Abstract
Repair of double-strand breaks by gene conversions between homologous sequences located on different Saccharomyces cerevisiae chromosomes or plasmids requires RAD51. When repair occurs between inverted repeats of the same plasmid, both RAD51-dependent and RAD51-independent repairs are found. Completion of RAD51-independent plasmid repair events requires RAD52, RAD50, RAD59, TID1 (RDH54), and SRS2 and appears to involve break-induced replication coupled to single-strand annealing. Surprisingly, RAD51-independent recombination requires much less homology (30 bp) for strand invasion than does RAD51-dependent repair (approximately 100 bp); in fact, the presence of Rad51p impairs recombination with short homology. The differences between the RAD51- and RAD50/RAD59-dependent pathways account for the distinct ways that two different recombination processes maintain yeast telomeres in the absence of telomerase.
Double-strand breaks (DSBs) are efficiently repaired by homologous recombination in Saccharomyces cerevisiae. The best-studied pathways of repair require the participation of Rad51p, the eukaryotic homologue of the bacterial strand exchange protein, RecA. Rad51p binds to single-stranded DNA (ssDNA) and forms a filament in which each subunit is bound to 3 nucleotides (nt) of ssDNA (30). Rad51p interacts with and is assisted in recombination by a number of other proteins, most notably Rad52p (42), Rad54p (5, 14, 27, 35, 38), Rad55p, and Rad57p (12, 48). Rad52p is essential for nearly all types of homologous recombination; however, there are a number of instances in which recombination can occur in the absence of Rad51p. In spontaneous recombination, a rad51Δ rad59Δ double mutant is more defective than a rad51Δ or rad59Δ single mutant (1).
In yeast cells lacking the telomerase EST1 or TLC1 gene, the maintenance of telomeres depends on homologous recombination, specifically on Rad52p (22). In fact, there are two independent telomere maintenance pathways, each yielding different outcomes. One pathway requires Rad52p, Rad51p, Rad54p, Rad57p, and presumably Rad55p (20, 50). This pathway yields telomeres in which there have been frequent recombination events involving subtelomeric Y′ sequences. A second pathway requires Rad52p, Rad50p, Mre11p, Xrs2p, Rad59p, and Sgs1p (4, 6, 16, 20, 49, 50). Here the TG1-3 telomere sequences themselves are elongated, either by intertelomere recombination or by an intrachromosomal event, such as rolling circle replication. Double mutants, rad51Δ rad50Δ and rad51Δ rad59Δ, fail to maintain telomeres without telomerase, as does rad52Δ.
A similar relationship between RAD51- and RAD50/RAD59-dependent mechanisms is found in a well-studied case where there is a single DSB in the middle of one chromosome in a diploid. The DSB, induced by the site-specific HO endonuclease, can be efficiently repaired by gene conversion in wild-type cells. In the absence of Rad51p, Rad54p, Rad55p, and Rad57p, gene conversions are eliminated but repair can still take place by break-induced replication (BIR). Here the centromere-proximal end of the chromosome can invade the intact homologous chromosome and promote replication to the chromosome end (23, 44). BIR, in the absence of Rad51p, depends on Rad50p (and presumably Mre11p and Xrs2p), Rad59p, and Tid1p. The double mutants rad51Δ rad50Δ, rad51Δ rad59Δ, and rad54Δ tid1Δ all eliminate 90% of homologous recombinational repair, although rad52Δ is still more severe (44). Without Rad51p, repair also depends on a special, distant chromosomal site, termed a facilitator of BIR, that is apparently needed to allow the initial steps of homologous recombination to occur (26). Thus, it appears that both repair of a single DSB in a diploid and maintenance of telomeres in telomerase-deficient haploid cells can occur by these two RAD52-dependent pathways.
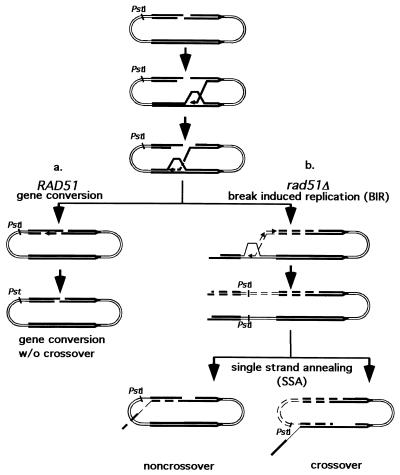
DSB-induced gene conversion events have also been extensively studied in centromere-containing plasmids carrying two inverted copies of Escherichia coli LacZ sequences, one of which harbors a recognition site for HO endonuclease. Here, too, it appears that there are both RAD51-dependent and RAD51-independent pathways of repair, both of which need RAD52 (1, 46). Recently it was suggested that Rad51-independent recombination on a plasmid may begin by BIR, in which repair is completed by single-strand annealing (BIR-SSA model) (Fig. 1) (17).
FIG. 1.
Models of RAD51-dependent and RAD51-independent intraplasmid recombination between inverted repeats (thick lines). One end invades and primes new DNA synthesis (dashed lines). (a) In the presence of Rad51p, DSB repair occurs by gene conversion, which in mitosis is mostly not associated with crossovers. (b) In the absence of Rad51p, DSB repair occurs by BIR-SSA. Strand invasion sets up a replication fork in which both leading and lagging strand synthesis occurs. If synthesis goes to the very end of the linearized plasmid, repair can be completed by SSA, in which the ends are resected by 5′ to 3′ exonucleases. Alternative annealing between repeats yields equal numbers of crossover and noncrossover products. The position of the PstI cut site is changed in the crossover product.
