Abstract
The c-fps/fes proto-oncogene encodes a 92-kDa protein tyrosine kinase that is preferentially expressed in myeloid and endothelial cells. Fes is believed to play a role in vascular development and myelopoiesis and in the inflammatory responses of granulocytes and macrophages. To help define the biological role of this kinase and identify its downstream targets, we have developed a gain-of-function allele of Fes that has potent biological activity in myeloid cell progenitors. Introduction of constitutively active Fes into bipotential U937 cells induced the appearance of fully differentiated macrophages within 6 to 12 days. The Fes-expressing differentiated cells became adherent, had distinctive macrophage morphology, and exhibited increased expression of myelomonocytic differentiation markers, including CD11b, CD11c, CD18, CD14, and the macrophage colony-stimulating factor receptor. These cells acquired phagocytic properties and exhibited NADPH oxidase and nonspecific esterase activities, confirming that they were functionally active macrophages. Concomitantly, there was downregulation of the granulocytic marker granulocyte colony-stimulating factor receptor, indicating that the biological activity of Fes was coordinated in a lineage-specific manner. A constitutively active Src did not induce macrophage morphology or upregulation of myelomonocytic markers in U937 cells, suggesting that the biological activity we observed was not a general consequence of expression of an activated nonreceptor tyrosine kinase. Analysis of possible downstream targets of Fes revealed that this kinase activated the ets family transcription factor PU.1, which is essential for macrophage development. Our results strongly implicate Fes as a key regulator of terminal macrophage differentiation and identify PU.1 as a transcription factor that may mediate some of its biological activities in myeloid cells.
The differentiation of myeloid progenitors into granulocytes and monocyte-macrophages is regulated by extracellular ligands such as colony-stimulating factors (CSFs) (i.e., granulocyte CSF [G-CSF], granulocyte-macrophage CSF [GM-CSF], and macrophage CSF [M-CSF]) (56) and adhesion molecules acting on specific receptors at the cell surface (43). These receptors encounter the inductive signals for differentiation in the stromal cell environment, in circulation, and during cell attachment and extravasation en route to inflammatory sites (55). The signaling cascades initiated by these extracellular stimuli culminate with the activation of lineage-specific transcription factors (11, 80), which sets in motion a specific program of gene expression. Very often this involves the sequential activation of tyrosine and serine/threonine protein kinases which carry the proliferation and/or differentiation instructions to the nucleus.
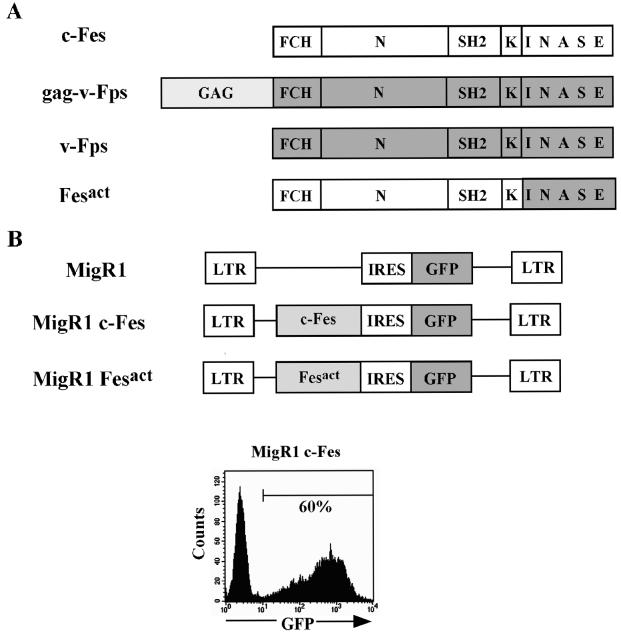
One of the tyrosine kinases that has been implicated in myelopoiesis is Fps/Fes. The c-fps/fes proto-oncogene encodes a 92-kDa cytoplasmic protein tyrosine kinase (17, 19, 53, 67) that has been repeatedly transduced by RNA tumor viruses (28). In the adult, c-fps/fes is preferentially expressed in endothelial cells (23, 26) and in hematopoietic cells of the myeloid lineage with the highest levels present in macrophages (17, 24, 34, 47, 69). This gene is known as fps in the avian system and fes in mammalian cells. The Fes tyrosine kinase contains a unique N-terminal region that harbors a Fes/CIP 4 homology domain (2), an SH2 domain, and a conserved tyrosine kinase domain (Fig. 1). Differently from Src kinases, Fes does not contain a myristylation site for membrane localization, an SH3 domain, or a negative regulatory phosphorylation site at the C terminus equivalent to tyrosine 527 in Src.
FIG. 1.
Construction of constitutively active Fes (Fesact) and retroviral-mediated gene transfer. (A) A constitutively active Fes molecule was constructed by making a chimera between human c-Fes and avian v-Fps as described in Materials and Methods and illustrated in the figure. FCH, Fes/CIP 4 homology domain. (B) c-Fes and Fesact were subcloned into the MigR1 vector upstream of the IRES, and the recombinant DNAs were used for retroviral-mediated gene transfer as described in Materials and Methods. The bottom panel shows a flow cytometry analysis of U937 cells infected with MigR1 c-Fes 48 h after infection. About 60% of the cells in the infected culture were GFP+.
There is much evidence that Fes kinase plays a role in the development and function of myeloid cells. Carmier and Samarut have shown that infection of chicken bone marrow myeloid progenitors with Fujinami sarcoma virus (FSV), which encodes a gag-v-fps oncogene (74), resulted in macrophage differentiation in the absence of M-CSF (10). Introduction of human c-fes into K562 erythroleukemia cells allowed their differentiation into macrophage-like cells, particularly in the presence of tetradecanoyl phorbol acetate (TPA) (85), which normally induces megakaryocytic differentiation in K562 cells (82). The suppression of Fes expression by antisense oligonucleotides in two myeloid cell lines, FDC-P1/MAC-11 and HL-60, abolished TPA-induced macrophage differentiation (50). Fes may also protect cells from apoptosis during differentiation. In HL-60 cells, antisense inhibition of Fes expression followed by treatment with the granulocytic differentiation inducer retinoic acid resulted in apoptosis (51). Fes has also been shown to be downstream of cytokine receptors. Treatment of myeloid cells with GM-CSF and other cytokines can elicit tyrosine phosphorylation of Fes and its association with receptor subunits (8, 29), linking Fes to cytokines that regulate the proliferation, differentiation, and functional responses of these cells.
There is also in vivo evidence that Fes is important for myelopoiesis. A kinase-inactivating mutation of the c-fes locus in mice showed reduced responses of bone marrow-derived macrophages to GM-CSF and lipopolysaccharide, and there were slight elevations of total leukocyte counts and splenomegaly (73). Definitive evidence that Fes plays a nonredundant role in the development of immunocompetent myeloid cells has been obtained from the analysis of Fes null mice (25), which exhibit a more pronounced phenotype. Fes−/− mice have defects in hematopoietic homeostasis and are compromised in their innate immunity. These mice have low numbers of B lymphocytes and increased numbers of granulocytes and monocyte-macrophages, but the latter appear to be functionally defective. Fes−/− macrophages are less adherent than their normal counterparts, and they exhibit hyperproliferative responses to GM-CSF which correlate with increased STAT3 and STAT5 activation. This hyperproliferative response to cytokines may explain the abnormally higher numbers of myeloid cells in Fes null mice. Thus, in the absence of Fes, cytokines can deliver an enhanced mitogenic signal, but full macrophage differentiation is not achieved. This incomplete macrophage maturation is likely to contribute to the defects in innate immunity observed in Fes−/− mice.
Since the physiological inducers of differentiation that activate Fes also activate other tyrosine kinases, it has been very difficult to study the specific role of this kinase in myeloid cell function. For instance, GM-CSF binding to its receptor activates Jak2 (64) and Src family kinases (13, 38, 79) in addition to Fes. Therefore, the downstream events under Fes control cannot be easily studied in a ligand-dependent conditional system. Another reason for the paucity of information on the mechanism of action of this kinase is that in hematopoietic cells, the activity of c-Fes is tightly regulated. Therefore, ectopically expressed c-Fes has little or no biological activity in myeloid cells, with the exception of K562 cells (85), where Fes is normally not expressed. To overcome these limitations, we sought to develop Fes reagents that would be biologically active in myeloid cells.
In this report we describe a gain-of-function allele of Fes that was made by replacement of part of the kinase domain of human c-fes with the corresponding region from avian v-fps. We found that this chimeric Fes molecule was constitutively active and exhibited dramatic biological activity in myeloid cells. Introduction of activated Fes into U937 monocytic precursors induced their differentiation into functional macrophages. We also identified PU.1 as a downstream target of Fes which may act in concert with this kinase to amplify signals for myeloid differentiation.
MATERIALS AND METHODS
Cells and transfection.
The human U937 and HL-60 cell lines were obtained from American Type Culture Collection (Manassas, Va.). These cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, 1% penicillin, and 1% streptomycin. The murine macrophage cell line RAW 264.7 was obtained from Keiko Ozato (National Institutes of Health, Bethesda, Md.) and was maintained in Dulbecco's modified Eagle's medium, 10% fetal bovine serum, and antibiotics. The amphotropic retroviral packaging cell line Phoenix ampho was obtained from Gary Nolan (Stanford University, Palo Alto, Calif.), and was maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, antibiotics, and 1% sodium pyruvate.
RAW 264.7 cells were transiently transfected with 2 μg of total plasmid DNA by using LipofectAmine Plus (Life Technologies, Gaithersburg, Md.) according to the manufacturer's instructions. The transfection efficiency was measured using a promoterless Renilla vector, pRL-0 (Promega, Madison, Wis.) (4). Transactivation of the promoters for tissue factor (TF) and other genes linked to luciferase reporters was assayed 15 h after transfection by using a dual luciferase kit from Promega. Phoenix ampho cells were transiently transfected with retroviral DNAs by the calcium phosphate method using 10 μg of plasmid DNA (22). The preparation of retroviral stocks is described below.
Plasmid DNA and construction of the constitutively active Fes molecule.
The v-fps viral DNA sequence of FSV without gag (pSRα-F36) (20) was obtained from Hidesaburo Hanafusa (The Rockefeller University, New York, N.Y.). The v-fps coding region from pSRα-F36 was excised with NcoI at the 5′ end and SmaI at the 3′ end and subcloned into the elongation factor 1α promoter-based pCEFL expression vector (3), which was obtained from J. Silvio Gutkind (National Institutes of Health, Bethesda, Md.). The murine stem cell virus-derived internal ribosome entry site (IRES)-containing retroviral vector MigR1 (61) was obtained from Warren Pear (University of Pennsylvania, Philadelphia, Pa.).
To construct the active Fes molecule (Fesact), we used a BamHI site in the kinase domain that is conserved between avian v-fps and human c-fes. A BamHI to EcoRI fragment encoding amino acids 610 to 822 in the kinase domain of human c-fes (19, 67) was replaced with the homologous region from v-fps (74), and the c-fes/v-fps chimera was cloned into pCEFL or a modified pCEFL vector that incorporates an in-frame hemagglutinin (HA) epitope tag at the N terminus. The coding sequences of untagged or HA-tagged c-Fes and Fesact were then inserted into MigR1. The mutations of Arg 483 to Leu (SH2 domain mutant, FesactR483L), Lys 590 to Arg (kinase-inactive mutant, FesactK590R), and Tyr 713 to Phe (autophosphorylation site mutant, FesactY713F) in the sequence of Fesact were performed by site-directed mutagenesis using the overlap extension method as described previously (35). These mutations were verified by DNA sequencing (70). pCA3CST-SrcE381G, a plasmid carrying a constitutively active mutant of human src (6), was obtained from Don Fujita (University of Calgary, Calgary, Canada). The coding sequence of srcE381G was subcloned into MigR1. Further details of these constructions are available upon request.
