Abstract
The mammalian SWI/SNF-like chromatin-remodeling BAF complex plays several important roles in controlling cell proliferation and differentiation. Interferons (IFNs) are key mediators of cellular antiviral and antiproliferative activities. In this report, we demonstrate that the BAF complex is required for the maximal induction of a subset of IFN target genes by alpha IFN (IFN-α). The BAF complex is constitutively associated with the IFITM3 promoter in vivo and facilitates the chromatin remodeling of the promoter upon IFN-α induction. Furthermore, we show that the ubiquitous transcription activator Sp1 interacts with the BAF complex in vivo and augments the BAF-mediated activation of the IFITM3 promoter. Sp1 binds constitutively to the IFITM3 promoter in the absence of the BAF complex, suggesting that it may recruit and/or stabilize the BAF complex binding to the IFITM3 promoter. Our results bring new mechanistic insights into the antiproliferative effects of the chromatin-remodeling BAF complex.
Interferons (IFNs) play several fundamental roles in cellular antiviral and antiproliferative activities (35, 40). The signal of alpha/beta IFNs (IFN-α/βs) is mainly mediated by IFN-stimulated gene factor 3 (ISGF3), a trimeric complex consisting of STAT1, STAT2, and p48 (14, 38, 44), which binds to IFN-stimulated regulatory elements (ISREs) to activate their target genes (6). Transcriptional activation of the IFN-induced genes is required for the actions of the IFNs (4, 40).
The genomic DNA in the nucleus is packaged into nucleosomes that are inhibitory to the access of transcription factors to their target sites. Modification of the nucleosomal template is thought necessary to allow transcriptional activation. This modification can be either covalent bond formation by acetylation, phosphorylation, and methylation at the histone tails and/or noncovalent action by ATP-utilizing remodeling enzymes (1, 13, 17, 19, 30, 36, 41, 43, 47). Whereas the mechanism by which ISGF3 overcomes the chromatin barrier for binding to its targeting sites is not clear, signal transducer and activator of transcription (STAT) proteins have been found to interact with the highly conserved and ubiquitously expressed cyclic AMP response element-binding proteins, CBP/p300 (3, 15, 34, 49, 52). Both CBP and p300 have histone acetyltransferase activity (33). Overexpression of E1A, which binds to CBP/p300 and inhibits its histone acetyltransferase activity, blocks the ISRE-mediated responses to IFN-α/β (3), suggesting that the modification of the chromatin structure might be an important step in the induction of IFN target genes.
It was reported recently that the reconstitution of the active BAF complex by transient expression of the essential ATPase subunit BRG1 in BRG1-deficient SW-13 cells up-regulates several IFN target genes (27), suggesting that the chromatin-remodeling activity of the BAF complex might be required for the induction of these genes by IFNs. In this report, we demonstrate that reconstitution of the BAF complex by transiently expressing BRG1 in SW-13 cells potentiates the maximal induction of a group of IFN target genes by IFN-α. The BAF complex facilitates chromatin remodeling of the IFITM3 promoter upon IFN-α induction. We also show that Sp1 interacts with the BAF complex in vivo and that the Sp1-binding site in the promoter is required for optimal activation of the promoter by the BAF complex.
MATERIALS AND METHODS
Constructs and antibodies.
pREP4-luc and pREP7-RL have been described previously (27). pGL3-TM3-luc and pREP4-TM3-luc were constructed by inserting the PCR-amplified IFITM3 promoter (−238 to −25) into the XhoI-HindIII sites of pGL3/basic or pREP4-luc. The constructs for promoter deletion analysis were prepared by inserting the corresponding fragments amplified by PCR into the XhoI-HindIII sites of pREP4-luc. Point mutation of the Sp1-binding site in pREP4-TM3-luc was performed with the QuikChange kit (Stratagene) following the manufacturer's instructions.
The BRG1 antibody has been described previously (21). The other antibodies were purchased from the following sources: Sp1 from Santa Cruz Biotechnology, Inc. (catalog no. SC-420 and SC-14027), STAT1 from Santa Cruz Biotechnology, Inc. (SC-346), p48 from Santa Cruz Biotechnology, Inc. (SC-496), and STAT2 from Transduction Laboratories (S21220).
Cell culture, transfection, luciferase assay, and RT-PCR.
SW-13 cells and HeLa cells were maintained in Dulbecco's modified Eagle's medium complemented with 10% fetal bovine serum, 1 mM glutamine, and penicillin-streptomycin. Transfection was done by using Superfect (Qiagen) transfection reagent according to the manufacturer's instructions. The luciferase assay was performed with the dual luciferase assay kit (Promega). Reverse transcription (RT)-PCR analysis of the mRNA levels was performed as described previously (27) with the following primers: IFITM2 forward, 5′ ATTCTGCTCGTCATCATCCCAG 3′, and reverse, 5′ TGATGCAAGACTCGGCTGTG 3′; IFITM3 forward, 5′ ATGAGTCACACTGTCCAAACCTTCTTC 3′, and reverse, 5′ AACAGGGACCAGACGACATGGTCG 3′; IFIT1 forward, 5′ ACAAGGTGGAGAACATTTGCAAG 3′, and reverse, 5′ AAGGAGAACCTTAATATATCC 3′; GBP forward, 5′ TGATGAACAAGCTGGCTGGAAG 3′, and reverse, 5′ TCTGTCACATAGTACAGTTG 3′; ISG15 forward, 5′ CAACGAATTCCAGGTGTC 3′, and reverse, 5′ TCACTTGCTGCTTCAGGTG 3′; and ISGF3G forward, 5′ TACAAGGTGTATCAGTTGCTG 3′, and reverse, 5′ AAAGTACCTGACCAAGTCTG 3′.
Restriction enzyme accessibility assay.