In the present study we show that HO endonuclease-induced intrachromosomal recombination in plasmids is different from equivalent recombination events between two plasmids, between a chromosome and a plasmid, or between two chromosomes, all of which only occur when RAD51 is present. We present further evidence that intrachromosomal RAD51-independent recombination on plasmids occurs by BIR-SSA. We report that intrachromosomal recombination in plasmids with as little as 33 bp on either side of the DSB is surprisingly efficient, but that repair of such short homologous sequences is almost exclusively carried out by a RAD51-independent, RAD50-dependent, and RAD59-dependent process that also requires TID1 and SRS2. We further show that Rad51p participates in a competing pathway that requires longer homology and promotes gene conversion without crossovers. This work provides an insight into the way that apparently analogous RAD51-dependent and RAD51-independent pathways can repair telomeres in the absence of telomerase.
MATERIALS AND METHODS
Strains and plasmids.
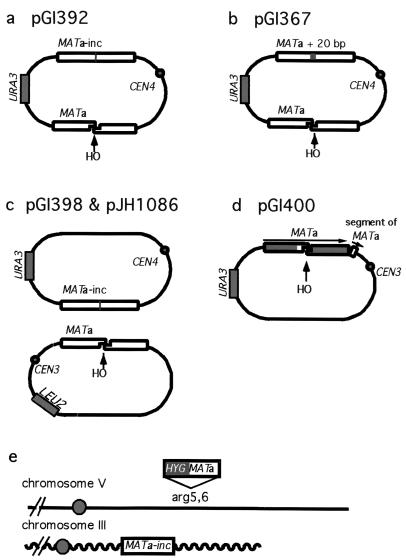
All plasmids analyzed in this study, described in Table 1, are derivatives of the centromeric plasmid YCp50 marked by URA3. They contain two copies of either yeast MATα, MATa, or E. coli LacZ sequences in inverted orientation. One copy, placed between EcoRI and HindIII restriction sites, carries an HO endonuclease recognition site and is the recipient in our recombination assays. The other copy inserted in the opposite direction at the SmaI site cannot be cut by HO endonuclease and is a donor of homologous sequences during DSB repair (Fig. 2). pGI36, pGI333, pGI343, pGI344, pGI345, pGI388, pGI389, pGI390, pGI391, and pGI392 (Fig. 2a and Table 1) carry inverted repeats of MATa and MATa-inc sequences. A 2.5-kb-long MATa-inc sequence was inserted in a SmaI site, and 46- to 2,067-bp-long fragments of the MATa sequence were inserted in the EcoRI-HindIII sites of YCp50. These constructs have the HO cut site in the middle of the repeat. The donor repeat of MATa sequences in pGI367, pGI364, pGI365, pGI366, and pGI368 have an insertion of 20 bp in the HO cut site so that the two 3′ ends of HO-cut DNA are perfectly matched to the donor MATa repeat (Fig. 2b and Table 1). The 16-bp gap has to be filled in during DSB repair. These constructs have 46 to 1,673 bp of MATa sequence inserted between the EcoRI and HindIII sites of YCp50 with the HO cut site in the middle. In the plasmid pGI348 (Table 1) the donor sequence MATα-inc has a single base pair substitution, C to T, placed 2 bp to the left of the HO cut site. The recipient repeat is a 70-bp MATα sequence with the HO cut site in the middle. Plasmid retention was tested with JKM146 (Δho Δhml::ADE1 Δhmr::ADE1 MATa-inc ade1 leu2-3,112 lys5 ura3-52 trp1::hisG ade3::GAL10::HO) or tNS1368 (Δho Δhmr HML mat::leu2::hisG ura3-52 thr-4 trp1 ade3::GAL-HO) or with derivatives of these two strains: tGI203 (tNS1368, rad51::KanMX2); tGI236 (JKM146, rad51::KanMX2); tGI235 (JKM146, rad51::KanMX2 rad50::hisG); YFP218 (JKM146, srs2::LEU2); YFP206 (JKM146, rad50::hisG); tNS1703 (JKM146, rad59::KanMX2); tGI52 (JKM146, rad59::KanMX2 srs2::LEU2); tGI223 (JKM146, tid1::KanMX2); YFP255 (JKM146, msh2::LEU2); and YFP259 (JKM146, msh3::leu2). Plasmid retention was tested in the wild type or rad51Δ and was the same in both genetic backgrounds, which means that the results are not strain specific.
TABLE 1.
Homology requirement for intraplasmid DSB repair in wild-type strains
| Plasmid name | Length of homologous repeats (bp) | Homology 5′ and 3′ of DSB (5′ bp/3′ bp) | % P plasmid retention (± SEM) |
|---|---|---|---|
| MATa repeatsa | |||
| pGI392 | 2,067 | 1,046/1,025 | 95.0 |
| pGI36 | 1,247 | 633/618 | 82.6 ± 7.45 |
| pGI391 | 812 | 408/408 | 82.0 |
| pGI390 | 412 | 208/208 | 77.0 |
| pGI389 | 302 | 153/153 | 74.2 ± 5.0 |
| pGI388 | 136 | 77/63 | 69.3 |
| pGI333 | 70 | 37/37 | 42.7 ± 10.4 |
| pGI343 | 62 | 33/33 | 26.3 ± 8.4 |
| pGI344 | 54 | 29/29 | 1.6 ± 0.6 |
| pGI345 | 46 | 25/25 | 0.2 ± 0.04 |
| MATa repeatsb | |||
| pGI367 | 1,673 | 1,353/324 | 83.9 ± 2.1 |
| pGI364 | 70 | 37/37 | 39.5 ± 1.4 |
| pGI365 | 62 | 33/33 | 24.6 ± 8.1 |
| pGI366 | 54 | 29/29 | 1.6 ± 0.7 |
| pGI368 | 46 | 25/25 | 0.3 ± 0.2 |
| MATα repeatsc | |||
| pGI348 | 70 | 37/37 | 36.9 ± 2.7 |
MATa repeats, donor contains a single base pair substitution 2 or 3 nt from the DSB.