The MigCD8t vector was constructed by replacing the green fluorescent protein (GFP) cassette from MigR1 with the human CD8 α chain devoid of its cytoplasmic domain (CD8t), which was obtained from Arthur Weiss (University of California at San Francisco, San Francisco, Calif.) (76). The CD8t sequence was amplified from pUC-CD8t by PCR using a 5′ primer that contained an NcoI site at the 5′ ATG initiation codon and a 3′ primer that had a SalI site at the 3′ end after the stop codon. The 5′ primer sequence was 5′-GAT CCC ATG GCC TTA CCA GTG ACC GCC TTG CTC CTG-3′, and the 3′ primer sequence was 5′-ACC GAA GAA GAT CTT AAT AGG GAT CCG TCG ACG ATC-3′. An NcoI-to-SalI fragment from the amplified product was then used to replace the GFP sequence from MigR1.
The PU.1 cDNA (pCB6-CMV-PU.1) was obtained from Michael Atchison (University of Pennsylvania) (57). The PU.1 coding region from pCB6-CMV-PU.1 was excised with BglII at the 5′ end and BamHI at the 3′ end and subcloned into the BglII site of the MigCD8t vector.
The TF promoter linked to a luciferase reporter was obtained from Nigel Mackman (Scripps Research Institute, San Diego, Calif.) (16, 49). Luciferase-linked promoters of CD11b and other myeloid genes were obtained from Daniel Tenen (Harvard Medical School, Boston, Mass.) (60).
Retrovirus-mediated gene transfer.
To produce viral supernatants, retroviral DNAs were transiently transfected into Phoenix ampho cells, the culture medium was replaced 24 h after transfection, and viral supernatant was harvested 24 h later. The indicated myeloid cell lines were infected with viral supernatants at a density of 1.5 × 106 cells/well of 24-well plates using spin infection at 2,000 × g for 1 h at room temperature in the presence of 6 μg of Polybrene (Sigma, St. Louis, Mo.)/ml. The spin infection was repeated three consecutive times.
Flow cytometry.
Flow cytometric analysis was done using a Becton Dickinson FACSort with Cell Quest software. Cells were stained with the indicated antibodies, and dead cells were excluded by staining with 7-aminoactinomycin D (7-AAD). For surface marker staining, we used the following antibodies purchased from BD Pharmingen (San Diego, Calif.): phycoerythrin (PE)-labeled antibodies against human CD11b, CD11c, CD14, CD18 (β2 integrin), CD29 (β1 integrin), CD32 (FcγRII), CD49d (α4 integrin), CD49e (α5 integrin), CD54 (intercellular adhesion molecule 1 [ICAM-1]), and CD64 (FcγR1). For the staining of human CD114 G-CSF receptors (G-CSFR), we used biotinylated antibodies against the receptors and PE-conjugated streptavidin. For the staining of CD8, we used CyChrome-labeled antibody against human CD8 α chain. TF was stained using monoclonal anti-human TF antibody (American Diagnostica Inc., Greenwich, Conn.) and secondary PE-conjugated anti-mouse immunoglobulin G1 (IgG1) (BD Pharmingen).
Phagocytosis and enzymatic staining of myeloid cells.
Next, 3 × 105 U937 cells infected with different viruses in 12-well plates were incubated with 1.0 μm fluorescein isothiocyanate-labeled latex beads (Fluoresbrite; Polysciences, Warrington, Pa.) for 2 h at 37°C. After washing five times with phosphate-buffered saline (PBS), the cells were collected by cytospin centrifugation (Shandon, Pittsburgh, Pa.) and were then stained with May-Grünwald Giemsa solution. Stained cells were examined using an Axioskop 2 fluorescence microscope (Zeiss Inc., Thornwood, N.J.). Nonspecific esterase (NSE) activity was assayed using α-naphthyl butyrate as a substrate as described previously (84). For NADPH oxidase activity, nitroblue tetrazolium reduction was assayed as described previously (7).
Immunofluorescence analysis.
Cells to be processed for immunofluorescence analysis were fixed with 4% paraformaldehyde. The fixed cells were permeabilized with 0.5% Triton X-100, blocked with 2% BSA in PBS containing 0.1% Tween 20, and then stained with the indicated antibodies for 1 h at room temperature. This was followed by incubation with appropriate secondary antibodies conjugated to fluorochrome. For HA epitope staining, we incubated with biotinylated anti-HA monoclonal antibody (Roche, Indianapolis, Ind.) followed by incubation with Alexa Fluor 594-conjugated streptavidin (Molecular Probes, Eugene, Oreg.). The nuclei were stained with 1 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI). Stained cells were observed with a Zeiss fluorescence microscope, and digital images were taken using a SPOT digital camera (Diagnostic Instruments Inc., Sterling Heights, Mich.).
Immunoprecipitation and Western blot analysis.
Cells were lysed in a buffer containing 50 mM HEPES-KOH (pH 7.4)-1% (vol/vol) Triton X-100-150 mM NaCl-2.5 mM EDTA-10% (vol/vol) glycerol-1 mM sodium orthovanadate-2% Trasylol (FBA Pharmaceuticals, New York, N.Y.). Total cell lysates containing 30 to 50 μg of protein or immunoprecipitates from 250 μg of protein extract were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblotting as previously described (1, 18). Monoclonal antibodies directed against Fes have been previously described (39). The antibody to phosphotyrosine (PY99) was purchased from Santa Cruz (Santa Cruz, Calif.). The anti-human macrophage colony-stimulating factor receptor (M-CSFR) antibodies used for immunoprecipitation and immunoblotting were purchased from Calbiochem (San Diego, Calif.) and Santa Cruz, respectively. Protein bands were visualized by using an ECL kit according to the manufacturer's instructions (Amersham Phamacia Biotech, Piscataway, N.J.).
Preparation of nuclear extract and EMSA.
Nuclear proteins were extracted essentially as previously described (71). Cells (107) were resuspended in 400 μl of ice-cold buffer A (10 mM HEPES-KOH [pH 7.8]; 10 mM KCl; 0.1 mM EDTA; 1 mM dithiothreitol [DTT]; 0.5 mM phenylmethylsulfonyl fluoride [PMSF]; aprotinin, leupeptin, and pepstatin A [5 μg/ml each]) and allowed to swell on ice for 15 min. After the addition of NP-40 to a final concentration of 0.5%, cells were lysed by passage through a 25-gauge-needle tuberculin syringe 10 times. Cell lysates were centrifuged, and the nuclear pellets were resuspended in 50 μl of ice-cold buffer C (50 mM HEPES [pH 7.8]; 420 mM KCl; 0.1 mM EDTA; 5 mM MgCl2; 1 mM DTT; 1 mM PMSF; aprotinin, leupeptin, pepstatin A [5 μg/ml each]; 1 mM sodium orthovanadate). For electrophoretic gel mobility shift assay (EMSA), double-stranded oligonucleotides were end-labeled with polynucleotide kinase and [γ-32P]ATP. For competition, nonspecific or specific unlabeled oligonucleotide at a 100-fold molar excess was added to the reaction mixture. For supershift assay, 2 μg of specific antibodies against human PU.1 (BD Pharmingen) was mixed with the nuclear extract for 15 min at 4°C before labeled oligonucleotides were added. The reaction mixtures were incubated for 15 min at 37°C, analyzed on 4% nondenaturing acrylamide gels, and exposed for autoradiography. The oligonucleotide sequences used were: PU.1 (CD11b) (60) sense, 5′-CTT CTG CCT CCT ACT TCT CCT TTT CTG GCC T-3′; PU.1 antisense, 5′-AGG CCA GAA AAG GAG AAG TAG GAG GCA GAA G-3′; Oct-1 (Ig heavy chain) (40) sense, 5′-GAT CGA ATG CAA ATC ACT AGC T-3′; Oct-1 antisense, 5′-AGC TAG TGA TTT GCA TTC GAT C-3′; Stat3-SIE (sis-inducible element) (75) sense, 5′-GTG CAT TTC CCG TAA ATC TTG TC-3′; and Stat3-SIE antisense, 5′-GAC AAG ATT TAC GGG AAA TGC AC-3′. All sequences were derived from humans.
RESULTS
Construction of Fesact.
To help elucidate the mechanism of action of Fes, we wanted to develop a gain-of-function allele of this kinase which would be biologically active in myeloid cells of mammalian origin. We first considered the feline v-Fes oncoproteins, because they are the only known activated mammalian Fes proteins. However, the Gag-v-Fes oncoproteins of Feline sarcoma viruses were not suitable for our purpose, because they contain large deletions at the N terminus (27, 66), which has been shown to be important for interactions of Fes with its substrates (52). Although fusion of Gag to the N terminus of avian and human c-Fps/Fes activates these kinases (19, 21), this mode of activation was also avoided because the presence of Gag might interfere with the ability of Fes to interact with its substrates and other myeloid-specific factors.
Since the transforming gene of FSV contains no deletions of cell-derived fps sequences (36, 74), is colinear with human c-fes, and still retains transforming activity in avian fibroblasts after the removal of gag (20), we thought that isogenic chimeras between human c-fes and the v-fps-specific sequence of FSV might be constitutively active. To screen for biological activity in myeloid cells of mammalian origin, candidate chimeric Fes molecules were tested for their ability to transactivate myeloid gene promoters in transient transfection assays in murine RAW 264.7 macrophages. We found that a c-Fes/v-Fps chimera (Fig. 1A) containing the N terminus, the SH2 domain, the first one-fifth portion of the kinase domain derived from human c-Fes, and the remaining four-fifths portion of the kinase domain derived from the v-Fps sequence of avian FSV was able to transactivate the promoter of TF and other myeloid genes (data not shown). In other words, human c-Fes was activated by replacing the C-terminal four-fifths of its kinase domain with the corresponding region from avian v-Fps. Fesact is isogenic with both human c-Fes and avian c-Fps. In the switched kinase region, avian c-Fps and FSV-v-Fps differ in six point mutations, and the percentage of amino acid homology between human c-Fes and avian v-Fps in this region is 88%, reflecting a high degree of conservation in the kinase domain of Fps/Fes across species (28). The reciprocal chimera containing most of v-Fps and four-fifths of the kinase domain from human c-Fes was inactive in promoter assays, as was normal human c-Fes. v-Fps, which differs from avian c-Fps in 25 point mutations (36), was also able to transactivate the TF promoter (data not shown), but it was not as effective as the chimeric Fesact molecule in mammalian cells. This suggests that the human N terminus and SH2 domain of the chimera were better suited than the corresponding avian regions to recognizing the targets of Fes in cells of mammalian origin.
The ability of Fesact to transactivate myeloid-specific genes suggested that this gain-of-function allele of Fes might mimic the physiological activation of c-Fes and enable us to identify downstream events under Fes regulation in myeloid cells.
Fesact kinase is constitutively active and has the same cytoplasmic localization as c-Fes.
To characterize the biological properties of Fesact in myeloid cells, we cloned this gene into the IRES-GFP retroviral vector MigR1 (Fig. 1B) and used retrovirus-mediated gene transfer to introduce Fes into target myeloid cells. This system allowed us to analyze a population of GFP-gated cells rather than selected clones. This is particularly important when the introduced genes are involved in regulation of cell proliferation or differentiation, since selected, stable clones may have to undergo substantial physiological changes in order to survive, compromising the experimental system.