The restriction enzyme accessibility assay was performed according to a published procedure (46) with the following modifications. Exponentially growing SW-13 and HeLa cells were treated with 1,000 U of IFN-α/ml (catalog no. PHC4814; Biosource) for 2 h prior to lysis in cell lysis buffer (10 mM Tris [pH 7.5], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, 0.15 mM spermine, 0.5 mM spermidine). After being washed once with 200 μl of nuclear buffer (10 mM Tris [pH 7.5], 50 mM NaCl, 10 mM MgCl2, 0.2 mM EDTA, 0.2 mM EGTA, 1 mM β-mercaptoethanol, 0.15 mM spermine, 0.5 mM spermidine), the nuclei were resuspended in 50 μl of restriction buffer (10 mM Tris [pH 7.9], 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol) and digested with 2 μl of HincII (20 U) for 10 min at 37°C. The reaction was stopped by adding 150 μl of stop buffer (10 mM Tris [pH 7.5], 10 mM EDTA, 0.4% sodium dodecyl sulfate [SDS], 0.6 mg of proteinase K/ml) and incubated at 50°C for 3 h. Following purification by phenol-chloroform extraction and precipitation, the genomic DNA (2 μg) was digested to completion with 1 μl of XmnI (10 U) in 50 μl of the restriction buffer for 2 h at 37°C. The DNA was purified by phenol-chloroform extraction and precipitation and resuspended in 20 μl of H2O. Linker ligation was carried out by adding 5 μl of 10× ligation buffer-20.5 μl of H2O-2.5 μl of 25 μM universal linker (32)-2 μl of T4 DNA ligase (New England Biolabs; 400 U/μl) and incubating at 14°C for 12 h. The DNA was purified as before and resuspended in 20 μl of H2O. For PCR amplification, 10 μl of DNA was mixed with 5 μl of 10× PCR buffer-5 μl of 25 mM MgCl2-5 μl of 1 mM deoxynucleoside triphosphates-2 μl of 2.5 μM the long universal primer-2 μl of 2.5 μM gene-specific primer IFITM3/445R (5′ TATGAAGCCCAGGCAGCAGGG 3′)-20 μl of H2O-1 μl of Taq polymerase (5 U) and cycled at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min and then 25 times at 72°C for 5 min. The PCR products were labeled by adding 32P-labeled IFITM3/402R (5′ AACAGGGACCAGACGACATGGTCG 3′) to 0.05 μM and two cycles of PCR (94°C for 2 min, 55°C for 2 min, 72°C for 2 min; repeated once). The labeled products were purified and resolved on an 8% acrylamide-6 M urea-1× Tris-borate-EDTA gel and visualized by autoradiography.
For transient expression of BRG1 in SW-13 cells, the cells were transfected with a cell selection kit (catalog no. 130-070-201; Miltenyi Biotec) by using pMACS Kk and pBJ5-BRG1. Following treatment with IFN-α for 2 h after 24 h of transfection, the transfected and nontransfected cells were sorted by using magnetic beads according to the manufacturer's instructions and processed for HincII accessibility as described above.
Nucleosome mapping with MNase.
The nucleosomal structure was mapped as described previously (51). Briefly, SW-13 cells and HeLa cells were lysed in the cell lysis buffer as described above. The nuclei were washed once with 1 ml of the nuclear buffer and resuspended in the same buffer at a concentration of about 6 million nuclei per 1,000 μl of buffer. The final Ca2+ concentration was adjusted to 1 mM with 1 M CaCl2. A total of 250 μl of the nuclei was digested with 0.05, 0.1, 0.3, and 0.6 U of MNase (Sigma), respectively, for 10 min at 30°C. The reaction was stopped, and the DNA was purified as in the restriction enzyme accessibility assay. The purified DNA (2 μg) was phosphorylated with T4 polynucleotide kinase, and double-stranded cleavages were detected by ligation-mediated PCR (LM-PCR) as described above.
The nucleosomal boundaries were mapped with mononucleosome-sized DNA by LM-PCR according to published protocol (28) by using the following IFITM3-specific primers: forward PCR primer (5′ GAGGGCTCACTGAGTAACCATC 3′), forward labeling primer F1 (5′ AGTAACCCGACCGCCGCTGGTCTT 3′), reverse PCR primer (5′ GGCTGGCCACTGTTGACAGGA 3′), and reverse labeling primer R1 (TGTGACTCATGGTGTCCAGCGAAG 3′).
In vivo DSP cross-linking.
In vivo cross-linking and immunoprecipitation (IP) were carried out as reported previously (49) with the following modifications. Exponentially growing Jurkat cells (108) were pelleted by centrifugation at 1,000 rpm in a Sorvall Biofuge for 5 min. The cells were resuspended in 2 ml of 1× phosphate-buffered saline and cross-linked with 0.6 mg of dithiobis(succinimidylpropionate) (DSP)/ml for 15 min at room temperature. The reaction was stopped by pelleting in a Sorvall Biofuge at 4,000 rpm for 1 min and washing once with 1× phosphate-buffered saline. Following lysis by Dounce homogenizing with a glass homogenizer in 1 ml of the lysis buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 1 mM phenylmethylsulfonyl fluoride), the nuclei were washed twice with 1 ml of the lysis buffer plus 0.2% Triton X-100. The nuclei were then extracted with 0.5 ml of the extraction buffer (20 mM HEPES [pH 7.9], 0.42 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride, proteinase inhibitor cocktail) for 30 min at 4°C with rotation. For the IP assay, 150 μl of the nuclear extracts, 250 μl of IP wash buffer (20 mM Tris [pH 7.5], 0.5 M NaCl, 1 mM EDTA, 0.2% Triton X-100), and 2 μg of the specific antibodies were added to 20 μl of protein A beads, which were then incubated for 4 h at 4°C with rotation. Following three 10-min washes with 1 ml each of the IP wash buffer, the immunoprecipitated proteins were eluted and reverse cross-linked by incubating the beads in 30 μl of 2× SDS loading buffer (0.1 M Tris [pH 6.8], 4% SDS, 10% glycerol, 0.2 M dithiothreitol) for 10 min at 95°C. The samples were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and detected by Western blot analysis.
ChIP assay and electrophoresic mobility shift assay (EMSA).
The chromatin IP (ChIP) assay was performed as described previously (27) with the following modifications. Following cross-linking with formaldehyde and sonication of the cells, the chromatin lysates were adjusted to a standard IP buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA [pH 8.0], 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS) plus 0.14 M NaCl. The soluble chromatin (500 μl), precleared by incubating with 50 μl of protein A-Sepharose beads for 1 h at room temperature, was added with 2 μg of purified specific antibodies or normal rabbit immunoglobulins and 50 μg of bovine serum albumin to 30 μl of protein A-Sepharose beads that were previously blocked by incubating with 300 μl of 1× phosphate-buffered saline containing 0.5% bovine serum albumin and salmon sperm DNA (0.1 μg/μl) for 1 h at room temperature. Following incubation for 16 h at 4°C with rotation, the beads were washed sequentially twice with IP buffer containing 0.3 M NaCl, twice with IP buffer containing 0.14 M NaCl, twice with L buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA [pH 8.0], 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate), and twice with 1× Tris-EDTA buffer. The immunoprecipitated DNA was reverse cross-linked and purified. One-tenth of the DNA was analyzed by multiplex PCR with the following primers: IFITM3/−251F, 5′ CAAGATGAGACTTGTGCTCCCTTGG 3′; IFITM3/+61R, 5′ CTCGTGCTCCTCCTTGAGCATCT 3′; IFITM2U/F2, 5′ CGGTCCTGTGACCCCTTAATGGT 3′; and IFITM2U/R1, 5′ CAAGACTTCATCTCCAACCGGAC 3′).