MATa repeats, donor is perfectly homologous to DSB ends, plus a 20-bp insertion.
MATα repeats, donor contains a single base pair substitution 2 or 3 nt from the DSB.
FIG. 2.
Plasmid and chromosomal constructs. Open rectangles represent homologous sequences. Plasmids carry two inverted or direct repeats of MATa. The recipient copy containing an HO cleavage site was inserted between the EcoRI and HindIII sites, and the donor copy with a mutated HO cut site was inserted at the SmaI site of YCp50. (a) In one set of plasmids the donor sequence cannot be cleaved because of a single C to T substitution, designated MATa-inc. (b) Plasmids in which the donor and recipient MATa repeats are perfectly homologous, but in which the donor cannot be cleaved by HO because of a 20-bp insertion at the HO cut site. (c) Interplasmid recombination system: MATa and MATa-inc sequences are on different plasmids, marked with URA3 or LEU2. (d) The MATa sequence and the first 33 bp of the MATa sequence upstream of the HO cut site were inserted in the same orientation between the EcoRI and HindIII sites and between the HindIII and BamHI sites of YCp50, respectively. Here, DSB can only be repaired by SSA. (e) Interchromosomal ectopic recombination system: the MATa-inc sequence is located on chromosome III, and the MATa sequence marked by an adjacent hygromycin resistance gene (HYG) was inserted in the ARG5,6 locus on chromosome V.
Measurement of DSB repair efficiency and crossover frequency.
Cells were grown in SC-URA (synthetic complete medium lacking uracil), YEPL (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto Peptone, 3.7% lactic acid, pH 5.5), YEPGal (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto Peptone, 2% galactose), or YEPD (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto Peptone, 2% [wt/vol] dextrose). Yeast strains containing a plasmid were grown overnight in SC-URA liquid minimal medium, washed once with water, resuspended in YEPL medium, grown again overnight to a density of 107 cells per ml, and plated on YEPGal plates and YEPD plates at about 100 cells per plate. Since these strains have a GAL10::HO construct, HO endonuclease was expressed on YEPGal plates so that the plasmid is cut at the HO cleavage site. The percentage of plasmid retention was calculated as the fraction of colonies retaining the repaired plasmid on SC-URA replica plated from YEPGal divided by the fraction of colonies retaining plasmid on SC-URA replica plated from YEPD. The percentage of crossing over was measured from Southern blots as a ratio between the intensity of the crossover product to the sum of the intensities of gene conversion and crossover bands. The efficiency of chromosomal DSB repair was calculated as the number of colonies on YEPGal divided by the number of colonies on YEPD.
RESULTS
RAD51 is required for DSB-induced interchromosomal gene conversion but not intrachromsomal DSB-induced recombination.
Gene conversions can be readily recovered from a centromeric plasmid containing inverted repeats of a homologous sequence, one of which can be cleaved by the HO endonuclease, even in the absence of RAD51 (46). In contrast, interchromosomal gene conversions between the same MAT sequences used in the plasmid studies require RAD51 (23, 44). To establish if the difference between these results was attributable to a difference between plasmid and chromosome or if it reflected the difference between an inter- and intrachromosomal recombination event, we constructed a series of strains to test interplasmid, interchromosomal, and plasmid-chromosome recombination. In each case a MATa sequence was the recipient while a MATa-inc sequence, containing a single base pair difference that prevents HO cleavage, was used as the donor (Fig. 2).
The plasmids were introduced into strains in which sequences homologous to the HO cleavage site at MAT, HML, and HMR had been deleted or mutated (see Materials and Methods). HO endonuclease was induced by using a galactose-inducible HO gene integrated at the ade3 locus. HO cleavage is essentially 100% when the gene is induced for at least 1 h; here HO was expressed continuously, since once the plasmid was repaired further cleavage was not possible. As shown in Table 2, the repair was readily obtained without Rad51p in plasmid pGI367, where recombination between inverted repeats was intrachromosomal.
TABLE 2.
All intermolecular DSB-induced gene conversions are RAD51 dependent
| MATa-inc donor sequence | MATa recipient sequence | Efficiency of DSB-induced gene conversion (%)a
|
|
|---|---|---|---|
| RAD51 | rad51Δ | ||
| Plasmid pGI398 | Plasmid pJH1086 | 52 | 0.3 |
| Chromosome III | Plasmid pGI387 | 18 | 0.6 |
| Plasmid pGI398 | Chromosome III | 55 | 0.4 |
| Chromosome III | Chromosome V | 82 | 0.4 |
The efficiency of DSB repair was calculated as described in Materials and Methods. No DSB-induced gene conversion was seen with intermolecular recombination. For intramolecular recombination (pGI367), DSB-induced gene conversion with RAD51 was 84%; with rad51Δ it was 67%.
When the MATa sequence was on one centromeric plasmid (pJH1086) marked with LEU2 and the MATa-inc donor was present on a different centromeric plasmid (pGI398) marked with URA3 (Fig. 2c), gene conversion in a wild-type strain was approximately 50% as efficient as it was in the intrachromosomal case. Here, crossings over would yield a dicentric plasmid so that only gene conversions without exchange were scored (Table 2). In the absence of Rad51p, the repair was inefficient on the level seen for nonhomologous end-joining of a DSB (28).
We also examined gene conversions in which HO-cleaved MATa was at its normal location on chromosome III and MATa-inc was present on a centromeric plasmid. Here, DSB repair was 55% efficient. Essentially all gene conversions required Rad51p (Table 2).
In the converse case, where MATa was on a plasmid and MATa-inc was on chromosome III, the efficiency of gene conversion was about 20%. The lower efficiency of these results may reflect the fact that extensive 5′ to 3′ degradation of the broken ends on a plasmid may cause plasmid loss before homology searching between different chromosomes is successful, whereas resection along the chromosome may not pose the same problem. Again, virtually all gene conversion events required Rad51p (Table 2).
