Abstract
The transcriptional coactivator p/CIP is a member of a family of nuclear receptor coactivator/steroid receptor coactivator (NCoA/SRC) proteins that mediate the transcriptional activities of nuclear hormone receptors. We have found that p/CIP is predominantly cytoplasmic in a large proportion of cells in various tissues of the developing mouse and in a number of established cell lines. In mouse embryonic fibroblasts, serum deprivation results in the redistribution of p/CIP to the cytoplasmic compartment and stimulation with growth factors or tumor-promoting phorbol esters promotes p/CIP shuttling into the nucleus. Cytoplasmic accumulation of p/CIP is also cell cycle dependent, occurring predominantly during the S and late M phases. Leptomycin B (LMB) treatment results in a marked nuclear accumulation, suggesting that p/CIP undergoes dynamic nuclear export as well as import. We have identified a strong nuclear import signal in the N terminus of p/CIP and two leucine-rich motifs in the C terminus that resemble CRM-1-dependent nuclear export sequences. When fused to green fluorescent protein, the nuclear export sequence region is cytoplasmic and is retained in the nucleus in an LMB-dependent manner. Disruption of the leucine-rich motifs prevents cytoplasmic accumulation. Furthermore, we demonstrate that cytoplasmic p/CIP associates with tubulin and that an intact microtubule network is required for intracellular shuttling of p/CIP. Immunoaffinity purification of p/CIP from nuclear and cytosolic extracts revealed that only nuclear p/CIP complexes possess histone acetyltransferase activity. Collectively, these results suggest that cellular compartmentalization of NCoA/SRC proteins could potentially regulate nuclear hormone receptor-mediated events as well as integrating signals in response to different environmental cues.
Nuclear receptors (NRs) are a superfamily of structurally related proteins that function as ligand-regulated transcription factors. Members of this family include receptors for estrogen, glucocorticoids, nonsteroidal ligands such as thyroid hormone, and retinoic acid, as well as receptors that bind by-products of lipid metabolism such as fatty acids and prostaglandins. These receptors control a complex array of genes involved in many biological functions including cell proliferation and differentiation, metabolism and growth, morphogenesis, programmed cell death, and homeostasis. In the absence of hormone, some NRs such as the thyroid hormone receptor and retinoic acid receptor function as transcriptional repressors by interacting with corepressor proteins. Hormone binding results in a conformational change in the receptor that results in the release of corepressor proteins and in the recruitment of coactivator proteins (17).
The nuclear receptor coactivator/steroid receptor coactivator (NCoA/SRC) proteins were among the first coactivators to be identified. This family includes steroid receptor coactivator 1 (SRC-1) (51) also designated nuclear receptor coactivator 1 (NCoA-1) (28); GRIP1 (25), also known as TIF2/NCoA-2 (65, 69), and the mouse p300/CBP-interacting protein (p/CIP) (65), which has been identified in humans as AIB1/ACTR/RAC3/SRC-3/TRAM-1 (2, 6, 38, 63, 64). Biochemical and functional studies have provided strong evidence to support the hypothesis that SRC proteins mediate the transcriptional effects of NRs. All SRC family members interact directly with NRs in a ligand- and AF-2-dependent manner (6, 35, 37, 45, 50, 65). This interaction is mediated by three leucine-rich motifs (referred to as LXXLL motifs or NR boxes) found within the NR interaction domain of the SRC proteins (23, 44, 65). Crystallographic studies have established that the LXXLL motif forms a short α-helix that makes direct contact with amino acids found in the AF-2 domain of all ligand-bound NRs (48, 72). Overexpression of individual SRC proteins can enhance the transcriptional activities of several NRs in response to their respective ligand in vivo and in vitro (2, 6, 51, 63). Single-cell microinjection of antibodies against specific SRC proteins blocks ligand-dependent stimulation of reporter genes containing NR response elements, suggesting that they are essential for some NR signaling events (65). It has also been shown that SRC proteins may function as coactivators for other classes of transcription factors such as AP-1 and NF-κB (48, 59, 65, 68). Chromatin immunoprecipitation assays have demonstrated that members of this class of coactivators are recruited to several endogenous NR target genes, such as cathepsin D and p21, in response to hormone (7, 58). More recently, in vitro transcription experiments using chromatinized templates have demonstrated that SRC proteins, in association with CBP/p300, enhance NR-mediated transcriptional initiation (31, 40).
The SRC proteins mediate their transcriptional effects primarily by functioning as bridging factors which, on binding to NRs, recruit other coactivator proteins important for regulating transcriptional events. Many of these interacting proteins possess enzymatic activity and include acetyltransferases such as p/CAF (33), GCN5 (1), and CBP/p300 (28), as well as CARM1 and PRMT1, which possess methyltransferase activity (5, 32). SRC proteins can also associate with several other coactivators whose mechanism is not entirely clear, such as the steroid receptor RNA coactivator (35) and ASC-I (30).
It has also been demonstrated that some members of the SRC family possess intrinsic histone acetyltransferase (HAT) activity, which is mediated by their respective carboxy termini (6, 61). This suggests that SRC proteins mediate their transcriptional response in part by directly acetylating histones or possibly other proteins. A large body of evidence has supported the proposal that reversible acetylation of core histones changes the nucleosomal conformation, which renders DNA more accessible to binding of the transcriptional machinery, thereby facilitating a transcriptional response (9, 36).
Although recruitment of coactivator proteins represents a major mechanism through which NRs mediate their effects, it has become increasingly clear that additional layers of complexity exist which can regulate transcriptional activity. For example, activation of phosphorylation pathways in response to intra- or extracellular signals can serve as an important mechanism. It has been shown that the AF-1 domain of some NRs can be phosphorylated by mitogen-activated protein kinase in response to various growth factors (20, 67). This phosphorylation may serve to target the SRC proteins to NRs independently of the hormone. Furthermore, all of the SRC proteins can be phosphorylated by several distinct protein kinases in vivo and in vitro, resulting in a concomitant change in activity (15, 41, 56).
Intracellular shuttling, in response to specific signals, constitutes another mechanism for regulating transcriptional activity. There are numerous examples of transcription factors whose activity is critically regulated by regulated nuclear import and export. For example, in unstimulated cells the glucocorticoid receptor (GR) is part of a multiprotein complex consisting mainly of heat shock proteins (55). The binding of GR to its ligand causes it to dissociate from the complex, allowing it to enter the nucleus and facilitate gene expression (19). The transcription factor NF-κB is maintained in an inactive form associated with IκBα in the cytoplasm (62). Following cellular activation, IκB is rapidly phosphorylated by a multisubunit kinase complex and is then degraded. In turn, the nuclear localization signal (NLS) on NF-κB is unmasked, allowing it to translocate to the nuclear compartment. More recently, it has been shown that the class II histone deacetylases (HDACs) shuttle into and out of the nuclei of skeletal muscle cells in response to differentiation signals (29, 46, 71).
Although small proteins can exit and enter the cell by passive diffusion, cells have evolved energy-dependent transport mechanisms which shuttle proteins between the cytoplasm and the nucleus (26). This shuttling is mediated primarily by members of the importin family of Ran GTP-binding proteins which recognize specific NLSs (18). Thus, the steady-state localization of a protein is determined by the relative rates of import and export, both of which can be targeted for regulation in response to intra- or extracellular stimuli. In addition, this type of mechanism may allow the coordination of events occuring in the cytoplasm and in the nucleus.
In the present study, we have examined the subcellular distribution of endogenous p/CIP in mouse tissue, and in various cell types. We have found that p/CIP expression is compartmentalized and exhibits a strong cytoplasmic component in addition to its nuclear localization. In untransformed mouse embryonic fibroblasts (MEFs), p/CIP undergoes a redistribution, in response to serum withdrawal, from a predominantly nuclear to a cytoplasmic localization. On exposure of serum- starved cells to epidermal growth factor (EGF), insulin, or phorbol esters, p/CIP redistributes back to the nucleus. Our results suggest that p/CIP redistribution is controlled by several novel mechanisms which may serve to control p/CIP levels in the nucleus and possibly integrate specific signals coming from the cell surface.
MATERIALS AND METHODS
Plasmids
The cDNAs encoding full-length p/CIP, p/CIP (amino acids [aa] 1-1085), and p/CIP(aa 1-636) were generated by restriction digestion from plasmid PCMXp/CIP (65), and the corresponding fragments were subcloned into pEGFPC1 (Clontech). The cDNA fragments corresponding to p/CIP (aa 1-214) and p/CIP (aa 1-150) were amplified by PCR and subcloned into pEGFPC2. Plasmid pHM829 p/CIP (aa 1 to 34) was generated by PCR amplifying the region of p/CIP corresponding to p/CIP (aa 1-34) and subcloning into pHM829 (60), which allows the simultaneous expression of cDNAs fused to both lacZ and the green fluorescent protein DNA (GFP).
Site-directed mutagenesis was performed using the Quickchange kit (Stratagene) as specified by the manufacturer.
Western blotting and antibody production
Subcellular fractions of cells were prepared by standard methods (13). Western blotting was performed as described previously (65). Normally, 20 μg of protein was fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and specific proteins were analyzed by Western blotting. Proteins were detected using enhanced chemiluminescence as recommended by the manufacturer (Amersham). The p/CIP rabbit polyclonal antiserum was raised against a His-tagged recombinant p/CIP (aa 591-803) protein. No significant cross-reactivity was observed when it was tested against homologous regions found in other SRC family members. The GRIP1 rabbit polyclonal antisera was raised against a recombinant His-tagged GRIP1 protein (aa 787-1100). Antibodies were purified by protein A-Sepharose chromatography by standard methods (22). For protein purification experiments, the immunoglobulin G (IgG)-purified antibodies were fractionated by affinity chromatography involving the corresponding His-tagged recombinant protein cross-linked to Sepharose 4B (Pharmacia). The α-tubulin monoclonal antibody (clone B-512) was obtained from Sigma.
