Abstract
Study of the mechanism(s) of genomic instability induced by the c-myc proto-oncogene has the potential to shed new light on its well-known oncogenic activity. However, an underlying mechanism(s) for this phenotype is largely unknown. In the present study, we investigated the effects of c-Myc overexpression on the DNA damage-induced G1/S checkpoint, in order to obtain mechanistic insights into how deregulated c-Myc destabilizes the cellular genome. The DNA damage-induced checkpoints are among the primary safeguard mechanisms for genomic stability, and alterations of cell cycle checkpoints are known to be crucial for certain types of genomic instability, such as gene amplification. The effects of c-Myc overexpression were studied in human mammary epithelial cells (HMEC) as one approach to understanding the c-Myc-induced genomic instability in the context of mammary tumorigenesis. Initially, flow-cytometric analyses were used with two c-Myc-overexpressing, nontransformed immortal lines (184A1N4 and MCF10A) to determine whether c-Myc overexpression leads to alteration of cell cycle arrest following ionizing radiation (IR). Inappropriate entry into S phase was then confirmed with a bromodeoxyuridine incorporation assay measuring de novo DNA synthesis following IR. Direct involvement of c-Myc overexpression in alteration of the G1/S checkpoint was then confirmed by utilizing the MycER construct, a regulatable c-Myc. A transient excess of c-Myc activity, provided by the activated MycER, was similarly able to induce the inappropriate de novo DNA synthesis following IR. Significantly, the transient expression of full-length c-Myc in normal mortal HMECs also facilitated entry into S phase and the inappropriate de novo DNA synthesis following IR. Furthermore, irradiated, c-Myc-infected, normal HMECs developed a sub-G1 population and a >4N population of cells. The c-Myc-induced alteration of the G1/S checkpoint was also compared to the effects of expression of MycS (N-terminally truncated c-Myc) and p53DD (a dominant negative p53) in the HMECs. We observed inappropriate hyperphosphorylation of retinoblastoma protein and then the reappearance of cyclin A, following IR, selectively in full-length c-Myc- and p53DD-overexpressing MCF10A cells. Based on these results, we propose that c-Myc attenuates a safeguard mechanism for genomic stability; this property may contribute to c-Myc-induced genomic instability and to the potent oncogenic activity of c-Myc.
The proto-oncogene c-myc has been implicated as a central switch in the onset or progression of many types of cancer. However, the underlying molecular mechanism(s) responsible for c-Myc-induced tumorigenesis remains elusive. Human c-Myc is a 439-amino-acid protein that when bound to its partner, Max, can interact with DNA and function as a transcription factor (4). Myc-Max heterodimerization, which occurs through the interaction of C-terminal basic helix-loop-helix and leucine zipper motifs, is required for virtually all well-known c-Myc-associated phenotypes: cell proliferation, growth, transformation, and apoptosis (9, 15). In addition to these well-established phenotypes, recent studies have demonstrated that c-Myc expression can cause gross chromosomal aberrations and gene amplifications (16, 35, 36). The onset and progression of tumors depend upon the stepwise accumulation of somatic mutations in proto-oncogenes and tumor suppressor genes. Therefore, destabilizing genomic integrity may accelerate the accumulation of mutations required for the aggressive growth and malignant progression of tumors (6, 29, 34). As such, the c-Myc-induced genomic instability phenotype may provide an informative paradigm for understanding oncogene-induced tumor progression in general.
One mechanism crucial to the maintenance of genomic stability is that of the DNA damage-induced checkpoints (12, 24, 44, 56). Cell cycle checkpoints ensure the proper execution of sequential events of the eukaryotic cell cycle; unprepared entry into either S phase or M phase can be fatal. As a cellular surveillance system for DNA damage, the DNA damage-induced checkpoint is a specific class of cell cycle checkpoints that ensure that cells containing damaged DNA do not proceed into S phase or into mitosis (1, 11, 32, 62). Many biochemical components of the G1/S checkpoint have been identified, such as the tumor suppressor p53 and the cyclin-dependent kinase inhibitor (CKI) p21/Cip1 (cyclin-dependent kinase inhibitor protein 1) (22, 61). When stabilized by a DNA damage signal, p53 is homotetramerized and acts as a transcription factor. Importantly, CKI p21/Cip1 links p53 and the DNA damage-induced inhibition of cell cycle progression (11, 13). These roles of p53 and p21/Cip1 in the DNA damage response have been firmly established through recent studies using gene knockout approaches (5, 10, 57, 58). Inhibition of cyclin/Cdk then leads to hypophosphorylation of the tumor suppressor retinoblastoma protein (Rb). The hypophosphorylated form of Rb (pRb) actively sequesters the E2F-1 transcription factor, preventing transactivation of E2F-1 targets. Since E2F-1-mediated transactivation is essential for progression into S phase, Rb-mediated inhibition of E2F-1 activity causes cell cycle arrest at G1/S. The crucial involvement of Rb in the DNA damage-induced G1/S arrest was demonstrated in a study employing Rb−/− nullizygous mouse fibroblasts.
Expression of c-Myc is highly regulated throughout the cell cycle (4), and deregulation of c-Myc leads to the alteration of cell cycle regulation, such as a shortened G1 and facilitation of G1/S progression (43). Therefore, we hypothesized that c-Myc overexpression may also override or alter the scheduled cellular response (cell cycle arrest) to DNA damage at the G1/S boundary. In our experiments with nontransformed immortal HMEC lines and normal mortal HMECs, we demonstrated that a transient excess of c-Myc activity was sufficient to alter the G1/S checkpoint. For comparison, two independent controls were also used: an N-terminally truncated c-Myc (c-MycS) (50) and a dominant negative p53 (p53DD) (47). Based on these findings, we suggest a model in which c-Myc can lead to an elimination of a crucial safeguard mechanism for genomic stability in human mammary epithelial cells (HMECs).
MATERIALS AND METHODS
Cell culture.
To study the effects of c-Myc overexpression in HMECs, both nontransformed immortal HMEC lines (184A1N4 and MCF10A) and normal mortal HMECs were used. The 184A1N4 clone (provided by M. Stampfer, Lawrence Berkeley National Laboratory and University of California, Berkeley, Calif.) was initially derived from an epithelial outgrowth of reduction mammoplasty tissue that was subjected to chemical immortalization but not to malignant transformation (51, 52). The 184A1N4-Myc line was originally established via retroviral transfection of 184A1N4 cells with a murine c-myc construct under the control of a constitutively active Moloney mouse leukemia virus (MoMuLV) long terminal repeat (LTR) promoter (54). Both 184A1N4 and 184A1N4-Myc were maintained in improved minimal essential medium (Life Technologies, Rockville, Md.) supplemented with 0.5% fetal calf serum, 10 ng of epidermal growth factor (EGF) per ml, 5 μg of insulin per ml, and 0.5 μg of hydrocortisone per ml. The wild-type genomic sequences of p53 genes in 184A1N4 and 184A1N4-Myc cells were confirmed by PCR sequencing of all exons at the Molecular Diagnostics Shared Resource of the Lombardi Cancer Center. MCF10A (obtained from the American Type Culture Collection, Manassas, Va.) was originally derived from a subcutaneous mastectomy, performed on a 36-year-old parous premenopausal woman; cells were immortalized spontaneously (49, 53). MCF10A was maintained in a 1:1 mixture of Dulbecco's modified Eagle medium and nutrient mixture F-12 medium (Life Technologies) supplemented with 5% horse serum, 20 ng of EGF per ml, 10 μg of insulin per ml, and 0.5 μg of hydrocortisone per ml. Normal, finite-life-span HMECs (1001-8, CC-2551) at passage 8, originally derived from reduction mammoplasty tissues, were purchased from Clonetics-BioWhittaker, Inc. (Walkersville, Md.), and cells between passages 8 and 10 were used for experiments (roughly three population doublings per passage). Population doubling times (PDTs) for normal HMECs were roughly 18 to 24 h under in vitro culture conditions. Normal HMECs were maintained according to the supplier's instructions in mammary epithelial cell growth medium (CC-3152), supplemented with 52 μg of bovine pituitary extract per ml, 10 ng of human EGF per ml, 5 μg of insulin per ml, and 0.5 μg of hydrocortisone per ml and were grown in 37°C incubators with low (0.1 to 0.2%) CO2 settings.
Transgene constructs and retroviral infection.
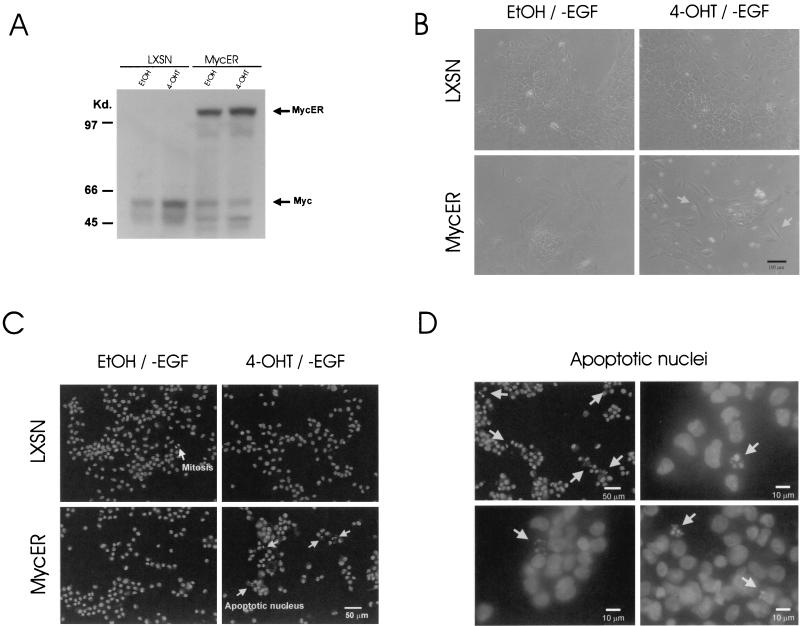
A full-length human c-myc cDNA construct, cloned in the pLXSN (LTR-gene X-simian virus 40-Neo) retroviral vector, was obtained from M. Stampfer and P. Yaswen (Lawrence Berkeley National Laboratory and University of California). In pLXSN, transgene expression and polyadenylation are facilitated by a Moloney murine sarcoma virus (MoMuSV) LTR promoter and a MoMuLV LTR polyadenylation signal (41). To create a retroviral construct expressing c-MycS, c-myc cDNA with a 5′ truncation was prepared through sequential recombinant DNA manipulations. Human c-myc cDNA has a unique recognition site for EcoRV between the first AUG (the second translation start codon) and the second AUG (see Fig. 2A), and pBluescript II SK(+/−) possesses a unique EcoRV site which is immediately behind the EcoRI site in the multiple cloning site (MCS) of the vector. Therefore, after the initial subcloning of human c-myc cDNA into the EcoRI site at the MCS of the cloning vector, EcoRV fragments containing the 5′-truncated form of the human c-myc cDNA (c-mycS) were isolated by digesting the initial construct with EcoRV and were then subcloned back into the HpaI site in the MCS of pLXSN retroviral vector by blunt-end ligation (see Fig. 2B). A subclone with c-mycS positioned in the correct direction was selected, and the predicted deletion of the 5′ region containing MB-I was confirmed by DNA sequencing. The pLXSN-p53DD construct was provided by M. Oren (Weizmann Institute of Science, Rehovot, Israel). All cloning junctions in each construct were confirmed by DNA sequencing. For the preparation of a high-titer retrovirus stock, PA317 retroviral packaging cells were transfected with each construct, and G418-resistant high-titer-producing clones were selected. To obtain stably transfected MCF10A pooled clones, cells were infected at a multiplicity of infection (MOI) of 5 in the presence of 8 μg of Polybrene per ml twice, sequentially, in order to increase the viral copy number in the infected cells (41). After a week of drug selection with 200 μg of G418 per ml, at least 1,000 independent colonies were pooled. The clones were pooled in order to overcome any effect of clonal variation and to examine the average effects of overexpression of transgenes. The pBabepuro-MycER constructs (encoding a fusion protein between N-terminal full-length human c-Myc and a C-terminal engineered ligand-binding domain of a murine estrogen receptor; its inactive c-Myc conformation changes into an active c-Myc form when it binds to 4-hydroxy-tamoxifen [4-OHT]) (19, 31) were obtained from T. Littlewood (Imperial Cancer Research Fund, London, United Kingdom) and L. Z. Penn (University of Toronto, Toronto, Canada). MycER was subcloned into an LXSN vector in order to have all constructs in a single type of retroviral backbone, and the L-MycER-SN retrovirus was used to infect MCF10A for the isolation of single clones. For the transient expression of transgenes in normal HMECs, cells were infected at an MOI of 5 for 12 h in the presence of 8 μg of Polybrene per ml. Cells were then grown for 24 or 48 h to allow transgene expression and harvested for experimentation.
