Abstract
The interaction between Hfq and RNA is central to multiple regulatory processes. Using site-directed mutagenesis, we have found a missense mutation in Hfq (V43R) which strongly affects2 the RNA binding capacity of the Hfq protein and its ability to stimulate poly(A) tail elongation by poly(A)-polymerase in vitro. In vivo, overexpression of this Hfq variant fails to stimulate rpoS–lacZ expression and does not restore a normal growth rate in hfq null mutant. Cells in which the wild-type gene has been replaced by the hfqV43R allele exhibit a phenotype intermediate between those of the wild-type and of the hfq minus or null strains. This missense mutation derepresses Hfq synthesis. However, not all Hfq functions are affected by this mutation. For example, HfqV43R represses OppA synthesis as strongly as the wild-type protein. The dominant negative effect of the V43R mutation over the wild-type allele suggests that hexamers containing variant and genuine subunits are presumably not functional. Finally, molecular dynamics studies indicate that the V43R substitution mainly changes the position of the K56 and Y55 side chains involved in the Hfq–RNA interaction but has probably no effect on the folding and the oligomerization of the protein.
INTRODUCTION
RNA transmits the genetic information encoded in DNA and also plays a variety of essential structural, catalytic and regulatory roles within the cell, often in association with specific proteins. One of the most intriguing examples of the global regulatory role of RNA–protein interactions in gene expression is provided by the Hfq protein [reviewed in (1–4)]. In Escherichia coli, inactivation of the hfq gene causes a variety of phenotypes and alters the expression of many proteins, indicating that Hfq acts as a pleiotropic regulator which controls expression of many genes positively or negatively, directly or indirectly (5–8).
Recently, this protein was reported to regulate poly(A)-dependent mRNA decay (9–11), to affect RNA stability (12–15), to modulate RNA processing by RNase E (13,16–19) and translation (20,21). Hfq binds to 30% of the small regulator noncoding RNAs (ncRNAs) identified in E.coli (22) and it is essential for the function exerted by the OxyS, DsrA, RyhB, Spot 42, MicA, SgrS ncRNAs on their mRNA targets rpoS, fhlA, sodB, sdhCDAB, galK, ompA, ptsG (15,23–27).
Although the multiple roles of the protein seem to be correlated with its capacity to bind RNA, the structural basis of the Hfq–RNA interaction has not been elucidated. Hfq recognizes A-U rich single-stranded region of RNA and was also reported to interact with a curved form of DNA (28). Moreover, the finding that Hfq activates poly(A)-polymerase (PAP) (11), copurifies with H-NS, PAP and polynucleotide phosphorylase (9,29), interacts with RNA polymerase (30), associates with RNase E through the C-terminal scaffold of the protein (19) and stimulates Qβ RNA replication (31) suggests that it probably has the capability to establish structural and functional interactions with several proteins. Finally, Hfq has been reported to exhibit an ATPase activity (30).
Several studies have aimed at identifying the functional regions of Hfq. It was shown that a truncated protein deprived of its 19 C-terminal residues is fully able to bind RNA (32) and that the hfq2::Ω mutation which leaves 79 N-terminal amino acids intact out of 101 residues does not affect its functions (7). Consistently, the Pseudomonas aeruginosa Hfq which comprises only 82 amino acids functionally replaces the E.coli protein (33). Hfq is a member of the Sm protein family involved in RNA splicing and degradation (23,24,34). These proteins form ring-shaped oligomeric structures which are homohexameric in bacteria (35) and heteroheptameric in eukaryotes. Comparison of the structure of the E.coli Hfq with that of Staphylococcus aureus crystallized in association with a short oligonucleotide suggests that D40, F42 and K56 residues of the E.coli protein may be implicated in the RNA binding site (34–36). Moreover, it was reported that in Azorhizobium caulinodans, a C to R missense mutation at position 43 altered the function of the Hfq paralog NrfA (37).
