Abstract
Eukaryotic translation initiation factor 4E (eIF4E) binds to the cap structure at the 5′ end of mRNAs and is a critical target for the control of protein synthesis. eIF4E is phosphorylated in many systems in response to extracellular stimuli, but biochemical evidence to date has been equivocal as to the biological significance of this modification. Here we use a genetic approach to this problem. We show that, in Drosophila melanogaster, homozygous eIF4E mutants arrest growth during larval development. In Drosophila eIF4EI, Ser251 corresponds to Ser209 of mammalian eIF4E, which is phosphorylated in response to extracellular signals. We find that, in vivo, eIF4EI Ser251 mutants cannot incorporate labeled phosphate. Furthermore, transgenic Drosophila organisms expressing eIF4ESer251Ala in an eIF4E mutant background have reduced viability. Escapers develop more slowly than control siblings and are smaller. These genetic data provide evidence that eIF4E phosphorylation is biologically significant and is essential for normal growth and development.
Eukaryotic translation initiation factor 4E (eIF4E) is a rate-limiting component of translation initiation, and its activity is tightly regulated in cells (13, 35). Regulation of eIF4E activity is critical to normal cell growth, as overexpression of eIF4E in rodent cells is oncogenic (22), while injection of eIF4E into quiescent NIH 3T3 cells induces DNA synthesis (38). In addition, Saccharomyces cerevisiae cells carrying the temperature-sensitive eIF4E allele cdc33ts4-2 arrest at the G1-to-S transition of the cell cycle (3) at the nonpermissive temperature, further implicating eIF4E in the regulation of proliferation.
eIF4E is a subunit of complex eIF4F, which associates with the 5′ end of the mRNA and facilitates the binding of the small ribosomal subunit and associated factors. In mammals, eIF4F consists of three subunits: eIF4E, eIF4A, and eIF4G (35). eIF4E binds to the 7-methyl-guanosine cap structure at the 5′ end of the mRNA. The activity of eIF4E is regulated by two known mechanisms: the inhibitory eIF4E-binding proteins (4E-BPs) control the availability of eIF4E by competing for its binding with eIF4G (15, 25), while phosphorylation of eIF4E at a conserved serine is hypothesized to control its mRNA cap-binding activity (35).
Unlike that of the 4E-BPs, the function of eIF4E phosphorylation is poorly understood. Unphosphorylated eIF4E can stimulate translation in vitro and can bind the mRNA cap or cap analogues, suggesting that phosphorylation is not strictly required for eIF4E function (35). However, when translation activity is altered by treatments with various extracellular stimuli, the phosphorylation state of eIF4E changes; in most cases, increased eIF4E phosphorylation correlates with increased translational activity. In contrast, eIF4E is hypophosphorylated during mitosis when the translation rate of mRNAs is low (1), and various cellular stresses such as heat shock and viral infection are correlated with reduced eIF4E phosphorylation (35). The major phosphorylation site of mammalian eIF4E is Ser209 (9, 43). Structural studies suggest that phosphorylation at Ser209 might allow tighter binding of the mRNA cap by formation of a salt bridge with a lysine residue on the other side of the postulated mRNA trajectory, thereby clamping the mRNA (26).
Work in invertebrate systems supports a link between eIF4E phosphorylation and translation efficiency. An antibody specific to the phosphorylated form of Aplysia californica eIF4E was used to show a significant correlation between translation rates and increases in eIF4E phosphorylation in ganglion preparations (8). In Drosophila melanogaster, a gene encoding eIF4E was identified and mapped to polytene chromosome region 67A on the left arm of chromosome 3 (17, 21). As for mammals, the phosphorylation of Drosophila eIF4E decreases upon heat shock concomitant with a decrease in translation rates (7).
These correlations between increased eIF4E phosphorylation and elevated growth rates suggest that phosphorylation is important for the regulation of eIF4E activity. To test this idea, it was critical to examine the relevance of eIF4E phosphorylation in a genetically tractable multicellular organism. To do this, we identified the major phosphorylation site of Drosophila eIF4E and found it to correspond to the site that is phosphorylated in the mammalian protein. By mutating this site, we demonstrated that phosphorylation of eIF4E and the integrity of the serine residue that corresponds to the major phosphorylation site in mammalian eIF4E are necessary for the efficient growth and development of Drosophila.
MATERIALS AND METHODS
Fly work.
Alleles l(3)589/11 (eIF4E589/11) and l(3)715/13 (eIF4E715/13) were provided by Kim Kaiser and originated in a screen for lines with lethal alleles on the third chromosome (6). All other strains were provided by the Bloomington Stock Center. Phenotypic characterization of the larval growth defect produced by eIF4E mutant alleles was performed as previously described (4, 12, 27) with some modifications. We used Tubby (Tb) on the TM6B Tb balancer to identify larvae with a wild-type copy of eIF4E, and for the larval growth assays we examined hemizygotes for an eIF4E allele and a deficiency that includes eIF4E to rule out the effects of unknown second-site recessive mutations. The deficiencies used were Df(3L)AC1/TM6B Tb and Df(3L)29A6/TM6B Tb. Embryos were collected on standard apple juice egg lay medium for 1 to 2 h at 24°C. The living Tb+ (eIF4E transheterozygotes) and Tb− (control siblings) flies were counted at 24-h intervals. Control experiments with wild-type strain Oregon-R (Ore-R) were performed in parallel, with results identical to those for the TM6B Tb control siblings.
P-element constructs and generation of transgenic flies by germ line transformation.
