Abstract
Organogenesis in plants depends upon the proper regulation of many genes, but how such necessary changes in gene expression are coordinated is largely unknown. The serrate (se) mutant of Arabidopsis displays defects in the initiation and elaboration of cotyledons and post-embryonic lateral organs. Cloning the SE gene revealed that it encodes a protein with a single, C2H2-type, zinc finger related to genes in other eukaryotes. Consistent with a role in organogenesis, the SE gene is transcribed in shoot meristems and in emerging organ primordia throughout development. Expression of the SE cDNA under the control of a heterologous promoter caused both accelerated and arrested plant growth, and these phenotypes were due to overexpression and co-suppression of the SE gene, respectively. Our analysis of the se mutant and the SE gene suggests a role for the SE gene product in regulating changes in gene expression via chromatin modification. Consistent with this proposed function, a synergistic double mutant phenotype was seen for plants mutant at both the SE locus and the locus encoding the largest subunit of chromatin assembly factor I.
INTRODUCTION
The aerial structures of a plant are produced by the shoot apical meristem, a collection of undifferentiated cells at growing shoot tips. Marked by drastically different rates of cell division, two zones within meristems can be distinguished (Steeves and Sussex, 1989). Slow cell divisions in the central zone serve to maintain this zone and to produce daughter cells for the peripheral zone. Cells in the peripheral zone rapidly proliferate to form organ primordia. Initiated reiteratively throughout plant development, organ primordia assume diverse fates depending on when and where they are initiated. Primordia initiated during the vegetative phase of development become leaves, while later primordia develop as floral meristems, which in turn produce sepal, petal, stamen, and carpel primordia. Furthermore, mature leaf structure differs depending on the developmental phase of the plant at the time of leaf initiation (Kerstetter and Poethig, 1998).
Along a cell lineage path from the central zone to the organ primordia, critical changes in gene expression occur. Knotted1-like homeobox (knox) genes are expressed throughout the meristem and downregulated in organ primordia (Reiser et al., 2000). Genes that are upregulated during organogenesis include the Arabidopsis AINTEGUMENTA, PIN-HEAD/ZWILLE, and ASYMMETRIC LEAVES1 (AS1) genes (Elliott et al., 1996; Klucher et al., 1996; Lynn et al., 1999; Byrne et al., 2000), the snapdragon PHANTASTICA (PHAN) gene (Waites et al., 1998), and the maize rough sheath2 (rs2) gene (Timmermans et al., 1999; Tsiantis et al., 1999). The AS1, PHAN, and rs2 genes encode highly similar MYB-like proteins and are required for maintaining knox gene repression in developing leaves (Waites et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000; Ori et al., 2000).
Given the sequential mechanism of organogenesis, genes that regulate widespread changes in gene expression are likely to be identified by mutations with pleiotropic effects on shoot and organ morphology. Here, we report the characterization of the Arabidopsis SERRATE (SE) gene. The se mutation has recently been shown to truncate the juvenile phase of vegetative development (Clarke et al., 1999; Serrano-Cartagena et al., 1999), increase plastochron lengths (Groot and Meicenheimer, 2000b), slow an early phase of leaf development (Groot and Meicenheimer, 2000a), and interact with as1 and as2 mutations with respect to knox gene repression in leaves (Ori et al., 2000). Our analyses of the se mutation revealed a pleiotropic phenotype affecting the positioning and the elaboration of leaves and floral organs. In addition, the se mutation affects cotyledon initiation in embryos when the maternal parent is homozygous for the mutation. SE encodes a zinc-finger protein whose mRNA accumulates in meristems and organ primordia. Transgenic plants expressing the SE cDNA with a heterologous promoter displayed a range of phenotypes consistent with a role for the SE gene in organogenesis but not with a direct role in the regulation of vegetative phase transitions. These results indicate that the SE gene product plays a role in coordinating gene expression in the meristem and lateral organ primordia, possibly by regulating chromatin structure.
RESULTS
Phenotypic Characterization of the se Mutant
The se mutation was first utilized as a genetic marker for linkage map construction (Rédei and Hirono, 1964). The se mutation segregates as a single, recessive Mendelian trait albeit at a slightly reduced transmission rate (Table 1). The reduced transmission is likely due to lethality of se/se embryos rather than poor transmission through either gametophyte because similar allele transmission frequencies were observed for both heterozygous parents in test crosses (Table 1). Complementation tests indicated that se is not allelic to other serrated leaf mutants—fas1, fas2, tousled, and leunig (data not shown; Leyser and Furner, 1992; Roe et al., 1993; Liu and Meyerowitz, 1995); nor is it allelic to mutants that have been mapped to a similar position on chromosome II—stunted plant1 and emb152 (data not shown; Baskin et al., 1995; Vernon and Meinke, 1995). Every mutation conferring se-like phenotypes that arose in mutagenized populations screened in our laboratory complemented the se mutation (unpublished observations), and mutant screens performed by two other laboratories also failed to identify alleles (Berná et al., 1999; Clarke et al., 1999). Finally, a directed approach failed to identify new se alleles. Approximately 9000 progeny were generated by pollinating se male sterile1-1 pistils with gamma- or UV-irradiated pollen from the Landsberg erecta (Ler-0) ecotype. (The male sterile1-1 mutation was included to prevent self-fertilization [van der Veen and Wirtz, 1968].) No progeny had a se-like phenotype and was heterozygous for Ler flanking sequences (data not shown). These results show that the se mutation identifies a novel locus and may represent a unique lesion.
Table 1.
Frequencies of Serrate Phenotype in Populations Segregating the se Mutation
| Cross (Female × Male) |
Serrate−a | Wild Typea | Frequencyb (%) |
χ2 | Pc |
|---|---|---|---|---|---|
| se/SE × se/SE | 277 | 948 | 22.6 | 3.72 | 0.05 |
| se/se × se/SE | 419 | 495 | 45.8 | 6.32 | 0.01 |
| se/SE × se/se | 407 | 504 | 44.7 | 10.3 | 0.001 |
a The phenotypes of progeny from the given crosses were scored after the expansion of the first two true leaves.
b Percentage of progeny with a Serrate− phenotype.
c The transmission frequency from each cross differed significantly from the expected Mendelian frequency (P ≤ 0.05).
All leaf margins of the se mutant are serrated as compared with wild-type plants in which only leaves produced late in development are serrated (Figures 1A and 1B). In addition, mutant leaf margins—like those of later-produced wild-type leaves—do not curl abaxially (downward). Further analysis revealed a complex mutant phenotype that extends beyond leaf margin development.
Figure 1.

Phenotype of se Plants.
(A) Outlines of all the rosette leaves of a Columbia ecotype (Col-1; wild type) plant (left) and a se mutant plant (right). Rosettes of se mutant plants contain fewer leaves and all the leaves are serrated, whereas only later-produced leaves of Col-1 plants are serrated.
(B) Rosette of a Col-1 (wild type) plant (left) and a se mutant plant (right).
(C) Inflorescences of a Col-1 (wild type) plant (left) and a se mutant plant (right). The arrow points to an abnormal cluster of flowers and siliques on the se mutant inflorescence.
WT, wild type.
Several se mutant phenotypes reflect defects in the initiation and elaboration of lateral organs. Normally, lateral structures such as leaves and flowers are produced in a spiral arrangement, or phyllotaxy, with nearly constant radial and vertical displacement between adjacent appendages (Leyser and Furner, 1992). Rosettes and inflorescences of se mutant plants often exhibit significant deviations from the normal radial positioning, and the internode lengths between adjacent flowers were more random in se relative to wild-type plants (Figure 1C; data not shown). The se mutation exerts a subtle effect on flower development—extra sepals and petals and fewer stamens were frequently present in the earliest formed flowers (Table 2). Throughout vegetative development, mutant plants produced visible leaves at a rate ∼70% of that observed for wild-type plants in the same growth conditions (Figure 2). This delay in leaf appearance may reflect a slower rate of leaf initiation or leaf elaboration, but two inflorescence phenotypes suggest that organ elaboration is primarily affected. First, because immature primordia in the shoot apex at the time of floral induction develop as either cauline leaves or as flowers, depending upon their developmental state (Hempel and Feldman, 1994), the increased number of cauline leaves on se mutant plants (Table 3) indicates that there are more immature leaf primordia at the time of floral induction. Second, se mutant inflorescences have an increased number of immature floral buds compared with those of wild-type inflorescences (Table 3), suggesting a slower rate of flower maturation.
Table 2.
Numbers of Floral Organs in Wild-Type and se Mutant Basal Flowers
| Wild Type (Col-1)
|
serrate
|
|||
|---|---|---|---|---|
| Whorl | Averagea | Abnormalb (%) |
Averagea | Abnormalb (%) |
| Sepal | 4.0 ± 0.0 | 0 | 4.3 ± 0.5 | 30 |
| Petal | 4.0 ± 0.0 | 0 | 4.3 ± 0.4 | 23 |
| Stamen | 5.5 ± 0.6 | 3 | 4.9 ± 0.8 | 37 |
| Carpel | 2.0 ± 0.0 | 0 | 2.0 ± 0.0 | 0 |
| All whorls | 15.5 ± 0.6 | 3 | 15.5 ± 1.0 | 63 |
a The number of floral organs in the first flower produced on 30 plants of both genotypes were counted and reported as the average ± twice the standard error.
b The percentage of deviation from the normal number of organs, which were four sepals, four petals, five or six stamens, and two carpels.
