Abstract
SUMOs are small ubiquitin-related polypeptides that are reversibly conjugated to many nuclear proteins. Although the number of identified substrates has grown rapidly, relatively little is still understood about when, where, and why most proteins are modified by SUMO. Here, we demonstrate that enzymes involved in the SUMO modification and demodification of proteins are components of the nuclear pore complex (NPC). We show that SENP2, a SUMO protease that is able to demodify both SUMO-1 and SUMO-2 or SUMO-3 protein conjugates, localizes to the nucleoplasmic face of the NPC. The unique amino-terminal domain of SENP2 interacts with the FG repeat domain of Nup153, indicating that SENP2 associates with the nucleoplasmic basket of the NPC. We also investigated the localization of the SUMO conjugating enzyme, Ubc9. Using immunogold labeling of isolated nuclear envelopes, we found that Ubc9 localizes to both the cytoplasmic and the nucleoplasmic filaments of the NPC. In vitro binding studies revealed that Ubc9 and SUMO-1-modified RanGAP1 bind synergistically to form a trimeric complex with a component of the cytoplasmic filaments of the NPC, Nup358. Our results indicate that both SUMO modification and demodification of proteins may occur at the NPC and suggest a connection between the SUMO modification pathway and nucleocytoplasmic transport.
SUMOs are small ubiquitin-like modifiers that are posttranslationally conjugated to a wide range of proteins in the cell (21). In vertebrates, there are three related SUMO proteins, SUMO-1, SUMO-2, and SUMO-3. All three SUMOs are linked to proteins through an isopeptide bond formed between their carboxyl-terminal glycine residues and a lysine(s) in the substrate. The majority of proteins modified by SUMOs reside in the nucleus. Known substrates include transcription factors, components of promyelocytic leukemia (PML) nuclear bodies, and chromatin-associated proteins (21). The specific effects of SUMO modification on these proteins are at least in part substrate dependent. SUMO-1 modification targets RanGAP1 to the nuclear pore complex (NPC) but targets PML to PML nuclear bodies (18, 20, 23). SUMO-1 modification also regulates the DNA binding activities of the heat shock transcription factors HSF1 and HSF2 (7, 9), and several examples indicate that it may function to stabilize proteins by acting as an antagonist to ubiquitin-mediated proteolysis (4). In a majority of instances, however, the specific effects of SUMO modification on targeted substrates are not known.
The mechanisms by which SUMOs are conjugated to their substrates closely parallel the mechanisms of ubiquitination. All three SUMOs appear to use the same heterodimeric E1 activating enzyme, Aos1/Uba2, and the same E2 conjugating enzyme, Ubc9 (21). Ubc9 is unique from ubiquitin E2 conjugating enzymes in that it is directly involved in substrate recognition and binding. Nearly all SUMO-modified proteins identified to date contain a conserved motif that surrounds the modified lysine. Structural and mutational analyses of this motif indicate that it is recognized directly by Ubc9 (2, 28, 34). The direct interaction between Ubc9 and SUMO substrates may preclude an absolute requirement for E3-like factors. However, several factors that enhance Ubc9-mediated SUMO modification have been identified, including the PIAS family of proteins and Nup358 (10, 12, 26, 30, 39). Nup358 is a component of the cytoplasmic filaments of the NPC, suggesting that SUMO substrates may be modified in connection with their nucleocytoplasmic transport (26). The majority of the SUMO E1 and E2 enzymes are localized in the nucleoplasm, however, indicating that SUMO modification is also likely to occur inside the nucleus (28).
The modification of substrates by SUMO is reversible and is mediated by a family of SUMO proteases. The hallmark of this family of proteases is a highly conserved carboxyl-terminal domain of ∼200 amino acids that contains the active site of the enzymes (16). The budding yeast Saccharomyces cerevisiae contains two members of this protease family, Ulp1 and Ulp2 (SMT4) (16, 17, 35), whereas vertebrates contain at least seven genes that encode related proteases (41). Each of the vertebrate proteases contains the highly conserved catalytic domain near the carboxyl terminus and extensive and divergent amino-terminal domains. Although the specific biological activities of the individual proteases remain to be determined, it is interesting that each of the vertebrate SUMO proteases characterized to date localizes to a distinct subcellular domain. SMT3IP1 (also known as SENP3) localizes to the nucleolus (25), SUSP1 (also known as SENP6), localizes predominantly to the cytoplasm (13), and SENP1 localizes to the nucleoplasm (6). The fourth vertebrate SUMO protease to be characterized, Axam (also known as SENP2), appears to exist as two different isoforms derived from alternatively spliced mRNAs (11, 24). The two isoforms differ in their amino-terminal domains, with one isoform containing an additional 47 amino acids at its extreme amino terminus (24). Both SENP2 isoforms interact with Axin, a regulator of the Wnt signaling pathway, and therefore have been implicated as possible regulators of β-catenin degradation (11, 24). The unique localizations of the known SUMO proteases suggest that these enzymes may have specialized functions and may regulate specific subsets of SUMO substrates.
A SUMO protease activity that copurifies with the NPC was previously detected (19). Here, we report the identification of this protease as the SUMO protease SENP2. We demonstrate that full-length SENP2 associates with the nucleoplasmic face of the NPC and that residues in its unique amino-terminal domain function to target it there. This domain of SENP2 interacts with the carboxyl-terminal FG repeat domain of Nup153, indicating that SENP2 associates with the nucleoplasmic basket of the NPC. Our results indicate that SENP2 functions at the NPC to process SUMO precursors and/or SUMO protein conjugates, possibly in connection with nucleocytoplasmic transport.
We have also characterized the association of the SUMO conjugating enzyme, Ubc9, with the NPC. Previous reports indicated that Ubc9 is concentrated at the nuclear envelope of cultured mammalian cells (15) and that it associates with Nup358 in Xenopus laevis egg extracts (32). However, the precise association of Ubc9 with the intact NPC has not been analyzed yet. We have localized Ubc9 by immunogold labeling of isolated nuclear envelopes and found that Ubc9 is associated with both the cytoplasmic and the nucleoplasmic filaments of the NPC. Using in vitro binding assays, we have demonstrated that Ubc9 is able to interact synergystically with SUMO-1-modified RanGAP1 to form a trimeric complex with Nup358, a component of the cytoplasmic filaments of the NPC. Together, our data indicate that both SUMO modification and demodification of substrates may occur at the NPC, possibly in association with nucleocytoplasmic transport.
MATERIALS AND METHODS
cDNA cloning and plasmid construction.
A human SENP2 cDNA clone was amplified from a fetal brain cDNA library (Clontech, Palo Alto, Calif.) by PCR. Isolated clones were sequenced and found to be identical to a previously reported sequence for SENP2 (GenBank accession number AK027599). The SENP2 cDNA was cloned into the mammalian expression vector pcDNA3 (Invitrogen, Carlsbad, Calif.), previously engineered to contain an in-frame amino-terminal myc epitope tag (37). Amino-terminal deletions of SENP2 were cloned into the same vector. Deletions were generated by using the full-length SENP2 cDNA as a template and unique site-specific primers for PCR. To generate plasmids for expressing pyruvate kinase-SENP2 fusion proteins, various domains of SENP2 were amplified by PCR and inserted into the previously described pcDNA3-myc-PK vector (37). The full-length SENP2 cDNA clone was also subcloned into the expression vector pQE30 (Qiagen, Inc., Valencia, Calif.). A fragment of SENP2 coding for the amino-terminal 63 amino acids was subcloned into the expression vector pGEX-4T-1 (Amersham Pharmacia Biotech, Piscataway, N.J.).
