Abstract
The structural basis for the association of eukaryotic and prokaryotic protein receptors and their triple-helical collagen ligand remains poorly understood. Here, we present the crystal structures of a high affinity subsegment of the Staphylococcus aureus collagen-binding CNA as an apo-protein and in complex with a synthetic collagen-like triple helical peptide. The apo-protein structure is composed of two subdomains (N1 and N2), each adopting a variant IgG-fold, and a long linker that connects N1 and N2. The structure is stabilized by hydrophobic inter-domain interactions and by the N2 C-terminal extension that complements a β-sheet on N1. In the ligand complex, the collagen-like peptide penetrates through a spherical hole formed by the two subdomains and the N1–N2 linker. Based on these two structures we propose a dynamic, multistep binding model, called the ‘Collagen Hug' that is uniquely designed to allow multidomain collagen binding proteins to bind their extended rope-like ligand.
Keywords: bacterial adhesion, collagen binding, protein–protein, surface proteins
Introduction
The collagens are the most abundant proteins in vertebrates. This class of proteins provides the structural support for tissues, serves as a scaffold for the assembly of extracellular matrices (ECM), and can also directly affect cell behavior through specific cellular receptors. Over 20 genetically different collagen types have been identified, in addition to a number of proteins that contain collagenous subdomains. The collagenous domains have a characteristic triple helix structure where each of the participating polypeptides are composed of repeating Gly-X-Y sequences that form hetero or homo-trimeric L-proline helices. The limited amino-acid sequence variations and the rope-like triple-helical structure of collagens present a unique challenge to the elucidation of the molecular and structural details of protein–collagen interactions. Currently, a plethora of proteins are known to interact with collagens; however, the mechanisms by which these interactions occur remain poorly understood.
Both eukaryotes and prokaryotes express collagen-binding proteins, which include noncollagenous components of the ECM, cellular receptors such as some integrins, and bacterial collagen adhesins. Collagen degrading enzymes such as the eukaryotic matrix metalloproteinases (MMP) and the bacterial collagenases also contain collagen-binding domains. So far the I domain of integrin α2 (α2I) is the only structure of a collagen binding protein in complex with a collagen-derived peptide ligand that has been solved (Emsley et al, 2000). In this structure, an Mg2+ ion facilitates the protein–protein interaction by coordinating residues in the metal ion-dependent adhesion site (MIDAS) of α2I and a Glu residue in the collagen peptide.
A family of structurally related collagen binding adhesins of the MSCRAMM (microbial surface component recognizing adhesive matrix molecules) type is found on Gram-positive pathogens such as Staphylococcus aureus (Patti et al, 1993), Enterococcus faecalis (Rich et al, 1999c), Enterococcus faecium (Nallapareddy et al, 2003), Streptococcus equi (Lannergard et al, 2003), Streptococcus mutans (Sato et al, 2004), Erysipelothrix rhusiopathiae (Shimoji et al, 2003), and Bacillus anthracis (Xu et al, 2004). These adhesive proteins from Gram-positive bacteria do not share sequence homology with the collagen binding I domains of integrins and do not require metal ions for collagen binding. Thus, these proteins appear to employ a collagen binding mechanism that maybe drastically different from that of the collagen binding integrins.
The collagen binding MSCRAMM on S. aureus is called CNA and is the prototype member of this family. CNA participates in the infectious process of pathogenic S. aureus and is shown to be a virulence factor in many different animal models of staphylococcal infections including arthritis, endocarditis, osteomyelitis, mastitis and keratitis (Patti et al, 1993, 1994; Hienz et al, 1996; Mamo et al, 2000; Rhem et al, 2000; Elasri et al, 2002), suggesting that the ability to interact with collagen provides a general advantage to the bacteria in pathogenesis. Furthermore, the recombinant CNA can be used as an effective vaccine component and antibodies raised against CNA are protective in a mouse model of S. aureus induced septic death (Nilsson et al, 1998).
CNA is composed of a so-called A-region and a varying number of B-repeats depending on the strain (Figure 1A). At the C-terminal end of CNA are features required for surface targeting and covalently anchoring to the peptidoglycan. The collagen binding activity is located in the A-region (Patti et al, 1993). Earlier studies from our laboratories have shown that a central segment of the A region contains the minimal collagen binding site and its crystal structure revealed a single domain representing a novel variant of the IgG-fold. Molecular modeling and docking experiments helped us to identify a grove on the surface of this domain as a putative collagen-binding site (Symersky et al, 1997). However, this central segment has a 10-fold lower affinity for collagen compared to the full length A-region, suggesting that regions flanking the central segment significantly participate in the collagen interaction (Patti et al, 1993; Xu et al, 2004).
Figure 1.
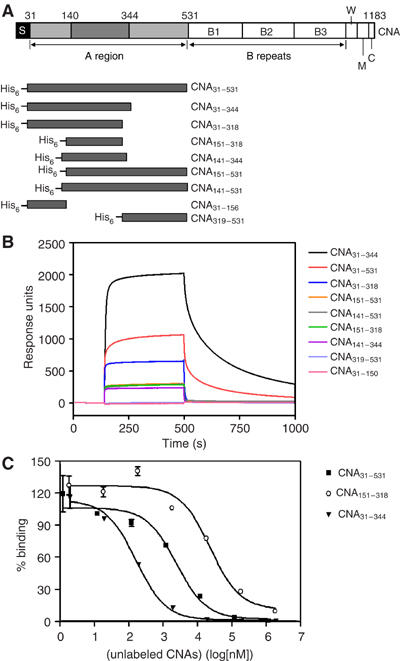
(A) The domain organization of S. aureus CNA and different constructs. The collagen binding A region is followed by B repeats. S, signal peptide; W, cell wall anchoring region containing the LPETG motif; M, transmembrane segment; and C, cytoplasmic tail. The three subdomains of A-region are from residues 31–140 (N1), 141–344 (N2), and 345–531 (N3). The previously identified minimum collagen-binding domain is from residues 151–318. CNA fragments constructed as N-terminal His-tag fusion proteins are illustrated. (B) Representative Biacore sensorgrams of different CNA fragments passed over collagen. The same concentration of purified CNA fragments (10 μM) was passed over a bovine type I collagen-coated surface. Injection started at ∼140 s and ended at ∼550 s. Responses from a blank surface were subtracted from the responses from the collagen-coated surface. (C) Inhibition of the binding of CNA31–344 to type I collagen by different CNA fragments. Biotin-labeled CNA31–344 (100 nM) was mixed with increasing concentrations of unlabeled CNA31-344 (inverted triangles), CNA31–531 (squares), and CNA151–318 (open circles), and then incubated in wells coated with bovine type I collagen.
We have identified a two-domain subregion of CNA that binds collagen with high affinity, crystallized this subregion and solved its crystal structure both as an apo-protein and in complex with a synthetic, collagen-like triple-helical peptide. Analyses of these structures point to an extraordinary multistep binding mechanism where the two subdomains cooperate to wrap around and ‘hug' the rope-like structure of a collagen monomer. The proposed binding mechanism, with some aspects of the Dock, Lock and Latch mechanism previously reported for MSCRAMM binding of linear peptides (Ponnuraj et al, 2003), demonstrates how bacteria by using similar building blocks, albeit with subtle modifications, can generate high affinity binding proteins that are specifically designed to adhere structurally diverse ligands. The possibility that eukaryotic multidomain collagen-binding proteins use similar ‘hugging' mechanisms to embrace the collagen triple helix is also discussed.