Finally, we examined ectopic interchromosomal gene conversion between MATa-inc on chromosome III and a MATa locus inserted at the arg5,6 locus on chromosome V (Fig. 2e). Gene conversions both with (4%) and without crossing over could be recovered (data not shown). In the absence of Rad51p, virtually all gene conversions were eliminated (Table 2).
Taken together these data suggest that intrachromosomal recombination on a plasmid is distinctly different from any of the interchromosomal or interplasmid situations. One explanation for such a difference is that intraplasmid repair of a DSB does not occur by gene conversion per se but by a pathway, such as BIR-SSA, as suggested by Kang and Symington (17) (Fig. 1).
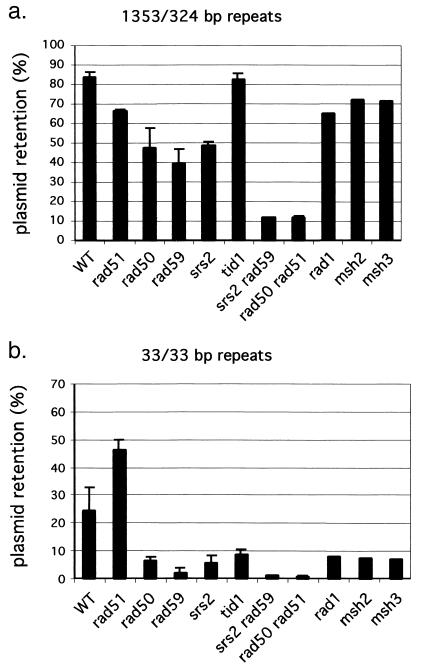
Two chromosomal situations in which there is repair of a DSB in the absence of Rad51p and in the absence of gene conversion have previously been described: in a diploid cell in which one chromosome III homologue is cleaved at MAT (23) and in the maintenance of telomeres lacking telomerase by RAD52-dependent recombination (20). In both cases these RAD51-independent events required RAD50 and RAD59. We found that intraplasmid recombination has the same requirements, as shown for a case in which a plasmid contains MATa and MATa-inc, modified to share approximately 1 kb of homology on either side of the DSB. Knocking out RAD51, RAD50, or RAD59 each reduced recombination from 90 to 50 to 70% of the wild-type value, respectively, but a rad51 rad50 double mutant eliminates approximately 90% of gene conversions (Fig. 3a). Thus, RAD51-independent repair in the plasmid strongly resembles BIR repair of chromosomal DSBs. On the basis of these results we decided to study the homology and genetic requirements of RAD50- and RAD59-dependent recombination pathways by examining different plasmid substrates and different mutant backgrounds.
FIG. 3.
DSB repair efficiency of two plasmids containing inverted repeats with intermediate lengths of homology (a) or short 33-bp inverted repeats (b). In both cases the DSB ends are perfectly homologous to those of the donor. The effect of deleting several genes important for recombination was determined. WT, wild type.
Homology requirements for intraplasmid DSB repair.
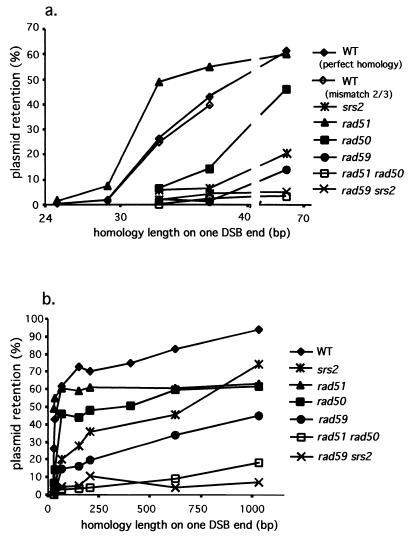
To study the effect of homology on DSB-mediated gene conversion on plasmids, we created a set of plasmids carrying inverted repeats of either MATa or MATα sequences (Table 1). The size of the DNA adjacent to the DSB that is homologous to the donor template was varied from 25 bp to >2 kb on either side of the break (Table 1). One series of plasmids contained MATa sequences and a donor carrying a 1-bp MATa-inc mutation (Fig. 4a). When homology was >150 bp on either side, successful recombination—measured by retention (and thus repair) of the HO-cleaved plasmids—occurred 74% of the time (Table 1 and Fig. 4). With less homology the efficiency decreased, but even with only 37 or 33 bp of homology, repair occurred nearly 43 and 26%, respectively, compared to that with much greater homology. With only 29 bp of homology on either side, recombination efficiency fell 15-fold to 1.6%. With homology of 25 bp, the proportion of cells able to retain the plasmid fell even further, to 0.2%, a portion of which is likely to be the result of nonhomologous end-joining. The sharp inflection at 29 bp suggests that the minimum length needed for efficient recombination is between 29 and 33 bp on one side of the DSB.
FIG. 4.
DSB repair efficiency for a set of plasmids carrying inverted MATa and MATa-inc repeats of various lengths. The effect of different homology lengths as well as the effects of deleting several genes involved in recombination is shown. DSB repair efficiency is plotted against the length of homology on each side of the DSB in a range of 25 to 70 bp (a) and in a range of 25 bp to 1 kb (b). WT, wild type.
The presence of the MATa-inc mutation creates mismatches 2 and 3 bp from the ends on each side of the DSB. This heterology did not impair recombination, as we obtained virtually identical results with a second series of plasmids in which the ends of the DNA are perfectly matched to a donor containing a modified HO cleavage site with a 16-bp insertion at its center, making it uncleavable (Fig. 4a). It is likely that these mismatches, located very close to the DSB ends, do not form heteroduplex DNA and are removed by the proofreading activity of repair DNA polymerases during DSB repair (32).
