Abstract
MLL-AFX is a fusion gene created by t(X;11) chromosomal translocations in a subset of acute leukemias of either myeloid or lymphoid derivation. It codes for a chimeric protein consisting of MLL fused to AFX, a forkhead transcription factor that normally regulates genes involved in apoptosis and cell cycle progression. We demonstrate here that forced expression of MLL-AFX enhances the self-renewal of hematopoietic progenitors in vitro and induces acute myeloid leukemias after long latencies in syngeneic recipient mice. MLL-AFX interacts with the transcriptional coactivator CBP, which is also a fusion partner for MLL in human leukemias. A potent minimal transactivation domain (CR3) at the C terminus of AFX mediates interactions with the KIX domain of CBP and is necessary for transformation of myeloid progenitors by MLL-AFX. However, CR3 alone is not sufficient, suggesting that simple acquisition of a transactivation domain per se does not activate the oncogenic potential of MLL. Rather, two conserved transcriptional effector domains (CR2 and CR3) of AFX are required for full oncogenicity of MLL-AFX and also endow it with the potential to competitively interfere with transcription and apoptosis mediated by wild-type forkhead proteins. Furthermore, a dominant-negative mutant of AFX containing CR2 and CR3 enhances the growth of myeloid progenitors in vitro, although considerably less effectively than does MLL-AFX. Taken together, these data suggest that recruitment of transcriptional cofactors utilized by forkhead proteins is a critical requirement for oncogenic action of MLL-AFX, which may impact both MLL- and forkhead-dependent transcriptional pathways.
Rearrangements of the mixed lineage leukemia (MLL) gene (also known as HRX, ALL-1, and Htrx) result from chromosomal translocations in a subset of acute leukemias with lymphoid, myeloid, or biphenotypic features (16, 25, 56). MLL codes for a 431-kDa protein that is a structural and functional homolog of Drosophila trithorax, a positive regulator of Hox genes during embryonic development (24, 26, 48, 61, 62). Remarkably, MLL undergoes fusion with more than 30 different partner proteins as a result of chromosomal translocations, to yield chimeric proteins containing amino-terminal portions of MLL fused in-frame to a carboxy-terminal partner. MLL fusion partners are highly diverse but appear to comprise two general categories of proteins, either nuclear factors with putative roles in transcriptional regulation or cytoplasmic proteins that may be involved in signal transduction (14). The wide array and diverse functions of MLL fusion partners have suggested several possible mechanisms for the oncogenic activation of MLL, including positive and negative gain-of-function, as well as forced oligomerization (17, 21).
Knockin mouse models (11) and retroviral transduction or transplantation assays (36) support a gain-of-function mechanism for a subset of MLL fusion proteins in leukemogenesis. For several nuclear partners, this appears to be the consequence of transcriptional effector domains contributed by the partner proteins. Indeed, structure-function analyses of MLL-ENL and MLL-ELL reveal a complete correlation between domains required for transformation and transcriptional activation (15, 50). These findings suggest the hypothesis that constitutive transcriptional activation by MLL may be a common pathway for its oncogenic conversion by nuclear fusion partners. However, such a mechanism does not exclude possible roles for MLL fusion proteins as transdominant inhibitors of wild-type partner protein function as observed for other chimeric proteins in human leukemias (37, 45).
Two MLL fusion partners, AFX (7, 40) and FKHRL1 (27), and the related FKHR, comprise a subclass of forkhead/winged helix transcription factors (1) that normally regulate genes involved in apoptosis and cell cycle progression (8, 41). Participation of forkhead proteins in these cellular processes suggests their critical roles as tumor suppressors, which may be attenuated after chromosomal translocations in leukemias (7, 27, 40), as well as solid tumors (3, 12, 20). Forkhead proteins bind their cognate DNA sites through a conserved forkhead domain and appear to regulate target gene transcription through recruitment of coactivators such as CBP, p300, or SRC-1, with which they physically interact (43). The coactivators CBP and p300 are also fusion partners for MLL in a subset of leukemias (23, 29, 53, 54).
In this report we demonstrate that transformation of hematopoietic progenitors by MLL-AFX requires the transactivation and CBP interaction domain of AFX. However, this domain alone does not confer oncogenic activity to MLL, demonstrating that simple fusion of a transactivation domain to MLL is not sufficient for transformation of hematopoietic progenitors. In addition to positive gain of function, we also observed a dominant-negative effect of MLL-AFX on transcription and apoptosis mediated by wild-type forkhead family proteins and demonstrate that antagonism of AFX function has a growth-enhancing effect on myeloid progenitors in vitro. Thus, MLL-AFX is capable of impacting MLL- and forkhead-dependent pathways, both of which may contribute to cellular transformation.
MATERIALS AND METHODS
DNA constructs.
Retroviral constructs were made by cloning of various AFX DNA fragments amplified by PCR into the NruI and XhoI sites of the MSCV-neo MLL 5′ cloning vector that encodes MLL amino acids (aa) 1 to 1396 (56), as previously described (15). A synthetic murine stem cell virus (MSCV) MLL-VP16 was made by cloning a fragment encoding the minimum transcriptional transactivation domain (MTD) (aa 413 to 453) of VP16 (22) into the MSCV-neo MLL 5′ retroviral vector. Gal4 fusion constructs consisted of various PCR fragments cloned into the KpnI and SacI sites of pSG424 that encodes the Gal4 DNA-binding domain (DBD; aa 1 to 147) (47). The full-length Gal4-AFX was reported previously (33). All constructs were sequenced to exclude mutations introduced by PCR. In vitro expression vectors encoding FKHR and FKHRL1 (1) and glutathione S-transferase (GST)-CBP (57) constructs have been previously described.
Hematopoietic progenitor transformation and tumorigenicity assays.
Hematopoietic progenitor transformation assays were performed as previously described (50) with modifications. Viral supernatants were collected 3 days after transfection of Phoenix cells and used to infect hematopoietic progenitors and stem cells (harvested from the bone marrow of 4- to 10-week-old C57BL/6 mice) that were positively selected for c-Kit expression by magnetic activated cell sorting. After spinoculation by centrifugation at 500 × g for 2 h at 32°C, transduced cells were cultured overnight in RPMI supplemented with 10% fetal calf serum (FCS), 20 ng of stem cell factor/ml, and 10 ng each of interleukin-3 (IL-3) and IL-6 (R&D Systems, Minneapolis, Minn.)/ml. Transduced cells were then plated in 1% methylcellulose (Stem Cell Technologies, Vancouver, British Columbia, Canada) supplemented with the same cytokines plus 10 ng of granulocyte-macrophage colony-stimulating factor (R&D Systems)/ml in the presence or absence of 1 mg of G418/ml. After 7 days of culture, colonies were counted to calculate the transduction efficiency. Single-cell suspensions (104 cells) of G418-resistant colonies were then replated in methylcellulose media supplemented with the same growth factors without G418. Plating was repeated every 7 days.
In each round of replating, single cell suspensions were also expanded in RPMI liquid culture containing 20% FCS plus 20% WEHI-conditioned medium. For tumorigenicity assays, 106 immortalized cells (MLL-AFX271-501 or AFX3′) were injected via tail vein into 6-week-old syngeneic C57BL/6 mice, which had received a sublethal dose of 5.25 Gy of total-body gamma irradiation (135Cs). Mice were maintained on antibiotic water to avoid infection and monitored for development of leukemia by complete blood count, blood smear, and fluorescence-activated cell sorting (FACS) analysis. Tissues were fixed in buffered formalin, sectioned, and stained with hematoxylin and eosin for histological analysis.
Phenotype analysis.