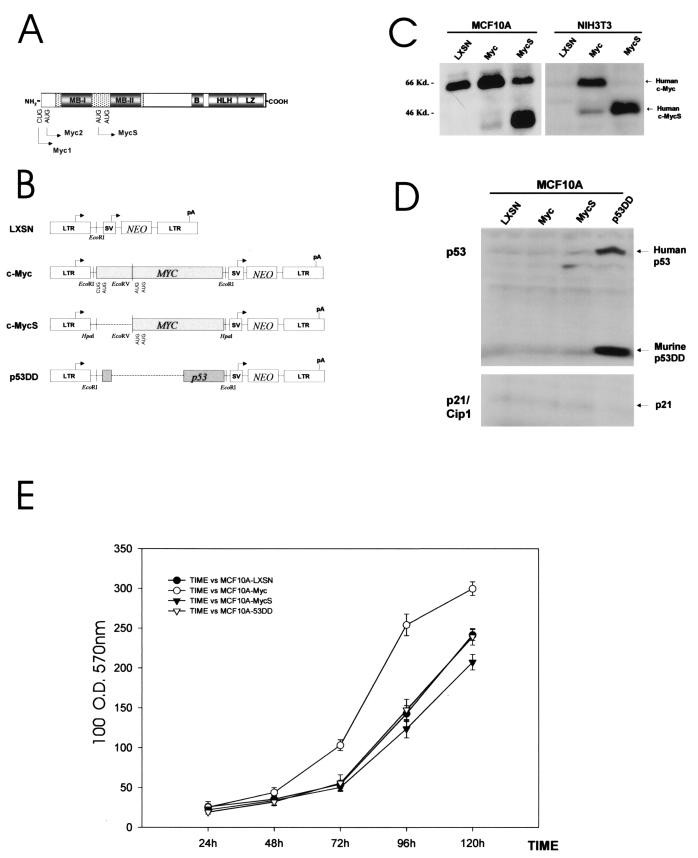
FIG. 2.
Retroviral constructs. (A) Protein structure of human c-Myc. c-Myc has well-separated functional domains in its amino terminus and its carboxy terminus. The C-terminal structure promotes the essential DNA binding activity through heterodimer formation with Max. Basic (B), helix-loop-helix (HLH), and leucine zipper (LZ) regions are essential to Myc-Max heterodimerization. The N-terminal structure determines the transcription-regulatory activity of c-Myc. This transcription-regulatory domain, represented by shaded regions in N terminus, contains highly conserved Myc box I (MB-I, amino acids 45 to 63) and Myc box II (MB-II, amino acids 129 to 141) sequences. Myc1 (67 kDa) and Myc2 (64 kDa) initiate their translation at the first start codon (CUG) and at the second start codon (AUG) located upstream of MB-I. However, when translation is initiated at the third start codon (the second AUG), it produces a truncated form of c-Myc (c-MycS, 46 kDa) without an MB-I sequence, since the internal start codon is located downstream of MB-I. (B) Schematic maps of retroviral constructs used in this study. The pLXSN retroviral vector has two promoters (the 5′ MoMuSV LTR directs transcription of gene X at the MCS, containing EcoRI and HpaI sites, and the simian virus 40 early promoter [SV] directs transcription of neomycin resistance gene [NEO]). The dotted lines in pLXSN-MycS and pLXSN-p53DD represent the deleted sequences in each construct. c-MycS was based on the full-length c-Myc construct (see Materials and Methods for details). Polyadenylation of the exogenous mRNA transcript is mediated by the polyadenylation signal (pA) in the 3′ MoMuLV LTR. (C) Immunoblot analysis for exogenous human c-Myc and c-MycS proteins in c-Myc- and c-MycS-infected MCF10A and NIH 3T3 cells. Anti-human c-Myc monoclonal antibody 9E10 detected endogenous human c-Myc in MCF10A-LXSN and failed to detect murine c-Myc in NIH 3T3 cells. Production of c-MycS appeared to be tightly regulated; it is apparent only in cells with deregulated c-Myc and c-MycS. Equal loading was confirmed by staining the membrane blot with amido black (data not shown). (D) Immunoblot analysis for p53 proteins in retrovirus-infected MCF10A cells.Overexpression of murine p53DD (∼19 kDa) in p53DD-infected MCF10A cells was clearly detected with an anti-pan-species p53 C-terminal monoclonal antibody, PAb421. The accompanying increase of endogenous full-length human p53 (53 kDa) suggests the efficient binding of p53DD to endogenous p53. The same blot was reprobed with anti-p21/WAF1 antibody to confirm the absence of p53 function in p53DD-infected cells. Equal loading was confirmed by staining the membrane blot with amido black (data not shown). (E) Proliferation of the infected MCF10A HMECs was measured using a 96-well-plate assay. The x axis represents incubation time after plating, and the y axis represents an arbitrary unit of optical density at 570 nm. After crystal violet staining of cells on the plates at each time point, stained total cell proteins were dissolved in the sodium citrate solutions, and the absorption at 570 nm was measured as readout for cell proliferation. Error bars show standard deviations from sextuplicate samples.
Cell proliferation assay.
On day 0, exponentially growing (∼70% confluence) cells were harvested and plated on flat-bottom, 96-well plates (Becton Dickinson Labware, Franklin Lakes, N.J.) at a density of 104 cells per well. After overnight attachment, the media were refreshed. For day 1 (24-h) samples, immediately after media were refreshed, cells were washed twice with phosphate-buffered saline (PBS) and stained for 15 min with 0.5% crystal violet solutions containing 25% methanol as a fixative. After staining, cells were washed three times with tap water. Next, plates were allowed to air dry. After the plates were dry, the stain was dissolved by adding 100 μl of 0.1 M sodium citrate-50% ethanol per well. The amount of stain was then quantitated and analyzed on a Vmax kinetic microplate reader (Molecular Devices, Menlo Park, Calif.) at 570 nm with the SOFTmax program. After the first medium change on day 1, culture media were not further refreshed, in order to prevent any disturbance of the growing cells.
γ-Irradiation for inducing DNA damage.
DNA damage was induced by ionizing radiation (IR) treatment, using a 137Cs source γ-irradiator (Mark-I irradiator; JL Shepherd, San Fernando, Calif.), at a rate of 3.6 Gy per min, until the specified absorbed dosage was reached. For irradiation of attached cells in culture dishes, cells were plated and irradiated on day 2. For irradiation of suspended cells in conical flasks, each sample was adjusted to the same cell density (∼3 × 106 cells per 5-ml culture medium in a 15-ml conical tube). All experimental samples, either in dishes or in flasks, were kept rotating on the turntable inside the irradiator during the irradiation procedure.
Flow-cytometric analysis of the cell cycle.
For flow-cytometric experiments to determine DNA content and the cell cycle profile, cells were plated at a density of 1 × 106 to 3 × 106 cells per T-75 flask. At various time points, cells were harvested and fixed with ice-cold 100% ethanol with vortexing at low speed; cells were then placed at −20°C for overnight. After fixation, cells were centrifuged and washed once with PBS containing 1% bovine serum albumin. For staining with DNA dye, cells were resuspended in 0.5 to 1 ml of propidium iodide (PI) solution containing RNase and incubated at 37°C for 30 min, followed by overnight incubation at 4°C. Cell cycle profiles were obtained with a FACScan flow cytometer (Becton Dickinson, San Jose, Calif.) and data were analyzed with ModFit software (Verity Software House, Inc.) for cell cycle analysis and ReproMan software (TruFacts Software Inc.) for region analysis of sub-G1 fractions.
BrdU labeling and immunofluorescence for de novo DNA synthesis.
DNA synthesis was monitored by measuring incorporation of the artificial thymidine nucleotide analog 5-bromo-2′-deoxyuridine (BrdU) (Sigma, St. Louis, Mo.) into newly synthesized DNA. After γ-irradiation, cells were plated on glass coverslips that had been prewarmed in media in 12-well culture plates. Cells (5 × 104 per well) were plated and allowed, for the first 24 h, to grow in complete medium without BrdU. Next, cells were refreshed with complete medium containing 10 μM (3-μg/ml) BrdU and incubated for an additional 6 or 24 h for BrdU incorporation. BrdU was added to the medium from a 10,000× stock solution in distilled water. After BrdU incubation, cells were washed three times with PBS (10 min each time) and fixed in 4% paraformaldehyde-PBS for 20 min. Next, the fixed cells were permeabilized with 0.5% Triton X-100-PBS. After the partial denaturation of DNA with 2 N HCl, cells were incubated with a fluorescein-conjugated anti-BrdU mouse monoclonal antibody (Roche/Boehringer Mannheim, Indianapolis, Ind.), at a 1:2 dilution (50 μg/ml) for 1 h in the dark. Cells were then washed with PBS three times and counterstained with DNA dyes (PI or DAPI [4′,6′-diamidino-2-phenylindole]) for 5 min. Finally, after several additional PBS washes, the cover slips were mounted on glass slides with Prolong mounting medium (Molecular Probes, Eugene, Oreg.) for fluorescence microscopy. Quantitative analysis of immunofluorescence data was carried out with Image-Pro Plus software (Media Cybernetics, Silver Spring, Md.).
Western blot analysis.