Preparation of mouse embryos and cell culture maintenance
Embryos were removed at 14.5 days postcoitum (dpc). The head, blood, and liver were removed after tryptic digestion (0.1% trypsin in EDTA) for 30 min at 37°C. MEFs were maintained in high-glucose Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS). The cells were used for experiments after approximately 10 passages.
HeLa, MDA-435, and NIH 3T3 cells were maintained in DMEM with high glucose supplemented with 10% (vol/vol) FBS. MCF7 cells were grown in Opti-MEM containing 5% FBS. BC3H-1 cells were maintained in DMEM containing 20% FBS. For the myogenic differentiation of BC3H-1 cells, normal growth medium was replaced with DMEM containing 2% horse serum for 4 days prior to immunostaining and detection with 3-amino-9-ethylcarbazole (AEC) as described below.
Immunohistochemistry on mouse tissues
Ovaries and uterina tissues were obtained from mice that were euthanized by cervical dislocation. After incubation overnight in 4% paraformaldehyde, the tissues were subjected to alcohol dehydration and embedding in paraffin, followed by sectioning and mounting. Endogenous peroxidase activity was quenched by treatment with 3% H2O2-methanol for 10 min. Nonspecific binding was blocked with 1:300 dilution of normal goat serum for 30 min. Rabbit anti-p/CIP IgG antibody was diluted (1:300) in phosphate-buffered saline (PBS) containing 3% bovine serum albumin, added to the sections, and incubated for 15 h at room temperature in a humidified chamber. After the mixture was washed, goat anti-rabbit IgG antibody conjugated with horseradish peroxidase (1:300) was added for 30 min at room temperature. The slides were washed with three changes of PBS for 5 min and dipped once in H2O after the addition of the chromagen AEC or NovaRED as specified by the manufacturer (Vector). The slides were then counterstained with Mayer's hematoxylin (Sigma) for 5 min. All images were acquired using an Olympus Provis AX70A microscope equipped with Sensicam and analyzed using Image Pro Plus 4.0.
Immunohistochemical and immunofluorescence analysis of cell lines
Cells were grown to approximately 80% confluence before being washed with PBS, trypsinized, and plated onto glass slides at a cellular density of 106 cells/ml. The cells were allowed to adhere to the slides for approximately 18 h and then washed with cold PBS and fixed at 4°C in 4% paraformaldehyde-PBS for 10 min. This was immediately followed by a 5-min incubation in PBS containing 0.1% Triton X-100. Nonspecific binding was blocked with a 1:300 dilution of normal goat serum in PBS for 30 min. Rabbit anti-p/CIP IgG (1:400) and/or mouse anti- α-tubulin (Sigma) (1:500) antibody was diluted in PBS-3% bovine serum albumin before being applied to the slides, which were then incubated overnight at 4°C in a humidified chamber. For immunohistochemical detection of p/CIP, goat anti-rabbit antibody conjugated to horseradish peroxidase (Bio-Rad) was incubated for 1 h at room temperature before being washed with PBS, and then the chromagen (NovaRED) was added as specified by the manufacturer. For coimmunofluorescence detection, slides were incubated for 1 h at room temperature with anti-mouse IgG conjugated to rhodamine (1:500 dilution) and anti-rabbit IgG conjugated to fluorescein (1:500 dilution) (Santa Cruz). After several washes in PBS, the slides were mounted in aqueous mounting medium for viewing.
For the redistribution experiments, MEFs were serum starved according to previously published methods (42, 66). Briefly, the cells were allowed to adhere to slides overnight and then incubated in DMEM-0.5% FBS serum for approximately 72 h prior to stimulation with 100 nM EGF, insulin, or phorbol myristate acetate (PMA) for a maximum of 4 h. For some experiments, the cells were pretreated with 10μM cyclohexamide or 1 μM colchicine prior to the addition of various stimuli. They were then washed twice with ice-cold PBS, fixed in paraformaldehyde, and stained for p/CIP, as described above.
For the nuclear export inhibition assay, HeLa cells were seeded onto slides overnight in growth medium containing 10% FBS. Individual slides were then incubated for 30 min to 4 h in duplicate, with or without 50 nM leptomycin B.
Cell cycle synchronization studies
HeLa cells were plated onto slides and allowed to adhere for approximately 18 h in DMEM-10% FBS prior to the addition of 2.5 mM thymidine. After a 36-h incubation period, the cells were washed twice with PBS and changed to fresh culture medium to release the cells from S-phase arrest. To determine the proportion of cells present in a particular cell cycle phase, the cells were analyzed by flow cytometry at 30-min intervals. The cells were washed in PBS, harvested, centrifuged for 5 min at 200 × g and resuspended in 1 ml of chilled PBS. A small aliquot of cells was taken for trypan blue exclusion and counting to determine cell viability and number. The cell suspension was added dropwise to 4.5 ml of chilled 70% ethanol while vortexing, left in the ethanol solution overnight, centrifuged, washed once in PBS, and resuspended in propidium iodide staining solution (0.1% Triton X-100, 20 mg of RNase A, and 2 mg of PI in 100 ml of PBS). After a 30-min incubation at room temperature, the DNA content was analyzed using a Beckman Coulter flow cytometer. Duplicate slides were processed concurrently by washing twice with PBS and fixing for 5 min in 4% paraformaldehyde. They were then incubated with anti-p/CIP antibody and processed for immunohistochemical analysis, as described above.
Green fluorescence microscopy of living cells
NIH 3T3 and HeLa cells were plated into six-well plates and transfected with plasmids expressing GFP fusion proteins using Effectene transfection reagent (Qiagen). At 16 h posttransfection, living cells were washed once with PBS and resuspended in DMEM. Expression of the GFP fusions was analyzed using an Olympus Provis AX70A inverted microscope equipped with Sensicam. Images were captured and and analyzed using Image Pro Plus 4.0. For some experiments, transfected cells were treated with 50 nM LMB for 4 h prior to analysis.
Purification of p/CIP
Cytoplasmic and nuclear extracts were prepared by standard methods (13). The cytosol was dialyzed against buffer A (20 mM Tris [pH 7.9], 0.5 mM EDTA, 0.5 mM EGTA, 10% glycerol, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 5 μg each of leupeptin, aprotinin, and pepstatin per ml) containing 20 mM KCl and was used as starting material. This was loaded onto a DE52 anion-exchange column, and the column was eluted in a stepwise fashion with buffer A containing 0.15 M, 0.3 M, and 0.5 M KCl. The majority of cytosolic p/CIP was found in the 0.15 M KCl fraction. This fraction was concentrated using a Millipore concentrator and then passed through a Sephadex 200 column. All fractions were analyzed by Western blotting using anti-p/CIP antibody (1:3,000 dilution), and the p/CIP-containing fractions were pooled prior to affinity purification.
To purify the nuclear p/CIP, the nuclear extract was dialyzed against buffer A containing 100 mM KCl and loaded onto a P11 phosphocellulose column preequilibrated in the same buffer. The flowthrough was collected, and the column was washed sequentially with buffer A containing increasing concentrations of KCl. The 0.1 M fraction containing p/CIP was precipitated with 20 to 60% ammonium sulfate, and the precipitated proteins were resuspended in 4 ml of buffer A containing 100 mM KCl. This was then dialyzed against the same buffer to remove residual ammonium sulfate before being applied to a Sephacryl S300 column. The column was then washed with buffer A at a flow rate of 0.4 ml/min. Fractions were collected, pooled, and analyzed for p/CIP by Western blotting. The p/CIP-containing fractions were pooled and dialyzed against buffer A containing 100 mM KCl.
For immunoaffinity purification of p/CIP, affinity-purified p/CIP antibody was cross-linked to protein A-Sepharose by using dimethylpalmilidate by standard procedures (22). Fractions from the gel filtration step were pooled and precleared by being passed through a control affinity column containing anti-rabbit IgG. The eluant was then loaded onto the anti-p/CIP affinity column at a flow rate of 0.2 to 0.5 ml/min. The flowthrough was collected and reloaded on the column five times prior to elution of the bound proteins with 100 mM glycine (pH 2.8). For mock purification experiments, samples from the gel filtration step were loaded onto protein A-Sepharose cross-linked to an irrelevant antibody.
Measurement of histone acetyltransferase activity
The HAT activity was measured using a previously described method (4). Typically, approximately 50 μg of protein from the pooled gel filtration fractions containing p/CIP was suspended in 250 μl of IPH buffer (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 5 mM EDTA, 0.1% NP-40, 0.1 mM phenylmethylsulfonyl fluoride, 5 μg each of aprotinin, leupeptin, and pepstatin per ml) and incubated with 50 μl (50% [vol/vol] slurry) of anti-p/CIP affinity resin for 1 h at 4°C. The immuncomplexes were pelleted by gentle centrifugation and washed three times with 1 ml of IPH buffer. After the final wash, the buffer was aspirated down to 30 μl, 25 μg of total histones or BSA was added, the reaction was initiated by the addition of 1 μl of [3H]acetyl coenzyme A (acetyl-CoA) (1.85 mBq, 7.7 Ci/mmol; Amersham), and the mixture was incubated at 30°C for 30 min. The reaction products were spotted onto P-81 phosphocellulose paper disks, which were soaked in 50 mM NaHCO3 buffer (pH 9.2). The disks was washed several times with acetone:methanol:chloroform (1:1:1, vol/vol/vol) and dried. The incorporation of acetyl-CoA was determined by liquid scintillation counting.