An 8.9-kb SpeI genomic fragment that includes eIF4E was obtained from a genomic clone (21) and was subcloned into pCaSpeR-4. This fragment contains approximately 4.9 kb of 5′ flanking DNA upstream of eIF4E and about 1.0 kb of 3′ flanking DNA. Codon 251 (TCC) was changed to GCC (Ser251Ala) and GAC (Ser251Asp) using Pfu high-fidelity polymerase, and the changes were verified by sequencing. The three constructs, referred to as P{eIF4Ewt}, P{eIF4ESer251Ala}, and P{eIF4ESer251Asp}, were transformed into yw flies by standard germ line transformation techniques (39) using the mini-white+ selection marker and the pTurbo helper plasmid as the source of transposase.
Antisera.
Protein blots were probed using a rabbit polyclonal antiserum generated against a peptide derived from the N-terminal sequence of eIF4EI (amino acid sequence MQSDFHRMKNFANPKSMF). The eIF4EI serum was affinity purified against the peptide and used at a dilution of 1:1,000 in all our experiments. An affinity-purified eIF4E antiserum, directed against the whole protein (37), was used at a 1:1,000 dilution.
In vivo metabolic labeling of eIF4E.
Twenty pairs of Drosophila ovaries were dissected into 1 ml of phosphate-free Schneider's cell culture medium (Biofluids) and were incubated for 2 h at room temperature in the presence of 0.1 mCi of [32P]orthophosphate. The ovaries were then washed three times with phosphate-buffered saline, homogenized in 1 ml of lysis buffer (10% glycerol, 50 mM Tris-HCl [pH 7.5], 60 mM KCl, 2 mM CDTA [trans-1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid], 1% Triton X-100, 2 mM dithiothreitol, 50 mM β-glycerophosphate, 5 mM NaF, 0.1 mM NaVO3), and extracts were frozen at −20°C until immunoprecipitations were performed. Products of immunoprecipitations using the eIF4EI antiserum were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Xymotech). Membranes were autoradiographed and analyzed by Western blotting.
Analysis of adult eyes.
Whole flies were dehydrated in an ethanol series. The ethanol was then gradually replaced with Freon-113 by incubation in Freon-113-ethanol mixtures containing increasing concentrations of Freon-113 in 24-h increments. Flies were mounted, and scanning electron microscopy was performed to obtain photographs of eyes for five individuals of each genotype and gender examined. All micrographs were obtained at identical magnifications (160×). Analysis of individual ommatidium areas was performed by scanning the micrographs into Adobe Photoshop and using the histogram function. The ommatidium size for each compound eye is an average of the areas for five ommatidia near the center of the eye. Ommatidia were counted manually from photographs of individual eyes, and the numbers were averaged (n = 5 for each genotype and gender examined).
Flow cytometry.
Larvae from 2-h collections were aged at 24°C for a total of 116 h after egg deposition. To compensate for the developmental delay observed with eIF4ESer251Ala, larvae of this genotype were aged longer and used when they reached the third-instar wandering stage. Wing imaginal disks were dissected in Schneider's medium (Gibco-BRL) and subjected to flow cytometry as previously described (29) using a Becton Dickinson FACScan. Data were analyzed using WinMDI, version 2.8, software.
RESULTS
Isolation and molecular characterization of eIF4E mutant alleles.
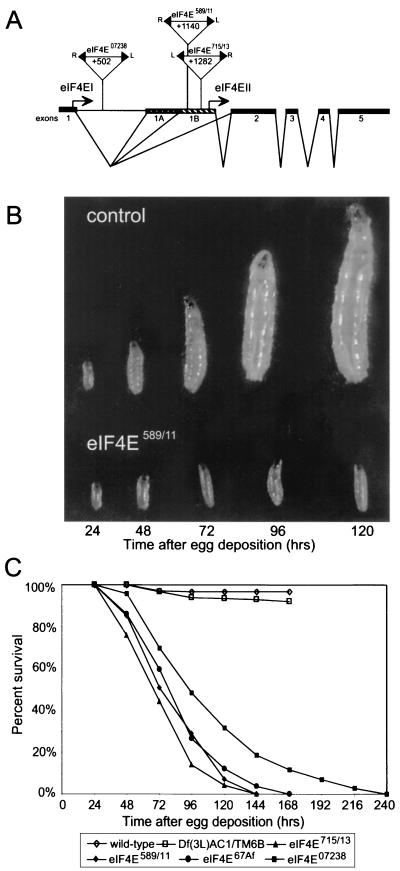
eIF4E mutants were identified by probing a plasmid rescue library generated from lines with lethal alleles on the third chromosome (14) with a radiolabeled eIF4EI cDNA (21). Plasmids corresponding to three P-element-lethal lines hybridized to the probe. By sequencing we determined that alleles eIF4E07238, eIF4E589/11, and eIF4E715/13 have P-element insertions at nucleotide positions +502, +1140, and +1282, respectively, all of which are within the large first intron of the gene (Fig. 1A). Nucleotide positions are in accordance with an earlier description of the eIF4E gene (21). These lines all failed to genetically complement one another. Recessive lethal mutant l(3)67Af, generated in a screen for ethyl methane sulfonate-induced lethal lines (24), was also found to be allelic to eIF4E and is referred to as eIF4E67Af. We completely restored viability and fertility to flies carrying the eIF4E alleles by introducing an 8.9-kb SpeI genomic fragment that includes the eIF4E gene (P{eIF4Ewt}).
FIG. 1.
Recessive lethal mutant alleles of eIF4E produce a larval growth arrest phenotype. (A) Schematic representation of the insertion site of the P elements in the eIF4E alleles. Insertion position with respect to the previously published eIF4E genomic sequence is indicated (21). (B) The growth of eIF4E589/11 larvae is arrested at the first-instar larval stage, whereas control siblings continue to develop. eIF4E589/11 and control larvae are shown at 24, 48, 72, 96, and 120 h after egg deposition. Similar growth arrests were observed for all eIF4E alleles examined. (C) Life spans of growth-arrested eIF4E mutants (solid symbols) carrying the different alleles of eIF4E. The survival rates of wild-type larvae and control siblings (open symbols) are also shown.
eIF4E mutants produce a larval growth arrest phenotype.