Cell lysates were prepared in 2× Laemmli sample buffer, containing 1% sodium dodecyl sulfate (SDS) and β-mercaptoethanol. After approximately 106 cells had been collected in 100 μl of sample buffer, whole-cell lysates were boiled for 10 min, cleared by centrifugation at 4°C, and stored at −80°C until use. For SDS-polyacrylamide gel electrophoresis, 20 μl of the cleared lysates was loaded into a well of the gel. Separated proteins were transferred to Immobilon-P polyvinylidene fluoride membranes (Millipore, Bedford, Mass.). After tank transfer, membranes were blocked for 1 h at room temperature in PBS-0.05% Tween 20 (PBS-T) containing 5% nonfat dried milk and then probed with primary antibody diluted in PBS-T overnight in a 4°C cold room. Antibodies and concentrations were as follows: for human c-Myc, 9E10, 2 μg/ml (4851A; Pharmingen, San Diego, Calif.); for human p53, PAb1801, 2.5 μg/ml (Ab-2; Oncogene Research Products, Boston, Mass.); pan-species p53 PAb 421, 10 μg/ml (Ab-1; Oncogene Research Products); for p21/WAF1, EA10, 1 μg/ml (Ab-1; Oncogene Research Products); for human Rb, 1 μg/ml (14001A; Pharmingen); for cyclin A, H-432, 1 μg/ml (sc-751; Santa Cruz Biotechnology, Santa Cruz, Calif.); for p14/ARF, 1 μg/ml (AHZ0472; Biosource, Camarillo, Calif.); and for α-tubulin, DM1A, 1:1,000 dilution (Ab-2; NeoMarkers, Fremont, Calif.). Blots were then washed with PBS-T three times for 10 min. All blots were next incubated in horseradish peroxidase-conjugated secondary goat anti-mouse or anti-rabbit antibody (1:3,000 dilution; Bio-Rad) for 1 h at room temperature for chemiluminescent detection.
Hoechst staining for microscopic analysis of apoptosis.
For the analysis of c-Myc-induced apoptosis with the MCF10A-MycER system, cells were starved of EGF for 6 days in the presence of ethanol (a vehicle control) or 1 μM 4-OHT. After harvest, cells were resuspended in 50 μl of PBS, and cell density was checked by phase-contrast microscopy. Next, the entire population of resuspended cells was added to 350 μl of Hoechst staining solution consisting of 10 μg of Hoechst 33258 (B-2883; Sigma) per ml, 1% NP-40, and 3.7% formaldehyde in PBS. Finally, the condensed nuclei of apoptotic cells were examined by fluorescent microscopy.
RESULTS
c-Myc-overexpressing 184A1N4 cells exhibited elimination of G1/S arrest following γ-irradiation.
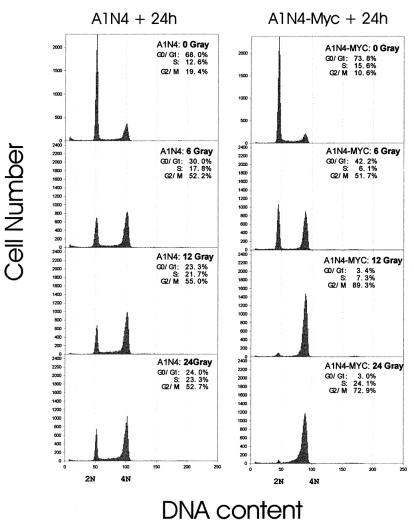
To determine if overexpression of c-Myc alters the DNA damage-induced checkpoints, we first investigated changes in cell cycle progression, following ionizing radiation (IR), of an isogenic pair of HMEC lines, 184A1N4 and 184A1N4-Myc. We chose γ-irradiation as a DNA-damaging agent because γ-irradiation clearly induces DNA strand breaks. The 184A1N4 and 184A1N4-Myc cells were first synchronized at G0/G1 phase by the EGF withdrawal technique. Previous studies with these lines have demonstrated that the EGF withdrawal technique allows a reversible G0/G1 synchronization (43). As expected, nonirradiated (0-Gy control) 184A1N4 and 184A1N4-Myc cells showed cell cycle progression and a gradual loss of synchrony during the 24-h post-IR incubation (roughly one cell cycle period of 184A1N4 and 184A1N4-Myc). In response to DNA damage induced by 6, 12, and 24 Gy of IR, 30, 23.3, and 24% of 184A1N4 cells, respectively, were present in G0/G1 phase (represented by 2N DNA content) and 52.2, 55, and 52.7%, of cells, respectively, were in G2/M phase (represented by 4N DNA content) (Fig. 1). In 184A1N4-Myc cells, surprisingly, 42.2% of cells were in G0/G1 and 51.7% of cells were in G2/M, following 6 Gy of IR. However, only about ∼3% of 184A1N4-Myc cells remained in G0/G1 phase, and most (89.3% after 12 Gy and 72.9% after 24 Gy) were in G2/M phase following 12 and 24 Gy of IR. We interpreted these results to suggest that in response to DNA damage a significant percentage (∼25 to 30%) of parental 184A1N4 cells arrested at the G1/S checkpoint. In contrast, a significant percentage (compared to 12- and 24-Gy profiles) of 184A1N4-Myc cells were able to repopulate G0/G1 following 6 Gy of IR, and cells were arrested at G2/M only following 12 and 24 Gy of IR. This result suggested that 184A1N4-Myc cells were altered in G1/S arrest following IR exposures of up to 24 Gy and in G2/M arrest following IR exposures of up to 6 Gy. In addition, our results also suggested that the strength of the G1/S arrest signal correlates with the amount of DNA damage. Considering 184A1N4-Myc results, 24-Gy-induced DNA damage retarded the S-to-G2/M progression more severely (S-phase percentage: 7.3% after 12 Gy of IR and 24.1% after 24 Gy of IR) than did 12-Gy-induced DNA damage.
FIG. 1.
Elimination of G1/S arrest following γ-irradiation in c-Myc-overexpressing 184A1N4 cells. Flow-cytometric profiles representing cell cycle changes of 184A1N4 or 184A1N4-Myc after 24 h of post-IR incubation. DNA content flow-cytometric analysis, measuring the amount of fluorescent dye (PI) intercalated into DNA, provides a readout for the distribution of cells with different DNA contents in the population. Before IR treatment, 184A1N4 and 184A1N4-Myc cells were synchronized in G0/G1 phase by an EGF withdrawal method. After postrelease recovery for 3 h in complete media containing EGF, HMECs were irradiated with 0-, 6-, 12-, and 24-Gy doses of IR to induce DNA damage. HMECs were then allowed to progress in the cell cycle during 24 h of post-IR incubation.
Retrovirus-mediated introduction of transgenes into MCF10A HMECs.
To further investigate DNA damage-induced G1/S arrest in the background of an independently derived nontransformed HMEC line, we examined the response of MCF10A cells to γ-irradiation; the MCF10A line has been reported to have wild-type, functional p53 (20, 39). For MCF10A and later studies, we irradiated an actively growing, asynchronous population of cells in order to avoid complications resulting from repeated growth factor withdrawal and stimulation. According to the flow-cytometric profile, MCF10A cells exhibited an increase in G0/G1 phase fraction (represented by 2N DNA content) and exhibited a significant reduction in S-phase fraction following IR with 4 to 12 Gy (data not shown). We next introduced a human c-myc cDNA construct into MCF10A cells by retroviral infection (Fig. 2A and C). In addition to c-myc, we also employed two independent controls, a construct encoding an N-terminally truncated c-Myc (c-mycS) and p53DD (Fig. 2B). c-MycS was previously known to be defective in transactivation of c-Myc targets (50). However, a recent study reported that some targets were transactivated by c-MycS (25). It served as an additional control for comparison, together with the empty retroviral vector (LXSN) control. p53DD, an N-terminally truncated, dominant negative (tetramerization domain only) form of murine p53 (2, 47), served initially as a potential positive control (Fig. 2B and D). p53DD was chosen in case the loss of p53 function provides an abolished-checkpoint phenotype in HMECs, as previously established in other cell types. Elimination of the G1/S checkpoint in the p53DD-overexpressing MCF10A cells was confirmed in subsequent experiments, and it served as a working positive control to be compared with the effects of c-Myc overexpression. After MCF10A cells had been infected with the recombinant retroviruses, we pooled all G418-resistant colonies from each infection procedure, in order to prevent clonal variations. In MCF10A-Myc cells (MCF10A cells infected with human c-myc), the level of c-Myc expression was greater than that of the MCF10A-LXSN control (MCF10A cells infected with empty retroviral vector only) (Fig. 2C). Since it is known that c-Myc downregulates its own expression through an autorepression mechanism (14), a majority of this signal may be due to expression of the introduced exogenous c-myc. The constitutive overexpression of exogenous c-myc was even more evident following γ-irradiation (see Fig. 4). We also infected the mouse embryonic fibroblast line NIH 3T3 with the same retroviruses and confirmed the specific expression of the exogenous human c-myc by retroviral infection (Fig. 2C). As expected, cells infected with c-mycS showed only the expression of a 46-kDa protein, a truncated version of human c-Myc. Expression of p53DD was also confirmed by Western analysis (Fig. 2D). Importantly, the overexpression of murine p53DD led to the increase of endogenous human full-length p53 proteins. This observation verified that the murine p53DD proteins bind to endogenous human p53 proteins to stabilize them and to inhibit their function. Hetero-oligomer formation between the p53DD miniproteins and the full-length p53 proteins has been known to impair the scheduled degradation of endogenous p53 and to lengthen its half-life (47). The level of p21/Cip1 proteins was also low in MCF10A-p53DD cells, further indicating that the increased amount of p53 proteins in these cells is not functional.
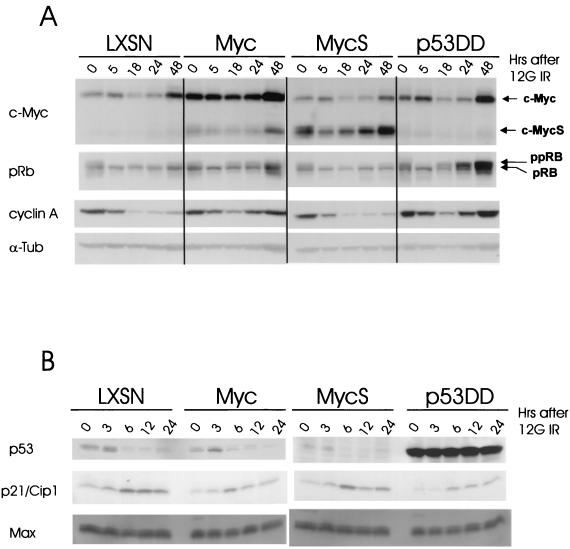
FIG. 4.
Time-course analysis of changes in cell cycle-related proteins following γ-irradiation. (A) Immunoblot analysis for c-Myc, Rb, or cyclin A in whole-cell lysates prepared at the indicated time points after 12 Gy of IR. All cells were irradiated, and lysates were prepared immediately following γ-irradiation. An equal loading control is provided by the α-tubulin (α-Tub) blot. The same membrane was reprobed repeatedly for each protein. ppRb in the slow-migrating bands and pRb in the fast-migrating bands are indicated. (B) Immunoblot analyses for p53 and p21/Cip1 in whole-cell lysates prepared at the indicated time points after 12 Gy of IR. An equal-loading control is provided by the Max blot. The same membrane was reprobed repeatedly for each protein.
