Abstract
Eukaryotic initiation factor eIF1 and the functional C-terminal domain of prokaryotic initiation factor IF3 maintain the fidelity of initiation codon selection in eukaryotes and prokaryotes, respectively, and bind to the same regions of small ribosomal subunits, between the platform and initiator tRNA. Here we report that these nonhomologous factors can bind to the same regions of heterologous subunits and perform their functions in heterologous systems in a reciprocal manner, discriminating against the formation of initiation complexes containing codon–anticodon mismatches. We also show that like IF3, eIF1 can influence initiator tRNA selection, which occurs at the stage of ribosomal subunit joining after eIF5-induced hydrolysis of eIF2-bound GTP. The mechanisms of initiation codon and initiator tRNA selection in prokaryotes and eukaryotes are therefore unexpectedly conserved and likely involve related conformational changes induced in the small ribosomal subunit by factor binding. YciH, a prokaryotic eIF1 homologue, could perform some of IF3's functions, which justifies the possibility that YciH and eIF1 might have a common evolutionary origin as initiation factors, and that IF3 functionally replaced YciH in prokaryotes.
Keywords: eIF1, IF3, ribosome, translation, YciH
Introduction
The fidelity of translation depends on accurate selection of the correct reading frame during initiation. In eukaryotes, this process involves at least 11 eukaryotic initiation factors (eIFs). Met-tRNAiMet forms a ternary complex with eIF2 and GTP, which together with eIF1, eIF1A and eIF3 binds to the 40S ribosomal subunit to form a 43S preinitiation complex. After loading onto the mRNA's 5′ end in a process requiring eIFs 4A, 4B and 4F, the 43S complex scans downstream until it encounters an AUG triplet in a favorable context GCC(A/G)CCAUGG (in which the nucleotides at the −3 and +4 positions are the most important; Kozak, 1991), stops and forms a stable 48S complex with established codon–anticodon base pairing in the P site. These two context nucleotides are important features of mammalian mRNAs but differ in sequence and importance in other eukaryotes. Subsequent joining of a 60S subunit is mediated by eIF5 (which induces hydrolysis of eIF2-bound GTP and dissociation of eIF2-GDP from the 40S subunit) and eIF5B (which mediates subunit joining and dissociation of other factors) (Pestova et al, 2000; Unbehaun et al, 2004).
eIF1 plays the key role in initiation codon selection. In an in vitro reconstituted system, eIF1 enables scanning 43S complexes to discriminate against non-AUG triplets and AUG triplets that have poor context or are located within 4 nt. of the 5′ end of an mRNA, and also promotes dissociation of ribosomal complexes aberrantly assembled at such triplets in its absence (Pestova et al, 1998; Pestova and Kolupaeva, 2002). Mutations in eIF1 permit initiation at noncognate initiation codons in vivo in yeast (e.g. Yoon and Donahue, 1992). eIF1 also prevents premature eIF5-induced hydrolysis of eIF2-bound GTP before codon–anticodon base pairing has been established (Unbehaun et al, 2004).
By contrast, initiation in prokaryotes requires only three initiation factors (IFs), does not involve scanning, and the 30S ribosomal subunit itself plays a direct role in initiation codon selection, by binding to the Shine–Dalgarno sequence upstream of the initiation codon (Laursen et al, 2005). IF3 increases the accuracy of initiation codon selection by promoting dissociation of pseudo-initiation complexes assembled either on noninitiation codons, or with noninitiator tRNA, particularly discriminating against mutations in three conserved G-C pairs in its anticodon stem, and also modulates the translation efficiency of leaderless mRNAs (Hartz et al, 1990; Meinnel et al, 1999; Dallas and Noller, 2001; Petrelli et al, 2001). IF3's proofreading function requires only its C-terminal domain (IF3-CTD) (Petrelli et al, 2001).
IF3 thus displays several activities that resemble those of eIF1 in ensuring the fidelity of initiation. Moreover, both eIF1 and IF3-CTD bind to the same region of the small ribosomal subunit, between the platform and initiator tRNA (Dallas and Noller, 2001; Lomakin et al, 2003). This location corresponds to a region of high homology between 16S and 18S rRNA. These observations suggest that eIF1 and IF3 may influence initiation codon selection by similar mechanisms. Ribosome-bound eIF1 and IF3-CTD are both out of reach of the initiation codon and the conserved G-C base pairs in the anticodon stem of initiator tRNA, so that any role for these factors in promoting the fidelity of initiation must be indirect. Binding of IF3 induces changes in the conformation of the 30S subunit and in its interaction with mRNA (Shapkina et al, 2000; Petrelli et al, 2001). It has been proposed that IF3-induced tilting of the 30S subunit head toward the platform and juxtaposition of GA1338–9 of 16S rRNA with the anticodon stem of initiator tRNA could constitute a means for checking the identity of tRNA in the P site (Dallas and Noller, 2001).
Despite similarities in function, size, shape and charge distribution (Lomakin et al, 2003), there is no sequence or structural homology between eIF1 and IF3-CTD. This is surprising in light of the homology between other prokaryotic and eukaryotic initiation factors, and between the regions of the small ribosomal subunit to which they both bind. However, all archaea and some prokaryotes encode homologues of eIF1 (Kyrpides and Woese, 1998), which in enteric bacteria is known as YciH (Cort et al, 1999). YciH is nonessential and its function is unknown.
These similarities between eIF1 and IF3 prompted us to test if they could perform some functions in heterologous systems in a reciprocal manner. We now report that eIF1 and IF3 bind to identical regions on homologous and heterologous ribosomal subunits, and can discriminate against the formation of initiation complexes with codon–anticodon mismatches in heterologous systems. Moreover, like IF3, eIF1 can influence initiator tRNA selection, but this activity is manifested only after hydrolysis of eIF2-bound GTP and dissociation of eIF2-GDP from initiator tRNA in the P site. Related conformational changes induced in the small ribosomal subunit may therefore be responsible for the selection of the initiation codon and initiator tRNA by eIF1 and IF3. The mechanisms of initiation codon and initiator tRNA selection in prokaryotes and eukaryotes are therefore unexpectedly conserved. YciH was able to perform some of the functions of IF3 in prokaryotic initiation, an observation that justifies the possibility that YciH and eIF1 have a common evolutionary origin as initiation factors, and that IF3 has functionally replaced YciH in prokaryotes.
Results
Binding of eIF1, IF3 and YciH to 40S and 30S ribosomal subunits
Although eIF1 and IF3-CTD have unrelated sequences and structures, their functions in ensuring the fidelity of initiation codon selection and positions on the small ribosomal subunit are similar. The homology of their respective binding sites on eukaryotic 40S and prokaryotic 30S subunits (Spahn et al, 2001) prompted us to test their binding to heterologous subunits. YciH was included in these studies. In pull-down experiments, T7-tagged IF3 and IF3-CTD bound 40S subunits as well as eIF1, whereas binding of YciH was lower (Figure 1A). Untagged eIF1 competed with YciH but not with IF3 for binding to 40S subunits (Figure 1B), indicating that YciH and eIF1 bind to the same or overlapping sites. Our failure to detect eIF1 in lanes 3 and 4 suggested that rather than binding to different sites on 40S subunits, eIF1 and IF3 bind to the same site but IF3's affinity is higher. To confirm this, we investigated competition between eIF1 and IF3 for binding to 40S subunits using sucrose density gradient centrifugation. Although we previously noted that this method is not very suitable for studying the eIF1/40S subunit interaction because these complexes are unstable under centrifugation conditions (Lomakin et al, 2003), loading of large amounts of eIF1/40S subunit complex on sucrose gradients permitted us to detect eIF1 and IF3 in ribosomal complexes. Incubation of preformed eIF1/40S subunit complexes with IF3 reduced binding of eIF1 to 40S subunits approximately four-fold (Figure 1C), confirming that eIF1 and IF3 bind to the same or overlapping sites. In pull-down experiments, eIF1 and YciH also bound to 30S subunits (Figure 1D), and again, the ribosome binding activity of YciH was lower.
Figure 1.