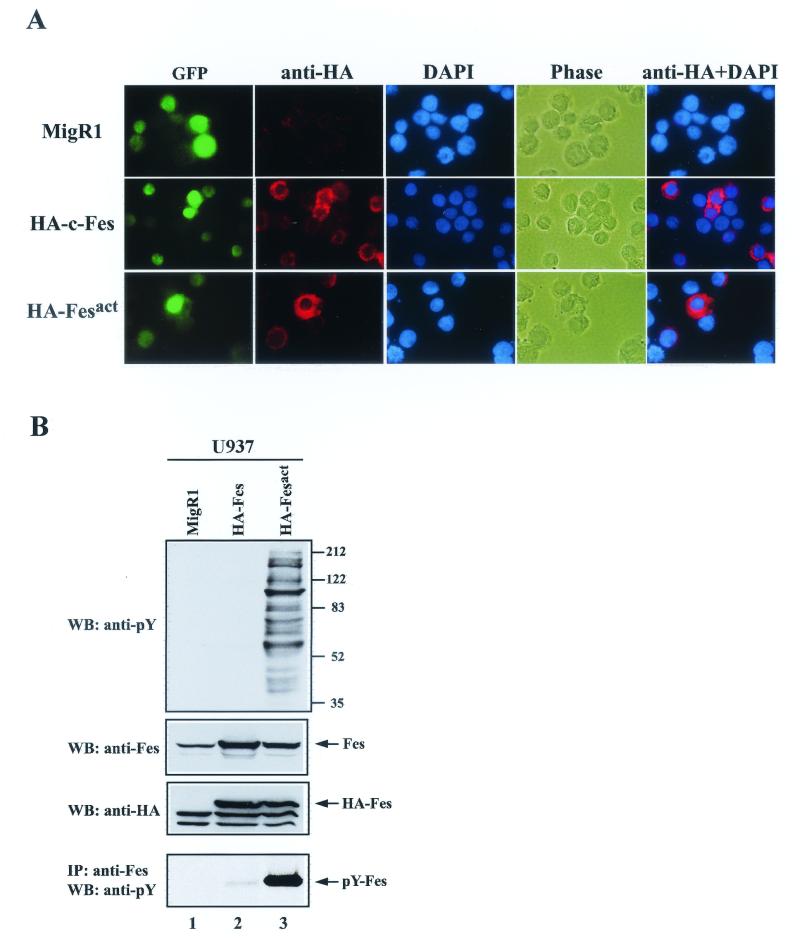
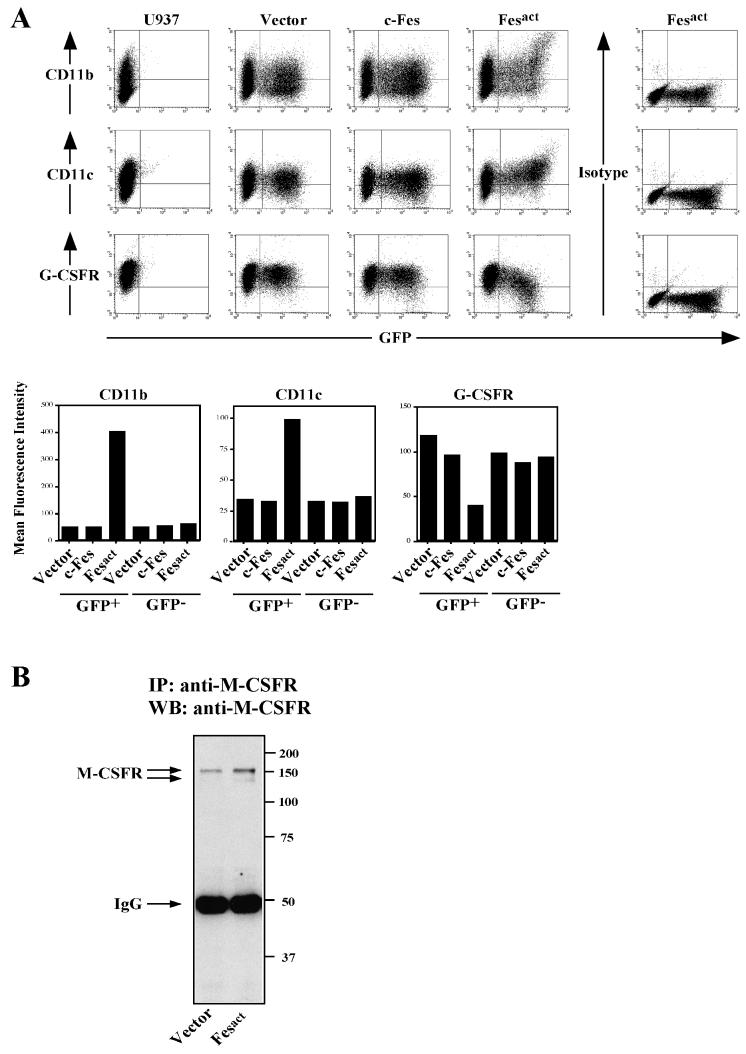
To begin our analysis we infected U937 cells with our recombinant viruses. U937 is a human bipotential promonocytic cell line that can be induced to differentiate into either macrophages or granulocytes by treatment with tetradecanoyl phorbol acetate (TPA) or all-trans retinoic acid (ATRA), respectively (65). The efficiency of infection ranged from 30 to 70%; most often, it was in the 40 to 60% range (Fig. 1B). After infection, HA-tagged Fes proteins were only expressed in GFP+ cells, and there was a close correlation between the expression levels of Fes and GFP (Fig. 2A). It should be noted that both c-Fes and Fesact were localized in the cytoplasm, especially in the perinuclear region, but not in the nucleus or at the plasma membrane. As shown in Fig. 2B, there was a considerable increase in the total levels of phosphotyrosine in Fesact-infected U937 cells compared to those in vector- or c-Fes-infected cells. The expression levels of retrovirally transduced Fes proteins were 5- to 10-fold higher than those of endogenous Fes in U937 cells. It is also apparent that Fesact but not c-Fes was phosphorylated on tyrosine in vivo.
FIG.2.
Coexpression of Fes and GFP in infected cells and tyrosine phosphorylation of cellular proteins by active Fes. (A) U937 cells infected with MigR1 vector, MigR1 HA-c-Fes, or MigR1 HA-Fesact were stained 4 days after infection by sequential incubation with biotinylated anti-HA antibody and Alexa Fluor 594-conjugated streptavidin. The nuclei were stained with DAPI. In the final column, the images from anti-HA antibody and DAPI staining were merged. Cells were observed by fluorescence microscopy (magnification, ×63). HA, hemagglutinin tag. (B) U937 cells infected with MigR1 vector (lane 1), MigR1 HA-c-Fes (lane 2), or MigR1 HA-Fesact (lane 3) were lysed 3 days after infection. The cell lysates were analyzed by SDS-PAGE in 8.5% gels followed by immunoblotting (WB) with the indicated antibodies, either directly or after immunoprecipitation (IP) with anti-Fes antibodies. The first, second, and third panels correspond to immunoblots of total cellular proteins detected with antiphosphotyrosine, anti-Fes, and anti-HA antibodies, respectively. In the fourth panel, the cell lysates were immunoprecipitated with anti-Fes antibody, and the immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting with antiphosphotyrosine antibodies. Numbers at the right indicate the positions of molecular weight (MW) markers. The arrows indicate the positions of Fes proteins, which have an apparent molecular mass of 92 kDa.
We conclude from these results that Fesact kinase is constitutively active in U937, that the enzymatic activity of c-Fes is tightly repressed even when this kinase is overexpressed, and that Fes autophosphorylation correlates with increased enzymatic activity in vivo. As described below, mutation of the Fes autophosphorylation site (Tyr 713) greatly diminished the enzymatic and biological activities of Fesact.
Introduction of active Fes into U937 results in the appearance of fully differentiated macrophages.
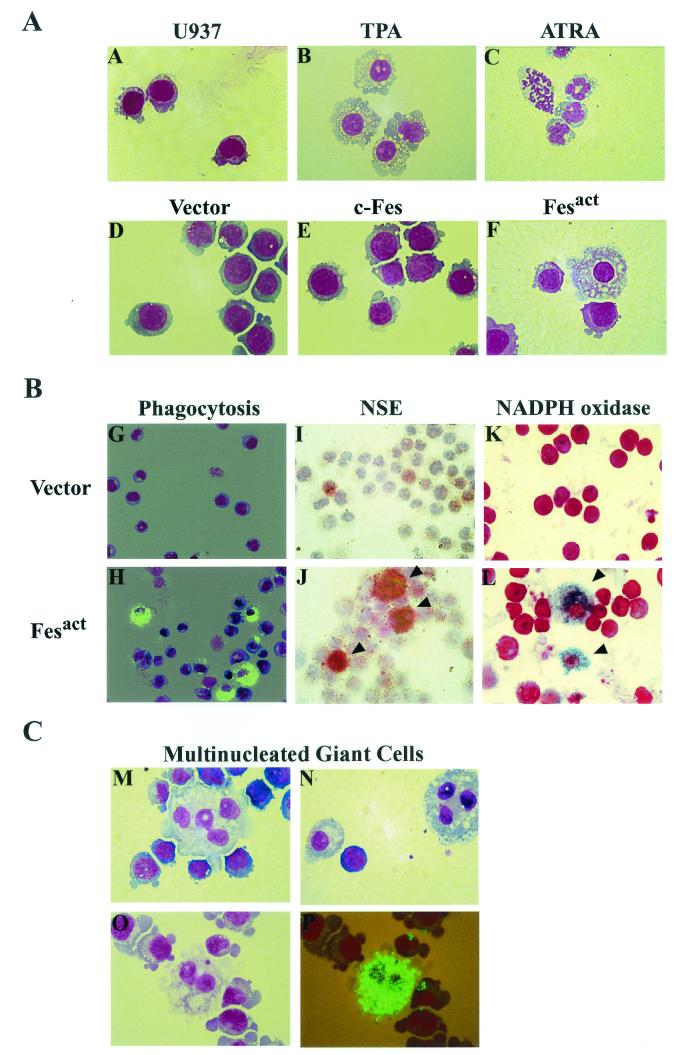
As shown in Fig. 3A, TPA treatment of U937 cells caused macrophage differentiation, whereas ATRA induced granulocytic differentiation, indicating that our stock of U937 cells was capable of both macrophage and granulocyte differentiation. Introduction of Fesact into U937 cells resulted in cell adhesion on substratum and, strikingly, this was followed by differentiation of these myeloid precursors into macrophages within 6 to 12 days (Fig. 3A). The differentiated cells showed characteristic macrophage morphology, namely larger cell size, increased membrane ruffles, and small nucleus and large cytoplasm filled with vacuoles. In contrast, vector- or c-Fes-infected U937 cells showed typical blast morphology, with large oblong nucleus and small cytoplasm, which is the phenotype observed in normal uninfected U937 cells (Fig. 3A). These results and those described in the following sections were obtained by using nontagged c-Fes or Fesact constructs, but the corresponding HA-tagged molecules had the same biological activity as the nontagged versions.
FIG.3.