We report here the construction of a set of E.coli Hfq variants at positions expected to participate in RNA binding and Hfq functions. These include single point mutants (D40A, F42A and K56A) and a double mutant (D40A–F42A) (35,36). The V43R variant was constructed to mimic the mutation previously shown to be deficient for Hfq function in A.caulinodans (37) as well as the V43C variant used as a control protein. We find that only one of them, in which valine at position 43 was substituted for arginine (V43R), was significantly affected for RNA binding and stimulation of poly(A) elongation. We also show that this Hfq variant (HfqV43R) failed to stimulate RpoS expression and that substitution of the wild-type gene by the mutant allele on the chromosome affects growth rate and abolishes autocontrol of Hfq synthesis. We present evidence that Hfq represses the expression of OppA, a protein whose synthesis was previously shown to be regulated by the GcvB ncRNA (38). Interestingly, the V43R mutation does not alter the repression of OppA synthesis. We also performed molecular dynamics (MD) simulations which indicate that the V43R substitution mainly affects side chain positions of K56 and Y55 within the monomer but not the overall structure of the hexamer. This set of data leads us to propose that introduction of arginine at position 43 perturbs binding of the RNA at the proximal site or interferes with the good positioning of some RNA molecules on the Hfq protein. This may impair formation of Hfq–RNA complexes and perturbates some functions of Hfq. Incidently, our data also provide evidence that Hfq, which is known to be involved in various stress situations, also ensures important functions during exponential growth in rich medium.
MATERIALS AND METHODS
Strains and plasmids
Plasmids
Mutant forms of the hfqHis6 gene were constructed either by using the PCR-based Stratagene QuickChange kit or by a two-step PCR amplification of the hfq gene from pTE607 (kindly provided by Thomas Elliott) using the reverse primer and T7term-GCTAGTTATTGCTCAGCGG primer and the corresponding forward primer and the pT7-TAATACGACTCACTATAGGG primer (Supplementary Table 1). Purified PCR fragments were used in a second PCR amplification without further addition of primers and template. Fragments were digested by NdeI and BamHI and cloned in the corresponding sites of pET11c. Hfq and Hfq variants were overproduced from BL21 λDE3hfq1 strain (Supplementary Table 2) transformed with these plasmids.
Mutagenesis of the hfq gene was performed on pTX381 carrying the hfq gene on a 1.5 kb fragment between the BamH1 and HindIII sites of pACYC184 (7), giving pTX381V43C and pTX381V43R plasmids. The BamHI–HindIII fragments of pTX381, pTX381V43C and pTX381V43R were inserted in the corresponding sites of pCL1921 (39) giving rise to plasmids pCLhfq+, pCLhfqV43C and pCLhfqV43R.
Construction of the strain with the hfq gene deleted
Deletion of the hfq gene was performed by a two-step PCR amplification. At the first step, two PCR fragments were generated on pTX367 plasmid (7) with two pairs of forward and reverse primers; ATGGCTAAGGGGCAATCTTTAGAAGAAACCGAATAAGGTTTCGGG and CACACAGGAAACAGCTATGACCATG (R primer), CCCGAAACCTTATTCGGTTTCTTCTAAAGATTGCCCCTTAGCCAT and CGACGTTGTAAAACGACGGCCAGT (U primer). At the second step, the two purified PCR products were mixed and amplified in the presence of R and U primers. The resulting fragment was cleaved with BamHI and HindIII, blunt ended at the HindIII site and inserted in the SmaI and BamHI sites of pKO3 plasmid (40), generating the pKO3hfqΔ22-294 plasmid. The pKO3hfqV43R was obtained by ligating the BamHI–HindIII fragment of pTX381V43R into the same sites of pKO3.
Chromosomal gene replacement
The pKO3hfqV43R and pKO3ΔhfqΔ22-294 plasmids were used to replace the hfq1::Ω allele from strain TX2808 (7) by the corresponding gene as described in (40), giving strain IBPC937 and IBPC946, respectively. Gene replacement was tested by loss of the kanamycin resistance. Candidates were validated by sequencing of the hfq gene. Other strains were constructed by generalized transduction with bacteriophage P1.