We also showed that different DNA sequences on one or both sides of the DSB yielded similar results. In one experiment both MATa repeats were replaced by MATα sequences in which the DNA sequences to the left of the cleavage site are completely different, beginning 8 bp from the 3′ end of the DSB. The donor sequence contained an inc mutation identical to that in the MATa-inc donor 2 and 3 bp from the two ends created by the DSB (Table 1). Plasmid pGI348, containing 37 bp of homology to MATα on either side of the DSB, was repaired as efficiently (36.9%) as the analogous plasmid with MATa sequences (pGI333, 42.7%). We conclude that the minimum effective processing segment for plasmid recombination is between 29 and 33 bp. Given that heterologies 2 and 3 nt from the ends of the DSB do not impair recombination, the size of perfectly matched homology needed for efficient recombination seems to be 30 or 31 bp on each side of the DSB.
Repair involving short homologies is RAD50 and RAD59 dependent.
In plasmids where the donor sequences shared long homologies and there are no nonhomologies at the DSB ends, knocking out either RAD51, RAD50, RAD59, or SRS2 causes relatively small (≤40%) reductions in repair efficiency (Fig. 3a). The more substantial defect for rad59Δ in HO-mediated intraplasmid recombination by Bai and Symington (1) can be attributed to an additional role of Rad59p in the removal of nonhomologous 3′-ended ssDNA tails when the HO cleavage site is present in the recipient as an insertion heterology (45). A rad51Δ rad50Δ double mutant decreased recombination eightfold, to 12%, compared to that of the wild-type strain (Fig. 3a). These results suggest that Rad51p and Rad50p independently contribute to the ability of plasmids to recombine, consistent with the idea that there may be two independent recombination pathways for DSB repair.
With plasmid pGI365 having 33 bp on each side of the DSB, the impact of different recombination mutants was quite different. Sixty to 80% of the recombination was lost in rad50Δ, rad59Δ, tid1Δ, and srs2Δ strains, with rad59Δ having the greatest impact. Most of the residual recombination was lost in a rad51Δ rad50Δ double mutant. Surprisingly, rad51Δ alone increased recombination from roughly 25 to 60% (Fig. 3b). We interpret these results to mean that recombination involving short homologies relies almost exclusively on a recombination process in which Rad59p, Rad50p (and presumably Mre11p and Xrs2p), Tid1p, and Srs2p are essential. Rad51p is able to carry out a very low level of recombination with short substrates; consequently, rad50 rad51 double mutants are more severely affected than rad50 alone.
The double mutant rad59Δ srs2Δ eliminates almost all recombination repair, even between long repeats. RAD51-dependent recombination between two chromosomes is reduced in rad59, rad50, and srs2 (data not shown). Thus, although these proteins are each essential for recombination between short repeats, Srs2 helicase, Rad59p, and Rad50p evidently play important but overlapping roles in RAD51-dependent recombination as well (Fig. 3 and 4).
If RAD51-independent recombination occurs by BIR coupled to SSA, then mutations that prevent SSA should also eliminate RAD51-independent events. We have previously shown that both Rad1-Rad10 nucleotide excision repair endonuclease and the Msh2-Msh3 mismatch repair heterodimer are needed for efficient removal of nonhomologous tails produced by SSA (11, 47). Indeed, rad1Δ, msh2Δ, and msh3Δ all severely impair repair of plasmid pGI365 with 33 bp of homology on either side of the DSB but have much less impact on a plasmid with longer homology (pGI367) where RAD51-dependent recombination can occur (Fig. 3).
Finally, we confirmed that RAD51-independent SSA events in the plasmid can efficiently arise from 33-bp repeats. Plasmid pGI400 carries two direct repeats, including both an intact copy of the MATa repeat and the first 33 bp to the left of the HO cut site, inserted on the right side of MATa in the same orientation (Fig. 2d). When this plasmid is cut it can form an intermediate which is analogous to that expected for BIR-SSA: an SSA event in which there is a single long, 3′ended nonhomologous tail. SSA between 33-bp repeats was tested in a rad51Δ strain. SSA is very efficient, as 82% of plasmids were retained. Again, this result supports the BIR-SSA model of intraplasmid repair. The lower efficiency of repair between 33-bp inverted repeats (59%) than between direct repeats (82%) appears to reflect additional constraints associated with the BIR step of repair in inverted substrates.
Different homology requirements for RAD51-dependent and -independent repair.
The fact that rad51Δ actually increases successful recombination of substrates with short homology suggests that Rad51p may compete with an alternative pathway by binding to ssDNA ends but that it cannot accomplish successful recombination with such short homology. To establish more precisely the homology requirements in these two pathways we examined the efficiency of repair of a series of short-homology plasmids in rad51Δ, rad50Δ, and rad59Δ strains. The results, shown in Fig. 4a, are consistent with the idea that the two processes have different homology requirements. We estimate that the RAD51-independent pathway, which must involve strand invasion and initiation of new DNA synthesis, occurs 10% of the time with as little as 29 bp of homology on either side of the DSB. To have a 10 to 20% level of recombination in the absence of Rad59p or Srs2p requires repeats with approximately 70 bp of homology on each side of the DSB (Fig. 4). An accurate measure of RAD51-dependent events is more difficult to obtain, because the results presented here indicate that Rad59p, Srs2p, and Rad50p all probably participate in that pathway as well.
RAD51- and RAD59-mediated recombination pathways have different outcomes.