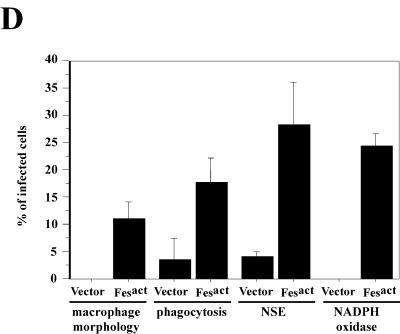
Fesact induces differentiation of U937 cells into macrophages. (A) U937 cells were either left untreated (A), treated with 16 nM TPA for 2 days (B) or 1 mM ATRA for 5 days (C), or infected with either MigR1 (D), MigR1 c-Fes (E), or MigR1 Fesact (F) viruses. Following the indicated TPA and ATRA treatment or at 9 days postinfection, cells were collected by cytospin centrifugation and stained with May-Grünwald Giemsa. Magnification, ×63. (B) U937 cells were infected with either MigR1 (panels G, I, and K) or MigR1 Fesact (panels H, J, and L) viruses, and the infected cells were assayed for phagocytosis and NSE and NADPH oxidase activities. For the phagocytosis assay (panels G and H), infected cells were incubated with fluorescein isothiocyanate-labeled latex beads for 2 h at 37°C, washed thoroughly with PBS, and collected by cytospin centrifugation. Cells were then stained with May-Grünwald Giemsa and observed by fluorescence microscopy. NSE (panels I and J) and NADPH oxidase (panels K and L) activities were assayed as described in Materials and Methods. After NSE and NADPH oxidase staining, cells were counterstained with hematoxylin and Safranin O, respectively. The phagocytosis and NSE assays were done 9 days after infection, and NADPH oxidase was assayed 6 days after infection. Magnification, ×40 for phagocytosis and NSE and ×63 for NADPH oxidase. Arrowheads in panels J and L indicate the positions of NSE- and nitroblue tetrazolium-stained cells, respectively. (C) U937 cells were infected with Fesact viruses (panels M, N, O, and P) and collected by cytospin centrifugation 9 days after infection. The infected cells were then stained with May-Grünwald Giemsa either directly (panels M and N) or after assaying for phagocytosis (panels O and P) as described above. In panels O and P, the same microscopic field was observed under light (O) or fluorescence microscopy (P). (D) U937 cells were infected with MigR1 or MigR1 Fesact, and 9 days after infection the cells were analyzed for macrophage morphology, phagocytosis, NSE, and NADPH oxidase activities as described in panel B, above. The results represent the percentages of positive cells in the infected populations. The error bars indicate standard deviations from three different microscopic fields that were randomly chosen and contained at least 100 cells per field. Cells that had characteristic macrophage morphology and a cytoplasmic/nuclear ratio greater than 1 were counted as macrophages. Cells that contained more than 10 fluorescent beads were counted as positive for the phagocytosis assay. In every case, P was <0.05 (Student's t test).
Full differentiation to macrophages was observed in about 10% of the Fesact-infected population (Fig. 3D), and the majority of the remaining 90% showed a partially differentiated phenotype, such as more irregular shape, increased membrane blebbing, and larger cell size compared to uninfected U937 cells. To assess the functional capacity of the Fesact-induced macrophages, we performed phagocytosis and enzymatic assays. As shown in Figs. 3B and 3D, about 15% of the Fesact-infected cells acquired strong phagocytic properties and 25 to 30% exhibited NSE and NADPH oxidase activities, confirming that these cells were functionally active macrophages. These results indicated that Fesact was capable of inducing terminal differentiation in a significant fraction of the infected population but the expression of Fesact alone was not sufficient to drive complete macrophage differentiation in all of the cells. Under physiological conditions, additional cooperating signals are likely to be required to induce full macrophage differentiation. At late stages after Fesact infection, we observed the appearance of highly phagocytic, multinucleated giant cells containing 2 to 5 nuclei (Fig. 3C). These cells are indicative of the final stages of macrophage differentiation. Multinucleated giant cells are also a phenotype of macrophages in tuberculosis lesions.
Consistent with the macrophage phenotype, flow cytometric analysis showed that Fesact upregulated the expression of myelomonocytic surface markers such as CD11b (αM), CD11c (αx), CD14, CD18 (β2), CD54 (intercellular adhesion molecule 1 [ICAM-1]), and CD64 (FcγRI). As shown in Fig. 4A, after Fesact expression there was a 7- to 8-fold increase in the mean fluorescence intensity (MFI) of CD11b and about 50% of the CD11b− population became CD11b+. The GFP− population had no change in MFI of CD11b in both the vector- and the Fesact-infected groups, indicating that upregulation of CD11b was not mediated by soluble secreted factors induced by Fesact. Fesact also caused an increase in the expression levels of CD11c by threefold, and 65% of the CD11c− population shifted to CD11c+ (Fig. 4A). By contrast with Fesact, c-Fes did not induce any significant changes in the level of expression of these markers (Fig. 4A). Other markers, including CD14, CD18 (β2), and CD54 (ICAM-1) were also upregulated in Fesact-infected cells (data not shown). We also found that, in Fesact-infected cells, there was slightly reduced expression of α5 and β1 integrins and no change in expression of α4 integrin (data not shown).
FIG. 4.
Coordinated upregulation of monocytic markers and downregulation of G-CSFR by Fesact. (A) U937 cells were infected with either MigR1, MigR1 c-Fes, or MigR1 Fesact viruses. Four days later, infected cells and uninfected controls were analyzed for surface expression of CD11b, CD11c, and G-CSFR by flow cytometry using specific antibodies or isotype-matched control antibodies as described in Materials and Methods. The lower panels represent the MFIs of each marker calculated from the dot plots shown in the upper panels. (B) U937 cells were infected with either MigR1 or MigR1 Fesact viruses, and cells were lysed 6 days after infection. The cell lysates were immunoprecipitated (IP) with anti-human M-CSF receptor (M-CSFR), and the immunoprecipitates were analyzed by SDS-PAGE in 8.5% gels followed by immunoblotting (WB) with anti-M-CSFR antibodies. Numbers at the right indicate the positions of MW markers. The arrows indicate the positions of M-CSFR and IgG.
Introduction of Fesact into U937 cells caused a twofold reduction in the overall expression level of the granulocytic marker G-CSFR, and about 40% of G-CSFR+ cells became G-CSFR− (Fig. 4A). On the other hand, Fesact upregulated the expression of M-CSF receptor by two- to threefold, as measured by immunoblotting (Fig. 4B). Since M-CSFR expression was analyzed in unselected populations containing both uninfected and infected cells, this probably represents an underestimate of the level of M-CSFR induction by Fesact. The reciprocal upregulation of monocytic markers and downregulation of granulocytic markers we observed indicated that the biological activity of Fesact was coordinated and specific for macrophage differentiation and not the result of a general induction of myeloid gene expression by Fes.
We conclude from these results that active Fes is a potent inducer of terminal macrophage differentiation of U937 myeloid precursors and that this may reflect a critical role of c-Fes kinase in signaling pathways leading to macrophage differentiation. We should point out that the differentiation activity of Fesact in myeloid cells was in sharp contrast to its transforming activity in fibroblasts (data not shown), cautioning that studies of Fes in cells that normally do not express this kinase can be quite misleading.
Fesact upregulates the expression of inflammatory molecules.
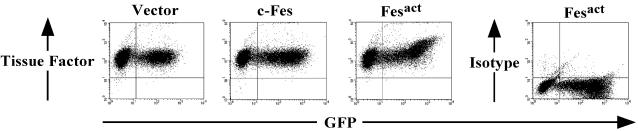
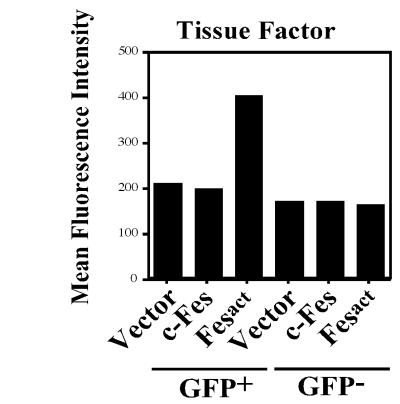
As shown in Fig. 3C, at late stages of Fesact infection in U937 cells we noticed the appearance of giant multinucleated cells, which were highly phagocytic. This suggested that Fes may be involved in macrophage activation and may also regulate the production of inflammatory mediators. TF is a transmembrane glycoprotein that initiates blood coagulation and acts as a cofactor of factor VIIa in the blood coagulation cascade (77). Since TF is induced during macrophage activation (15, 48), we examined whether Fesact would be able to induce TF. As shown in Fig. 5, Fesact upregulated the expression of TF in U937 cells by about twofold.
FIG. 5.
Fesact expression upregulates TF. U937 cells were infected with either MigR1, MigR1 c-Fes, or MigR1 Fesact viruses. Four days later, cells were analyzed for surface expression of TF by flow cytometry using specific antibodies or isotype-matched control antibodies as described in Materials and Methods. The dot plots and mean fluorescence intensities of TF are shown in the upper and lower panels, respectively.
The ability of Fesact to induce the appearance of highly phagocytic multinucleated giant cells and to upregulate inflammatory mediators suggests that Fes may also play a role in macrophage activation and in the inflammatory responses of these cells.
Fesact induces myeloid-specific markers in HL-60 myeloid leukemia cells.
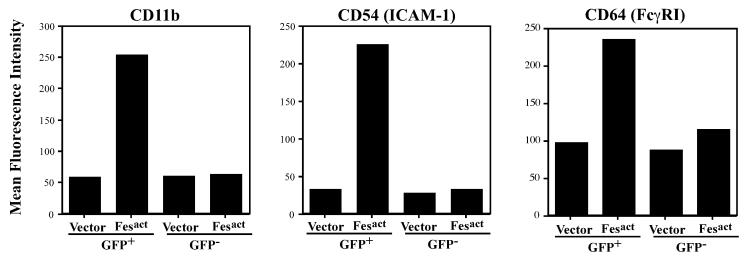
The biological activity of Fes was tested in another bipotential myeloid cell line, HL-60. In these cells Fesact also upregulated the expression of myeloid-specific markers, including CD11b, CD11c, CD18, CD54, and CD64. The expression levels of CD11b, CD54, and CD64 were increased about four- to fivefold, six- to sevenfold, and two- to threefold, respectively, by Fesact in the GFP+ population only (Fig. 6). However, Fesact-expressing HL-60 cells did not acquire definitive macrophage or granulocyte morphology as assessed by May-Grünwald Giemsa staining, suggesting that these cells are missing some cellular factors required for full macrophage differentiation by Fesact.
FIG. 6.
Fesact upregulates CD11b, CD54 (ICAM-1), and CD64 (FcγR1) in HL-60 cells. HL-60 cells were infected with MigR1 or MigR1 Fesact, and 4 days later cells were stained with anti-CD11b, anti-CD54, or anti-CD64 antibodies. Stained cells were analyzed by flow cytometry, and the mean fluorescence intensity of each marker was plotted.
Active Src does not induce macrophage differentiation in U937 cells.
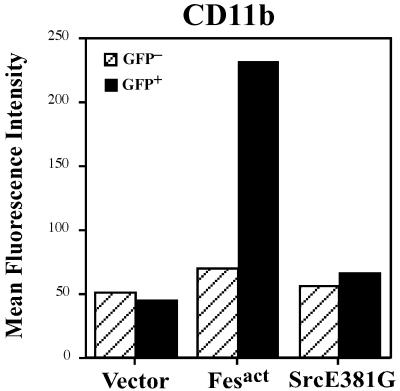
To address the question of specificity of Fes kinase, we tested the activity of a constitutively active mutant of human src, namely srcE381G (6), which was cloned into the same retroviral vector. Active Src did not induce significant changes in the expression of myeloid differentiation markers like CD11b (Fig. 7), and we could not find any typical macrophage morphology by staining. However, active Src induced adhesion to plastic and aberrant morphology, which is consistent with previously described roles of Src in cell adhesion and cytoskeletal remodeling (58, 78, 81). These results suggest that the macrophage differentiation activity we observed was not a general consequence of overexpressing an activated nonreceptor tyrosine kinase but reflected a specific property of Fesact.