RESULTS
Transient expression of BRG1 in SW-13 cells potentiates the maximal induction of a group of IFN target genes by IFN-α.
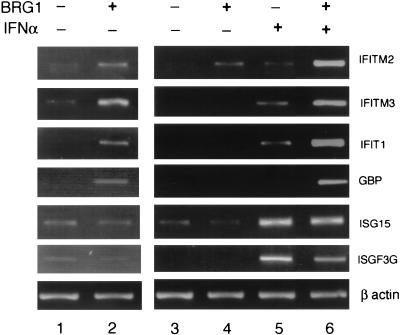
The BAF complex elevates the basal-level expression of 80 genes in SW-13 cells, including a number of IFN-inducible genes (27). Since chromatin remodeling is considered an important step in the transcriptional activation of a gene, we hypothesized that the presence of the chromatin-remodeling BAF complex may enhance the induction of these IFN target genes by IFN. RT-PCR analysis confirmed the conclusion from the cDNA microarray results that the basal-level expression of four IFN target genes including IFITM2, IFITM3, IFIT1, and GBP was augmented by the transient expression of BRG1 in SW-13 cells, while no difference in expression was observed for ISG15 and ISGF3G (Fig. 1, compare lanes 1 and 2). IFN-α alone induced only weakly the first four genes. However, the presence of BRG1 in SW-13 cells dramatically enhanced the induction of these genes by IFN-α (Fig. 1, compare lanes 5 and 6). In contrast, ISG15 and ISGF3G were induced strongly by IFN-α alone and no significant difference in induction was observed in the presence of BRG1.
FIG. 1.
Reconstitution of the BAF complex potentiates the maximal induction of a group of IFN target genes by IFN-α. SW-13 cells transfected with BRG1 expression construct for 24 h were treated with 500 U of IFN-α/ml for 10 h. Total RNA was extracted from the cells, reverse transcribed, and used for RT-PCR analysis with primers for the genes shown on the right. The samples in lanes 1 and 2 were amplified five more cycles than the samples in lanes 3 to 6 to visualize the difference caused by the presence of BRG1. β actin was used as a negative control.
To rule out the possibility that the IFN-α signaling pathway is defective in SW-13 cells, we examined the phosphorylation status of STAT1 and STAT2 upon IFN-α treatment. We found that IFN-α treatment efficiently induced the phosphorylation of STAT1 and STAT2 in both SW-13 cells and HeLa cells (data not shown). Therefore, the defect in the induction of IFITM2, IFITM3, IFIT1, and GBP by IFN-α in SW-13 cells might be caused by the absence of the functional BAF complex, suggesting that the maximal induction of these genes may require chromatin remodeling by the BAF complex.
Chromatin remodeling at the IFITM3 promoter is compromised in SW-13 cells upon IFN-α induction.
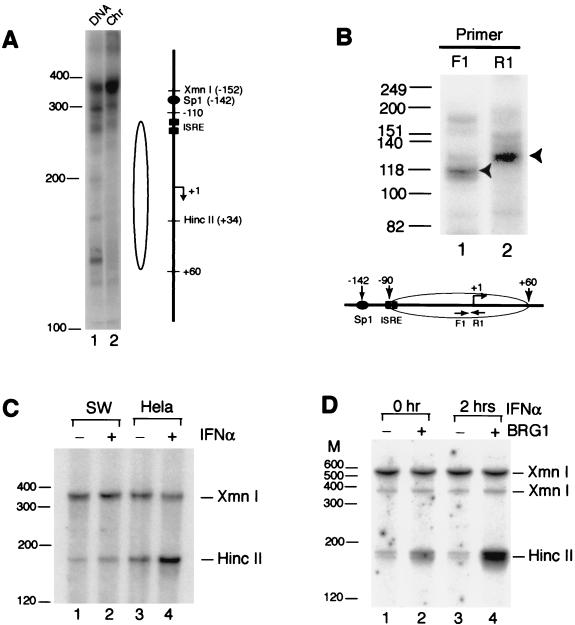
The IFITM2 (1-8D) and IFITM3 (1-8U) genes are located in the same chromosome locus and share significant homology in their coding and 5′ promoter regions (26). They are highly induced by IFN-α in HeLa cells (12). The observation that the maximal induction of these genes by IFN-α requires the presence of BRG1 suggests that the chromatin structure at their promoters may not be efficiently remodeled in the absence of a functional BAF complex upon IFN-α induction. We examined whether there is a nucleosome-like structure at the IFITM3 promoter in SW-13 cells by MNase digestion and LM-PCR. Nuclei isolated from exponentially growing cells were treated with MNase, which generates double-stranded cleavages in the linker region. To map the double-stranded cleavage sites, a unidirectional linker was added to the blunt ends generated by the double-stranded cleavages by MNase. The linker primer and an IFITM3 promoter-specific primer were then used to amplify the DNA and then to label it with a nested 32P-labeled IFITM3 promoter-specific primer. The purified genomic DNA was treated the same way as the control. As shown in Fig. 2A, about 160 bp of DNA from +60 to −100 relative to the transcription start site was protected from MNase digestion, suggesting the presence of a nucleosome-like structure. The same MNase digestion pattern was observed in HeLa cells (data not shown). In order to map more precisely the boundaries of the possible nucleosomal structure, the mononucleosome-sized DNA isolated from SW-13 nuclei digested with MNase was analyzed by LM-PCR with primers within the protected region. As indicated in Fig. 2B, the labeled F1 primer (−35 to −12) detected a major band of 118 bases that included 25 bases of the universal linker DNA, which was ligated to the mononucleosomes, indicating that the 3′ boundary is at +60 relative to the transcription start site. By using the labeled R1 primer (+10 to −14), the 5′ boundary of the nucleosome was determined to be at −90 (Fig. 2B, lane 2). Therefore, taken together, the data shown in Fig. 2A and B indicate the existence of a positioned nucleosome from −90 to +60. The majority of the two ISREs (−102 to −72) in the promoter are located within the nucleosomal DNA.
FIG. 2.