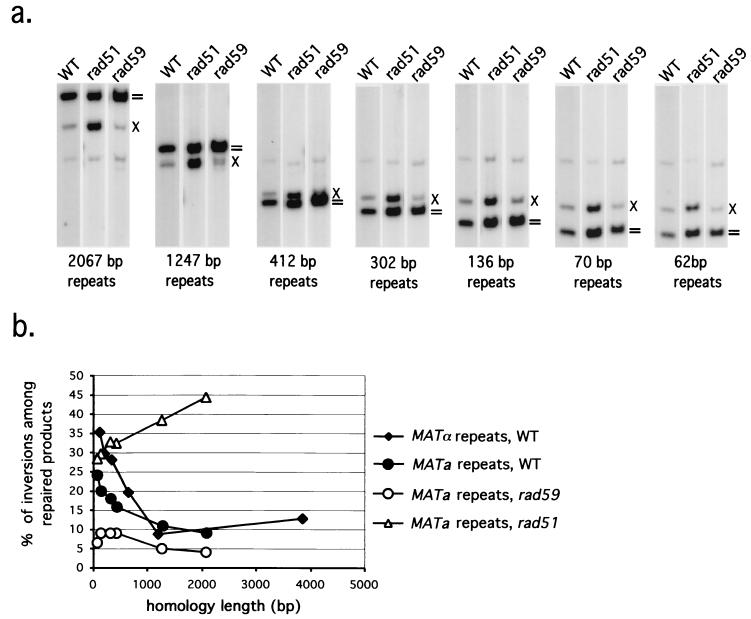
The percentage of crossover events was determined by densitometry from Southern blots for two series of plasmids carrying either MATα or MATa inverted repeats of various lengths (Fig. 5). Previous studies of HO-induced mitotic recombination between chromosomes have shown that mitotic RAD51-dependent gene conversion is associated with crossing over less than 10% of the time (24, 31). In contrast, if RAD51-independent repair occurs by BIR-SSA, then there should be equal numbers of crossover and noncrossover products (17) (see Fig. 1). If DSBs within short homologous sequences are repaired only by BIR-SSA, then we should observe a higher level of crossing over when repeats are shorter. As shown in Fig. 5, crossing over accompanied gene conversion only 10% of the time when homology was greater than approximately 500 bp on either side of the DSB, but the percentage of exchanges went up progressively as the length of homology decreased, culminating in approximately 25 to 35% crossovers when there was less than 100 bp on each side of the break. The increase in crossing over appears to correlate with the proportion of recombination events that rely on the RAD59-, RAD50-mediated pathway. This is also seen with mutants that eliminate one or the other process. In rad59Δ the level of crossover is very low (5 to 9%) for the entire set of plasmids, and in rad51Δ the crossover level is substantially higher (28 to 45%) (Fig. 4). These data strongly support a BIR-SSA model for RAD51-independent repair between inverted repeats on plasmids.
FIG. 5.
The effect of homology length on the frequency of crossing over accompanying gene conversion. (a) Crossover frequencies were determined by densitometry of Southern blots for a set of strains with plasmids carrying inverted repeats of MATa and MATa-inc. Wild type (WT), rad59Δ, and rad51Δ mutants were analyzed. Crossover product bands are indicated by multiplication signs, and gene conversion without crossover product bands are indicated by equal signs. (b) The percentage of crossover products was plotted against the length of homology for the plasmids shown in panel a and a set of plasmids containing inverted repeats of MATα and MATα-inc.
DISCUSSION
Rad51 is a key protein in recombination in all eukaryotic organisms, but previous studies had suggested that DSB repair could occur without Rad51p when the two repeats were on a plasmid (46). It has been shown that essentially all intermolecular DSB-induced gene conversions in yeast—between two chromosomes or between two plasmids—depend on Rad51p (Table 2), and it is therefore likely that bona fide gene conversions on a plasmid also require RAD51 (2, 10). It was previously reported that recombination between a chromosome and a plasmid could occur without Rad51 (46). We now believe that this occurred by SSA and not by gene conversion, because the break on the chromosome could be repaired by Rad51p-independent annealing to the intact donor sequence on a linearized plasmid.
There are, however, RAD51-independent repair events that can occur between chromosomes, including both BIR of a DSB in the middle of a chromosome and the related process of recombination-dependent maintenance of telomeres in the absence of telomerase.
Our data strongly support the idea that the RAD51-independent events occurring within a plasmid with inverted repeats also involves BIR followed by SSA (Fig. 1) (17). It is known that SSA and BIR do not require Rad51p. Here we demonstrate that RAD51-independent intraplasmid recombination relies on the same genes that were previously shown to be involved in RAD51-independent telomere maintenance and in RAD51-independent BIR repair of a DSB in the middle of a chromosome: RAD50, RAD59, and TID1. Consistent with this mechanism, when plasmid substrates have very short homology so that only the RAD50/RAD59-dependent pathway is functional, recovery of recombined plasmids was strongly dependent on Rad1, Msh2, and Msh3 proteins previously shown to be required to remove long 3′-ended ssDNA tails produced during SSA. Rad1p is also needed in the absence of Rad51p when homologous repeats are long (17). RAD51-dependent recombination occurs by a process that does not require SSA, and thus Rad1p is not necessary. This process appears to be best explained by gene conversion (31).
The idea that intraplasmid repair is frequently accomplished by BIR-SSA is also supported by Southern blot analysis of the kinetics of HO-induced DSB repair (7, 40), where the two restriction fragments characteristic of reciprocal crossing over appear asynchronously, with one crossover band visible an hour before the second. If all crossovers arose by resolution of a Holliday junction intermediate, the two products should appear simultaneously. A restriction fragment identical to one of the two crossover products would be produced if there was strand invasion preferentially from one of the two DSB ends, leading to the establishment of a replication fork (Fig. 1).
Consistent with the idea that BIR would be followed by SSA is the observation that the ratio of crossover to noncrossover products is much more equivalent in the RAD50/RAD59-dependent pathway. If all BIR events succeeded in reaching the end of the template before dissociating, SSA would produce an equal number of the two outcomes. However, it is quite likely that some BIR events dissociate before reaching the end of the template (19, 33), in which case there will be more noncrossover than crossover products, as we did in fact observe (Fig. 5). In addition, when the homologies are short, 5′ to 3′ resection of the DSB ends will leave a template in which the BIR replication fork will stop before it copies the homologous region; SSA will then favor noncrossover products. The very low level of crossing over seen with RAD51-mediated events is consistent with many observations of the level of crossing over associated with DSB-mediated interchromosomal gene conversions (which require RAD51) (13, 24, 31).