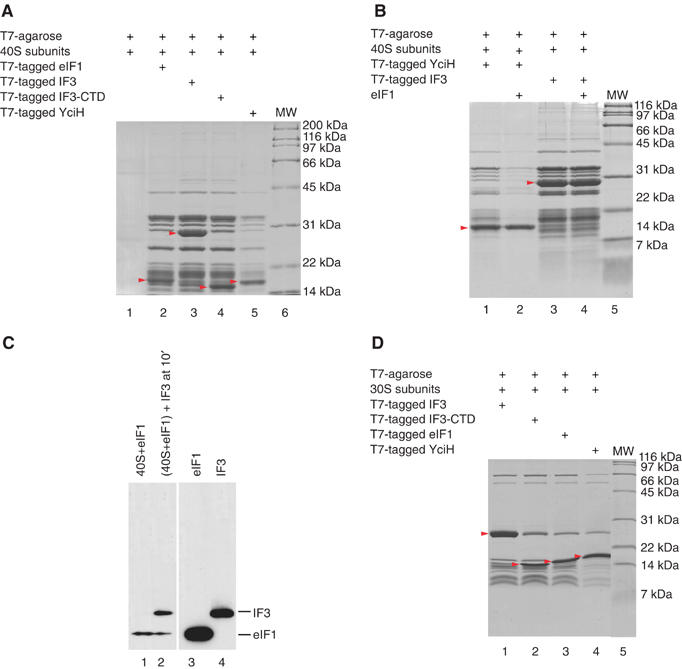
Binding of IF3, YciH and eIF1 to (A–C) 40S subunits and (D) 30S subunits. (A, B, D) Interaction of ribosomal subunits with T7-tag antibody agarose-immobilized eIF1, IF3, IF3-CTD or YciH (as indicated), and (B) in the presence of recombinant untagged eIF1 (as indicated) in in vitro binding assays. Ribosomal proteins and initiation factors were stained with Coomassie blue. Initiation factors are indicated by red arrows. (C) Presence of T7-tagged eIF1 and IF3 in ribosomal complexes isolated from sucrose density gradients (lanes 1 and 2), and eIF1 and IF3 markers (lanes 3 and 4) visualized by Western blotting.
The eIF1-binding site on the 30S subunit was determined by specific cleavage of 16S rRNA by hydroxyl radicals generated at Fe(II) site, specifically tethered to unique cysteine residues on the surface of 30S-bound eIF1, after treatment with ascorbic acid and H2O2. Hydroxyl radicals have a small radius of action (∼20 Å), which allows precise localization of eIF1. To determine its orientation on 30S subunits, we used four eIF1 mutants with single surface-exposed cysteine residues (Figure 2A) previously used to locate eIF1 on 40S subunits (Lomakin et al, 2003). Cleavage of 16S rRNA at nt. 694–696 (helix 23b) from Cys75, at nt. 1400 (helix 44) from Cys38 and Cys42, and at nt. 784–789 (helix 24a) from Cys38, Cys42, Cys61 and Cys75 (Figures 2B–H) was identical to cleavage of 18S rRNA from these residues in eIF1/40S subunit complexes (Lomakin et al, 2003). These cleavage sites in the 16S rRNA and eIF1's position on the 40S subunit were modeled onto a Thermus thermophilus 30S subunit (Supplementary Figure 1A). In addition to these cleavage sites, which are consistent with eIF1 binding to identical positions on 40S and 30S subunits, Cys61 and Cys75 also cleaved 16S rRNA at nt. 722–723 (helix 23a) and nt. 830–832 (helix 26) (Figures 2B, C, D, I and J). This second set of cleavage sites on the solvent side of the 30S subunit (Supplementary Figure 1B) would be consistent with eIF1 binding to a position like that of IF3-CTD in IF3-CTD/30S subunit complexes as determined by X-ray crystallography of IF3-CTD soaked into 30S subunit crystals (Pioletti et al, 2001). We therefore cannot exclude the possibility of two eIF1-binding sites on a 30S subunit with potentially different affinities.
Figure 2.
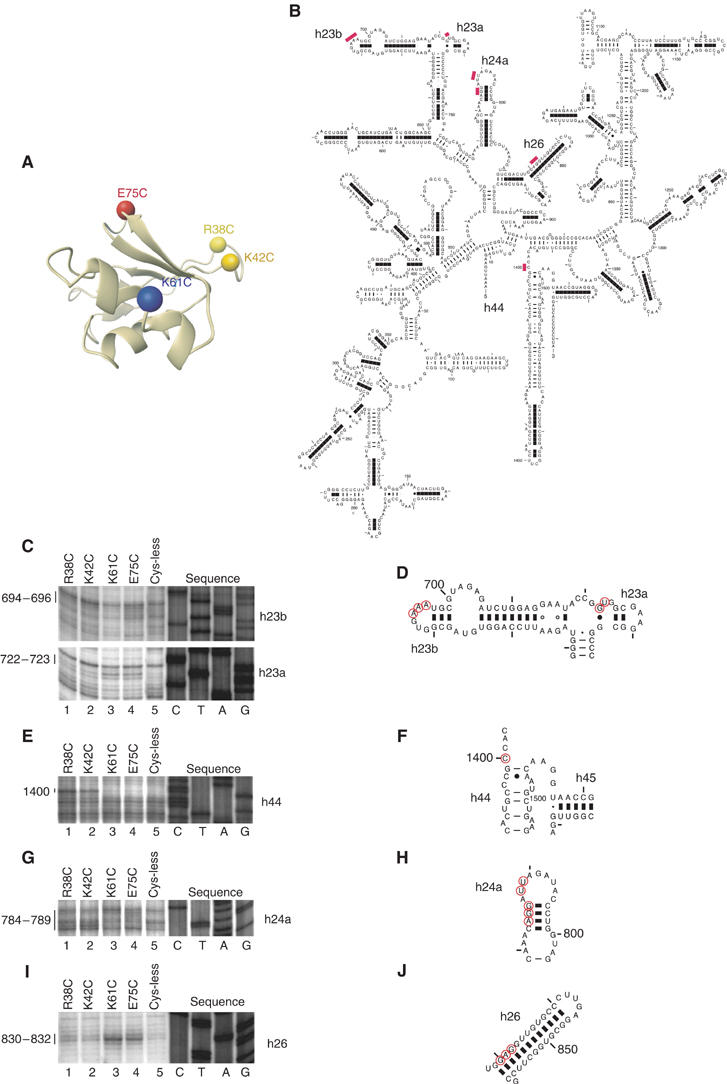
Directed hydroxyl radical cleavage of 16S rRNA in 30S/eIF1 complexes from Fe(II) tethered to different positions on eIF1. (A) Ribbon diagram of the structured domain of eIF1. Colored spheres indicate the positions of cysteines introduced to tether Fe(II)-BABE. (B) Secondary structure of Escherichia coli 16S rRNA. Sites of directed hydroxyl radical cleavage are shown as red bars. (C, E, G, I) Primer extension analysis of directed hydroxyl radical cleavage of 16S rRNA (in helices 23, 44, 24 and 26, respectively) from Fe(II) tethered to positions on eIF1 as indicated. Cleavage sites are indicated to the left of each panel. Lanes marked ‘Cys-less' correspond to reaction mixtures that contained the cysteine-less eIF1 mutant. Lanes C, T, A and G depict 16S rRNA sequence generated from the same primer. (D, F, H, J) Elements of helices 23, 44, 24 and 26 of 16S rRNA with hydroxyl radical cleavage sites (red circles).
As the results of pull-down and sucrose density gradient centrifugation experiments, and detection of hydroxyl radical cleavage in helices 44, 23b and 24a of 16S rRNA in eIF1/30S subunit complexes both suggest that eIF1, IF3-CTD and YciH bound to similar regions on 40S and 30S subunits, we compared their activities in eukaryotic and prokaryotic translation initiation.
Activities of eIF1, IF3 and YciH in ribosomal dissociation and subunit antiassociation
Unlike IF3, eIF1 alone has no ribosome dissociation or antiassociation activity, but consistent with its position on 40S subunits (which would block access of 60S subunits to 18S rRNA elements that form B2b and B2d intersubunit bridges; Spahn et al, 2001), it strongly enhances the ribosome dissociation/antiassociation activity of eIF3 (Kolupaeva et al, 2005). That IF3-CTD can dissociate 70S ribosomes while eIF1 cannot dissociate 80S ribosomes despite binding to the same area of 30S/40S subunits could be because of differences in the affinity of these factors to 30S and 40S subunits or in intersubunit interactions of prokaryotic and eukaryotic ribosomes (and thus in mechanisms of subunit association). We therefore tested the dissociation activity of eIF1 on 70S ribosomes, of IF3 on 80S ribosomes and of YciH on both. eIF1 did not dissociate 80S or 70S ribosomes and did not protect 30S subunits from association with 50S subunits (Figures 3A and B; data not shown). IF3-CTD, capable of dissociating 70S ribosomes, could not dissociate 80S ribosomes or prevent association of 40S and 60S subunits, and YciH had no dissociation/anti-association activity in either system (Figures 3A and B; data not shown). Although IF3, eIF1 and YciH bind to identical/overlapping regions of 70S and 80S ribosomes, only IF3-CTD had ribosome dissociation activity and it was specific for 70S ribosomes. This activity of IF3 in the prokaryotic system could be because only IF3-CTD binds sufficiently avidly to 30S subunits, or because the architecture of IF3-CTD/30S subunit complexes causes bound IF3 to inhibit subunit association at an early stage when it cannot be displaced by 50S subunits.
Figure 3.