Chromatin remodeling at the IFITM3 promoter requires the activity of the BAF complex. (A) MNase digestion defines a nucleosome positioned over the IFITM3 promoter. The nuclei or genomic DNA isolated from SW-13 cells were digested with MNase. The double-stranded cleavages in the nucleosomal linker region were detected by LM-PCR. The open oval on the right side of the lanes indicates the protected region of DNA. The filled oval represents the Sp1-binding site. The filled squares represent ISREs. “+1” and an arrow indicate the position relative to the transcription start site and direction of transcription, respectively. The HincII and XmnI restriction sites are also indicated. Chr, nuclei (28). (B) A map of the boundaries of the protected region is shown. The mononucleosome-sized DNA isolated from SW-13 nuclei digested with MNase was subjected to LM-PCR analysis (28). The nested 32P-labeled primers are indicated F1 (−35 to −12) and R1 (+10 to −14). The arrowhead indicates the major band. The nucleosomal boundaries indicated by the open oval were determined after deduction of the DNA sequence (25 bases) that was ligated to the mononucleosomes as a universal linker (adaptor). (C) Chromatin remodeling at the IFITM3 promoter is defective in SW-13 cells. Nuclei isolated from SW-13 and HeLa cells with or without IFN-α treatment were briefly digested with HincII, followed by complete digestion of the purified genomic DNA with XmnI. The cleavage products were detected by LM-PCR. (D) Transient expression of BRG1 in SW-13 cells restored the chromatin remodeling at the IFITM3 promoter upon IFN-α stimulation. SW-13 cells were transiently transfected with pMACS Kk and pBJ5-BRG1 for 24 h. Following treatment with IFN-α for 2 h, the transfected and nontransfected cells were sorted by magnetic beads and processed for HincII accessibility as described for panel C. The digestion of the XmnI site at −152 was somehow incomplete. The band of 520 bp was derived from complete digestion of the XmnI site at −330. Size markers are indicated on the left of each panel.
MNase digestion was not able to reveal any differences in the chromatin structure at the IFITM3 promoter between SW-13 cells and HeLa cells with or without IFN-α treatment (data not shown). We then used the restriction enzyme accessibility assay to monitor the chromatin-remodeling event at the IFITM3 promoter upon IFN-α treatment. There is a HincII site (+34) located in the nucleosome which is downstream of the two ISREs, as shown in Fig. 2A. Nuclei from SW-13 cells and HeLa cells with or without IFN-α treatment were briefly digested with HincII. The genomic DNA was purified and digested completely with XmnI, which recognizes a site at −152. The DNA cleavage pattern was analyzed by LM-PCR. Cleavage at the HincII site would generate a fragment of 181 bp, and cleavage at the XmnI site would generate a fragment of 367 bp. The nucleosome remodeling at the promoter would increase HincII digestion and therefore generate more 181-bp fragment. As shown in Fig. 2B, the HincII site became significantly more accessible after 2 h of treatment with IFN-α in HeLa cells (lanes 3 and 4). However, no significant changes were observed in SW-13 cells upon IFN-α stimulation (lanes 1 and 2).
In order to demonstrate that the defect in remodeling the IFITM3 promoter in SW-13 cells was caused by the absence of the BRG1 protein, we transiently transfected the cells with pMACS Kk and pBJ5-BRG1. Following treatment with IFN-α for 2 h after 24 h of transfection, the transfected and nontransfected cells were sorted with magnetic beads and processed for HincII accessibility as described in the legend to Fig. 2C. The results showed that BRG1 expression alone slightly increased HincII digestion (Fig. 2D, compare lanes 1 and 2) and that chromatin remodeling was fully restored in SW-13 cells expressing BRG1 protein upon treatment with IFN-α (Fig. 2D, compare lanes 3 and 4). The control XmnI site at −152 was somehow inefficiently digested, and the band of 520 bp was derived from the XmnI site at −320.
These results indicate that the BAF complex is required for chromatin remodeling at the IFITM3 promoter upon induction with IFN-α.
Activation of the IFITM3 promoter by the BAF complex is dependent on chromatin formation.
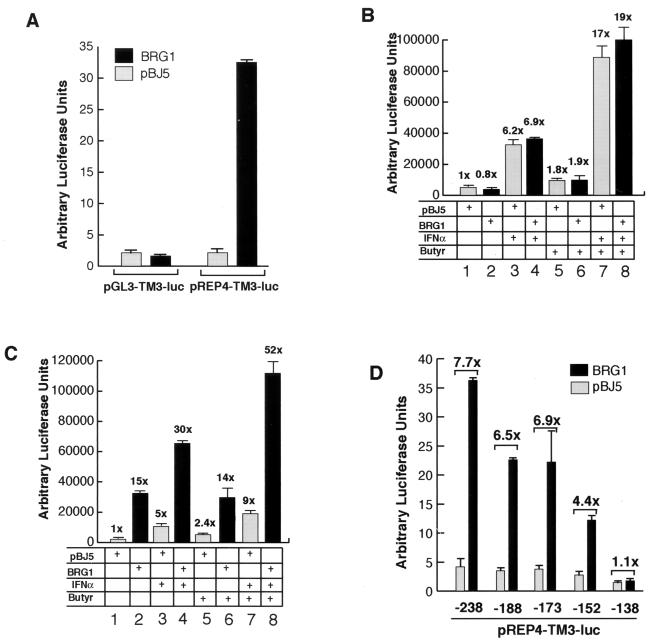
In order to identify the DNA elements and the corresponding protein factors that mediate the activity of the BAF complex, we cloned 238 bp of the IFITM3 promoter sequence into the pGL3-basic vector and tested the promoter activity in SW-13 cells by cotransfection with the BRG1 expression vector. As shown in Fig. 3A, cotransfection with BRG1 did not activate the promoter. Since the BAF complex is a chromatin-remodeling complex, the activation of the IFITM3 promoter by the complex might require the formation of proper chromatin structure. Therefore, we cloned the promoter into a pREP4-based episomal reporter vector that forms proper chromatin structure when transiently transfected into cells (27, 42). The IFITM3 promoter in the pREP4 vector was significantly activated by BRG1 (Fig. 3A). Interestingly, transient expression of the BRG1 homologue, hBRM, also efficiently activated the IFITM3 promoter in the pREP4 vector (data not shown).
FIG. 3.
Synergistic activation of the IFITM3 promoter by BRG1 and IFN-α is dependent on the formation of proper chromatin structure. (A) Activation of the IFITM3 promoter by the BAF complex requires formation of proper chromatin structure. The IFITM3 promoter was cloned into the pGL3/basic or pREP4-luc reporter vector. The constructs were cotransfected with either pBJ5 or BRG1 into SW-13 cells. Luciferase activity was determined with the dual luciferase assay kit from Promega. The bar graph was derived from the average of two experiments, with the error bar indicating the range from two experiments. (B and C) The IFITM3 promoter in pGL3 vector (panel B) or pREP4 vector (panel C) was cotransfected with pBJ5 or BRG1 into SW-13 cells for 24 h, followed by treatment with 500 U/ml and/or 5 mM sodium butyrate for 12 h. The luciferase activity was analyzed as described for panel A. The fold activation is indicated above each column. (D) Deletion analysis of the IFITM3 promoter is illustrated. The pREP4-TM3-luc constructs with 5′ deletions were analyzed as described for panel A. The numbers below the graph represent the base pairs from the transcription start site. The numbers above the bars represent the fold difference in activation by BRG1 compared to that by the pBJ5 vector.