Cytocentrifugated cells were stained with May-Grünwald-Giemsa (MGG) to assess cell morphology. Immunophenotypic analysis was performed by FACS with fluorochrome-conjugated monoclonal antibodies to Sca-1 (D7 clone), c-Kit (2B8 clone), CD43 (S7 clone), Mac-1 (M1/70 clone), and B220 (RA3-6B2 clone; Pharmingen, Inc., San Diego, Calif.), respectively. Staining was generally performed on ice for 15 min, and then cells were washed twice in staining medium and resuspended in 1 μg of propidium iodine (PI)/ml before analysis with a Moflops (a modified triple laser Cytomation/Becton Dickinson hybrid FACS apparatus). Dead cells were gated out by high PI staining and forward light scatter.
Transcriptional transactivation assays.
293, NIH 3T3, or COS7 cells (5 × 104) were seeded overnight in 24-well plates before transfection with Fugene (Roche Molecular Biochemicals). Gal4 fusion constructs (0.1 μg) were cotransfected with pcDNA3.1/LacZ internal control plasmid (0.2 μg) and a luciferase reporter construct (0.2 μg), which contained two tandem copies of Gal4 consensus binding sites and the luciferase gene driven by either a herpes simplex virus thymidine kinase (TK), adenovirus E1b (46), or myelomonocytic growth factor promoter (18). U937 cells (5 × 106) were electorphorated with 0.8 μg of each Gal4-DBD fusion construct and luciferase reporter construct, together with 0.4 μg of pcDNA3.1/LacZ, in RPMI containing 10 μg of DEAE-dextran at 300 V and 960 μF in a 0.4-mm cuvette (Bio-Rad electroporator). At 24 h after transfection, luciferase and β-galactosidase activities were analyzed by using commercially prepared reagents. Similar procedures were employed for transactivation studies by using the FHRE (8) or p27kip (41) Luc reporters except that 0.4 μg of FKHRL1 expression vector was cotransfected with 0.2 μg of pcDNA3.1/LacZ control plasmid in combination with 0.4 μg of luciferase reporter construct and 0.4 μg of pcDNA expression constructs. Luciferase activities were normalized based on β-galactosidase levels. Means and standard deviations were determined from at least two independent experiments performed in duplicate.
In vitro binding assays.
GST pull-down and electrophoretic mobility shift assays were performed as previously described (51, 52). Briefly, [35S]methionine-labeled FKHR, FKHRL1, AFX, and MLL-AFX fusion proteins were generated by in vitro transcription and translation by using the TNT-coupled reticulocyte lysate system according to the manufacturer's instructions (Promega). GST fusion protein (1 μg) was preincubated with glutathionine-Sepharose beads (Sigma) in NETN buffer (0.5% [vol/vol] Nonidet P-40, 20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA]) for 60 min at 4°C before [35S]methionine-labeled proteins were added. After 90 min incubation at 4°C, the beads were washed five times in buffer H (20 mM HEPES [pH 7.7], 50 mM KCl, 20% [vol/vol] glycerol, 0.1% [vol/vol] Nonidet P40, 0.007% β-mercaptoethanol). Bound proteins were eluted by boiling in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, resolved by electrophoresis, and detected by autoradiography. For electrophoretic mobility shift assay, AFX and MLL-AFX fusion proteins were synthesized by in vitro transcription and translation as described above except in the absence of [35S]methionine. In vitro-translated proteins were preincubated for 15 min at room temperature in buffer B [20 mM HEPES, pH 7.4; 50 mM KCl; 1 mM β-mercaptoethanol; 10% glycerol; 1 μg of poly(dI-dC); 100 μg of bovine serum albumin] to allow proper protein folding before they were incubated with γ-32P-end-labeled complementary oligonucleotides corresponding to either Fas ligand site encompassing the IGFBP1 IRS (Fas ligand promoter, 5′-ATTAGATCTTAAATAAATAGATCTTTA-3′) (8) or a mutated IRS (Am2Bm2, 5′-CACTAGCAACCATGCCATGGTTGAACAC-3′) of the IGFBP-1 (33). Binding reactions were preformed for 30 min at room temperature and 4°C, respectively. Protein-DNA complexes were resolved in 6% polyacrylamide nondenaturing gel equilibrated with 0.5× Tris-borate-EDTA and visualized by autoradiography.
Apoptosis assays.
Ba/F3 cells stably expressing FKHRL1 (A3):ER∗ have been described previously (13). For transient transfection, Ba/F3 cells were electrophorated with 5 μg of enhanced green fluorescent protein (EGFP) construct, together with 15 μg of empty MSCV-puro vector or DNA expression constructs encoding MLL fusion protein, truncated 5′ MLL/3′ AFX or cyclin E. At 2 h after electroporation, dead cells were removed by Ficoll gradient centrifugation. Cells were then cultured in RPMI supplemented with 10% FCS and 20% WEHI-conditioned medium in the absence or presence of 0.1 μM 4-hydroxy tamoxifen (4-OHT) for 24 h. Cells were washed twice in binding buffer (10 mM HEPES, pH 7.4; 140 mM NaCl; 2.5 mM CaCl2) before staining with phycoerythrin (PE)-conjugated Annexin V (Pharmingen). After incubation at room temperature for 15 min, cells were washed and resuspended in binding buffer with 1 μg of PI/ml, and immediately analyzed by FACS. Dead cells were gated out by PI staining, and 5,000 GFP-positive cells were analyzed for PE staining.
IP and Western blots.
Both Western blots and immunoprecipitation (IP) were performed as previously described (30). Briefly, transfected cells were lysed and denatured in lysis buffer (50 mM Tris, pH 6.8; 2% SDS; 10% glycerol) at 100°C for 5 min. Genomic DNA was sheared by passage of samples through a 28-gauge needle. After polyacrylamide gel electrophoresis, proteins were transferred to ECL (Amersham) or polyvinylidene difluoride membranes (Bio-Rad) and blotted with monoclonal MLL antibody (N4.4), monoclonal p27kip-1 antibody (BD Transduction Laboratories), monoclonal Gal4 DBD antibody (Santa Cruz Biotechnology), or rabbit polyclonal FKHRL1 antibody (Upstate Biotechnology). For IP-Western blotting, COS7 cells were transfected in 90-mm dishes with 3 μg each of CMV-CBP and pcDNA MLL-AFX or Gal4-AFX constructs and then washed in detachment buffer (40 mM Tris, pH 7.6; 150 mM NaCl; 1 mM EDTA) before lysis in buffer A (20 mM HEPES, pH 7.9; 10 mM KCl; 1 mM EDTA; 1 mM dithiothreitol [DTT]; 1 mM phenylmethylsulfonyl fluoride; 0.1% NP-40) on ice for 1 min. Nuclear pellets were recovered after centrifugation and resuspended in buffer B (20 mM HEPES, pH 7.9; 0.4 mM NaCl; 1 mM EDTA; 1 mM DTT; 1 mM phenylmethylsulfonyl fluoride) for 20 min incubation at 4°C. Nuclear extracts were obtained by centrifugation at 12,000 × g for 15 min and brought to 20% glycerol for IP. Then, 3 μg of αMLL-N4.4 or αGal4 DBD antibodies were added to the nuclear extracts and incubated at 4°C for 3 h. Next, 20 μl of protein G-Sepharose beads (Pharmacia) were then added to the extracts and incubated for additional 2 h at 4°C before they were washed five times in buffer D (0.5 M KCl, 0.1% Tween 20, 0.5 mM DTT, 0.2 mM EDTA). Precipitated proteins were then denatured at 100°C for 5 min and separated by PAGE before transfer to polyvinylidene difluoride membranes and blotting with polyclonal CBP antibody (Upstate Biotechnology).
RESULTS
MLL-AFX enhances the self-renewal of primary myeloid progenitors.