To confirm the proproliferation activity of c-Myc, we performed a 96-well-plate proliferation assay (Fig. 2E). Although we plated exponentially growing (∼70% confluence) cells on day 0, it appeared that the initial PDT is much longer than the later PDT. Since we usually plated the exponentially growing cells on the day before experiments, the initial PDT was more relevant for most experiments. The estimated initial PDTs (based on a 48- to 72-h period) are 31.4 h for MCF10A-LXSN, 20.4 h for MCF10A-Myc, 32.2 h for MCF10A-MycS, and 27.6 h for MCF10A-p53DD cells. The growth rate of MCF10A-Myc was highest; this is consistent with proproliferation effects of c-Myc. The growth rate of MCF10A-MycS was lower than that of MCF10A-LXSN. MCF10A-p53DD grew marginally faster than the MCF10A-LXSN control in vitro.
Overexpression of c-Myc attenuated G1/S arrest following IR and induced the inappropriate DNA synthesis in MCF10A cells.
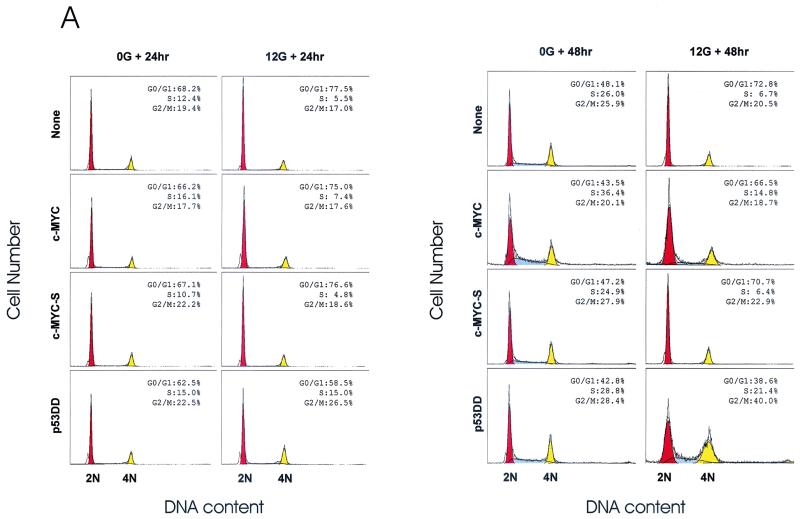
The MCF10A cells harboring the aforementioned transgenes were irradiated in suspension and were then replated in culture flasks. Following 24 or 48 h of post-IR incubation, cell cycle profiles were analyzed by flow cytometry. According to the computer generated, best-fit cell cycle modeling by ModFit software, irradiated c-Myc- or p53DD-overexpressing cells consistently showed a lower percentage of G0/G1 phase than the irradiated LXSN- or c-MycS-overexpressing cells (Fig. 3A). Interestingly, irradiated MCF10A cells did not show a prominent accumulation of cells at G2/M, suggesting an attenuated G2/M checkpoint in this HMEC line. However, it was not easy to interpret the changes because nonirradiated controls also changed their cell cycle profile after being replated in the culture flask. In terms of changes in S fraction, while c-Myc did not provide a dramatic effect on changes in S-phase fraction at 24 h post-IR, its effect was clear at 48 h post-IR, compared to those of LXSN and MycS. The effects of c-Myc overexpression could be clearly perceived when the higher S-phase fraction in nonirradiated MCF10A-Myc than in nonirradiated MCF10A-LXSN and MCF10A-MycS cells at 48 h post-IR was taken into account. Following IR, MCF10A-Myc cells exhibited a degree of reduction in S phase (to ∼46% of nonirradiated control: 16.1% S phase in nonirradiated and 7.4% S phase in irradiated cells) similar to those of MCF10A-LXSN and MCF10A-MycS cells at 24 h post-IR. However, at 48 h post-IR, irradiated MCF10A-Myc cells maintained 40.7% of the S-phase fraction of the nonirradiated control (36.4% S phase in nonirradiated and 14.8% S phase in irradiated cells), while MCF10A-LXSN cells displayed 25.7% of the S-phase fraction of nonirradiated controls (26.0% S phase in nonirradiated and 6.7% S phase in irradiated cells). Effects of loss of p53 function by p53DD were evident in that MCF10A-p53DD cells clearly exhibited the abolished G1/S arrest following IR (100% of S fraction in nonirradiated cells at 24 h post-IR and 74.3% of S fraction in nonirradiated cells at 48 h post-IR). Therefore, these results suggested the possibility that overexpression of c-Myc attenuates the maintenance, rather than the initial set-up, of the G1/S arrest following IR.
FIG. 3.
c-Myc attenuates IR-induced G1/S arrest in MCF10A cells. (A) Flow-cytometric profiles representing cell cycle changes of MCF10A-LXSN, MCF10A-Myc, MCF10A-MycS, and MCF10A-p53DD cells following IR. Asynchronous, exponentially growing MCF10A HMEC populations were harvested and irradiated in suspension with 0 or 12 Gy of IR. Next, infected cells were allowed to progress in the cell cycle for 24 or 48 h of post-IR incubation. Primary histogram data are displayed as fine line graphs, and the computer-generated best fit is shown with the orange (G0/G1), yellow (G2/M), and dashed (S) areas. (B) BrdU incorporation assay for de novo DNA synthesis following 0 or 12 Gy of IR. After irradiation of suspended cells, cells were replated and incubated for 24 h before an additional 24-h incubation with BrdU. After immunofluorescence staining with anti-BrdU monoclonal antibody (αBrdU) for nuclei that had undergone de novo DNA synthesis, all nuclei were counterstained with the nonspecific DNA-intercalating dye PI to allow quantification of the total number of nuclei in the same microscopic field. At least 300 PI-stained nuclei were analyzed for each infected population with image analysis software.
While the flow-cytometric data provided valuable information on the global changes in cell cycle progression in irradiated cells, they did not necessarily provide evidence for entry into S phase following IR. Therefore, in order to obtain more direct evidence for the inappropriate entry into S phase, we performed a BrdU incorporation assay, measuring de novo DNA synthesis and representing all cells encountering S phase following IR. MCF10A cells were preincubated for 24 h following IR before the addition of BrdU, in order to provide sufficient time for preexisting S-phase cells to move from S phase to the next cell cycle phase. Next, the culture media were changed to BrdU-containing complete media, and cells were incubated for an additional 24 h for BrdU incorporation. In all four groups of sample populations, although a 24-h BrdU incubation was allowed, nonirradiated controls failed to exhibit 100% saturation of BrdU incorporation (Fig. 3B) (LXSN, 45.4%; c-Myc, 53.2%; c-MycS, 50.7%; and p53DD, 54.8%). In these results, MCF10A-Myc and MCF10A-p53DD cells had more entries into S phase per unit of time; a greater percentage of nonirradiated cells incorporated BrdU for 24 h. As expected, while the irradiated MCF10A-LXSN and MCF10A-MycS cells demonstrated a significant loss of BrdU labeling (LXSN, 4.9%, and MycS, 1.5%) (Fig. 3B), both c-Myc- and p53DD-overexpressing cells exhibited a higher percentage of BrdU incorporation following IR (c-Myc, 29.2%, and p53DD, 46.8%) (Fig. 3B). Therefore, the flow-cytometric analysis provided valuable information on the post-IR changes in cell cycle progression, and the BrdU incorporation assay clearly demonstrated the effects of c-Myc on the inappropriate entry into S phase following IR. One consideration for the interpretation of our BrdU incorporation data might be that BrdU could be additionally incorporated into the damaged chromosomes during DNA repair following IR, regardless of checkpoint control. However, it is very unlikely that only c-Myc- or p53DD-infected cells undergo a massive DNA repair following the same dosage of IR.
Inappropriate hyperphosphorylation of Rb and reappearance of cyclin A following IR in c-Myc-overexpressing MCF10A cells.
To gain initial insight into the underlying molecular basis of the altered G1/S checkpoint in c-Myc-overexpressing cells, we measured post-IR time course changes in the levels of some important cell cycle-related proteins. We started with c-Myc itself. Interestingly, the oncoprotein itself was significantly downregulated, as early as 18 h post-IR, in the irradiated LXSN-, c-MycS-, and p53DD-infected MCF10A cells (Fig. 4A). However, the level of c-Myc stayed relatively high and was unaffected by IR treatment in MCF10A-Myc. Rb is a prominent target for Cdk2-cyclin E and Cdk2-cyclin A during scheduled G1/S progression, and we wanted to assess any post-IR changes in Rb phosphorylation status and Cdk2 holoenzyme status in each transfected cell population. Reprobing the same blot with an anti-Rb antibody demonstrated inappropriate hyperphosphorylation of Rb, following IR, selectively in c-Myc- and p53DD-infected cells. We irradiated exponentially growing, asynchronous cells, and various levels of phosphorylation in Rb were detected at the 0-h time point. In all sample groups, the majority of hyperphosphorylated Rb (ppRb) disappeared as early as 5 h post-IR. pRb dominated until 24 h post-IR in LXSN- and c-MycS-infected cells. In contrast, the irradiated c-Myc- and p53DD-infected cells showed a rapid reappearance of ppRb (slower-migrating bands) as early as 18 h post-IR. Although the effects of c-Myc on post-IR Rb phosphorylation appeared to be moderate and delayed with respect to the effects of p53DD, they were significant compared to those of LXSN and MycS controls. The strength of the ppRb signal of c-Myc at 48 h post-IR was comparable to that of p53DD at 24 h post-IR. Cyclin A is known to be a positive regulatory subunit for Cdk2 and abundant both in late-G1- and in S-phase cells; the levels of cyclins mainly determine the activities of Cdk holoenzymes. While the levels of cyclin A were dramatically suppressed in the irradiated LXSN- and c-MycS-infected cells 18 h post-IR, irradiated MCF10A-Myc and MCF10A-p53DD cells exhibited reappearance of cyclin A 24 h post-IR. The amount of cyclin A at 48 h post-IR in irradiated MCF10A-Myc cells was comparable to that of cyclin A at 24 h post-IR in irradiated MCF10A-p53DD cells.
To test the p53-dependent molecular changes to IR, we performed a time course Western blot analysis of p53 and its downstream effector, p21/Cip1. As expected, the endogenous p53 protein was upregulated in MCF10A-LXSN, MCF10A-Myc, and MCF10A-MycS cells at 3 h post-IR (Fig. 4B). Stabilized, but nonfunctional, endogenous, full-length p53 in MCF10A-p53DD was also apparent. Next, upregulation of p21/Cip1 appeared as early as 6 h post-IR following the upregulation of p53. Max, the c-Myc partner, is known to maintain a constant level throughout the cell cycle; it served as a loading control in the Western analysis.
A transient excess of c-Myc activity was sufficient to induce the inappropriate entry into S phase and DNA synthesis following IR.