FIG. 7.
Specific induction of CD11b by Fesact but not by active Src. U937 cells were infected with MigR1, MigR1 Fesact, or MigR1 SrcE381G viruses, and 5 days later cells were stained with anti-CD11b antibody. Stained cells were analyzed by flow cytometry.
Fes-dependent differentiation requires an enzymatically active Fesact, autophosphorylation of Tyr 713, and a functional SH2 domain.
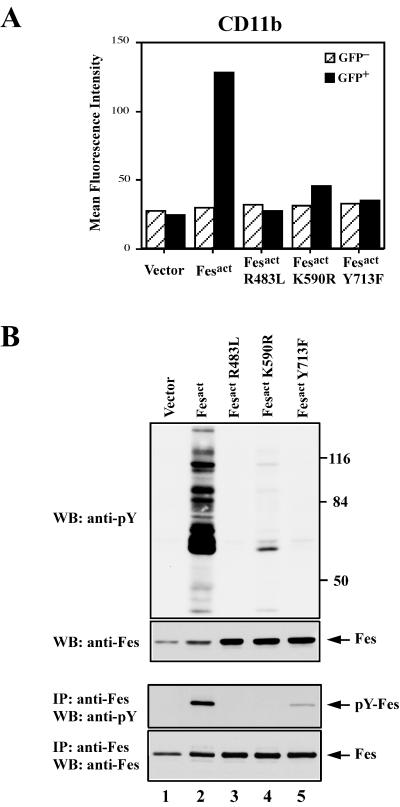
To assess whether the enzymatic activity, SH2 domain, and autophosphorylation site of Fesact were required for its macrophage differentiation activity, we generated three Fesact mutants: FesactK590R has a mutation in the ATP binding site and is kinase deficient, FesactY713F has a Tyr to Phe mutation at the conserved autophosphorylation site, and FesactR483L has a point mutation in the SH2 domain that renders it unable to bind phosphotyrosine residues (5). None of these Fes mutants were able to induce any morphological changes in infected U937 cells compared to uninfected or vector-infected cells. As shown in Fig. 8A, while Fesact strongly induced CD11b expression, all three Fesact mutants had greatly diminished biological activity. Mutation of the Fes SH2 domain completely abolished the ability of Fesact to upregulate CD11b, whereas the kinase-deficient and autophosphorylation mutants still had some residual activity. The loss of biological activity in these mutants correlated with greatly diminished levels of tyrosine phosphorylation of cellular proteins (Fig. 8). As shown in Fig. 8B, all three mutant proteins were stably expressed in infected cells, indicating that the absence of biological activity was not due to lower expression levels or instability of these mutants. Phosphotyrosine analysis of Fes immunoprecipitates showed that, while Fesact was highly phosphorylated on tyrosine in infected cells, the kinase-deficient and SH2 mutants were not, and very little autophosphorylation was observed in the Tyr 713 mutant. The low level of phosphorylation observed in FesactY713F suggests that there may be a secondary tyrosine phosphorylation site in Fesact (68).
FIG. 8.
Fesact requires its kinase activity, a functional SH2 domain, and Tyr 713 for biological activity. (A) U937 cells were infected with MigR1, MigR1 Fesact, MigR1 FesactR483L, MigR1 FesactK590R, and MigR1 FesactY713F viruses. Five days after infection, cells were stained with anti-CD11b antibody and analyzed by flow cytometry. (B) Portions of the U937 cells infected with MigR1 (lane 1), MigR1 Fesact (lane 2), MigR1 FesactR483L (lane 3), MigR1 FesactK590R (lane 4), and MigR1 FesactY713F (lane 5) viruses described in the panel A legend, above, were lysed 5 days after infection. The cell lysates were analyzed by SDS-PAGE in 8.5% gels followed by immunoblotting (WB) with the indicated antibodies either directly or after immunoprecipitation (IP) with anti-Fes antibodies. Top and upper-middle panels, immunoblotting of total cellular proteins with antiphosphotyrosine or anti-Fes antibodies, respectively. Lower-middle and bottom panels, immunoprecipitates of Fes were immunoblotted with anti-phosphotyrosine or anti-Fes antibodies, respectively. Numbers at the right indicate the positions of MW markers. Arrows indicate the positions of Fes proteins.
We conclude from these results that induction of myeloid gene expression by Fesact requires its enzymatic activity, the presence of the conserved autophosphorylation site at Tyr 713, and a functional SH2 domain.
Fesact activates the ets transcription factor PU.1.
Myeloid differentiation is critically dependent on the combinatorial action of general and lineage-restricted transcription factors activated in response to extracellular stimuli. Since Fesact was able to induce macrophage differentiation, we thought that this kinase might upregulate or activate one or more transcription factors known to be involved in macrophage differentiation, including PU.1, Sp-1, interferon consensus sequence binding protein, early growth response gene 1, and interferon regulatory factor 1. In initial experiments we did not detect any changes in mRNA levels of any of these transcription factors after Fesact expression (data not shown).
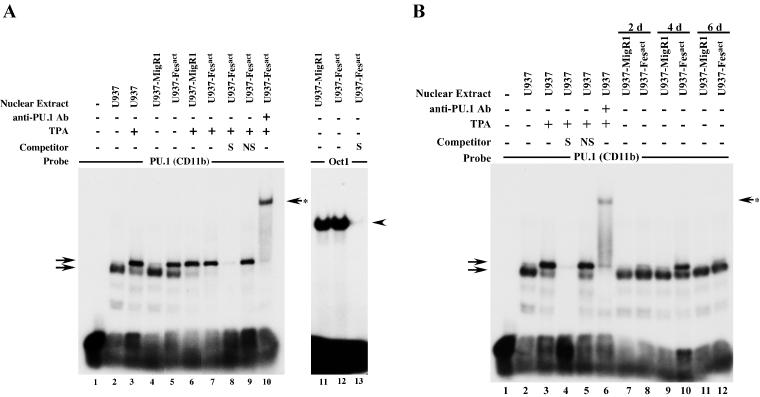
Since PU.1 is essential for lymphoid and macrophage development (31, 54, 72), we examined the possibility that this transcription factor was activated by Fesact. To investigate whether Fesact would be able to induce DNA-binding activity of PU.1, we carried out EMSA analysis of Fesact-infected U937 cells. As shown in Fig. 9A, lane 5, the expression of Fesact in U937 cells resulted in the activation of PU.1, as evidenced by the appearance of a new slow-migrating band, which comigrated with a band induced by TPA (Fig. 9A, lane 3), a known activator of PU.1 (9). Although the shift appeared to be weaker in the case of Fesact compared to that of TPA, it should be pointed out that the EMSA was carried out in an unselected population containing both uninfected and Fesact-infected cells. Infection with vector alone did not cause any PU.1 activation (Fig. 9A, lane 4), and TPA treatment of vector- and Fesact-infected U937 cells (Fig. 9A, lanes 6 and 7) resulted in similar patterns of PU.1 activation. The PU.1 bands were competed out by excess unlabeled specific oligonucleotide but not by a nonspecific STAT3-SIE oligonucleotide (Fig. 9A, lanes 8 and 9). The addition of anti-PU.1 antibodies to the EMSA reaction resulted in a band supershift, confirming that the Fesact- and TPA-induced bands contained PU.1 (Fig. 9A, lane 10). Fesact did not induce changes in the DNA-binding activity of Oct1, which was used as a control (Fig. 9A, lanes 11 to 13). PU.1 activation was detectable 2 days after Fesact infection, reached a peak level on day 4, and decreased by day 6 (Fig. 9B, lanes 8, 10, and 12).
FIG. 9.
Fesact expression results in activation of PU.1. (A) U937 cells were infected with MigR1 (lanes 4, 6, and 11) or MigR1 Fesact (lanes 5, 7, 8, 9, 10, 12, and 13) viruses, and 4 days after infection, the infected cells and uninfected controls (lanes 2 and 3) were either left untreated (lanes 2, 4, 5, 11, 12, and 13) or were treated with 16 nM TPA for 24 h (lanes 3, 6, 7, 8, 9, and 10) before cell lysis. Nuclear extracts were prepared and PU.1 DNA-binding activity was analyzed by EMSA as described in Materials and Methods. Lane 1 had no nuclear extract. Unlabeled, specific (S) PU.1 (lane 8) or nonspecific (NS) Stat3-SIE (lane 9) oligonucleotides were included in the EMSA as indicated. Anti-PU.1 antibody was used for supershift (lane 10). In the right panel, portions of the nuclear extracts of U937 cells infected with MigR1 (lane 11) or MigR1 Fesact (lanes 12 and 13) were assayed for DNA-binding activity of Oct1 in the absence (lanes 11 and 12) or presence (lane 13) of unlabeled, specific (S) Oct1 oligonucleotide. Arrows at the left indicate the positions of PU.1-specific bands. The upper left arrow indicates the position of the slower migrating PU.1 complex. The arrow with the asterisk at the right indicates the supershifted PU.1 complex obtained after incubation with specific anti-PU.1 antibody. Arrowhead indicates the position of the Oct1 complex. (B) U937 cells were infected with MigR1 (lanes 7, 9, and 11) or MigR1 Fesact viruses (lanes 8, 10, and 12), and nuclear extracts were prepared at days 2, 4, and 6 postinfection. Nuclear extracts were also prepared from uninfected U937 cells that were either left untreated (lane 2) or treated with 16 nM TPA for 24 h (lanes 3 to 6). PU.1 DNA-binding activity in nuclear extracts was assayed as described for panel A, above. Lane 1 has no nuclear extract. Unlabeled, specific (S) PU.1 (lane 4) or nonspecific (NS) Stat3-SIE (lane 5) oligonucleotides were included in the EMSA as indicated. Anti-PU.1 antibody was used for supershift (lane 6). Arrows at the left indicate PU.1 complexes. The arrow with the asterisk at the right shows the supershifted PU.1 complex obtained after incubation with specific anti-PU.1 antibody.
We conclude from these results that in U937 cells, PU.1 is a direct or indirect downstream target of Fesact.
PU.1 cooperates with Fes in the induction of CD11b expression.
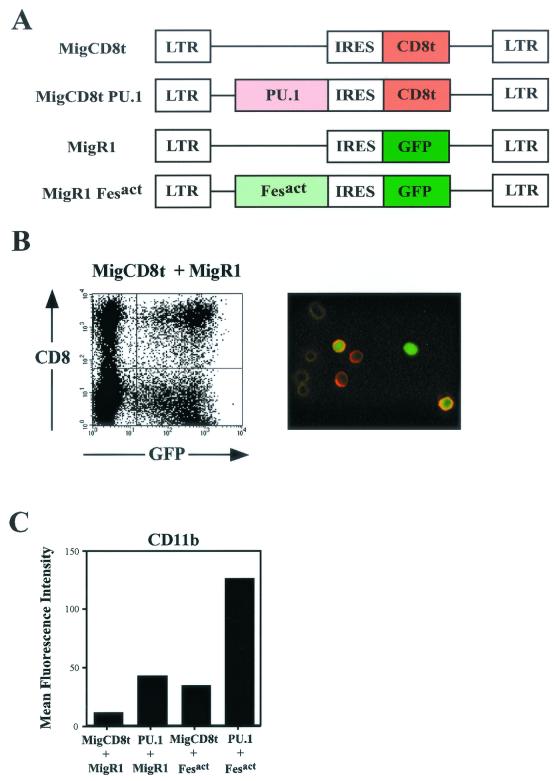
To further examine the role of PU.1 activation by Fesact in upregulation of myeloid gene expression, we wanted to determine whether PU.1 was able to cooperate with this kinase in inducing gene expression. In order to coexpress Fesact and PU.1, we constructed a new retroviral vector in which the GFP portion of MigR1 was replaced by a truncated CD8 (CD8t), and we cloned PU.1 into this new IRES-CD8t vector (Fig. 10A). Retroviral supernatants containing Fesact or PU.1 viruses were generated from separate transfections in Phoenix ampho cells, and we then used a mixture of the two viruses to coinfect U937. Cells expressing Fesact or PU.1 were discriminated by their respective GFP or CD8t expression (Fig. 10B). As shown in Fig. 10C, after 3 days, cells infected with Fesact or PU.1 alone exhibited similar levels of CD11b upregulation of about threefold, but the simultaneous expression of Fesact and PU.1 in U937 cells cooperatively upregulated CD11b by 12-fold.