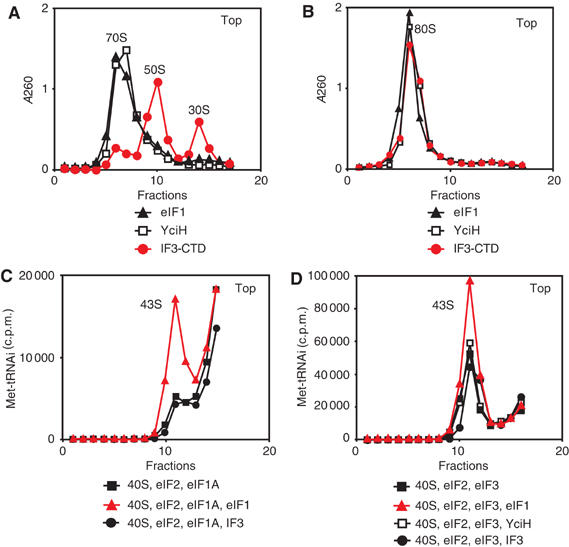
Activities of eIF1, IF3, IF3-CTD and YciH in promoting ribosomal dissociation and formation of 43S complexes. Dissociation of (A) 70S and (B) 80S ribosomes in the presence of factors as indicated. The optical density of ribosomal profiles was measured after centrifugation through 10–30% sucrose density gradients. The positions of ribosomes and ribosomal subunits are indicated. (C, D) 43S complex formation in reaction mixtures containing [35S]Met-tRNAiMet, 40S subunits and factors as indicated was assayed by centrifugation through 10–30% sucrose density gradients. Aliquots of gradient fractions were analyzed by scintillation counting. The position of 43S complexes is indicated. Fractions from upper parts of gradients were omitted for clarity.
Activities of eIF1, IF3 and YciH in 43S complex formation
eIF1 stimulates binding of eIF2-ternary complexes to 40S subunits strongly in the presence of eIF1A and relatively less so in the presence of eIF3, although the overall binding of eIF2-ternary complexes to 40S subunits is higher in the presence of eIF3 (Kolupaeva et al, 2005). Neither IF3/IF3-CTD nor YciH enhanced 43S complex formation (Figures 3C and D; data not shown). The mechanism by which eIF1 stimulates this process is not known, but it is likely indirect, and IF3 and YciH may not induce the necessary conformational changes in 40S subunits. The relative activity of eIF1 in stimulating 43S complex formation in the presence of both eIF3 and eIF1A is very low (Kolupaeva et al, 2005), and the activities of IF3 and YciH were therefore not tested in this combination.
Activities of IF3 and YciH in dissociating aberrant eukaryotic 48S complexes
We also investigated if IF3 and YciH can play any of eIF1's roles in maintaining the fidelity of initiation codon selection in eukaryotes. Toe-printing analysis was used to test if IF3, IF3-CTD and YciH can dissociate complex I formed at the 5′-end of native capped β-globin mRNA and aberrant ribosomal complexes formed on AUG triplets located 1 nt. from the 5′-end of mRNA, on near-cognate initiation codons, or on AUG triplets in bad nucleotide context using mRNAs (Figure 4A) that are derivatives of (CAA)n-GUS mRNA containing a GUS reporter gene and an unstructured 5′-UTR lacking potential near-cognate initiation codons (Figure 6B).
Figure 4.
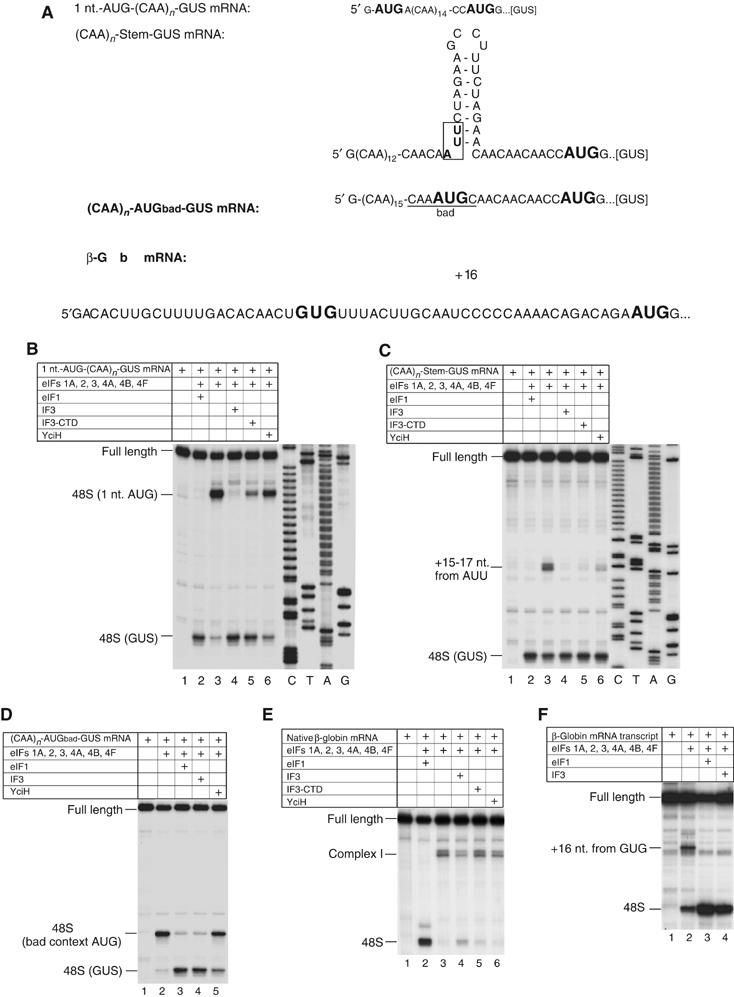
Activities of eIF1, IF3, IF3-CTD and YciH in dissociating aberrant eukaryotic 48S complexes. (A) Sequences of 5′-UTRs of β-globin mRNA and (CAA)n-GUS mRNA derivatives with initiation codons in bold. (B–F) Toe-printing analysis of 48S complexes assembled on mRNAs as indicated. Reaction mixtures contained 40S subunits, Met-tRNAiMet and factors as indicated. The positions of toe-prints caused by assembled 48S complexes are shown on the left. Full-length cDNAs are labeled. (E) ‘Complex I' indicates the position of toe-prints caused by ribosomal complexes whose leading edge was 21–24 nt. from the 5′-end of β-globin mRNA. (B, C) Lanes marked C, T, A and G show cDNA sequences derived using the same primer as that for toe-printing.
Figure 6.
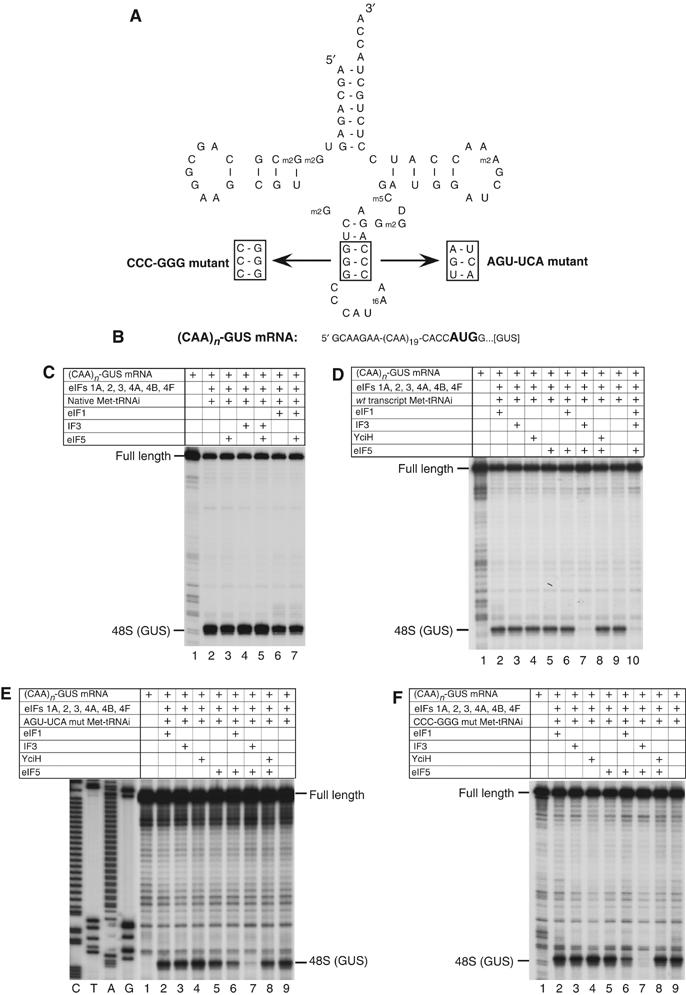
The activities of eIF1, IF3 and YciH in discriminating between wt and mutant forms of Met-tRNAiMet with mutations in the anticodon stem during 48S complex formation on (CAA)n-GUS mRNA. (A) The structure of wt tRNAiMet showing mutated nucleotides and the sequences of mutations in the anticodon stems of CCC-GGG and AGU-UCA mutant transcripts. (B) The sequence of 5′-UTR of (CAA)n-GUS mRNA showing the initiation codon in bold. (C–F) Toe-printing analysis of 48S complex formation on (CAA)n-GUS mRNA in reaction mixtures containing 40S subunits, factors, native eukaryotic Met-tRNAiMet, wt eukaryotic transcript Met-tRNAiMet, AGU-UCA or CCC-GGG mutant transcript Met-tRNAiMet, as indicated. Full-length cDNAs are labeled. The label ‘48S (GUS)' indicates the position of toe-prints caused by 48S complexes assembled on the GUS gene AUG. Lanes (C, T, A and G) show cDNA sequences derived using the same primer.