RESULTS
p/CIP is found in the cytoplasm and in the nucleus
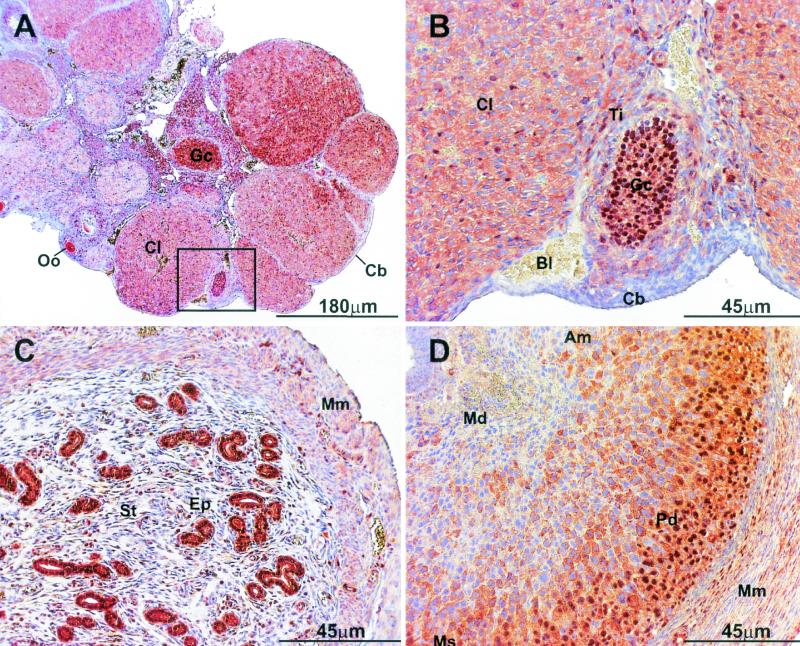
To characterize the subcellular distribution of p/CIP, we used a polyclonal antibody generated against a recombinant p/CIP protein. This antibody recognizes both the human and mouse forms of p/CIP, as determined by Western blotting and immunohistochemistry. Surprisingly, immunohistochemical analysis for p/CIP revealed a significant cytoplasmic component in many tissues of the developing mouse including the pancreas, lungs, and muscle and within the reproductive organs (data not shown and Fig. 1). A representative section of the female reproductive organs obtained from 6-week-old mice highlights the heterogenous distribution of endogenous p/CIP in vivo (Fig. 1). Examination of ovaries and uteri taken from adult virgin and pregnant mice revealed nucleocytoplasmic staining for p/CIP in a variety of different cell types. In the ovaries, intense nuclear staining was observed in the granulosa cells of the follicle surrounding the oocyte (Fig. 1A and B). In contrast, the surrounding thecal cells displayed mainly cytoplasmic staining for p/CIP. During the follicular stage of the ovarian cycle, granulosa cells respond to gonadotropins secreted by the anterior pituitary, which is essential for the proliferation and maturation of the follicle. This hormonal surge ultimately leads to the release of the oocyte during ovulation. Ovulation marks the beginning of the luteal stage, in which the newly formed corpus luteum secretes a large amount of progesterone and estradiol. Within the highly vascularized corpora lutea, which is composed mainly of terminally differentiated thecal and granulosa cells, p/CIP staining was predominantly cytoplasmic, although some sporadic nuclear staining was observed. In contrast, the cuboidal cells, which envelop the entire ovary, showed little, if any, staining for p/CIP.
FIG. 1.
Immunohistological analysis of p/CIP expression in mouse female reproductive organs. (A and B) Low magnification (A) and high magnification (B) of the same section of an ovary taken from a 6-week-old mouse. Predominantly nuclear staining was observed within the granulosa cells (Gc) of the tertiary follicle, and diffuse p/CIP staining was also observed within the cytoplasm. In the surrounding theca interna (Ti) and corpora lutea (Cl), mainly cytoplasmic staining was observed. The cubiodal cells (Cb) enveloping the entire ovary also displayed diffuse cytoplasmic staining. (C) Cross section of a uterine horn taken from an adult virgin mouse. Intense nuclear and cytoplasmic staining can be observed within the uterina epithelial cells (Ep), whereas in the surrounding stroma (St) and smooth muscle myometrium (Mm), cytoplasmic staining predominates. (D) Sagittal section of a uterus taken 7.5 d.p.c. Mature nonproliferating decidua (Md), close to the embryo proper, demonstrates little to no staining for p/CIP. Decidual cells in between the myometrium and the embryo show mainly diffuse cytoplasmic staining for p/CIP. In contrast, the outer layer of highly proliferative deciduas (Pd) demonstrates intense nuclear staining. Each of these tissues were isolated from approximately the same age and strain of mice. Control experiments were performed using nonimmune IgG or p/CIP IgG preincubated with antigen. In both cases, no staining was observed (data not shown). Oo, oocyte; Bl, blood; Am, antimesometrium; Ms, mesometrium.
In the uteri isolated from virgin female mice, intense p/CIP staining was observed in both the cytoplasmic and nuclear compartments of the uterine secretory epithelium (Fig. 1C). Cytoplasmic immunoreactivity was observed in the cells of the enveloping smooth muscle myometrium. The uteri isolated from 7.5 d.p.c. mice demonstrated little to no staining within differentiated (mature) nonproliferating decidua (Fig. 1D). However, increased cytoplasmic immunoreactivity was observed in the more proliferative decidua until it became intensely nuclear in the highly proliferative cells bordering the uteri (Fig. 1D). Overall, this type of compartmentalization was not restricted to the reproductive organs and was observed in many mouse tissues at various stages of differentiation and/or proliferation (data not shown) and suggests that the localization of p/CIP may be developmentally regulated.
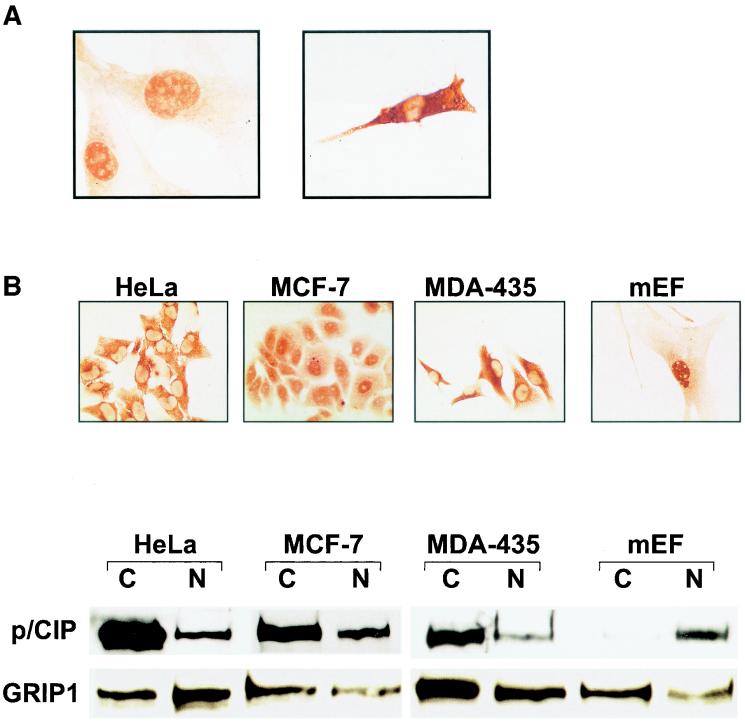
To investigate further the relationship between differentiation and p/CIP localization, we examined the localization of p/CIP during differentiation of the BC3H-1 nonfusing myocytic cell line (Fig. 2A). In proliferating undifferentiated BC3H-1 myoblasts, p/CIP was found predominantly in the nuclear compartment. However, when BC3H-1 cells were grown in differentiation medium, p/CIP became localized in the cytoplasm, although some residual nuclear staining was evident. These results suggest that cytoplasmic localization of p/CIP may be coupled to the muscle differentiation program.
FIG. 2.
Intracellular localization of p/CIP varies between cell types. (A) Differentiation of BC3H-1 cells stimulates cytoplasmic accumulation of p/CIP. Cells were placed in differentiation medium for 4 days (right panel) prior to immunostaining using anti-p/CIP antibody. (B) (Top) Immunohistochemical staining for p/CIP in various cell lines. Significant cytoplasmic staining was observed in the HeLa S3, MCF-7, and MDA-435 cell lines. In the MEFs, p/CIP was localized mainly to the nuclei of the cells. (Bottom) Western blot analysis of nuclear and cytosolic fractions isolated from the corresponding cell lines used in panel A. Approximately 20 μg of protein was loaded into each well prior to SDS-PAGE and Western blotting. The antibodies used are indicated on the left. C, cytosolic extract; N, nuclear extract;
We also examined the subcellular localization of endogenous p/CIP in a number of different cell lines that included HeLa cells, the MCF-7 and MDA-435 breast cancer cell lines, and MEFs isolated from 13-dpc embryos (Fig. 2B). All of the cell lines were asynchronous and tumor derived, with the exception of the fibroblasts. In the cell lines, the amount of p/CIP staining found in the nucleus was highly variable, although a prominent cytosolic component was always detected. For example, in the HeLa S3 cell line, we detected p/CIP predominantly in the cytoplasmic compartment, whereas in the MCF-7 cells, a strong nuclear signal, as well as a cytoplasmic signal, was detected. In contrast, in the MDA-435 cells, an estrogen receptor-negative cell line, p/CIP was found predominantly within the cytoplasmic compartment. In the MEFs, p/CIP was found in the nucleus although some cytoplasmic staining was also detected.
The subcellular localization of p/CIP was also examined by Western blotting of cytosolic and nuclear fractions isolated from the same cell types, which again indicated that p/CIP was relatively abundant in the cytoplasm (Fig. 2B). Identical results were obtained using a second antibody which recognizes a different epitope (data not shown). Finally, we examined the subcellular distribution of the related family member GRIP-1. Western blotting using a GRIP-1-specific antibody demonstrated that it was relatively abundant in both the cytosolic and nuclear fractions. Collectively, these data support the immunohistochemical observations that p/CIP and related family members are differentially distributed between the nucleus and cytoplasm depending on the cell type.