Results and discussion
A CNA segment with high affinity for collagen
We previously reported that the fibrinogen binding staphylococcal MSCRAMMs contain three subdomains called N1, N2 and N3 (Perkins et al, 2001; Ponnuraj et al, 2003) in their ligand-binding regions. By comparing the amino-acid sequences of CNA with these fibrinogen-binding proteins, we propose that the A-region of CNA is also composed of three subdomains; N1 corresponding to residues 31–140, N2 to residues 141–344 and N3 to residues 345–531. The previously identified minimal collagen-binding domain of CNA (res. 151–318) would, in this model, correspond to a truncated form of the N2 domain. The affinity of CNA151–318 for type I collagen is about 10-fold lower than that of the full-length CNA A-region (Patti et al, 1993). In our attempts to identify a subsegment of the CNA A-region with full binding activity, we constructed a set of seven recombinant proteins (Figure 1A, Supplementary Table 1) based on the putative subdomains and determined their relative affinity for type 1 collagen by surface plasmon resonance (SPR). Each protein at 10 μM was run over a BIA-core chip containing immobilized collagen (Figure 1B). Remarkably, the protein construct corresponding to the predicted N1N2 domains (CNA31–344) bound collagen with an affinity that appears to be substantially higher than that of even the full-length A-region (CNA31–531). Recombinant proteins corresponding to the predicted N2 domain (CNA141–344) and the N2N3 domains (CNA141–531) bound collagen with affinities similar to that of the previously examined CNA151–318, lower than that of CNA31–531 and much lower than that of CNA31–344. Recombinant proteins representing the predicted independent N1 or N3 domains did not show any measurable affinity for collagen. Further analyses using solid phase ELISA-type binding assays (Table I) or SPR analyses (data not shown) with multiple concentrations of CNA proteins confirmed the relative affinities indicated in Figure 1B. We further characterized the binding of the different recombinant CNA proteins to type 1 collagen in inhibition-ELISAs. Each of the CNA31–344, CNA31–531 and CNA151–318 proteins could completely inhibit the binding of biotin derivatives of the individual proteins to type I collagen (e.g., see Figure 1C), suggesting that the different CNA constructs have the same binding specificity but differ in their binding affinity.
Table 1.
Summary of the apparent collagen binding affinities of the different recombinant fragments of CNA
| Construct | KDapp (μM)a |
|---|---|
| CNA31–531 | 2.2±0.4 |
| CNA31–344 | 0.2±0.02 |
| CNA31–318 | 1.5±0.5 |
| CNA151–318 | 31.6±4.4 |
| CNA141–344 | 69.6±6.1 |
| CNA151–531 | 28.3±3.3 |
| CNA141–531 | 38.1±11.8 |
| CNA31–156 | NDb |
| CNA319–531 |
NDb |
| aThe apparent dissociation constants (KDapp) were determined from ELISA assays as described in Materials and methods. Recombinant fragments of the different constructs were added to wells coated with bovine type I collagen. Bound proteins were detected with mouse anti-His antibodies and goat-anti-mouse IgG-AP conjugants. Data analysis was performed using the nonlinear regression method (GraphPad Prism). | |
| bCNA31–156 and CNA319–531 did not show any binding in the ELISA assays and their KDapp values were not determined. | |
Identification of a collagen peptide with high affinity for CNA31–344
To identify binding sites in collagen for CNA31–344, we initially examined the binding of this MSCRAMM to fragments of bovine type I collagen generated by cyanogen bromide cleavage. CNA31–344 bound all fragments tested but with varying apparent affinities (P Speziale, Y Xu and M Höök, unpublished observation). Rotary shadowing coupled with electron microscopy was used to locate CNA binding sites; however, we could not identify any particularly ‘hot spots' and the binding sites appeared scattered over the type I collagen molecule (D Keene, Y Xu, and M Höök, unpublished observation). It seems that CNA31–344 can bind many sites along the collagen molecule, in agreement with previous SPR analysis of the binding characteristics of CNA31–531 (Rich et al, 1999b).
We screened a panel of 16 synthetic collagen peptides that were generated based on sequences from the α1 chain of bovine or chicken type I collagen. To facilitate the formation of collagen-like triple helices, the specific sequences in individual peptides were flanked by three or four GPO (O being Hydroxyproline) triplets at either side. In addition to this available library, we synthesized two generic peptides consisting of 11 GPO triplets or 11 GPP triplets.
In SPR binding analysis, some of the collagen peptides exhibited a high affinity for CNA31–344, some had an intermediate affinity, and others had a low affinity. An example of the binding of four of the peptides, DBS4, (GPO)11, (GPP)11 and DBS3 are shown in Figure 2A. The CD spectra of these peptides indicated that DBS4, (GPO)11, and (GPP)11 formed triple helices at both 4°C and room temperature, while DBS3 does not (Supplementary Figure 1). The binding of DBS4, (GPO)11, and (GPP)11 to CNA31–344 was further analyzed using SPR by passing increasing concentrations of the three peptides over a surface with immobilized CNA31–344. The dissociation constants (KD) of the interactions between CNA and DBS4, (GPO)11 and (GPP)11 were determined to be ∼3.0, ∼140, and ∼7.5 nM, respectively. In solid phase inhibition assays, DBS4 and (GPP)11 completely inhibited the binding of CNA31–344 to type I collagen with IC50 values of 5.3±1.3 and 11.8±1.5 μM, respectively. (GPO)11 and DBS3 were less potent inhibitors and only caused ∼50% inhibition at 100 μM (Figure 2B). Taken together, these results suggest that DBS4 and (GPP)11 contain at least one high-affinity binding site for CNA31–344.
Figure 2.
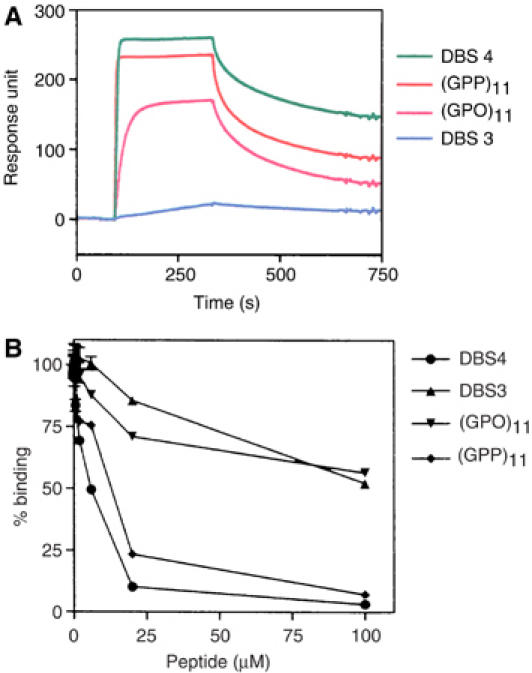
(A) Representative sensorgrams of the synthetic collagen peptides passed over CNA31–344. The same concentrations of four collagen peptides (1 μM) were passed over a CNA31–344-coated surface. Injection started at ∼95 s and ended at ∼335 s. Responses from a blank surface were subtracted from responses from collagen-coated surface. (B) Inhibition of the binding of CNA31–344 to type I collagen by synthetic collagen peptides. CNA31–344 (10 nM) was preincubated with increasing amounts of synthetic collagen peptides DBS4 (•), DBS3 (▴), (GPO)11 (▾), and (GPP)11 (⧫), and then added to wells coated with bovine type I collagen.