Since activation of the IFITM3 promoter by the BAF complex is dependent upon the formation of proper chromatin structure, we hypothesized that the synergistic effect of the BAF complex and IFN-α on the induction of the IFITM3 gene might also require the proper chromatin structure. As shown in Fig. 3B, the IFITM3 promoter in pGL3 vector was activated 6.2 times by IFN-α alone. The presence of BRG1 did not cause a significant increase in the activation (compare columns 3 and 4). However, the IFITM3 promoter in the pREP4 vector was significantly more activated by IFN-α in the presence of BRG1 (30 times) than in the absence of BRG1 (5 times) (Fig. 3C, compare columns 3 and 4), indicating that the proper chromatin structure is a critical mediator of synergy.
Since the histone acetyltransferase activity of CBP/p300 is required for the induction of IFN-α/β target genes (3), we tested the effect of inhibiting histone deacetylase activity by butyrate treatment on activation of the IFITM3 promoter. As shown for the pGL3 vector in Fig. 3B, butyrate activated the promoter 1.8-fold (column 5). Both IFN-α and butyrate synergistically activated the promoter about 17-fold (column 7). No further activation was observed in the presence of BRG1 (compare columns 7 and 8). In contrast, the presence of BRG1 dramatically increased activation of the IFITM3 promoter in the pREP4 vector from 9-fold to 52-fold (columns 7 and 8 in Fig. 3C), consistent with the idea that histone acetylation stabilizes the binding of the SWI/SNF complex to the chromatin template (18).
A 5′ deletion analysis of the promoter revealed that deletion to −173 slightly reduced the fold activation by BRG1 (Fig. 3D). A more significant decrease in the fold activation by BRG1 was observed when the promoter sequence was deleted to −152. Further deletion to −138 abolished activation by BRG1. Therefore, at least two major elements may be mediating the activity of the BAF complex, one located between −173 and −152 and the other between −152 and −138.
Sp1-binding site contributes to activation of the IFITM3 promoter by the BAF complex.
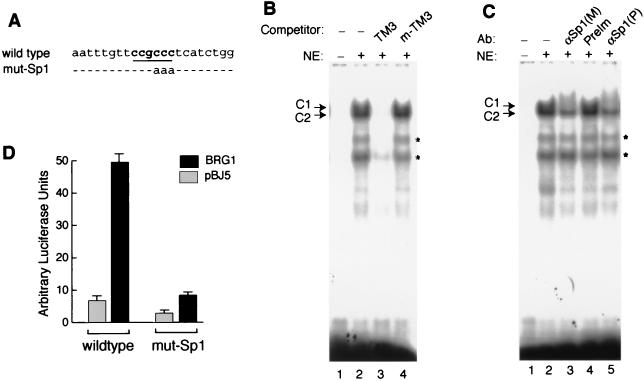
While examination of the DNA sequence between −173 and −152 did not reveal any apparent binding sites for known protein factors, the region between −152 and −138 contains one Sp1-binding motif (Fig. 4A). EMSA showed that the SW-13 nuclear extracts produced two reproducible shifts (labeled C1 and C2) of the 22-mer probe covering the Sp1-binding site from the IFITM3 promoter (Fig. 4B, lane 2). The shifts were inhibited with a 100-fold excess of the unlabeled wild-type probe (lane 3) but not with the mutant-binding site (lanes 4). Both of the monoclonal and polyclonal antibodies against the Sp1 protein induced a very faint supershifted band and at the same time completely inhibited the upper shift, C1, while the C2 shift was only partially inhibited (Fig. 4C, lanes 3 and 5). These data indicate that the C1 shift and part of the C2 shift were generated by Sp1 binding (Fig. 4C, lanes 2 and 3).
FIG. 4.
Sp1 contributes to the activation of the IFITM3 promoter by the BAF complex. (A) The sequence of the oligonucleotide probe surrounding the Sp1-binding site (underlined) from the wild-type IFITM3 promoter used for the EMSA is shown. The mutated bases used in the EMSA and in the reporter construct for transfection are indicated. (B) EMSA was performed with the wild-type probe labeled with 32P, 1 μg of SW-13 nuclear extracts, and 500 ng of poly(dI-dC) nonspecific competitor as described previously (27). Where applicable, 100× excess of unlabeled oligonucleotide was used as competitor. TM3, wild-type oligonucleotide from the IFITM3 promoter; m-TM3, Sp1 site-mutated oligonucleotide from the IFITM3 promoter. (C) The EMSA reaction included 1 μg of Sp1 antibody. M, monoclonal antibody; P, polyclonal antibody; PreIM, normal rabbit serum; ∗, band not observed reproducibly. (D) The IFITM3 promoter was mutated as mut-Sp1 as described for panel A. The mutant construct was analyzed by luciferase assay as described in the legend to Fig. 3A.
In order to determine if the Sp1-binding site contributes to the regulation of the IFITM3 promoter by the BAF complex, we tested the pREP4-TM3-luc construct with the same mutation as shown in Fig. 4A in transfection experiments. As shown in Fig. 4D, mutation of the Sp1-binding site reduced dramatically the activation of the IFITM3 promoter by the BAF complex while it caused a modest decrease in the basal-level activity of the promoter.
These results suggest that Sp1 binds to the IFITM3 promoter and augments activation by the BAF complex.
BAF complex and Sp1 are constitutively associated with the IFITM3 promoter in vivo.
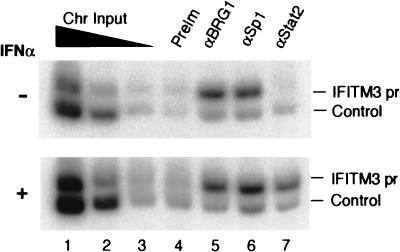
Previous studies suggest that the chromatin-remodeling complexes are recruited to their target promoters upon induction (2, 5, 37). In order to determine if the SWI/SNF-like BAF complex and Sp1 are recruited to the IFITM3 promoter upon induction by IFN-α, we performed the ChIP assay with HeLa cells. Chromatin fractions prepared from formaldehyde-cross-linked HeLa cells with or without prior IFN-α treatment were immunoprecipitated with antibodies against Sp1, BRG1 (which recognizes both BRG1 and hBRM), and STAT2. The immunoprecipitated DNA was reverse cross-linked and analyzed by multiplex PCR, with the DNA sequence downstream of the 3′ untranslated region of the IFITM2 gene as the negative control. As expected, STAT2 became bound to the promoter upon treatment with IFN-α, while no binding was detected in the absence of IFN-α treatment (Fig. 5). However, antibodies against BRG1 and Sp1 enriched the promoter sequence relative to the control sequence in both the presence and absence of IFN-α treatment, indicating that the BAF complex and Sp1 are constitutively associated with the promoter in vivo. These data are consistent with the observation that transient expression of BRG1 in SW-13 cells remodeled and activated the IFITM3 promoter even in the absence of IFN-α treatment (Fig. 1, 2D, and 3).