Localization of p/CIP is dynamically regulated
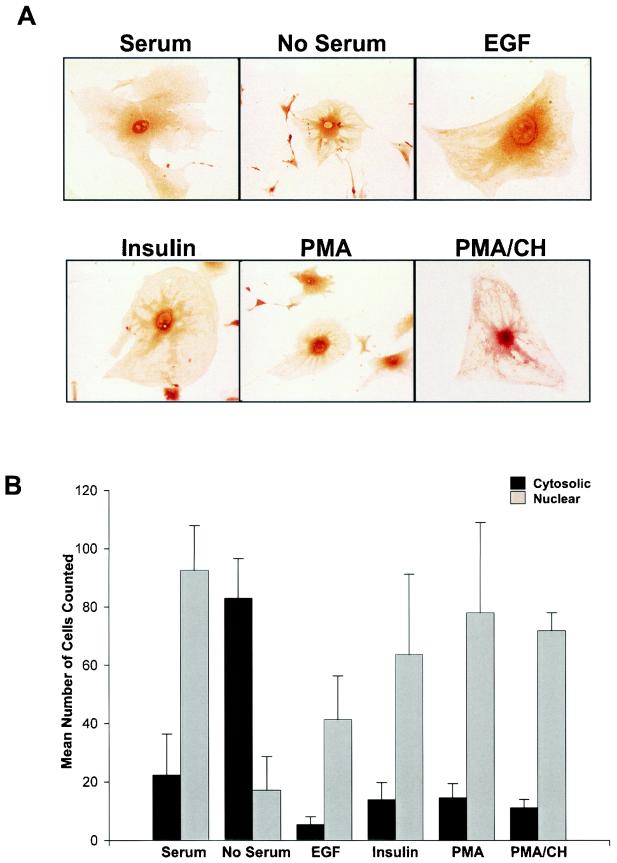
The distribution of p/CIP, both in vivo and in a number of cell types, suggests that it may undergo regulated intracellular trafficking. To assess whether components found in serum could stimulate p/CIP redistribution, MEFs were serum deprived and p/CIP localization was examined by immunohistochemical analysis. In the presence of serum-containing medium, p/CIP was found mainly in the nuclear compartment (Fig. 3). However, serum deprivation stimulated a dramatic shift in p/CIP localization to the cytoplasmic compartment. Interestingly, immunoreactivity for p/CIP was particularly intense in the perinuclear region. Similar results were obtained with NIH 3T3 cells (data not shown). To define specific factors responsible for mediating p/CIP redistribution, serum-starved cells were stimulated with various growth factors which included EGF and insulin as well as the tumor-promoting phorbol ester PMA. In the majority of cells, treatment with these agents resulted in a redistribution of p/CIP back into the nucleus (Fig. 3B). The effect of PMA was particularly dramatic, with significant redistribution occuring as early as 30 min following its addition. This redistribution was not dependent on changes in protein levels, since preincubation of cells with cycloheximide had no effect on p/CIP redistribution in response to PMA.
FIG. 3.
p/CIP undergoes redistribution in MEFs. (A) Immunohistochemical analysis of p/CIP in MEFs grown in the presence or absence of serum, or in serum-starved cells treated with either 100 nM EGF, insulin, PMA, or PMA in the presence of cycloheximide (PMA/CH). (B) Graphical representation of the results of the experiment in panel A, showing the mean number of cells counted in 10 random fields of cells stained for p/CIP in the presence or absence of serum, EGF, insulin, PMA, or PMA with cycloheximide. Error bars indicate standard errors of the means.
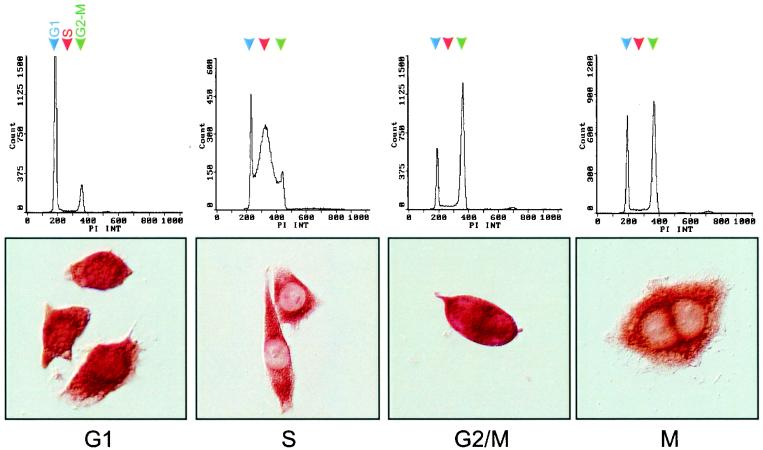
We also determined if p/CIP localization was dependent on the cell cycle by synchronizing a population of HeLa S3 cells at the beginning of S phase with a thymidine block, releasing the cells, and examining p/CIP distribution throughout the cell cycle (Fig. 4). Throughout the G1 phase, p/CIP was found in both the cytoplasmic and nuclear compartments, although a greater percentage of the cells showed higher levels of cytoplasmic staining. This changed dramatically in cells which were predominantly in S phase. In these cells, p/CIP accumulation was predominantly cytoplasmic and excluded from the nucleus. During late G2 phase, just prior to entry into mitosis and before nuclear envelope breakdown, a rapid nuclear accumulation of p/CIP was observed. This was again followed by redistribution back into the cytoplasm during late M phase. These results suggest that the subcellular localization of p/CIP can occur in a cell cycle-dependent manner.
FIG. 4.
Redistribution of p/CIP is cell cycle dependent. Subcellular compartmentalization of p/CIP was determined by immunohistochemistry of synchronized HeLa cells advancing through the cell cycle. HeLa cells were arrested in S phase on treatment with 2.5 mM thymidine for 36 h. Cells were collected for flow cytometry at 30-min intervals after removal of the blocker, and the proportions of cells in G1 (77.4%), S (68.8%), G2/M (66.2%), and M (57.5%) were determined by flow cytometry (top) or by immunostaining for p/CIP (bottom).
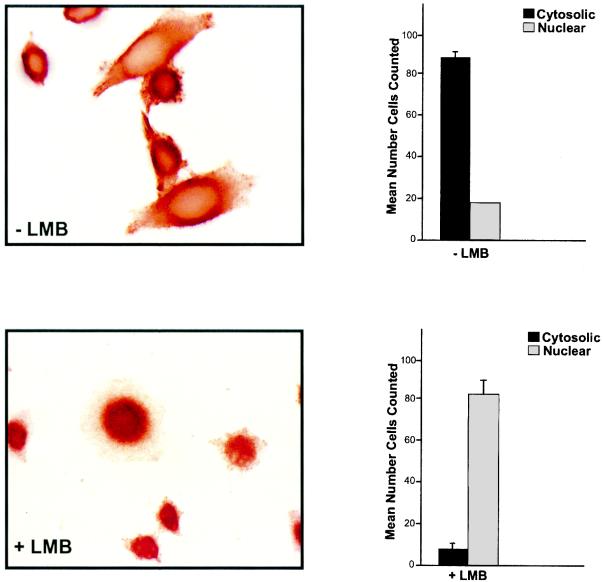
It is well known that the distribution of many proteins depends on their export as well as their import. Exportin/CRM1, the best-characterized export receptor, recognizes leucine-rich nuclear export sequence (NES) found within many proteins and is specifically inhibited by the antibiotic LMB. Therefore, to determine if the cytoplasmic accumulation of p/CIP depends on an active nuclear export mechanism, HeLa cells were treated with LMB for various periods, fixed, and stained with anti-p/CIP antibody. In cells treated with vehicle alone, p/CIP was both cytoplasmic and nuclear, consistent with our previous observations. However, in response to LMB, p/CIP was found predominantly within the nuclear compartment (Fig. 5). This suggests that the cytoplasmic accumulation of p/CIP reflects its continuous nuclear export as well as import.
FIG. 5.
p/CIP distribution is determined by nuclear export as well as nuclear import. (Bottom) Immunohistochemical analysis of HeLa cells treated with 50 nM LMB resulted in accumulation of p/CIP in the nucleus. (Top) In the absence of LBM, endogenous p/CIP expression was predominantly cytoplasmic with some nuclear staining. The corresponding graphs on the right represent the mean numbers of cells in six random fields of cells stained for p/CIP in the absence or presence of LMB. Data are representative of four separate experiments. Error bars indicate standard errors of the means.
p/CIP localization is mediated by nuclear import and export signals
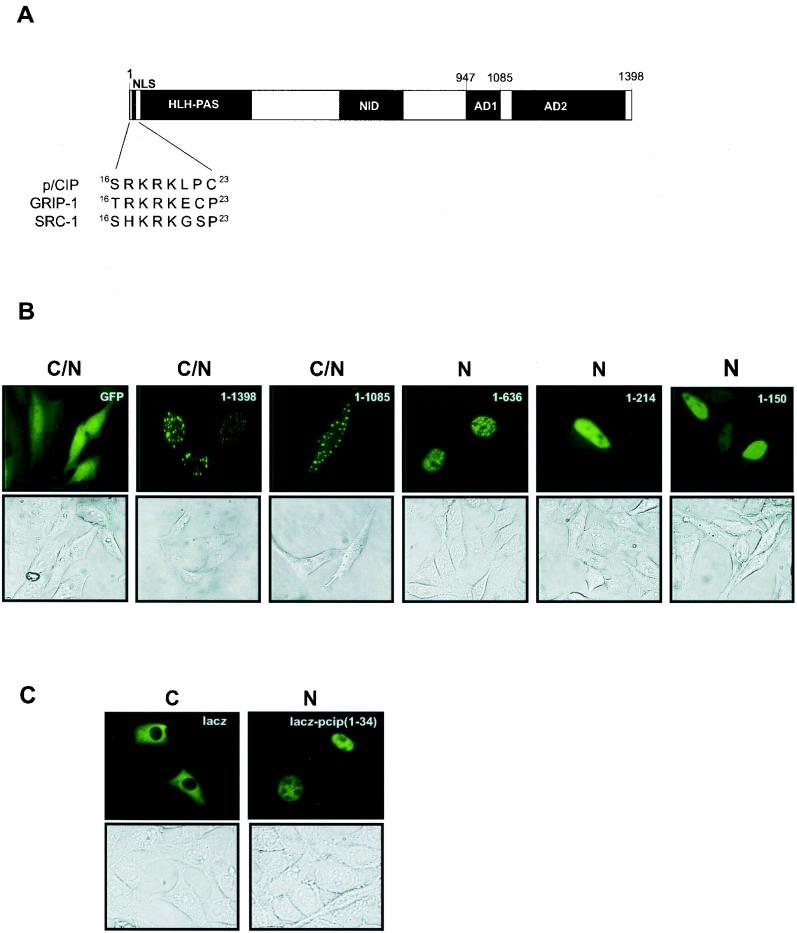
To define specific sequences which may mediate cellular localization, we generated a panel of p/CIP truncation mutants fused to the carboxy terminus of GFP and expressed them in living cells. The expression of full-length p/CIP (GFP-p/CIP aa 1-1398) or a truncation mutant in which the C-terminal activation domain (AD2) was deleted (GFP-p/CIP aa 1-1085) resulted in a strong cytoplasmic as well as nuclear signal when analyzed by fluorescence microscopy (Fig. 6B). Interestingly, the expression of the GFP fusions was not diffuse but was concentrated in discrete foci throughout the cell. Further truncation mutations were localized exclusively to the nucleus, which indicated that the amino terminus of p/CIP harbors determinants responsible for nuclear localization. Inspection of the primary amino acid sequence revealed a motif, within the first 30 aa, which resembles the NLS of the simian virus 40 large T antigen (Fig. 6A). To confirm that this motif represents a bona fide NLS, we tested its ability to target an expression vector containing GFP fused to lacZ, which is normally expressed in the cytoplasm. The addition of p/CIP (aa 1-34) containing the KKRK motif resulted in the accumulation of the lacZ-GFP fusion exclusively in the nucleus of all cells examined (Fig. 6C).