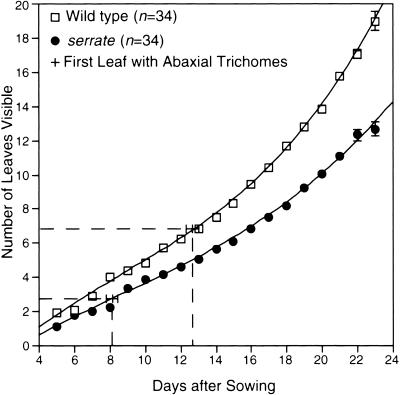
Figure 2.
The se Mutation Affects the Rate of Leaf Production.
The number of leaves visible with the aid of a dissecting microscope were recorded daily, and the mean leaf number was plotted relative to the number of days since sowing. Error bars indicate ± twice the standard error. Col-1 (wild type) data points are indicated with open boxes, and se data points are indicated with filled circles. Superimposed onto this graph are the average leaf positions of the first adult leaves plotted against the average day on which these leaves were first visible.
Table 3.
Developmental Phase Transitions in Wild-Type and se Mutant Plants
| Number of Leavesa
|
|||||
|---|---|---|---|---|---|
| Genotype | Juvenile | Adult | Cauline | Flowering Timeb | Flower Budsc |
| Wild type (Col-1) | 5.8 ± 0.4 | 10.1 ± 0.6 | 3.9 ± 0.3 | 22.4 ± 0.5 | 16.9 ± 1.1 |
| serrate | 1.8 ± 0.3 | 8.4 ± 0.8 | 5.2 ± 0.6 | 22.6 ± 1.0 | 26.8 ± 2.6 |
a Adult rosette leaves were defined as those with trichomes on the abaxial leaf surface, whereas juvenile rosette leaves lacked abaxial trichomes. Only cauline leaves on primary inflorescence were included; n = 30; mean ± twice the standard error.
b Days until flower buds were visible; n = 30; mean ± twice the standard error.
c When the primary inflorescences had between 10 and 25 post-anthesis flowers, the number of developing flower buds (between stages 6 and 12) were counted; mean ± twice the standard error. Wild type, n = 35; serrate, n = 25.
In wild-type plants, the distribution of trichomes is developmentally regulated (Martínez-Zapater et al., 1995; Chien and Sussex, 1996; Telfer et al., 1997). Juvenile leaves produce trichomes only on the adaxial (upper) surface, whereas adult leaves produce trichomes on both the adaxial and abaxial surfaces. Under our growth conditions, wild-type plants generally produced six leaves lacking abaxial trichomes; however, se mutant plants produced only one or two leaves without abaxial trichomes (Table 3 and Figure 2). Furthermore, the first leaves bearing abaxial trichomes were visible nearly 5 days earlier on mutant plants than on wild-type plants, suggesting a defect in the regulation of this developmental phase transition (Figure 2). In addition to leaf margin serration and trichome distribution, other characteristics such as leaf shape and the number of hydathodes differ between early and late leaves and were similarly affected by the se mutation (Clarke et al., 1999; data not shown).
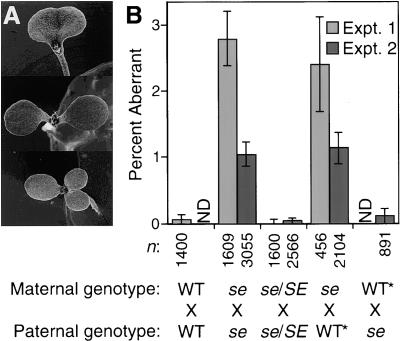
The SE gene is maternally required for normal embryo development. Whereas wild-type Arabidopsis embryos produced two cotyledons, progeny of selfed se plants often had either a single fused cotyledon or extra cotyledons (Figure 3). After reciprocal crosses using SE/SE or se/SE and se/se plants as parents, progeny cotyledon numbers were abnormal only when se/se plants were used as the maternal parent (Figure 3). These data indicate that the se mutation exerts its effect on embryo patterning from the surrounding maternal tissue and not from the gametophyte, endosperm, or embryonic cells. Consistent with this, seven out of thirteen aberrant progeny from the cross involving se/se (maternal parent) and se/SE (pollen donor) were genotypically heterozygous at the SE locus and displayed otherwise normal post-embryonic development (data not shown).
Figure 3.
Maternal Effects of the se Mutation.
(A) Variation in cotyledon number in week-old progeny from se mutant ovules.
(B) After germination, the number of seedlings with abnormal cotyledon numbers were counted. Error bars indicate ±sd of the binomial distribution. Experiments (Expt.) 1 and 2 differ only in the growth conditions of the parents: Experiment 2 was performed in a greenhouse during the winter. Supplemented artificial light was provided from 8:00 A.M. to midnight, and the daytime temperatures were held at less than ∼25°C (nighttime temperatures were cooler). Asterisks denote that wild-type (Col-1) plants were used in this cross for experiment 1 and se/SE plants in experiment 2. n, total number of seedlings scored; ND, not determined.
Similar to the rate of leaf production, the rate of root growth was also slower in se mutants. When the changes in primary root length during day 7 of growth were compared, se roots elongated at a rate ∼70% of that observed in wild-type roots (se, 5.5 ± 0.4 mm; wild type, 8.2 ± 0.7 mm; mean ± twice the standard error).
The SE Locus Encodes a Zinc-Finger Protein
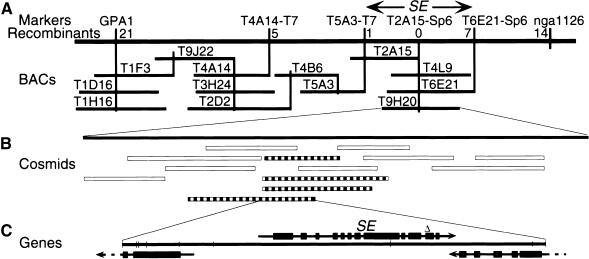
The complex se mutant phenotype suggests that the SE gene product regulates the activity of a number of developmental pathways. To help decipher its role in development, we isolated the SE gene by positional cloning (Figure 4). Genetic linkage to the middle of chromosome 2 had previously been reported (Rédei and Hirono, 1964), and this was confirmed using the GPA1 cleaved amplified polymorphic sequence (CAPS) marker (Konieczny and Ausubel, 1993). Using mapping lines with meiotic recombination breakpoints near the SE locus, SE was determined to be between the GPA1 and the nga1126 (http://thale.salk.edu/) loci. A bacterial artificial chromosome (BAC) contig was assembled extending from the GPA1 locus. During contig assembly, restriction fragment length polymorphism (RFLP) markers derived from BAC end sequences were mapped relative to SE until it was found that the T6E21-Sp6 RFLP marker mapped distally from the SE locus. BAC T9H20 spanned most of the region between flanking RFLP markers genetically separable from SE and was subcloned for plant transformation. Of 13 cosmids introduced into se mutant plants, four conferred a wild-type phenotype. Within the 10-kb region shared by the complementing cosmids, a single complete gene and two partial genes were found by DNA sequence analysis. Flanking cosmids that deleted portions of the complete gene but spanned either of the partial genes did not rescue the mutant phenotype. Furthermore, the complete gene contained a 7-bp deletion in the corresponding region from the se mutant genome, confirming that this was the SE gene.
Figure 4.
Positional Isolation of the SE Locus.
(A) Position of SE relative to polymorphic markers in the middle of chromosome 2 (long, thick horizontal line) and the contig of BACs (horizontal bars). The vertical lines represent the positions of DNA fragments used for mapping (those named across the top) or those used only for constructing the BAC contig (unnamed). The number of meiotic recombination events identified between a marker and the se mutation are indicated below the marker names.
(B) Cosmid subclones of the BAC T9H20 (solid bar). Striped bars indicate cosmid subclones that complemented the se mutant phenotype upon transformation, and open bars indicate those that did not.
(C) Schematic representation of the gene structures in the region that complemented the se mutant phenotype. Filled boxes represent exons, lines designate noncoding regions, arrowheads show the direction of transcription, and dashed lines indicate that the genes extend beyond the region sequenced. The position of the 7-bp deletion of the se mutant genome is indicated (open triangle).
The SE open reading frame encodes a 720–amino acid protein predicted to contain a single C2H2-type zinc finger and multiple bipartite nuclear localization motifs (Figure 5A). The se mutation would cause a frameshift altering the last 27 amino acids. Two expressed sequence tags (ESTs) from sugarcane that were highly similar to the C terminus of the predicted SE protein were present in public databases and were used to isolate two distinct SE homologs from another monocot, maize. The amino acid sequences encoded by three partial monocot cDNAs (from sugarcane and maize) share ∼90% identity with one another and 60% identity with the dicot-derived SE protein. Currently, the public databases contain homologous ESTs from maize (six accessions), rice (two), sorghum (eleven), tomato (three), potato (one), soybean (one), and hybrid aspen (one). In addition, several genomic survey sequences (four BAC end sequences and the RG365 RFLP marker sequences) from rice contain sequence similarity to the SE gene. The maize genome contains at least four SE-related genes because the ESTs identify genes distinct from those isolated using the sugarcane cDNA sequence (Figure 5A). The rice and sorghum genomes, however, appear to have only two expressed SE-related genes. An alignment of SE-related sequences from plants is shown in Figure 5A.