To assay for SUMO carboxyl-terminal hydrolase activity, the SUMO-1 and SUMO-3 precursor proteins were engineered to have carboxyl-terminal extensions consisting of 10 additional alanine residues. These alanines were added by cloning a DNA fragment to the 3′ ends of the SUMO-1 and SUMO-3 precursor cDNAs by PCR. The PCR products were cloned into the expression vector pQE30 for bacterial expression. Plasmids for expressing glutathione S-transferase (GST)-NΔ419 (amino acids 420 to 589 of mouse RanGAP1), mature SUMO-1, Ubc9, GST-Nup358 (amino acids 2596 to 2836 of human Nup358), and Nup98 were previously described (5, 31, 34). Plasmids for expressing human Nup153 and human Tpr were kind gifts from Brian Burke (University of Florida, Gainesville) and Larry Gerace (Scripps Research Institute, La Jolla, Calif.). Plasmids for in vitro translation of the amino-terminal domain (amino acids 1 to 609), the zinc finger domain (amino acids 610 to 869), and the carboxyl-terminal domain (amino acids 870 to 1475) of Nup153 were produced by cloning PCR-generated DNA fragments into the vector pCR2.1 TOPO (Invitrogen).
Protein expression and purification.
Recombinant full-length SENP2 was produced in bacteria by inducing expression with 0.5 mM isopropyl-β-d-galactopyranoside (IPTG) at 16°C for 12 h and was purified by nickel affinity chromatography. SUMO-1 and SUMO-3 precursor proteins were expressed by induction with 0.5 mM IPTG at 37°C for 4 h and were purified by nickel affinity chromatography under standard conditions. GST, a GST-SENP2 fusion protein containing just the amino-terminal 63 amino acids of SENP2 [GST-SENP2(1-63)], GST-Nup358 (amino acids 2596 to 2836), GST-NΔ419, and GST-Ubc9 were expressed in bacteria under standard conditions, purified by affinity chromatography on glutathione-Sepharose beads (Amersham Pharmacia Biotech), and eluted from the beads with 10 mM reduced glutathione or by cleavage with the appropriate protease.
Recombinant SUMO-modified NΔ419 was generated in vitro by using recombinant NΔ419 and SUMO-1 or SUMO-3 and a modification reaction containing recombinant SUMO E1 and E2 enzymes as previously described (2). SUMO-modified NΔ419 was purified from the reaction by separation on a Mono-Q column (Amersham Pharmacia Biotech).
In vitro transcription and translation of Nup153, Nup98, Tpr, and domains of Nup153 were performed with rabbit reticulocyte lysates in the presence of [35S]methionine as described by the manufacturer (Promega Corp., Madison, Wis.).
In vitro SENP2 enzyme assays and protein binding assays.
To assay for carboxyl-terminal hydrolase activity, the SUMO-1 or SUMO-3 precursor protein (10 μM) was incubated with purified SENP2 (0.2 μM) at 30°C for 1 h in buffer containing 10 mM Tris (pH 8.0), 150 mM NaCl, and 1 mM dithiothreitol. Reactions were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) followed by Coomassie blue staining. The processing of SUMO-modified RanGAP1 was similarly assayed by incubating SUMO-1- or SUMO-3-modified NΔ419 (4 μM) with purified SENP2 (0.1 μM) for 1 h at 30οC and then analyzing the reactions by SDS-PAGE.
To identify nucleoporins that interact with SENP2, GST-SENP2(1-63) or GST alone was bound to 20 μl of glutathione-Sepharose beads (1 mg of protein/ml of beads) in phosphate-buffered saline (PBS) containing 1 mM dithiothreitol. Nonspecific protein binding sites were blocked by incubation with 2% bovine serum albumin for 20 min at 4°C. Equivalent amounts of each in vitro-translated protein were incubated with the beads in 100 μl of binding buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 0.1% Tween 20) for 30 min at room temperature. The beads were washed three times with binding buffer, followed by elution of the bound proteins with SDS sample buffer.
To characterize interactions among Ubc9, RanGAP1 NΔ419, and Nup358, GST-Nup358 (amino acids 2596 to 2836) or GST as a control was bound to 20 μl of glutathione beads (0.2 mg of protein/ml of beads) in binding buffer. Nonspecific protein binding sites were blocked as described above. After two washes with binding buffer, 10 μg of each recombinant protein was incubated with the beads in 100 μl of binding buffer for 30 min at room temperature. The beads were washed three times with binding buffer, followed by elution with SDS sample buffer. Binding reactions were analyzed by SDS-PAGE.
Cell culturing, transfection, and immunofluorescence microscopy.
HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Cells were grown on glass coverslips in 35-mm dishes and transfected with 2 μg of plasmid by using Lipofectamine (GIBCO BRL, Gaithersburg, Md.) as described by the manufacturer. At 36 h posttransfection, the cells were washed twice with PBS, fixed for 30 min at room temperature with 2% formaldehyde in PBS, and permeabilized with −20°C acetone for 3 min or with 10 μg of digitonin (Aldrich Chemical Co., Milwaukee, Wis.)/ml in PBS for 10 min. After three rinses with PBS, the cells were incubated with the anti-myc monoclonal antibody 9E10 for 1 h at room temperature, followed by three washes with PBS. The cells were subsequently incubated with Alexa-488-conjugated goat anti-mouse antibody (Molecular Probes, Eugene, Oreg.) for 30 min at room temperature. Alternatively, the cells were incubated with a rabbit anti-myc antibody (Cell Signaling Technology) followed by Alexa-594-conjugated goat anti-rabbit antibody (Molecular Probes). After three washes with PBS, coverslips were mounted and analyzed with a Zeiss Axiovert fluorescence microscope or a Zeiss LSM410 confocal microscope. For double-labeling experiments, the indicated primary antibodies were coincubated with the anti-myc antibodies, followed by coincubation with the appropriate secondary antibodies. RanGAP1 was detected by using the monoclonal antibody 19C7 (19). Lamin B was detected by using a rabbit polyclonal antibody (22).
Immunogold electron microscopy.
Rat liver nuclear envelopes were prepared and processed for immunogold labeling and analysis as previously described (19). Antibodies were generated by injecting rabbits with full-length human Ubc9 purified from bacteria.
RESULTS
SENP2 is an NPC-associated SUMO protease.
A SUMO protease activity that fractionates with mammalian NPCs and that is able to remove SUMO-1 from RanGAP1 was previously detected. This activity was sensitive to N-ethylmaleimide, suggesting that the protease could be a member of the Ulp1-related cysteine protease family (19). Consistent with this prediction, SENP2 was identified in a proteomic analysis of a highly enriched NPC fraction by mass spectrometry (3a). SENP2 is a 590-amino-acid protein that contains an ∼200-amino-acid carboxyl-terminal domain that shares significant sequence homology with the catalytic domains of Ulp1 and other characterized SUMO proteases (41). The amino-terminal 390 amino acids of SENP2 are unrelated to other proteins in the currently available databases.
To provide further evidence that SENP2 is associated with NPCs, we analyzed its localization in cells transiently transfected with a plasmid coding for a myc epitope-tagged fusion protein. The localization of SENP2 was determined by indirect immunofluorescence with an anti-myc antibody. When expressed at low to moderate levels, SENP2 localized predominantly to the nuclear envelope (intranuclear foci appeared in cells expressing high levels of SENP2). When we focused on the midplane of the nucleus, the SENP2 signal appeared as a discontinuous ring around the periphery of the nucleus (Fig. 1a), whereas the signal appeared as punctate dots when the focal plane was adjusted to view the surface of the nucleus (Fig. 1b). These specific localization patterns are consistent with the finding that SENP2 copurifies with isolated NPCs and indicate that SENP2 localizes to NPCs in vivo.
FIG. 1.
SENP2 localizes to the NPC. (a) SENP2 localized to a discontinuous ring around the nuclear periphery, as evident when we focused on the midplane of the nucleus. (b) When we focused on the surface of the nucleus, the SENP2 signal appeared as punctate dots, indicative of localization to the NPC. Bar, 5 μm.