Since individuals homozygous for lethal alleles of eIF4A are deficient in growth and arrest during larval development (12), we examined individuals carrying our eIF4E alleles for similar phenotypes. The growth defect phenotype differs from simple larval lethality in that growth-defective larvae never reach the normal third-instar larval size, or else reach it after a substantially longer time than wild-type larvae, yet survive for a minimum of 4 days after hatching from the egg (12). Wild-type larvae reach the second larval instar in approximately 24 h and reach the third instar in another 24 h. eIF4E mutants have a larval growth arrest phenotype (Fig. 1B and C). Three alleles (eIF4E715/13, eIF4E589/11, and eIF4E67Af) arrest development in the first-instar larval stage but survive for several days, while eIF4E07238 arrests growth in the second instar. For the weakest allele, eIF4E07238, second-instar-arrested larvae can live up to 10 days after egg deposition (Fig. 1C). In addition, we observed some embryonic lethality with allele eIF4E715/13 (18% of embryos failed to hatch). No other morphological defects were observed for any of the eIF4E alleles.
Drosophila eIF4EI Ser251 mutants cannot be phosphorylated in vivo.
In mammalian cells, Ser209 of eIF4E is the major site of phosphorylation (9, 43). A serine residue in a similar sequence context (Ser251 in eIF4EI) is present near the C terminus of Drosophila eIF4E. To study whether Drosophila eIF4E is phosphorylated on this site in vivo and to study the function of this residue in development, we generated transgenic Drosophila lines expressing eIF4E under the control of its own promoter; in these lines Ser251 was altered to Ala (P{eIF4ESer251Ala}) or Asp (P{eIF4ESer251Asp}).
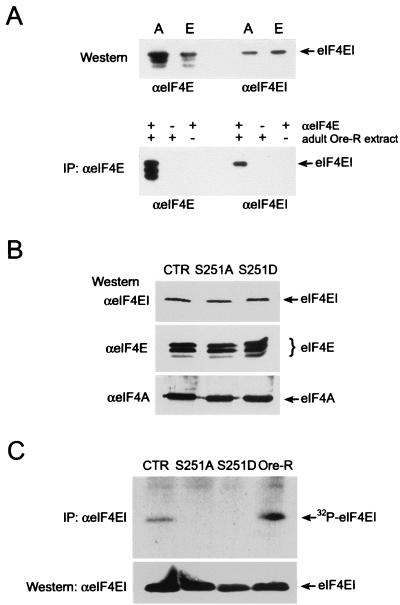
To analyze eIF4E, we used an eIF4EI antiserum that was generated against a peptide limited to the unique N-terminal sequence of this isoform, a region that is highly variable in all eIF4E cognates (20). This antibody binds to the slowest-migrating eIF4E isoform detected by an antiserum against all forms of eIF4E (37) (Fig. 2A). Thus, the eIF4EI antiserum is specific for this isoform. We compared the levels of expression of eIF4E in our transgenic lines and found that P{eIF4ESer251Ala} and P{eIF4ESer251Asp} express eIF4E at levels similar to that for the wild type (Fig. 2B). A Western blot using anti-eIF4A (37) confirms that equivalent amounts of protein are present in lines with each genotype. Thus the phenotypes we describe below cannot be attributed to a dosage effect.
FIG. 2.
Drosophila eIF4EI is phosphorylated on Ser251. (A) Characterization of the affinity-purified antiserum raised and purified against eIF4EI N-terminal peptide MQSDFHRMKNFANPKSMF. (Top) Western blot containing Drosophila adult (lane A) and embryo (lane E) extracts was probed with a general eIF4E antiserum (αeIF4E) or the eIF4EI peptide serum (αeIF4EI). Both antisera were used at 1:1,000 dilutions. (Bottom) Products of immunoprecipitations (IP) performed with αeIF4E were transferred to a nitrocellulose membrane and probed with αeIF4E or αeIF4EI. These data indicate that the eIF4EI antiserum is specific for the slowest-migrating form of eIF4E. (B) Western blot examining the levels of eIF4E (detected by αeIF4E or αeIF4EI) and, as loading control, of eIF4A (detected by αeIF4A [40]) in adult extracts from control siblings (CTR, P{eIF4ESer251Ala} eIF4E589/11/TM3 Sb) and from the eIF4E Ser251 mutants (S251A, P{eIF4ESer251Ala} eIF4E589/11; S251D, P{eIF4ESer251Asp} eIF4E589/11). (C) Immunoprecipitation of eIF4EI from extracts of Drosophila ovaries metabolically labeled with [32P]orthophosphate (top, autoradiography; bottom, Western blot with αeIF4EI). Immunoprecipitations from control sibling (CTR), eIF4ESer251Ala (S251A), eIF4ESer251Asp (S251D), and wild-type (Ore-R) ovary extracts are shown (genotypes are as indicated for panel B).
To assess whether eIF4EI was phosphorylated when Ser251 was replaced by Ala or Asp, we immunoprecipitated eIF4EI from Drosophila ovaries that were metabolically labeled with [32P]orthophosphate (Fig. 2C). While eIF4EI was immunoprecipitated from ovaries from flies of all genotypes examined (Fig. 2C, bottom), eIF4EI was labeled with [32P]orthophosphate only in Ore-R ovaries and in transgenic ovaries expressing wild-type eIF4EI (Fig. 2C, top, lanes Ore-R and CTR, respectively). eIF4EI immunoprecipitated from mutant ovaries, in which the only source of eIF4EI is either the P{eIF4ESer251Ala}or P{eIF4ESer251Asp} transgene, does not detectably incorporate [32P]orthophosphate. These results are most readily interpreted as indicating that Drosophila eIF4EI is phosphorylated in vivo on Ser251, although the alternative, that Ser, and not Ala, at position 251 is required for phosphorylation of eIF4E at a different site, cannot be ruled out.
eIF4ESer251Ala mutants are delayed in development and are small.