Crystal structure of apo-CNA31–344
The crystal structure of CNA31–344 was determined by molecular replacement methods, using the CNA151–318 crystal structure (Symersky et al, 1997) as a search model, and refined to an R factor of 19.1% (Rfree of 23.3%) using diffraction data to 1.95 Å resolution. Crystallographic, diffraction data collection, and model refinement details are presented in Table II. The crystal structure of CNA31–344 (Figure 3A) exhibits two distinct domains (called N1 and N2). The N-terminal N1 domain (residues 31–163) exhibits a DeV-IgG fold, which contains two additional strands D1′ and D1″ (Figure 3B) compared to the conventional IgG fold (Deivanayagam et al, 2002; Ponnuraj et al, 2003). The N2 domain corresponding to 174–329 residues is very similar to the previously determined crystal structure of CNA151–318 (Symersky et al, 1997), which contains in addition to the D2′ and D2″ strands, an extra D2‴ strand and a two-turn α-helix (Figure 3B).
Table 2.
Data collection and refinement statistics
| Crystal | CNA31–344 | CNA31–344–collagen peptide complex |
|---|---|---|
| Unit cell dimensions | ||
| a (Å) | 41.98 | 90.55 |
| b (Å) | 106.43 | 193.82 |
| c (Å) | 44.08 | 205.19 |
| β (deg) | 116.45 | |
| Space group | P2 (1) | C222 (1) |
| Resolution limits (Å) | 1.95–50.0 | 3.30–50.0 |
| Total reflections | 159 629 | 289 690 |
| Unique reflections | 19 783 | 24 313 |
| Completeness (%)a | 95.8 (93.6) | 94.3 (83.1) |
| Rsymm (%)a | 3.0 (6.1) | 16.8 (60.7) |
| <I/S(I)>a | 36.0 (19.5) | 3.5 (1.2) |
| Refinement | ||
| R | 19.11 | 25.10 |
| Rfree | 23.33 | 30.21 |
| R.m.s. deviation | ||
| Bond length (Å) | 0.0051 | 0.0101 |
| Bond angle (deg) | 1.220 | 1.825 |
| Water molecules | 230 | 0 |
| Ramachandran plot | ||
| Core | 93.5% | 71.3% |
| Allowed regions | 4.5% | 25.5% |
| Generously allowed | 2.0% | 2.5% |
| Disallowed regions | 0.0% | 0.6% |
| Over all B-factor | 24.2% | 39.4 |
| R.m.s. deviation | ||
| Main chain bond B values | 0.98 | 1.75 |
| Main chain angle B values | 1.35 | 2.81 |
| Side chain bond B values | 2.62 | 3.93 |
| Side chain angle B values |
4.82 |
6.10 |
| aNumbers in parentheses refer to the highest resolution shell. | ||
Figure 3.
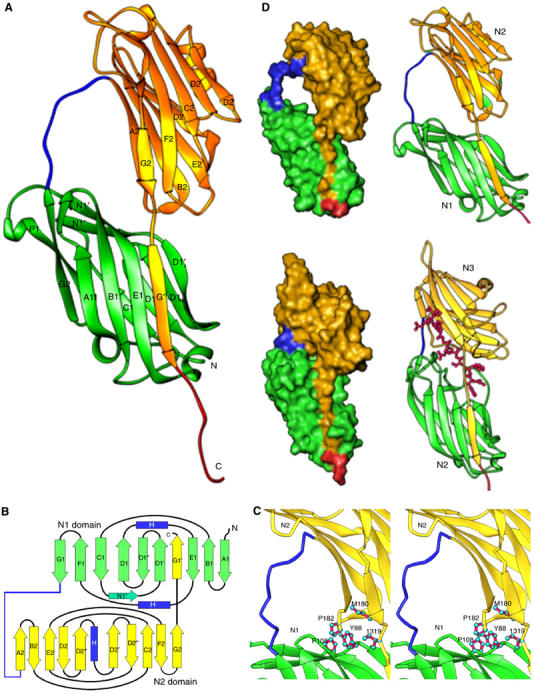
Crystal structure of the apo-CNA31–344. (A) Apo-CNA31–-344 consists two domains, N1 (Green) and N2 (Yellow). Residues 164–173 form a linker (blue in color) that joins the N1 and N2 domains. (B) Cartoon representation of the apo-CNA31–344 crystal structure. Strands are represented as arrows (green for the N1 domain and yellow for the N2 domain), and helices in blue color. The linker joining the N1 and N2 domains is represented in blue and all other loop regions in black. Images of the molecular structures were prepared using RIBBONS program (Carson, 1997). (C) The inter domain hole in apo-CNA31–344 is formed by the linker that connects N1 and N2 domains and hydrophobic residues contributed by both N1 and N2 domains. (D) Comparison of apo-CNA31–344 and SdrGN2N3 crystal structures. The crystal structure of apo-CNA31–344 (top) is compared to the ligand bound SdrGN2N3 crystal structure (bottom) to illustrate the common C-terminal extension ‘donor strand' mode and the length differences in the inter domain linker region (blue).
The N1 and N2 domains of CNA are connected by a long linker region (residues 164–173; colored blue in Figures 3A and B). This organization creates a two-domain structure with a distinct hole between the two domains (Figure 3D). The electron density for the main chain atoms of the 164–173 linker is poorly defined suggesting some flexibility in this linker segment. The N2 domain ends at residue 318 and the C-terminal extension of this domain (residues 319–329) stretches towards the N1 domain and forms a β strand (G′) that complements one of the β-sheets of the N1 domain (Figure 3A), while the following residues 332–344 are disordered and extend into the solvent region. This intra-molecular donor strand observed in CNA31–344 is analogous to the ‘latch' observed in the crystal structure of SdrGN2N3 in complex with a ligand peptide (Ponnuraj et al, 2003) (Figure 3D). In the structure of the apo-form of SdrGN2N3, the C-terminal ‘latch' points to the solvent, resulting in an open conformation that allows the ligand to dock onto its binding site. Upon ligand binding, due to hydrophobic interactions with the ligand peptide, the C-terminal extension of the SdrG N3 subdomain is redirected and latched into a trench present in the N2 subdomain, resulting in a closed conformation. However, in the apo-CNA31–344 structure, a closed conformation is observed with the analogous latch (residues 319–330) inserted into its N1 domain. The back of the latching trench on N1 contains a TYTFTDYVD motif, which is found in a number of MSCRAMMs (McCrea et al, 2000) and was predicted to contribute to the structuring of the ‘latching cleft' in SdrG (Ponnuraj et al, 2003). It is interesting to note that the inter-domain N2 C-terminal extension is only three residues (319–321) long in CNA31–344, compared to six residue length of N3 C-terminal extension in SdrGN2N3, resulting in a close contact between N1 and N2 domains of CNA. The two CNA31–344 domains interact through hydrophobic residues Tyr88 and Pro108 of N1, Pro182 and Met180 of N2, and Ile319 of the C-terminal N2 extension (Figure 3C). In this closed conformation, the combined buried surface area between the N1 and N2 domains of CNA31–344 is 1493 Å2. The inter-domain N2 C-terminal extension and the hydrophobic residues present between the two domains complete the hole created by the linker 164–173 (Figure 3D) in the apo-CNA31–344 crystal structure.
The overall structure of CNA31–344 in complex with a collagen peptide
Crystals of CNA31–344 in complex with DBS4 [(GPO)4GPRGRT(GPO)4], a synthetic triple-helical collagen peptide, belong to the C2221 space group and diffracted anisotropically to 3.2 Å resolution at best, and 3.5 Å at worst. The asymmetric unit contains two copies of complex, each one composed of two molecules of CNA31–344 bound to one collagen-like peptide. The crystal structure of this complex was determined by molecular replacement methods using the apo-CNA31–344 crystal structure as a search model. Electron density for the collagen peptide developed gradually, and the collagen peptide model was not included in phase and map calculations until the final cycles of refinement (Figure 4A). The final model was refined to an R factor of 26.5% (Rfree of 33.5%) using diffraction data to 3.3 Å resolution. The two complex molecules present in one asymmetric unit are not related by a two-fold noncrystallographic symmetry.
Figure 4.