FIG. 5.
BAF complex and Sp1 are constitutively associated with the IFITM3 promoter in vivo. Chromatin was prepared from HeLa cells without (−) or with (+) IFN-α treatment for 30 min prior to formaldehyde cross-linking. The DNA was purified from immunoprecipitates with the antibodies indicated above the panel and analyzed by multiplex PCR with IFITM3 promoter primers (−251 to +61) and a pair of negative control primers (IFITM2U/F2 and IFITM2U/R1) that recognized the DNA sequence downstream of the 3′ untranslated region of the IFITM2 gene. PCR was performed in the presence of 5 μCi of [α-32P]dCTP, and the products were analyzed in 5% polyacrylamide gels as described previously (39). Chromatin input was diluted 3 times at each step.
Sp1 is bound to the IFITM3 promoter in the absence of the active BAF complex in SW-13 cells.
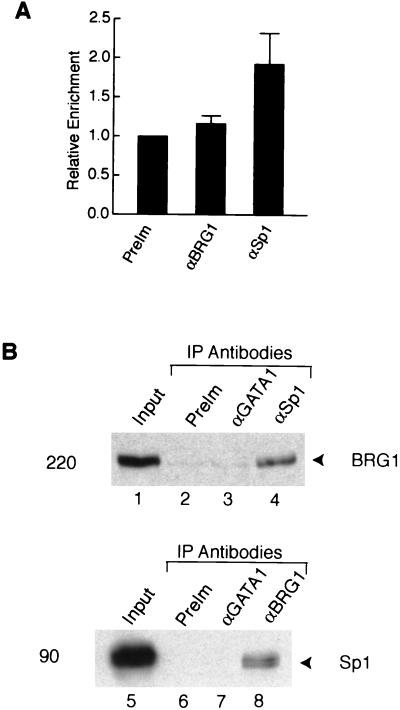
The Sp1-binding site in the IFITM3 promoter appears to be located outside of the positioned nucleosome (Fig. 2A). Therefore, Sp1 binding to the promoter might not require the activity of the BAF complex. To test this hypothesis, we performed a ChIP assay with SW-13 cells. As quantified by real-time PCR in Fig. 6A, Sp1 antibodies modestly enriched the promoter sequence in the immunoprecipitates while no enrichment was observed with antibodies against BRG1, which does not have detectable levels of expression in the cells. Similar results were obtained with SW-13 cells treated with IFN-α (data not shown). These results suggest that Sp1 is constitutively associated with the IFITM3 promoter, even in the absence of the BAF complex.
FIG. 6.
Sp1 interacts with the BAF complex in vivo. (A) Association of Sp1 with the IFITM3 promoter does not require the activity of the BAF complex. The immunoprecipitated chromatin from SW-13 cells with each antibody was analyzed, respectively, with the IFITM3 promoter primers and the negative control primers by real-time PCR. The graph shows the relative enrichment of the IFITM3 promoter sequence after normalization to the negative control sequence. The error bar indicates the range from two experiments. (B) Sp1 interacts with the BAF complex in vivo. Nuclear extracts prepared from DSP-cross-linked cells were immunoprecipitated with the indicated antibodies. The immunoprecipitates were resolved by SDS-PAGE and blotted with BRG1 or Sp1 antibodies. One percent of the nuclear extracts used for IP was loaded in lanes 1 and 5 as input. The size of the band is indicated on the left of the panels.
Sp1 interacts directly with the BAF complex in vivo.
Since the Sp1-binding site contributed to activation of the promoter by the BAF complex (Fig. 4) and the zinc finger DNA-binding domain of Sp1 has been shown to interact with BRG1 and BAF155 in vitro in a GST pull-down assay (20), we decided to test the direct in vivo interaction between Sp1 and the BAF complex. While coimunoprecipitation from the nuclear extracts of Jurkat cells did not yield conclusive results, we used an in vivo cross-linking strategy to stabilize any possible interaction between them. Exponentially growing Jurkat cells were cross-linked briefly with the cell membrane permeable and reversible cross-linker DSP, followed by nuclear extract preparation as described previously (50). The cross-linked nuclear extracts were immunoprecipitated with anti-Sp1 and anti-BRG1 antibodies in high-salt buffer, and the coimmunoprecipitated proteins were detected by Western blot analysis. As shown in Fig. 6B, the Sp1 and BRG1 antibodies could each coprecipitate the other (lanes 4 and 8). The preimmune serum and anti-GATA1 antibodies did not pull down either the Sp1 or BRG1 proteins (lanes 2, 3, 6, and 7). Since the cross-linker, DSP, does not cross-link protein to DNA, which rules out the possibility that the coimmunoprecipitation of the BAF complex with Sp1 resulted from the BAF complex and Sp1 being cross-linked to the same DNA fragment, and the mild conditions we used favor the cross-linking of directly interacting proteins (50), these data strongly suggest that Sp1 and the BAF complex interact directly in vivo.
DISCUSSION
BAF complex is involved in IFN signaling pathways.
IFNs mediate a broad range of cellular activities. In particular, they suppress cancer development by inhibiting cell growth and controlling apoptosis. It is not known how IFN-induced gene products control the antiproliferative activity. However, at least in some cases, factors involved in cell cycle control, such as c-Myc, pRB, etc., are targeted (24, 29). IFNs can either promote or inhibit apoptosis, depending on the developmental status of a cell (40). The activities of IFNs are mainly mediated by the STAT proteins. Upon IFN binding, the IFN receptors oligomerize and lead to the phosphorylation and activation of Janus kinases, which leads to the dimerization and nuclear translocation of the STAT proteins (6, 25). The STAT proteins in the nucleus bind to their target sites and activate transcription of their target genes. However, very little is known about how the nuclear STAT complexes overcome the chromatin barrier to activate their target genes. We compared the chromatin-remodeling activity at the IFITM3 promoter upon IFN-α induction in HeLa cells and SW-13 cells. Our results indicate that chromatin was efficiently remodeled in HeLa cells upon IFN-α treatment. However, chromatin remodeling was defective in SW-13 cells (Fig. 2C), even though IFN-α signaling efficiently activated STAT proteins (data not shown). Significantly, transient expression of BRG1 in SW-13 cells fully restored the chromatin remodeling of the IFITM3 promoter upon IFN-α treatment (Fig. 2D) and the induction of a group of IFN-α target genes (Fig. 1). These data indicate that the BAF complex mediated chromatin remodeling for the transcriptional activation of these genes.