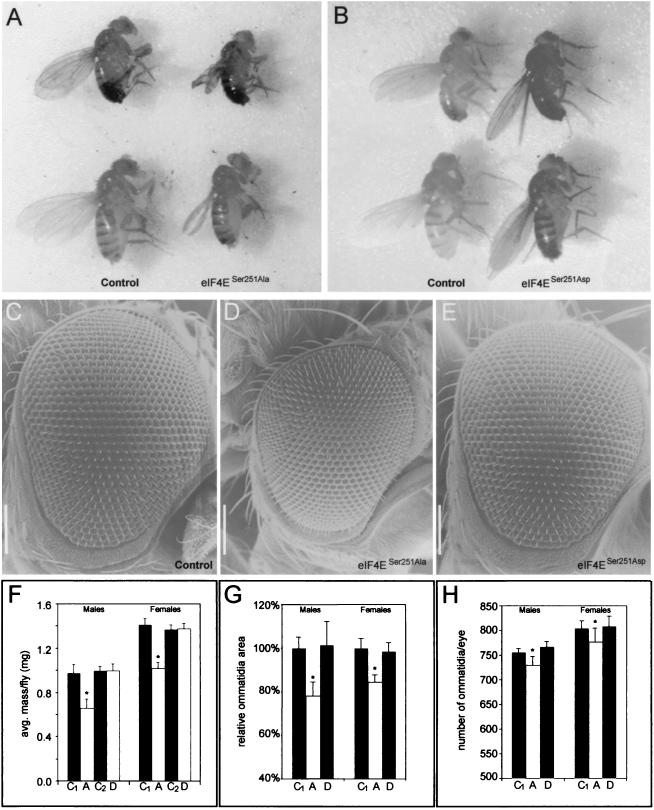
Flies carrying the P{eIF4ESer251Ala} transgene in the background of eIF4E mutant alleles have reduced viability (35% lethality), take longer to develop to adulthood, and have blistered wings (Fig. 3A). The viability of eIF4ESer251Ala-expressing mutants is lower for males than females, and the developmental delay varies depending on the strength of the eIF4E alleles used as genetic background, varying from 1 to 2 additional days for weaker alleles (eIF4E07238 and eIF4E67Af) to 3 to 4 additional days for stronger alleles (eIF4E589/11 and eIF4E715/13). Strikingly, when P{eIF4ESer251Ala} is placed in the background of stronger eIF4E alleles, such as eIF4E589/11, surviving females and males are smaller and their weights are reduced by 29 (one-tailed t test; P < 0.001) and 32% (one-tailed t test; P < 0.01), respectively, compared to those of control siblings grown under identical conditions (Fig. 3F). Although smaller, the body parts of eIF4ESer251Ala flies are appropriately proportioned, and no patterning defects are observed except for the wings. Wings of eIF4ESer251Ala flies are blistered, smaller than those of the wild type, and occasionally clipped.
FIG. 3.
eIF4ESer251Ala mutant flies have reduced growth. Rescue of the eIF4E589/11 allele with the P{eIF4ESer251Ala} transgene results in adult flies with small body size and aberrant wing growth (A), while flies carrying the P{eIF4ESer251Asp} transgene in the background of the eIF4E589/11 allele are similar to controls (B). Males (top row) and females (bottom row) are shown. The control flies, which were grown under conditions identical to those for the transgenic mutants, are siblings of eIF4ESer251Ala (P{eIF4ESer251Ala} eIF4E589/11/TM3 Sb) (A) and eIF4ESer251Asp (P{eIF4ESer251Asp} eIF4E589/11/TM3 Sb) (B) flies. Adult compound eyes of control (C), eIF4ESer251Ala (D), and eIF4ESer251Asp (E) male flies in the background of the eIF4E589/11 allele are also shown. Note the reduction in size of eIF4ESer251Ala mutant eyes compared to those of control and eIF4ESer251Asp flies. The control shown is from a male of genotype P{eIF4ESer251Ala} eIF4E589/11/TM3 Sb, which was grown under conditions identical to those for the eIF4ESer251Ala and eIF4ESer251Asp mutants. Similar results were obtained with female fly eyes (data not shown). Magnification for all electron micrographs, ×160. Bar = 100 μm. (F) Average masses of eIF4ESer251Ala and eIF4ESer251Asp mutants compared to those of their respective control siblings (n = 60). (G and H) Relative areas of individual ommatidia (G) and average numbers of ommatidia per compound eye (H) of males and females (n = 5 individuals for each genotype). C1, control sibling of eIF4ESer251Ala (P{eIF4ESer251Ala} eIF4E589/11/TM3 Sb) mutant; C2, control sibling of eIF4ESer251Asp (P{eIF4ESer251Asp} eIF4E589/11/TM3 Sb) mutant; A, P{eIF4ESer251Ala} eIF4E589/11 mutant; D, P{eIF4ESer251Asp} eIF4E589/11 mutant. ∗, significant P values, as calculated using a one-tailed Student t test comparing the average masses of male and female mutants to those of their control siblings. Data represent means ± standard deviations.
Transgenic lines in which Ser251 was mutated to Asp were generated to test the effects of mimicking constitutive phosphorylation. The P{eIF4ESer251Asp} transgene can fully rescue the lethality of all eIF4E transheterozygote combinations tested (Fig. 3B). No morphological defects or changes in size or weight were observed in whole animals or dissected wings from any of the eIF4ESer251Asp lines (Fig. 3F and data not shown).
Adult eyes of eIF4ESer251Ala mutants have smaller and fewer ommatidia.