In the presence of eIF1, 48S complexes formed only on the GUS start codon of 1 nt.-AUG-(CAA)n-GUS mRNA, but in its absence, formed almost exclusively on the 5′-proximal AUG (Figure 4B, lanes 2 and 3; Pestova and Kolupaeva, 2002). Like eIF1, IF3 also promoted 48S complex formation predominantly on the GUS initiation codon (Figure 4B, lane 4). IF3-CTD dissociated ∼70% of 48S complexes from the 5′-proximal AUG and enhanced 48S complex formation on the GUS initiation codon accordingly, whereas YciH was less active and dissociated only ∼25% of 5′-proximal 48S complexes and formed fewer 48S complexes on the GUS initiation codon (Figure 4B, lanes 5 and 6).
A stem inserted into the 5′-UTR of (CAA)n-GUS mRNA contained a near-cognate AUU initiation codon (Figure 4A). In the absence of eIF1, 48S complexes formed efficiently on this triplet (Figure 5C, lanes 2 and 3; Pestova and Kolupaeva, 2002). Like eIF1, IF3 promoted exclusive 48S complex formation on the GUS initiation codon (Figure 4C, lane 4). IF3-CTD and YciH reduced 48S complex formation on the AUU triplet by ∼90 and ∼75%, respectively (Figure 4C, lanes 5 and 6).
Figure 5.
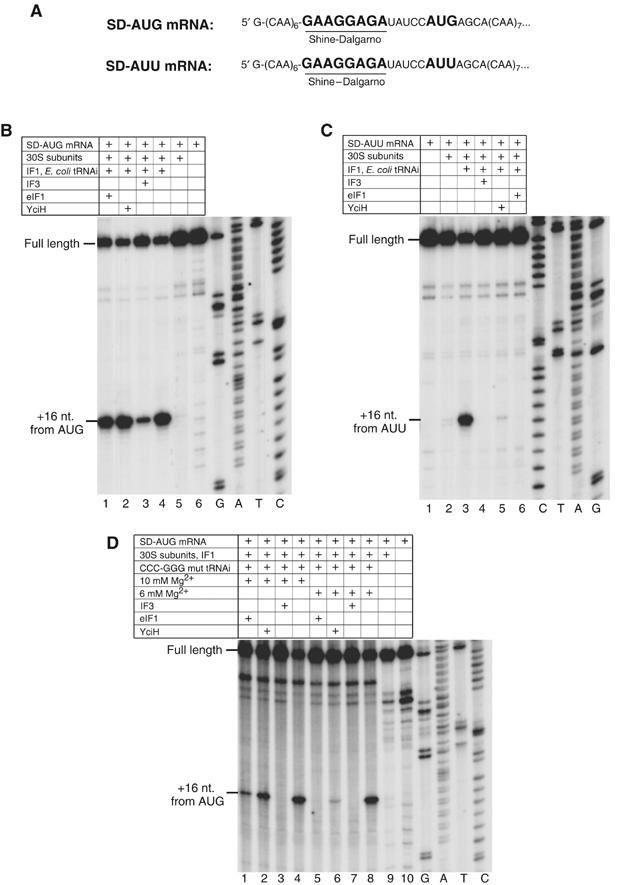
Activities of eIF1, IF3 and YciH in initiation codon and initiator tRNA selection in prokaryotes. (A) The sequences of the 5′UTRs of SD-AUG and SD-AUU mRNAs. The Shine–Dalgarno sequence is bold and underlined; initiation codons are bold. (B–D) Toe-printing analysis of prokaryotic initiation complexes assembled on (B, D) SD-AUG mRNA and (C) SD-AUU mRNA. Reaction mixtures contained 30S subunits, IF1 and wild-type E.coli tRNAiMet or CCC-GGG mutant transcript tRNAiMet (Figure 6A) as well as IF3, eIF1 and YciH as indicated. Assembly reactions in (D) were carried out at 6 or 10 mM Mg2+ as indicated. Full-length cDNAs are labeled; cDNAs labeled ‘+16 nt. from AUG' or ‘+16 nt. from AUU' correspond to toe-prints caused by initiation complexes. Lanes marked G, A, T and C show cDNA sequences derived using the same primer as that for toe-printing.
Discrimination of initiation codon context by IF3 and YciH was investigated using mRNA with an AUG triplet in ‘bad' context upstream of the GUS initiation codon (Figure 4A). In the presence of eIF1, ∼90% of 43S complexes scanned to the GUS initiation codon, whereas in the absence of eIF1, 48S complexes assembled mostly on the first AUG triplet despite its bad context (Figure 4D, lanes 2 and 3; Pestova and Kolupaeva, 2002). IF3 also caused 48S complexes to form almost exclusively on the GUS initiation codon, whereas YciH reduced 48S complex formation on the bad context AUG by only 15% (Figure 4D, lanes 4 and 5).
In the absence of eIF1, an aberrant ribosomal complex I forms at the 5′-end of native β-globin mRNA (Figure 5E, lanes 2 and 3; Pestova et al, 1998). Its formation was reduced by 60% by IF3 and by 15–20% by YciH or IF3-CTD (Figure 4E, lanes 3–6). However, in contrast to eIF1, neither IF3/IF3-CTD nor YciH promoted efficient 48S complex formation on the correct initiation codon (Figure 4E, lanes 4–6). Formation of complex I is dependent on the eIF4F–cap interaction. Thus consistent with our previous data (Pestova and Kolupaeva, 2002), complex I did not form on uncapped β-globin mRNA in the absence of eIF1, but instead small amounts of 48S complex were formed at the AUG codon of β-globin mRNA, and an additional aberrant ribosomal complex was formed at a near-cognate GUG initiation codon in the β-globin 5′-UTR (Figure 4F, lane 2). Like eIF1, IF3 promoted exclusive 48S complex formation on the initiation codon of β-globin mRNA, but with an efficiency of only half that of eIF1 (Figure 4F, lanes 3 and 4).
In conclusion, IF3, IF3-CTD and YciH all discriminated against 48S complex formation on a near-cognate initiation codons, on AUG triplets at the 5′-end of mRNA or with bad context, and against formation of aberrant ribosomal complex I on β-globin mRNA. Full-length IF3 was most and YciH least active.
Dissociation of prokaryotic initiation complexes assembled on AUU triplets by eIF1 and YciH
We next examined the ability of eIF1 and YciH to dissociate prokaryotic initiation complexes containing codon–anticodon mismatches using mRNAs with AUG or AUU codons and canonical Shine–Dalgarno (S–D) sequences (Figure 5A). To increase initiation efficiency, these elements were flanked by multiple CAA triplets to minimize secondary structure. Binding of initiator tRNA to 30S subunits does not require IF2 (e.g. Hartz et al, 1989). This allowed us to assemble initiation complexes from only 30S subunits, mRNA, Escherichia coli tRNAiMet and IF1. Inclusion of eIF1 or YciH did not influence initiation complex formation on mRNA containing an AUG triplet, but N-terminally tagged IF3 surprisingly reduced its formation by 60% (Figure 5B, lanes 1–4). Since native untagged IF3 also reduced initiation complex formation on mRNA containing an AUG triplet (data not shown) and showed the same activities as N-terminally tagged IF3 in all assays presented in Figure 4 (data not shown), we conclude that an N-terminal tag did not alter IF3's activity. Ribosomal complexes did not form at all on SD-AUU mRNA in the presence of either IF3 or eIF1, and YciH reduced complex formation 20-fold (Figure 5C, lanes 3–6). In conclusion, like IF3, eIF1 and YciH discriminated against prokaryotic initiation complexes with codon–anticodon mismatches.