BRG1 protein is the essential ATPase subunit of the BAF complex. It is not expressed at any detectable levels in the SW-13 cells derived from pheochromocytoma (11, 31). Interestingly, transient expression of BRG1 induces cell differentiation and growth arrest by an unknown mechanism (11). In order to understand the mechanism by which the BAF complex induces differentiation and growth arrest, we used the cDNA microarray approach to identify target genes of the BAF complex. Among the 80 genes activated by the BAF complex were several IFN-induced genes, including IFITM1, IFITM2, IFITM3, and IFIT1 (27). Furthermore, we demonstrated in this report that the BAF complex is required for the maximal induction of these IFN target genes by IFN-α (Fig. 1). Since the IFN system mediates cellular antiproliferative activity, up-regulation of the IFN-induced genes might have a negative effect on cell growth and, in combination with other unidentified effector genes, result in the observed cell growth arrest of SW-13 cells.
Recent data suggest that the BAF complex may function as a tumor suppressor and play an essential role in cell proliferation and cell differentiation in various experimental and pathological systems. For example, the homozygous mutation of INI1 or BAF47 is linked to rhabdomyosarcoma in early childhood (45). Mice with the heterozygous mutation of INI1 are predisposed to cancer formation (16, 22). Various mutations in the BAF complexes have been identified in a broad range of cancer cell lines (7). Expression of a dominant negative form of BRG1 or hBRM prevents MyoD-mediated muscle differentiation (8). However, it is not clear what mediates the tumor suppressor activity of the BAF complex. The present hypothesis asserts that the SWI/SNF or BAF complex cooperates with the retinoblastoma protein to induce cell cycle arrest (11, 48). Based on our discovery that the BAF complex is involved in the IFN signaling processes, we hypothesize that mutations in the BAF complex reduce the efficiency of the IFN system in their antiproliferative activities and therefore result in the disruption of cell growth control.
Recruiting mechanism of the BAF complex to the IFITM3 promoter.
In this report, we demonstrated that a subset of the IFN-α target genes required the BAF complex for maximal induction, being consistent with previous reports that the BAF complex is often required for only a subset of genes involved in a particular pathway (9, 10, 23). In order to elucidate the molecular mechanisms by which the BAF complex is targeted to the IFN-α target genes, we analyzed the IFITM3 promoter. The major recognition sequences for the BAF complex in the IFITM3 promoter are located between −138 and −173 relative to the transcription start site. Deletion of the sequence between −152 and −173 reduced the activation by the BAF complex to about 50%, while the basal-level activity was not significantly changed. However, mutation of the Sp1-binding site between −140 and −145 dramatically inhibited activation by the BAF complex while the basal activity decreased moderately. Therefore, the sequence from −152 to −173 and the binding site for the ubiquitous transcription activator Sp1 synergistically mediate the activity of the BAF complex on the IFITM3 promoter. Sp1 is associated with the IFITM3 promoter even in the absence of the BAF complex. Furthermore, we find that Sp1 interacts with the BAF complex in vivo, suggesting that Sp1 protein might recruit the complex and/or stabilize the binding of the complex to the promoter in collaboration with another unidentified activity located between −152 and −173.
Analysis of the DNA sequence of several IFN-α target promoters, including 6-16, ISG15, ISG20, ISGF3G, etc., which do not respond to the BAF complex, revealed the existence of Sp1-binding sites in many of the promoters, suggesting that Sp1 alone may not be sufficient for recruiting the BAF complex. The DNA sequence from −148 to −172 in the IFITM3 promoter is well conserved among IFITM1, IFITM2, and IFITM3, which could serve as the primary recognition sequence for the BAF complex and is under further investigation.
Several reports suggest that the yeast SWI/SNF complex and the mammalian BAF complex are actively recruited to promoters upon activation of genes (2, 5, 37). However, our data indicate that the BAF complex is associated with the IFITM3 promoter constitutively in HeLa cells in the absence or presence of IFN stimuli (Fig. 5). It remodels the chromatin structure of the IFITM3 promoter and activates the IFN-α target genes in the absence of IFN-α treatment (Fig. 1, 2D, and 3). It was also reported recently that the BAF complex is associated constitutively with the CSF1 (colony-stimulating factor 1) promoter that is inducible in the primary human cell line, the WI-38 cell line, upon tumor necrosis factor alpha treatment (27). In both cases, the binding of the BAF complex to the promoter up-regulated the basal-level expression of the genes, which could have important functional consequences in their cellular activities. Chromatin remodeling is a highly energy-consuming and, consequently, rate-limiting step. Therefore, constitutive binding of the chromatin-remodeling complex to the promoter might keep the promoter at a remodeled state that would allow rapid induction of the promoter by a stimulus.
Acknowledgments
We thank the members of the Leonard and Zhao laboratories for stimulating discussions and Warren J. Leonard for critical reading of the manuscript. We thank Mei Huang and Zilong Wen for help with the STAT phosphorylation experiments. K.Z. thanks Warren J. Leonard for continuous encouragement and support.
This work was supported by intramural grants to NHLBI, NIH.