In experiments with the permanently transfected MCF10A line, we pooled the G418-resistant colonies to provide an average phenotype without clonal variations. However, it was demonstrated in a fibroblast model that deregulated c-Myc activity could induce various genomic instabilities in a reasonably short period, 2 days (22). Therefore, to rule out any hypothetical effects potentially resulting from secondary genetic alterations acquired during the G418 selection period, it was necessary to determine whether the transient excess of c-Myc activity was sufficient to alter the G1/S arrest following IR. To address this question, we established a regulatable c-Myc system with MCF10A by utilizing the MycER construct. MycER is a fusion protein consisting of full-length human c-Myc (at the N terminus) and the hormone-binding domain of the murine estrogen receptor (ER) (at the C terminus). Only in the presence of an ER-binding ligand, such as 4-OHT, does the C-terminal hormone-binding domain fail to interfere with the function of c-Myc in the N terminus, enabling the activation of c-Myc in a posttranslational manner. First, we confirmed the expression of MycER in MCF10A cells, at the expected size of ∼110 kDa (Fig. 5A). In addition, Western analysis demonstrated that 4-OHT-treatment downregulated the endogenous c-Myc, in comparison to mock treatment with ethyl alcohol (EtOH) (vehicle control). This is consistent with negative feedback regulation at the c-myc promoter by active Myc-Max transcription factors. For the verification of a functional MCF10A-MycER system, we measured c-Myc-induced apoptosis in the absence of growth factor; a Hoechst 33258 dye assay was used to identify apoptotic nuclei. c-Myc is well known to promote apoptosis, particularly in the absence of growth and survival factors. Therefore, we starved cells of EGF for up to 6 days in the presence of EtOH or 4-OHT (Fig. 5B). After 4 days of this treatment, the morphological difference between EtOH-treated cells and 4-OHT-treated cells was identifiable. In contrast to the flattened cells obtained with EtOH treatment and EGF starvation, 4-OHT treatment with EGF starvation induced slender cells (Fig. 5B) and significant amounts of floating bodies. Using Hoechst 33258 dye staining, we were also able to identify fragmented and condensed apoptotic nuclei selectively in 4-OHT-stimulated MCF10A-MycER cells (∼3% of roughly 400 nuclei observed in samples of 4-OHT-treated, EGF-starved cells) (Fig. 5C and D). These observations confirmed that c-Myc activity could be regulated by 4-OHT in the MCF10A-MycER system.
FIG. 5.
Regulatable c-Myc system in MCF10A cells. (A) Immunoblot analysis confirms protein expression of MycER in MCF10A HMECs. After 48 h of EtOH or 1 μM 4-OHT treatment, total cell lysates were prepared and separated by SDS-polyacrylamide gel electrophoresis. (B) Phase-contrast microscopy of MCF10A-LXSN and MCF10A-MycER cells after 6 days of EGF starvation in the presence of EtOH or 1 μM 4-OHT. EtOH-treated MCF10A-MycER cells were severely flattened in the absence of EGF. 4-OHT-treated MCF10A-MycER cells demonstrated a marked increase of floating bodies and slender cells (arrows). (C) A Hoechst 33258 staining assay demonstrated EGF starvation-induced apoptosis only in cells with excess c-Myc activity. MCF10A-LXSN and MCF10A-MycER cells were EGF starved for 6 days in the presence of EtOH or 1 μM 4-OHT. Mitotic chromosomes were clearly distinguished from apoptotic nuclei. (D) Fragmented and condensed nuclei (arrows) resulting from c-Myc-induced apoptosis in the EGF-starved HMECs.
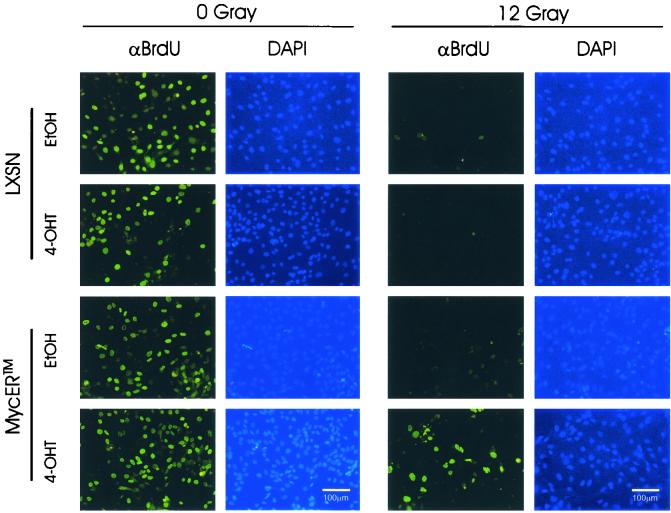
To determine directly if a transient excess of c-Myc activity is sufficient to attenuate DNA damage-induced G1/S arrest, we performed a BrdU incorporation assay, following 0 or 12 Gy of IR, in the presence of 4-OHT or EtOH (Fig. 6). In the absence of IR, 4-OHT-treated MycER cells showed a higher BrdU incorporation (37.1% of total nuclei) than EtOH vehicle-treated ones (33.6% of total nuclei). In irradiated MCF10A-MycER cells, only those stimulated with 4-OHT demonstrated a suppressed, but significant amount (28.2% of total nuclei) of BrdU-labeled nuclei, consistent with results with permanently transfected MCF10A cells. In addition, significant amounts of multimicronucleated nuclei displaying BrdU incorporation were selectively identified in γ-irradiated cells (Fig. 6) (our unpublished data). These BrdU-labeled micronuclei had morphologies distinct from those of the condensed apoptotic nuclei in these experiments. Therefore, although further in-depth analyses are required, it is unlikely that the BrdU incorporation occurred directly as a part of apoptosis.
FIG. 6.
A transient excess of c-Myc activity induced inappropriate entry into S phase following IR in MCF10A cells. BrdU incorporation was assayed for de novo DNA synthesis of MCF10A-MycER cells following IR. IR treatment was essentially the same as that in the stably transfected MCF10A cells. Following the harvest of asynchronous exponentially growing cells, cells were irradiated in suspension and replated for 24 h of incubation in the presence of EtOH or 1 μM 4-OHT before an additional 24-h incubation with BrdU under the same treatment conditions. After immunofluorescence staining with a fluorescein-linked anti-BrdU monoclonal antibody (αBrdU) to detect nuclei that had undergone de novo DNA synthesis, all nuclei were counterstained with the nonspecific DNA-intercalating dye DAPI to quantify the total number of nuclei in the same microscopic field. At least 150 DAPI-stained nuclei were analyzed for each treated population with image analysis software.
Transient overexpression of c-Myc induced inappropriate entry into S phase following IR in normal HMECs.
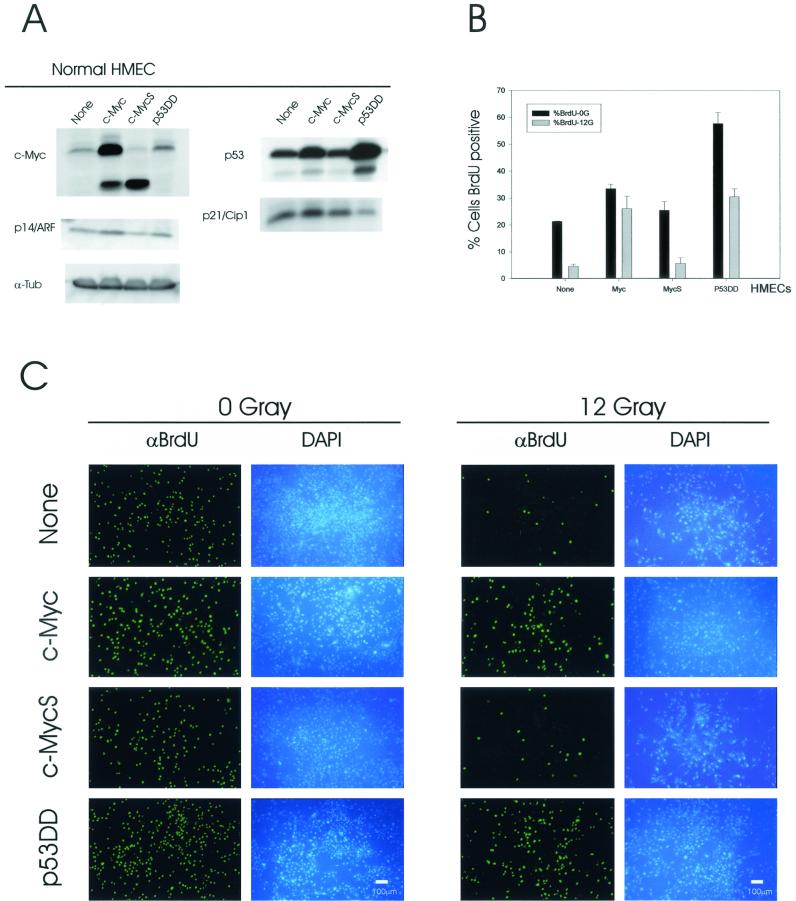
Establishment of immortal cell lines from primary human cells may involve mutations in known and/or unknown genes, complicating interpretation of cell cycle control and DNA damage response studies. Therefore, it was necessary to investigate whether overexpression of c-Myc attenuates the DNA damage-induced cell cycle checkpoints in normal mortal HMECs. For experiments with normal HMECs, we employed a transient-infection strategy (no selection with G418) in order to study the immediate effects of transient overexpression of c-Myc on the DNA damage responses. Normal HMECs were infected at a MOI of 5, theoretically increasing the chance of retroviral infection of most of target cells. For one of our controls, we used noninfected normal HMECs instead of empty retroviral vector-infected cells. It is generally known that recombinant retroviruses do not disturb the cell cycle, because they do not contain viral genes (A. Dusty Miller, The Fred Hutchinson Cancer Research Center, Seattle, Wash., personal communication). However, we wanted a control for any kind of potential effects of retroviral infection on cell cycle progression, at least at the moment of infection, so that we could study changes in cell cycle progression of undisturbed, normal HMECs following IR. We also used the c-MycS retrovirus as an additional control for the effects of c-Myc, based on the failure of c-MycS to induce attenuation of G1/S arrest in MCF10A. We confirmed the expression of transgenes in normal HMECs, following transient infection (Fig. 7A). At 48 h postinfection, levels of c-Myc and c-MycS were clearly elevated in c-Myc- and c-MycS-infected normal HMECs. c-Myc-infected HMECs demonstrated slight increases in levels of p53 and of p14/ARF, which are known transcriptional targets of Myc-Max transcription factors in some cell types (46). The p53DD-infected normal HMECs consistently demonstrated an increased level of endogenous p53 proteins that are probably stabilized by murine p53DD. Consistent with data from MCF10A-p53DD cells (Fig. 2D), p53DD-infected cells exhibited a decreased level of p21/Cip1 proteins.
FIG. 7.