Figure 5.
SE Is a Member of a Protein Family Conserved in Eukaryotes.
(A) Alignment of SE-related sequences from plants. Conserved residues are shaded in black, and conservative changes are shaded in gray. Gaps (indicated by dots) were introduced to optimize alignment, and tildes (∼) were added where sequence information is lacking. The sources of the sequences are described in Methods. The positions of the four domains used in the alignment shown in (B) are designated with brackets and labeled, and the positions of se-1 mutation and putative bipartite nuclear localization motifs are indicated with a triangle and hatched bars, respectively. Genes designated ZmSExx are derived from maize. C-term, C terminus.
(B) Alignments of four domains (A, B, zinc finger, and C terminus) of the predicted SE protein with comparable domains of the S. pombe SPBC725.08 protein; the Asr2-related homologs from Caenorhabditis elegans, Drosophila, and human; and the Chinese hamster Asr2 protein. Shading is as described for (A), and residues conserved in all homologs are highlighted with an asterisk. The number of residues in front of and between domains are indicated. The positions comparable to the two β strands and the α helix of C2H2-zinc-finger domains of known structure are indicated with arrows and a coil above the respective sequences.
Significant homology is also shared with the SPBC725.08 zinc-finger protein of unknown function from Schizosaccharomyces pombe and a protein family from animals that includes the hamster arsenite resistance2 (Asr2) protein (Rossman and Wang, 1999). Although the overall similarity between the plant, fungal, and animal proteins is modest (11 to 15% amino-acid identity), the sequences and relative orders of four domains are conserved (Figure 5B). Significantly, two of these domains correspond to the zinc-finger domain and the C-terminal sequences affected by the se mutation. Interestingly, the animal homologs lack the second conserved cysteine but retain high similarity in the region corresponding to the DNA binding helix of other zinc fingers (Figure 5B; Klug and Schwabe, 1995; Berg and Shi, 1996). Although no biochemical function has been ascribed to any of these proteins, overexpression of the asr2 cDNA—which is likely to encode only a portion of the full-length protein—confers arsenite resistance to hamster cell lines (Rossman and Wang, 1999).
Expression Pattern of the SE mRNA
The se mutant phenotype suggested that the wild-type SE gene product is required for normal development of shoot and floral meristems, developing leaves, and embryos. SE mRNA was detected in all of these tissue types by use of in situ hybridization (Figure 6; data not shown). In the developing embryo, SE expression is present in the shoot and root meristems and in the adaxial portion of cotyledons of torpedo-stage embryos, but expression is reduced in walking-stick stage embryos and absent from mature embryos (Figures 6A to 6C). Similar to cotyledons, the adaxial portion of newly emerging leaf primordia exhibits the highest level of SE expression, but more mature leaves retain some expression only in localized regions (Figure 6E). The downregulation of SE is rapid during floral organ development: for example, SE mRNA is detected in sepal primordia of stage 4 flowers, but by stage 7, expression is absent from sepals and is present in the emerging stamen and carpel primordia (Figure 6F; stages as defined in Smyth et al., 1990). SE mRNA is also detected throughout vegetative, inflorescence, and floral meristems (Figures 6E and 6F). SE mRNA accumulation in se mutant apices appears to be reduced in parallel in situ hybridization experiments, suggesting either that the C-terminal sequences may be required for autoregulation or that the small deletion adversely affects mRNA stability (Figure 6G).
Figure 6.
In Situ Localization of SE mRNA in Wild-Type and se Mutant Tissue.
(A) to (C) Torpedo-stage (A), walking-stick stage (B), and nearly mature (C) wild-type embryos hybridized with an antisense SE riboprobe.
(D) Torpedo stage wild-type embryo hybridized with a sense (control) SE riboprobe.
(E) Apex of the wild-type shoot apex prior to flower production but after hybridization with an antisense SE riboprobe.
(F) and (G) Wild-type (F) and se mutant (G) inflorescences hybridized with an antisense SE riboprobe. Numbers indicate stage of flower development.
c, cotyledon; f, floral meristem; g, developing gynoecium; l, leaf primordia; m, shoot apical meristem; r, root meristem; se, developing sepal; sf, developing stamen. Bars = 40 μm.
Phenotypic Effects of SE Misexpression
The SE locus was identified based upon a single mutation affecting only the C-terminal tail of the predicted protein. To observe the effects of altered levels of SE activity, wild-type and se mutant plants were transformed with the SE cDNA under the control of the cauliflower mosaic virus 35S RNA (35S) promoter (Odell et al., 1985). A range of phenotypes arose in the T1 plants, but a significant proportion of these lines (29/67) could not be recovered due to seedling lethality or sterility. The phenotypes of the T2 generation were studied in greater detail, and two classes emerged (Table 4, and Figures 7A and 7B). Class I lines displayed an essentially wild-type phenotype whether in a wild-type or se mutant background, and class II lines displayed a range of developmental phenotypes. Interestingly, lines carrying the se mutation tended to have more severe phenotypes than did those transformed into a wild-type background (Table 4). Furthermore, the severity of the class II lines frequently increased with successive generations regardless of the background. For example, the sbla3 T1 plant (in se background) was fertile, but every T2 plant was completely sterile (n > 20). Given the phenotypic instability of class II lines, and the class I and class II phenotypes (below), class I lines are likely to be overexpressing the SE gene and class II lines are likely to have suppressed SE function.
Table 4.
Phenotypic Categories of 35S::SE Linesa
| Number of Independent Lines per Class (%)
|
||||
|---|---|---|---|---|
| Background | Class I | Class IIa | Class IIb | Class IIc |
| serrate | 3 (11%) | 1 (4%) | 10 (36%) | 14 (50%) |
| Wild type (Col-1) | 4 (36%) | 3 (27%) | 3 (27%) | 1 (9%) |
The 35S::SE construct was transformed into se and wild-type plants, and the T2 transgenic lines were grown for ∼2 weeks before scoring the phenotypes. The phenotypes were classified based on the most severe segregants as described in the text: class I, wild type; IIa, mild; IIb, intermediate; and IIc, severe. The probability that the observed distribution of phenotypes was independent of genetic background was <0.02 using a 2 × 4 William's corrected G test of independence.
Figure 7.
Phenotypes of Plants Harboring the 35S::SE Transgene.
(A) A class I (sbla11) T4 plant is compared with the wild type (Col-1) and the se mutant after ∼25 days of growth. The class I plant's inflorescence is taller due to the early flowering phenotype.
(B) The phenotypes of classes IIa (sbla6), IIb (sbla1), and IIc (sala3) T3 plants are compared after ∼30 days of growth. Note the aberrant flower positions of the classes IIa and IIb plants and the aberrant leaf positioning of the class IIc plant.
(C) Ten-day-old wild-type seedling with two cotyledons and four true leaves.
(D) Ten-day-old class IIc (sbla17) plant that arrested after cotyledon expansion.
(E) Scanning electron micrograph of an arrested apex of a class IIc line. The arrowhead points to the meristem, which is flanked by arrested primordia. Magnification bar is ∼100 μm.
(F) to (H) Examples of the various inflorescence and flower patterning defects seen in class IIc lines. Note the wide range in floral organ numbers and organ morphologies. The mature “flower” shown in (H) failed to terminate after the production of carpels. Arrows point to radially symmetric filaments.
(I) Detection of SE mRNA in SE misexpression lines. Total leaf RNA was prepared from 10 T1 plants of the given phenotypic class, and SE mRNA was detected by semi-quantitative RT-PCR. The lines represented include cbla1, cala2, cala3, cala4, and cala7 in a wild-type background, and sbla2, sbla3, sbla4, sbla10, and sbla11 in a se mutant background. For control reactions, cDNA was prepared from wild-type leaves (WT Leaf), wild-type inflorescences including pre-anthesis flower buds (WT Inf.), and se leaves (se Leaf). The gel at top shows the detection of SE mRNA, whereas the bottom two gels show detection of the ubiquitously expressed β–ATPase and ROC1 genes (Boutry and Chua, 1985; Kelly et al., 1990; Lippuner et al. 1994). Note that the SE RT-PCR product from SE leaves is slightly smaller than that from the wild type due to the mutation.
One class I line in the se mutant background, sbla11, was chosen for detailed analysis because the transgene stably complemented all of the se mutant phenotypes (Figure 7A and Table 5) and exhibited high levels of SE mRNA expression (Figure 7I). In contrast to the se mutant, the sbla11 plant line displayed an increased rate of leaf production and had a decreased number of developing floral buds on inflorescences relative to wild-type plants (Table 5). Unexpectedly, the transgene also affected both the number of days and the number of leaves produced prior to flowering (Table 5). Each vegetative phase is shortened relative to those of wild-type plants, suggesting that development in general is accelerated by SE overexpression.