FIG. 10.
Coexpression of PU.1 with Fesact cooperatively upregulates CD11b. (A) Schematic diagram of retroviral vectors used to establish double infection with Fesact and PU.1 recombinant viruses. The MigCD8t vector was constructed by replacing the GFP cassette of MigR1 with a truncated CD8 α chain that is devoid of its cytoplasmic tail, as described in Materials and Methods. (B) Double infection of U937 cells with MigCD8t and MigR1 retroviral vectors. Left panel: U937 cells were infected with a mixture of MigCD8t and MigR1 retroviral supernatants. Two days after infection, cells were stained with CyChrome-conjugated anti-CD8 antibody and analyzed by flow cytometry. Right panel: Stained cells represented in the left panel were observed under fluorescence microscopy. MigR1-infected cells are green fluorescent. MigCD8t-infected cells exhibit orange-red staining on the cell surface. Double-infected cells appear green inside and bright orange-red on the surface. At the left side of the right panel, a few uninfected cells can be observed. (C) U937 cells were double infected with MigCD8t + MigR1, MigCD8t PU.1 + MigR1, MigCD8t + MigR1 Fesact, or MigCD8t PU.1 + MigR1 Fesact viruses. Three days later, the cells were stained with PE-conjugated anti-CD11b antibody and CyChrome-conjugated anti-CD8 antibody and analyzed by flow cytometry. The double-infected population (CD8+GFP+) from each combination of infections was gated and analyzed for CD11b surface expression, and mean fluorescence intensities of CD11b were plotted.
Taken together, the activation of PU.1 by Fesact, the cooperation between these two molecules in marker induction, and the correlation between the time course of PU.1 activation and appearance of the differentiated phenotype strongly support the idea that PU.1 activation by Fesact may be part of the mechanism by which this kinase induces macrophage differentiation in U937 cells.
DISCUSSION
Using a constitutively active Fes molecule, we have obtained genetic evidence that Fes kinase may be involved in regulation of terminal macrophage differentiation. We also identified the ets transcription factor PU.1 as a downstream target of Fes, suggesting a potential mechanism by which Fes may regulate lineage-specific gene expression.
Fesact regulates myeloid differentiation in a lineage-specific manner.
In U937 cells, Fesact induced the development of functionally mature macrophages within 6 to 12 days of infection, which is similar to the time course for development of macrophages from bone marrow precursors under physiological conditions (37). This suggests that, in contrast to TPA, which can induce macrophage differentiation within 2 days, Fes may utilize a physiological pathway to drive differentiation. Full macrophage differentiation was observed in about 10% of the Fesact-expressing cells, while the majority of the rest showed intermediate phenotypic changes. Therefore, macrophage differentiation under physiological conditions is likely to require other signaling events in addition to the activation of c-Fes, but the expression of a constitutively active Fes molecule can partially compensate for their absence. Fes-induced macrophages were functionally mature by several criteria. They had the typical morphology of macrophages, they acquired phagocytic activity, and they expressed markers of macrophage differentiation as determined by flow cytometry and cytochemical analysis. This remarkable activity was not the result of a general induction of myeloid gene expression by Fes, because concomitantly with the upregulation of macrophage markers, there was downregulation of granulocytic markers. The observation that active Src was not able to upregulate myeloid-specific genes suggests that the differentiation activity of Fes was not merely the result of introducing an activated nonreceptor tyrosine kinase but that it was a specific biological property of Fes. The late appearance of multinucleated giant cells in some Fesact-expressing U937 cells showed that continuous Fes activity can drive these cells into the ultimate stages of macrophage differentiation (37), suggesting that Fes kinase may also be involved in macrophage activation. This idea is supported by the observation that Fes null mice have compromised innate immunity, which relies to a large extent on the inflammatory responses of activated macrophages.
Fesact can override the differentiation block of leukemic cells.
Our results showed that Fes activation can override the differentiation block in U937 and HL60 cells, and we also observed that this was coupled to progressive loss of proliferative potential. Stable expression of c-Fes in K562 cells, which contain a bcr-abl oncogene, also causes a reduction in proliferative potential compared to that of parental cells (85), and this led to the proposal that c-Fes might be able to antagonize bcr-abl transformation in chronic myeloid leukemia cells (45).
This ability of Fesact to restore the capacity of human leukemic cells to terminally differentiate seems to be unique among constitutively active nonreceptor tyrosine kinases. Expression of constitutively active Src (41) or Abl (12, 14) in factor-dependent cell lines does not cause myeloid differentiation but factor independence and tumorigenicity. Similarly, constitutively active versions of Jak kinases resulting from chromosomal translocations have been implicated in human leukemogenesis (42), and gain-of-function alleles of hopscotch, the Drosophila homolog of Jak kinases, can cause hematopoietic neoplasms in this organism (30). Our results suggest that elucidation of the pathway used by Fes to induce differentiation may be of therapeutic importance for acute myeloid leukemia or chronic myeloid leukemia.
Functional requirements of Fesact for induction of myeloid gene expression.
When c-Fes was ectopically expressed in U937 cells, no autophosphorylation or tyrosine phosphorylation of cellular proteins above control levels was observed, and this correlated with the absence of biological activity. On the other hand, P92c-fes immunoprecipitated from U937 cells exhibits strong in vitro kinase activity that results in P92c-fes autophosphorylation (17). Thus, the enzymatic activity of c-Fes appears to be tightly regulated within the cell, perhaps by binding to a cellular inhibitor that may be washed off during immunoprecipitation. This is a plausible mechanism, since c-Abl, another nonreceptor tyrosine kinase that does not have a negative regulatory site equivalent to tyrosine 527 in c-Src, is negatively regulated by F-actin (83). By contrast with c-Fes, Fesact was autophosphorylated in vivo, induced tyrosine phosphorylation of cellular proteins, and had potent biological activity in U937 cells, indicating that the ability of Fesact to induce myeloid gene expression requires an active kinase capable of phosphorylating its cellular substrates. Our mutational analysis provided further evidence that autophosphorylation at tyrosine 713 and a functional SH2 domain are essential for both enzymatic and biological activity by Fesact.
Fes activation of PU.1 and macrophage differentiation.
Our results showed that Fesact induced a novel slow-migrating DNA-binding complex that contained PU.1, a transcription factor that is absolutely required for myelopoiesis, and that Fesact and PU.1 cooperated to induce myeloid gene expression. PU.1 null mice do not produce lymphoid or myeloid cells and succumb before or shortly after birth (54, 72). Fetal liver or embryonic stem cells from PU.1-deficient mice can be induced to express early markers of monocytic differentiation such as NSE and GM-CSF receptors, but late markers such as CD11b and CD64 are not expressed (59). Thus, PU.1 appears to be dispensable for myeloid lineage commitment but is required for terminal differentiation, an observation consistent with the presence of PU.1 sites in the promoters of a number of late myeloid genes like CD11b, CD18, CD64, and M-CSFR, all of which were upregulated by Fesact. Since PU.1 is a downstream target of Fes, it is tempting to speculate that PU.1 may be one of the transcription factors that mediate the macrophage differentiation activity of Fes, and that this kinase might regulate terminal differentiation rather than a lineage commitment step. This idea is supported by the observation that, in Fes−/− mice, macrophages are produced but their maturation appears to be defective (25).
It has been shown that the Fes promoter has binding sites for PU.1, Sp1, and a novel transcription factor called c-fes expression factor (32, 33). Ectopic expression of PU.1 transactivates the c-fes promoter in Cos cells (33), and myeloid cells derived from PU.1−/− embryonic stem cells show greatly reduced expression of Fes (59), indicating that PU.1 regulates the expression of Fes. Our finding that Fes can activate PU.1 suggests that these two molecules may be part of a positive regulatory loop that operates during myeloid differentiation. In this scenario, differentiation signals acting on myeloid precursors would activate PU.1, leading to the induction of Fes expression. Subsequently, Fes kinase activation by similar or different signals would in turn lead to the activation of PU.1. This positive feedback mechanism would result in the rapid amplification of signals for the induction of genes under PU.1 control and would also explain the upregulation of Fes observed during myeloid differentiation (44, 59).
An interesting question that arises from our studies concerns the mechanism by which Fesact induced PU.1 DNA-binding activity. The binding of PU.1 to other transcription factors to form active complexes is known to be mediated by phosphorylation of PU.1 by kinases such as casein kinase II (46, 63). In B cells, phosphorylation of PU.1 can induce recruitment to the DNA complex of Pip, a member of the interferon regulatory factor family, or C/EBPδ (57, 62). In U937 cells, TPA treatment induces the phosphorylation of PU.1 and the appearance of a slow-migrating band in the EMSA, which was coupled to the differentiation-inducing ability of TPA (9). In our studies, both Fesact-transduced and TPA-treated U937 cells exhibited a similar pattern of PU.1 activation in the EMSA. This raises the possibility that the band shift observed in the EMSA results from Fesact-dependent phosphorylation of PU.1, triggering its association with other transcription factors to form a transcriptionally active complex. Alternatively, Fes may activate another transcription factor, which then associates with PU.1 in a complex capable of binding PU.1 sites on target genes. Future analysis should clarify the nature of the PU.1 complex induced by Fes and the precise mechanisms leading to formation of this complex.
Loss-of-function and gain-of-function approaches to study the biological role and mechanism of action of Fes.
The potent biological activity of Fesact we describe here lends strong support to the idea that c-Fes is an important component of the myeloid differentiation machinery and that it may also play a role in modulating inflammatory responses of mature macrophages. Although the biological properties of Fesact do not fully correspond to the phenotype of Fes−/− mice, this may be due to the existence of overlapping pathways for myeloid differentiation, as exemplified by the large number of physiological and nonphysiological agents (i.e., colony-stimulating factors, TPA, ATRA, dimethyl sulfoxide) capable of inducing myeloid differentiation. Because of this functional redundancy, elucidation of the role and mechanism of action of Fes kinase in its target tissues will require integrating information obtained by using both gain-of-function and loss-of-function approaches.
In conclusion, we have constructed a constitutively active Fes kinase that allows direct identification of downstream events under Fes control. Using this new reagent, we obtained direct evidence that Fes can regulate lineage-specific gene expression in a highly coordinated manner leading to terminal macrophage differentiation, and we identified PU.1 as a possible downstream target of Fes.
Acknowledgments
We thank Hidesaburo Hanafusa (Rockefeller University) for generously providing us with v-Fps DNA, Don Fujita (University of Calgary) for a constitutively active Src mutant, Michael Atchison (University of Pennsylvania) for PU.1 cDNA, Warren Pear (University of Pennsylvania) for the retroviral vector MigR1, J. Silvio Gutkind (NIH) for pCEFL vector, Nigel Mackman (Scripps Research Institute) for TF promoters, Daniel Tenen (Harvard University) for myeloid gene promoters, and Arthur Weiss (University of California, San Francisco) for CD8t cDNA. Excellent technical assistance by Humayra Ali is greatly appreciated.