FIG. 6.
p/CIP contains an NLS in its amino terminus. (A) Schematic representation highlighting the NLS in p/CIP and the homologous regions found in GRIP-1 and mouse SRC-1. (B) Representative green fluorescent images of living cells expressing GFP-p/CIP fusion constructs. HeLa cells were transfected with expression plasmids containing GFP fused to p/CIP deletion mutants as indicated in the upper right-hand corner. The lower images represent the corresponding bright-field images. (C) Representative green fluorescent images of living cells expressing lacZ fused to GFP (left) or lacZ-GFP fused to p/CIP (aa 1-34). C, predominantly cytosolic localization; N, predominantly nuclear localization; C/N, cytosolic and nuclear localization observed in more than 75% of the cells in a random field of view consisting of approximately 50 cells.
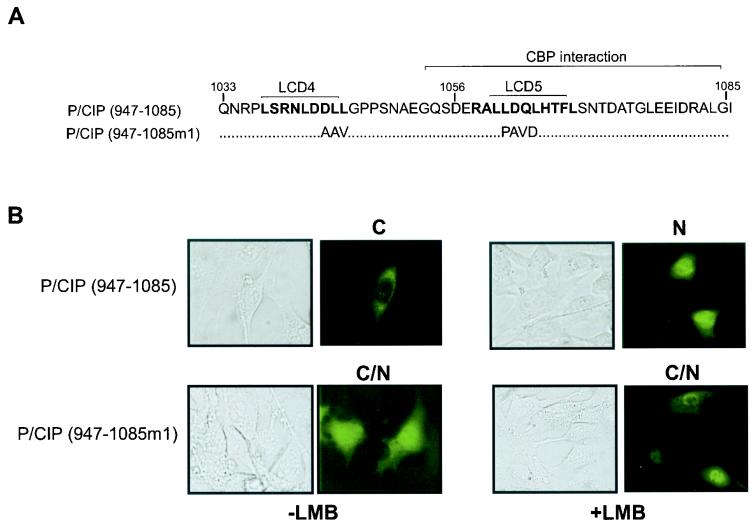
The observation that LMB causes nuclear accumulation of endogenous p/CIP suggests that p/CIP may also contain a CRM1-dependent NES which, based on the results of the above experiments, may be localized in the carboxy terminus. The region of p/CIP corresponding to aa 1033 to 1085 contains two leucine-rich regions (designated LCD4 and LCD5) which resemble CRM1-dependent nuclear export consensus sites (Fig. 7A). These generally include a series of spatially positioned leucine or similar hydrophobic residues, although many CRM1-dependent NESs diverge from this consensus site (24). To determine whether this region of p/CIP is sufficient to confer a cytoplasmic localization, we expressed a GFP-p/CIP (aa 947-1085) fusion in living cells. Remarkably, this GFP fusion was found predominantly in the cytoplasmic compartment in approximately 75% of the cells examined and treatment of cells with LMB resulted in a shift to the nuclear compartment in the majority of cells (Fig. 7B). In contrast, when amino acids within the LCD4 and LCD5 region were mutated, the expression of this fusion protein was much more diffuse and no significant redistribution was observed in response to LMB. Collectively, these results support the hypothesis that p/CIP distribution is mediated by intrinsic nuclear import and export signals.
FIG. 7.
The CBP/p300 interaction domain contains an NES region. (A) Amino acid sequence of the CBP/p300 interaction domain containing the two leucine-rich regions. The NES consensus sites are highlighted in bold. (B) Representative green fluorescent images of living cells expressing GFP-p/CIP fusion constructs. HeLa cells were transfected with expression plasmids containing GFP fused to p/CIP (aa 947-1085) or GFP-p/CIP (aa 947-1085m1) in which the leucine-rich regions were mutated as indicated in panel A. The cells were then treated with vehicle (−LMB) or 50 nM LMB (+ LMB) for 4 h. The images on the left represent the corresponding bright-field images. C, predominantly cytosolic localization; N, predominantly nuclear localization; C/N, cytosolic and nuclear localization observed in more than 75% of the cells in a random field of view consisting of approximately 50 cells.
Cytoplasmic p/CIP is associated with the microtubule network
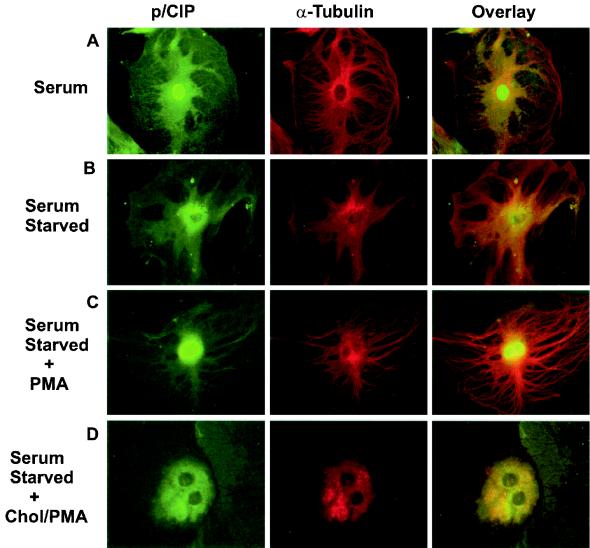
In several of the cell types we examined, the staining pattern of p/CIP following serum withdrawal is highly consistent with the reticular pattern observed for the cytoskeletal microtubule network. Based on this observation, we hypothesized that a component of p/CIP may somehow be tethered to microtubules in the cytoplasm and, consequently, sequestered from transcriptional complexes. On stimulation, this pool can be rapidly sequestered to the nucleus in response to the specific needs of the cell. To test this hypothesis, we used serum-starved MEFs and visualized microtubules by immunofluorescence using a monoclonal antibody directed against α-tubulin, which stained exclusively cytoplasmic (Fig. 8A). Comparison of the immunofluorescence staining patterns of p/CIP and α-tubulin clearly demonstrate similar reticular patterns. Importantly, colocalization of p/CIP and α-tubulin was confirmed by the yellow color that is apparent in the overlay image. These observations indicate that a component of cytosolic p/CIP is indeed associated with cellular microtubules. To determine if the interaction of p/CIP with the microtubule network is a necessary requirement for p/CIP shuttling, we preincubated cells with colchicine, which disrupts the microtubule network (Fig. 8D). This resulted in significantly diminished nuclear accumulation of p/CIP in response to PMA, suggesting that a functional microtubule network is required for intracellular trafficking of p/CIP.
FIG. 8.
Colocalization of p/CIP and α-tubulin in MEFs. p/CIP staining (green) and α-tubulin (red) are colocalized in the cytoplasm, as indicated by the yellow color in the overlay images. (A) Cells grown in the presence of serum. (B) Immunofluorescence of serum-starved cells. (C) Immunofluorescence of serum-starved cells which have been stimulated for 4 h with 100 nM PMA. (D) Immunofluorescence of serum-starved cells which have been stimulated for 4 h with 100 nM PMA after colchicine (Chol) treatment. No p/CIP shuttling was observed, in response to PMA, when microtubules were disrupted.
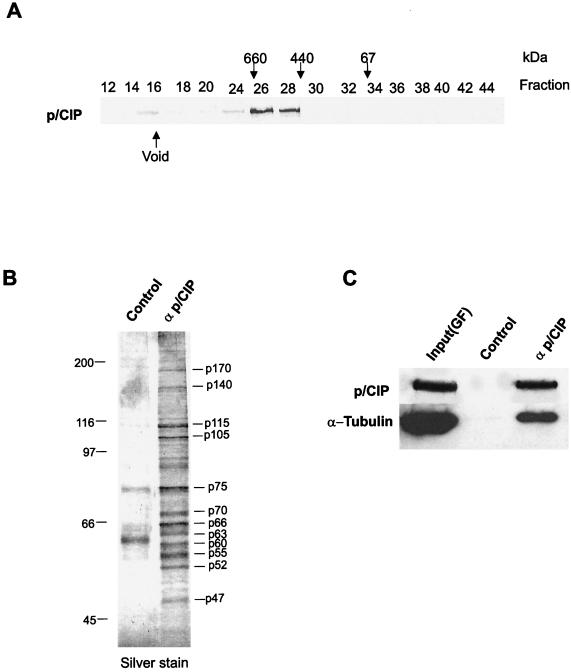
To confirm the association between endogenous p/CIP and microtubules biochemically, cytosolic extracts were isolated from HeLa cells by using conventional chromatography in combination with immunoaffinity purification (Fig. 9). When analyzed by gel filtration chromatography, cytosolic p/CIP consistently eluted as a single major peak corresponding to a molecular mass of approximately 700 kDa (Fig 9A). The p/CIP-containing fractions isolated by gel filtration chromatography were pooled and then fractionated by affinity chromatography using anti-p/CIP antibody cross-linked to protein A-Sepharose. Analysis of the affinity purified proteins by Western blotting demonstrated that both p/CIP and α-tubulin were indeed present (Fig. 9C). In addition, silver staining of the affinity-purified complex identified several other proteins that copurify with p/CIP, suggesting that this complex is heterogenous (Fig. 9B).