Table 5.
Phenotypic Analysis of the sbla11 Class I 35S::SE Line
| Number of Leaves Visiblea
|
Number of Leavesb
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Day 6 | Day 11 | Day 16 | Juvenile | Adult | Cauline | Flowering Timec | Flower Budsd |
| 35S::SE se | 2.0 ± 0.0 | 5.8 ± 0.2e | 11.6 ± 0.3f | 4.2 ± 0.2f | 5.8 ± 0.3f | 2.7 ± 0.1f | 18.6 ± 0.3f | 15.1 ± 1.9e |
| Wild type (Col-1) | 2.0 ± 0.1 | 5.5 ± 0.2 | 11.0 ± 0.3 | 4.6 ± 0.3 | 7.9 ± 0.4 | 3.7 ± 0.2 | 20.9 ± 0.4 | 17.0 ± 1.0 |
| serrate | 0.0 ± 0.0f | 4.0 ± 0.1f | 7.8 ± 0.2f | 1.9 ± 0.2f | 5.8 ± 0.4f | 4.6 ± 0.4f | 21.1 ± 0.5 | 30.7 ± 1.4f |
a On given days after sowing, the number of leaves visible without dissection and using a dissecting microscope were recorded; mean ± twice the standard error; n = 45 to 48.
b Adult rosette leaves were defined as those with trichomes on the abaxial leaf surface, whereas juvenile rosette leaves did not have abaxial trichomes. Only cauline leaves on primary inflorescence were included. Mean ± twice the standard error; n = 45 to 48.
c Days until flower buds were visible; mean ± twice the standard error; n = 45 to 48.
d When the primary inflorescences had between 10 and 25 post-anthesis flowers, the number of developing flower buds (between stages 6 and 12) were counted; mean ± twice the standard error; n = 25 to 35.
e Significantly different from the wild type (P < 0.05).
f Significantly different from the wild type (P ≤ 0.001).
Class II lines displayed strikingly variable phenotypes both within and between lines. The class was further subdivided based upon the more severe phenotypes appearing in T2 families (Figure 7B). Mildly affected (class IIa) lines produced relatively normal leaves but consistently developed inflorescence phyllotaxy defects similar to those seen in se mutants. The intermediate class of transformants (class IIb) produced leaves with adaxially curled lamina and reduced inflorescence internode elongation. Growth of the severely affected (class IIc) lines intermittently arrested after the production of a variable number of leaves (most often after 0, one, or two leaves; Figure 7D). Microscopic analysis of arrested shoot apices revealed protrusions with outgrowths emerging along their sides (Figure 7E). On the basis of their positions and morphology, the tips of these protrusions are likely to be apical meristems, and the outgrowths appear to be arrested organ primordia. Eventually, growth continued, with new leaf primordia emerging both from the original meristem's location and from positions on the side of the apex. The leaves produced by class IIc lines were similar to those of the se mutant but were often asymmetric in shape (data not shown). The inflorescences of these plants were severely affected: there was little internode elongation between cauline leaves, and few flowers were produced (Figure 7F; data not shown). The flowers that did form were disorganized and had extremely variable numbers of floral organs and radially symmetric filaments (Figures 7F to 7H). In several class IIc lines, floral meristems failed to terminate properly after the production of the carpel whorl, resulting in sterile masses predominately composed of unfused carpels and exposed ovules (Figure 7H).
The range of phenotypes seen in the 35S::SE transformants suggested that expression from the transgene might be variable. After the T1 plants were phenotypically scored, gene expression was analyzed by semi-quantitative reverse transcriptase–polymerase chain reaction (RT-PCR). As shown in Figure 7I, expression levels were variable, but there was only a slight correlation between phenotypic class and SE mRNA accumulation. This poor correlation suggests either that a post-transcriptional mechanism is responsible for the phenotypic variability or that transcript accumulation is spatially and/or temporally variable and was not adequately assayed by our analysis of mature leaf tissue.
Synergistic se fas1 Double Mutant Phenotype
Mutations in the FAS1 gene cause pleiotropic phenotypes that overlap the phenotypes observed for se mutants: mutations in either gene affect leaf serration, phyllotaxy, and the numbers of floral organs in the outer three whorls (Figure 8A; Leyser and Furner, 1992). Unlike se, however, fas1 mutants have narrow leaves and enlarged meristems that often lead to stem fasciation. To determine whether the SE and FAS1 genes function in a common pathway or in converging pathways, double mutants were constructed. Two fas1 alleles from the En ecotype background were used in these analyses. The inflorescence phenotypes are similar, but the fas1-11 allele caused more deeply serrated leaves and less fertile flowers than did the fas1-1 mutant allele (Serrano-Cartagena et al., 1999; data not shown). Despite having a less severe phenotype, molecular analysis suggests that fas1-1 is a null allele (Kaya et al., 2001). The se fas1-1 and se fas1-11 double mutants produced a variable number of extremely narrow lateral organs before the meristems degenerate into masses of callous-like cells (Figures 8B to 8E). Out of 59 se fas1-1 double mutants, 42 (71%) arrested prior to stem elongation, seven (12%) arrested after inflorescence elongation but prior to flower formation, and 10 (17%) arrested only after the initiation of a few misshapen flowers (Figure 8B). The last leaves produced prior to meristem degeneration often had radial symmetry (data not shown). Similar phenotypes were previously reported for the mgo1-1 fas1-1 double mutant (Laufs et al., 1998). This synergistic genetic interaction suggests that the SE and FAS1 genes function in separate pathways that converge during organogenesis, but a common-pathway model cannot be completely dismissed because the se mutation is not likely null.
Figure 8.
se fas1 Double Mutant Phenotype.
(A) fas1-1 single mutant.
(B) Range of phenotypes displayed by ∼28-day-old se fas1-1 double mutants. Note that all leaves are extremely narrow and that the shoot apex has arrested growth in each individual albeit at different developmental times.
(C) se fas1-11 double mutant.
(D) and (E) Scanning electron micrographs of the arrested shoot apex of an ∼30-day-old se fas1-1 double mutant plant. The meristem has degenerated into a mass of callous-like tissue. Bar in (D) = ∼500 μm; bar in (E) = 20 μm.
DISCUSSION
The phenotypes of the se mutant indicate a role for the SE protein in organogenesis. The slower rate of leaf production, the displacement of more leaves onto the inflorescence stem, and the presence of more immature flower buds are all consistent with a primary defect in the early steps of organ elaboration. This interpretation is also consistent with the analyses by Groot and Meicenheimer (2000a) that revealed that growth of se leaves is biphasic, with slower growth early (until ∼2 mm in length) and normal growth thereafter. Detailed mutational analysis of the SE gene was handicapped by the lack of additional mutant alleles. Allelic mutants have not been identified based on leaf phenotypes in our laboratory or in screens by other laboratories (data not shown; Berná et al., 1999; Serrano-Cartagena et al., 1999; Clarke et al., 1999). Furthermore, a directed pollen-mutagenesis approach also failed to identify new alleles. One explanation for the failure to identify new alleles is that the original se allele retains some function that is essential for development. This is consistent with molecular analysis of the existing se-1 mutant allele, which only affects the C-terminal 27 amino acids (Figures 5A). Despite the uniqueness of the se-1 allele, it is unlikely that the phenotype results from the gain of a novel function because the mutation acts recessively.
Analysis of transgenic plants misexpressing the SE gene revealed two classes of phenotypes. The nearly wild-type phenotype of class I lines suggests that SE activity is permissive for normal development because ectopic SE expression rescues the mutant phenotypes with few additional consequences. These lines produce leaves at a slightly faster rate than do wild-type plants, but the most profound effect was on flowering time, suggesting that the SE protein may be involved in the vegetative-to-floral phase transition as well as the transitions from meristematic to organogenetic growth. Given the appearance of phenotypes similar to those found in the se mutant, class II lines appear to have reduced SE function due to co-suppression (Matzke and Matzke, 1998). Although the increase in phenotypic severity in several of the class II lines in successive generations suggests an epigenetic mechanism, there was little correlation between phenotypic severity and transgene mRNA expression (Figure 7I). One explanation for this is that the SE gene is essential and that stably silenced plant lines were lost due to selection of viable transformants. In this scenario, SE expression in the class II lines is unstable and therefore detectable in total leaf RNA preparations. Alternatively, SE function may be suppressed by a “squelching” mechanism. Genetic squelching may result when a protein is overexpressed and dilutes the pool of required co-factors (Ptashne, 1992; Larkin et al., 1994).