This work was supported by grant CA55293 from the National Institutes of Health.
REFERENCES
- 1.Areces, L. B., P. Dello Sbarba, M. Jücker, E. Richard Stanley, and R. A. Feldman. 1994. Functional specificity of cytoplasmic and transmembrane tyrosine kinases: identification of 130- and 75-kilodalton substrates of c-fps/fes tyrosine kinase in macrophages. Mol. Cell. Biol. 14:4606-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aspenstrom, P. 1997. A Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr. Biol. 7:479-487. [DOI] [PubMed] [Google Scholar]
- 3.Bais, C., B. Santomasso, O. Coso, L. Arvanitakis, E. G. Raaka, J. S. Gutkind, A. S. Asch, E. Cesarman, M. C. Gershengorn, E. A. Mesri, and M. C. Gerhengorn. 1998. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature 391:86-89. [DOI] [PubMed] [Google Scholar]
- 4.Behre, G., L. T. Smith, and D. G. Tenen. 1999. Use of a promoterless Renilla luciferase vector as an internal control plasmid for transient co-transfection assays of Ras-mediated transcription activation. BioTechniques 26:24-26, 28. [DOI] [PubMed]
- 5.Bibbins, K. B., H. Boeuf, and H. E. Varmus. 1993. Binding of the Src SH2 domain to phosphopeptides is determined by residues in both the SH2 domain and the phosphopeptides. Mol. Cell. Biol. 13:7278-7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorge, J. D., C. Bellagamba, H. C. Cheng, A. Tanaka, J. H. Wang, and D. J. Fujita. 1995. Characterization of two activated mutants of human pp60c-src that escape c-Src kinase regulation by distinct mechanisms. J. Biol. Chem. 270:24222-24228. [DOI] [PubMed] [Google Scholar]
- 7.Breitman, T. R., S. E. Selonick, and S. J. Collins. 1980. Induction of differentiation of the human promyelocytic leukemia cell line (HL-60) by retinoic acid. Proc. Natl. Acad. Sci. USA 77:2936-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brizzi, M. F., M. G. Aronica, A. Rosso, G. P. Bagnara, Y. Yarden, and L. Pegoraro. 1996. Granulocyte-macrophage colony-stimulating factor stimulates JAK2 signaling pathway and rapidly activates p93fes, STAT1 p91, and STAT3 p92 in polymorphonuclear leukocytes. J. Biol. Chem. 271:3562-3567. [DOI] [PubMed] [Google Scholar]
- 9.Carey, J. O., K. J. Posekany, J. E. deVente, G. R. Pettit, and D. K. Ways. 1996. Phorbol ester-stimulated phosphorylation of PU.1: association with leukemic cell growth inhibition. Blood 87:4316-4324. [PubMed] [Google Scholar]
- 10.Carmier, J. F., and J. Samarut. 1986. Chicken myeloid stem cells infected by retroviruses carrying the v-fps oncogene do not require exogenous growth factors to differentiate in vitro. Cell 44:159-165. [DOI] [PubMed] [Google Scholar]
- 11.Clarke, S., and S. Gordon. 1998. Myeloid-specific gene expression. J. Leukoc. Biol. 63:153-168. [DOI] [PubMed] [Google Scholar]
- 12.Cleveland, J. L., M. Dean, N. Rosenberg, J. Y. Wang, and U. R. Rapp. 1989. Tyrosine kinase oncogenes abrogate interleukin-3 dependence of murine myeloid cells through signaling pathways involving c-myc: conditional regulation of c-myc transcription by temperature-sensitive v-abl. Mol. Cell. Biol. 9:5685-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corey, S., A. Eguinoa, K. Puyana-Theall, J. B. Bolen, L. Cantley, F. Mollinedo, T. R. Jackson, P. T. Hawkins, and L. R. Stephens. 1993. Granulocyte macrophage-colony stimulating factor stimulates both association and activation of phosphoinositide 3OH-kinase and src-related tyrosine kinase(s) in human myeloid derived cells. EMBO J. 12:2681-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortez, D., G. Reuther, and A. M. Pendergast. 1997. The Bcr-Abl tyrosine kinase activates mitogenic signaling pathways and stimulates G1-to-S phase transition in hematopoietic cells. Oncogene 15:2333-2342. [DOI] [PubMed] [Google Scholar]
- 15.Cunningham, M. A., P. Romas, P. Hutchinson, S. R. Holdsworth, and P. G. Tipping. 1999. Tissue factor and factor VIIa receptor/ligand interactions induce proinflammatory effects in macrophages. Blood 94:3413-3420. [PubMed] [Google Scholar]
- 16.Fan, S. T., N. Mackman, M. Z. Cui, and T. S. Edgington. 1995. Integrin regulation of an inflammatory effector gene. Direct induction of the tissue factor promoter by engagement of beta 1 or alpha 4 integrin chains. J. Immunol. 154:3266-3274. [PubMed] [Google Scholar]
- 17.Feldman, R. A., J. L. Gabrilove, J. P. Tam, M. A. Moore, and H. Hanafusa. 1985. Specific expression of the human cellular fps/fes-encoded protein NCP92 in normal and leukemic myeloid cells. Proc. Natl. Acad. Sci. USA 82:2379-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldman, R. A., D. R. Lowy, and W. C. Vass. 1990. Selective potentiation of c-fps/fes transforming activity by a phosphatase inhibitor. Oncogene Res. 5:187-197. [PubMed] [Google Scholar]
- 19.Feldman, R. A., W. C. Vass, and P. E. Tambourin. 1987. Human cellular fps/fes cDNA rescued via retroviral shuttle vector encodes myeloid cell NCP92 and has transforming potential. Oncogene Res. 1:441-458. [PubMed] [Google Scholar]
- 20.Foster, D. A., and H. Hanafusa. 1983. A fps gene without gag gene sequences transforms cells in culture and induces tumors in chickens. J. Virol. 48:744-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster, D. A., M. Shibuya, and H. Hanafusa. 1985. Activation of the transformation potential of the cellular fps gene. Cell 42:105-115. [DOI] [PubMed] [Google Scholar]
- 22.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 23.Greer, P., J. Haigh, G. Mbamalu, W. Khoo, A. Bernstein, and T. Pawson. 1994. The Fps/Fes protein-tyrosine kinase promotes angiogenesis in transgenic mice. Mol. Cell. Biol. 14:6755-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greer, P., V. Maltby, J. Rossant, A. Bernstein, and T. Pawson. 1990. Myeloid expression of the human c-fps/fes proto-oncogene in transgenic mice. Mol. Cell. Biol. 10:2521-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hackenmiller, R., J. Kim, R. A. Feldman, and M. C. Simon. 2000. Abnormal Stat activation, hematopoietic homeostasis, and innate immunity in c-fes−/− mice. Immunity 13:397-407. [DOI] [PubMed] [Google Scholar]
- 26.Haigh, J., J. McVeigh, and P. Greer. 1996. The fps/fes tyrosine kinase is expressed in myeloid, vascular endothelial, epithelial, and neuronal cells and is localized in the trans-golgi network. Cell Growth Differ. 7:931-944. [PubMed] [Google Scholar]
- 27.Hampe, A., I. Laprevotte, F. Galibert, L. A. Fedele, and C. J. Sherr. 1982. Nucleotide sequences of feline retroviral oncogenes (v-fes) provide evidence for a family of tyrosine-specific protein kinase genes. Cell 30:775-785. [DOI] [PubMed] [Google Scholar]
- 28.Hanafusa, H. 1988. The fps/fes oncogene, p. 39-57. In E. P. Reddy, A. M. Skalka, and T. Curran (ed.), The oncogene handbook. Elsevier Science Publishers B. V., Amsterdam, The Netherlands.
- 29.Hanazono, Y., S. Chiba, K. Sasaki, H. Mano, A. Miyajima, K. Arai, Y. Yazaki, and H. Hirai. 1993. c-fps/fes protein-tyrosine kinase is implicated in a signaling pathway triggered by granulocyte-macrophage colony-stimulating factor and interleukin-3. EMBO J. 12:1641-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrison, D. A., R. Binari, T. S. Nahreini, M. Gilman, and N. Perrimon. 1995. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 14:2857-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henkel, G. W., S. R. McKercher, H. Yamamoto, K. L. Anderson, R. G. Oshima, and R. A. Maki. 1996. PU.1 but not ets-2 is essential for macrophage development from embryonic stem cells. Blood 88:2917-2926. [PubMed] [Google Scholar]
- 32.Heydemann, A., J. H. Boehmler, and M. C. Simon. 1997. Expression of two myeloid cell-specific genes requires the novel transcription factor, c-fes expression factor. J. Biol. Chem. 272:29527-29537. [DOI] [PubMed] [Google Scholar]
- 33.Heydemann, A., G. Juang, K. Hennessy, M. S. Parmacek, and M. C. Simon. 1996. The myeloid-cell-specific c-fes promoter is regulated by Sp1, PU.1, and a novel transcription factor. Mol. Cell. Biol. 16:1676-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heydemann, A., S. Warming, C. Clendenin, K. Sigrist, J. P. Hjorth, and M. C. Simon. 2000. A minimal c-fes cassette directs myeloid-specific expression in transgenic mice. Blood 96:3040-3048. [PubMed] [Google Scholar]
- 35.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 36.Huang, C. C., C. Hammond, and J. M. Bishop. 1985. Nucleotide sequence and topography of chicken c-fps. Genesis of a retroviral oncogene encoding a tyrosine-specific protein kinase. J. Mol. Biol. 181:175-186. [DOI] [PubMed] [Google Scholar]
- 37.Johnston, R. B. 1988. Current concepts: immunology. Monocytes and macrophages. N. Engl. J. Med. 318:747-752. [DOI] [PubMed] [Google Scholar]
- 38.Jücker, M., and R. A. Feldman. 1995. Identification of a new adapter protein that may link the common beta subunit of the receptor for granulocyte/macrophage colony-stimulating factor, interleukin (IL)-3, and IL-5 to phosphatidylinositol 3-kinase. J. Biol. Chem. 270:27817-27822. [DOI] [PubMed] [Google Scholar]
- 39.Jücker, M., K. McKenna, A. J. da Silva, C. E. Rudd, and R. A. Feldman. 1997. The Fes protein-tyrosine kinase phosphorylates a subset of macrophage proteins that are involved in cell adhesion and cell-cell signaling. J. Biol. Chem. 272:2104-2109. [DOI] [PubMed] [Google Scholar]
- 40.Kemler, I., E. Bucher, K. Seipel, M. M. Muller-Immergluck, and W. Schaffner. 1991. Promoters with the octamer DNA motif (ATGCAAAT) can be ubiquitous or cell type-specific depending on binding affinity of the octamer site and Oct-factor concentration. Nucleic Acids Res. 19:237-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kruger, A., and S. M. Anderson. 1991. The v-src oncogene blocks the differentiation of a murine myeloid progenitor cell line and induces a tumorigenic phenotype. Oncogene 6:245-256. [PubMed] [Google Scholar]
- 42.Lacronique, V., A. Boureux, V. D. Valle, H. Poirel, C. T. Quang, M. Mauchauffe, C. Berthou, M. Lessard, R. Berger, J. Ghysdael, and O. A. Bernard. 1997. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science 278:1309-1312. [DOI] [PubMed] [Google Scholar]