A full-length MLL-AFX fusion cDNA was expressed under control of the long terminal repeat (LTR) of the MSCV after transduction into murine bone marrow cells enriched for hematopioetic progenitor and stem cells. MSCV-neo vector alone and retroviruses expressing 5′ MLL or 3′ AFX cDNAs encoding the respective portions of the MLL-AFX fusion protein were also tested (Fig. 1A). Colonies were observed for all constructs in the initial round of plating, although bone marrow cells transduced with MLL-AFX consistently gave rise to fewer colonies, which may reflect possible toxic effects of the fusion protein or a lower viral titer. MLL-AFX transduced cells continued to replate and yielded increasing numbers of colonies in the second and subsequent platings. In contrast, cells transduced with empty vector or MLL-5′ exhausted their self-renewal potential by the third round. MLL-AFX colonies typically displayed a distinctive compact CFU-GEMM morphology (Fig. 1B). Unexpectedly, cells transduced with AFX-3′ (Fig. 1A) replated into the third round of methylcellulose culture. The colonies displayed a more diffuse morphology (Fig. 1B), suggesting enhanced growth of cells with some capacity for myeloid differentiation. Although these cells did not form colonies in the fourth plating, they adapted to growth in liquid medium containing IL-3, as did cells transduced with MLL-AFX. MLL-AFX transduced cells expressed c-Kit, CD43, Gr-1, and Mac-1 but were negative for Sca-1, CD3, and B220 (Fig. 1C), a surface antigen profile consistent with early myeloid derivation. AFX-3′-expressing cells displayed a similar but more differentiated phenotype (CD43+, Gr-1+, Mac-1+, and c-Kit+/−) with a majority lacking c-Kit expression (data not shown), a finding consistent with their diffuse colony morphologies in methylcellulose media. Therefore, MLL-AFX induces enhanced self-renewal of myeloid progenitors in vitro. AFX-3′ displayed substantially less potent effects, which nevertheless suggest that antagonism of AFX-dependent pathways also promotes the self-renewal of myeloid progenitors.
FIG. 1.
Transformation of myeloid progenitors by MLL-AFX. (A) Schematic diagram of MLL-AFX and the retroviral constructs used in hematopoietic progenitor transformation assays (left). MTase, DNA methyltransferase homology region; AT hook, AT hook DBD; FK, forkhead DBD; S, consensus serine phosphorylation sites; PFK, post-forkhead homology region (1); CR2, conserved region 2, which contains three alpha helices shown as thin vertical bars; CR3, conserved region 3. The bar graph (right) represents corresponding numbers of colonies after each round of plating in methylcellulose (average of three independent assays). (B) Typical morphology of methylcellulose colonies generated from bone marrow cells transduced with retroviruses expressing the indicated constructs. (C) Phenotypic analysis of cells transduced by MLL-AFX. Red lines represent FACS staining obtained with antibodies specific for the indicated cell surface antigens. Blue lines represent staining obtained with isotype control antibodies.
Progenitors immortalized by MLL-AFX induce myeloid leukemias after long latencies.
To investigate their leukemogenic potential, cells (106) immortalized by MLL-AFX or AFX-3′ were injected intravenously into sublethally irradiated syngeneic C57BL/6 mice. A total of 90% of the animals that received MLL-AFX cells succumbed to acute leukemias within 15 months (Fig. 2). Histological and cytological studies demonstrated that >30% of bone marrow cells were comprised of leukemic blasts, which were also present in the peripheral blood (Fig. 2). Both the spleen and the liver were infiltrated with leukemic cells, which were particularly prominent in periportal zones of the liver and effaced the normal splenic architecture. The leukemic blasts were c-Kit+ CD43+ Mac-1+ Sca-1− B220−, a phenotype consistent with the early myeloid phenotype of the injected transduced cells (data not shown). To date (9 month observation period), none of the mice injected with cells immortalized by MLL-3′ have developed leukemia. Therefore, MLL-AFX was capable of inducing the leukemic transformation of myeloid progenitors, but AFX-3′ was not in spite of its ability to alter the growth properties of progenitors in vitro.
FIG. 2.
MLL-AFX transformed cells induce acute myeloid leukemias. (A) Representative histology is shown for control and MLL-AFX mice. Paraffin sections were stained with hematoxylin and eosin; blood smears were stained with MGG. In MLL-AFX mice, the spleen and liver were infiltrated with leukemic blasts. Bone marrow was densely packed with a homogeneous population of blasts. Leukemic cells are present in the peripheral blood. (B) Survival curves are shown for cohorts (n = 10) of sublethally irradiated C57BL/6 mice that were injected with MLL-AFX immortalized cells (MLL-AFX), AFX-3′ immortalized cells, or mock injected (control).
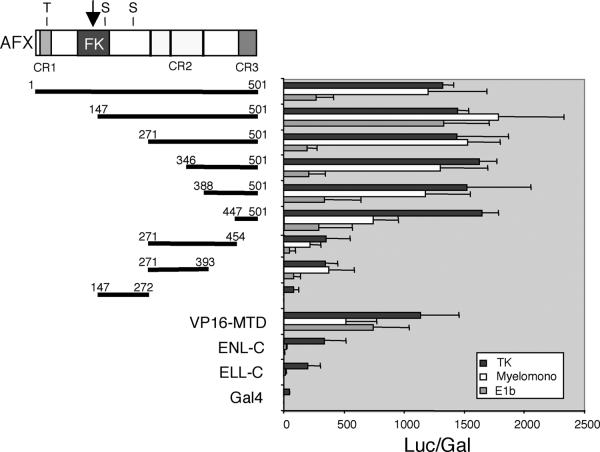
A strong transcriptional activation domain maps to the CR3 region of AFX.
Members of the mammalian FKHR-related transcription factor subfamily (AFX, FKHR, and FKHRL1) display three conserved regions of sequence similarity, which we will refer to as conserved regions 1 to 3 (CR1 to CR3), in addition to the forkhead DBD (Fig. 3). To determine whether these or other regions of AFX mediate transcriptional effector activity, various portions of AFX were fused to the GAL4-DBD and tested for transcriptional activation potential in transient transfection assays. These studies employed luciferase reporter gene constructs driven by three different promoters (HSV-TK, adenovirus E1b, and myelomonocytic growth factor) in four different cell lines (293 human kidney, NIH 3T3 mouse fibroblast, U937 early myeloid, and COS7 monkey kidney cells). Since similar results were obtained with all cell lines, representative data are only shown for studies with 293 cells (Fig. 3). GAL4 fusion constructs containing the minimal oncogenic domains of MLL fusion partners ENL (ENL-C) (50) and ELL (ELL-C) (15) were also evaluated for comparison. AFX displayed strong transcriptional activation, which was comparable to that of the VP16 minimal transactivation domain (VP16-MTD). By comparison, ENL-C and ELL-C activated transcription only on the TK promoter, and their activities were significantly less than that displayed by AFX. Further mapping studies identified two activation domains in the portion of AFX retained in MLL-AFX fusion proteins. A strong transactivation domain localized to CR3 at the carboxy terminus of AFX (447 to 501 aa). Although considerably less potent than CR3, a second transactivation domain was localized to CR2, which displayed activation potential comparable in magnitude to that observed for ENL-C and ELL-C on the TK promoter (Fig. 3).
FIG. 3.
Mapping of transcriptional activation domains in AFX. Schematic diagram (top left) illustrates the conserved domains of AFX. The arrow indicates the fusion site with MLL in human leukemias. Thick horizontal black lines with amino acid numbers represent the AFX fragments fused to the Gal4-DBD for transactivation assays. ENL-C (477 to 559 aa) and ELL-C (496 to 621 aa) correspond to the minimal regions sufficient for immortalization of myeloid progenitors by MLL-ENL and MLL-ELL fusion proteins, respectively (15, 50). VP16-MTD encodes the minimal transcriptional activation domain (aa 413 to 453) of herpes simplex virus VP-16. Expression constructs were cotransfected with pcDNA-LacZ into 293 cells with a luciferase reporter gene under the control of the indicated promoters. TK, herpes simplex virus TK promoter; E1b, adenovirus E1b promoter; Myelomono, myelomonocytic growth factor promoter (described in Materials and Methods). Luciferase values were normalized for β-galactosidase expression from the internal LacZ control construct. Similar expression level of the Gal4 constructs was confirmed by Western blot (data not shown).