SENP2 localizes to the nucleoplasmic side of the NPC.
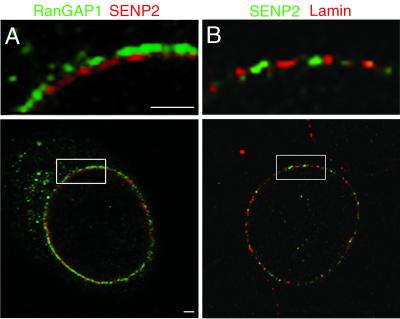
NPCs are composed of a highly symmetric central domain but also contain unique sets of protein filaments that localize asymmetrically to either the cytoplasmic or the nucleoplasmic face of the NPC (40). To further refine the localization of SENP2 at the NPC, we performed double-label immunofluorescence confocal microscopy and determined the localization of SENP2 relative to those of RanGAP1 and lamin B. RanGAP1 is associated with the distal tips of the cytoplasmic filaments of the NPC (19) and therefore provides a marker for the cytoplasmic face of the NPC. When RanGAP1 and myc-tagged SENP2 were colocalized in transiently transfected cells, both proteins could be detected as a discontinuous ring around the nuclear periphery (Fig. 2A, bottom panel). When viewed at a high magnification, the signals for the proteins could be seen to be nonoverlapping in many regions, with the SENP2 signal consistently present in a ring internal to the RanGAP1 signal (Fig. 2A, top panel). We also examined the relative localizations of SENP2 and a component of the nuclear lamina, the lamin B protein. When colocalized in transiently transfected cells, the SENP2 and lamin B signals appeared nearly coincident (Fig. 2B). Together, these data suggest that SENP2 localizes to the nucleoplasmic face of the NPC.
FIG. 2.
SENP2 and RanGAP1 localize to distinct domains of the NPC. (A) Cells were probed simultaneously with a rabbit anti-myc antibody and a mouse monoclonal antibody specific for RanGAP1. RanGAP1 (green) localizes to the filaments on the cytoplasmic face of the NPC (19). The signal for SENP2 (red) is distinct from the RanGAP1 signal and localizes toward the nucleoplasmic side of the NPC. The highlighted box is shown at a fourfold-higher magnification in the top panel. (B) Cells were probed simultaneously with a mouse anti-myc monoclonal antibody and a rabbit antibody specific for lamin B. The SENP2 signal (green) and the lamin signal (red) are coincident at the nucleoplasmic face of the nuclear envelope. The highlighted box is shown at a fourfold-higher magnification in the top panel. Bars, 1 μm.
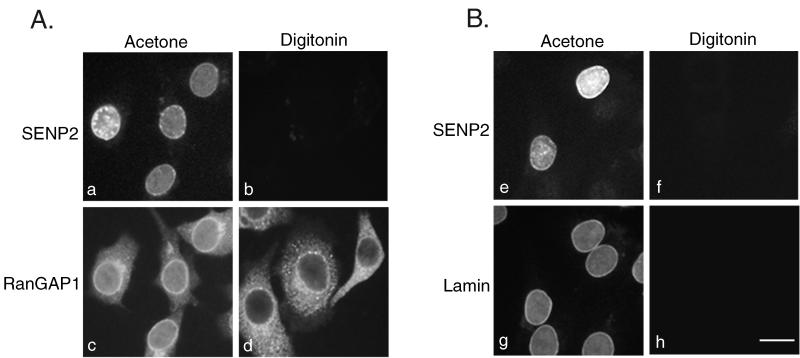
To provide further evidence that SENP2 is asymmetrically localized to the nucleoplasmic face of the NPC, we also performed immunofluorescence analysis on transiently transfected cells selectively permeabilized with digitonin or permeabilized with acetone. Under the specific conditions used, digitonin permeabilizes the plasma membrane but not the nuclear envelope, whereas acetone permeabilizes all the membranes. When digitonin-permeabilized cells were probed with an anti-myc antibody to detect SENP2, no signal could be detected in any cells (Fig. 3A, panel b). In contrast, nuclear envelope localization could be detected in transfected cells permeabilized with acetone (Fig. 3A, panel a). As a control for plasma membrane permeabilization, the same cells were also probed with an antibody specific for RanGAP1. Because RanGAP1 localizes to the cytoplasm and to the cytoplasmic filaments of the NPC, it could be detected in both digitonin-permeabilized cells (Fig. 3A, panel d) and acetone-permeabilized cells (Fig. 3A, panel c). As an additional control, we also probed cells with an antibody specific for lamin B. Because of its nuclear localization, lamin B was not detected in cells permeabilized with digitonin (Fig. 3B, panel h) but was readily detected in cells permeabilized with acetone (Fig. 3B, panel g). SENP2 accessibility mirrored that of lamin B (Fig. 3B, panels e and f). These data provide further evidence that SENP2 localizes to the nucleoplasmic face of the NPC.
FIG. 3.
SENP2 localizes to the nucleoplasmic face of the NPC. (A) Cells were fixed, permeabilized with either acetone (panels a and c) or digitonin (panels b and d), and probed simultaneously with a rabbit anti-myc antibody and a mouse monoclonal antibody specific for RanGAP1. RanGAP1 (panels c and d) localizes to the cytoplasm and to the filaments on the cytoplasmic face of the NPC (19) and is detectable in cells permeabilized with both acetone and digitonin. SENP2 (panels a and b) localizes to the nucleoplasmic face of the NPC and is detectable only in cells permeabilized with acetone. (B) Cells were fixed, permeabilized with either acetone (panels a and c) or digitonin (panels b and d), and probed simultaneously with a mouse anti-myc monoclonal antibody and a rabbit antibody specific for lamin B. SENP2 (panels e and f) and lamin B (panels g and h) are detectable only in cells permeabilized with acetone. Bar, 10 μm.
The amino terminus of SENP2 contains nuclear localization and NPC localization signals.
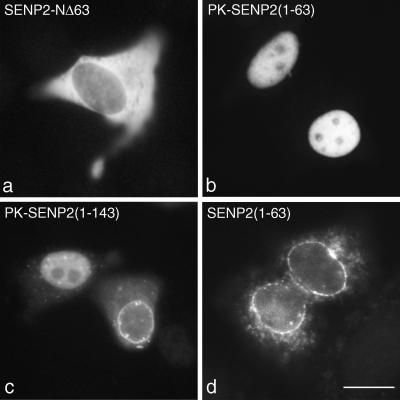
Remarkably, each of the different SUMO proteases characterized to date localizes to a unique subcellular domain (6, 13, 25). To identify the region of SENP2 that is responsible for its specific localization to the nucleoplasmic face of the NPC, we created a series of constructs expressing proteins with amino-terminal deletions and determined the subcellular localizations of the truncated SENP2 proteins in vivo. Deletion of just the first 63 amino acids of SENP2 resulted in a protein that no longer localized to the NPC and that was restricted to the cytoplasm (Fig. 4a). This finding suggests that the first 63 amino acids of SENP2 contain a nuclear localization signal (NLS). To test this notion, we fused these 63 residues to pyruvate kinase, a reporter protein that normally localizes to the cytosol (data not shown). Upon transient transfection of HeLa cells, the resulting fusion protein, PK-SENP2(1-63), localized exclusively to the nucleus (Fig. 4b), demonstrating that the amino-terminal 63 amino acids of SENP2 contain an NLS.
FIG. 4.