The phenotypes of eIF4ESer251Ala mutants suggest that phosphorylation of eIF4E is important for the normal growth of Drosophila. The eIF4ESer251Ala phenotypes are similar to previously described phenotypes produced by genes that influence growth, such as Dmyc and Dras1, and by genes of the insulin signaling pathway (42). These genes influence final body size by affecting the size and number of cells in specific tissues. We thus examined whether cell growth is affected in flies carrying eIF4E alleles rescued by the phosphorylation mutant transgenes.
The Drosophila compound eye is a highly precise hexagonal array of units termed ommatidia. The female wild-type eye is composed of approximately 800 ommatidia, while the male counterpart has on average 50 fewer ommatidia (44). Due to the readily quantifiable architecture and size of the eye, we opted to use this adult structure to examine whether cell size and number were affected in our various eIF4E transgenic lines. While control and eIF4ESer251Asp flies have normal-size eyes (Fig. 3C and E), the compound eyes of transgenic eIF4ESer251Ala mutants are markedly reduced in size (Fig. 3D). The reduction in size of the eIF4ESer251Ala mutant compound eye is caused mostly by a reduction in the area of individual ommatidia and only slightly by a reduction in their number. The average areas of individual ommatidia in the center of the eye in both male and female eIF4ESer251Ala mutants are significantly reduced (males, 206.51 ± 16.16 μm2; females, 246.60 ± 10.80 μm2) compared to those of the ommatidia of control siblings (males, 265.00 ± 13.17 μm2; females, 293.24 ± 12.46 μm2) (Fig. 3G), a difference of 22.1% for males (one-tailed t test; P < 0.001) and 15.9% for females (one-tailed t test; P < 0.001). The areas of individual eIF4ESer251Asp mutant ommatidia (males, 269.25 ± 28.50 μm2; females, 289.02 ± 11.51 μm2) are essentially the same as those of the controls (Fig. 3G). A slight reduction in ommatidium number is also observed in eIF4ESer251Ala mutants (Fig. 3H). This reduction is small but statistically significant for males (729 ± 19 ommatidia in the mutant compared to 759 ± 8 in controls; one-tailed t test; P < 0.01) and females (777 ± 28 ommatidia compared to 805 ± 14 in controls; one-tailed t test; P < 0.05). Again, no difference between the number of ommatidia in eIF4ESer251Asp mutants (males, 766 ± 12 ommatidia; females, 808 ± 21 ommatidia) and in the controls is observed. These data argue that the overall reduction in size of the compound eye observed in eIF4ESer251Ala mutants mostly results from reduced cell size, with a minor contribution from a reduction in cell number.
Wing imaginal disk cells of eIF4ESer251Ala mutants are smaller than wild-type cells.
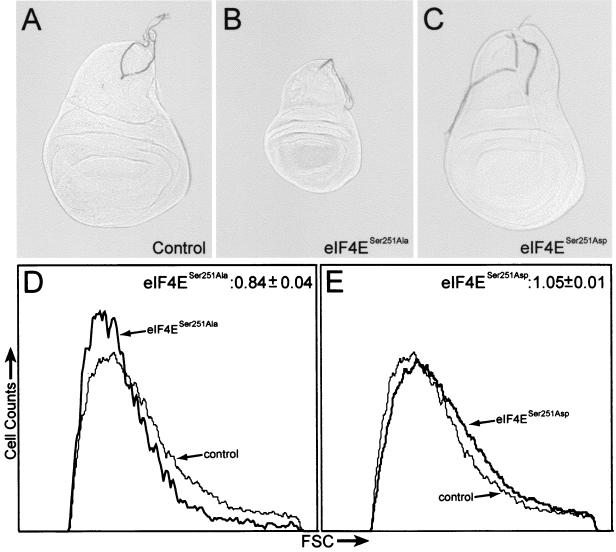
The larval precursors of Drosophila adult structures are the imaginal disks. We examined wing imaginal disks from late-stage third-instar eIF4ESer251Ala larvae and found that they are markedly reduced in size compared to disks from controls and eIF4ESer251Asp mutants (Fig. 4A to C). The decrease in size of the eIF4ESer251Ala wing disk suggests that growth may be reduced at the level of individual wing disk cells. To examine the relative sizes of individual cells from wing disks, imaginal disks were dissociated and their cells were analyzed by flow cytometry (Fig. 4D and E). The forward light scatter value obtained by flow cytometry is a measure of cell size. The mean forward light scatter value of cells from eIF4ESer251Ala mutant imaginal disks is decreased by 16% compared to that of cells from control disks (Fig. 4D). Also, a slight increase in size of 5% was observed for cells from eIF4ESer251Asp mutant disks (Fig. 4E), suggesting that, despite our inability to detect a difference in adult tissues, a mutation that mimics constitutive phosphorylation of eIF4E has a detectable effect on growth during the development of Drosophila wing imaginal disks. We also assessed wing imaginal disks from wild-type, eIF4ESer251Ala, and eIF4ESer251Asp larvae for apoptotic cells and observed no differences among the three genotypes (data not shown). This is consistent with our conclusion that the primary cause for the reduced size of eIF4ESer251Ala wing disks is a reduction in cell size.
FIG. 4.
Phenotypic analysis of wing imaginal disks in the eIF4E phosphorylation mutants. Dissected wing imaginal disks from control sibling P{eIF4ESer251Ala} eIF4E589/11/TM6 Tb (A), P{eIF4ESer251Ala} eIF4E589/11 mutant (B), and the P{eIF4ESer251Asp} eIF4E589/11 mutant larvae. Note the reduction in size of eIF4ESer251Ala mutant imaginal disks compared to control and eIF4ESer251Asp mutant disks. (D and E) Forward light scatter (FSC) comparison of cells dissociated from eIF4ESer251Ala (D) and eIF4ESer251Asp (E) mutants with cells from control disks. Numbers at the top right corner represent ratios of mean FSC values of wing disk cells from phosphorylation mutants compared to the control cells. Control sibling animals (P{eIF4ESer251Ala} eIF4E589/11/TM6 Tb) have wild-type eIF4E and were raised under conditions identical to those for the phosphorylation mutants.