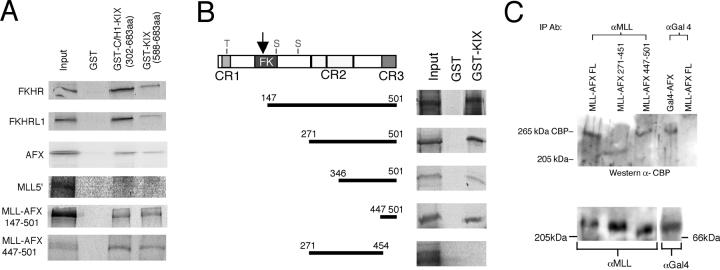
The AFX CR3 domain interacts with the KIX domain of CBP.
FKHR-related subfamily proteins interact with CBP (43), which itself is a fusion partner for MLL in acute leukemias (53, 54). To determine whether MLL-AFX retains an ability to interact with CBP, in vitro binding assays were conducted with GST-CBP fusion proteins that spanned the KIX domain, which mediates interactions with forkhead family proteins. MLL-AFX bound to GST-C/H1-KIX (encoding the C/H1 and KIX domains of CBP) and GST-KIX, but not GST, under conditions that wild-type forkhead family proteins AFX, FKHR, and FKHRL1 also bound to CBP (Fig. 4A). In contrast, 5′ MLL did not interact with GST-CBP proteins, indicating that the CBP interacting domain resided within AFX. To map the region of AFX required for CBP interaction, various AFX deletion mutants were tested for their ability to bind GST-KIX. All AFX proteins that retained CR3 interacted with the CBP-KIX domain, whereas a construct spanning CR2 but lacking CR3 (aa 271 to 454) failed to interact with GST-KIX (Fig. 4B). Consistent with the mapping studies, MLL-AFX447-501, consisting of MLL fused to CR3, interacted with CBP-KIX/C/H1 and CBP-KIX in a GST pull-down assay (Fig. 4A). Taken together, these results indicate that CR3 is sufficient for transcriptional activation and interaction with CBP in vitro.
FIG. 4.
The AFX CR3 interacts in vitro with the KIX domain of CBP. (A) GST pull-down experiments were conducted with in vitro translated [35S]methionine-labeled proteins indicated on the left. GST constructs are indicated at the top of gel lanes. GST-CBP constructs have been previously described (58). (B) Mapping of KIX interacting domain to CR3 of AFX. Various AFX fragments (indicated by horizontal bars on left) were in vitro translated with [35S]methionine and incubated with GST (lane 2) or GST-KIX (lane 3) fusion proteins. Bound proteins were washed, eluted, and subjected to SDS-PAGE after autoradiography. Protein inputs are shown in lane 1. (C) In vivo interaction between CBP and AFX proteins. Constructs used in cell transfection are shown at the top of each lane and their expression was confirmed by Western blots (bottom panel). Cell lysates were precipitated with antibodies to MLL or Gal4 DBD and then blotted with anti-CBP antibody. Specific 265-kDa CBP bands are indicated in the top panel.
MLL-AFX interacts with CBP in vivo.
MLL-AFX was immunoprecipitated from nuclear extracts of cotransfected COS7 cells, and the immune complexes were assessed for the presence of CBP by Western blot analysis. A specific 265-kDa band corresponding to CBP was detected in the anti-MLL precipitates but not in control precipitations employing anti-Gal4 antibodies, which were capable of coprecipitating CBP from cells transfected with a Gal4-AFX construct (Fig. 4C). CBP coprecipitated with MLL-AFX447-501, but not with MLL-AFX271-451, confirming the requirement for CR3 consistent with the in vitro binding assays. The integrity of input MLL proteins was confirmed by Western blot (Fig. 4C). These data further demonstrate that CR3 is a minimal transactivation domain that recruits CBP.
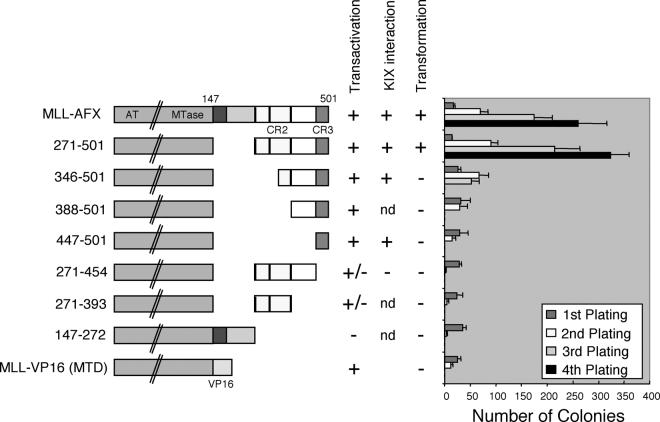
The AFX CR3 domain is necessary but not sufficient for transformation of hematopoietic progenitors by MLL-AFX.
To determine which portions of AFX were necessary for the oncogenic activity of MLL-AFX, various MSCV MLL-AFX constructs were transduced into primary hematopoietic progenitors, whose self-renewal potentials were then evaluated. Retroviral constructs used for these assays displayed similar transduction efficiencies as determined by enumeration of colony numbers in methylcellulose medium with or without G418 selection in the first round of plating. Expression of MLL-AFX constructs was confirmed in COS7 cells and in primary transduced hematopoietic progenitors by Western blot and reverse transcription-PCR analyses, respectively (data not shown). Replating assays demonstrated that the forkhead DBD and consensus PKB phosphorylation sites of AFX were not necessary for myeloid immortalization since a construct (MLL-AFX271-501) with deletion of these domains immortalized hematopoietic progenitors (Fig. 5). Conversely, constructs lacking CR3 (MLL-AFX271-454 and MLL-AFX271-393) were unable to enhance the self-renewal of progenitors under these conditions. These data indicated that CR3, the transcriptional activation and CBP interaction domain, was necessary for transformation. However, fusion of CR3 alone to MLL (MLL-AFX447-501) was not sufficient for hematopoietic cell transformation, failing to give rise to colonies after the second round of plating. This suggested that CR2 may also be necessary for transformation, prompting analysis of constructs with partial or complete CR2 deletion (MLL-AFX346-501 and MLL-AFX388-501, respectively). Although MLL-AFX346-501 with partial deletion of CR2 yielded significant numbers of colonies in the second round of plating, the colony number dropped dramatically in the third plating. Both constructs resulted in no colonies in the fourth round, indicating an inability to immortalize hematopoietic progenitors. These results suggested that fusion of a transcriptional activation domain per se to MLL was not sufficient for transformation of hematopoietic progenitors. To further test this conclusion, a synthetic construct containing MLL fused with the VP16 MTD (aa 413 to 453) was evaluated for its transformation abilities. MLL-VP16 (MTD) was unable to immortalize hematopoietic progenitors and failed to form colonies in the third plating (Fig. 5). Therefore, the strong transcriptional activation domain (CR3) of AFX is necessary but not sufficient for transformation by MLL-AFX, which appears to also require contributions from CR2.
FIG. 5.
CR2 and CR3 are both necessary for MLL-AFX-mediated transformation. Schematic diagram (left column) shows the deletion mutants of MLL-AFX used for hematopoietic transformation assays. A synthetic MLL-VP16 (MTD) construct was also included. Transformation abilities, transactivation properties, and in vitro binding to the CBP KIX domain are summarized in the middle. Bar graph (right) represents the number of colonies generated by the respective constructs in replating assays (mean ± the standard derivation, n = 3).
MLL-AFX antagonizes FKHRL1-mediated transcriptional activation.