The amino-terminal domain of SENP2 contains an NLS and an NPC localization signal. (a) Cells were transfected with a plasmid coding for a form of SENP2 lacking the first 63 amino acids (SENP2-NΔ63). Deletion of this amino-terminal region of SENP2 caused the remaining protein to mislocalize to the cytoplasm. (b) Cells were transfected with a plasmid coding for the first 63 amino acids of SENP2 fused to the carboxyl terminus of pyruvate kinase [PK-SENP2(1-63)]. This region of SENP2 targeted pyruvate kinase to the nucleoplasm but not to the NPC. (c) Cells were transfected with a plasmid coding for the first 143 amino acids of SENP2 fused to the carboxyl terminus of pyruvate kinase [PK-SENP2(1-143)]. The expressed fusion protein localized to both the nucleoplasm and the NPC. (d) Cells were transfected with a plasmid coding for the first 63 amino acids of SENP2 fused to a myc tag [SENP2(1-63)]. The expressed fusion protein localized to the NPC. Bar, 10 μm.
PK-SENP2(1-63) did not appear to localize to the NPC, suggesting that additional residues of SENP2 are required for proper NPC targeting. In order to further define the region of SENP2 that determines NPC localization, we fused progressively longer amino-terminal regions of SENP2 to pyruvate kinase. As expected, a pyruvate kinase fusion protein containing the amino-terminal 143 residues of SENP2 localized to the nucleus (Fig. 4c). However, this fusion protein also clearly localized to the NPC in approximately 30% of the transfected cells (Fig. 4c). We reasoned that the pyruvate kinase domain could be interfering with NPC targeting, and we therefore determined the localizations of amino-terminal domains of SENP2 not fused to pyruvate kinase. When expressed alone with an amino-terminal myc epitope tag, residues 1 to 63 of SENP2 localized exclusively to the NPC (Fig. 4d). These data demonstrate that the amino-terminal 63 amino acids of SENP2 contain both an NLS and an NPC-targeting signal.
The SENP2 NPC-targeting signal interacts with the carboxyl-terminal domain of Nup153.
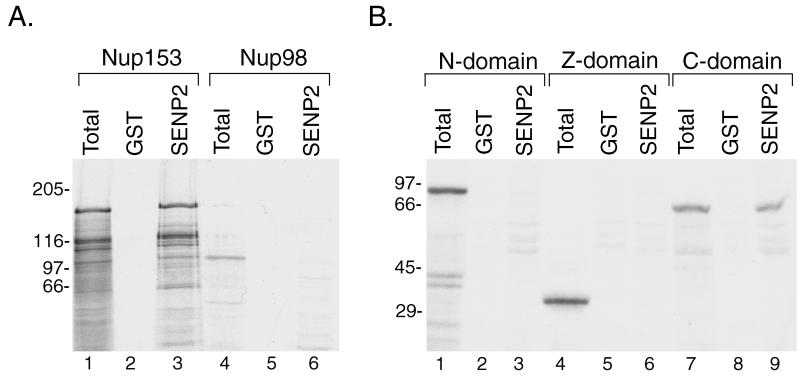
Several nucleoporins that localize specifically to the nucleoplasmic face of the NPC have been characterized, including Nup153 (38), Tpr (3), and Nup98 (27). To determine whether any of these proteins may play a role in the targeting of SENP2 to the NPC, we expressed them in rabbit reticulocyte lysates in the presence of [35S]methionine and assayed for their ability to interact with the NPC localization signal of SENP2. GST-SENP2(1-63) was immobilized on glutathione-Sepharose beads and incubated with the in vitro-translated proteins. Following incubation and washes, bound proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE. Total translation reactions (Fig. 5A, lanes 1 and 4) showed that full-length Nup153 and Nup98 were produced in vitro. During incubation with immobilized GST-SENP2(1-63), significant binding of Nup153 was detected (Fig. 5A, lane 3), but no binding of Nup98 was observed (Fig. 5A, lane 6). No binding was observed to beads containing GST alone (Fig. 5A, lanes 2 and 5). We also performed similar binding assays with full-length Tpr and observed no interaction with SENP2 (data not shown).
FIG. 5.
The NPC localization signal of SENP2 interacts with the carboxyl-terminal domain of Nup153. (A) The amino-terminal 63 amino acids of SENP2 were fused to GST, and the expressed fusion protein or GST alone was immobilized on glutathione-Sepharose beads. Full-length Nup153 and full-length Nup98 were translated in vitro in the presence of [35S]methionine, and the translation products (lanes 1 and 4) were incubated with immobilized GST-SENP2(1-63) (lanes 3 and 6) or with immobilized GST as a control (lanes 2 and 5). Bound proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE. Only Nup153 was able to bind to the NPC localization signal of SENP2. (B) Three separate domains of Nup153, corresponding to the amino terminus (amino acids 1 to 609) (N-domain), the zinc finger region (amino acids 610 to 869) (Z-domain), and the carboxyl terminus (amino acids 870 to 1475) (C-domain), were expressed in vitro in the presence of [35S]methionine. The translation products (lanes 1, 4, and 7) were incubated with immobilized GST-SENP2(1-63) (lanes 3, 6, and 9) or GST as a control (lanes 2, 5, and 8). Bound proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE. Only the carboxyl-terminal domain of Nup153 was retained by the NPC localization signal of SENP2. Molecular mass standards (kilodaltons) are indicated to the left of each gel.
Nup153 can be divided into three separate domains corresponding to an amino-terminal region that contains information for NPC localization (1), a zinc finger region (amino acids 610 to 869), and a carboxyl-terminal region containing FG repeats (amino acids 870 to 1475). To determine which of these domains interacts with SENP2, we created plasmids that would allow us to express the amino-terminal domain (amino acids 1 to 609), the zinc finger domain (amino acids 610 to 869), and the carboxyl-terminal domain (amino acids 870 to 1475) separately by in vitro translation (Fig. 5B, lanes 1, 4, and 7). We performed binding experiments similar to those outlined above and found that the amino-terminal and zinc finger domains of Nup153 (Fig. 5B, lanes 2, 3, 5, and 6) showed no interactions with the NPC localization domain of SENP2. However, the carboxyl-terminal domain of Nup153 was specifically retained on beads containing immobilized GST-SENP2(1-63) (Fig. 5B, lane 9) but not on beads containing GST alone (Fig. 5B, lane 8). These findings demonstrate that SENP2 binds specifically to the carboxyl-terminal FG-repeat domain of Nup153 and indicate that Nup153 plays a role in localizing SENP2 to the nucleoplasmic face of the NPC.
SENP2 has carboxyl-terminal hydrolase activity and is able to process both SUMO-1 and SUMO-3 conjugates in vitro.
SUMO proteases can potentially perform two different types of cleavage reactions in vivo. First, all SUMOs are synthesized as precursor proteins with carboxyl-terminal extensions that must be recognized and proteolytically removed. In addition, SUMO proteases are required to reverse SUMO modifications by cleaving the isopeptide bonds formed between the carboxyl-terminal glycine of SUMO and the lysine residue of a covalently modified substrate (21). The murine SENP2-related protein Axam2 is able to process both SUMO precursors and SUMO substrates in vitro (24). To determine whether SENP2 has either of these activities, we expressed it in bacteria and performed in vitro protease assays with SUMO precursors and protein conjugates.
To detect carboxyl-terminal hydrolase activity, we engineered SUMO-1 and SUMO-3 precursor proteins with extended carboxyl termini (each contained an additional 10 alanine residues following the natural carboxyl-terminal extensions) that would allow us to distinguish between cleaved and uncleaved proteins by SDS-PAGE. We expressed both SUMO-1 and SUMO-3 precursor proteins (Fig. 6A, lanes 1 and 3) and incubated them with purified, recombinant SENP2. As evident by the increased electrophoretic mobility (Fig. 6A, lanes 2 and 4), both SUMO-1 and SUMO-3 precursor proteins were recognized and processed by SENP2.
FIG. 6.