Activities of eIF1, IF3 and YciH in dissociation of eukaryotic 48S complexes assembled with initiator tRNAs containing mutations in the anticodon stem
Both eIF1 and IF3 monitor the fidelity of initiation codon selection but IF3 also participates in initiator tRNA selection, discriminating against initiator tRNA with mutations in three conserved GC pairs in its anticodon stem (Hartz et al, 1989, 1990). IF3 has been proposed to mediate selection indirectly by inducing conformational changes in the 30S subunit, tilting the head towards the platform and enabling GA1338–9 of 16S rRNA in the head to inspect the minor groove of the anticodon stem in the region of these GC pairs (Dallas and Noller, 2001). Eukaryotic tRNAiMet also contains these GC pairs and although it is stringently selected by eIF2, eIF1 might also contribute to selection if it induces conformational changes in 40S subunits similar to those induced in 30S subunits by IF3. To investigate this potential role of eIF1, we used tRNAiMet mutants in which three GC pairs were reversed (CCC-GGG mutant) or in which two were substituted by AU pairs (AGU-UCA mutant) (Figure 6A). Both tRNAs were as active as wt tRNAiMet transcripts or native tRNAiMet in eIF2/GTP/Met-tRNAi ternary complex and 43S complex formation (data not shown). We therefore assayed their activity in 48S complex formation with or without eIF1 on (CAA)n-GUS mRNA (Figure 6B), which has no potential near-cognate initiation codons in its 5′-UTR and which allows 48S complex assembly on the GUS initiation codon in the absence of eIF1 (Pestova and Kolupaeva, 2002). We also determined the influence on 48S complex formation of eIF5-induced hydrolysis of eIF2-bound GTP, after which tRNAiMet remains on the 40S subunit but is no longer bound to eIF2. IF3 and YciH were also assayed.
In the absence of eIF5-stimulated hydrolysis of eIF2-bound GTP, eIF1, IF3 and YciH did not affect 48S complex formation on (CAA)n-GUS mRNA with wt native or in vitro transcribed tRNAiMet (Figure 6C, lanes 2, 4 and 6; Figure 6D, lanes 2–4 and 9). However, in the presence of eIF5, IF3 dissociated ∼90% of 48S complexes assembled with tRNAiMet transcripts but not with native wt tRNAiMet (Figure 6C, lane 5; Figure 6D, lane 7). Neither eIF1 nor YciH dissociated 48S complexes in the presence of eIF5, and eIF1 did not protect 48S complexes containing wt tRNAiMet transcripts from dissociation by IF3 (Figure 6D, lanes 6, 8 and 10). eIF1, IF3 or YciH did not discriminate against 48S complex formation with CCC-GGG or AGU-UCA mutant tRNAs in the absence of eIF5-induced hydrolysis of eIF2-bound GTP (Figures 6E and F, lanes 2–4), but these complexes were dissociated almost completely by IF3 and by 70% by eIF1 (Figures 6E and F, lanes 6 and 7) following hydrolysis of eIF2-bound GTP. YciH did not dissociate 48S complexes containing mutant tRNAs in the presence of eIF5 (Figures 6E and F, lane 8). The activities of eIF1, IF3 and YciH in the dissociation of initiation complexes assembled with wt or mutant in vitro transcribed initiator tRNAs did not depend on the order of mixing and incubation of components. Thus, identical results were obtained if eIF5 and eIF1, IF3 or YciH were added simultaneously with all other translational components, or if initiation complexes were first assembled without these factors, then incubated for 15 min with eIF5 to promote complete hydrolysis of eIF2-bound GTP and after that for 10 min with eIF1, IF3 or YciH (data not shown). The fact that eIF1 dissociated a large proportion of 48S complexes assembled with mutant tRNAiMet after hydrolysis of eIF2-bound GTP suggests that initiator tRNA selection occurs at two stages. Initial selection occurs during eIF2-ternary complex formation and involves only eIF2. A second selection step involves eIF1 and occurs after eIF5-stimulated hydrolysis of eIF2-bound GTP and release of eIF2/GDP from initiation complexes. eIF1 therefore ensures the fidelity of initiation codon selection during 48S complex formation and ensures the fidelity of initiator tRNA selection at the later ribosomal subunit joining stage.
Activities of eIF1 and YciH in dissociating prokaryotic initiation complexes assembled with initiator tRNAs containing mutations in the anticodon stem
The activity of eIF1 in initiator tRNA selection in eukaryotes and its ability to discriminate against prokaryotic initiation complexes containing codon–anticodon mismatches prompted us to test initiator tRNA selection by eIF1 and YciH in prokaryotes. The fact that the efficiency of initiation complex formation and the activities of IF3, eIF1 and YciH in the presence of native E. coli initiator tRNA (Figure 5B) and human tRNAiMet transcripts (data not shown) were identical justifies the use of human CCC-GGG mutant tRNAiMet (see above) to investigate discrimination by eIF1 and YciH against prokaryotic initiation complexes assembled with initiator tRNA with mutations in the anticodon stem. At 6 mM Mg2+, eIF1 and YciH discriminated as well as IF3 against initiation complexes formed with mutant tRNA (Figure 5D, lanes 5–8). At 10 mM Mg2+, discrimination by eIF1 and YciH was reduced: initiation complexes formed in the presence of eIF1 and YciH at ∼20 and ∼60%, respectively, of the level in their absence (Figure 5D, lanes 1–4). YciH and eIF1 can therefore monitor the fidelity of initiator tRNA selection in prokaryotes, although their activity was reduced at 10 mM Mg2+.
Activity of eIF1 in dissociating eukaryotic 48S complexes assembled using initiator tRNA with a mutated anticodon loop
To determine if eIF1 can discriminate against perfectly matched codon–anticodon base pairs that differ from canonical AUG/CAU pairs, we introduced a CAU-CCU mutation into the anticodon loop of initiator tRNA (Figure 7A) and assayed its ability to promote 48S complex formation on the complementary AGG triplets of (CAA)n-AGGgood-GUS and (CAA)n-AUGbad-GUS mRNAs (Figure 7B) in the presence and absence of eIF1. Although methionyl-tRNA synthetase recognizes the anticodon loop of tRNAiMet, it can acylate the (CAU-CCU) anticodon loop mutant tRNAiMet with methionine, and this tRNA supports initiation in vivo from an AGG codon (Cigan et al, 1988). Despite the in vivo activity of this mutant codon–anticodon pair, eIF1 might still reduce initiation from the AGG codon. In fact, eIF1 enhanced 48S complex formation on the good context AGG triplet and reduced 48S complex formation only on the bad context AGG triplet (Figures 7C and D). We conclude that eIF1 does not discriminate against perfectly matched noncanonical codon–anticodon pairs and even enables 43S complexes containing the mutant tRNA to sense the nucleotide context of the AGG codon. In the absence of eIF1, this mutant tRNA also formed 48S complexes on the AUG triplet of (CAA)n-GUS mRNA (Figure 7E), yielding an initiation complex with an unusual mismatch in the second position.
Figure 7.
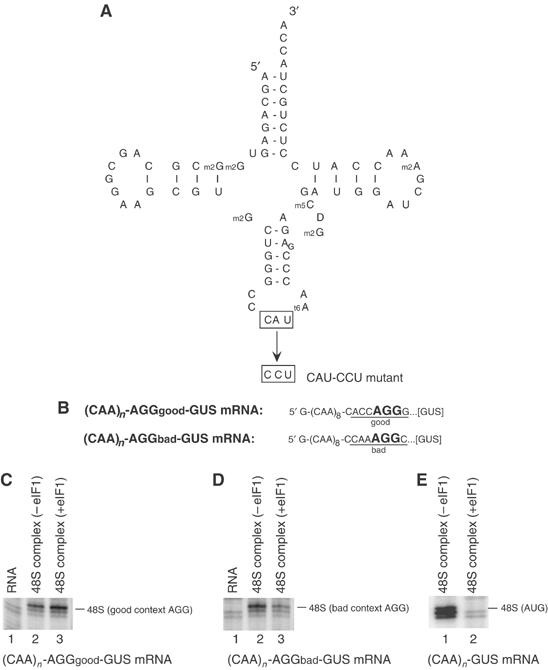
eIF1 does not discriminate against noncanonical complementary codon–anticodon base pairs. (A) Structure of wt tRNAiMet showing mutated nucleotides and the sequences of mutations in the anticodon loop of AGU-UCA mutant transcripts. (B) The sequence of the 5′-UTR of (CAA)n-AGGgood-GUS and (CAA)n-AGGbad-GUS mRNAs with initiation codons in bold. Context residues from −3 to +4 positions are underlined. (C–E) Toe-printing analysis of 48S complex formation on (CAA)n-AGGgood-GUS, (CAA)n-AGGbad-GUS and (CAA)n-GUS mRNAs in reaction mixtures containing 40S subunits, AGU-UCA mutant Met-tRNAiMet, eIFs 2, 3, 1A, 4B, 4B, 4F and eIF1, as indicated. The labels to the right of each panel show the position of toe-prints caused by 48S complexes assembled on AGG or AUG codons, as indicated.