Construction of translational fusions with the lacZ gene on the chromosome
To generate the hfq–lacZ translation fusion, the hfq translation initiation region (TIR) covering positions from −95 to +42 (where +1 is A of the start ATG) was amplified from genomic DNA by PCR and inserted between BamHI and HindIII sites of pEMBLΔ46 in frame with the lacZ coding sequence under the control of lac-promoter/operator, as described earlier for lacZ translational fusions with exogenic TIRs (41). BamHI site was included in a forward primer (Hfq-for 5′-GCAGGATCCGCAGGCTGAATGTGTAC) and HindIII site in a reverse primer for PCR (Hfq-rev 5′-CGCAAGCTTGCGTTCAGGAACGGATC). The resulting plasmid pEHfq95 was then used to transfer the hfq–lacZ fusion onto the chromosome of the ENSO strain [former name HfrG6Δ12, see (42)] by homologous recombination so that β-galactosidase activity was measured from a single-copy lacZ gene, giving the IBhfq95 strain. Supplementary Table 2 listed the strains used in this work.
Protein purification
Cells from the induced cultures were resuspended in 10 ml of buffer containing 20 mM Tris–HCl, pH 7.8, 0.5 M NaCl, 10% (v/v) glycerol, 0.1% (v/v) Triton X-100, 10 µl DNase I (10 mg/ml) and a protease inhibitor cocktail tablet (Complete mini, EDTA-free, Roche Diagnostic) at 4°C. The suspension was passed through a French press (1200 bar, 20 000 psi) and centrifugated for 30 min at 15 000 g. The supernatant was heated 80°C for 15 min and the insoluble material was removed by centrifugation for 30 min at 15 000 g. Imidazole-HCl, pH 7.8, was added to the supernatant to give a final concentration of 1 mM and the resultant suspension was applied to a 1 ml Ni2+-NTA column (Qiagen). The resin was then sequentially washed with about 15 column volumes of (i) 20 mM Tris–HCl, pH 7.8, 0.3 M NaCl, 20 mM imidazole and (ii) 50 mM sodium phosphate, pH 6.0, 0.3 M NaCl. Hfq protein was eluted in buffer containing 20 mM Tris–HCl, pH 7.8, 0.3 M NaCl and 0.5 M imidazole. The fractions containing Hfq were dialysed in buffer A (50 mM Tris–HCl, pH 7.5, 1 mM EDTA, 5% glycerol) containing 50 mM NH4Cl. After centrifugation, the supernatant was applied to a 5 ml poly(A) Sepharose (Amersham Biosciences). The resin was washed with buffer A containing 1 M NH4Cl and the Hfq protein was eluted in buffer A containing 1 M NH4Cl and 8 M urea. The fractions containing Hfq were then dialysed in buffer A containing 50 mM NH4Cl and 0.1 % Triton X-100. The protein was conserved at 4°C. Protein concentrations were determined by using the Bradford protein assay with BSA as a standard. Hfq molar concentration was calculated on the basis of the monomer form. These proteins were used in in vitro studies.
RNA preparation and labelling
The 3′-rpsO RNA fragment corresponds to the last 97 nt of the rpsO transcript, with a GGG sequence at the 5′ end, the 3′-rpsO-(A)18 RNA fragment corresponds to the same RNA with 18 A residues at its 3′ end (17). RNA corresponding to the 5′-untranslated region of the hfq gene with a GG sequence at the 5′ end was transcribed form a DNA fragment obtained by PCR amplification from genomic DNA using the T7 RNA polymerase promoter containing oligonucleotide 5′-TAATACGACTCACTATAGGGTATCGTGCGCAATTTTTTC as a forward primer and the 5′-GAACGGATCTTGTAAAGATTG as a reverse primer. These RNAs were synthesised by T7 RNA polymerase yielding uniformly labelled RNA with [α-32P]UTP as tracer (17).
Electrophoretic mobility shift assays
Increasing concentrations of His-tagged Hfq protein and variants were incubated with 5′ [32P]RNA in 20 µl buffer containing 10 mM Tris–HCl (pH 8), 1 mM EDTA, 80 mM NaCl and 1% glycerol (v/v). Reactions were incubated at 37°C for 30 min and complexes were separated on native polyacrylamide gels. A PhosphoImager and the ImageQuant software (MD) were used to view the separated components and to quantify results. The apparent dissociation constant (Kd) was determined by incubating increasing concentrations of protein ranging from 5 pM to 100 nM with 5 pM RNA. The data were plotted using KALEIDAGRAPH 3.0.4 (Abelbeck Software, Reading, PA) and the generated curves were fitted by non-linear least squares regression assuming a bimolecular model such that the Kd values represent the protein concentration at half-maximal RNA binding (17).