DISCUSSION
In mammals, increased phosphorylation of eIF4E has been correlated with increased cellular proliferation; nevertheless, unphosphorylated eIF4E or bacterially expressed eIF4E can still bind the mRNA cap and initiate translation (28, 35). Thus, biochemical analyses have not resolved the question of the biological importance of eIF4E phosphorylation. Increased phosphorylation of mammalian eIF4E is reported to increase its affinity for mRNA caps in vitro (28). However, the eIF4E preparations used in this study were not recombinant proteins and may not have been homogeneous, in that the unphosphorylated eIF4E might have been associated with 4E-BPs. Also, it was originally believed that the phosphorylation of mammalian eIF4E occurred on Ser53. Mutations in Ser53 were generated to examine the role of eIF4E phosphorylation in mediating the oncogenic transformation of mammalian cells (reviewed in reference [35]). Later studies showed unambiguously that the phosphorylation of eIF4E occurs on Ser209 in vivo (41, 43). The cocrystal structure of eIF4E with m7GDP is consistent with the idea that phosphorylation of eIF4E on Ser209 enhances mRNA binding by forming a salt bridge with Lys159, thereby clamping the mRNA into the cap-binding slot, and does not enhance a direct interaction with the cap structure itself, which is located distantly from Ser209 (26). The structure of eIF4E also shows that Ser53 resides in the hydrophobic core of the protein, and thus alterations in this residue probably affect protein folding rather than phosphorylation (26). Since Ser209 resides on the surface of eIF4E and since the eIF4ESer251Asp mutant can fully replace wild-type eIF4E, we believe that the mutations in Ser251 of the Drosophila protein that we generated do not alter the three-dimensional structure of eIF4E.
In this work, we demonstrate that flies in which eIF4E can no longer be phosphorylated on Ser251 are viable. However, the flies are delayed in development and are smaller than control animals. This genetic work thus provides evidence that Ser251 of eIF4E is important for the normal growth and development of a multicellular organism. We also show that lethal eIF4E mutations result in growth arrest during larval development. Similar phenotypes were previously described for mutations in eIF4A (12). In addition to mutations in eIF4A, mutations in several genes implicated in biosynthesis of proteins and nucleic acids have been shown to produce a larval-growth deficiency. These include mutations in the ribosomal protein S15 gene bonsai, the Myc-regulated DEAD-box RNA helicase gene pitchoune, and the DNA replication regulator gene peter pan (12, 27, 45). eIF4E thus appears to be part of a growing class of genes which when mutated produce a larval growth defect phenotype and which regulate macromolecular synthesis.
Although phenotypes affecting viability and growth were observed for eIF4ESer251Ala, we did not obtain strong phenotypes suggestive of overgrowth in an eIF4E mutant by mimicking phosphorylation by conversion of Ser251 to Asp. The only phenotype that we observed in eIF4ESer251Asp individuals was a small increase in cell size in wing imaginal disks from third-instar larvae, as detected by flow cytometry. We also attempted to increase eIF4E activity by overexpressing the gene using the GAL4-upstream activation sequence system (2) and various drivers. Overexpression of eIF4E in cultured mammalian cells results in increased growth (22, 38). Since in mammalian cells a negative-feedback loop between increased eIF4E activity and 4E-BP hypophosphorylation has been observed (18), overexpression of eIF4EIwt was also performed in the background of a Drosophila 4E-BP (d4E-BP) null mutant allele. Alteration of wild-type d4E-BP expression does not influence growth as flies carrying d4E-BP null alleles (M. Miron and N. Sonenberg, unpublished data) and flies in which d4E-BPwt is overexpressed (29) are indistinguishable from the wild type. We did not observe any overgrowth phenotypes upon overexpression of eIF4EIwt in various tissues in the background of the d4E-BP null allele (P. E. D. Lachance and P. Lasko, unpublished results). The absence of phenotypes upon overexpression of Drosophila eIF4E was also reported elsewhere (46). Since eIF4ESer251Asp or eIF4EIwt overexpression did not result in increased growth, we conclude that Drosophila can physiologically withstand increases in eIF4E activity more readily than decreases. Consistent with this, ectopic expression of a highly active form of d4E-BP has been shown to decrease growth (29). Our inability to obtain an overproliferation phenotype may be explained if our treatments did not increase the activity of eIF4E to a level at which a phenotype can be observed. However, the explanation that we favor is that elevated eIF4E activity results in increased growth and proliferation only if growth-promoting pathways are simultaneously activated. In primary mammalian cell cultures, coexpression of other proto-oncogenes is necessary for eIF4E-mediated transformation (23).
In mammals, the best candidate for the eIF4E kinase is mitogen-activated protein kinase (MAPK)-interacting protein kinase 1 (MNK1), which phosphorylates eIF4E on Ser209 upon activation by either the p44/42 or p38 MAPKs (11, 41). MNK1 physically interacts with eIF4G, bringing it in close proximity to eIF4E in vivo (31, 34). The Drosophila protein most similar to MNK1 is MAPK Lk6 (19). It will be of interest to determine whether Lk6 interacts with Drosophila eIF4G (16) and if disruption of that interaction results in a phenotype similar to that described here for eIF4ESer251Ala. A role for the Ras/MAPK signaling cascade in eIF4E phosphorylation is consistent with the finding that mammalian cells transformed by ras or src have increased eIF4E phosphorylation (10, 36). In this respect, ras has been shown to regulate cellular growth in the Drosophila wing (33). Although ras has never been shown to be upstream of eIF4E phosphorylation in Drosophila, it is possible that a portion of the effect on growth exhibited by Drosophila ras is mediated through changes in eIF4E phosphorylation. Nevertheless, the effects of Drosophila ras are likely to be pleiotropic since genetic manipulations of ras lead to changes in the activity of the Drosophila homologues of myc and cyclin E (33).