Dissociation of binary CrPV IRES/40S subunit complexes by IF3
Initiation on a few mRNAs is 5′-end independent and is instead mediated by an internal ribosomal entry site (IRES). Translation of the second cistron in the dicistronic Cricket paralysis virus (CrPV) genome is mediated by a ∼190 nt. long intergenic region (IGR) IRES that comprises three pseudoknots (Jan and Sarnow, 2002). Initiation on it does not require initiation factors or initiator tRNA (Wilson et al, 2000; Pestova and Hellen, 2003). The IRES binds directly to 40S subunits, so that the P site is occupied by the part of the IRES containing pseudoknot (PK) I that mimics codon–anticodon base pairs (Figure 8D). Most of the IRES occupies the ribosomal E and P sites (Pestova et al, 2004; Spahn et al, 2004). IRES/40S subunit complexes yield toe-prints at AG6228–6229 (15–16 nt. downstream of the P site CCU codon) and AA6161–6162 (caused by interaction of the PK III-containing domain with 40S subunits) (Figures 8A and D; Wilson et al, 2000). Although initiation on this IRES does not require initiation factors, eIF1 stabilizes IRES binding to the 40S subunit in the region of the P site CCU triplet, which is evident by enhancement of the AG6228–6229 toe-prints and the consequent reduction of those at AA6161–6162 (Figure 8A, lanes 2 and 3; Pestova et al, 2004). The ribosomal location of the IRES, and eIF1's enhancement of IRES/40S subunit interaction prompted us to investigate the influence of IF3 and YciH on this interaction. IF3 strongly destabilized IRES binding to the 40S subunit at the P site, nearly abrogating the AG6228–6229 toe-prints and slightly increasing those at AA6161–6162 (Figure 8A, lane 4). IF3-CTD had a weaker destabilizing effect on the IRES/40S subunit interaction in the P site area, and YciH had no effect (Figure 8A, lanes 5 and 6). IRES/40S subunit complexes were slightly protected by eIF1 from destabilization by IF3 (data not shown), confirming that eIF1 and IF3-CTD bind to identical or overlapping ribosomal sites. In sucrose density gradient centrifugation experiments, IF3 decreased IRES/40S subunit complex formation by only 10–15% (data not shown). We conclude that IF3 does not significantly influence binding of the IRES to 40S subunits, but mainly disrupts its local interaction at the P site. IF3 did not destabilize the IRES' interaction with 80S ribosomes, and addition of 60S subunits to reaction mixtures prevented IF3-dependent weakening of the AG6228–6229 toe-prints (Figure 8B, lanes 2–5). This result is expected, because the IRES can bind productively to 80S ribosomes as well as to 40S subunits (Pestova et al, 2004) and because IF3 cannot prevent subunit association (see above). IRES/80S ribosome complexes formed in the presence of IF3 were elongation-competent and were translocated in the presence of cognate Ala-tRNAAla and eEFs 1 and 2 (Figure 8B, lane 6). The IRES bound only to eukaryotic ribosomes; neither 30S subunits nor 70S ribosomes formed complexes with it that could be detected by toe-printing or sucrose density gradient centrifugation (Figure 8B, lanes 7 and 8; data not shown). The opposing effects of eIF1 and IF3 on the IRES/40S subunit interaction could be explained either by differences in potential conformational changes in 40S subunits induced by these factors, or by differences in the shapes and size of eIF1 and IF3, so that simultaneous binding of IF3 and the IRES at the P site may be sterically impossible.
Figure 8.
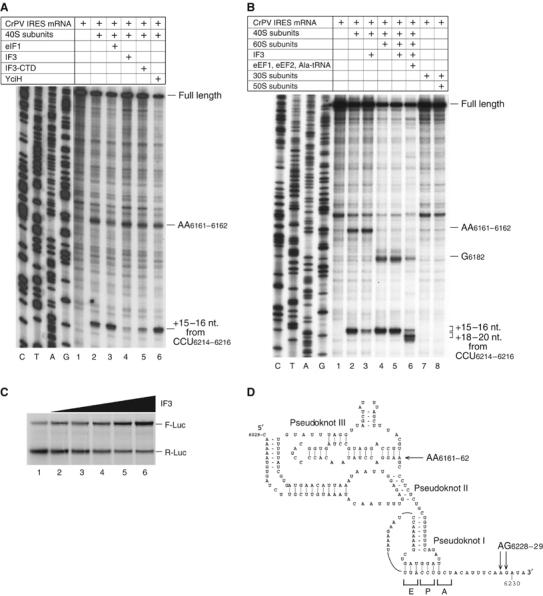
Activities of eIF1, IF3, IF3-CTD and YciH in discriminating against initiation on the CrPV IGR IRES. (A, B) Toe-printing analysis of binding of 40S, 30S subunits, 80S and 70S ribosomes to this IRES in the presence of eIF1, IF3, IF3-CTD, YciH, and eEF1, eEF2 and Ala-tRNAAla as indicated. The 40S/80S-dependent toe-prints and toe-prints that appear following ribosomal translocation are indicated to the right. Full-length cDNAs are labeled. (C) Influence of IF3 on translation mediated by the CrPV IRES. In total, 0.2 μg of bicistronic RLuc-CrPV IRES-FLuc mRNA were translated for 60 min at 30°C in 15 μl of reticulocyte lysate that had been preincubated for 10 min with increasing amounts (0.2–1.5 μg) of IF3. Samples were analyzed by SDS–PAGE and autoradiography. (D) Structure of the CrPV IRES (Jan and Sarnow, 2002) showing the three pseudoknots, and toe-prints (black arrows) due to binding of 40S subunits (Wilson et al, 2000). mRNA triplets located in E, P and A sites are indicated.
The effect of IF3 on IRES/40S subunit binding led us to expect that IF3 might influence translation driven by the IRES. We therefore assayed translation in vitro of dicistronic mRNA (Wilson et al, 2000) in which the IRES mediated translation of the second firefly luciferase (Fluc) cistron and translation of the first Renilla luciferase (Rluc) cistron was 5′-end dependent. Surprisingly, IF3 inhibited 5′-end-dependent translation of Rluc (Figure 8C) and simultaneously stimulated IRES-mediated translation of Fluc, which might be a secondary consequence of the inhibitory effect of IF3 on canonical initiation that could be explained by IF3's destabilizing effect on 48S complexes formed on the AUG of the first cistron. An increase in 40S subunits, temporarily or permanently free of eIF2-ternary complexes, would increase the efficiency of initiation on the IRES.
Discussion
Ribosomal binding and dissociation/antiassociation activities of eIF1, IF3 and YciH
The ability of the nonhomologous factors eIF1/YciH and IF3-CTD to bind to the same site on the platform of heterologous 30S/40S subunits may be rationalized by the homology of these sites and between eIF1 and YciH. eIF1 may also bind to a second site on 30S subunits that corresponds to the secondary IF3-binding site revealed by crystallography (Pioletti et al, 2001) through binding of one of the negatively charged patches on eIF1 to 16S rRNA (Fletcher et al, 1999). eIF1 may not bind the equivalent region on 40S subunits because of differences in protein and rRNA composition (e.g. in helix 26) between those areas of 30S and 40S subunits.
One role of IF3 in initiation is to provide free 30S subunits by preventing subunit association. This activity is specific for prokaryotic ribosomes. eIF1 and YciH cannot dissociate either prokaryotic or eukaryotic ribosomes, and ribosomal dissociation in eukaryotes instead requires eIF3 and involves eIF1 in an accessory role (Kolupaeva et al, 2005). It possibly occurs by a mechanism that differs significantly from that of prokaryotes, which may be because of differences in intersubunit interactions between prokaryotic and eukaryotic ribosomes (Spahn et al, 2001).
Initiation codon selection
A major function of eIF1 and IF3 in homologous translation systems is to ensure the fidelity of initiation codon selection by dissociating aberrant initiation complexes. We identified complete intrakingdom overlap in this function: eIF1, YciH and IF3 all dissociated prokaryotic initiation complexes with codon–anticodon mismatches and eukaryotic 48S ribosomal complexes aberrantly assembled on for example, AUG triplets with bad context or which contained codon–anticodon mismatches. YciH was least active in all assays, possibly because of its lower ribosome-binding activity; it discriminated most strongly against complexes containing codon–anticodon mismatches, which are possibly the least stable of those that we assayed. The fact that initiation factors can discriminate against the assembly of heterologous ribosomes into aberrant initiation complexes is consistent with a common mechanism of action, most likely involving the same set of induced conformational changes in the small subunit.