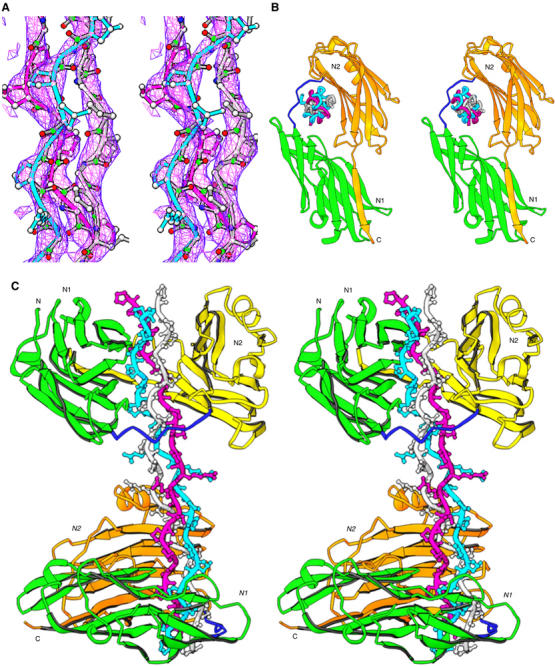
The crystal structure of (CNA31–344)2–collagen-like peptide complex. (A) The representative simulated annealing omit map for the collagen-like peptide is viewed at 1.5σ. (B) The triple helical collagen peptide is seen parked in the hole between N1 and N2 domains. The three chains of the collagen peptide are named as leading (magenta), middle (cyan) and trailing (white) as viewed from their N-termini. (C) Two CNA31–344 molecules interact with one collagen peptide in an identical manner. The leading and trailing chains peptide interact with N2 domain β sheet, and the middle chain with N1. The N1–N2 linker covers the leading and trailing chains, and holds the rope like ligand in place.
The structure of the collagen-like peptide
Figures 4B and C present a schematic representation of interactions between the CNA31–344 molecules and the collagen peptide. The three (GPO)4GPRGRT(GPO)4 strands form a right handed triple helix that is about 90 Å long. We named the three chains of the collagen peptide as leading, middle and trailing when viewed from their N-termini (Emsley et al, 2000). The conformation and structure of the collagen triple helix (Figure 4C) in this structure is similar to that observed for the collagen peptide (POG)4POA(POG)5 reported in 1994 (Bella et al, 1994). No structural water molecules are identified due to the low resolution of the present structure. However, the inter chain hydrogen bonds essential for the stabilization of the supercoiled structure are present similar to those observed in the (POG)4POA(POG)5 triple helical peptide. The N- and C-terminal tips of the collagen peptide are disordered and no electron density is seen for either of the termini. The peptide is slightly bent before and after the central RGRT sequence. The side chains of Arg and Thr residues in the middle of the collagen peptide are pointing to the solvent region and do not participate in intra-collagen interactions. Different chains have specific interactions with different parts of CNA31–344 in the (CNA31–344)2–collagen peptide complex, which we will discuss below.
The structure of (CNA31–344)2–collagen peptide complex
The crystal structure of the (CNA31–344)2–collagen peptide complex looks like a dumbbell, with two CNA31–344 molecules bound at each end of the collagen peptide (Figure 4C). The collagen triple helix is seen penetrating through the hole between the N1 and N2 domains in CNA31–344 (Figure 4B). This hole is about 12 to 15 Å in diameter. The overall diameter of the (GPO)4 region of the collagen peptide is about 11 Å, increasing to 15–19 Å around the central RGRT sequence and at termini. The GPO repeating regions of the collagen peptide precisely fit into this inter-domain hole and there is no extra space to accommodate any larger side chains (Figure 5A).
Figure 5.
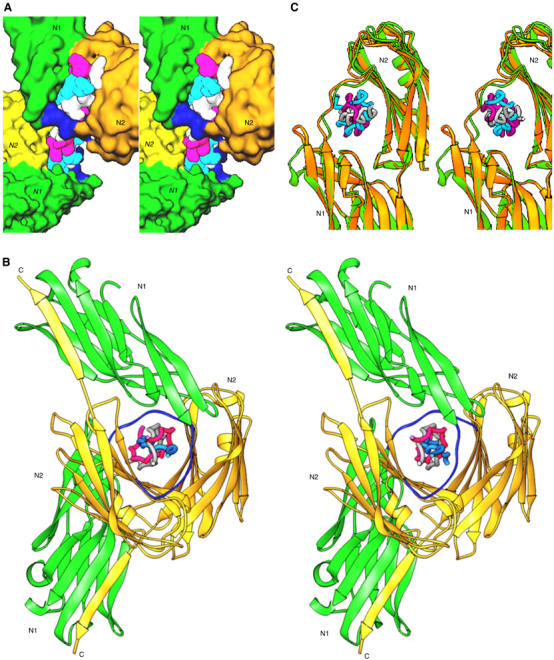
(A) A surface plot of the collagen peptide with the two CNA31–344 molecules. The N1–N2 linker regions are shown in blue. (B) Two CNA31–344 molecules in one complex molecule have a rotational difference of 120° when viewed down the collagen peptide from N-terminus. (C) The conformational switch of a loop region (residue 138–148) between apo-CNA31–344(green) and ligand bound CNA31–344 (yellow) for stabilizing the bound ligand.
In the crystal structure, a turn in the super coil of the collagen is calculated to require approximately 7.3 GXY (X and Y represent any amino acids) repeats, giving an average 49° rotational difference between the consecutive GXY repeats. However, the helical pitch between the GPO repeats varies among the various regions of the (GPO)4GPRGRT(GPO)4 peptide, suggesting a significant helical twist relaxation, which may be the result of various amino acid substitutions in the X and Y positions (Bella et al, 1994). Hence, the approximate 75 triple-helical symmetry observed for (GPO)4GPRGRT(GPO)4 is different from the ideal 107 symmetry calculated from fiber diffraction experiments of native collagen (Fraser et al, 1979; Okuyama et al, 1981). The first GPO repeat that is bound by one CNA31–344 molecule is five repeats apart from the first GPO repeat bound by the second CNA31–344 molecule. The rotational difference between the two CNA31–344 molecules is around −245° (115°; Figure 5C), close to the expected 255° value for five repeats in a collagen peptide with 75 screw symmetry. Viewing the collagen peptide from the N-terminus down its axis, the two bound CNA31–344 molecules are aligned in the same orientation and face the same direction, relative to the collagen axis, with the predicted rotational difference of ∼115°.
A closer examination reveals that the two CNA31–344 molecules bind to the (GPO)4 repeats in an identical manner. At either end of the collagen peptide, the N2 domain of each CNA31–344 molecule interacts with the leading and trailing chains, while the N1 domain interacts with the middle chain of the ligand peptide. The 164–173 inter-domain linker directly covers the leading chain of the collagen peptide (Figure 5A). The linker region is generally hydrophobic in nature, and does not seems to have specific contacts with GPO residues on the ‘leading' and ‘trailing' chains of the collagen peptide. However, the linker is essential for the confinement of collagen super coil, in the absence of which the ligand affinity drops to CNA318–344 level (Symersky et al, 1997).