The FKHR-related subclass of forkhead/winged helix transcription factors induce cell cycle arrest and apoptosis by activating expression of the p27kip-1 (41) and Fas ligand (8) genes, respectively. To evaluate whether MLL-AFX has the potential to dominantly interfere with these normal processes, we first determined whether MLL-AFX binds to forkhead DNA recognition sites. As determined by band shift assay, wild-type AFX efficiently bound the consensus forkhead DNA site (Fas ligand promoter) but not a mutated form (Am2Bm2) of the site. Conversely, MLL-AFX displayed minimal binding consistent with the fact that it retains only half of the forkhead DBD (Fig. 6A). Since MLL-AFX retains the transcriptional activation and CBP interaction domains of AFX, we tested its effect on FKHRL1-mediated transcriptional activation in transient transfection assays (Fig. 6B). As expected, wild-type FKHRL1 stimulated expression of the luciferase reporter gene driven by the p27kip-1 or Fas ligand promoters by more than 10-fold above the basal level. However, coexpression of MLL-AFX or MLL-AFX271-501 impaired FKHRL1-mediated transactivation (Fig. 6B). The observed reduction was specific since coexpression of wild-type MLL or Gal4-AFX with FKHRL1 did not compromise FKHRL1-mediated transactivation. The reduction is unlikely to be an artifact of transient hyperexpression since the level of MLL-AFX expression in transiently transfected cells was comparable to that observed in leukemic cells (Fig. 6C). Since FKHRL1 levels were considerably lower in the latter, the ratio of MLL-AFX to FKHRL1 expression was actually greater in leukemic cells. These results suggest that MLL-AFX has the potential to exert a dominant-negative effect on the FKHRL1 transcriptional pathway.
FIG. 6.
Potential dominant-negative effects of MLL-AFX fusion proteins. (A) MLL-AFX is unable to bind consensus forkhead DNA-binding sites. In vitro-translated proteins (indicated at tops of gel lanes) were incubated with 32P-labeled oligonucleotides encompassing the Fas ligand site corresponding to IGFBP1 IRS (Fas ligand promoter) or a mutated IRS (Am2Bm2) of the IGFBP-1. Reticulocyte lysate (lane 1) was used as a control for nonspecific binding. The arrow indicates specific DNA-binding complexes. (B) MLL-AFX antagonizes FKHRL1-mediated transcriptional activation. An expression construct encoding FKHRL1 was cotransfected into 293 cells with FKHRL1, MLL-AFX, or AFX expression constructs and a reporter gene driven by the Fas ligand promoter (FHRE promoter) or the p27kip-1 promoter (p27kip promoter). The fold induction was corrected for β-galactosidase activity from an internal lacZ control construct in each transfection. (C) Comparable expression levels of MLL-AFX in leukemic (lane 2) and transfected (lane 3) cells. The control (lane 1) consisted of the REH pre-B cell line, which expressed wild-type MLL but not MLL-AFX. Specific bands corresponding to wild-type MLL, MLL-AFX, and FKHRL1 are indicated to the left.
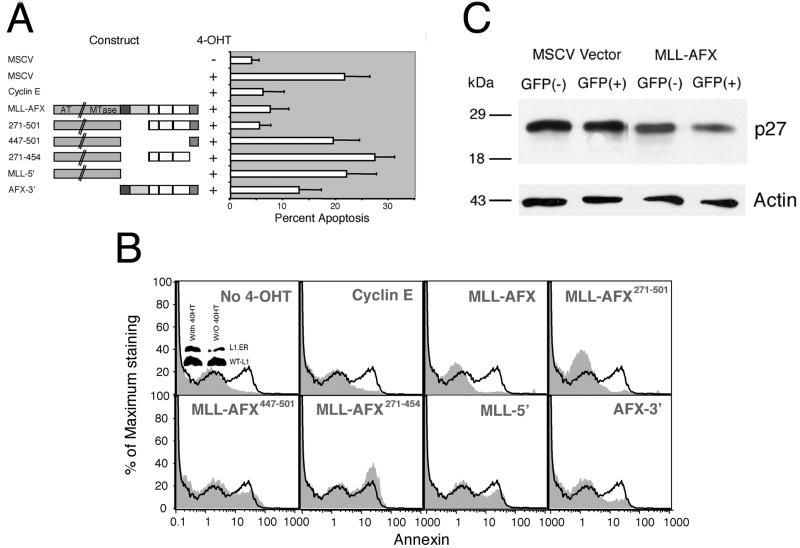
MLL-AFX suppresses FKHRL1-mediated apoptosis in Ba/F3 cells.
The dominant-interfering transcriptional potential of MLL-AFX was evaluated for possible functional consequences on apoptosis mediated by forkhead family proteins. These studies employed a Ba/F3 pro-B cell line that stably expresses FKHRL1 (A3):ER∗, an estrogen receptor fusion protein that is activated to induce apoptosis by upregulation of p27kip-1 upon addition of 4-hydroxy tamoxifen (4-OHT) (13). These cells were cotransfected with various MLL-AFX constructs and a CMV EGFP vector and then induced to undergo apoptosis by treatment with 4-OHT. EGFP-positive cells were sorted and analyzed. Typically, ca. 20% of Ba/F3 cells undergo apoptosis after 4-OHT treatment (Fig. 7A). Transient expression of MLL-AFX or MLL-AFX271-501 reduced the percentage of apoptotic cells to ca. 7%, which was slightly higher than the basal level of 3% (Fig. 7A and B). This reduction was similar to that observed after expression of cyclin E, which served as a positive control. Conversely, expression of MLL-5′, MLL-AFX271-454, or MLL-AFX447-501, which were incapable of immortalizing hematopoietic progenitors, did not significantly impair apoptosis, suggesting that both CR2 and CR3 of AFX were required for the observed antiapoptotic effects. Expression of AFX-3′, which enhances self-renewal of myeloid progenitors, reduced apoptosis to ca. 13%. Under the conditions of this experiment, Ba/F3 cells transfected with MLL-AFX consistently displayed downregulation of endogenous p27 levels by Western blot analysis in comparision to nontransfected or vector-transfected cells (Fig. 7C). Taken together, these studies indicate that MLL-AFX has the potential to dominantly interfere with the apoptotic functions of wild-type forkhead family proteins through the AFX moiety.
FIG. 7.
MLL-AFX suppresses FKHRL1-mediated apoptosis in Ba/F3 cells. (A) Ba/F3 cells stably expressing FKHRL1 (A3): ER∗ were cotransfected with EGFP and various expression constructs (left). Cells were treated with or without 0.1 μM 4-OHT for 24 h before analysis of apoptosis by Annexin V staining. EGFP was used as a marker to identify transfected cells for apoptosis analysis. Bars (right) represent the percentage change in apoptosis after the induction of FKHRL1 (A3) expression in the absence or presence of various DNA constructs. (B) FACS data of Ba/F3 cells from a representative transfection experiment. The results are shown for EGFP-positive, PI-negative gated cells. Black lines represent controls transfected with EGFP only and treated with 4-OHT for 24 h. Shadow profiles represent either the control without 4-OHT treatment or cells transfected with various constructs and treated with 4-OHT. Expression of FKHRL1 (A3)∗ER was detected by Western blot, as shown by the small insert in the left top panel. (C) MLL-AFX- or MSCV-transfected cells were sorted into GFP-positive and -negative populations. Cell lysates from 105 cells were analyzed by Western blotting with antibodies specific for p27kip-1 or actin.
DISCUSSION
Our studies demonstrate that MLL-AFX enhances the self-renewal of primary hematopoietic progenitors and arrests their maturation at a developmental stage similar to that of human myeloid leukemias expressing this fusion protein (7). Progenitors immortalized by MLL-AFX were also leukemogenic after transplantation into sublethally irradiated, syngeneic recipients, albeit with long latencies, suggesting the need for secondary mutations. A presumed requirement for secondary mutations is typical of myeloid leukemias associated with other MLL fusion proteins in the transduction and transplantation model employed here (36) and in MLL-AF9 knockin mice (11). However, the latency periods required for the development of overt leukemias vary substantially for different MLL fusion proteins, despite their induction of similar immortalized phenotypes on primary myeloid progenitors in vitro (2). With a latency of at least 4 to 15 months, MLL-AFX clearly falls into the long-latency group. Our observations support the hypothesis that MLL fusion partners serve important, although currently undefined, roles in regulating the rate of acquisition of critical secondary mutations that affect progression to acute leukemia in vivo.