IF3 promoted dissociation of aberrant ribosomal complexes containing codon–anticodon mismatches and formation of 48S complexes on the AUG initiation codons of uncapped β-globin mRNA and of an uncapped derivative of (CAA)n-GUS mRNA, but did not promote efficient 48S complex formation on the initiation codon of capped native β-globin mRNA. The molecular mechanism of initial ribosomal attachment to the 5′-terminal region of mRNA is unknown. However, it is likely that ribosomal loading on capped and uncapped mRNAs occurs by fundamentally different mechanisms, which for capped mRNAs involves the eIF4E–cap interaction. It is not clear how the transition from ribosomal loading to scanning occurs, and what eIF1's role is in this process, but IF3 may be unable to induce this transition, or may even impair correct loading of 43S complexes onto capped mRNA. The fact that in contrast to (CAA)n-GUS mRNA, on which 48S complexes formed equally efficiently in the presence of either eIF1 or IF3, 48S complex formation on uncapped β-globin mRNA in the presence of IF3 was about half as efficient as that in the presence of eIF1 might also indicate that even though IF3 promotes dissociation of aberrant ribosomal complexes, it does not fully support scanning, which can occur on the unstructured 5′UTR of (CAA)n-GUS mRNA in the absence of eIF1; but on the structured 5′-UTR of β-globin mRNA might require this factor. The suggested inability of IF3 to support scanning might be because of its inability either to put the 43S complex in the necessary conformation, or to interact productively with other components of the 43S complex that bind eIF1, such as eIF3c (Fletcher et al, 1999).
Initiator tRNA selection
Another major function of IF3 is selection of initiator tRNA, which is particularly important because 30S subunits can efficiently bind any tRNA directly. In contrast, 40S subunits cannot bind tRNA directly, and tRNAiMet binds to 40S subunits only in complex with eIF2, which ensures its stringent selection. IF3 discriminates particularly against mutations in three conserved G-C pairs in the anticodon stem (Hartz et al, 1990). Eukaryotic tRNA also contains these conserved, functionally important G-C pairs (Drabkin et al, 1993). Although their role could be simply to maintain a specific conformation of the anticodon loop, there could also be a selection mechanism that requires them.
We found that after hydrolysis of eIF2-bound GTP, IF3 and, to a lesser extent, eIF1 could discriminate against 48S complexes containing tRNAs with mutations in these G-C pairs. It is not known if eIF2 protects tRNAiMet from a challenge by IF3, or if dissociation of eIF2 alters the position of tRNAiMet on the 40S subunit and enhances its sensitivity to the challenge. eIF1 caused only partial rejection of mutant tRNAs under these circumstances, which suggests that this mechanism may be an evolutionary relic intrinsic to the ribosome rather than an efficient proof-reading activity. Initiator tRNA and initiation codon selection in prokaryotes may both rely on the same integrated set of nonseparable conformational changes in the small subunit. The fact that eukaryotes rely on the same mechanism as prokaryotes for initiation codon selection possibly resulted in the conservation of the structure of initiator tRNA in order to preserve the integrity of 48S complexes during initiation codon selection. Initiator tRNA selection on the eukaryotic ribosome may therefore simply be an inevitable consequence of retaining the prokaryotic mechanism of initiation codon selection. Curiously, IF3 destabilized these complexes more strongly than eIF1, and even discriminated against wild-type (unmodified) tRNAiMet transcripts. It is possible that IF3 induces greater conformational changes in the 40S subunit than eIF1, or that its shape is less compatible with the presence of tRNAiMet on the ribosome.
Consistent with the universality of the mechanism for ensuring the fidelity of initiation codon and initiator tRNA selection that we have revealed, we also found that eIF1 and YciH both dissociated prokaryotic initiation complexes formed with mutant tRNAiMet. Surprisingly, eIF1 appeared to be more active than YciH. Although eIF1 may cause similar conformational changes in 30S and 40S subunits, the structure of the former may differ from that of the latter in such a way that these changes discriminate more effectively against mutant tRNAiMet.
It may be significant that eIF1 did not discriminate against tRNAiMet with a mutated (CCU) anticodon loop when it was base-paired with a complementary (AGG) triplet; IF3 also does not discriminate against perfectly base-paired non-AUG triplets in the P site (e.g. Meinnel et al, 1999) unless additional negative determinants are present (Van Etten and Janssen, 1998).
Initiation on the CrPV IGR IRES
The CrPV IRES occupies P and E sites, mimicking initiator tRNA and the base-paired initiation codon/anticodon. The fact that eIF1 stabilized this interaction (Pestova et al, 2004) suggests that the IRES may have adapted to eIF1's presence on 40S subunits. IF3 strongly destabilized binding of the IRES to the 40S subunit in the P site, further suggesting that IF3 and eIF1 bind to the same region of 40S subunits. The different outcomes of binding could be because of the different, induced conformational changes in the 40S subunit, or simply because eIF1 and IF3 differ in size.
In conclusion, we have found that eIF1, IF3-CTD and YciH all bind to the same part of 30S and 40S subunits and can perform many of their functions in discriminating against the formation of aberrant initiation complexes in heterologous systems. These functions are all relevant to the selection of either the initiation codon or the initiator tRNA, which are thus unexpectedly highly conserved and possibly involve a conserved set of conformational changes in the small ribosomal subunit induced by factor binding.
Materials and methods
Plasmids
(CAA)n-GUS, 1 nt.-AUG-(CAA)n-GUS, (CAA)n-Stem-GUS, (CAA)n-AUGbad-GUS and β-globin (Pestova and Kolupaeva, 2002), ptRNA-Ala1 (Pestova and Hellen, 2003), pTRM-1 (Pestova and Hellen, 2001) and monocistronic and bicistronic CrPV IGR IRES-containing (Wilson et al, 2000) transcription vectors and vectors for the expression of recombinant wt eIFs 1, 1A, 4A, 4B and 5 (Pestova et al, 1996, 1998, 2000), and cysteine eIF1 mutants (Lomakin et al, 2003) have been described. CCC-GGG, AGU-UCA and CAU-CCU tRNAiMet mutants were generated by PCR and cloned after a T7 promoter between Ssp1 and HindIII sites of pBluescript SK (Stratagene). Coding regions for IF3, IF3-CTD (aa 80–180) (infC, acc.#V00291) and YciH (acc.#P08245) genes were amplified by PCR from E. coli DH5α DNA and cloned between BamH1 and HindIII sites of pET28b (Novagen). pMetRS plasmid containing both N-terminal (fragment RS1) and C-terminal (fragment RS2) coding regions of E. coli methionyl-tRNA synthetase gene (metG, acc.#K02671) was made by cloning PCR fragments amplified from E. coli DH5α DNA between NcoI and XhoI sites of pET28a.
To construct transcription vectors for SD-AUG and SD-AUU mRNAs, oligonucleotides containing sequences for T7 promoter, 5′-UTRs and coding regions were inserted between BamHI and EagI sites of pBR322. CAA-AGGgood-GUS and CAA-AGGbad-GUS mRNAs were transcribed using mutant PCR fragments derived from (CAA)n-GUS DNA as a template. All RNAs were transcribed using T7 RNA polymerase.
Purification of factors and ribosomal subunits
40S and 60S subunits, eIF2, eIF3, eIF4F, eEF1 and eEF2 were purified from rabbit reticulocyte lysate (Green Hectares), and recombinant eIFs 1, 1A, 4A, 4B and 5 were expressed and purified as described (Pestova et al, 1996, 1998, 2000; Pestova and Hellen, 2003). Recombinant IF3, IF3-CTD, YciH, IF1 and methionyl-tRNA synthetase were expressed in E. coli BL21(DE3) and purified on Ni2+-NTA (QIAGEN) and MonoS (the first three factors) or MonoQ (the last two proteins). 70S ribosomes, 30S and 50S subunits were purified as described (Yusupov and Spirin, 1988). Native E. coli tRNAiMet was from Sigma. Native tRNAiMet was purified as described (Unbehaun et al, 2004). Aminoacylation of native and in vitro transcribed wt and mutant tRNAiMet by recombinant methionyl-tRNA synthetase and of tRNAAla by aminoacyl-tRNA synthetases from RRL was carried out as described (Pestova and Hellen, 2001, 2003). For assaying 43S complex formation (Figure 4C and D), tRNAiMet was aminoacylated with [35S]methionine (spec. act. 400 000 c.p.m./pmol).
In vitro translation
CrPV IGR IRES-containing bicistronic mRNA was translated in RRL with [35S]methionine under standard conditions with or without IF3. Translation products were resolved by electrophoresis in 12% SDS–PAAG and visualized by autoradiography.
Directed hydroxyl radical cleavage
Fe(II)-BABE modification of eIF1 mutants was done as described (Lomakin et al, 2003). Ribosomal complexes were assembled by incubating 500 pmol Fe(II)-eIF1 with 50 pmol 30S subunits in 100 μl buffer (80 mM Hepes, pH 7.6, 140 mM NH4Cl, 10 mM MgCl2 and 10% glycerol) at 37°C for 15 min. Directed hydroxyl radical cleavage and analysis of 16S rRNA were carried out as described (Lomakin et al, 2003).