SENP2 has carboxyl-terminal hydrolase activity and is able to process SUMO-1 and SUMO-3 conjugates. (A) SUMO-1 and SUMO-3 precursors were engineered to contain an additional 10 amino acids at their carboxyl termini and were expressed and purified from bacteria. Following a 30-min incubation without (lanes 1 and 3) or with (lanes 2 and 4) SENP2, proteins were resolved by SDS-PAGE and visualized by Coomassie blue staining. Both SUMO-1 and SUMO-3 precursor proteins were cleaved by SENP2, as evident by a shift in the molecular mass following incubation with SENP2. The arrowheads indicate cleaved protein products. (B) The carboxyl-terminal domain of RanGAP1 (NΔ419) (lane 1) was modified with SUMO-1 (lane 2) or SUMO-3 (lane 4) in vitro, and the resulting conjugates were incubated with SENP2 (lanes 3 and 5). Both SUMO-1 and SUMO-3 conjugates were recognized and deconjugated by SENP2. Molecular mass standards (kilodaltons) are indicated on the left.
To determine whether SENP2 was able to process SUMO substrates, we incubated purified SENP2 with SUMO-modified RanGAP1. The carboxyl-terminal domain of RanGAP1 (NΔ419; Fig. 6B, lane 1) was modified in vitro with either SUMO-1 or SUMO-3 (Fig. 6B, lanes 2 and 4). The conjugates were purified and incubated with SENP2, and then the reactions were analyzed by SDS-PAGE. Both SUMO-1- and SUMO-3-modified forms of NΔ419 were converted to free NΔ419 and SUMO (Fig. 6B, lanes 3 and 5), indicating that SENP2 is able to process both SUMO-1 and SUMO-3 protein conjugates in vitro. These data demonstrate that SENP2 is able to recognize and process SUMO precursors and SUMO substrates and that it has no apparent specificity for either SUMO-1 or SUMO-3 under the in vitro conditions tested.
Ubc9 localizes to the cytoplasmic and nucleoplasmic filaments of the NPC.
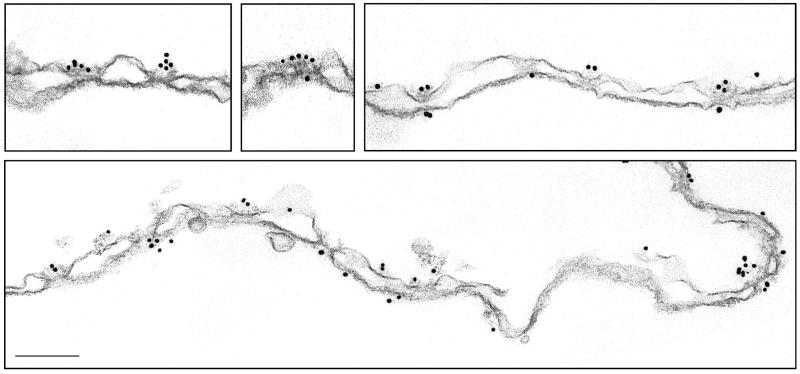
The above findings indicating that SUMO demodification may occur at the NPC are particularly intriguing in light of the finding that SUMO modification of substrates may also occur there (26). It was previously demonstrated by immunofluorescence microscopy that Ubc9, the SUMO E2 enzyme, partially localizes to the nuclear envelope (15). Like SENP2, Ubc9 was also detected as a factor that copurifies with vertebrate nucleoporins (3a), suggesting that it associates with intact NPCs. To more precisely determine the localization of Ubc9, we probed isolated rat liver nuclear envelopes with a Ubc9-specific antibody. The antibody was produced by immunizing rabbits with full-length Ubc9, and the specificity was assayed by immunoblot analysis (data not shown). Following labeling with a secondary antibody conjugated to 10-nm gold particles, the localization of Ubc9 was detected by electron microscopy. Heavy labeling of the cytoplasmic fibrils of the NPC was detected, with four or five gold particles often being present at a single NPC (Fig. 7). Of 425 gold particles clearly labeling NPCs, 70% were associated with the cytoplasmic fibrils. The remaining label was detected on the nucleoplasmic face of the NPC, possibly associated with the filaments that form the nucleoplasmic basket. The cytoplasmic and nucleoplasmic sides of the nuclear envelopes were distinguished by the occasional blebs in the membrane that are found on the cytoplasmic side. These blebs are not found on the nucleoplasmic side due to the presence of the nuclear lamina. Control experiments with preimmune serum revealed no specific labeling of NPCs (data not shown). These data demonstrate that Ubc9 localizes to the nuclear envelope through interactions with the nucleoplasmic and cytoplasmic filaments of the NPC.
FIG. 7.
Ubc9 localizes to the cytoplasmic and nucleoplasmic filaments of the NPC. Nuclear envelopes were prepared from rat liver nuclei and labeled with a rabbit polyclonal antibody specific for Ubc9. Following incubation with a secondary antibody conjugated to 10-nm gold particles, the nuclear envelopes were processed for thin sectioning and analyzed by electron microscopy. Several sections of nuclear envelopes demonstrating the typical labeling pattern are shown. The cytoplasmic faces of the nuclear envelopes (evidenced by occasional blebs in the membrane) are oriented toward the top of the figure. Bar, 0.15 μm.
Ubc9 and SUMO-1-modified RanGAP1 bind synergistically to form a trimeric complex with Nup358.
Several previous studies indicated that Ubc9 can associate with Nup358, a large nucleoporin that localizes to the cytoplasmic filaments of the NPC (26, 31, 33). These studies demonstrated that Ubc9 binds to a carboxyl-terminal region of Nup358 (amino acids 2596 to 2836) that coincides with the region found to interact with SUMO-1-modified RanGAP1 (18, 20, 31). It was recently demonstrated that this carboxyl-terminal domain of Nup358 can bind to SUMO-1-modified RanGAP1 and to Ubc9 simultaneously (26); however, the specific interactions within this trimeric complex have not been fully analyzed.
Notably, direct interactions between SUMO-1-modified RanGAP1 and the carboxyl-terminal domain of Nup358 have not been tested, as previous studies involved proteins expressed in rabbit reticulocyte lysates (18, 20). Furthermore, it was shown that SUMO-1-modified RanGAP1 forms a stable complex with Ubc9, indicating that Ubc9 could act to tether RanGAP1 to Nup358 (34). To more precisely investigate the interactions among SUMO-1-modified RanGAP1, Ubc9, and Nup358, we performed in vitro binding assays with recombinant, bacterially expressed proteins.
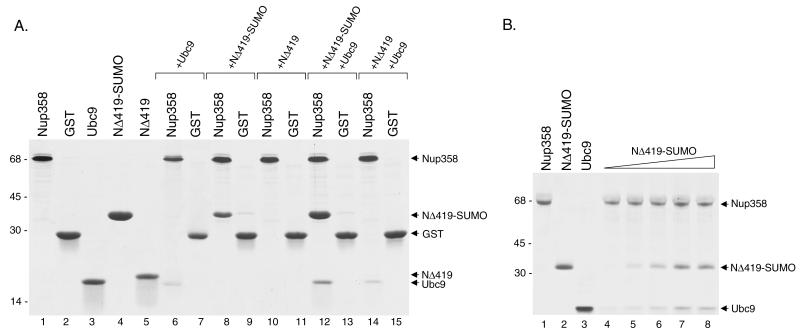
The carboxyl-terminal region of human Nup358 (amino acids 2596 to 2836) was expressed as a GST fusion protein (Fig. 8A, lane 1) and immobilized on glutathione-Sepharose beads. The beads were subsequently incubated with bacterially expressed, purified Ubc9 (Fig. 8A, lane 3). Ubc9 was specifically retained on the beads containing the GST-Nup358 fusion protein (Fig. 8A, lanes 6 and 7), confirming the finding that Ubc9 interacts directly with Nup358. We next performed binding experiments with purified SUMO-1-modified RanGAP1 to determine whether it could also interact directly with this same domain of Nup358. The carboxyl-terminal domain of RanGAP1 (NΔ419) was expressed in bacteria, modified with SUMO-1 in an in vitro modification reaction, and subsequently purified. When expressed in rabbit reticulocyte lysates, this domain of RanGAP1 interacts with Nup358, but only when modified by SUMO-1 (20). SUMO-1-modified NΔ419 (Fig. 8A, lane 4) and unmodified NΔ419 (Fig. 8A, lane 5) were incubated with immobilized GST-Nup358 and with GST alone as a negative control. Significantly higher levels of SUMO-1-modified NΔ419 bound to GST-Nup358 than to GST alone (Fig. 8A, lanes 8 and 9), indicating that SUMO-1-modified NΔ419 also binds directly to Nup358. No interaction was detected between Nup358 and unmodified NΔ419 (Fig. 8A, lanes 10 and 11), consistent with previous findings obtained with proteins expressed in reticulocyte lysates.