An unexpected observation in the current study is that fusion of a strong transcriptional activation domain to MLL is not sufficient to activate its oncogenic properties. We localized a strong transactivation domain (CR3) to the C terminus of AFX and showed that it interacts with the KIX domain of coactivator CBP. CR3, which has features of an acidic activation domain, was required but not sufficient for in vitro immortalization of myeloid progenitors by MLL-AFX. Further evidence that acquisition of an acidic transactivation domain per se is not sufficient for MLL-mediated transformation is provided by MLL-VP16, a synthetic construct encoding the potent MTD of VP16, which failed to transform hematopoietic progenitors. This may not necessarily be predicted to be effective since different transcriptional scaffolds are organized by different proteins, which may result in activation in different ways. Nevertheless, this contrasts with MLL fusion partners ENL and ELL, whose minimal domains required for transcriptional activation correlate with those necessary and sufficient for oncogenic transformation (15, 50). The C-terminal activation domain of ELL has been shown to interact with EAF1, which itself is capable of conferring oncogenic potential when fused with MLL (38). Our current findings suggest that there are qualitative differences in the transcriptional effector domains that activate the oncogenic properties of MLL, which are not correlated with quantitative levels of activation measured on experimental reporter genes.
Two conserved AFX domains (CR3 and CR2) are required to achieve full oncogenicity of MLL-AFX. CR2 displays weak transactivation properties, equivalent to those of ELL and ENL, but nevertheless is not sufficient for transformation. CR2 displays primary sequence and predicted secondary structure conservation (three small conserved α-helical segments) with other mammalian forkhead subfamily proteins, but its molecular function is unknown. Prior studies of PAX-FKHR fusion proteins are consistent with an oncogenic requirement for CR2. As a result of chromosomal translocations in rhabdomyosarcomas, FKHR is fused with the paired box transcription factors PAX3 or PAX7 at an analogous point in the forkhead DBD as in MLL-AFX fusions (3, 12, 20). Structure-function analysis of PAX3-FKHR indicates that, like MLL-AFX, transactivation ability does not directly correlate with transformation, which also requires undefined FKHR sequences upstream of CR3 for efficient transformation (32, 34). Thus, the contributions of forkhead family proteins may be similar in MLL and PAX fusion proteins. Recent studies have revealed that fusion with FKHR deregulates normal transcriptional control by the N-terminal repression domain of PAX, suggesting that override of repression rather than strength of transactivation is critical for transformation by PAX-FKHR (4-6, 28). By analogy, the portion of MLL retained in all fusion proteins also contains a repression domain that interacts with histone deacetylases HDAC1 and HDAC2, which may antagonize the activation properties of MLL fusion partners (60). It remains to be determined whether override of MLL-mediated repression is a critical factor in transformation.
Covalent fusion of MLL with CBP in MLL-CBP is sufficient for in vitro immortalization and leukemic transformation of myeloid progenitors (35). This contrasts with our current results that a CBP interaction domain of AFX is not capable of activating the oncogenic potential of MLL in a similar myeloid transformation assay. There are several possible explanations for this discrepancy. Covalent fusion of CBP with MLL, which deletes amino-terminal portions of CBP, may remove regulatory regions of CBP that negatively modulate one or more of its effector properties resulting in its “activation.” Another possibility is that the CR3 of AFX does not functionally interact with CBP on MLL target genes, despite its ability to interact with the CBP KIX domain in vitro and function as a potent transactivation domain in transient transfection assays. Alternatively, CR3 may not efficiently recruit CBP or result in a sufficiently stable interaction in the targeted transformation cell types. Indeed, previous studies have suggested that the mode of recruitment of a forkhead activation domain may be crucial for activation (43). One possibility for the requirement of CR2 is that it stabilizes or otherwise enhances the AFX-CBP interaction. Precedent for essential dual sites of CBP interaction is provided by the human T-cell leukemia virus type 1 Tax protein. It contains a KIX-C/H1 interaction domain, as well as an amphipathic helical region that interacts with the C-terminal region of CBP to mediate transcriptional activation (49). Mutation of the latter domain significantly compromises Tax-mediated transactivation despite an intact KIX-C/H1 interaction domain.
The possibility that disruption of MLL partner protein functions may contribute to leukemogenesis is based in part on their putative growth suppressive properties (Abi-1, AF6, AFX, FKHRL1, and GAS7) and involvement in other malignances (AF10, GRAF, CBP, and p300). In the present study, we observed that MLL-AFX suppressed transcription and apoptosis mediated by the forkhead protein FKHRL1. Structure-function studies indicated that these interfering effects required the same portions of AFX (i.e., CR2 and CR3) necessary for transformation of hematopoietic progenitors. Furthermore, the proliferation of myeloid progenitors in vitro was enhanced by their transduction with a truncated form of AFX (AFX-3′). Although the mechanisms that mediate this hyperproliferative capability are not known, the absence of an intact forkhead DBD in AFX-3′ and MLL-AFX suggests that they may act as dominant-negative mutants to compete for limiting amounts of transcriptional cofactors utilized by endogenous forkhead family proteins. Independent evidence that disruption of AFX-dependent pathways perturbs growth control is provided by studies of a dominant-negative form of AFX containing only the forkhead DBD, which induced a significant increase in colony formation by A14 cells and a decrease in the percentage of cells in G1 (41).
A dominant interfering function for MLL-AFX on AFX-dependent pathways does not exclude a gain-of-function effect on MLL target genes. Previous studies of PML-RARα in acute promyelocytic leukemia demonstrate that it antagonizes PML-mediated apoptosis and the retinoic acid signaling pathway by sequestering the respective wild-type proteins and their interacting partners (e.g., RXR) (31, 44), in addition to its role as a transcriptional repressor on retinoid target genes. Similar effects have been reported for the variant PLZF-RARα and NPM-RARα proteins in acute promyelocytic leukemia (10, 51). Analogously, the AML1-ETO fusion protein resulting from t(8;21) inhibits the functions of AML1 and ETO by sequestering the respective wild-type proteins and their interacting partners (9, 55), in addition to directly repressing AML1 target genes (19, 39, 42, 58). In our studies, the dominant-negative AFX-3′ was considerably less effective compared to MLL-AFX in perturbing the growth of myeloid progenitors, suggesting that disruption of MLL-dependent pathways has a greater role in leukemogenesis.
In summary, our data indicate that the strength of transcriptional activation is not directly correlated with MLL-AFX-mediated transformation, which requires additional functions contributed by CR2. This region is also critical for transformation mediated by PAX3-FKHR fusion proteins that maintain a constitutive transactivation status by subverting normal transcriptional repression of the PAX protein (28, 59). Although the actual function of CR2 remains to be determined, the presence of weak transactivation activity and conserved helical structures suggest that it may interact with the transcriptional machinery. CR2 along with CR3 is also necessary for the dominant-negative effect of MLL-AFX on normal forkhead protein function. This suggests a model in which MLL-AFX requires and competes for accessory proteins utilized by wild-type forkhead transcription factors to facilitate its positive and negative effects on MLL and forkhead-dependent pathways, respectively.
Acknowledgments
We thank B. Burgering, P. Coffer, M. Greenberg, K. Arden, J. Nyborg, J. Lipsick, and P. Jackson for providing essential reagents to conduct these studies. We are particularly grateful to P. Ayton for insightful critique of the manuscript, A. Yokoyoma for technical advice, and other members of the Cleary lab for numerous helpful discussions. We are also indebted to Caroline Tudor and Erica So for assistance with artwork.