FIG. 9.
α-Tubulin is present in the affinity-purified cytosolic p/CIP complex. (A) Gel filtration chromatography of HeLa cell cytosolic extracts. (B) Silver stain SDS-PAGE gel of affinity-purified p/CIP-associated proteins isolated from HeLa cell cytosolic extracts. (C) Western blot of affinity- purified p/CIP isolated from HeLa cell cytosolic extract. p/CIP and its associated proteins were isolated from cytosolic extract by affinity chromatography using an anti-p/CIP affinity column or control affinity column as indicated above the lanes. Affinity-purified proteins were Western blotted and probed for either α-tubulin or p/CIP as indicated on the left. Input(GF) indicates an aliquot of the pooled p/CIP-containing fractions obtained from the gel filtration chromatography.
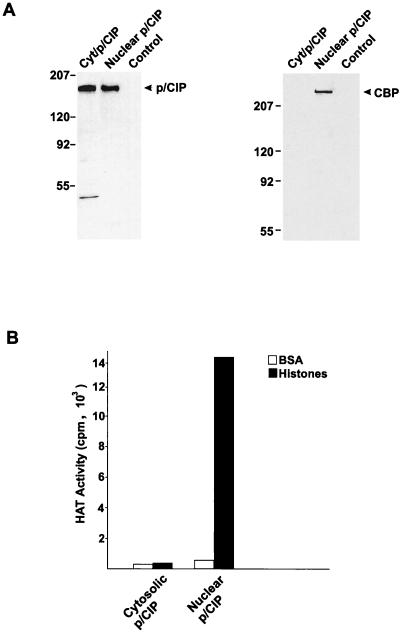
To provide further evidence that p/CIP associates with different proteins depending on its location, we also examined the colocalization of the transcriptional coactivator CBP, which can associate with p/CIP, as well as other SRC proteins, in vivo and in vitro (31, 39, 65, 68, 73). This association appears to be critical for mediating some of the transcriptional effects of NRs. CBP was found only in the immunopurified p/CIP fraction isolated from the nuclear extract and was absent from the cytosolic complex (Fig. 10A).
FIG. 10.
Only nuclear p/CIP possesses HAT activity. (A) Western blots of p/CIP isolated from either cytosolic (Cyt/p/CIP) or nuclear (Nuclear p/CIP) extracts using an anti-p/CIP affinity column or an anti-IgG column as a control. Aliquots of the affinity-purified nuclear and cytoplasmic p/CIP complexes were separated by SDS-PAGE, transferred to nitrocellulose, and probed with anti-p/CIP or anti-CBP antibodies indicated on the right of each blot. (B) HAT activity of affinity-purified complexes. P/CIP was affinity purified from the cytosolic and nuclear HeLa cell extracts, and aliquots of each were incubated with either BSA or free histones in a buffer containing [3H]acetyl-CoA. Histones were isolated, and the incorporation of [3H]acetyl-CoA was determined by liquid scintillation counting. The data are representative of two separate purifications.
Finally, we tested the ability of cytoplasmic and nuclear p/CIP complexes to acetylate free histones in vitro. Consistent with the presence of CBP, immunopurified nuclear p/CIP complexes possess a very robust HAT activity in vitro (Fig. 10B). In contrast, no HAT activity was detected in the immunopurified cytoplasmic p/CIP component. Collectively, these results indicate that cytoplasmic and nuclear p/CIP are associated with different proteins and, furthermore, that there are functional differences associated with the cytoplasmic and nuclear p/CIP complexes that are reflected in their ability to acetylate histones and possibly other proteins.
DISCUSSION
In this study, we have found that endogenous p/CIP is cytoplasmic and undergoes nucleocytoplasmic shuttling in response to various stimuli. A predominant cytoplasmic localization was also detected for GRIP-1, based on Western blotting of subcellular fractions, suggesting that a similar mechanism may regulate the distribution of other SRC proteins. We have also demonstrated that a substantial fraction of cytoplasmic p/CIP associates with the microtubule network and that this association plays a role in the mechanism of nuclear import of p/CIP.
Immunohistochemical analysis of p/CIP localization in the female reproductive organs of mice clearly showed a segregated pattern of localization depending on the cell type. Intense nuclear staining was observed in the highly proliferative follicular cells, whereas in the corpus luteum, which is composed mainly of terminally differentiated thecal and granulosa cells, p/CIP was found predominantly in the cytoplasmic compartment. Within the uteri of pregnant female mice, we consistently observed a “gradient-like” nuclear localization of p/CIP, with increasing nuclear localization observed in cells directly adjacent to the myometrium. Taken together, these results suggest a possible correlation between the proliferative and/or differentiation state of a particular cell type, and p/CIP localization in mouse tissues. This hypothesis is supported by additional correlative evidence. For example, in synchronized HeLa cells, cytoplasmic localization of p/CIP is cell cycle dependent. Surprisingly, in these cells, p/CIP is absent from the nucleus during S phase and translocates to the nucleus at late G2 phase in a manner similar to cyclin B1 (54). This occurs prior to nuclear envelope breakdown and chromosomal condensation, supporting a possible role for p/CIP in mitotic events (data not shown). In addition, factors which promote cell cycle progression, such as insulin, EGF, or PMA, triggered a dramatic redistribution of p/CIP from the cytoplasmic to the nuclear compartment in serum-starved cells. Finally, we have shown that the induction of differentiation of BC3H-1 myoblasts stimulates the translocation of p/CIP to the cytoplasm. In contrast, a recent report has shown that the induction of differentiation of C2C12 myoblasts by serum withdrawal stimulates the translocation of GRIP1 to the nucleus (8). Collectively, these results suggest that there may be functional differences between various SRC factors, depending on the cell type.
While our results demonstrate that p/CIP undergoes both import and export, they do not address the status of p/CIP during transport. However, one plausible explanation is that the shuttling mechanism involves the phosphorylation of p/CIP, either directly or indirectly, by protein kinase C (PKC). The PKC family of serine/threonine kinases consists of 12 distinct isoforms which can be classified according to their cofactor requirements (43). All of the isoforms have a common requirement for phosphatidylserine. Class I, which consists of three isoforms (α, β, and γ), also requires Ca2+ and diacylglycerol (DAG) for activity. The remaining isoforms show various degrees of dependence on DAG or Ca2+ (10, 11). Phorbol esters such as PMA are potent activators of PKC and mimic the effects of DAG, resulting in a sustained activation of PKC. Interestingly, it has been shown in several cell types that both EGF and insulin can activate phospholipase C to release DAG, which, in turn, activates several distinct isoforms of PKC (3, 10, 11).
Our results are in agreement with previous results demonstrating that p/CIP undergoes rapid nuclear import in Rat 1 cells in response to insulin (70). This response is inhibited by okadiac acid, suggesting that the mechanism of nuclear import may be phosphorylation dependent, although it remains to be determined whether phosphorylation is directly involved in regulating p/CIP distribution. Alternatively, because p/CIP redistribution involves a nuclear export pathway as well as an import pathway, changes in the relative activity of either, as a result of PKC activation, could alter the distribution of p/CIP in a given cell type.
p/CIP contains nuclear import and export signals
We have identified a novel NLS in the N terminus of p/CIP which exhibits strong nuclear targeting activity when fused to GFP-lacZ. Interestingly, the p/CIP NLS, which is conserved in other members of the SRC family, resembles the simian virus 40 T antigen class of NLSs, which are arginine and lysine rich, suggesting that the SRC proteins may interact with the importin-α family of proteins.
The NES region contains two lysine-rich domains which conform to the consensus NESs found in several other proteins that exhibit CRM-1-dependent nuclear export. Overexpression of a GFP fusion containing the NES region is retained in the cytoplasm and is LMB sensitive. Importantly, a mutant version of the GFP fusion is no longer restricted to the cytoplasm and is no longer responsive to LMB. Thus, nuclear export of p/CIP is likely to be achieved through its direct interaction with CRM-1. Interestingly, amino acids which participate in nuclear export are also critical for the coactivator activity of p/CIP (Fig. 7 and data not shown). In addition, the recent nuclear magnetic resonance spectroscopic structural determination of the ACTR/CBP interface indicates that the NES region partially overlaps with the CBP interaction domain (12). Collectively, these results support a model in which the binding of CBP and of CRM-1 to p/CIP are mutually exclusive. Consequently, during transcriptional activation when p/CIP is bound to CBP, the NES may be masked and incapable of interacting with CRM-1. The physiological stimuli which regulate the interaction between CBP/p300 and p/CIP are not known, although it has been demonstrated that mitogen-activated protein kinase activation stimulates the recruitment of p300, suggesting that the mechanism involved may be phosphorylation dependent (15).
Cytoplasmic p/CIP associates with microtubules
Using both immunohistochemical localization and biochemical purification, we have also demonstrated that endogenous p/CIP associates with the microtubule network. The association of proteins with the microtubule network is rapidly emerging as a potentially important pathway for regulating transcription factor activity. Microtubules are composed of polar polymerized tubulin tracks along which kinesin and dynein motor proteins transport a wide variety of cargos in response to various stimuli. Studies carried out on the glucocorticoid and androgen receptors have demonstrated that an intact cytoskeleton is necessary for nucleocytoplasmic shuttling of the receptor and that disruption of the cytoskeletal components renders the receptor unable to shuttle between the cytoplasm and the nucleus (52, 53). The SMAD proteins, which mediate transcriptional activation of transforming growth factor β-responsive genes, bind microtubules, and microtubule-destabilizing drugs disrupt the SMAD-microtubule interaction, resulting in the nuclear import of SMAD proteins (14). More recently, it has been shown that p53 is transported along microtubules in a dynein-dependent manner and that this facilitates its accumulation in the nucleus after DNA damage (16).