The relationship of the organ elaboration phenotypes with the leaf margin and the trichome distribution phenotypes is unclear. One possibility—corroborated by SE mRNA expression pattern (Figure 6) and the SE misexpression phenotypes (Figure 7)—is that there is an associated defect in leaf polarity in lateral organs of the se mutant. Organogenesis and organ polarity defects are linked in other mutants. The phan mutants of snapdragon fail to initiate organ primordia at low temperatures and produce partially or completely abaxialized organs at higher temperatures (Waites and Hudson, 1995; Waites et al., 1998). Strong mutant alleles of the Arabidopsis ARGONAUTE1 (AGO1) gene produce organs at a much slower rate than do wild-type plants, and many organ types are abaxialized in the mutant (Bohmert et al., 1998; Lynn et al., 1999). Weaker ago1 mutants display a delay in organ production and allele-specific defects in organ patterning: one class of alleles produces narrow organs and another class produces serrated leaves that are frequently trumpet shaped and have outgrowths on the abaxial surfaces—both likely due to a partial adaxialization of organ primordia (Champagne, 1998). Although trumpet-shaped leaves have never been observed in se mutants, the phenotypes of the latter class of weak ago1 mutants and the se mutant are remarkably similar and suggest that SE and AGO1 act in overlapping developmental pathways. Both mutants display slow rates of organ production, similar leaf margin morphologies, and flower-positioning defects.
The embryo's dependence on SE activity for proper cotyledon initiation is interesting, and similar maternal control over plant embryo patterning has only previously been observed for the Arabidopsis sin1-2 mutant and transgenic tobacco plants overexpressing the oat phyA gene (Ray et al., 1996; Emmler and Schäfer, 1997). Given the presence of a haploid gametophyte generation between diploid sporophyte generations, maternal effects on plant embryo development are paradoxical because the embryo is isolated both spatially (by the endosperm) and mitotically (during megagametophyte development) from diploid maternal tissue. One possibility is that the SE and SIN1 proteins may be involved in establishing the expression state of one or more maternal genes prior to gametogenesis. The MEDEA/FIS1 gene of Arabidopsis is required for proper embryo and endosperm development and was recently found to be imprinted such that only maternally inherited copies are expressed during endosperm development (Chaudhury et al., 1997; Grossniklaus et al., 1998; Kinoshita et al., 1999; Kiyosue et al., 1999; Luo et al., 1999; Vielle-Calzada et al., 1999). In another possibility, the SE and SIN1 genes may be involved in signaling to the early embryo positional information required for the proper induction of bilateral symmetry. A third possibility is that sufficient maternally contributed wild-type protein or mRNA persists through gametogenesis and early embryogenesis and that zygotic expression appears too late to rescue the defect in embryo symmetry.
The putative zinc-finger and nuclear localization motifs suggest that the SE protein regulates transcription, possibly by altering chromatin structure. Unlike conventional zinc-finger transcription factors (Klug and Schwabe, 1995; Berg and Shi, 1996), SE contains a single zinc finger—rather than tandem repeats—and is, consequently, unlikely to bind alone with high sequence specificity. Having lower levels of sequence specificity would allow SE to coordinate the expression of many genes by altering chromatin structure in a region-specific manner. Previous analyses of leaf-shape mutants have already established a link between gene silencing and leaf development. Mutations in the CURLY LEAF gene of Arabidopsis, encoding an E(z)-like polycomb-group protein, cause the leaf lamina to roll up as a result of ectopic expression of the floral homeotic gene AGAMOUS (Goodrich et al., 1997). The leaf curling phenotype of class IIb 35S::SE plants is very similar to that of curly leaf mutants, suggesting that SE might also be involved in repressing the AGAMOUS gene, and preliminary RT-PCR results agree with this interpretation (M.J. Prigge and D.R. Wagner, unpublished results).
A role for SE in gene regulation during organogenesis was recently identified by analysis of the se as1 and se as2 double mutants. The se mutation synergistically enhanced the as1 and as2 phenotypes, resulting in phenotypes very similar to those of plants overexpressing the KNAT1 knox gene—including the appearance of ectopic stipules in the sinuses of leaf lobes (Ori et al., 2000). Although as1 and as2 single mutants ectopically express the KNAT1 and KNAT2 knox genes in leaf tissue, only rarely do ectopic stipules develop (Byrne et al., 2000; Ori et al., 2000). Although se single mutants do not express knox genes ectopically, se as1 and se as2 double mutants exhibit slightly increased knox gene expression in the sinuses but reduced expression in other parts of leaves relative to as single mutants (Ori et al., 2000). This suggests that the se mutation either allows knox gene expression to rise above a critical threshold specifically in leaf margins, lowers the knox gene expression threshold required to activate downstream genes, or a combination of both possibilities. A role for the SE protein in regulating chromatin structure would be consistent with both of these possibilities.
Two other proteins with single C2H2 zinc fingers have been implicated in regulating chromatin structure: FIS2 from Arabidopsis (Luo et al., 1999) and the GAGA factor (GAF) encoded by the Trithorax-like gene of Drosophila (Farkas et al., 1994; Pedone et al., 1996). Although no target genes have been identified, mutant analysis suggests that FIS2 is required for repressive chromatin structure during megagametophyte development in the same pathway as two polycomb-group genes: FIS1/MEDEA and FIE, which are homologous to the Drosophila E(z) and Esc proteins, respectively (Chaudhury et al., 1997; Grossniklaus et al., 1998; Kiyosue et al., 1999; Luo et al., 1999; Ohad et al., 1999). In contrast to the proposed role for FIS2, GAF antagonizes silent chromatin maintenance, resulting in timely derepression of several developmentally and environmentally regulated genes (reviewed in Wilkins and Lis, 1997). In a manner analogous to the FIS2 and GAF proteins, SE may coordinate the expression of multiple genes during development by modifying the chromatin structure surrounding these genes.
The synergistic genetic interaction between se and fas1 mutations is most easily explained as a convergence of two pathways regulating chromatin structure. FAS1 encodes the largest subunit of chromatin assembly factor I (Kaya et al., 2001), which directs assembly of histones onto newly replicated DNA (Smith and Stillman, 1989; Adams and Kamakaka, 1999). Mutations in the yeast homolog CAC1/RLF2 impair the maintenance of gene silencing at the telomeres and at the silent mating loci (Enomoto et al., 1997; Kaufman et al., 1997; Enomoto and Berman, 1998). Reminiscent of the fas1 interaction with se, cac1/rlf2 mutants interact synergistically with mutations in the SIR1 gene (Enomoto and Berman, 1998). Because SIR1p is required for the establishment of gene silencing at the silent mating loci (Pillus and Rine, 1989), it was proposed that SIR1p cooperates with chromatin properly assembled by CAC1/RLF2p function (Enomoto and Berman, 1998). An intriguing possibility is that SE protein may downregulate genes during organogenesis in a manner analogous to SIR1p.
In an alternative hypothesis, SE may be directly regulating the cell cycle. Such a hypothesis is consistent with the SE mRNA accumulation pattern (the highest expression in tissue with the shortest cell-cycle lengths) and with the SE overexpression phenotypes because similar phenotypes were seen when an Arabidopsis D-type cyclin was overexpressed in tobacco (Cockcroft et al., 2000). A role for SE in cell-cycle regulation is, however, less easily reconciled with the phenotype of the se fas1 double mutant in which cell division continued after organogenesis had arrested (Figure 8D). Further investigations of the SE protein function should reveal the mechanism by which SE regulates so many aspects of plant development.
METHODS
Plant Materials, Culture, and Analysis
Arabidopsis thaliana seed for the se, fas1-1, fas1-11, and phyB-10 mutant strains and the Columbia (Col-1) and Wassilewskija (Ws-2) wild-type strains were obtained from the Arabidopsis Biological Resource Center (ABRC, Ohio State University, Columbus). Eva Sundberg and George Coupland (John Innes Centre, Norwich, UK) supplied Tn28 and alb3-1 seed (Long et al., 1993).
Seeds were imbibed at 4°C for 3 to 5 days before or after sowing to 6-cm square pots containing coarse vermiculite overlaid with a 1- to 2-cm layer of soil mix comprised of a 2:1 mixture of Premier Pro potting soil (Red Hill, PA) and fine vermiculite. Plants were grown either in a long-day growth room (18-hr-light:6-hr-dark cycle) or in a greenhouse during the winter with supplemented light. Temperatures in the growth room were maintained between 18 and 22°C, but the greenhouse temperatures were very variable with cooler nighttime temperatures. Peters Professional (20-20-20) fertilizer was administered upon sowing and again after ∼3 weeks of growth. An in planta Agrobacterium-mediated transformation protocol was used to introduce constructs into Arabidopsis (Bechtold et al., 1993; Clough and Bent, 1998). Selection of kanamycin- or hygromycin-resistant seedlings was performed on aseptic solid media containing 0.5 × MS salts (Murashige and Skoog, 1962; Sigma), 2% sucrose, and 1% purified agar (Sigma).
The se fas1-1 double mutants were identified in the second generation after backcrossing se × fas1-1 F2 individuals with a Fas phenotype to se. Roughly one-quarter of the progeny from selfed Se backcross progeny displayed the described double mutant phenotype. The se fas1-11 double mutants were identified among the F2 progeny from a cross between se and fas1-11. Differences between the Columbia (Col-1) and Enkeim (En) ecotype backgrounds of the parental strains may have contributed to the broad range of observed phenotypes; however, very similar ranges of double mutant severities were seen in both the se × fas1-1 SE/se F2 (3:1 Col-1:En) and the original se × fas1-1 F2 (1:1 Col-1:En), suggesting that other factors also contributed to the variability (data not shown).