C. W. So is the recipient of a Croucher Foundation Fellowship. This work was supported by a grant from the National Institutes of Health (CA55209) and in part by a Croucher Foundation Research grant to C. W. So.
REFERENCES
- 1.Anderson, M. J., C. S. Viars, S. Czekay, W. K. Cavenee, and K. C. Arden. 1998. Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics 47:187-199. [DOI] [PubMed] [Google Scholar]
- 2.Ayton, P. M., and M. L. Cleary. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20:5695-5707. [DOI] [PubMed] [Google Scholar]
- 3.Barr, F. G., N. Galili, J. Holick, J. A. Biegel, G. Rovera, and B. S. Emanuel. 1993. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 3:113-117. [DOI] [PubMed] [Google Scholar]
- 4.Bennicelli, J. L., S. Advani, B. W. Schafer, and F. G. Barr. 1999. PAX3 and PAX7 exhibit conserved cis-acting transcription repression domains and utilize a common gain of function mechanism in alveolar rhabdomyosarcoma. Oncogene 18:4348-4356. [DOI] [PubMed] [Google Scholar]
- 5.Bennicelli, J. L., R. H. Edwards, and F. G. Barr. 1996. Mechanism for transcriptional gain of function resulting from chromosomal translocation in alveolar rhabdomyosarcoma. Proc. Natl. Acad. Sci. USA 93:5455-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennicelli, J. L., W. J. Fredericks, R. B. Wilson, F. J. Rauscher III, and F. G. Barr. 1995. Wild-type PAX3 protein and the PAX3-FKHR fusion protein of alveolar rhabdomyosarcoma contain potent, structurally distinct transcriptional activation domains. Oncogene 11:119-130. [PubMed] [Google Scholar]
- 7.Borkhardt, A., R. Repp, O. A. Haas, T. Leis, J. Harbott, J. Kreuder, J. Hammermann, T. Henn, and F. Lampert. 1997. Cloning and characterization of AFX, the gene that fuses to MLL in acute leukemias with a t(X;11)(q13;q23). Oncogene 14:195-202. [DOI] [PubMed] [Google Scholar]
- 8.Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857-868. [DOI] [PubMed] [Google Scholar]
- 9.Chen, L. F., K. Ito, Y. Murakami, and Y. Ito. 1998. The capacity of polyomavirus enhancer binding protein 2αB (AML1/Cbfa2) to stimulate polyomavirus DNA replication is related to its affinity for the nuclear matrix. Mol. Cell. Biol. 18:4165-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, G. X., X. H. Zhu, X. Q. Men, L. Wang, Q. H. Huang, X. L. Jin, S. M. Xiong, J. Zhu, W. M. Guo, J. Q. Chen, S. F. Xu, C. W. So, L. C. Chan, S. Waxman, A. Zelent, G. Q. Chen, S. Dong, J. X. Liu, and S. J. Chen. 1999. Distinct leukemia phenotypes in transgenic mice and different corepressor interactions generated by promyelocytic leukemia variant fusion genes PLZF-RARα and NPM-RARα. Proc. Natl. Acad. Sci. USA 96:6318-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corral, J., I. Lavenir, H. Impey, A. J. Warren, A. Forster, T. A. Larson, S. Bell, A. N. McKenzie, G. King, and T. H. Rabbitts. 1996. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell 85:853-861. [DOI] [PubMed] [Google Scholar]
- 12.Davis, R. J., C. M. D'Cruz, M. A. Lovell, J. A. Biegel, and F. G. Barr. 1994. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 54:2869-2872. [PubMed] [Google Scholar]
- 13.Dijkers, P. F., R. H. Medema, C. Pals, L. Banerji, N. S. Thomas, E. W. Lam, B. M. Burgering, J. A. Raaijmakers, J. W. Lammers, L. Koenderman, and P. J. Coffer. 2000. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27KIP1. Mol. Cell. Biol. 20:9138-9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiMartino, J. F., and M. L. Cleary. 1999. MLL rearrangements in haematological malignancies: lessons from clinical and biological studies. Br. J. Haematol. 106:614-626. [DOI] [PubMed] [Google Scholar]
- 15.DiMartino, J. F., T. Miller, P. M. Ayton, T. Landewe, J. L. Hess, M. L. Cleary, and A. Shilatifard. 2000. A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL. Blood 96:3887-3893. [PubMed] [Google Scholar]
- 16.Djabali, M., L. Selleri, P. Parry, M. Bower, B. D. Young, and G. A. Evans. 1992. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat. Genet. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 17.Dobson, C. L., A. J. Warren, R. Pannell, A. Forster, and T. H. Rabbitts. 2000. Tumorigenesis in mice with a fusion of the leukaemia oncogene Mll and the bacterial lacZ gene. EMBO J. 19:843-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firestein, R., and M. L. Cleary. 2000. Set domain-dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3-9. Mol. Cell. Biol. 20:4900-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank, R., J. Zhang, H. Uchida, S. Meyers, S. W. Hiebert, and S. D. Nimer. 1995. The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene 11:2667-2674. [PubMed] [Google Scholar]
- 20.Galili, N., R. J. Davis, W. J. Fredericks, S. Mukhopadhyay, R. J. Rauscher III, B. S. Emanuel, G. Rovera, and F. G. Barr. 1993. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 5:230-235. [DOI] [PubMed] [Google Scholar]
- 21.Galoian, K., T. Milne, H. Brock, A. Shilatifard, R. Slany, and J. L. Hess. 2000. Deregulation of c-myc by leukemogenic MLL fusion proteins. Blood 96:1967. [Google Scholar]
- 22.Ghosh, S., C. Toth, B. M. Peterlin, and E. Seto. 1996. Synergistic activation of transcription by the mutant and wild-type minimal transcriptional activation domain of VP16. J. Biol. Chem. 271:9911-9918. [DOI] [PubMed] [Google Scholar]
- 23.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 24.Gould, A. 1997. Functions of mammalian Polycomb group and trithorax group related genes. Curr. Opin. Genet. Dev. 7:488-494. [DOI] [PubMed] [Google Scholar]
- 25.Gu, Y., T. Nakamura, H. Alder, R. Prasad, O. Canaani, G. Cimino, C. M. Croce, and E. Canaani. 1992. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell 71:701-708. [DOI] [PubMed] [Google Scholar]
- 26.Hanson, R. D., J. L. Hess, B. D. Yu, P. Ernst, M. van Lohuizen, A. Berns, N. M. van der Lugt, C. S. Shashikant, F. H. Ruddle, M. Seto, and S. J. Korsmeyer. 1999. Mammalian trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc. Natl. Acad. Sci. USA 96:14372-14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hillion, J., M. Le Coniat, P. Jonveaux, R. Berger, and O. A. Bernard. 1997. AF6q21, a novel partner of the MLL gene in t(6;11)(q21;q23), defines a forkhead transcriptional factor subfamily. Blood 90:3714-3719. [PubMed] [Google Scholar]
- 28.Hollenbach, A. D., J. E. Sublett, C. J. McPherson, and G. Grosveld. 1999. The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. EMBO J. 18:3702-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ida, K., I. Kitabayashi, T. Taki, M. Taniwaki, K. Noro, M. Yamamoto, M. Ohki, and Y. Hayashi. 1997. Adenoviral E1A-associated protein p300 is involved in acute myeloid leukemia with t(11;22)(q23;q13). Blood 90:4699-4704. [PubMed] [Google Scholar]
- 30.Jacobs, Y., C. A. Schnabel, and M. L. Cleary. 1999. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol. Cell. Biol. 19:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jansen, J. H., A. Mahfoudi, S. Rambaud, C. Lavau, W. Wahli, and A. Dejean. 1995. Multimeric complexes of the PML-retinoic acid receptor alpha fusion protein in acute promyelocytic leukemia cells and interference with retinoid and peroxisome-proliferator signaling pathways. Proc. Natl. Acad. Sci. USA 92:7401-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kempf, B. E., and P. K. Vogt. 1999. A genetic analysis of PAX3-FKHR, the oncogene of alveolar rhabdomyosarcoma. Cell Growth Differ. 10:813-818. [PubMed] [Google Scholar]