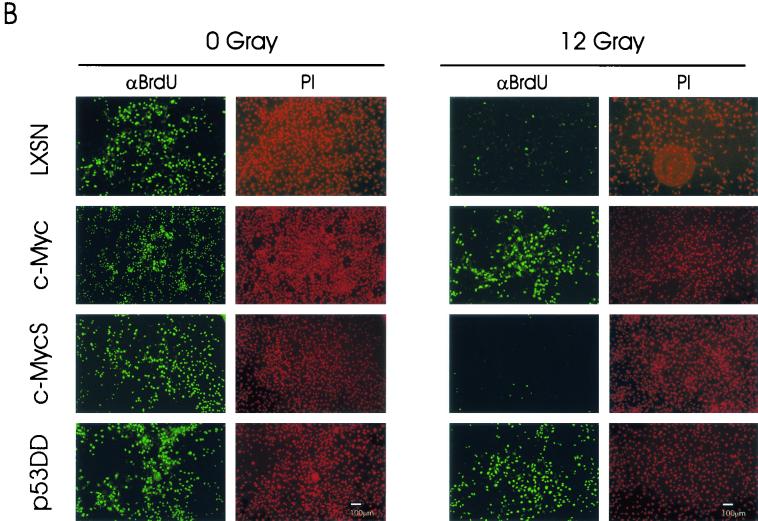
Transient overexpression of c-Myc attenuates IR-induced G1/S arrest and induces inappropriate entry into S phase in normal HMECs. (A) Transient infection of normal HMECs with recombinant retroviruses. At 48 h postinfection, total cell lysates were prepared and probed with each specific antibody. An equal loading control is provided by the α-tubulin blot. (B) BrdU incorporation assay for de novo DNA synthesis of normal HMECs after IR. Retroviral infection and IR were applied directly to the attached cells on the glass coverslip (see Material and Methods). At least 300 DAPI-stained nuclei were analyzed for each population with image analysis software. Error bars show standard deviations for three samples. (C) Representative images of BrdU incorporation following 0 or 12 Gy of IR in normal HMECs. After BrdU staining, all nuclei were counterstained with DAPI to determine the total number of nuclei in the same microscopic field.
We first examined the response of normal HMECs to γ-irradiation by phase-contrast microscopy (data not shown). In this experiment, transiently infected cells were irradiated with 0 or 12 Gy of IR, and then morphological changes were observed. Flattened cellular morphologies were apparent in noninfected and c-MycS-infected HMECs. However, following prolonged post-IR incubation, c-Myc- and p53DD-infected cells frequently demonstrated rounded or floating bodies. We next performed a BrdU incorporation assay, following 0 or 12 Gy of IR (Fig. 7B and C). In this series of experiments, we used a 6-h BrdU pulse, in addition to a 24-h BrdU incubation, in order to avoid potential complications resulting from a prolonged BrdU incubation. All four nonirradiated groups of samples exhibited significant incorporation of BrdU after a 6-h BrdU pulse (Fig. 7B) (noninfected, 21.3% of total nuclei; c-Myc, 33.5%; c-MycS, 25.4%; and p53DD, 57.7%). These results indicate that transient overexpression of c-Myc or p53DD significantly enhanced the number of cellular entries into S phase per unit of time (for 6 h in this study) in normal HMECs. Following 12 Gy of IR, noninfected and c-MycS-infected HMECs demonstrated a detectable but significantly reduced amount of BrdU incorporation (4.6% in noninfected and 5.6% in c-MycS cells). In contrast, both c-Myc-infected and p53DD-infected cells maintained a significant BrdU incorporation following IR (26.1% in c-Myc and 30.5% in p53DD cells). Taking into account the more extensive G1/S passage in nonirradiated p53DD-infected cells, overexpression of c-Myc attenuated the G1/S arrest more aggressively than did p53DD in normal HMECs.
Overexpression of c-Myc induced a sub-G1 population and a >4N population following IR.
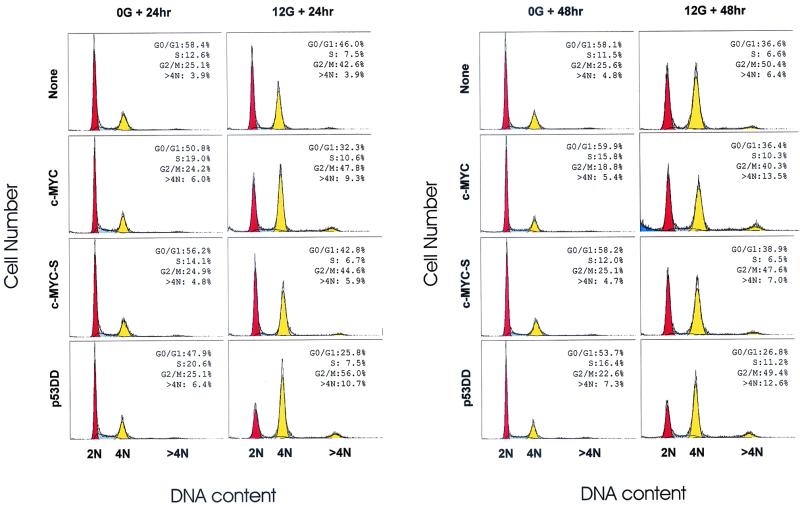
We investigated global changes of cell cycle progression in irradiated normal HMECs. Notable changes in the cell cycle profile of normal HMECs were the decreases in G0/G1 fractions (78.8% of G0/G1 fraction in nonirradiated control) and the increases in G2/M fractions (169.7% of G2/M fraction in nonirradiated control) after 24 h of post-IR incubation (Fig. 8). Furthermore, at 48 h post-IR, noninfected normal HMECs demonstrated a lower G0/G1 fraction and a higher G2/M fraction than those at 24 h post-IR, implicating a suppressed but progressive movement of irradiated cells from G0/G1 to G2/M. These results suggested an incomplete G1/S arrest and a functional G2/M arrest following IR in normal HMECs. c-MycS-infected normal HMECs showed similar changes in the cell cycle profile following IR. However, despite the intrinsically relaxed G1/S arrest in parental normal HMECs, effects of full-length c-Myc and p53DD were clearly demonstrated in the flow-cytometric analyses. Overexpression of c-Myc induced an even lower G0/G1 fraction (63.6% of G0/G1 fraction in nonirradiated c-Myc-expressing cells) and a comparatively higher S-phase fraction at 24 h post-IR. Furthermore, significant development of a sub-G1 population and a >4N population in irradiated c-Myc-infected HMECs was shown at 48 h post-IR. The changes also coincided with decreases in the G2/M fraction at 48 h post-IR. In p53DD-infected cells, p53DD greatly facilitated G1/S passage immediately and demonstrated dramatic decreases in G0/G1 fraction following IR. Even with a defective p53 function, irradiated p53DD-infected HMECs accumulated at G2/M, at least temporarily, at 24 h post-IR and began to display decreases in G2/M fraction at 48 h post-IR. In a region analysis of the sub-G1 fraction, c-Myc-infected HMECs displayed a moderately higher sub-G1 fraction, even in the absence of γ-irradiation, than other populations (Fig. 8) (noninfected, 0.4%; c-Myc, 1.6%; c-MycS, 0.4%; and p53DD, 0.6%). However, IR significantly induced the sub-G1 population in c-Myc-infected HMECs (Fig. 8) (noninfected, 0.5%; c-Myc, 4.5%; c-MycS, 0.4%; and p53DD, 1.9%). While the sub-G1 populations represent apoptosis in some situations, it has not yet been determined by other independent methods whether the sub-G1 population in irradiated c-Myc-infected HMECs truly represent apoptosis.
FIG. 8.
Overexpression of c-Myc led to an increase in the sub-G1 population and >4N population following IR in normal HMECs. Flow-cytometric profiles show cell cycle changes of each infected normal HMEC population following IR. Following 48 h of postinfection incubation for transgene expression, asynchronously growing cell populations were irradiated with 0 or 12 Gy of IR. Next, cells were allowed to progress for 24 or 48 h. Primary histogram data are displayed as fine line graphs, and the computer-generated best fit (ModFit) is shown by the orange (G0/G1), dashed (S), yellow (G2/M and >4N), and blue (sub-G1) areas.
DISCUSSION
According to previous studies, overexpression of c-myc induces aneuploidy, polyploidy, and gene amplifications in cultured fibroblasts (16, 35, 36). Chromosome translocations, trisomy, and dicentric chromosomes in mouse mammary carcinoma cells derived from c-myc-transgenic mice have also been identified (33, 37, 59). However, the underlying mechanism(s) for these dramatic effects of deregulated c-Myc on the destabilization of the cellular genome is largely unknown. As an approach to identify a possible mechanism(s) for this c-Myc-induced genomic instability, we examined the effects of c-Myc overexpression on the DNA damage-induced cell cycle checkpoints in the present study.
Overexpression of c-Myc alters DNA damage-induced G1/S arrest.
We have demonstrated that overexpression of c-Myc attenuated DNA damage-induced G1/S arrest, both in immortal HMEC lines and in mortal normal HMECs. Following 12 Gy of IR, c-Myc-overexpressing 184A1N4-Myc cells showed an absence of G1/S arrest and accumulation of most cells at G2/M, while the irradiated parental 184A1N4 cells significantly arrested both at G1/S and at G2/M (Fig. 1). In experiments with MCF10A, despite the absence of G2/M arrest, irradiated cells apparently arrested at the G1/S boundary and effects of c-Myc overexpression on the G1/S arrest were clearly demonstrated (Fig. 3). Furthermore, we clearly demonstrated that a transient excess of c-Myc activity was sufficient to alter the DNA damage-induced G1/S arrest and induced the inappropriate entry into S phase (Fig. 6). However, according to a previous study performed in an immortal 32D murine myeloid cell line (61a), c-Myc expression itself had no effects on the cell cycle arrest following IR. On the other hand, overexpression of c-Myc abolished another type of cell cycle checkpoint (previously known as a spindle assembly checkpoint) and accelerated the appearance of tetraploidy in 32D cells following nocodazole (a mitotic spindle poison) treatment. The effects of c-Myc expression on this spindle assembly checkpoint in 184A1N4 HMECs were also demonstrated in an independent study (30). It has not yet been determined whether this c-Myc-mediated alteration of cellular response to a mitotic spindle poison is potentially linked, at the molecular level, to the altered DNA damage-induced G1/S checkpoint phenotype in c-Myc-overexpressing HMECs.