Leaf outlines were traced using Deneba Canvas (Miami, FL) after the series of rosette leaves were scanned using a flatbed scanner. Scanning electron microscopy was performed as previously described (Pickett et al., 1996; Yu et al., 2000).
Molecular Biology Techniques
Plant genomic DNA was prepared in one of two ways. Large-scale preparations were isolated from lyophilized material by CTAB precipitation (Reiter et al., 1992). A scaled-down version of a protocol by Dean et al. (1992) was used for mini-preparations suitable for polymerase chain reaction (PCR) and DNA gel blot analyses and is described briefly. One or two leaves (∼5 cm2 total leaf area) were transferred to microcentrifuge tubes, frozen in liquid nitrogen, spun briefly in a microcentrifuge, and re-frozen. Using a polypropylene mini-pestle attached to a drill, the tissue was ground until it started to thaw. After adding 500 μL of extraction buffer (140 mM sorbitol, 220 mM Tris, pH 8, 22 mM EDTA, pH 8, 0.8 M NaCl, 1% sarkosyl, and 0.8% CTAB), the contents were mixed with the pestle. Samples were incubated at 60 to 65°C for 20 to 40 min with occasional mixing. Cooled samples were extracted with chloroform and ethanol precipitated. For PCR analysis, the pellet was washed with 70% ethanol, dried, and resuspended in 50 μL of TE (10 mM Tris and 0.5 mM EDTA, pH 8). For DNA gel blot analysis, the samples were resuspended in 100 μL of 300 mM NaCl and further extracted at least once with 1:1 phenol:chloroform then chloroform, precipitated, washed, dried, and resuspended in 20 μL of TE; the yield was determined using a DyNA Quant 200 fluorometer (Amersham Pharmacia Biotech).
To facilitate higher throughput PCR mapping, loading dyes and sucrose were included in the reaction mixtures (Hoppe et al., 1992). PCR reactions contained 50 mM KCl, 10 Tris-HCl, pH 8.3, 1 mM MgCl2, 0.1 g/L BSA, 12% sucrose, 0.1% Triton X-100, 0.2 mM cresol red, 0.3% yellow food coloring (McCormick and Co., Inc., Hunt Valley, MD), 125 μM each dNTP, 250 nM each primer, and 0.1 U/μL Taq polymerase.
The non-radioactive Genius System (Boehringer Mannheim, Mannheim, Germany) was used—essentially as prescribed—for DNA gel blot analysis and for screening bacterial artificial chromosome (BAC) and phage libraries. Between 0.3 and 2 μg of restricted Arabidopsis genomic DNA was fractionated and blotted to nylon membranes for DNA gel blot analysis. Probe detection using alkaline phosphatase– and horseradish peroxidase–conjugated anti-digoxigenin antibodies was performed essentially as described by the manufacturer except that casein (U.S. Biochemical) was substituted for Boehringer Blocking powder and an extra post-antibody binding wash was added. CDP-Star was used for alkaline phosphatase detection, and luminol-based chemiluminescence was used for horseradish peroxidase detection.
Mapping the SE Locus
SE was initially mapped using a se (Col-1 ecotype) × Ws F2 population, the GPA1 and PHYB CAPS markers (Konieczny and Ausubel, 1993; Boerjan et al., 1995), and the nga168 and AthGPA1 SSLP markers (Bell and Ecker, 1994). The locus was finely mapped using mapping lines with meiotic recombination near the SE locus derived from three different crosses: se × phyB-10 (Ws ecotype), se × Tn28 (Landsberg erecta [Ler] ecotype), and se × alb3-1 (Ler ecotype). Lines with breakpoints above of SE were identified as kanamycin-resistant (phyB-10/PHYB) or hygromycin-resistant (Tn28/+) se/se plants. Lines with breakpoints between SE and ALB3 were identified as hygromycin-resistant (alb3-1/ALB3) se/se plants. Finally, lines with breakpoints between ER and ALB3 were identified as non-albino er/er plants. These mapping lines were genotyped using the B68 restriction fragment length polymorphism (RFLP) marker (Ellen Wisman, Max-Planck-Institut, Cologne, Germany), HY1 CAPS marker (Muramoto et al., 1999), nga1126 SSLP marker (http://thale.salk.edu/), and RFLP markers derived from BACs isolated during contig assembly (below).
Isolating the SE Locus
To minimize wear on individual BAC library filters, each of the 11 plates was prescreened by DNA gel blot analysis. Each of the first 11 plates of the TAMU BAC library (Choi et al., 1995) was stamped onto solid media and grown overnight, and the pooled colonies from each plate were collected for DNA preparation. HindIII-restricted plate-pool DNA (3 μg) were fractionated and blotted. Because the BACs were constructed using HindIII-restricted genomic DNA (Choi et al., 1995), diagnostically sized fragments were detected in positive plate pools, thus reducing the chances of false positives. Individual library plates were then stamped onto nylon membranes, placed on solid media, and grown overnight, and the filters were prepared for hybridization (Sambrook et al., 1989). After identification, positive BACs were compared by HindIII restriction and DNA gel blot analysis. Digoxigenin-labeled BAC end probes were generated directly from the EcoRV-digested BACs using T7 RNA polymerase or by labeling subcloned end fragments. The BAC end subclones were isolated by end rescuing: religation after BamHI or SphI digestion (Sp6 ends) or after BstBI or BspDI digestion (both ends). In the latter case, the T7 ends were usually isolated by subcloning the appropriate BstBI- HindIII or HindIII fragment.
The BAC T9H20 was partially digested with HindIII and size-fractionated using low-melting-point agarose. Two size ranges (12 to 23 kb and 23 to 50 kb) were purified using β-agarase (New England Biolabs, Beverly, MA) and phenol:chloroform extraction (necessary to remove residual agarose). Both size-ranges were ligated into HindIII-digested and shrimp alkaline phosphatase–treated pOCA28 plant transformation vector, a derivative of pOCA18 obtained from Neil Olszewski (University of Minnesota, St. Paul; Olszewski et al., 1988). Ligation reactions were used in transforming ultra-competent Escherichia coli DH5αF′ cells (Inoue et al., 1990; Tang et al., 1994). DNA from 105 individual cosmids was prepared, digested with HindIII, run on a 1% agarose gel, and blotted to duplicate filters. The relative position of each cosmid was determined by comparing the restriction patterns and the hybridization patterns of several probes. Overlapping subclones were selected and transformed individually into se mutant plants. After selection on kanamycin-containing solid media, the phenotypes were noted. All transformants had a se-like phenotype except for those transformed with four overlapping subclones, which rescued the mutant phenotypes. The region of overlap between these cosmids was sequenced from cosmids directly or after subcloning. The region containing the SE gene was PCR-amplified from se mutant DNA, and pooled products from 10 independent reactions were purified and sequenced. Prior to publication, the region containing the SE gene was also sequenced by the Arabidopsis Genome Initiative (Lin et al., 1999; GenBank accession number AC005623). The determined genomic sequences were identical, although our SE cDNA sequence differed from the predicted At2g27100 mRNA sequence at the position of the splice-site acceptor site of the sixth intron.
cDNA Isolation
Expressed sequence tags (ESTs) corresponding to the SE gene (accession numbers T42213 and AA585883) and from a closely related gene from sugarcane (accession numbers AA080638 and AA080660) were obtained from ABRC and from D. Carson (South African Sugar Association Experiment Station, Mount Edgecombe, South Africa), respectively. These cDNAs were labeled and used as probes for screening λZAPII-based cDNA libraries derived from either Arabidopsis hypocotyls (Kieber et al., 1993) or maize leaves (Fisk et al., 1999) provided by ABRC and Alice Barkan (University of Oregon, Eugene), respectively. Ten positive Arabidopsis cDNA clones and seven maize cDNA clones were excised according to the manual provided by Stratagene (La Jolla, CA) and compared on agarose gels after XhoI-XbaI double digestion. The longest Arabidopsis cDNA was sequenced, and the 5′ end of each maize cDNA was sequenced initially. Two classes of maize clone sequences emerged after sequence comparisons, and the longest representative from each locus was used to finish sequencing. Simultaneously, the sugarcane cDNA was also sequenced. To indicate their paralogous nature, the names ZmSE1A and ZmSE1B were used for the maize loci. The GenBank accession numbers for the cDNA sequences are AF311221 through AF311224.
Protein Sequence Alignments
The GenBank accession numbers for the maize EST sequences used in Figure 5A are AI987373 and AW056145 for ZmSE2A; and AW330612 for ZmSE2B. Amino acid sequences were extracted from consensus EST, and a frameshift was introduced to the ZmSE2B sequence to preserve the open reading frame. The GenBank accession numbers (and references) for the Schizosaccharomyces pombe, Drosophila, and hamster Asr2 protein sequences are CAA22180 (S. pombe Sequencing Group at the Sanger Centre), AAF57281 (Adams et al., 2000), and AAA83777 (Rossman and Wang, 1999), respectively. The Caenorhabditis elegans protein sequence was deduced after manually splicing the genomic sequence (AC006627; Genome Sequencing Center, Washington University School of Medicine) on the basis of EST sequences, and the human protein sequence was deduced after manually splicing the chromosome 7 working draft sequences (NT007969; International Human Genome Sequencing Consortium, 2001) on the basis of the DKFZp564H2023 cDNA sequence (AL096723).