FIG. 8.
Ubc9 and SUMO-1-modified RanGAP1 both interact directly with Nup358 to form a trimeric complex. (A) A carboxyl-terminal region of Nup358 (amino acids 2596 to 2836) was expressed as a GST fusion protein in bacteria and purified (lane 1). Other proteins, including GST (lane 2), Ubc9 (lane 3), and the carboxyl-terminal domain of RanGAP1 (NΔ419) (lane 5), were also expressed in bacteria and purified. NΔ419 was modified by SUMO-1 in an in vitro assay and purified from reaction components (lane 4). To assay for interactions with Nup358, the GST-Nup358 fusion protein was immobilized on glutathione-Sepharose beads and incubated with Ubc9 alone (lane 6), SUMO-1-modified NΔ419 (NΔ419-SUMO) alone (lane 8), NΔ419 alone (lane 10), NΔ419-SUMO plus Ubc9 (lane 12), and NΔ419 plus Ubc9 (lane 14). Following washes, bound proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE. As controls for binding specificity, similar binding assays were also performed with glutathione-Sepharose beads containing GST alone (lanes 7, 9, 11, 13, and 15). Molecular mass standards (kilodaltons) are indicated on the left. (B) The synergistic effect of SUMO-1-modified RanGAP1 on the binding of Ubc9 to Nup358 was investigated. A fixed amount of Ubc9 (5 μg) was incubated with immobilized Nup358 in the presence of increasing amounts of SUMO-1-modified RanGAP1 (lane 4, 0 μg; lane 5, 1 μg; lane 6, 2 μg; lane 7, 4 μg; and lane 8, 8 μg).
We next investigated the formation of the trimeric complex among Ubc9, SUMO-1-modified RanGAP1, and Nup358. For these experiments, GST-Nup358 was again immobilized on glutathione-Sepharose beads and incubated with SUMO-1-modified NΔ419 in the presence of equimolar amounts of Ubc9. Under these conditions, we observed a significant enhancement in the binding of SUMO-1-modified NΔ419 to Nup358 relative to binding in the absence of Ubc9 (Fig. 8A, lanes 8 and 12). The binding of Ubc9 to Nup358 was also significantly enhanced relative to binding in the absence of SUMO-1-modified NΔ419. No significant interactions with GST alone were detected (Fig. 8A, lane 13). These results indicate that SUMO-1-modified RanGAP1 and Ubc9 bind synergistically to Nup358. To test this notion more directly, we performed similar binding assays with a fixed, excess amount of Ubc9 and increasing concentrations of SUMO-1-modified RanGAP1. Under these conditions, the amount of Ubc9 bound to Nup358 was found to increase proportionally with the amount of SUMO-1-modified RanGAP1 added to the binding assays (Fig. 8B). These experiments provide further evidence that Ubc9 and SUMO-1-modified RanGAP1 bind synergistically to Nup358 to form a trimeric complex. The ability to form a trimeric complex was also confirmed by gel filtration chromatography (data not shown) (26).
It was previously shown that Ubc9 forms a very stable complex with both SUMO-1-modified NΔ419 and unmodified NΔ419 (2, 34). We therefore also examined whether Ubc9 could influence the ability of unmodified NΔ419 to interact with Nup358. Ubc9 had no effect on the ability of unmodified NΔ419 to interact with Nup358 (Fig. 8A, lane 14). These results indicate that interactions between Ubc9 and SUMO-1-modified RanGAP1 promote binding to Nup358, whereas interactions between Ubc9 and unmodified RanGAP1 do not. Together with our immunolocalization data, these findings indicate that Ubc9 likely exists in a complex with SUMO-1-modified RanGAP1 and Nup358 on the cytoplasmic filaments of the NPC.
SUMO-1-modified RanGAP1 is resistant to SENP2 protease activity when complexed with Ubc9 and Nup358.
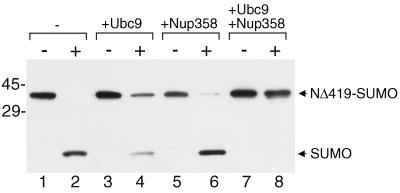
RanGAP1 is the most abundant SUMO-1 conjugate in the cell, with approximately 50% of the total RanGAP1 accumulating in the SUMO-modified form (19). Moreover, SUMO-1-modified RanGAP1 is relatively stable, even after cell lysis, when most SUMO-1 conjugates become accessible to endogenous SUMO proteases. One possible explanation for the unusual stability of the SUMO-1-RanGAP1 conjugate is that it is protected from SUMO proteases when complexed with Nup358 and Ubc9. To test this notion experimentally, we made preformed complexes of SUMO-1-modified RanGAP1, Ubc9, and Nup358 and assayed them for SENP2 protease activity by using immunoblot analysis to detect the release of free SUMO-1. As previously observed, SENP2 was able to quantitatively demodify SUMO-1-modified RanGAP1 when incubated with it alone (Fig. 9, lanes 1 and 2). When SENP2 was incubated with SUMO-1-modified RanGAP1 that had been preincubated with excess Ubc9, we observed a partial reduction in the demodification of RanGAP1 (Fig. 9, lanes 3 and 4). Preincubation of SUMO-1-modified RanGAP1 with excess Nup358 had very little effect on SENP2 activity (Fig. 9, lanes 5 and 6). Most significantly, when SUMO-1-modified RanGAP1 was allowed to form a complex with both Nup358 and Ubc9 prior to incubation with SENP2, complete protection from protease activity was observed (Fig. 9, lanes 7 and 8). These experiments demonstrate that SUMO-1-modified RanGAP1 is protected from SUMO protease activity when complexed with Nup358 and Ubc9.
FIG. 9.
SUMO-1-modified RanGAP1 is protected from SENP2 protease activity when complexed with Nup358 and Ubc9. The carboxyl-terminal domain of RanGAP1 (amino acids 420 to 589) was modified by SUMO-1 in vitro and purified from reaction components. The purified product (NΔ419-SUMO) was mixed with buffer (lanes 1 and 2), an eightfold molar excess of Ubc9 (lanes 3 and 4), a sixfold molar excess of Nup358 (amino acids 2596 to 2836) (lanes 5 and 6), or an eightfold molar excess of Ubc9 plus a sixfold molar excess of Nup358 (amino acids 2596 to 2836) (lanes 7 and 8). Following preincubation to allow for complex formation, reactions were split into two tubes; SENP2 was added to one tube (lanes marked with a plus sign), and buffer was added to the other (lanes marked with a minus sign). Following further incubation, the reactions were stopped with SDS sample buffer and analyzed by SDS-PAGE and immunoblot analysis with an antibody specific for SUMO-1. Molecular mass standards (kilodaltons) are indicated on the left, and the positions of SUMO-1-modified NΔ419 and free SUMO-1 are indicated on the right.