The association of p/CIP with the microtubule network may serve multiple functions. First, compartmentalization of p/CIP within the cytoplasm may represent an active retention mechanism, limiting the accessibility of p/CIP to the transport machinary and consequently preventing it from entering the nucleus. In other words, this type of association may serve as a negative regulator of transcription. This is also supported by the finding that immunopurified p/CIP from cytosolic extracts is devoid of any HAT activity, based on the in vitro acetylation of core histones. In contrast, nuclear p/CIP, in association with CBP, possesses a robust HAT activity. Our inability to detect HAT activity associated with cytoplasmic p/CIP was surprising in light of recent reports demonstrating that both SRC-1 and ACTR possess intrinsic HAT activity that is mediated by their respective carboxy termini (6, 61). It is possible, however, that p/CIP HAT activity may be inhibited, perhaps as a result of its interaction with other proteins which may be present in the cytoplasmic complex. Second, our results suggest that the association of p/CIP with the cytoskeleton is required for its translocation. This is based on the observation that disruption of the microtubule network with colchicine prevents shuttling of p/CIP in response to PMA. Third, a shuttling mechanism which serves to control entry into the nucleus might allow p/CIP to interact with a repertoire of transcriptional proteins depending on the specific needs of the cell.
Intracellular shuttling between the cytoplasm and the nucleus often acts as switch to regulate the activity of many proteins. For example, transcription factors containing a conserved Per-Arnt-Sim(PAS) domain, such as the dioxin receptor and HIF1α, are found in the cytoplasm in unstimulated cells and are translocated to the nucleus in response to specific signals, where they activate the transcription of target genes (21, 27). More recently, it has been shown that differentiation of muscle cells is dependent on the regulated nuclear export of the class II HDACs HDAC4 and HDAC5 (46, 71). In undifferentiated muscle cells, HDAC4 and HDAC5 interact with members of the myocyte enhancer factor 2 (MEF-2) family of transcription factors, resulting in repression of MEF-2-dependent genes. Upon activation of the Ca/calmodulin-dependent signalling pathway, HDAC4 and HDAC5 are phosphorylated and are exported from the nucleus. This relieves repression of the transcription factor MEF-2, which, in turn, associates with MyoD to activate the expression of muscle-specific genes.
Amplified in breast cancer I (AIB1), the human homologue of p/CIP, was identified within a region on chromosome 20, which is often amplified in breast and ovarian cancer (2). Preliminary results from our laboratory have shown that cytoplasmic localization of p/CIP is a characteristic feature in specific types of breast cancers (data not shown), suggesting that compartmentalization of p/CIP could have important implications in breast cancer development. For example, it is conceivable that normal cell growth is dependent on an equilibrium between the cytoplasmic and nuclear SRC proteins. Consequently, the “oncogenic” behavior of p/CIP may be linked not only to its overexpression but also to a shift in equilibrium between the cytoplasmic and nuclear components, resulting in abnormal growth. This type of mechanism has also been postulated for p53, which relies on a regulated transport mechanism to shuttle into and out of the nucleus (47, 57). Under normal conditions, p53 activity is regulated by preventing its import to the nucleus, in part through its association with microtubules. In response to specific extracellular stimuli, p53 relocalizes to the nucleus to regulate transcriptional processes. However, in a percentage of p53-related tumors, particularly in breast cancer and neuroblastoma, p53 is predominantly cytoplasmic and is inactivated and may contribute to uncontrolled cell proliferation. Based on the observations presented in this study, we believe that the cytoplasmic and nuclear localization of p/CIP may be an important regulatory mechanism that serves to control the availability of coactivator proteins in the nucleus and possibly to limit the activity of NRs at specific stages of the cell cycle or during normal development. In addition, this type of regulation could serve to integrate diverse signaling pathways at the level of coactivator proteins and could have important implications for understanding cell growth in breast cancer. Further delineation of the mechanism of nucleocytoplasmic shuttling of p/CIP is necessary to understand its complex mode of regulation.
ADDENDUM IN PROOF
While this paper was in revision, a report by Wu et al. describing the translocation of SRC-3 in response to tumor necrosis factor was published (R.-C. Wu, J. Qin, Y. Hashimoto, J. Wong, J. Xu, S. Y. Tsai, M.-J. Tsai, and B. W. O’Malley, Mol. Cell. Biol. 22: 3549-3561, 2002.)
Acknowledgments
We are grateful to R. Evans for the ACTR antibody and to Minoru Yoshida, The University of Tokyo, for kindly providing LMB. Special thanks go to Caroline Underhill and Lynne Cayer for technical assistance. We thank Alan Tuck and Nancy Kerkvleit for providing immunohistochemical sections of the human breast cancer tissue biopsy specimens, Michael Keeney for use of the flow cytometer, and Barb Lyons for animal husbandry. We thank M. Tini for critical reading of the manuscript.
This work was supported by an operating grant from the National Cancer Institute of Canada (grant 012298).
REFERENCES
- 1.Anafi, M., Y. F. Yang, N. A. Barlev, M. V. Govindan, S. L. Berger, T. R. Butt, and P. G. Walfish. 2000. GCN5 and ADA adaptor proteins regulate triiodothyronine/GRIP1 and SRC-1 coactivator-dependent gene activation by the human thyroid hormone receptor. Mol. Endocrinol. 14:718-732. [DOI] [PubMed] [Google Scholar]
- 2.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. [DOI] [PubMed] [Google Scholar]
- 3.Braiman, L., L. Sheffi-Friedman, A. Bak, T. Tennenbaum, and S. R. Sampson. 1999. Tyrosine phosphorylation of specific protein kinase C isoenzymes participates in insulin stimulation of glucose transport in primary cultures of rat skeletal muscle. Diabetes 48:1922-1929. [DOI] [PubMed] [Google Scholar]
- 4.Brownell, J. E., and C. D. Allis. 1995. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc. Natl. Acad. Sci. USA 92:6364-6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. [DOI] [PubMed] [Google Scholar]
- 6.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 8.Chen, S. L., S. C. Wang, B. Hosking, and G. E. Muscat. 2001. Subcellular localization of the steroid receptor coactivators (srcs) and mef2 in muscle and rhabdomyosarcoma cells. Mol. Endocrinol. 15:783-796. [DOI] [PubMed] [Google Scholar]
- 9.Collingwood, T. N., F. D. Urnov, and A. P. Wolffe. 1999. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J. Mol. Endocrinol. 23:255-275. [DOI] [PubMed] [Google Scholar]
- 10.Corbit, K. C., D. A. Foster, and M. R. Rosner. 1999. Protein kinase Cδ mediates neurogenic but not mitogenic activation of mitogen-activated protein kinase in neuronal cells. Mol. Cell. Biol. 19:4209-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbit, K. C., J.-W. Soh, K. Yoshida, E. M. Eves, I. B. Weinstein, and M. R. Rosner. 2000. Different protein kinase C isoforms determine growth factor specificity in neuronal cells. Mol. Cell. Biol. 20:5392-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demarest, S. J., M. Martinez-Yamout, J. Chung, H. Chen, W. Xu, H. J. Dyson, R. M. Evans, and P. E. Wright. 2002. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature 415:549-553. [DOI] [PubMed] [Google Scholar]
- 13.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong, C., Z. Li, R. Alvarez, Jr., X. H. Feng, and P. J. Goldschmidt-Clermont. 2000. Microtubule binding to Smads may regulate TGF beta activity. Mol. Cell 5:27-34. [DOI] [PubMed] [Google Scholar]
- 15.Font de Mora, J., and M. Brown. 2000. AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol. Cell. Biol. 20:5041-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannakakou, P., D. L. Sackett, Y. Ward, K. R. Webster, M. V. Blagosklonny, and T. Fojo. 2000. p53 is associated with cellular microtubules and is transported to the nucleus by dynein. Nat. Cell Biol. 2:709-717. [DOI] [PubMed] [Google Scholar]
- 17.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 18.Gorlich, D., M. Dabrowski, F. R. Bischoff, U. Kutay, P. Bork, E. Hartmann, S. Prehn, and E. Izaurralde. 1997. A novel class of RanGTP binding proteins. J. Cell Biol. 138:65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hache, R. J., R. Tse, T. Reich, J. G. Savory, and Y. A. Lefebvre. 1999. Nucleocytoplasmic trafficking of steroid-free glucocorticoid receptor. J. Biol. Chem. 274:1432-1439. [DOI] [PubMed] [Google Scholar]
- 20.Hammer, G. D., I. Krylova, Y. Zhang, B. D. Darimont, K. Simpson, N. L. Weigel, and H. A. Ingraham. 1999. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. Mol. Cell 3:521-526. [DOI] [PubMed] [Google Scholar]
- 21.Hankinson, O. 1995. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 35:307-340. [DOI] [PubMed] [Google Scholar]
- 22.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 24.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256:213-224. [DOI] [PubMed] [Google Scholar]
- 25.Hong, H., K. Kohli, A. Trivedi, D. L. Johnson, and M. R. Stallcup. 1996. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl. Acad. Sci. USA 93:4948-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaffman, A., and E. K. O’Shea. 1999. Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol. 15:291-339. [DOI] [PubMed] [Google Scholar]