Protein sequences were initially aligned using the PILEUP program (Genetics Computer Group, Madison, WI) and then manually adjusted. Figures were generated using MacBoxshade (Michael D. Baron, Institute for Animal Health, UK) and modified using Deneba Canvas (Miami, FL).
In Situ Detection of the SE mRNA
The tissue preparation and detection protocols followed for the in situ detection of SE mRNA were based on those described previously (Long and Barton, 1998; http://www.wisc.edu/genetics/CATG/barton/index.html) except that a 3-hr prehybridization step replaced the dehydration and drying step (Larkin et al., 1993). Digoxigenin-labeled riboprobes were synthesized from each strand of the full-length SE cDNA insert by in vitro transcription by the T7 (anti-sense strand) and T3 (sense strand) RNA polymerases. Probe yield was determined by spot test (relative to a labeled DNA standard), and the probe was diluted to 50 ng/mL for hybridization.
Misexpressing the SE Gene
To replace the β-glucuronidase coding sequence of the pBI121 plant expression vector (Clontech Co., Palo Alto, CA) with the SE coding sequence, a BamHI restriction site was added 5′ from the SE initiation codon. A primer was designed that replaced nucleotides aac-gaaATGgcc of the cDNA (initiation codon in capital letters) to ggatcc- ATGgcc, creating a BamHI (as well as a NcoI) site. A PCR fragment containing this mutated sequence was cloned into a pBluescript II vector (Stratagene, La Jolla, CA) and sequenced. The full-length coding sequence was reconstituted using a BsaBI site 11 nucleotides 3′ from the initiation codon. The resulting cDNA was inserted into the pBI121 vector for introduction into plants. Transformed plants were identified by selection on kanamycin-containing media.
Reverse Transcriptase–PCR
Mini-preparations of total leaf RNA were performed as previously described (Carpenter and Simon, 1998). M-MLV reverse transcriptase (RT) (Promega, Madison, WI) and a poly (dT) primer were used to generate the cDNA templates for PCR. PCR conditions were the same as described above except that only 10 cycles were performed prior to gel fractionation. To control for differences in loading, two ubiquitously expressed genes were detected using the same cDNA preparations: the ROC1 cyclophilin gene (Lippuner et al., 1994) and the Arabidopsis ortholog of the tobacco β-ATPase gene (Boutry and Chua, 1985; Kelly et al., 1990). The PCR primers used were the following: SE, ctgttgtctccggcctttag and ctctagccctgtcttgtctac; ROC1, gatctacgggagcaagttcg and ttctcgatggcctttaccac; and β-ATPase, gtg-cccgtaagatcc-agaga and tcttctctgcctttgcaacc.
Acknowledgments
We thank Kris Soebroto for assistance in mapping, Yanling Wang of the Institute of Molecular Biology Sequencing Facility for sequencing, and Karen Hicks, Steve Clark, and Sang Ho Jeong for helpful comments on the manuscript. We thank Hidetaka Kaya and Takashi Araki for sharing unpublished results and the Arabidopsis Biological Resource Center, Eva Sundberg and George Coupland, Deborah Lee Carson, and Alice Barkan for supplying materials. We are indebted to George Rédei for sharing his mutant collection. This research was supported by National Science Foundation Grant MCB-9808208 and the U.S.-Israel Binational Agricultural Research and Development (BARD) Agency Grant US-2964-97. M.J.P. was supported by predoctoral training grants from the National Science Foundation (DBI-9413223) and National Institutes of Health (GM07413).
References
- Adams, C.R., and Kamakaka, R.T. (1999). Chromatin assembly: Biochemical identities and genetic redundancy. Curr. Opin. Genet. Dev. 9, 185–190. [DOI] [PubMed] [Google Scholar]
- Adams, M.D., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I., Cork, A., Williamson, R.W., and Gorst, J.R. (1995). STUNTED PLANT 1, a gene required for expansion in rapidly elongating but not in dividing cells and mediating root growth responses to applied cytokinin. Plant Physiol. 107, 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1195–1197. [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Berg, J.M., and Shi, Y. (1996). The galvanization of biology: A growing appreciation for the roles of zinc. Science 271, 1081–1085. [DOI] [PubMed] [Google Scholar]
- Berná, G., Robles, P., and Micol, J.L. (1999). A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152, 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan, W., Cervera, M.T., Delarue, M., Beeckman, T., Dewitte, W., Bellini, C., Caboche, M., Van Onckelen, H., Van Montagu, M., and Inze, D. (1995). superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7, 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., and Benning, C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17, 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutry, M., and Chua, N.-H. (1985). A nuclear gene encoding the beta subunit of the mitochondrial ATP synthase in Nicotiana plumbaginifolia. EMBO J. 4, 2159–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. [DOI] [PubMed] [Google Scholar]
- Carpenter, C.D., and Simon, A.E. (1998). Preparation of RNA. In Arabidopsis Protocols, J.M. Martínez-Zapater and J. Salinas, eds (Totowa, NJ: Humana Press, Inc.), pp. 85–89.
- Champagne, M.M. (1998). Genetic Regulation of Shoot Development in Arabidopsis thaliana. Ph.D. Dissertation (Eugene, OR: University of Oregon).
- Chaudhury, A.M., Ming, L., Miller, C., Craig, S., Dennis, E.S., and Peacock, W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94, 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, J.C., and Sussex, I.M. (1996). Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 111, 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S., Creelman, R.A., Mullet, J.E., and Wing, R.A. (1995). Construction and characterization of a bacterial artificial chromosome library of Arabidopsis thaliana. Plant Mol. Biol. Rep. 13, 124–128. [Google Scholar]
- Clarke, J.H., Tack, D., Findlay, K., Van Montagu, M., and Van Lijsebettens, M. (1999). The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J. 20, 493–501. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–744. [DOI] [PubMed] [Google Scholar]
- Cockcroft, C.E., den Boer, B.G., Healy, J.M., and Murray, J.A. (2000). Cyclin D control of growth rate in plants. Nature 405, 575–579. [DOI] [PubMed] [Google Scholar]
- Dean, C., Sjodin, C., Page, T., Jones, J., and Lister, C. (1992). Behavior of the maize transposable element Ac in Arabidopsis thaliana. Plant J. 1, 69–81. [Google Scholar]
- Elliott, R.C., Betzner, A.S., Huttner, E., Oakes, M.P., Tucker, W.Q., Gerentes, D., Perez, P., and Smyth, D.R. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmler, K., and Schäfer, E. (1997). Maternal effect on embryogenesis in tobacco overexpressing rice phytochrome A. Bot. Acta 110, 1–8. [Google Scholar]
- Enomoto, S., and Berman, J. (1998). Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 12, 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto, S., McCune-Zierath, P.D., Gerami-Nejad, M., Sanders, M.A., and Berman, J. (1997). RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 11, 358–370. [DOI] [PubMed] [Google Scholar]
- Farkas, G., Gausz, J., Galloni, M., Reuter, G., Gyurkovics, H., and Karch, F. (1994). The Trithorax-like gene encodes the Drosophila GAGA factor. Nature 371, 806–808. [DOI] [PubMed] [Google Scholar]
- Fisk, D.G., Walker, M.B., and Barkan, A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18, 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E.M., and Coupland, G. (1997). A polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51. [DOI] [PubMed] [Google Scholar]
- Groot, E.P., and Meicenheimer, R.D. (2000. a). Comparison of leaf plastochron index and allometric analyses of tooth development in Arabidopsis thaliana. J. Plant Growth Regul. 19, 77–89. [DOI] [PubMed] [Google Scholar]
- Groot, E.P., and Meicenheimer, R.D. (2000. b). Short-day-grown Arabidopsis thaliana satisfies the assumptions of the plastochron index as a time variable in development. Int. J. Plant Sci. 161, 749–756. [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280, 446–450. [DOI] [PubMed] [Google Scholar]
- Hempel, F.D., and Feldman, L.J. (1994). Bi-directional inflorescence development in Arabidopsis thaliana: Acropetal initiation of flowers and basipetal initiation of paraclades. Planta 192, 276–286. [Google Scholar]
- Hoppe, B.L., Conti-Tronconi, B.M., and Horton, R.M. (1992). Gel-loading dyes compatible with PCR. Biotechniques 12, 679–680. [PubMed] [Google Scholar]
- Inoue, H., Nojima, H., and Okayama, H. (1990). High efficiency transformation of Escherichia coli with plasmids. Gene 96, 23–28. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Kaufman, P.D., Kobayashi, R., and Stillman, B. (1997). Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 11, 345–357. [DOI] [PubMed] [Google Scholar]
- Kaya, H., Shibahara, K., Taoka, K., Iwabuchi, M., Stillman, B., and Araki, T. (2001). FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104, 131–142. [DOI] [PubMed] [Google Scholar]