Although the N1 and N2 domains of CNA31–344 are rigid and identical in all four molecules, their inter-domain association is noticeably different among the four crystallographically independent molecules (r.m.s. deviation ranging between 0.65 and 1.42 Å). If we superimpose the N2 domain of each molecule, the N1 domain shifts by about 5–10°. The r.m.s. deviation between two collagen molecules in one asymmetric unit is 0.88 Å and range between 0.52 and 0.74 Å for the corresponding individual chains. Taking the torsional variations at different segments of the collagen peptide into consideration, we can suggest that the relative orientations of N1 and N2 domain are possibly dependent on the helical twist flexibility of the binding sites in collagen. When the apo- and CNA31–344–collagen peptide complex crystal structures are compared, besides the orientation difference of the N1/N2 domain association, the ligand bound CNA31–344 has undergone an interesting conformational change in the 138–148 loop region of the N1 domain. A single turn 310 helix preceding this segment observed in the apo-CNA31–344 was transformed into a β-strand upon ligand binding (Figure 5C). This newly formed β-strand facilitates the extension of the β-strand preceding the 164–173 linker, which results in shortening and tightening of the 164–173 linker around the bound ligand. Thus, these conformational changes not only stabilize the linker around the ligand but also help in shrinking the hole, similar to a vise, around the bound ligand.
Specific interactions between CNA31–344 and the collagen-like peptide
Each CNA31–344 molecule interacts with three and half GPO repeats at each end of the collagen peptide. The interactions involved are primarily hydrophobic in nature, and also include some hydrogen bonds and van der Waals contacts. Most of the direct interactions involve residues in the concave surface (the trench) on the N2 domain of CNA31–344, which is holding the leading and trailing chains of the collagen peptide. Although the direction of the bound ligand is about 30° rotated from the previously proposed ‘trench-docking' model (Symersky et al, 1997), the collagen peptide still covers most of the trench area. All the trench residues previously implicated in collagen binding (Symersky et al, 1997) are in fact contacting the collagen peptide in this crystal structure and the interactions are predominantly between residues in the N2 domain trench region and the leading and trailing chains of collagen (Figure 6A). We identify two additional CNA hydrophobic residues, Val172 and Leu181, that may be important for locking the ligand in place. Thus, the hydrophobic residues Tyr175 and Phe191 present in the N2 domain trench region are stacked against the hydrophobic Pro11L and Pro8L of the leading chain. Similarly, Tyr233 of the trench region is stacked against Pro5T of the trailing chain while the polar residues Asn193, Asn223 and Asn278 in the trench region are hydrogen bonded to trailing chain Hyp6T and Hyp9T residues. The N1 domain of CNA31–344 displays limited interactions with the collagen peptide. However, the Asp50 and Arg136 residues of the N1 domain do interact with Hyp6M of the middle chain and Hyp9T of the trailing chain, respectively (Figure 6B). The164–173 linker seems to be playing an important role in holding the ligand in place by interacting with proline residues in the leading and trailing chains (Figures 6A and B). Significantly, the Val172 residue from the 164–173 linker and Tyr175 from N2 domain are seen sandwiching the same proline Pro11L from the leading chain. The importance of this interaction can be gauged from the observation that the electron density was missing for the Val172 side chain in the apo-CNA31–344, crystal structure, suggesting a prominent role for Val172 in the conformational changes leading to sequestration of the triple helical collagen.
Figure 6.
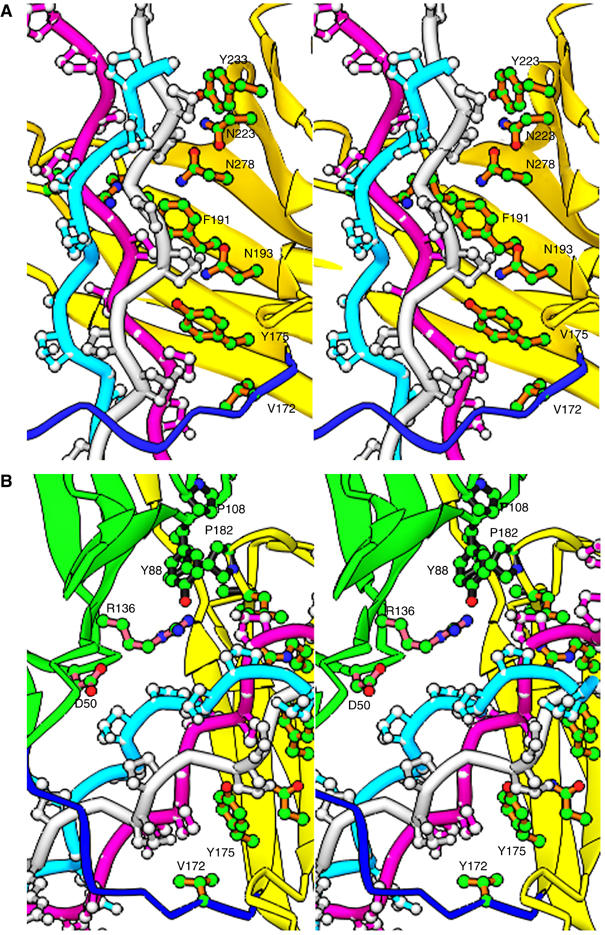
Specific interactions between CNA31–344 and the collagen peptide ligand. (A) The triple helical collagen peptide mainly interacts with residues in the trench of the N2 domain (yellow). Stacking hydrophobic interactions between the proline residues of the ligand and the bulky hydrophobic Tyr and Phe residues of the N2 domain are observed. Val172 from the N1–N2 linker (blue) also participates in such hydrophobic interactions. (B) The minimal ligand interactions contributed by the N1 domain (green) are presented. The hydrophobic inter-domain ‘locking' arrangement can be seen between residues Pro108 and Tyr88 of N1 domain, and Pro182 and Met180 from N2 domain, and the Ile319 from the C-terminal extension.
Most importantly, the interactions between proline residues in the collagen peptide and residues in the trench region on N2 are conserved. For example, theTyr175 and Tyr233 residues in one CNA31–344 molecule of the ligand complex and the Tyr175 and Tyr233 residues in the second CNA31–344 molecule exhibit identical interactions. The specific prolines targeted in collagen peptide are 15 residues apart along their corresponding helical chains. Thus, the leading and trailing chains of the collagen peptide interact with the two CNA31–344 molecules separated by five GXY repeats. The CNA31–344 molecule appears to bind and recognize the collagen peptide only in one direction. If we insert the collagen peptide in the opposite direction, it is obvious that most of the hydrogen bond and hydrophobic interactions observed between the protein and the ligand in the (CNA31–344)2-collagen peptide complex would be lost.
The Collagen Hug—a dynamic binding model for the CNA–collagen interaction
Figure 7 presents a cartoon form of the hypothetical multistep ‘Collagen Hug' model of CNA binding to its rope-like ligand. We propose that the apo-form of CNA exists in equilibrium between an open and closed conformation. Only the open conformation can bind collagen. The binding is initiated by a low-affinity interaction between the collagen triple helix and the complementary shallow trench on the N2 domain and stabilized by polar and hydrophobic interactions (Symersky et al, 1997) (Figure 7A). The N1 domain is not involved at this stage. In the next step, the Val172 residue from the interdomain linker interacts with a proximal proline residue from the leading chain and facilitates the repositioning of the N1 domain. The repositioned N1 domain is stabilized by the interactions between the critical hydrophobic residues Tyr88 and Pro108 of the N1 domain, and Pro182 and Met180 of the N2 domain (Figure 3C). This reorientation and stabilization of the N1 domain is the significant step in CNA wrapping around the bound collagen (Figure 7B). Now the crucial ‘locking' of ligand takes place in two steps. We suggest that after ligand binding, the C-terminal extension of N2 reorients itself due to the insertion of Ile319 present at the end of the G2 strand of N2 into the recently formed hydrophobic cluster of Tyr88, Pro108, Pro182 and Met180 residues present between the N1 and the N2 domains. This helps to ‘prime' the N2 domain extension latch for the second step; the insertion of the 322–330 segment into the conserved latching trench (Ponnuraj et al, 2003) on the N1 domain (Figure 7C). The collagen peptide ligand is further secured by conformational changes observed in the ‘wrapped' around linker region, by which the inter-domain ‘hole' shrinks to snuggly fit around the collagen (Figure 5F). As a result, the collagen triple helix is seen seamlessly enclosed in the ‘hole' created by the two domains and the linker joining them, and held tightly in place by the surrounding steric hindrance.