A caveat for studying cell cycle checkpoints with immortalized lines is that the checkpoint alteration phenotypes could be contributed by additional genetic mutations acquired during the immortalization process or during repeated passaging of the cell lines. In particular, it has been shown that in mouse embryonic fibroblasts (MEFs), the vast majority (reportedly ∼80%) of rare, immortal fibroblasts were hypotetraploid and harbored mutant p53 alleles. Those that remain near-diploid frequently sustain biallelic loss of the p19/ARF locus (the mouse homolog of human p14/ARF) (48). The importance of p19/ARF in constituting a fully functional G1/S checkpoint has been suggested in a previous study of p19/ARF−/− nullizygous MEFs which showed that loss of p19/ARF alters the DNA damage-induced G1/S arrest (27). Furthermore, c-Myc has been proposed to upregulate p53 and ARF in some cell types (63). Therefore, to resolve these controversial ideas, it was necessary to employ a primary cell system to study effects of c-Myc expression. Furthermore, the variable G2/M responses following IR in two immortal HMEC lines (184A1N4 and MCF10A) also prompted us to employ normal mortal HMECs in the checkpoint assay. We performed a series of experiments with normal HMECs and consistently observed that c-Myc significantly attenuated G1/S arrest following 12 Gy of IR (Fig. 7 and 8). Flow-cytometric analyses also demonstrated a functional G2/M arrest following DNA damage in normal HMECs. Therefore, the absence of IR-induced G2/M arrest in MCF10A may reflect the immortalization history of this specific cell line. Interestingly, irradiated normal HMECs also demonstrated a relaxed G1/S arrest following IR (Fig. 8). According to a previous study on the DNA damage responses in normal HMECs (40), HMECs demonstrated a lack of G1/S arrest following a limited dosage of 4 Gy of IR. We also identified an absence of G1/S arrest following 4 Gy of IR in our parental normal HMECs (data not shown). It is currently not known whether this relaxed G1/S arrest phenotype may be related to the fact that normal HMECs have an extended proliferative potential and the elimination of either p53 or p14/ARF is not necessary for their immortalization (28).
In terms of effects of c-Myc expression in primary cells, we demonstrated that a transient overexpression of c-Myc in normal HMECs led to slightly elevated levels of p53 and p14/ARF proteins (Fig. 7A). However, it did not dramatically induce either cell cycle arrest or apoptosis in normal HMECs (Fig. 8). A transient activation of MycER also failed to induce G2/M arrest in the MCF10A system (data not shown). These results are in striking contrast with a previous report on the effects of c-Myc in normal human fibroblasts (NHFs) (17). In that study, a transient excess of c-Myc activity in NHFs dramatically induced cell cycle arrest at G2 phase within 48 h. Therefore, further investigations are required to elucidate a potential difference between the responses of these two cell types to deregulated c-Myc. Interestingly, a recent study also emphasized the importance of cellular background. NHFs failed to upregulate ARF following the introduction of a Ras oncogene, suggesting a difference between human fibroblasts and murine cells in responding to an activated oncogene (60).
How does c-Myc override the p53-dependent pathway? Downstream targets for the c-Myc-induced checkpoint alteration.
Since its discovery some 20 years ago, c-Myc has been one of the most paradoxical oncogenes. It is well known that c-Myc facilitates two very important but seemingly opposite processes in cells, proliferation and apoptosis. The c-Myc-induced checkpoint alteration phenotype is not exempt from this type of paradox. p53 and ARF, two recently identified transcriptional targets of c-Myc (46), make c-Myc's action on the cell cycle even more intriguing. Although a significant increase of p53 protein was not detected following c-Myc infection of MCF10A, an IR-induced upregulation of p53 and p21/Cip1 protein was confirmed both in LXSN- and in c-Myc-infected HMECs over the same time course (Fig. 4B). According to several independent reports, c-Myc transrepresses p21, a universal Cdk inhibitor (7, 8, 42). Since p21 is a critical downstream effector of p53-mediated G1/S arrest, it would be a plausible target for c-Myc-induced override of this G1/S checkpoint. Our results however demonstrate only a subtle decrease of p21/Cip1 proteins in c-Myc-infected cells, following its initial upregulation at 6 h post-IR (Fig. 4B). Therefore, further molecular analyses following IR-induced DNA damage are needed to address the possible role of p21/Cip1 in the c-Myc-induced alteration of the G1/S checkpoint. Another candidate downstream target of c-Myc for override of the G1/S checkpoint could be Rb. Rb is a major downstream effector of the p53-p21-Rb pathway for DNA damage-induced G1/S arrest. According to previous studies, the absence of Rb in the Rb−/− nullizygous MEFs leads to an inappropriate entry into the S-phase following IR treatment (23). The reappearance of ppRb in irradiated c-Myc-infected MCF10A cells suggests the involvement of Rb inactivation in c-Myc-induced G1/S checkpoint alteration. However, because cells phosphorylate Rb whenever they prepare and initiate a scheduled entry into S phase, it is not clear yet whether c-Myc directly targets Rb phosphorylation or whether this effect is simply a reflection of entry into S. Expression of cyclin A is likely to have returned coincident with c-Myc- or p53DD-infected cells progressing again into S phase following IR. Reportedly, c-Myc transactivates cyclin E, and this results in deregulated activation of Cdk2-cyclin E and in hyperphosphorylation of Rb (3, 21, 26, 45). Any possible change of cyclin E levels following IR has not yet been determined. We employed c-MycS initially to probe a potential distinction between the transactivation and the transrepression mediated by c-Myc. However, a recent study suggested the potential of c-MycS in transactivating endogenous target genes in NHFs and in immortalized rat fibroblasts (25). This report also demonstrated that c-MycS has undetectable activity as an inducer of S phase in quiescent human fibroblasts.
Irradiated c-Myc-overexpressing HMECs develop micronuclei, sub-G1 populations, and >4N populations.
An unexpected, intriguing finding of the present G1/S study is an induction of micronuclei and a development of the sub-G1 and the >4N populations in irradiated c-Myc-overexpressing HMECs. According to a previous study of c-Myc-overexpressing rat embryonic cells (REC:Myc) using a computerized video time-lapse method (18, 55), these REC:Myc were susceptible to IR-induced apoptosis after 4 to 9.5 Gy of IR. Cell death occurred primarily by postmitotic apoptosis, following several divisions of the irradiated cells. These results suggest the possibility of an altered G2/M entry in these cells, because the irradiated c-Myc-overexpressing REC were allowed to undergo several mitotic divisions. That study also reported that 60% of the progeny of 4-Gy-irradiated REC:Myc either had micronuclei or were sisters of cells with micronuclei. The formation of micronuclei was correlated to apoptosis, as cells with micronuclei were more likely to undergo apoptosis during the generation in which the micronuclei were observed (18). Additionally, a study by McKenna et al. (38) reported a minimal delay in G2/M of Myc-expressing fibroblasts following IR. These studies provide interesting clues to stimulate further, rigorous investigations of the effects of deregulated c-Myc on DNA damage-induced arrest at the G2/M boundary. Interestingly, irradiated c-Myc-infected normal HMECs developed sub-G1 and >4N populations (Fig. 8). These results suggest possible effects of c-Myc on uncoupling of cell cycle progression following DNA damage and on the sensitization of cells to DNA-damaging agents, a phenotype previously demonstrated in p53- and p21-deficient cells (5, 58).
Based on the present study, we propose that c-Myc alters the DNA damage-induced G1/S checkpoint, a molecular safeguard mechanism for genomic stability. This effect of c-Myc may provide a mechanistic link between c-Myc-induced cell cycle alteration, genomic instability, and its role as a potent oncogene.
ADDENDUM IN PROOF
G. M. Wahl and colleagues have independently shown that c-Myc expression in normal human fibroblasts can attenuate G1/S arrest following DNA damage (personal communication).
Acknowledgments
We appreciate the materials and critical advice from Martha Stampfer, Paul Yaswen, Moshe Oren, Trevor Littlewood, Chi Dang, Linda Penn, and Dusty Miller. We also thank Todd Waldman for helpful discussions throughout this study and Geoffrey M. Wahl for sharing experimental information before publication. We gratefully received technical assistance from the microscopic imaging core facilities and the flow cytometry core facilities at the Lombardi Cancer Center, Georgetown University Medical Center. We thank Tim Jorgensen and members of our laboratory for comments on the manuscript.
This study was supported in part by a predoctoral training grant of the DOD breast cancer research program (DAMD17-99-1-9205) to J.-H.S. and NIH grant R01AG1496 to R.B.D.
REFERENCES
- 1.Agarwal, M. L., A. Agarwal, W. R. Taylor, and G. R. Stark. 1995. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc. Natl. Acad. Sci. USA 92:8493-8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacus, S. S., Y. Yarden, M. Oren, D. M. Chin, L. Lyass, C. R. Zelnick, A. Kazarov, W. Toyofuku, J. Gray-Bablin, R. R. Beerli, N. E. Hynes, M. Nikiforov, R. Haffner, A. Gudkov, and K. Keyomarsi. 1996. Neu differentiation factor (heregulin) activates a p53-dependent pathway in cancer cells. Oncogene 12:2535-2547. [PubMed] [Google Scholar]
- 3.Berns, K., E. M. Hijmans, and R. Bernards. 1997. Repression of c-Myc responsive genes in cycling cells causes G1 arrest through reduction of cyclin E/CDK2 kinase activity. Oncogene 15:1347-1356. [DOI] [PubMed] [Google Scholar]
- 4.Blackwood, E. M., B. Luscher, L. Kretzner, and R. N. Eisenman. 1991. The Myc:Max protein complex and cell growth regulation. Cold Spring Harbor Symp. Quant. Biol. 56:109-117. [DOI] [PubMed] [Google Scholar]
- 5.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497-1501. [DOI] [PubMed] [Google Scholar]
- 6.Cahill, D. P., K. W. Kinzler, B. Vogelstein, and C. Lengauer. 1999. Genetic instability and Darwinian selection in tumours. Trends Cell Biol. 9:M57-M60. [PubMed]
- 7.Claassen, G. F., and S. R. Hann. 2000. A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta-induced cell-cycle arrest. Proc. Natl. Acad. Sci. USA 97:9498-9503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coller, H. A., C. Grandori, P. Tamayo, T. Colbert, E. S. Lander, R. N. Eisenman, and T. R. Golub. 2000. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA 97:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang, C. V., L. M. Resar, E. Emison, S. Kim, Q. Li, J. E. Prescott, D. Wonsey, and K. Zeller. 1999. Function of the c-Myc oncogenic transcription factor. Exp. Cell Res. 253:63-77. [DOI] [PubMed] [Google Scholar]