REFERENCES
- 1.Aalfs, J. D., and R. E. Kingston. 2000. What does “chromatin remodeling” mean? Trends Biochem. Sci. 25:548-555. [DOI] [PubMed] [Google Scholar]
- 2.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya, S., R. Eckner, S. Grossman, E. Oldread, Z. Arany, A. D'Andrea, and D. M. Livingston. 1996. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature 383:344-347. [DOI] [PubMed] [Google Scholar]
- 4.Boehm, U., T. Klamp, M. Groot, and J. C. Howard. 1997. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 15:749-795. [DOI] [PubMed] [Google Scholar]
- 5.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 6.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 7.Decristofaro, M. F., B. L. Betz, C. J. Rorie, D. N. Reisman, W. Wang, and B. E. Weissman. 2001. Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J. Cell. Physiol. 186:136-145. [DOI] [PubMed] [Google Scholar]
- 8.de la Serna, I. L., K. A. Carlson, and A. N. Imbalzano. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27:187-190. [DOI] [PubMed] [Google Scholar]
- 9.de la Serna, I. L., K. A. Carlson, D. A. Hill, C. J. Guidi, R. O. Stephenson, S. Sif, R. E. Kingston, and A. N. Imbalzano. 2000. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 20:2839-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Serna, I. L., K. Roy, K. A. Carlson, and A. N. Imbalzano. 2001. MyoD can induce cell cycle arrest but not muscle differentiation in the presence of dominant negative SWI/SNF chromatin remodeling enzymes. J. Biol. Chem. 276:41486-41491. [DOI] [PubMed] [Google Scholar]
- 11.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 12.Friedman, R. L., S. P. Manly, M. McMahon, I. M. Kerr, and G. R. Stark. 1984. Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell 38:745-755. [DOI] [PubMed] [Google Scholar]
- 13.Fry, C. J., and C. L. Peterson. 2001. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11:R185-R197. [DOI] [PubMed] [Google Scholar]
- 14.Fu, X. Y., C. Schindler, T. Improta, R. Aebersold, and J. E. Darnell, Jr. 1992. The proteins of ISGF-3, the interferon alpha-induced transcriptional activator, define a gene family involved in signal transduction. Proc. Natl. Acad. Sci. USA 89:7840-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gingras, S., J. Simard, B. Groner, and E. Pfitzner. 1999. p300/CBP is required for transcriptional induction by interleukin-4 and interacts with Stat6. Nucleic Acids Res. 27:2722-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guidi, C. J., A. T. Sands, B. P. Zambrowicz, T. K. Turner, D. A. Demers, W. Webster, T. W. Smith, A. N. Imbalzano, and S. N. Jones. 2001. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol. 21:3598-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassan, A. H., K. E. Neely, M. Vignali, J. C. Reese, and J. L. Workman. 2001. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 6:D1054-D1064. [DOI] [PubMed] [Google Scholar]
- 18.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104:817-827. [DOI] [PubMed] [Google Scholar]
- 19.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 20.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 14:2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khavari, P. A., C. L. Peterson, J. W. Tamkun, D. B. Mendel, and G. R. Crabtree. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170-174. [DOI] [PubMed] [Google Scholar]
- 22.Klochendler-Yeivin, A., L. Fiette, J. Barra, C. Muchardt, C. Babinet, and M. Yaniv. 2000. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kowenz-Leutz, E., and A. Leutz. 1999. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4:735-743. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, R., and I. Atlas. 1992. Interferon alpha induces the expression of retinoblastoma gene product in human Burkitt lymphoma Daudi cells: role in growth regulation. Proc. Natl. Acad. Sci. USA 89:6599-6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonard, W. J., and J. J. O'Shea. 1998. Jaks and STATs: biological implications. Annu. Rev. Immunol. 16:293-322. [DOI] [PubMed] [Google Scholar]
- 26.Lewin, A. R., L. E. Reid, M. McMahon, G. R. Stark, and I. M. Kerr. 1991. Molecular analysis of a human interferon-inducible gene family. Eur. J. Biochem. 199:417-423. [DOI] [PubMed] [Google Scholar]
- 27.Liu, R., H. Liu, X. Chen, M. Kirby, P. O. Brown, and K. Zhao. 2001. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106:309-318. [DOI] [PubMed] [Google Scholar]
- 28.McPherson, C. E., E. Y. Shim, D. S. Friedman, and K. S. Zaret. 1993. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell 75:387-398. [DOI] [PubMed] [Google Scholar]
- 29.Melamed, D., N. Tiefenbrun, A. Yarden, and A. Kimchi. 1993. Interferons and interleukin-6 suppress the DNA-binding activity of E2F in growth-sensitive hematopoietic cells. Mol. Cell. Biol. 13:5255-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muchardt, C., and M. Yaniv. 1999. ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job. J. Mol. Biol. 293:187-198. [DOI] [PubMed] [Google Scholar]
- 31.Muchardt, C., and M. Yaniv. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mueller, P. R., and B. Wold. 1989. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science 246:780-786. [DOI] [PubMed] [Google Scholar]
- 33.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 34.Paulson, M., S. Pisharody, L. Pan, S. Guadagno, A. L. Mui, and D. E. Levy. 1999. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 274:25343-25349. [DOI] [PubMed] [Google Scholar]
- 35.Pestka, S., J. A. Langer, K. C. Zoon, and C. E. Samuel. 1987. Interferons and their actions. Annu. Rev. Biochem. 56:727-777. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 37.Reinke, H., P. D. Gregory, and W. Horz. 2001. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell 7:529-538. [DOI] [PubMed] [Google Scholar]
- 38.Schindler, C., X. Y. Fu, T. Improta, R. Aebersold, and J. E. Darnell, Jr. 1992. Proteins of transcription factor ISGF-3: one gene encodes the 91- and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc. Natl. Acad. Sci. USA 89:7836-7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 40.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 41.Sudarsanam, P., and F. Winston. 2000. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 16:345-351. [DOI] [PubMed] [Google Scholar]
- 42.van der Vlag, J., J. L. den Blaauwen, R. G. Sewalt, R. van Driel, and A. P. Otte. 2000. Transcriptional repression mediated by polycomb group proteins and other chromatin-associated repressors is selectively blocked by insulators. J. Biol. Chem. 275:697-704. [DOI] [PubMed] [Google Scholar]
- 43.Varga-Weisz, P. 2001. ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene 20:3076-3085. [DOI] [PubMed] [Google Scholar]
- 44.Veals, S. A., C. Schindler, D. Leonard, X. Y. Fu, R. Aebersold, J. E. Darnell, Jr., and D. E. Levy. 1992. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol. Cell. Biol. 12:3315-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Versteege, I., N. Sevenet, J. Lange, M. F. Rousseau-Merck, P. Ambros, R. Handgretinger, A. Aurias, and O. Delattre. 1998. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394:203-206. [DOI] [PubMed] [Google Scholar]
- 46.Weinmann, A. S., S. E. Plevy, and S. T. Smale. 1999. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11:665-675. [DOI] [PubMed] [Google Scholar]
- 47.Wu, J., and M. Grunstein. 2000. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 25:619-623. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79-89. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, J. J., U. Vinkemeier, W. Gu, D. Chakravarti, C. M. Horvath, and J. E. Darnell, Jr. 1996. Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc. Natl. Acad. Sci. USA 93:15092-15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao, K., W. Wang, O. J. Rando, Y. Xue, K. Swiderek, A. Kuo, and G. R. Crabtree. 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95:625-636. [DOI] [PubMed] [Google Scholar]
- 51.Zhao, X., P. S. Pendergrast, and N. Hernandez. 2001. A positioned nucleosome on the human U6 promoter allows recruitment of SNAPc by the Oct-1 POU domain. Mol. Cell 7:539-549. [DOI] [PubMed] [Google Scholar]
- 52.Zhu, M., S. John, M. Berg, and W. J. Leonard. 1999. Functional association of Nmi with Stat5 and Stat1 in IL-2- and IFNgamma-mediated signaling. Cell 96:121-130. [DOI] [PubMed] [Google Scholar]