Figure 7.
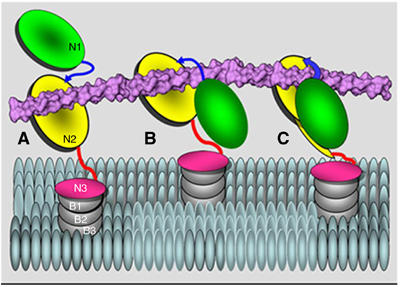
A hypothetical ‘Collagen Hug' model shown in a cartoon representation. (A) Collagen triple helix is initially associated with the N2 domain. (B) The collagen is then wrapped by the N1–N2 linker and the N1 domain. (C) The N1 domain interacts with the N2 domain via multiple hydrophobic interactions and finally the C-terminal latch is introduced in the N1 domain to secure the ligand in place.
Based on the observed binding of CNA31–344 to GPO repeats in the crystal structure, we suggest that CNA preferentially binds to GPO repeats. The observed higher affinity of CNA31–344 for (GPO)4 segments in DBS4 compared to the same sites in (GPO)11 can be due to a possible increased stability (Persikov et al, 2005) and torsional flexibility of the DBS4 collagen-like peptide, due to similar imino acid substitutions in all its three chain central parts (Persikov et al, 2000), as evident by its 75 symmetry in the crystal structure. Variations in helix twist may serve as a recognition feature or facilitate triple helical orientation suitable for binding (Brodsky and Persikov, 2005). Specially, the substituted Arg at X and Y positions imparts higher stability to the triple helical peptide through its dual, polar and hydrophobic side chain (Kramer et al, 2001). In addition, the Thr side-chain at Y position could participate in water mediated inter-chain hydrogen bonds as observed for Hyp side chains (Kramer et al, 2001). The higher binding affinities of (GPP)11 compared to (GPO)11 could be due to the dominance of hydrophobic interactions between the proline rings and the hydrophobic residues of the ‘collagen binding trench', compared to minimal polar interactions between the peptide and the protein. In summary, a synergic combination of helical twist relaxation, conformational flexibility due to Arg and Thr substitutions at X and Y positions of G-X-Y repeats, and the hydrophobic and hydrophilic interactions of Pro and Hyp residues can explain why CNA31–344 binds with different affinities to the collagen-like peptides DBS4>(GPP)11>(GPO)11.
We believe that the Collagen Hug model is the only model consistent with the structures we have determined for CNA31–344 as an apo-protein and in complex with a collagen peptide. This model requires that the closed conformation of CNA31–344, seen in the crystals of the apo-protein, in solution, is in equilibrium with a postulated open conformation. Only an open conformation is capable of binding collagen. In light of the suggested flexible orientations of the N1 domain, perhaps a closed conformation is favored by the conditions used to crystallize CNA31–344 or perhaps the closed form preferentially crystallizes. However, the ‘Collagen Hug' remains a hypothetical binding model and we are currently testing experimentally the different steps predicted by this model.
Concluding remarks
The Collagen Hug binding mechanism proposed in this communication is distinct from the metal ion-dependent collagen-binding mechanism employed by the integrin I domains. In addition to confirming that the collagen triple-helix itself is a major recognition element of CNA (Speziale et al, 1996), the present model also reveals that the binding site is versatile. The unique features of the Collagen Hug include: (A) Collagen binding is initiated primarily through hydrophobic interactions between residues in a shallow trench on CNA31–344 and the GPO repeats of the ligand; (B) A long linker that connects the N1 and N2 domains of CNA and allows the two domains to wrap around the triple helical ligand; (C) the ability of CNA to shrink its inter-domain hole to fit snuggly around the bound collagen; and (D) the insertion of the latch, representing the extension of the N2 subdomain, into a trench formed on the surface of the N1 domain, and thus securing the CAN–collagen complex. The later step in the Collagen Hug mechanism proposed for CNA is similar to that of the Dock, Lock and Latch mechanism used by MSCRAMMs binding linear peptides. The latching event that involves an interdomain β-strand complementation is observed in the ligand binding processes of both MSCRAMM types, although different subdomains are participating. This similarity, combined with similarities in the overall structural organization and amino acid sequence, suggests that the two MSCRAMM types have evolved from a common ancestor.
The Collagen Hug and the Dock, Lock and Latch binding mechanisms result in a secure and tightly held ligand. However, the mode and steps involved to reach such a secure state by the various MSCRAMMs are different. In the Dock, Lock and Latch mechanism, the ligand peptide docks into a well-defined binding pocket formed between the N2 and N3 domains of the interacting MSCRAMM. Only peptides with specific primary sequences can fit into the binding pocket. In the Collagen Hug model, the shallow trench on N2 positions the rope-like ligand to be embraced by the inter-domain linker and N1 domain. Although there are specific interactions between residues in the collagen and the MSCRAMM, the rope-like triple helical shape of the collagen is the structural determinant for binding. In fact, CNA dose not bind to gelatin or denatured collagen. It is fascinating to note the structural evolution of MSCRAMMs and how similar building blocks (i.e. IgG-like domains) are being used to accommodate structurally different ligands.
The Collagen Hug model predicts that CNA can only bind to monomeric forms of collagen. In the tissue, most of the collagen triple helix molecules assemble into fibrillar or fiber structures. CNA would not mediate staphylococcal adherence to these higher ordered structures but only allow bacteria to bind to triple helix monomers that are naturally occurring (Liotta et al, 1980; Kehrel, 1995) or generated through tissue injury.
CNA is a member of a family of collagen binding MSCRAMMs present on Gram -positive bacteria. A comparison of the amino-acid sequences of the A-regions of the proteins in this family indicate that CNA from S. aureus, ACE from E. faecalis (Rich et al, 1999c), ACM from E. faecium (Nallapareddy et al, 2003), CNE from S. equi (Lannergard et al, 2003), and CMN from S. mutans (Sato et al, 2004) are closely related. These adhesins all contain a region resembling the now crystallized N1N2 domain of CNA that exhibit a high degree of amino acid sequence similarities (data not shown). We therefore predict that they have a similar N1N2 structure and that these two domains cooperate to bind collagen using the ‘Collagen Hug' mechanism described herein.
Although many proteins are shown to bind collagen with a high affinity, the detailed mechanisms involved are often poorly understood. In both the previously reported integrin (Emsley et al, 2000) and the MSCRAMM, a shallow trench on the binding protein can accommodate the collagen triple helix. For the integrin, a divalent metal-ion coordinates residues in both the I domain and the collagen molecule; metal-ion coordination is required for a high affinity interaction. However, in addition to the bacterial collagen adhesins described above, many mammalian collagen binding proteins are not known to require metal ions for ligand binding, but appear to require multiple subdomains for high affinity binding (Clemetson and Clemetson, 2001). In the MSCRAMMs, we show a collagen-binding mechanism that is initiated mainly through hydrophobic interactions between one subdomain and the ligand. High affinity binding of collagen by the MSCRAMM requires a structural reorganization involving several subdomains in the binding protein resulting in this embracing the collagen triple helix structure. A recent modeling study with human proMMP-1 suggested that processing of the pro-domain caused structural changes that resulted in a conformation in which collagen could bind at the interface formed between the catalytic domain and the hemopexin domain of MMP-1 (Jozic et al, 2004, 2005). It is tempting to speculate that some aspects of the Collagen Hug mechanism proposed here may be shared by several eukaryotic protein–collagen interaction systems.