- 43.Laouar, A., F. R. Collart, C. B. Chubb, B. Xie, and E. Huberman. 1999. Interaction between alpha 5 beta 1 integrin and secreted fibronectin is involved in macrophage differentiation of human HL-60 myeloid leukemia cells. J. Immunol. 162:407-414. [PubMed] [Google Scholar]
- 44.Liebermann, D. A., and B. Hoffman-Liebermann. 1989. Proto-oncogene expression and dissection of the myeloid growth to differentiation developmental cascade. Oncogene 4:583-592. [PubMed] [Google Scholar]
- 45.Lionberger, J. M., and T. E. Smithgall. 2000. The c-Fes protein-tyrosine kinase suppresses cytokine-independent outgrowth of myeloid leukemia cells induced by Bcr-Abl. Cancer Res. 60:1097-1103. [PubMed] [Google Scholar]
- 46.Lodie, T. A., R. Savedra, D. T. Golenbock, C. P. Van Beveren, R. A. Maki, and M. J. Fenton. 1997. Stimulation of macrophages by lipopolysaccharide alters the phosphorylation state, conformation, and function of PU.1 via activation of casein kinase II. J. Immunol. 158:1848-1856. [PubMed] [Google Scholar]
- 47.MacDonald, I., J. Levy, and T. Pawson. 1985. Expression of the mammalian c-fes protein in hematopoietic cells and identification of a distinct fes-related protein. Mol. Cell. Biol. 5:2543-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mach, F., U. Schonbeck, J. Y. Bonnefoy, J. S. Pober, and P. Libby. 1997. Activation of monocyte/macrophage functions related to acute atheroma complication by ligation of CD40: induction of collagenase, stromelysin, and tissue factor. Circulation 96:396-399. [DOI] [PubMed] [Google Scholar]
- 49.Mackman, N., K. Brand, and T. S. Edgington. 1991. Lipopolysaccharide-mediated transcriptional activation of the human tissue factor gene in THP-1 monocytic cells requires both activator protein 1 and nuclear factor kappa B binding sites. J. Exp. Med. 174:1517-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manfredini, R., R. Balestri, E. Tagliafico, F. Trevisan, M. Pizzanelli, A. Grande, D. Barbieri, P. Zucchini, G. Citro, C. Franceschi, and S. Ferrari. 1997. Antisense inhibition of c-fes proto-oncogene blocks PMA-induced macrophage differentiation in HL60 and in FDC-P1/MAC-11 cells. Blood 89:135-145. [PubMed] [Google Scholar]
- 51.Manfredini, R., A. Grande, E. Tagliafico, D. Barbieri, P. Zucchini, G. Citro, G. Zupi, C. Franceschi, U. Torelli, and S. Ferrari. 1993. Inhibition of c-fes expression by an antisense oligomer causes apoptosis of HL60 cells induced to granulocytic differentiation. J. Exp. Med. 178:381-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maru, Y., K. L. Peters, D. E. Afar, M. Shibuya, O. N. Witte, and T. E. Smithgall. 1995. Tyrosine phosphorylation of BCR by FPS/FES protein-tyrosine kinases induces association of BCR with GRB-2/SOS. Mol. Cell. Biol. 15:835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mathey-Prevot, B., H. Hanafusa, and S. Kawai. 1982. A cellular protein is immunologically crossreactive with and functionally homologous to the Fujinami sarcoma virus transforming protein. Cell 28:897-906. [DOI] [PubMed] [Google Scholar]
- 54.McKercher, S. R., B. E. Torbett, K. L. Anderson, G. W. Henkel, D. J. Vestal, H. Baribault, M. Klemsz, A. J. Feeney, G. E. Wu, C. J. Paige, and R. A. Maki. 1996. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 15:5647-5658. [PMC free article] [PubMed] [Google Scholar]
- 55.Metcalf, D. 1991. Control of granulocytes and macrophages: molecular, cellular, and clinical aspects. Science 254:529-533. [DOI] [PubMed] [Google Scholar]
- 56.Metcalf, D. 1989. The molecular control of cell division, differentiation commitment and maturation in haemopoietic cells. Nature 339:27-30. [DOI] [PubMed] [Google Scholar]
- 57.Nagulapalli, S., J. M. Pongubala, and M. L. Atchison. 1995. Multiple proteins physically interact with PU.1. Transcriptional synergy with NF-IL6 beta (C/EBP delta, CRP3). J. Immunol. 155:4330-4338. [PubMed] [Google Scholar]
- 58.Ojaniemi, M., S. S. Martin, F. Dolfi, J. M. Olefsky, and K. Vuori. 1997. The proto-oncogene product p120(cbl) links c-Src and phosphatidylinositol 3′-kinase to the integrin signaling pathway. J. Biol. Chem. 272:3780-3787. [DOI] [PubMed] [Google Scholar]
- 59.Olson, M. C., E. W. Scott, A. A. Hack, G. H. Su, D. G. Tenen, H. Singh, and M. C. Simon. 1995. PU.1 is not essential for early myeloid gene expression but is required for terminal myeloid differentiation. Immunity 3:703-714. [DOI] [PubMed] [Google Scholar]
- 60.Pahl, H. L., R. J. Scheibe, D. E. Zhang, H. M. Chen, D. L. Galson, R. A. Maki, and D. G. Tenen. 1993. The proto-oncogene PU.1 regulates expression of the myeloid-specific CD11b promoter. J. Biol. Chem. 268:5014-5020. [PubMed] [Google Scholar]
- 61.Pear, W. S., J. P. Miller, L. Xu, J. C. Pui, B. Soffer, R. C. Quackenbush, A. M. Pendergast, R. Bronson, J. C. Aster, M. L. Scott, and D. Baltimore. 1998. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210bcr/abl-transduced bone marrow. Blood 92:3780-3792. [PubMed] [Google Scholar]
- 62.Perkel, J. M., and M. L. Atchison. 1998. A two-step mechanism for recruitment of Pip by PU.1. J. Immunol. 160:241-252. [PubMed] [Google Scholar]
- 63.Pongubala, J. M., C. Van Beveren, S. Nagulapalli, M. J. Klemsz, S. R. McKercher, R. A. Maki, and M. L. Atchison. 1993. Effect of PU.1 phosphorylation on interaction with NF-EM5 and transcriptional activation. Science 259:1622-1625. [DOI] [PubMed] [Google Scholar]
- 64.Quelle, F. W., N. Sato, B. A. Witthuhn, R. C. Inhorn, M. Eder, A. Miyajima, J. D. Griffin, and J. N. Ihle. 1994. JAK2 associates with the βc chain of the receptor for granulocyte-macrophage colony-stimulating factor, and its activation requires the membrane-proximal region. Mol. Cell. Biol. 14:4335-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Radomska, H. S., C. S. Huettner, P. Zhang, T. Cheng, D. T. Scadden, and D. G. Tenen. 1998. CCAAT/enhancer binding protein α is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol. Cell. Biol. 18:4301-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roebroek, A. J., J. A. Schalken, C. Onnekink, H. P. Bloemers, and W. J. Van de Ven. 1987. Structure of the feline c-fes/fps proto-oncogene: genesis of a retroviral oncogene. J. Virol. 61:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roebroek, A. J., J. A. Schalken, J. S. Verbeek, A. M. Van den Ouweland, C. Onnekink, H. P. Bloemers, and W. J. Van de Ven. 1985. The structure of the human c-fes/fps proto-oncogene. EMBO J. 4:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rogers, J. A., R. D. Read, J. Li, K. L. Peters, and T. E. Smithgall. 1996. Autophosphorylation of the Fes tyrosine kinase. Evidence for an intermolecular mechanism involving two kinase domain tyrosine residues. J. Biol. Chem. 271:17519-17525. [DOI] [PubMed] [Google Scholar]
- 69.Samarut, J., B. Mathey-Prevot, and H. Hanafusa. 1985. Preferential expression of the c-fps protein in chicken macrophages and granulocytic cells. Mol. Cell. Biol. 5:1067-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ′mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 17:6419.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scott, E. W., M. C. Simon, J. Anastasi, and H. Singh. 1994. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265:1573-1577. [DOI] [PubMed] [Google Scholar]
- 73.Senis, Y., R. Zirngibl, J. McVeigh, A. Haman, T. Hoang, and P. A. Greer. 1999. Targeted disruption of the murine fps/fes proto-oncogene reveals that Fps/Fes kinase activity is dispensable for hematopoiesis. Mol. Cell. Biol. 19:7436-7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shibuya, M., and H. Hanafusa. 1982. Nucleotide sequence of Fujinami sarcoma virus: evolutionary relationship of its transforming gene with transforming genes of other sarcoma viruses. Cell 30:787-795. [DOI] [PubMed] [Google Scholar]
- 75.Shimozaki, K., K. Nakajima, T. Hirano, and S. Nagata. 1997. Involvement of STAT3 in the granulocyte colony-stimulating factor-induced differentiation of myeloid cells. J. Biol. Chem. 272:25184-25189. [DOI] [PubMed] [Google Scholar]
- 76.Stein, P. H., J. D. Fraser, and A. Weiss. 1994. The cytoplasmic domain of CD28 is both necessary and sufficient for costimulation of interleukin-2 secretion and association with phosphatidylinositol 3′-kinase. Mol. Cell. Biol. 14:3392-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taubman, M. B., J. T. Fallon, A. D. Schecter, P. Giesen, M. Mendlowitz, B. S. Fyfe, J. D. Marmur, and Y. Nemerson. 1997. Tissue factor in the pathogenesis of atherosclerosis. Thromb. Haemostasis 78:200-204. [PubMed] [Google Scholar]
- 78.Thomas, S. M., P. Soriano, and A. Imamoto. 1995. Specific and redundant roles of Src and Fyn in organizing the cytoskeleton. Nature 376:267-271. [DOI] [PubMed] [Google Scholar]
- 79.Torigoe, T., R. O'Connor, D. Santoli, and J. C. Reed. 1992. Interleukin-3 regulates the activity of the LYN protein-tyrosine kinase in myeloid-committed leukemic cell lines. Blood 80:617-624. [PubMed] [Google Scholar]
- 80.Valledor, A. F., F. E. Borras, M. Cullell-Young, and A. Celada. 1998. Transcription factors that regulate monocyte/macrophage differentiation. J. Leukoc. Biol. 63:405-417. [DOI] [PubMed] [Google Scholar]
- 81.Vuori, K., H. Hirai, S. Aizawa, and E. Ruoslahti. 1996. Introduction of p130cas signaling complex formation upon integrin-mediated cell adhesion: a role for Src family kinases. Mol. Cell. Biol. 16:2606-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Whalen, A. M., S. C. Galasinski, P. S. Shapiro, T. S. Nahreini, and N. G. Ahn. 1997. Megakaryocytic differentiation induced by constitutive activation of mitogen-activated protein kinase kinase. Mol. Cell. Biol. 17:1947-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Woodring, P. J., T. Hunter, and J. Y. Wang. 2001. Inhibition of c-Abl tyrosine kinase activity by filamentous actin. J. Biol. Chem. 276:27104-27110. [DOI] [PubMed] [Google Scholar]
- 84.Yam, L. T., C. Y. Li, and W. H. Crosby. 1971. Cytochemical identification of monocytes and granulocytes. Am. J. Clin. Pathol. 55:283-290. [DOI] [PubMed] [Google Scholar]
- 85.Yu, G., T. E. Smithgall, and R. I. Glazer. 1989. K562 leukemia cells transfected with the human c-fes gene acquire the ability to undergo myeloid differentiation. J. Biol. Chem. 264:10276-10281. [PubMed] [Google Scholar]