Ribosome dissociation activity of IF3, eIF1 and YciH
80S ribosomes were assembled by incubating 50 pmol each of 40S and 60S subunits in 200 μl buffer A (20 mM Tris–HCl, 100 mM KCl, 2.5 mM MgCl2, 2 mM DTT, 0.25 mM spermidine). 70S ribosomes were assembled by incubating 50 pmol each of 30S and 50S subunits in buffer B (20 mM Tris–HCl, 100 mM NH4Cl, 6 mM mercaptoethanol) with 10 mM MgCl2. Ribosomes were incubated with 400 pmol IF3-CTD, eIF1 or YciH at 37°C for 10 min and analyzed by centrifugation in a Beckman SW55 rotor for 1 h and 40 min at 4°C and 50 000 r.p.m. in 10–30% sucrose density gradients.
Analysis of 43S complexes
Ribosomal complexes were assembled by incubating 10 pmol 40S subunits with 20 pmol eIF2, 15 pmol [35S]Met-tRNAiMet (spec. act. 400 000 c.p.m./pmol) and different combinations of 20 pmol eIF3, 40 pmol eIF1A and 100 pmol eIF1, IF3 or YciH in 150 μl buffer A containing 0.2 mM GTP for 10 min at 37°C and analyzed by sucrose density gradient centrifugation as described above. The presence of [35S]Met-tRNAiMet in ribosomal fractions was monitored by scintillation counting.
In vitro binding assay
Pull-down analysis was carried out essentially as described (Lomakin et al, 2003). In all, ∼10 μg T7-tagged eIF1, IF3, IF3-CTD or YciH were immobilized on 10 μl T7-antibody agarose by incubation in 100 μl buffer A containing 0.5% Triton X-100 at 25°C for 3 min followed by the addition of 20 mg BSA (Roche) and incubation for 10 min. Beads were washed 3 × with 300 μl of the same buffer. Then, 30 pmol 40S subunits with or without 300 × molar excess of untagged eIF1 were added to immobilized eIF1 and incubation was continued for 15 min. Beads were washed again and bound material was analyzed by SDS–PAGE electrophoresis. For experiments with 30S subunits, buffer A was replaced by buffer B containing 10 mM MgCl2. To analyze eIF1- and IF3-40S subunit interaction by sucrose density gradient centrifugation, 100 pmol 40S subunits and 10 000 pmol T7-tagged eIF1, or eIF1 and IF3 were incubated in 150 μl buffer A for 15 min at 37°C. Fractions corresponding to 40S subunits were analyzed for the presence of eIF1 or IF3 by Western blotting using T7-tag antibodies (QIAGEN).
Toe-printing analysis of initiation complexes
Eukaryotic 48S complexes were assembled on native β-globin mRNA (Invitrogen), CrPV IRES-containing mRNA or (CAA)n-GUS mRNA and its derivatives, and were analyzed by primer extension using AMV reverse transcriptase and primers as described (Pestova et al, 1998; Wilson et al, 2000; Pestova and Kolupaeva, 2002). Reaction mixtures (40 μl) containing 3 pmol mRNA, 3 pmol 40S subunits, 3 pmol 60S subunits, 9 pmol eIF2, 9 pmol eIF3, 5 pmol eIF4F, 10 pmol eIF4A, 10 pmol eIF4B, 50 pmol eIF1 (or IF3, IF3-CTD, or YciH), 10 pmol eIF1A, 25 pmol eIF5, 5 pmol eEF1, 5 pmol eEF2 and 5 pmol aminoacylated tRNAAla or wt or mutant tRNAiMet (as indicated in figures) were incubated for 10 min at 37°C in buffer A with 1 mM ATP and 0.2 mM GTP. cDNA products were analyzed in 6% polyacrylamide sequencing gels. Prokaryotic initiation complexes were assembled on SD-AUG and SD-AUU mRNAs. Reaction mixtures (40 μl) containing 3 pmol mRNA, 3 pmol 30S subunits, 10 pmol IF1, 50 pmol IF3, eIF1 or YciH and 5 pmol wt or mutant tRNAiMet (as indicated in figures) were incubated for 10 min at 37°C in buffer B containing 6 or 10 mM MgCl2. cDNA products were analyzed using a primer complementary to nt 1014–1031 of pBR322. PhosphorImager analysis was used to quantify the efficiency of initiation complex formation. All values presented in Results are the average of at least three independent experiments. Values obtained in independent experiments and using the same factor preparations differed by less than 10%.
Supplementary Material
Supplementary Figure S1
Acknowledgments
We are indebted to A Dahlberg and P Wollenzien for their generous gifts of the 16S rRNA-encoding plasmid pKK3535 and of an untagged IF3 expression vector, respectively. This work was supported by NIH Grants R01 GM59660 and R01 AI51340 to TVP and CUTH, respectively.
References
- Cigan AM, Feng L, Donahue TF (1988) tRNAi(Met) functions in directing the scanning ribosome to the start site of translation. Science 242: 93–97 [DOI] [PubMed] [Google Scholar]
- Cort JR, Koonin EV, Bash PA, Kennedy MA (1999) A phylogenetic approach to target selection for structural genomics: solution structure of YciH. Nucleic Acids Res 27: 4018–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas A, Noller HF (2001) Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell 8: 855–864 [DOI] [PubMed] [Google Scholar]
- Drabkin HJ, Helk B, RajBhandary UL (1993) The role of nucleotides conserved in eukaryotic initiator methionine tRNAs in initiation of protein synthesis. J Biol Chem 268: 25221–25228 [PubMed] [Google Scholar]
- Fletcher CM, Pestova TV, Hellen CU, Wagner G (1999) Structure and interactions of the translation initiation factor eIF1. EMBO J 18: 2631–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D, Binkley J, Hollingsworth T, Gold L (1990) Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes Dev 4: 1790–1800 [DOI] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Gold L (1989) Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev 3: 1899–1912 [DOI] [PubMed] [Google Scholar]
- Jan E, Sarnow P (2002) Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol 324: 889–902 [DOI] [PubMed] [Google Scholar]
- Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CUT, Pestova TV (2005) Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA 11: 470–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1991) Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem 266: 19867–19870 [PubMed] [Google Scholar]
- Kyrpides NC, Woese CR (1998) Universally conserved translation initiation factors. Proc Natl Acad Sci USA 95: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU (2005) Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev 69: 101–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV (2003) Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev 17: 2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T, Sacerdot C, Graffe M, Blanquet S, Springer M (1999) Discrimination by Escherichia coli initiation factor IF3 against initiation on non-canonical codons relies on complementarity rules. J Mol Biol 29: 825–837 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CUT (1998) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854–859 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CUT (2001) Preparation and activity of synthetic unmodified mammalian tRNAiMet in initiation of translation in vitro. RNA 7: 1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CUT (2003) Factor requirements for translation elongation after initiation at the A site on the Cricket paralysis virus internal ribosomal entry site. Genes Dev 17: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CUT, Shatsky IN (1996) Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol 16: 6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG (2002) The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 16: 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Hellen CU (2004) Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep 5: 906–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CUT (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335 [DOI] [PubMed] [Google Scholar]
- Petrelli D, LaTeana A, Garofalo C, Spurio R, Pon CL, Gualerzi CO (2001) Translation initiation factor IF3: two domains, five functions, one mechanism? EMBO J 20: 4560–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioletti M, Schlunzen F, Harms J, Zarivach R, Gluhmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, Hartsch T, Yonath A, Franceschi F (2001) Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J 20: 1829–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapkina TG, Dolan MA, Babin P, Wollenzien P (2000) Initiation factor 3-induced structural changes in the 30S ribosomal subunit and in complexes containing tRNAfMet and mRNA. J Mol Biol 299: 615–628 [DOI] [PubMed] [Google Scholar]
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J (2001) Structure of the 80S ribosome from Saccharomyces cerevisiae-tRNA-ribosome and subunit–subunit interactions. Cell 107: 373–386 [DOI] [PubMed] [Google Scholar]
- Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J (2004) Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell 118: 465–475 [DOI] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CUT, Pestova TV (2004) Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev 18: 3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten WJ, Janssen GR (1998) An AUG initiation codon, not codon–anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli. Mol Microbiol 27: 987–1001 [DOI] [PubMed] [Google Scholar]
- Wilson JE, Pestova TV, Hellen CUT, Sarnow P (2000) Initiation of protein synthesis from the A site of the ribosome. Cell 102: 511–520 [DOI] [PubMed] [Google Scholar]
- Yoon H, Donahue TF (1992) The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mo Cell Biol 12: 248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Spirin AS (1988) Hot tritium bombardment technique for ribosome surface topography. Meth Enzymol 167: 426–439 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1