Materials and methods
Protein purification, crystallization and data collection
DNA fragments encoding different regions of the CNA A region were PCR amplified from a CNA31–531 construct generated previously (Rich et al, 1999a), using primers listed in Supplementary Table 1. The PCR products were purified, digested with BamHI and SalI, and ligated into the pQE30 expression vector as described previously (Xu et al, 2000). Each construct was confirmed by DNA sequencing. Expression and large-scale purification of the recombinant fragments were as described previously (Visai et al, 2000). The masses of the purified proteins were verified by mass spectrometry.
The purified CNA31–344 protein was concentrated to 40 mg/ml before crystallization trials. Single crystals were obtained using 3.2 M ammonium sulfate, with 0.1 M MES buffer, pH 6.5 and trace amounts of PEG1000. The CNA31–344 crystals diffracted to 1.95 Å and diffraction data were collected at 100 K using an in house X-ray source.
The synthesized collagen peptide, DBS4, was dissolved and incubated with CNA31–344 overnight at 10:1 ratio. The CNA31–344–collagen complex was then purified using gel filtration chromatography and concentrated to 5 mg/ml. The crystals of the CNA31–344–collagen complex were grown at 22°C and we obtained two crystal forms at two different crystallization conditions. Hexagonal crystals, grown using 1.6 M ammonium sulfate, and 50 mM HEPES buffer, at pH 7.4, were of poor diffraction quality, and the crystals obtained using 0.8 M ammonium sulfate, 0.5 M sodium acetate, pH 6.7, belonged to orthorhombic system and diffracted to 3.5 Å resolution. The diffraction data were collected at 100 K using APS SERCAT 22ID beamline, and processed using HKL2000 (Otwinowski, 1997).
Phase determination, model building and refinement
The crystal structure of CNA31–344 was solved by molecular replacement using the AmoRe program (Navaza, 2001) and the crystal structure of CNA151–318 (1AMX) (Symersky et al, 1997) as a search model. The crystal structure was refined to an R factor of 19.1% (Rfree of 23.3%) to 1.95 Å resolution with the help of the CNS program (Brunger et al, 1998).
The initial molecular replacement solution for the CNA31–344–collagen complexes was obtained with the help of the CNS program while using the apo-CNA31–344 crystal structure as a search model. Only positions for two out of four CNA31–344 molecules in the asymmetric unit were obtained initially, and the remaining two CNA31–344 molecules were positioned manually into the electron density domain by domain as the phases improved during refinement. The CNA-collagen structure was first refined by rigid-body refinement and simulated annealing function, then it was refined by Refmac5 of CCP4 package with NCS restrains (i.e. the four N-terminal domains and the four C-terminal domains of CNA were applied by a local NCS restrains, respectively). The Refmac5 refined structure was further refined by a grouped-B-factor function and finally two cycles of energy minimization function of CNS.
Many rounds of positional refinement and model building with the program O (Jones et al, 1991) were performed gradually in steps of increasing resolution. All through these steps the electron density for the two collagen peptides in the asymmetric unit improved gradually. During the process of manual adjustment and refinement of individual domains and atoms of the four CNA31–344 molecules, the collagen peptide coordinates were not included in the phase calculations until the final rounds. The final R factor was reduced to 23.3% and Rfree 30.2%, using diffraction to 3.3 Å resolution.
Collagen peptide synthesis and characterization
Peptides were synthesized by a solid phase method on a TentaGel R RAM resin (RAPP Polymere GmbH, Tubingen, Germany) using Fmoc chemistry and a model 396 MBS Multiple Peptide Synthesizer from Advanced ChemTech Inc. (Louisville, KY) as described previously (Xu et al, 2000). Synthesized peptides were purified by reverse phase high pressure liquid chromatography as described (Xu et al, 2000).
Synthetic collagen peptides were analyzed by circular dicroism (CD) spectroscopy on a Jasco J720 spectropolarimeter as described previously (Xu et al, 2000).
SPR spectroscopy
SPR analysis was performed on a Biacore 3000 system (Biacore) as previously described (Xu et al, 2000). For analysis of the binding of recombinant CNA fragments to collagen, bovine type I collagen (Vitrogen) was immobilized onto the cells in a CM5 sensor chip. Various concentrations of CNA fragments in HBS buffer (10 mM HEPEs, 150 mM NaCl, 3.4 mM EDTA, pH 7.4) were run over the collagen surface. Response from a blank cell was subtracted. For analysis of the binding of collagen peptide to CNA31–344, recombinant CNA31–344 was immobilized onto the cells of a CM5 chip and various concentrations of peptides in HBS buffer were run over the surface. BiaEvaluation software (Biacore) as well as GraphPad Prism 4.0 (GraphPad) was used to analyze the data as described (Xu et al, 2000).
Solid-phase binding assays
These assays were performed as described previously (Xu et al, 2000, #200) with slight modifications. For direct binding of recombinant CNA fragments to collagen, bovine type I collagen was coated onto the wells of a 96-well microtiter plate at the concentration of 1 μg/well. Bound MSCRAMM proteins were detected with mouse anti-His mAb, followed by goat-anti-mouse IgG-alkaline phosphotase conjugant (BioRad). For competition assays, a fixed concentration of biotin-labeled CNA fragment (100 nM CNA31–344, 1 μM CNA31–531, and 20 μM CNA151–318, respectively) was mixed with increasing concentrations of the indicated unlabeled CNA fragments, and then incubated in wells coated with bovine type I collagen. Streptavidin-AP conjugant was used for the detection of bound labeled proteins. For peptide inhibition assays, CNA31–344 at 10 nM was mixed with increasing concentrations of the indicated synthetic collagen peptide, and added to wells coated with bovine type I collagen. Bound CNA31–344 was detected by anti-His mAb.
Supplementary Material
Supplementary Figure
Supplementary Materials
Acknowledgments
This study was supported by NIH Grants AR44415 and AI20624 to M Höök, AI061555 to Y Xu and NASA Cooperative agreement NCC8-246 and NIH Grant AI064815 to SVLN.