In addition to the Ras pathway, the insulin-like signaling pathway in Drosophila, which involves phosphatidylinositol 3-kinase and protein kinase B as intermediates, has been implicated in several studies as a key regulator of organism size (5, 42). In mammalian cells, this signal transduction pathway leads to the activation of kinase FRAP/mTOR, which in turn leads to the activation of translation via at least two mechanisms: the phosphorylation of S6K and of 4E-BP (35). dS6K is a critical component of this pathway in Drosophila as its overexpression can rescue the lethality of dTOR mutants (30, 46). The phenotype that we observe in eIF4E mutants encoding the S251A change most closely resembles that of dS6K mutants, in that the growth defect is predominantly manifested in a reduction in cell size rather than cell number. Moreover, the effects on growth that result from mutations in genes of the insulin pathway appear to be at least in part mediated by the regulation of eIF4E availability, as regulated by 4E-BP, although these proteins lie on a branch of the pathway independent of dS6K (29). Interestingly, insulin signaling may promote cell growth also by activating the Ras/MAPK pathway in Drosophila (5). Therefore, there may be a link in Drosophila between insulin signaling and the effects on growth that are linked to eIF4E phosphorylation.
Acknowledgments
We thank Francis Poulin for critically reading the manuscript.
This work was supported by a research grant from the Canadian Institutes of Health Research (CIHR) to P.L. and N.S., by graduate fellowships from the Natural Sciences and Engineering Research Council of Canada and McGill University to P.E.D.L. and from les Fonds de la recherche en santé du Québec and the Cancer Research Society to M.M. P.L. is a CIHR Investigator. N.S. is a CIHR Distinguished Scientist and a Howard Hughes Medical Institute International Scholar.
REFERENCES
- 1.Bonneau, A. M., and N. Sonenberg. 1987. Involvement of the 24-kDa cap-binding protein in regulation of protein synthesis in mitosis. J. Biol. Chem. 262:11134-11139. [PubMed] [Google Scholar]
- 2.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 3.Brenner, C., N. Nakayama, M. Goebl, K. Tanaka, A. Toh-e, and K. Matsumoto. 1988. CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol. Cell. Biol. 8:3556-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britton, J. S., and B. A. Edgar. 1998. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development 125:2149-2158. [DOI] [PubMed] [Google Scholar]
- 5.Brogiolo, W., H. Stocker, T. Ikeya, F. Rintelen, R. Fernandez, and E. Hafen. 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11:213-221. [DOI] [PubMed] [Google Scholar]
- 6.Deak, P., M. M. Omar, R. D. Saunders, M. Pal, O. Komonyi, J. Szidonya, P. Maroy, Y. Zhang, M. Ashburner, P. Benos, C. Savakis, I. Siden-Kiamos, C. Louis, V. N. Bolshakov, F. C. Kafatos, E. Madueno, J. Modolell, and D. M. Glover. 1997. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogaster: correlation of physical and cytogenetic maps in chromosomal region 86E-87F. Genetics 147:1697-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duncan, R. F., D. R. Cavener, and S. Qu. 1995. Heat shock effects on phosphorylation of protein synthesis initiation factor proteins eIF-4E and eIF-2 alpha in Drosophila. Biochemistry 34:2985-2997. [DOI] [PubMed] [Google Scholar]
- 8.Dyer, J. R., and W. S. Sossin. 2000. Regulation of eukaryotic initiation factor 4E phosphorylation in the nervous system of Aplysia californica. J. Neurochem. 75:872-881. [DOI] [PubMed] [Google Scholar]
- 9.Flynn, A., and C. G. Proud. 1995. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J. Biol. Chem. 270:21684-21688. [DOI] [PubMed] [Google Scholar]
- 10.Frederickson, R. M., K. S. Montine, and N. Sonenberg. 1991. Phosphorylation of eukaryotic translation initiation factor 4E is increased in Src-transformed cell lines. Mol. Cell. Biol. 11:2896-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukunaga, R., and T. Hunter. 1997. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 16:1921-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galloni, M., and B. A. Edgar. 1999. Cell-autonomous and non-autonomous growth-defective mutants of Drosophila melanogaster. Development 126:2365-2375. [DOI] [PubMed] [Google Scholar]
- 13.Gingras, A.-C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 14.Guo, Y., A. Gillan, T. Torok, I. Kiss, J. A. Dow, and K. Kaiser. 1996. Site-selected mutagenesis of the Drosophila second chromosome via plasmid rescue of lethal P-element insertions. Genome Res. 6:972-979. [DOI] [PubMed] [Google Scholar]
- 15.Haghighat, A., S. Mader, A. Pause, and N. Sonenberg. 1995. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 14:5701-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez, G., M. del Mar Castellano, M. Agudo, and J. M. Sierra. 1998. Isolation and characterization of the cDNA and the gene for eukaryotic translation initiation factor 4G from Drosophila melanogaster. Eur. J. Biochem. 253:27-35. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez, G., R. Diez del Corral, J. Santoyo, S. Campuzano, and J. M. Sierra. 1997. Localization, structure and expression of the gene for translation initiation factor eIF-4E from Drosophila melanogaster. Mol. Gen. Genet. 253:624-633. [DOI] [PubMed] [Google Scholar]
- 18.Khaleghpour, K., S. Pyronnet, A. C. Gingras, and N. Sonenberg. 1999. Translational homeostasis: eukaryotic translation initiation factor 4E control of 4E-binding protein 1 and p70 S6 kinase activities. Mol. Cell. Biol. 19:4302-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kidd, D., and J. W. Raff. 1997. LK6, a short lived protein kinase in Drosophila that can associate with microtubules and centrosomes. J. Cell Sci. 110:209-219. [DOI] [PubMed] [Google Scholar]