DISCUSSION
SUMO modification is believed to regulate a wide range of cellular activities, many associated with the nucleus. Consistent with a prominent role in the nucleus, a significant number of the known SUMO substrates are nuclear proteins (21), and the E1 and E2 enzymes that mediate SUMO modification localize to the nucleoplasm (28). These findings have suggested that SUMO modification itself is predominantly an intranuclear event. Here, we have analyzed the localizations of two factors that control SUMO modification, Ubc9 and SENP2, and have found that they localize to filaments on the cytoplasmic and nucleoplasmic faces of the NPC. The implication of these findings is that SUMO modification and demodification of proteins can occur at the NPC, possibly in connection with nucleocytoplasmic transport.
SENP2 is one of at least seven related SUMO proteases present in vertebrates. We have shown that it is able to cleave the carboxyl-terminal extensions of SUMO-1 and SUMO-3 precursor proteins and reverse the modification of SUMO-1 and SUMO-3 substrates. Although we were unable to observe any obvious substrate specificity for SENP2 in our in vitro studies, the localization of SENP2 at the NPC makes it unique from previously characterized SENPs that have been found to localize to the cytoplasm (13), nucleoplasm (6), and nucleolus (25). The specific functions of the individual SENPs are currently not known; however, it is intriguing to speculate that the unique localizations of these enzymes may contribute to distinct substrate specificities and functions. SENP2 was previously isolated as a protein that interacts with Axin, a regulator of the Wnt signaling pathway (11). In this study, a truncated version of SENP2 (lacking the fist 71 amino acids) was shown to induce the degradation of β-catenin in cultured cells and to inhibit Wnt-dependent axis duplication in Xenopus embryos. Consistent with our finding that the first 63 amino acids of SENP2 are required for the targeting of SENP2 to the NPC, the truncated protein used in this study localized exclusively to the cytoplasm (11). The mislocalization of SENP2 in this study raises questions about the relevance of the effects on the Wnt pathway that were observed. However, another study identified what appears to be an alternatively spliced form of SENP2 that lacks the amino-terminal 47 amino acids and that localizes to the cytoplasm (24). Because the activation of the Wnt pathway specifically involves the stabilization of β-catenin and its translocation from the cytoplasm to the nucleus (two processes that could involve SUMO modification), it will be particularly important to further investigate the possible roles of SENP2 in regulating the Wnt signaling pathway.
While this manuscript was in preparation, Hang and Dasso also reported that full-length SENP2 localizes to the NPC (8). Our findings complement theirs in establishing that SENP2 localizes to the nucleoplasmic face of the NPC, likely through interactions with Nup153. One notable difference between their work and ours is the localization of SENP2 containing amino-terminal deletions. Whereas we found that the deletion of 63 amino acids from the amino terminus of SENP2 caused it to localize almost exclusively to the cytoplasm, Hang and Dasso found that similar deletions caused SENP2 to localize to the nucleoplasm. A likely explanation for this difference is the use of different protein tags (myc in our study and green fluorescent protein in the study of Hang and Dasso). The use of the myc epitope tag allowed us to determine that the first 63 amino acids of SENP2 contain signals for both nuclear localization and NPC targeting. We have also extended the findings of Hang and Dasso by showing that SENP2 specifically interacts with the carboxyl-terminal FG repeat domain of Nup153. Interestingly, this domain of Nup153 plays important roles in NLS-mediated nuclear import (36) and mRNA export (1). The functions of the Nup153 FG repeat domain have been attributed to its ability to bind to nuclear import and nuclear export receptors. Given our finding that this domain also interacts with SENP2, it will be important to evaluate the potential roles of SENP2 in facilitating protein import and/or RNA export.
Ubc9, the E2 conjugating enzyme for SUMO, plays direct roles in substrate recognition and substrate modification. Ubc9 is unusual in comparison to ubiquitin E2 conjugating enzymes in that it binds directly to a consensus sequence found in most SUMO-modified proteins (2, 34). Because of this direct recognition of substrates by Ubc9, the role of E3-like factors in SUMO modification remains unclear. Nonetheless, several E3-like factors that stimulate SUMO modification have been identified, including Nup358 (26). Consistent with Nup358 having a role in SUMO modification, others (26, 31) and we have shown that Nup358 is able to interact directly with Ubc9. Using immunogold labeling of isolated nuclear envelopes, we have shown, for the first time, that Ubc9 localizes to the cytoplasmic filaments of intact NPCs, a finding that is consistent with Ubc9 interacting with Nup358. We have also shown, for the first time, that SUMO-1-modified RanGAP1 binds directly to Nup358 in the absence of other factors. Although SUMO-1-modified RanGAP1 and Ubc9 interact independently with the same 240-amino-acid domain of Nup358, a synergistic effect on binding was observed when they were incubated with Nup358 together. A likely explanation for the observed synergy is that Ubc9 and SUMO-1-modified RanGAP1 interact simultaneously with each other and with Nup358 to form a stable trimeric complex, a model that is consistent with previous observations that SUMO-1-modified RanGAP1 and Ubc9 form stable interactions (2, 34). The finding that SUMO-1-modified RanGAP1 is resistant to SENP2 only when complexed with both Nup358 and Ubc9 is also consistent with these proteins forming a stable trimeric complex. Overall, our localization data and protein-protein interaction studies indicate that Ubc9 localizes to the cytoplasmic filaments of the NPC, where it forms a complex with both Nup358 and SUMO-1-modified RanGAP1. These findings are likely to have important implications for the function of Ubc9 at the NPC and for the role of Nup358 as an E3 ligase. We are currently investigating the E3 ligase activity of Nup358 and how it may be affected by interactions with Ubc9 and SUMO-1-modified RanGAP1.
Immunogold labeling of isolated nuclear envelopes also revealed that Ubc9 may interact with the nucleoplasmic filaments of the NPC. Initial assays for interactions between Ubc9 and nucleoporins known to localize to the nucleoplasmic face of the NPC (Tpr, Nup153, and Nup98) revealed no direct binding. Because a significant fraction of Ubc9 localizes to the nucleoplasm, we cannot rule out the possibility that the labeling observed on the nucleoplasmic face of the NPC was nonspecific. However, labeling was clearly specific for NPCs and was not observed on the inner nuclear membrane or lamina. In addition, similar results were obtained with two independently produced antibodies (data not shown).
NPCs are large multiprotein complexes that span the double membrane of the nuclear envelope and mediate the trafficking of macromolecules between the nucleus and the cytoplasm. In general, the NPC is believed to have a relatively passive role in the trafficking process, with transport receptors and their cargos transiently interacting with nucleoporins and translocating across the pore by facilitated diffusion (29). SENP2 and Ubc9 are unusual in that they are among a very limited number of proteins that are associated with the NPC and that have known biochemical activities. Taken together, our results indicate that SUMO modification and demodification of proteins can occur at the distal ends of the NPC. Proteins associated with the cytoplasmic and nucleoplasmic filaments of the NPC have been proposed to have important roles in determining the directionality of transport through the NPC because of their unique asymmetric localizations (29). At this point, we can only speculate about the exact functions that Ubc9 and SENP2 may have at the NPC. In the simplest model, SUMO modification and demodification at the NPC may function to alter the activities of specific substrates in a manner that is directly linked to their import or export from the nucleus. Alternatively, the modification and demodification of proteins may be directly related to their vectorial transport through the NPC. The modification of proteins on the cytoplasmic filaments could, for example, serve as a signal that initiates nuclear import. SUMO modification at the cytoplasmic filaments of the NPC could mediate the interaction of substrates with a nuclear import receptor or could allow modified proteins to interact directly with components of the NPC. SUMO-1 modification of RanGAP1, for example, regulates its interaction with Nup358 (18, 20). Translocation of modified substrates through the NPC would be terminated by their demodification by SENP2 at the nucleoplasmic basket. It is also conceivable that the SUMO modification of nuclear import receptors themselves functions to regulate their interactions with the NPC or with their cargo.