References
- Bella J, Eaton M, Brodsky B, Berman HM (1994) Crystal and molecular structure of a collagen-like peptide at 1.9 Å resolution [see comment]. Science 266: 75–81 [DOI] [PubMed] [Google Scholar]
- Brodsky B, Persikov AV (2005) Molecular structure of the collagen triple helix. Adv Protein Chem 70: 301–339 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren G (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54 (Part 5): 905–921 [DOI] [PubMed] [Google Scholar]
- Carson M (1997) Ribbons. Methods Enzymol 277: 493–505 [PubMed] [Google Scholar]
- Clemetson KJ, Clemetson JM (2001) Platelet collagen receptors. Thromb Haemost 86: 189–197 [PubMed] [Google Scholar]
- Deivanayagam CC, Wann ER, Chen W, Carson M, Rajashankar KR, Hook M, Narayana SV (2002) A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. EMBO J 21: 6660–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elasri MO, Thomas JR, Skinner RA, Blevins JS, Beenken KE, Nelson CL, Smelter MS (2002) Staphylococcus aureus collagen adhesin contributes to the pathogenesis of osteomyelitis. Bone 30: 275–280 [DOI] [PubMed] [Google Scholar]
- Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC (2000) Structural basis of collagen recognition by integrin alpha2beta1. Cell 101: 47–56 [DOI] [PubMed] [Google Scholar]
- Fraser RD, MacRae TP, Suzuki E (1979) Chain conformation in the collagen molecule. J Mol Biol 129: 463–481 [DOI] [PubMed] [Google Scholar]
- Hienz SA, Schennings T, Heimdahl A, Flock JI (1996) Collagen binding of Staphylococcus aureus is a virulence factor in experimental endocarditis. J Infect Dis 174: 83–88 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallograph A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Jozic D, Bourenkov G, Lim NH, Visse R, Nagase H, Bode W, Maskos K (2004) X-ray structure of human proMMP-1: new insights into procollagenase activation and collagen binding. J Biol Chem 280: 9578–9585 [DOI] [PubMed] [Google Scholar]
- Jozic D, Bourenkov G, Lim NH, Visse R, Nagase H, Bode W, Maskos K (2005) X-ray structure of human proMMP-1: new insights into procollagenase activation and collagen binding. J BiolChem 280: 9578–9585 [DOI] [PubMed] [Google Scholar]
- Kehrel B (1995) Platelet–collagen interactions. Sem Thromb Hemost 21: 123–129 [DOI] [PubMed] [Google Scholar]
- Kramer RZ, Bella J, Brodsky B, Berman HM (2001) The crystal and molecular structure of a collagen-like peptide with a biologically relevant sequence. J Mol Biol 311: 131–147 [DOI] [PubMed] [Google Scholar]
- Lannergard J, Frykberg L, Guss B (2003) CNE, a collagen-binding protein of Streptococcus equi. FEMS Microbiol Lett 222: 69–74 [DOI] [PubMed] [Google Scholar]
- Liotta LA, Tryggvason K, Garbisa S, Hart I, Foltz CM, Shafie S (1980) Metastatic potential correlates with enzymatic degradation of basement membrane collagen. Nature 284: 67–68 [DOI] [PubMed] [Google Scholar]
- Mamo W, Froman G, Muller HP (2000) Protection induced in mice vaccinated with recombinant collagen-binding protein (CnBP) and alpha-toxoid against intramammary infection with Staphylococcus aureus. Microbiol Immunol 44: 381–384 [DOI] [PubMed] [Google Scholar]
- McCrea KW, Hartford O, Davis S, Eidhin DN, Lina G, Speziale P, Foster TJ, Hook M (2000) The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology 146 (Part 7): 1535–1546 [DOI] [PubMed] [Google Scholar]
- Nallapareddy SR, Weinstock GM, Murray BE (2003) Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol Microbiol 47: 1733–1747 [DOI] [PubMed] [Google Scholar]
- Navaza J (2001) Implementation of molecular replacement in AMoRe. Acta Crystallogr D 57: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Nilsson IM, Patti JM, Bremell T, Hook M, Tarkowski A (1998) Vaccination with a recombinant fragment of collagen adhesin provides protection against Staphylococcus aureus-mediated septic death. J Clin Invest 101: 2640–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama K, Okuyama K, Arnott S, Takayanagi M, Kakudo M (1981) Crystal and molecular structure of a collagen-like polypeptide (Pro-Pro-Gly)10. J Mol Biol 152: 427–443 [DOI] [PubMed] [Google Scholar]
- Otwinowski ZaMW (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol: Macromol Crystallogr A 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Patti JM, Allen BL, McGavin MJ, Hook M (1994) MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol 48: 585–617 [DOI] [PubMed] [Google Scholar]
- Patti JM, Boles JO, Hook M (1993) Identification and biochemical characterization of the ligand binding domain of the collagen adhesin from Staphylococcus aureus. Biochemistry 32: 11428–11435 [DOI] [PubMed] [Google Scholar]
- Perkins S, Walsh EJ, Deivanayagam CC, Narayana SV, Foster TJ, Hook M (2001) Structural organization of the fibrinogen-binding region of the clumping factor B MSCRAMM of Staphylococcus aureus. J Biol Chem 276: 44721–44728 [DOI] [PubMed] [Google Scholar]
- Persikov AV, Ramshaw JA, Brodsky B (2005) Prediction of collagen stability from amino acid sequence. J Biol Chem 280: 19343–19349 [DOI] [PubMed] [Google Scholar]
- Persikov AV, Ramshaw JA, Kirkpatrick A, Brodsky B (2000) Amino acid propensities for the collagen triple-helix. Biochemistry 39: 14960–14967 [DOI] [PubMed] [Google Scholar]
- Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, Xu Y, Hook M, Narayana SV (2003) A ‘dock, lock, and latch' structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115: 217–228 [DOI] [PubMed] [Google Scholar]
- Rhem MN, Lech EM, Patti JM, McDevitt D, Hook M, Jones DB, Wilhelmus KR (2000) The collagen-binding adhesin is a virulence factor in Staphylococcus aureus keratitis. Infect Immun 68: 3776–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich RL, Deivanayagam CC, Owens RT, Carson M, Hook A, Moore D, Symersky J, Yang VW, Narayana SV, Hook M (1999a) Trench-shaped binding sites promote multiple classes of interactions between collagen and the adherence receptors, alpha(1)beta(1) integrin and Staphylococcus aureus cna MSCRAMM. J Biol Chem 274: 24906–24913 [DOI] [PubMed] [Google Scholar]
- Rich RL, Deivanayagam CC, Owens RT, Carson M, Hook A, Moore D, Symersky J, Yang VW, Narayana SV, Hook M (1999b) Trench-shaped binding sites promote multiple classes of interactions between collagen and the adherence receptors, alpha(1)beta(1) integrin and Staphylococcus aureus cna MSCRAMM. [erratum appears in J Biol Chem 1999 Oct 1;274(40):28836]. J Biol Chem 274: 24906–24913 [DOI] [PubMed] [Google Scholar]
- Rich RL, Kreikemeyer B, Owens RT, LaBrenz S, Narayana SV, Weinstock GM, Murray BE, Hook M (1999c) Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J Biol Chem 274: 26939–26945 [DOI] [PubMed] [Google Scholar]
- Sato Y, Okamoto K, Kagami A, Yamamoto Y, Igarashi T, Kizaki H (2004) Streptococcus mutans strains harboring collagen-binding adhesin. J Dent Res 83: 534–539 [DOI] [PubMed] [Google Scholar]
- Shimoji Y, Ogawa Y, Osaki M, Kabeya H, Maruyama S, Mikami T, Sekizaki T (2003) Adhesive surface proteins of Erysipelothrix rhusiopathiae bind to polystyrene, fibronectin, and type I and IV collagens. J Bacteriol 185: 2739–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speziale P, Joh D, Visai L, Bozzini S, House-Pompeo K, Lindberg M, Hook M (1996) A monoclonal antibody enhances ligand binding of fibronectin MSCRAMM (adhesin) from Streptococcus dysgalactiae. J Biol Chem 271: 1371–1378 [DOI] [PubMed] [Google Scholar]
- Symersky J, Patti JM, Carson M, House-Pompeo K, Teale M, Moore D, Jin L, Schneider A, DeLucas LJ, Hook M, Narayana SV (1997) Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat Struct Biol 4: 833–838 [DOI] [PubMed] [Google Scholar]
- Visai L, Xu Y, Casolini F, Rindi S, Hook M, Speziale P (2000) Monoclonal antibodies to CNA, a collagen-binding microbial surface component recognizing adhesive matrix molecules, detach Staphylococcus aureus from a collagen substrate. J Biol Chem 275: 39837–39845 [DOI] [PubMed] [Google Scholar]
- Xu Y, Gurusiddappa S, Rich RL, Owens RT, Keene DR, Mayne R, Hook A, Hook M (2000) Multiple binding sites in collagen type I for the integrins alpha1beta1 and alpha2beta1. J Biol Chem 275: 38981–38989 [DOI] [PubMed] [Google Scholar]
- Xu Y, Liang X, Chen Y, Koehler TM, Hook M (2004) Identification and biochemical characterization of two novel collagen binding MSCRAMMs of Bacillus anthracis. J Biol Chem 279: 51760–51768 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure
Supplementary Materials