- 20.Lasko, P. 2000. The Drosophila melanogaster genome: translation factors and RNA binding proteins. J. Cell Biol. 150:F51-F56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavoie, C. A., P. E. D. Lachance, N. Sonenberg, and P. Lasko. 1996. Alternatively spliced transcripts from the Drosophila eIF4E gene produce two different cap-binding proteins. J. Biol. Chem. 271:16393-16398. [DOI] [PubMed] [Google Scholar]
- 22.Lazaris-Karatzas, A., K. S. Montine, and N. Sonenberg. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345:544-547. [DOI] [PubMed] [Google Scholar]
- 23.Lazaris-Karatzas, A., and N. Sonenberg. 1992. The mRNA 5′ cap-binding protein, eIF-4E, cooperates with v-myc or E1A in the transformation of primary rodent fibroblasts. Mol. Cell. Biol. 12:1234-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leicht, B. G., and J. J. Bonner. 1988. Genetic analysis of chromosomal region 67A-D of Drosophila melanogaster. Genetics 119:579-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mader, S., H. Lee, A. Pause, and N. Sonenberg. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15:4990-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcotrigiano, J., A.-C. Gingras, N. Sonenberg, and S. K. Burley. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89:951-961. [DOI] [PubMed] [Google Scholar]
- 27.Migeon, J. C., M. S. Garfinkel, and B. A. Edgar. 1999. Cloning and characterization of peter pan, a novel Drosophila gene required for larval growth. Mol. Biol. Cell 10:1733-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minich, W. B., M. L. Balasta, D. J. Goss, and R. E. Rhoads. 1994. Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc. Natl. Acad. Sci. USA 91:7668-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miron, M., J. Verdu, P. E. D. Lachance, M. J. Birnbaum, P. F. Lasko, and N. Sonenberg. 2001. The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat. Cell Biol. 3:596-601. [DOI] [PubMed] [Google Scholar]
- 30.Montagne, J., M. J. Stewart, H. Stocker, E. Hafen, S. C. Kozma, and G. Thomas. 1999. Drosophila S6 kinase, a regulator of cell size. Science 285:2126-2129. [DOI] [PubMed] [Google Scholar]
- 31.Morino, S., H. Imataka, Y. V. Svitkin, T. V. Pestova, and N. Sonenberg. 2000. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20:468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neufeld, T. P., A. F. de la Cruz, L. A. Johnston, and B. A. Edgar. 1998. Coordination of growth and cell division in the Drosophila wing. Cell 93:1183-1193. [DOI] [PubMed] [Google Scholar]
- 33.Prober, D. A., and B. A. Edgar. 2000. Ras1 promotes cellular growth in the Drosophila wing. Cell 100:435-446. [DOI] [PubMed] [Google Scholar]
- 34.Pyronnet, S., H. Imataka, A.-C. Gingras, R. Fukunaga, T. Hunter, and N. Sonenberg. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 18:270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raught, B., A.-C. Gingras, and N. Sonenberg. 2000. Regulation of ribosomal recruitment in eukaryotes, p. 245-293. In N. Sonenberg, J. Hershey, and M. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Rinker-Schaeffer, C. W., V. Austin, S. Zimmer, and R. E. Rhoads. 1992. Ras transformation of cloned rat embryo fibroblasts results in increased rates of protein synthesis and phosphorylation of eukaryotic initiation factor 4E. J. Biol. Chem. 267:10659-10664. [PubMed] [Google Scholar]
- 37.Sigrist, S. J., P. R. Thiel, D. F. Reiff, P. E. Lachance, P. Lasko, and C. M. Schuster. 2000. Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature 405:1062-1065. [DOI] [PubMed] [Google Scholar]
- 38.Smith, M. R., M. Jaramillo, Y. L. Liu, T. E. Dever, W. C. Merrick, H. F. Kung, and N. Sonenberg. 1990. Translation initiation factors induce DNA synthesis and transform NIH 3T3 cells. New Biol. 2:648-654. [PubMed] [Google Scholar]
- 39.Spradling, A. C., and G. M. Rubin. 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218:341-347. [DOI] [PubMed] [Google Scholar]
- 40.Styhler, S., A. Nakamura, A. Swan, B. Suter, and P. Lasko. 1998. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 125:1569-1578. [DOI] [PubMed] [Google Scholar]
- 41.Waskiewicz, A. J., A. Flynn, C. G. Proud, and J. A. Cooper. 1997. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 16:1909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinkove, D., and S. J. Leevers. 2000. The genetic control of organ growth: insights from Drosophila. Curr. Opin. Genet. Dev. 10:75-80. [DOI] [PubMed] [Google Scholar]
- 43.Whalen, S. G., A. C. Gingras, L. Amankwa, S. Mader, P. E. Branton, R. Aebersold, and N. Sonenberg. 1996. Phosphorylation of eIF-4E on serine 209 by protein kinase C is inhibited by the translational repressors, 4E-binding proteins. J. Biol. Chem. 271:11831-11837. [DOI] [PubMed] [Google Scholar]
- 44.Wolff, T., and D. Ready. 1993. Pattern formation in the Drosophila retina, p. 1277-1325. In M. Bate and A. Martinez-Arias (ed.), The development of Drosophila melanogaster, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Zaffran, S., A. Chartier, P. Gallant, M. Astier, N. Arquier, D. Doherty, D. Gratecos, and M. Semeriva. 1998. A Drosophila RNA helicase gene, pitchoune, is required for cell growth and proliferation and is a potential target of d-Myc. Development 125:3571-3584. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, H., J. P. Stallock, J. C. Ng, C. Reinhard, and T. P. Neufeld. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14:2712-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]