β-Galactosidase assay
Strains RQ91 (hfq+) and MCM11 (hfq10::cat) contain the rpoS-lacZ chromosomal fusion. These strains were transformed with the pCL series of plasmids described in the text. Cells were grown at 37°C in Luria–Bertani (LB) medium with the appropriate antibiotic. Strains IBhfq95 and IBhfq95-hfqV43R containing the hfq–lacZ chromosomal fusion under the control of the lac-promoter/operator region were grown in LB medium supplemented with isopropyl-β-d-thiogalactopiranoside (IPTG; 0.2 mM). Cells were harvested in exponential or prestationary phase, disrupted by sonication and β-galactosidase activities were measured in clarified cell extracts. Specific β-galactosidase activity was expressed as nmol ONPG (o-nitrophenyl-β-d-galactopiranoside) hydrolyzed/min/mg of total soluble cell proteins (41).
Western blotting
Frozen cells were resuspended in 0.1 M Tris–HCl, pH 8, 0.4 M NaCl, 0.1 mM EDTA, 1 mM 2-β-mercaptoethanol and lysed by sonication on ice. Protein concentrations were determined by the BCA protein assay (Pearce). Cellular proteins (20 µg) were separated on a 15% SDS–PAGE gel and transferred to Hybond C-super (Amersham) by electroblotting. The membrane was incubated successively with polyclonal rabbit antibodies raised against Hfq and with radiolabelled protein A. Quantification was carried out by PhosphorImager (MD) and results were normalized relative to the bands of E.coli proteins that cross-hybridized with the antibody.
Polyadenylation in vitro
5′ Radiolabelled polyadenylated RNA (250 fmol) was incubated with tRNA (0.5 µg), ATP (2 mM), Hfq variant (the amount is specified in each experiment) and PAP (1.4 pmol) (11,43) The reaction was stopped by addition of phenol/chloroform and samples were precipitated with ethanol. Pellets were counted, resuspended in 8 M urea, 0.025% bromophenol blue (w/v), 0.025% xylene cyanol blue (w/v) and identical amounts of radioactivity were analysed on the sequencing gel.
Theory section
MD simulations were carried out at pH 7 using the GROMACS program and the all-atom force field GROMOS96 (44). The initial conformation for wild-type E.coli Hfq was taken from its crystal structure (PDB entry 1HK9), free of RNA and containing the atomic positions of residues 6–69 (36). The starting points for the V43R and V43C mutants were constructed using the SWISS-MODEL server (45). The hexameric forms of wild-type E.coli Hfq and its variants (V43R and V43C) were then solvated in a dodecahedric box of 8.2 nm side containing ∼11 000 simple point charge water molecules. MD simulations were performed with periodic boundary conditions at constant temperature (300 K) and constant pressure (1 atm), with coupling constants of 0.1 and 0.5 ps, respectively. Bond lengths were constrained with the SHAKE algorithm and the time step for dynamics was 2 fs. The Particle Mesh Ewald method was used with a cutoff distance of 12 Å for the electrostatic interactions and the nonbonded interactions were updated every five time steps. The three Hfq proteins were equilibrated at 300 K for 50 ps and then subject to MD analysis for 1.2 ns. Each simulation was repeated using different initial velocities to verify that different simulations on the same sequence produce equivalent results on a nanosecond time scale. All simulations covering 7.5 ns took 2 months on a cluster of four 1.2 GHz processors.