- 10.Deng, C., P. Zhang, J. W. Harper, S. J. Elledge, and P. Leder. 1995. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82:675-684. [DOI] [PubMed] [Google Scholar]
- 11.Di Leonardo, A., S. P. Linke, K. Clarkin, and G. M. Wahl. 1994. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 8:2540-2551. [DOI] [PubMed] [Google Scholar]
- 12.Di Leonardo, A., S. P. Linke, Y. Yin, and G. M. Wahl. 1993. Cell cycle regulation of gene amplification. Cold Spring Harbor Symp. Quant. Biol. 58:655-667. [DOI] [PubMed] [Google Scholar]
- 13.el-Deiry, W. S., J. W. Harper, P. M. O'Connor, V. E. Velculescu, C. E. Canman, J. Jackman, J. A. Pietenpol, M. Burrell, D. E. Hill, Y. Wang, K. G. Wiman, W. E. Mercer, M. B. Kastan, K. W. Kohn, S. J. Elledge, K. W. Kinzler, and B. Vogelstein. 1994. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 54:1169-1174. [PubMed] [Google Scholar]
- 14.Facchini, L. M., S. Chen, W. W. Marhin, J. N. Lear, and L. Z. Penn. 1997. The Myc negative autoregulation mechanism requires Myc-Max association and involves the c-myc P2 minimal promoter. Mol. Cell. Biol. 17:100-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Facchini, L. M., and L. Z. Penn. 1998. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 12:633-651. [PubMed] [Google Scholar]
- 16.Felsher, D. W., and J. M. Bishop. 1999. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc. Natl. Acad. Sci. USA 96:3940-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsher, D. W., A. Zetterberg, J. Zhu, T. Tlsty, and J. M. Bishop. 2000. Overexpression of MYC causes p53-dependent G2 arrest of normal fibroblasts. Proc. Natl. Acad. Sci. USA 97:10544-10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forrester, H. B., N. Albright, C. C. Ling, and W. C. Dewey. 2000. Computerized video time-lapse analysis of apoptosis of REC:Myc cells X-irradiated in different phases of the cell cycle. Radiat. Res. 154:625-639. [DOI] [PubMed] [Google Scholar]
- 19.Gandarillas, A., and F. M. Watt. 1997. c-Myc promotes differentiation of human epidermal stem cells. Genes Dev. 11:2869-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudas, J., H. Nguyen, T. Li, D. Hill, and K. H. Cowan. 1995. Effects of cell cycle, wild-type p53 and DNA damage on p21CIP1/Waf1 expression in human breast epithelial cells. Oncogene 11:253-261. [PubMed] [Google Scholar]
- 21.Hanson, K. D., M. Shichiri, M. R. Follansbee, and J. M. Sedivy. 1994. Effects of c-myc expression on cell cycle progression. Mol. Cell. Biol. 14:5748-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 23.Harrington, E. A., J. L. Bruce, E. Harlow, and N. Dyson. 1998. pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc. Natl. Acad. Sci. USA 95:11945-11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartwell, L. 1992. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell 71:543-546. [DOI] [PubMed] [Google Scholar]
- 25.Hirst, S. K., and C. Grandori. 2000. Differential activity of conditional MYC and its variant MYC-S in human mortal fibroblasts. Oncogene 19:5189-5197. [DOI] [PubMed] [Google Scholar]
- 26.Jansen-Durr, P., A. Meichle, P. Steiner, M. Pagano, K. Finke, J. Botz, J. Wessbecher, G. Draetta, and M. Eilers. 1993. Differential modulation of cyclin gene expression by MYC. Proc. Natl. Acad. Sci. USA 90:3685-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan, S. H., J. Moritsugu, and G. M. Wahl. 2000. Differential requirement for p19ARF in the p53-dependent arrest induced by DNA damage, microtubule disruption, and ribonucleotide depletion. Proc. Natl. Acad. Sci. USA 97:3266-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiyono, T., S. A. Foster, J. I. Koop, J. K. McDougall, D. A. Galloway, and A. J. Klingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396:84-88. [DOI] [PubMed] [Google Scholar]
- 29.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 30.Li, Q., and C. V. Dang. 1999. c-Myc overexpression uncouples DNA replication from mitosis. Mol. Cell. Biol. 19:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Littlewood, T. D., D. C. Hancock, P. S. Danielian, M. G. Parker, and G. I. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23:1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livingstone, L. R., A. White, J. Sprouse, E. Livanos, T. Jacks, and T. D. Tlsty. 1992. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell 70:923-935. [DOI] [PubMed] [Google Scholar]
- 33.Liyanage, M., A. Coleman, S. du Manoir, T. Veldman, S. McCormack, R. B. Dickson, C. Barlow, A. Wynshaw-Boris, S. Janz, J. Wienberg, M. A. Ferguson-Smith, E. Schrock, and T. Ried. 1996. Multicolour spectral karyotyping of mouse chromosomes. Nat. Genet. 14:312-315. [DOI] [PubMed] [Google Scholar]
- 34.Loeb, L. A. 1991. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 51:3075-3079. [PubMed] [Google Scholar]
- 35.Mai, S. 1994. Overexpression of c-myc precedes amplification of the gene encoding dihydrofolate reductase. Gene 148:253-260. [DOI] [PubMed] [Google Scholar]
- 36.Mai, S., M. Fluri, D. Siwarski, and K. Huppi. 1996. Genomic instability in MycER-activated Rat1A-MycER cells. Chromosome Res. 4:365-371. [DOI] [PubMed] [Google Scholar]
- 37.McCormack, S. J., Z. Weaver, S. Deming, G. Natarajan, J. Torri, M. D. Johnson, M. Liyanage, T. Ried, and R. B. Dickson. 1998. Myc/p53 interactions in transgenic mouse mammary development, tumorigenesis and chromosomal instability. Oncogene 16:2755-2766. [DOI] [PubMed] [Google Scholar]
- 38.McKenna, W. G., G. Iliakis, M. C. Weiss, E. J. Bernhard, and R. J. Muschel. 1991. Increased G2 delay in radiation-resistant cells obtained by transformation of primary rat embryo cells with the oncogenes H-ras and v-myc. Radiat. Res. 125:283-287. [PubMed] [Google Scholar]
- 39.Merlo, G. R., F. Basolo, L. Fiore, L. Duboc, and N. E. Hynes. 1995. p53-dependent and p53-independent activation of apoptosis in mammary epithelial cells reveals a survival function of EGF and insulin. J. Cell Biol. 128:1185-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer, K. M., S. M. Hess, T. D. Tlsty, and S. A. Leadon. 1999. Human mammary epithelial cells exhibit a differential p53-mediated response following exposure to ionizing radiation or UV light. Oncogene 18:5795-5805. [DOI] [PubMed] [Google Scholar]
- 41.Miller, A. D., and G. J. Rosman. 1989. Improved retroviral vectors for gene transfer and expression. BioTechniques 7:980-986, 989. [PMC free article] [PubMed]
- 42.Mitchell, K. O., and W. S. El-Deiry. 1999. Overexpression of c-Myc inhibits p21WAF1/CIP1 expression and induces S-phase entry in 12-O-tetradecanoylphorbol-13-acetate (TPA)-sensitive human cancer cells. Cell Growth Differ. 10:223-230. [PubMed] [Google Scholar]
- 43.Nass, S. J., and R. B. Dickson. 1998. Epidermal growth factor-dependent cell cycle progression is altered in mammary epithelial cells that overexpress c-myc. Clin. Cancer Res. 4:1813-1822. [PubMed] [Google Scholar]
- 44.Paulovich, A. G., D. P. Toczyski, and L. H. Hartwell. 1997. When checkpoints fail. Cell 88:315-321. [DOI] [PubMed] [Google Scholar]
- 45.Perez-Roger, I., D. L. Solomon, A. Sewing, and H. Land. 1997. Myc activation of cyclin E/Cdk2 kinase involves induction of cyclin E gene transcription and inhibition of p27(Kip1) binding to newly formed complexes. Oncogene 14:2373-2381. [DOI] [PubMed] [Google Scholar]
- 46.Reisman, D., N. B. Elkind, B. Roy, J. Beamon, and V. Rotter. 1993. c-Myc trans-activates the p53 promoter through a required downstream CACGTG motif. Cell Growth Differ. 4:57-65. [PubMed] [Google Scholar]
- 47.Shaulian, E., A. Zauberman, D. Ginsberg, and M. Oren. 1992. Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol. Cell. Biol. 12:5581-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherr, C. J., and R. A. DePinho. 2000. Cellular senescence: mitotic clock or culture shock? Cell 102:407-410. [DOI] [PubMed] [Google Scholar]
- 49.Soule, H. D., T. M. Maloney, S. R. Wolman, W. D. J. Peterson, R. Brenz, C. M. McGrath, J. Russo, R. J. Pauley, R. F. Jones, and S. C. Brooks. 1990. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50:6075-6086. [PubMed] [Google Scholar]
- 50.Spotts, G. D., S. V. Patel, Q. Xiao, and S. R. Hann. 1997. Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol. Cell. Biol. 17:1459-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stampfer, M. R., and J. C. Bartley. 1985. Induction of transformation and continuous cell lines from normal human mammary epithelial cells after exposure to benzo[a]pyrene. Proc. Natl. Acad. Sci. USA 82:2394-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stampfer, M. R., C. H. Pan, J. Hosoda, J. Bartholomew, J. Mendelsohn, and P. Yaswen. 1993. Blockage of EGF receptor signal transduction causes reversible arrest of normal and immortal human mammary epithelial cells with synchronous reentry into the cell cycle. Exp. Cell Res. 208:175-188. [DOI] [PubMed] [Google Scholar]
- 53.Tait, L., H. D. Soule, and J. Russo. 1990. Ultrastructural and immunocytochemical characterization of an immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50:6087-6094. [PubMed] [Google Scholar]
- 54.Valverius, E. M., F. Ciardiello, N. E. Heldin, B. Blondel, G. Merlo, G. Smith, M. R. Stampfer, M. E. Lippman, R. B. Dickson, and D. S. Salomon. 1990. Stromal influences on transformation of human mammary epithelial cells overexpressing c-myc and SV40T. J. Cell Physiol. 145:207-216. [DOI] [PubMed] [Google Scholar]
- 55.Vidair, C. A., C. H. Chen, C. C. Ling, and W. C. Dewey. 1996. Apoptosis induced by X-irradiation of rec-myc cells is postmitotic and not predicted by the time after irradiation or behavior of sister cells. Cancer Res. 56:4116-4118. [PubMed] [Google Scholar]
- 56.Wahl, G. M., S. P. Linke, T. G. Paulson, and L. C. Huang. 1997. Maintaining genetic stability through TP53 mediated checkpoint control. Cancer Surv. 29:183-219. [PubMed] [Google Scholar]
- 57.Waldman, T., K. W. Kinzler, and B. Vogelstein. 1995. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 55:5187-5190. [PubMed] [Google Scholar]
- 58.Waldman, T., C. Lengauer, K. W. Kinzler, and B. Vogelstein. 1996. Uncoupling of S phase and mitosis induced by anticancer agents in cells lacking p21. Nature 381:713-716. [DOI] [PubMed] [Google Scholar]
- 59.Weaver, Z. A., S. J. McCormack, M. Liyanage, S. du Manoir, A. Coleman, E. Schrock, R. B. Dickson, and T. Ried. 1999. A recurring pattern of chromosomal aberrations in mammary gland tumors of MMTV-cmyc transgenic mice. Genes Chromosomes Cancer 25:251-260. [PubMed] [Google Scholar]
- 60.Wei, W., R. M. Hemmer, and J. M. Sedivy. 2001. Role of p14(ARF) in replicative and induced senescence of human fibroblasts. Mol. Cell. Biol. 21:6748-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiong, Y., G. J. Hannon, H. Zhang, D. Casso, R. Kobayashi, and D. Beach. 1993. p21 is a universal inhibitor of cyclin kinases. Nature 366:701-704. [DOI] [PubMed] [Google Scholar]
- 61a.Yin, X. Y., L. Grove, N. S. Datta, M. W. Long, and E. V. Prochownik. 1999. c-myc overexpression and p53 loss cooperate to promote genomic instability. Oncogene 18:1177-1184. [DOI] [PubMed] [Google Scholar]
- 62.Yin, Y., M. A. Tainsky, F. Z. Bischoff, L. C. Strong, and G. M. Wahl. 1992. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell 70:937-948. [DOI] [PubMed] [Google Scholar]
- 63.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]