- 33.Kops, G. J., N. D. de Ruiter, A. M. De Vries-Smits, D. R. Powell, J. L. Bos, and B. M. Burgering. 1999. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398:630-634. [DOI] [PubMed] [Google Scholar]
- 34.Lam, P. Y., J. E. Sublett, A. D. Hollenbach, and M. F. Roussel. 1999. The oncogenic potential of the Pax3-FKHR fusion protein requires the Pax3 homeodomain recognition helix but not the Pax3 paired-box DNA binding domain. Mol. Cell. Biol. 19:594-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavau, C., C. Du, M. Thirman, and N. Zeleznik-Le. 2000. Chromatin-related properties of CBP fused to MLL generate a myelodysplastic-like syndrome that evolves into myeloid leukemia. EMBO J. 19:4655-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavau, C., S. J. Szilvassy, R. Slany, and M. L. Cleary. 1997. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 16:4226-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Licht, J. D. 2001. AML1 and the AML1-ETO fusion protein in the pathogenesis of t(8;21) AML. Oncogene 20:5660-5679. [DOI] [PubMed] [Google Scholar]
- 38.Luo, R. T., C. Lavau, C. Du, F. Simone, P. E. Polak, S. Kawamata, and M. J. Thirman. 2001. The elongation domain of ELL is dispensable but its ELL-associated factor 1 interaction domain is essential for MLL-ELL-induced leukemogenesis. Mol. Cell. Biol. 21:5678-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lutterbach, B., D. Sun, J. Schuetz, and S. W. Hiebert. 1998. The MYND motif is required for repression of basal transcription from the multidrug resistance 1 promoter by the t(8;21) fusion protein. Mol. Cell. Biol. 18:3604-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCabe, N. R., M. Kipiniak, H. Kobayashi, M. Thirman, H. Gill, J. D. Rowley, and M. O. Diaz. 1994. DNA rearrangements and altered transcripts of the MLL gene in a human T-ALL cell line Karpas 45 with a t(X;11) (q13;q23) translocation. Genes Chromosomes Cancer 9:221-224. [DOI] [PubMed] [Google Scholar]
- 41.Medema, R. H., G. J. Kops, J. L. Bos, and B. M. Burgering. 2000. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404:782-787. [DOI] [PubMed] [Google Scholar]
- 42.Meyers, S., N. Lenny, and S. W. Hiebert. 1995. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol. Cell. Biol. 15:1974-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nasrin, N., S. Ogg, C. M. Cahill, W. Biggs, S. Nui, J. Dore, D. Calvo, Y. Shi, G. Ruvkun, M. C. Alexander-Bridges. 2000. DAF-16 recruits the CREB-binding protein coactivator complex to the insulin-like growth factor binding protein 1 promoter in HepG2 cells. Proc. Natl. Acad. Sci. USA 97:10412-10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez, A., P. Kastner, S. Sethi, Y. Lutz, C. Reibel, and P. Chambon. 1993. PMLRAR homodimers: distinct DNA binding properties and heteromeric interactions with RXR. EMBO J. 12:3171-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rego, E. M., and P. P. Pandolfi. 2001. Analysis of the molecular genetics of acute promyelocytic leukemia in mouse models. Semin. Hematol. 38:54-70. [DOI] [PubMed] [Google Scholar]
- 46.Rubnitz, J. E., J. Morrissey, P. A. Savage, and M. L. Cleary. 1994. ENL, the gene fused with HRX in t(11;19) leukemias, encodes a nuclear protein with transcriptional activation potential in lymphoid and myeloid cells. Blood 84:1747-1752. [PubMed] [Google Scholar]
- 47.Sadowski, I., and M. Ptashne. 1989. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 17:7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schumacher, A., and T. Magnuson. 1997. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 13:167-170. [PubMed] [Google Scholar]
- 49.Scoggin, K. E., A. Ulloa, and J. K. Nyborg. 2001. The oncoprotein Tax binds the SRC-1-interacting domain of CBP/p300 to mediate transcriptional activation. Mol. Cell. Biol. 21:5520-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slany, R. K., C. Lavau, and M. L. Cleary. 1998. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol. Cell. Biol. 18:122-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.So, C. W., S. Dong, C. K. So, G. X. Cheng, Q. H. Huang, S. J. Chen, and L. C. Chan. 2000. The impact of differential binding of wild-type RARα, PML-, PLZF-, and NPM-RARα fusion proteins towards transcriptional co-activator, RIP-140, on retinoic acid responses in acute promyelocytic leukemia. Leukemia 14:77-83. [DOI] [PubMed] [Google Scholar]
- 52.So, C. W., C. K. C. So, N Cheung, S. L. Chew, M H Sham, and L. C. Chan. 2000. The interaction between EEN and Abi, two MLL fusion partners, and synaptojanin and dynamin: implications for leukaemogenesis. Leukemia 14:594-601. [DOI] [PubMed] [Google Scholar]
- 53.Sobulo, O. M., J. Borrow, R. Tomek, S. Reshmi, A. Harden, B. Schlegelberger, D. Housman, N. A. Doggett, J. D. Rowley, and N. J. Zeleznik-Le. 1997. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3). Proc. Natl. Acad. Sci. USA 94:8732-8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taki, T., M. Sako, M. Tsuchida, and Y. Hayashi. 1997. The t(11;16)(q23;p13) translocation in myelodysplastic syndrome fuses the MLL gene to the CBP gene. Blood 89:3945-3950. [PubMed] [Google Scholar]
- 55.Tanaka, K., T. Tanaka, M. Kurokawa, Y. Imai, S. Ogawa, K. Mitani, Y. Yazaki, and H. Hirai. 1998. The AML1/ETO(MTG8) and AML1/Evi-1 leukemia-associated chimeric oncoproteins accumulate PEBP2beta(CBFbeta) in the nucleus more efficiently than wild-type AML1. Blood 91:1688-1699. [PubMed] [Google Scholar]
- 56.Tkachuk, D. C., S. Kohler, and M. L. Cleary. 1992. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71:691-700. [DOI] [PubMed] [Google Scholar]
- 57.Van Orden, K., H. A. Giebler, I. Lemasson, M. Gonzales, and J. K. Nyborg. 1999. Binding of p53 to the KIX domain of CREB binding protein. A potential link to human T-cell leukemia virus, type I-associated leukemogenesis. J. Biol. Chem. 274:26321-26328. [DOI] [PubMed] [Google Scholar]
- 58.Westendorf, J. J., C. M. Yamamoto, N. Lenny, J. R. Downing, M. E. Selsted, and S. W. Hiebert. 1998. The t(8;21) fusion product, AML-1-ETO, associates with C/EBP-alpha, inhibits C/EBP-alpha-dependent transcription, and blocks granulocytic differentiation. Mol. Cell. Biol. 18:322-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiggan, O., A. Taniguchi-Sidle, and P. A. Hamel. 1998. Interaction of the pRB-family proteins with factors containing paired-like homeodomains. Oncogene 16:227-236. [DOI] [PubMed] [Google Scholar]
- 60.Xia, Z. B., and N. J. Zeleznik-Le. 2000. MLL repression domain interacts with histone deacetylases. Blood 96:1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu, B. D., R. D. Hanson, J. L. Hess, S. E. Horning, and S. J. Korsmeyer. 1998. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl. Acad. Sci. USA 95:10632-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu, B. D., J. L. Hess, S. E. Horning, G. A. Brown, and S. J. Korsmeyer. 1995. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378:505-508. [DOI] [PubMed] [Google Scholar]