RESULTS
Search for Hfq mutants affected in their ability to bind RNA and to activate PAP
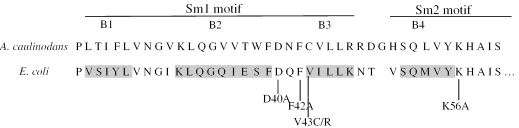
To provide insight into Hfq functions, and based on the crystal structure of Hfq protein, we replaced strategic residues expected to alter the RNA binding capacity of the protein. The conserved residues D40, F42 and K56 were individually changed to alanine (D40A, F42A and K56A). Alanine substitution eliminates the side chain beyond the [beta] carbon and yet neither does it alter the main-chain conformation nor does it impose extreme electrostatic or steric effects. In addition, we constructed a D40A–F42A double mutant. We also generated the V43R mutation that mimics in E.coli the C43R substitution which inactivates the Hfq homolog of A.caulinodans (37) and a control protein where valine 43 is replaced by a cystein as in A.caulinodans (Figure 1). Mutations were made on a plasmid-borne wild-type hfq gene harbouring a C-terminus His-tagged to overproduce variants in a strain bearing an inactivated hfq gene (Supplementary Table 2). Furthermore, Hfq mutant proteins retained on Ni2+-NTA column were further purified on poly(A) sepharose column to eliminate contaminating RNAs.
Figure 1.
Localization in the Hfq sequence of the amino acids that were mutated. Sequence alignment of Hfqs of A.caulinodans and E.coli. Secondary structural elements are highlighted in grey. B indicate beta sheet. The amino acid replacements are indicated (by bars).
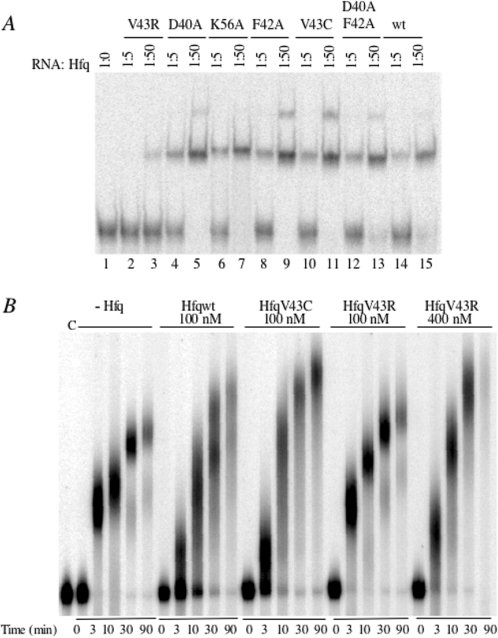
RNA binding capacity of Hfq variants was evaluated by mobility shift assay using a polyadenylated rpsO RNA fragment [3′-rpsO-(A)18] that was previously shown to be very strongly bound by wild-type Hfq (Hfqwt) (17). All mutant and wild-type proteins bound the RNA with similar affinities except HfqV43R whose affinity was dramatically lower (Figure 2). Precise measurements show that HfqV43C bound the 3′-rpsO-(A)18 with the same affinity (Kd = 360 ± 54 pM) as Hfqwt (Kd = 280 ± 51 pM), and that substitution of V43 by R increased the Kd value to 10 ± 2.6 nM, i.e. by 35-fold [Table 1, column (a)]. Because preincubation of HfqV43R for 10 min at 90°C before mixing with the RNA did not change the affinity of HfqV43R for the polyadenylated rpsO RNA compared with non-preincubated protein, we assumed that the lower affinity of HfqV43R for RNA does not result from a significant thermodynamic destabilization of the protein (data not shown).
Figure 2.
Effects of amino acid substitutions in the Hfq protein on RNA binding and stimulation of poly(A)polymerase elongation. (A) Relative interaction of wild-type and mutant Hfq proteins with polyadenylated 3′-rpsO RNA fragment assayed by gel retardation. Radiolabelled 3′-rpsO-(A)18 RNA (10 pM) was incubated without (lane 1) or with 50 pM (lanes 2, 4, 6, 8, 10, 12 and 14) and 500 pM (lanes 3, 5, 7, 9, 11, 13 and 15) of either wild-type Hfq protein (wt) or of the mutant protein indicated at the top. (B) Poly(A) polymerase activation by wild-type and Hfq variant proteins was assayed on the 3′-rpsO RNA fragment. Radiolabelled RNA fragment was incubated with the Hfq protein indicated at the top of the gel at the concentration specified, in the presence of tRNA and ATP. PAP was added at time 0. Samples were removed and reaction stopped at the times indicated at the bottom of the picture. c indicates RNA incubated without PAP.
Table 1.