- 27.Kallio, P. J., K. Okamoto, S. O'Brien, P. Carrero, Y. Makino, H. Tanaka, and L. Poellinger. 1998. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1α. EMBO J. 17:6573-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamei, Y., L. Xu, T. Heinzel, J. Torchia, R. Kurokawa, B. Gloss, S. C. Lin, R. A. Heyman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1996. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85:403-414. [DOI] [PubMed] [Google Scholar]
- 29.Kao, V., A. Tsai, C. C. Simon, C. Juguilon, H. S. Khochbin. 2001. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276:47496-47507. [DOI] [PubMed] [Google Scholar]
- 30.Kim, H.-J., J.-Y. Yi, H.-S. Sung, D. D. Moore, B. H. Jhun, Y. C. Lee, and J. W. Lee. 1999. Activating signal cointegrator 1, a novel transcription coactivator of nuclear receptors, and its cytosolic localization under conditions of serum deprivation. Mol. Cell. Biol. 19:6323-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, M. Y., S. J. Hsiao, and W. L. Kraus. 2001. A role for coactivators and histone acetylation in estrogen receptor alpha-mediated transcription initiation. EMBO J. 20:6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koh, S. S., D. Chen, Y. H. Lee, and M. R. Stallcup. 2001. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J. Biol. Chem. 276:1089-1098. [DOI] [PubMed] [Google Scholar]
- 33.Korzus, E., J. Torchia, D. W. Rose, L. Xu, R. Kurokawa, E. M. McInerney, T. M. Mullen,C. K. Glass, and M. G. Rosenfeld. 1998. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science 279:703-707. [DOI] [PubMed] [Google Scholar]
- 34.Kurokawa, R., D. Kalafus, M. H. Ogliastro, C. Kioussi, L. Xu, J. Torchia, M. G. Rosenfeld, and C. K. Glass. 1998. Differential use of CREB binding protein-coactivator complexes. Science 279:700-703. [DOI] [PubMed] [Google Scholar]
- 35.Lanz, R. B., N. J. McKenna, S. A. Onate, U. Albrecht, J. Wong, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1999. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97:17-27. [DOI] [PubMed] [Google Scholar]
- 36.Lee, D. Y., J. J. Hayes, D. Pruss, and A. P. Wolffe. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72:73-84. [DOI] [PubMed] [Google Scholar]
- 37.Leers, J., E. Treuter, and J.-Å. Gustafsson. 1998. Mechanistic principles in NR box-dependent interaction between nuclear hormone receptors and the coactivator TIF2. Mol. Cell. Biol. 18:6001-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, H., P. J. Gomes, and J. D. Chen. 1997. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc. Natl. Acad. Sci. USA 94:8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, J., B. W. O'Malley, and J. Wong. 2000. p300 requires its histone acetyltransferase activity and SRC-1 interaction domain to facilitate thyroid hormone receptor activation in chromatin. Mol. Cell. Biol. 20:2031-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, Z., J. Wong, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2001. Sequential recruitment of steroid receptor coactivator-1 (SRC-1) and p300 enhances progesterone receptor-dependent initiation and reinitiation of transcription from chromatin. Proc. Natl. Acad. Sci. USA 98:12426-12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez, G. N., C. W. Turek, F. Schaufele, M. R. Stallcup, and P. J. Kushner. 2001. Growth factors signal to steroid receptors through mitogen-activated protein kinase regulation of p160 coactivator activity. J. Biol. Chem. 276:22177-22182. [DOI] [PubMed] [Google Scholar]
- 42.Mangiarotti, R., M. G. Bottone, M. Danova, and C. Pellicciari. 1998. Bivariate flow cytometric analysis of DNA content versus immunopositivity for ribonucleotide reductase M1 subunit in the cell cycle. Cytometry 32:78-85. [DOI] [PubMed] [Google Scholar]
- 43.Martelli, A. M., N. Sang, P. Borgatti, S. Capitani, and L. M. Neri. 1999. Multiple biological responses activated by nuclear protein kinase C. J. Cell. Biochem. 74:499-521. [PubMed] [Google Scholar]
- 44.McInerney, E. M., D. W. Rose, S. E. Flynn, S. Westin, T. M. Mullen, A. Krones, J. Inostroza, J. Torchia, R. T. Nolte, N. Assa-Munt, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McInerney, E. M., M. J. Tsai, B. W. O'Malley, and B. S. Katzenellenbogen. 1996. Analysis of estrogen receptor transcriptional enhancement by a nuclear hormone receptor coactivator. Proc. Natl. Acad. Sci. USA 93:10069-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKinsey, T. A., C. L. Zhang, J. Lu, and E. N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moll, U. M., M. LaQuaglia, J. Benard, and G. Riou. 1995. Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc. Natl. Acad. Sci. USA 92:4407-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Na, S. Y., S. K. Lee, S. J. Han, H. S. Choi, S. Y. Im, and J. W. Lee. 1998. Steroid receptor coactivator-1 interacts with the p50 subunit and coactivates nuclear factor κB-mediated transactivations. J. Biol. Chem. 273:10831-10834. [DOI] [PubMed] [Google Scholar]
- 49.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, M. G. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and co- activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395:137-43. [DOI] [PubMed]
- 50.Onate, S. A., V. Boonyaratanakornkit, T. E. Spencer, S. Y. Tsai, M. J. Tsai, D. P. Edwards, and B. W. O'Malley. 1998. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J. Biol. Chem. 273:12101-12108. [DOI] [PubMed] [Google Scholar]
- 51.Onate, S. A., S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 52.Ozanne, D. M., M. E. Brady, S. Cook, L. Gaughan, D. E. Neal, and C. N. Robson. 2000. Androgen receptor nuclear translocation is facilitated by the f-actin cross-linking protein filamin. Mol. Endocrinol. 14:1618-1626. [DOI] [PubMed] [Google Scholar]
- 53.Perrot-Applanat, M., P. Lescop, and E. Milgrom. 1992. The cytoskeleton and the cellular traffic of the progesterone receptor. J. Cell Biol. 119:337-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pines, J., and T. Hunter. 1991. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J. Cell Biol. 115:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pratt, W. B., and D. O. Toft. 1997. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 18:306-360. [DOI] [PubMed] [Google Scholar]
- 56.Rowan, B. G., N. L. Weigel, and B. W. O'Malley. 2000. Phosphorylation of steroid receptor coactivator-1. Identification of the phosphorylation sites and phosphorylation through the mitogen-activated protein kinase pathway. J. Biol. Chem. 275:4475-4483. [DOI] [PubMed] [Google Scholar]
- 57.Schlamp, C. L., G. L. Poulsen, T. M. Nork, and R. W. Nickells. 1997. Nuclear exclusion of wild-type p53 in immortalized human retinoblastoma cells. J. Natl. Cancer Inst. 89:1530-1536. [DOI] [PubMed] [Google Scholar]
- 58.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 59.Sheppard, K. A., K. M. Phelps, A. J. Williams, D. Thanos, C. K. Glass, M. G. Rosenfeld, M. E. Gerritsen, and T. Collins. 1998. Nuclear integration of glucocorticoid receptor and nuclear factor-κB signaling by CREB-binding protein and steroid receptor coactivator-1. J. Biol. Chem. 273:29291-29294. [DOI] [PubMed] [Google Scholar]
- 60.Sorg, G., and T. Stamminger. 1999. Mapping of nuclear localization signals by simultaneous fusion to green fluorescent protein and to beta-galactosidase. BioTechniques 26:858-862. [DOI] [PubMed] [Google Scholar]
- 61.Spencer, T. E., G. Jenster, M. M. Burcin, C. D. Allis, J. Zhou, C. A. Mizzen, N. J. McKenna, S. A. Onate, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1997. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194-198. [DOI] [PubMed] [Google Scholar]
- 62.Stancovski, I., and D. Baltimore. 1997. NF-κB activation: the I κB kinase revealed? Cell 91:299-302. [DOI] [PubMed] [Google Scholar]
- 63.Suen, C. S., T. J. Berrodin, R. Mastroeni, B. J. Cheskis, C. R. Lyttle, and D. E. Frail. 1998. A transcriptional coactivator, steroid receptor coactivator-3, selectively augments steroid receptor transcriptional activity. J. Biol. Chem. 273:27645-27653. [DOI] [PubMed] [Google Scholar]
- 64.Takeshita, A., G. R. Cardona, N. Koibuchi, C. S. Suen, and W. W. Chin. 1997. TRAM-1, A novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J. Biol. Chem. 272:27629-2734. [DOI] [PubMed] [Google Scholar]
- 65.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 66.Traub, P., C. Bauer, R. Hartig, S. Grub, and J. Stahl. 1998. Colocalization of single ribosomes with intermediate filaments in puromycin-treated and serum-starved mouse embryo fibroblasts. Biol. Cell. 90:319-337. [PubMed] [Google Scholar]
- 67.Tremblay, A., G. B. Tremblay, F. Labrie, and V. Giguere. 1999. Ligand-independent recruitment of SRC-1 to estrogen receptor beta through phosphorylation of activation function AF-1. Mol. Cell 3:513-519. [DOI] [PubMed] [Google Scholar]
- 68.Voegel, J. J., M. J. Heine, M. Tini, V. Vivat, P. Chambon, and H. Gronemeyer. 1998. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 17:507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voegel, J. J., M. J. Heine, C. Zechel, P. Chambon, and H. Gronemeyer. 1996. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 15:3667-3675. [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, Z., D. W. Rose, O. Hermanson, F. Liu, T. Herman, W. Wu, D. Szeto, A. Gleiberman, A. Krones, K. Pratt, R. Rosenfeld, C. K. Glass, and M. G. Rosenfeld. 2000. Regulation of somatic growth by the p160 coactivator p/CIP. Proc. Natl. Acad. Sci. USA 97:13549-13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, A. H., and X.-J. Yang. 2001. Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell. Biol. 21:5992-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wurtz, J. M., U. Egner, N. Heinrich, D. Moras, and A. Mueller-Fahrnow. 1998. Three-dimensional models of estrogen receptor ligand binding domain complexes, based on related crystal structures and mutational and structure-activity relationship data. J. Med. Chem. 41:1803-1814. [DOI] [PubMed] [Google Scholar]
- 73.Yao, T. P., G. Ku, N. Zhou, R. Scully, and D. M. Livingston. 1996. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl. Acad. Sci. USA 93:10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]