- Kelly, A.J., Zagotta, M.T., White, R.A., Chang, C., and Meeks-Wagner, D.R. (1990). Identification of genes expressed in the tobacco shoot apex during the floral transition. Plant Cell 2, 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter, R.A., and Poethig, R.S. (1998). The specification of leaf identity during shoot development. Annu. Rev. Cell Dev. Biol. 14, 373–398. [DOI] [PubMed] [Google Scholar]
- Kieber, J.J., Rothenberg, M., Roman, G., Feldmann, K.A., and Ecker, J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72, 427–441. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Yadegari, R., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell 11, 1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue, T., Ohad, N., Yadegari, R., Hannon, M., Dinneny, J., Wells, D., Katz, A., Margossian, L., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher, K.M., Chow, H., Reiser, L., and Fischer, R.L. (1996). The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug, A., and Schwabe, J.W. (1995). Protein motifs 5. Zinc fingers. FASEB J. 9, 597–604. [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Larkin, J.C., Oppenheimer, D.G., Pollock, S., and Marks, M.D. (1993). The Arabidopsis GLABROUS1 gene requires downstream sequences for function. Plant Cell 5, 1739–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, J.C., Oppenheimer, D.G., Lloyd, A.M., Paparozzi, E.T., and Marks, M.D. (1994). Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA genes in Arabidopsis trichome development. Plant Cell 6, 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs, P., Dockx, J., Kronenberger, J., and Traas, J. (1998). MGOUN1 and MGOUN2: Two genes required for primordium initiation at the shoot apical and floral meristems in Arabidopsis thaliana. Development 125, 1253–1260. [DOI] [PubMed] [Google Scholar]
- Leyser, H.M.O., and Furner, I.J. (1992). Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development 116, 397–403. [Google Scholar]
- Lin, X., et al. (1999). Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402, 761–768. [DOI] [PubMed] [Google Scholar]
- Lippuner, V., Chou, I.T., Scott, S.V., Ettinger, W.F., Theg, S.M., and Gasser, C.S. (1994). Cloning and characterization of chloroplast and cytosolic forms of cyclophilin from Arabidopsis thaliana. J. Biol. Chem. 269, 7863–7868. [PubMed] [Google Scholar]
- Liu, Z., and Meyerowitz, E.M. (1995). LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121, 975–991. [DOI] [PubMed] [Google Scholar]
- Long, D., Martin, M., Sundberg, E., Swinburne, J., Puangsomlee, P., and Coupland, G. (1993). The maize transposable element system Ac/Ds as a mutagen in Arabidopsis: Identification of an albino mutation induced by Ds insertion. Proc. Natl. Acad. Sci. USA 90, 10370–10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., and Barton, M.K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125, 3027–3035. [DOI] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Koltunow, A., Dennis, E.S., Peacock, W.J., and Chaudhury, A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn, K., Fernandez, A., Aida, M., Sedbrook, J., Tasaka, M., Masson, P., and Barton, M.K. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126, 469–481. [DOI] [PubMed] [Google Scholar]
- Martínez-Zapater, J.M., Jarillo, J.A., Cruz-Alverez, M., Roldán, M., and Salinas, J. (1995). Arabidopsis late-flowering fve mutants are affected in both vegetative and reproductive development. Plant J. 7, 543–551. [Google Scholar]
- Matzke, A.J., and Matzke, M.A. (1998). Position effects and epigenetic silencing of plant transgenes. Curr. Opin. Plant Biol. 1, 142–148. [DOI] [PubMed] [Google Scholar]
- Muramoto, T., Kohchi, T., Yokota, A., Hwang, I., and Goodman, H.M. (1999). The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 11, 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant 15, 473–497. [Google Scholar]
- Odell, J.T., Nagy, F., and Chua, N.H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313, 810–812. [DOI] [PubMed] [Google Scholar]
- Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11, 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski, N.E., Martin, F.B., and Ausubel, F.M. (1988). Specialized binary vector for plant transformation: Expression of the Arabidopsis thaliana AHAS gene in Nicotiana tabacum. Nucleic Acids Res. 16, 10765–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532. [DOI] [PubMed] [Google Scholar]
- Pedone, P.V., Ghirlando, R., Clore, G.M., Gronenborn, A.M., Felsenfeld, G., and Omichinski, J.G. (1996). The single Cys2-His2 zinc finger domain of the GAGA protein flanked by basic residues is sufficient for high-affinity specific DNA binding. Proc. Natl. Acad. Sci. USA 93, 2822–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett, F.B., Champagne, M.M., and Meeks-Wagner, D.R. (1996). Temperature-sensitive mutations that arrest Arabidopsis shoot development. Development 122, 3799–3807. [DOI] [PubMed] [Google Scholar]
- Pillus, L., and Rine, J. (1989). Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell 59, 637–647. [DOI] [PubMed] [Google Scholar]
- Ptashne, M. (1992). A Genetic Switch: Phage λ and Higher Organisms (Cambridge, MA: Cell Press, Blackwell Scientific Publications).
- Ray, S., Golden, T., and Ray, A. (1996). Maternal effects of the short integument mutation on embryo development in Arabidopsis. Dev. Biol. 180, 365–369. [DOI] [PubMed] [Google Scholar]
- Rédei, G.P., and Hirono, Y. (1964). Linkage studies. Arabidopsis Info. Serv. 1, 9–10. [Google Scholar]
- Reiser, L., Sánchez-Baracaldo, P., and Hake, S. (2000). Knots in the family tree: Evolutionary relationships and functions of knox homeobox genes. Plant Mol. Biol. 42, 151–166. [PubMed] [Google Scholar]
- Reiter, R.S., Young, R.M., and Scolnik, P.A. (1992). Genetic linkage of the Arabidopsis genome: Methods for mapping with recombinant inbreds and random amplified polymorphic DNAs (RAPDs). In Methods in Arabidopsis Research, C. Koncz, N.-H. Chua, and J. Schell, eds (River Edge, NJ: World Scientific Press, Inc.), pp. 170–190.
- Roe, J.L., Rivin, C.J., Sessions, R.A., Feldmann, K.A., and Zambryski, P.C. (1993). The Tousled gene in A. thaliana encodes a protein kinase homolog that is required for leaf and flower development. Cell 75, 939–950. [DOI] [PubMed] [Google Scholar]
- Rossman, T.G., and Wang, Z. (1999). Expression cloning for arsenite-resistance resulted in isolation of tumor-suppressor fau cDNA: Possible involvement of the ubiquitin system in arsenic carcinogenesis. Carcinogenesis 20, 311–316. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsh, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Press).
- Serrano-Cartagena, J., Robles, P., Ponce, M.R., and Micol, J.L. (1999). Genetic analysis of leaf form mutants from the Arabidopsis Information Service collection. Mol. Gen. Genet. 261, 725–739. [DOI] [PubMed] [Google Scholar]
- Smith, S., and Stillman, B. (1989). Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58, 15–25. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves, T.A., and Sussex, I.M. (1989). Patterns in Plant Development, 2nd ed. (Cambridge, UK: Cambridge University Press).
- Tang, X., Nakata, Y., Li, H.-O., Zhang, M., Gao, H., Fujita, A., Sakatsume, O., Ohta, T., and Yokayama, K. (1994). The optimization of preparations of competent cells for transformation of E. coli. Nucleic Acids Res. 22, 2857–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer, A., Bollman, K.M., and Poethig, R.S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124, 645–654. [DOI] [PubMed] [Google Scholar]
- Timmermans, M.C., Hudson, A., Becraft, P.W., and Nelson, T. (1999). ROUGH SHEATH2: A myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284, 151–153. [DOI] [PubMed] [Google Scholar]
- Tsiantis, M., Schneeberger, R., Golz, J.F., Freeling, M., and Langdale, J.A. (1999). The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284, 154–156. [DOI] [PubMed] [Google Scholar]
- van der Veen, J.H., and Wirtz, P. (1968). EMS-induced genic male sterility in Arabidopsis thaliana: A model selection experiment. Euphytica 17, 371–377. [Google Scholar]
- Vernon, D.M., and Meinke, D.W. (1995). Late embryo-defective Mutants of Arabidopsis. Dev. Genet. 16, 311–320. [Google Scholar]
- Vielle-Calzada, J.P., Thomas, J., Spillane, C., Coluccio, A., Hoeppner, M.A., and Grossniklaus, U. (1999). Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13, 2971–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites, R., and Hudson, A. (1995). phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121, 2143–2154. [Google Scholar]
- Waites, R., Selvadurai, H.R., Oliver, I.R., and Hudson, A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93, 779–789. [DOI] [PubMed] [Google Scholar]
- Wilkins, R.C., and Lis, J.T. (1997). Dynamics of potentiation and activation: GAGA factor and its role in heat shock gene regulation. Nucleic Acids Res. 25, 3963–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L.P., Simon, E.J., Trotochaud, A.E., and Clark, S.E. (2000). POLTERGEIST functions to regulate meristem development downstream of the CLAVATA loci. Development 127, 1661–1670. [DOI] [PubMed] [Google Scholar]