Homology and genetic requirements for gene conversion and BIR.
Given the above results, we used the inverted-repeat plasmid system to examine the homology and genetic requirements for RAD51-dependent (presumably gene conversion) and RAD51-independent (presumably BIR-SSA) events. One of the most striking conclusions is that recombination of short inverted repeats (29 to 37 bp on each DSB side) is preferentially carried out by a recombination process that employs Rad52p, Rad59p, Rad50p, Mre11p, Xrs2p, and Tid1p. In fact, with substrates of this size the presence of Rad51p is surprisingly inhibitory, so that a rad51Δ strain allows the plasmid with 33 bp on either side of the DSB to recombine 60% as efficiently as one with more than 1 kb on either side.
We note that SSA itself is highly efficient even with 33 bp of homology. Our previous estimation of homology necessary for SSA on a chromosome showed low efficiency of repair with such small repeats (45). This difference might be due to the presence of two nonhomologous tails in the chromosomal version of this experiment, whereas there is only one on the plasmid. Another possibility is that annealing of such short repeats can be facilitated when they are on one molecule.
As shown in Fig. 3, RAD51-mediated (i.e., RAD59- and SRS2-independent) recombination requires approximately 70 to 100 bp of homology on each side of the DSB. We interpret this result to suggest that Rad51p forms a filament on ssDNA with 30 nt (10 monomers of Rad51p), but Rad51p and its auxiliary proteins—Rad52p, Rad54p, Rad55p, and Rad57p—are incapable of carrying out strand invasion on substrates that are smaller than 70 to 100 nt (23 to 33 Rad51p monomers). To our knowledge these are the first measurements for mitotic, DSB-induced recombination of the minimum size of homology (sometimes referred to as the minimum effective processing segment [41]). It is difficult to compare this result with a previous estimate of 250 bp of homology that is required for spontaneous intermolecular recombination (15), because we do not know the nature and position of the initiating lesion.
RAD51-dependent and -independent telomere maintenance without telomerase.
Our findings here allow us to explain why there are two recombination-dependent pathways of telomere lengthening in the absence of telomerase. The type I events require RAD51 and most often lead to the proliferation of long (5.2 or 6.7 kb) Y′ subtelomeric regions, whereas type II events seem to involve recombination of the irregular TG1-3 sequences themselves. We suggest that Rad51p-bound single-stranded TG1-3 regions cannot locate sufficient patches of near-perfect homology within the telomere sequences themselves; hence, Rad51p-mediated telomere maintenance will depend on recombination between adjacent, longer homologous sequences, including Y′, X, and other subtelomeric sequences. In a strain with very few Y′ sequences there are virtually no type I events even in the presence of Rad51p (21). In contrast, the Rad59p-, Rad50p-dependent recombination machinery is better suited to finding short, sufficiently homologous regions within TG1-3 of telomere sequence to permit the initiation of recombination. In both instances recombination proceeds by BIR, as the resected chromosome ends have only one end with which to initiate repair. In support of this idea Bucholc et al. (3) have found that collapse of very long telomere regions by recombination (presumably within TG1-3 repeats) also depends on RAD50.
Molecular activities of the Rad50, Rad59 pathway.
How exactly recombination takes place in the absence of a bona fide strand exchange protein such as Rad51p is not yet known. Three proteins or complexes in this study have ssDNA annealing activity (Rad52p, Rad59p, and the Mre11p-Rad50p-Xrs2p [M-R-X] complex) (8, 9, 29, 36, 43). In addition, the mammalian equivalent of the M-R-X complex has at least limited DNA unwinding activity (34), as does the 3′ to 5′ helicase Srs2p (18, 39). Tid1p, related to the SWI2/SNF2 family of chromatin remodeling proteins, also may unwind DNA (37). Thus, this set of proteins may be able to unwind a template and allow the assimilation of a 3′-ended ssDNA strand to set up the initial stages of homologous recombination.
It is undoubtedly simplistic to think that the components of the RAD50/RAD59-dependent DSB repair pathway act only in this regard and have no role in Rad51p-mediated activities. Certainly this is the case for both the M-R-X complex and Srs2p. When a DSB is made on a chromosome and is repaired with a homologous chromosome as the donor, gene conversions (as opposed to BIR) can occur only by a Rad51p-dependent process. Deleting RAD50 reduces the efficiency of gene conversion by 30% and alters the size of gene conversion tracts (25). In contrast, tid1Δ does not affect gene conversion repair in this assay. Moreover, when the impact of rad50Δ on intraplasmid recombination was studied (Fig. 4), both Rad59p- and Rad51p-mediated recombination appear to be affected. By these same criteria, Srs2p also seems to have a role in both pathways facilitating Rad51p-dependent and -independent recombination between short repeats.
Acknowledgments
We thank E. Coïc for the kind gift of the arg5,6::MATa-HYG cassette. N. Sugawara and A. Malkova offered insightful comments on the paper.
This research was supported by National Institutes of Health grant GM 20056.