The SUMO modification and demodification of proteins at the NPC could also have a role in regulating RNA and protein export. One possible role for SUMO modification and demodification at the nucleoplasmic basket is to remodel messenger ribonucleoprotein complexes as they are translocated through the NPC or even to alter the conformation of the NPC itself. Analysis of the nuclear export of Balbiani ring transcripts, for example, has revealed dramatic changes in messenger ribonucleoprotein and NPC structures that are coincident with translocation through the pore (14). Intriguingly, one of the two SUMO proteases in S. cerevisiae (Ulp1) also localizes to NPCs (16, 17, 35), suggesting that the role of SUMO modification and demodification at the NPC is evolutionarily conserved.
Acknowledgments
We thank Michelle Bowman and Diane Grove for initial efforts in identifying and characterizing SENP2, Helen Shio for assistance with preparing samples for electron microscopy, and Larry Gerace and Brian Burke for Tpr and Nup153 cDNA clones. We also acknowledge Janet Cronshaw, Richard Rogers, and Maria Vasilleva for comments and suggestions made during the course of this work.
Support for this work was provided by grant GM60980 from the National Institutes of Health and grant RSG-01-064-01-CSM from the American Cancer Society (to M.J.M.).
REFERENCES
- 1.Bastos, R., A. Lin, M. Enarson, and B. Burke. 1996. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernier-Villamor, V., D. A. Sampson, M. J. Matunis, and C. D. Lima. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345-356. [DOI] [PubMed] [Google Scholar]
- 3.Cordes, V. C., S. Reidenbach, H. R. Rackwitz, and W. W. Franke. 1997. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J. Cell Biol. 136:515-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Cronshaw, J. M., A. N. Krutchinsky, W. Zhang, B. T. Chait, and M. J. Matunis. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol., in press. [DOI] [PMC free article] [PubMed]
- 4.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 5.Fontoura, B. M., G. Blobel, and M. J. Matunis. 1999. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 144:1097-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong, L., S. Millas, G. G. Maul, and E. T. Yeh. 2000. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275:3355-3359. [DOI] [PubMed] [Google Scholar]
- 7.Goodson, M. L., Y. Hong, R. Rogers, M. J. Matunis, O. K. Park-Sarge, and K. D. Sarge. 2001. Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J. Biol. Chem. 276:18513-18518. [DOI] [PubMed] [Google Scholar]
- 8.Hang, J., and M. Dasso. 2002. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277:19961-19966. [DOI] [PubMed] [Google Scholar]
- 9.Hong, Y., R. Rogers, M. J. Matunis, C. N. Mayhew, M. Goodson, O. K. Park-Sarge, and K. D. Sarge. 2001. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 276:40263-40267. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, E. S., and A. A. Gupta. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106:735-744. [DOI] [PubMed] [Google Scholar]
- 11.Kadoya, T., S. Kishida, A. Fukui, T. Hinoi, T. Michiue, M. Asashima, and A. Kikuchi. 2000. Inhibition of Wnt signaling pathway by a novel axin-binding protein. J. Biol. Chem. 275:37030-37037. [DOI] [PubMed] [Google Scholar]
- 12.Kahyo, T., T. Nishida, and H. Yasuda. 2001. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8:713-718. [DOI] [PubMed] [Google Scholar]
- 13.Kim, K. I., S. H. Baek, Y. J. Jeon, S. Nishimori, T. Suzuki, S. Uchida, N. Shimbara, H. Saitoh, K. Tanaka, and C. H. Chung. 2000. A new SUMO-1-specific protease, SUSP1, that is highly expressed in reproductive organs. J. Biol. Chem. 275:14102-14106. [DOI] [PubMed] [Google Scholar]
- 14.Kiseleva, E., M. W. Goldberg, B. Daneholt, and T. D. Allen. 1996. RNP export is mediated by structural reorganization of the nuclear pore basket. J. Mol. Biol. 260:304-311. [DOI] [PubMed] [Google Scholar]
- 15.Lee, G. W., F. Melchior, M. J. Matunis, R. Mahajan, Q. Tian, and P. Anderson. 1998. Modification of Ran GTPase-activating protein by the small ubiquitin-related modifier SUMO-1 requires Ubc9, an E2-type ubiquitin-conjugating enzyme homologue. J. Biol. Chem. 273:6503-6507. [DOI] [PubMed] [Google Scholar]
- 16.Li, S. J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246-251. [DOI] [PubMed] [Google Scholar]
- 17.Li, S. J., and M. Hochstrasser. 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20:2367-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahajan, R., L. Gerace, and F. Melchior. 1998. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J. Cell Biol. 140:259-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matunis, M. J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matunis, M. J., J. Wu, and G. Blobel. 1998. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140:499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 22.Moss, S. F., V. Krivosheyev, A. de Souza, K. Chin, H. P. Gaetz, N. Chaudhary, H. J. Worman, and P. R. Holt. 1999. Decreased and aberrant nuclear lamin expression in gastrointestinal tract neoplasms. Gut 45:723-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishida, T., F. Kaneko, M. Kitagawa, and H. Yasuda. 2001. Characterization of a novel mammalian SUMO-1/Smt3-specific isopeptidase, a homologue of rat axam, which is an axin-binding protein promoting beta-catenin degradation. J. Biol. Chem. 276:39060-39066. [DOI] [PubMed] [Google Scholar]
- 25.Nishida, T., H. Tanaka, and H. Yasuda. 2000. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur. J. Biochem. 267:6423-6427. [DOI] [PubMed] [Google Scholar]
- 26.Pichler, A., A. Gast, J. S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109-120. [DOI] [PubMed] [Google Scholar]
- 27.Radu, A., M. S. Moore, and G. Blobel. 1995. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell 81:215-222. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez, M. S., C. Dargemont, and R. T. Hay. 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276:12654-12659. [DOI] [PubMed] [Google Scholar]
- 29.Rout, M. P., and J. D. Aitchison. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593-16596. [DOI] [PubMed] [Google Scholar]
- 30.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saitoh, H., M. D. Pizzi, and J. Wang. 2002. Perturbation of SUMOlation enzyme Ubc9 by distinct domain within nucleoporin RanBP2/Nup358. J. Biol. Chem. 277:4755-4763. [DOI] [PubMed] [Google Scholar]
- 32.Saitoh, H., R. Pu, M. Cavenagh, and M. Dasso. 1997. RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc. Natl. Acad. Sci. USA 94:3736-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitoh, H., D. B. Sparrow, T. Shiomi, R. T. Pu, T. Nishimoto, T. J. Mohun, and M. Dasso. 1998. Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr. Biol. 8:121-124. [DOI] [PubMed] [Google Scholar]
- 34.Sampson, D. A., M. Wang, and M. J. Matunis. 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276:21664-21669. [DOI] [PubMed] [Google Scholar]
- 35.Schwienhorst, I., E. S. Johnson, and R. J. Dohmen. 2000. SUMO conjugation and deconjugation. Mol. Gen. Genet. 263:771-786. [DOI] [PubMed] [Google Scholar]
- 36.Shah, S., and D. J. Forbes. 1998. Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors. Curr. Biol. 8:1376-1386. [DOI] [PubMed] [Google Scholar]
- 37.Siomi, H., and G. Dreyfuss. 1995. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sukegawa, J., and G. Blobel. 1993. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell 72:29-38. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi, Y., A. Toh-e, and Y. Kikuchi. 2001. A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene 275:223-231. [DOI] [PubMed] [Google Scholar]
- 40.Vasu, S. K., and D. J. Forbes. 2001. Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 13:363-375. [DOI] [PubMed] [Google Scholar]
- 41.Yeh, E. T., L. Gong, and T. Kamitani. 2000. Ubiquitin-like proteins: new wines in new bottles. Gene 248:1-14. [DOI] [PubMed] [Google Scholar]