REFERENCES
- 1.Bai, Y., and L. S. Symington. 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10:2025-2037. [DOI] [PubMed] [Google Scholar]
- 2.Bartsch, S., L. E. Kang, and L. S. Symington. 2000. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol. Cell. Biol. 20:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucholc, M., Y. Park, and A. J. Lustig. 2001. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:6559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Q., A. Ijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clever, B., H. Interthal, J. Schmuckli-Maurer, J. King, M. Sigrist, and W. D. Heyer. 1997. Recombinational repair in yeast: functional interactions between Rad51 and Rad54 proteins. EMBO J. 16:2535-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, H., and D. A. Sinclair. 2001. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl. Acad. Sci. USA 98:3174-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colaiacovo, M. P., F. Paques, and J. E. Haber. 1999. Removal of one nonhomologous DNA end during gene conversion by a RAD1- and MSH2-independent pathway. Genetics 151:1409-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, A. P., and L. S. Symington. 2001. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics 159:515-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Jager, M., M. L. Dronkert, M. Modesti, C. E. Beerens, R. Kanaar, and D. C. van Gent. 2001. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res. 29:1317-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elias-Arnanz, M., A. A. Firmenich, and P. Berg. 1996. Saccharomyces cerevisiae mutants defective in plasmid-chromosome recombination. Mol. Gen. Genet. 252:530-538. [DOI] [PubMed] [Google Scholar]
- 11.Fishman-Lobell, J., and J. E. Haber. 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258:480-484. [DOI] [PubMed] [Google Scholar]
- 12.Hays, S. L., A. A. Firmenich, and P. Berg. 1995. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl. Acad. Sci. USA 92:6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inbar, O., B. Liefshitz, G. Bitan, and M. Kupiec. 2000. The relationship between homology length and crossing over during the repair of a broken chromosome. J. Biol. Chem. 275:30833-30838. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, H., Y. Xie, P. Houston, K. Stemke-Hale, U. H. Mortensen, R. Rothstein, and T. Kodadek. 1996. Direct association between the yeast Rad51 and Rad54 recombination proteins. J. Biol. Chem. 271:33181-33186. [DOI] [PubMed] [Google Scholar]
- 15.Jinks-Robertson, S., M. Michelitch, and S. Ramcharan. 1993. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:3937-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, F. B., R. A. Marciniak, M. McVey, S. A. Stewart, W. C. Hahn, and L. Guarente. 2001. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 20:905-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang, L. E., and L. S. Symington. 2000. Aberrant double-strand break repair in rad51 mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 20:9162-9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein, H. L. 1997. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics 147:1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraus, E., W. Y. Leung, and J. E. Haber. 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louis, E. J., and J. E. Haber. 1990. The subtelomeric Y' repeat family in Saccharomyces cerevisiae: an experimental system for repeated sequence evolution. Genetics 124:533-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 23.Malkova, A., E. L. Ivanov, and J. E. Haber. 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93:7131-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malkova, A., F. Klein, W. Y. Leung, and J. E. Haber. 2000. HO endonuclease-induced recombination in yeast meiosis resembles Spo11-induced events. Proc. Natl. Acad. Sci. USA 97:14500-14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malkova, A., L. Ross, D. Dawson, M. F. Hoekstra, and J. E. Haber. 1996. Meiotic recombination initiated by a double-strand break in rad50 delta yeast cells otherwise unable to initiate meiotic recombination. Genetics 143:741-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malkova, A., L. Signon, C. B. Schaefer, M. L. Naylor, J. F. Theis, C. S. Newlon, and J. E. Haber. 2001. RAD51-independent break-induced replication to repair a broken chromosome depends on a distant enhancer site. Genes Dev. 15:1055-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazin, A. V., C. J. Bornarth, J. A. Solinger, W. D. Heyer, and S. C. Kowalczykowski. 2000. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell 6:583-592. [DOI] [PubMed] [Google Scholar]
- 28.Moore, J. K., and J. E. Haber. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortensen, U. H., C. Bendixen, I. Sunjevaric, and R. Rothstein. 1996. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. USA 93:10729-10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogawa, T., X. Yu, A. Shinohara, and E. H. Egelman. 1993. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science 259:1896-1899. [DOI] [PubMed] [Google Scholar]
- 31.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paques, F., and J. E. Haber. 1997. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6765-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paques, F., W. Y. Leung, and J. E. Haber. 1998. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol. 18:2045-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paull, T. T., and M. Gellert. 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13:1276-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petukhova, G., S. Stratton, and P. Sung. 1998. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature 393:91-94. [DOI] [PubMed] [Google Scholar]
- 36.Petukhova, G., S. A. Stratton, and P. Sung. 1999. Single strand DNA binding and annealing activities in the yeast recombination factor Rad59. J. Biol. Chem. 274:33839-33842. [DOI] [PubMed] [Google Scholar]
- 37.Petukhova, G., P. Sung, and H. Klein. 2000. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 14:2206-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petukhova, G., S. Van Komen, S. Vergano, H. Klein, and P. Sung. 1999. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem. 274:29453-29462. [DOI] [PubMed] [Google Scholar]
- 39.Rong, L., and H. L. Klein. 1993. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 268:1252-1259. [PubMed] [Google Scholar]
- 40.Rudin, N., E. Sugarman, and J. E. Haber. 1989. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics 122:519-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen, P., and H. V. Huang. 1986. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112:441-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shinohara, A., and T. Ogawa. 1998. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391:404-407. [DOI] [PubMed] [Google Scholar]
- 43.Shinohara, A., M. Shinohara, T. Ohta, S. Matsuda, and T. Ogawa. 1998. Rad52 forms ring structures and cooperates with RPA in single-strand DNA annealing. Genes Cells 3:145-156. [DOI] [PubMed] [Google Scholar]
- 44.Signon, L., A. Malkova, M. L. Naylor, H. Klein, and J. E. Haber. 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 21:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugawara, N., G. Ira, and J. E. Haber. 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugawara, N., E. L. Ivanov, J. Fishman-Lobell, B. L. Ray, X. Wu, and J. E. Haber. 1995. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature 373:84-86. [DOI] [PubMed] [Google Scholar]
- 47.Sugawara, N., F. Paques, M. Colaiacovo, and J. E. Haber. 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA 94:9214-9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sung, P. 1997. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11:1111-1121. [DOI] [PubMed] [Google Scholar]
- 49.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 50.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]