RNA binding of Hfqwt and variant proteins
| RNA | (a) | (b) |
|---|---|---|
| Protein | 3′-rpsO-(A)18 | hfq (1-101) |
| Hfqwt | 280 ± 51 pM (×3) | 2 ± 0.6 nM (×4) |
| HfqV43R | 10 ± 2.6 nM (×2) | 7.2 ± 1.2 nM (×4) |
| HfqV43C | 360 ± 54 pM (×1) | 1.1 ± 0.2 nM (×1) |
5′ End labelled RNA (5 pM) indicated at the top of each column was mixed with increasing concentrations of Hfq ranging from 5 pM to 100 nM. Complexes were separated on native polyacrylamide gels. The data were plotted using KALEIDAGRAPH 3.0.4 (Abelbeck Software, Reading, PA) and the generated curves were fitted by non-linear least squares regression assuming a bimolecular model such that the Kd values represent the protein concentration at half-maximal RNA binding. The apparent Kd values were calculated from the number of independent experiments as indicated between brackets. Predicted structures of the RNA are presented in Supplementary Figure 1.
We then assayed whether the Hfq deficiency in binding the 3′-rpsO-(A)18 RNA impairs elongation by PAP as proposed earlier (43). PAP is a distributive enzyme; at the beginning of the reaction, the 3′-rpsO RNA substrate rapidly disappeared and all molecules were elongated at approximately the same rate (Figure 2B, left part). Addition of Hfqwt caused partition of the RNA substrate into elongated and non-elongated molecules demonstrating that Hfq switches PAP from a distributive to a processive mode of polyadenylation as previously reported (Figure 2B) (11). Moreover, these kinetics confirm that Hfq stimulates the elongation of molecules already harbouring poly(A) tails (30 and 90 min of incubation). Figure 2B shows that HfqV43R is impaired in these functions, whereas the HfqV43C protein that binds this RNA normally, does have an effect on poly(A) synthesis. HfqV43R is able to provide the same stimulating effect only at increased concentration (compare 30 min lanes for HfqV43R 100 and 400 nM).
Effect of HfqV43C and HfqV43R expression in hfq and wild-type strains
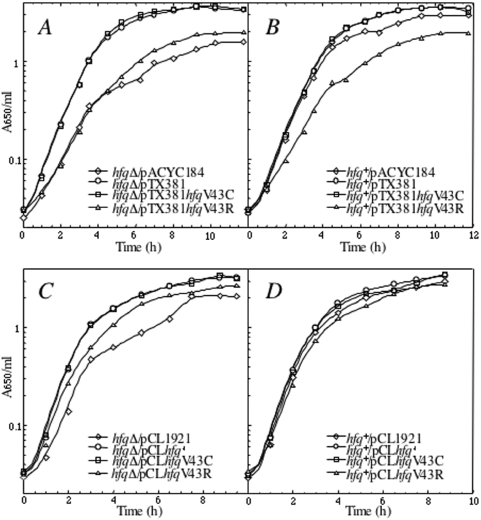
We then examined whether the HfqV43R mutation impairs Hfq functions in vivo. For that purpose, we first constructed an in-frame deleted hfq gene in order to avoid polar effects that affected expression of downstream genes in the available hfq1::Ω mutant (7). Then, we attempted to complement this hfqΔ strain with hfq mutant genes cloned in pACYC184 (pTX381 derivatives, about 15 copies per cell) and in pCL1921 (pCL derivatives, about 5 copies per cell). We verified that the amount of Hfq synthesized from pTX381 is two times higher than that synthesized from pCLhfq+ in an hfqΔ strain (data not shown). Strikingly, while HfqV43C and Hfqwt correct the growth defect of the hfqΔ, HfqV43R, faulty for RNA binding, either does not complement at all (when expressed from the higher copy number plasmid, Figure 3A) or only partly complements (when expressed from the lower copy number plasmid, Figure 3C) this growth deficiency, suggesting that higher amount of HfqV43R synthesized from pTX381 is deleterious for the cell. It is also interesting to point out that the hfq+ strain transformed with pTX381hfqV43R exhibited a growth curve similar to that of the hfqΔ strain transformed with the empty vector (Figure 3A and B), which suggests that the hfqV43R allele exerts a dominant negative effect on the wild-type gene when overexpressed from pTX381hfqV43R.
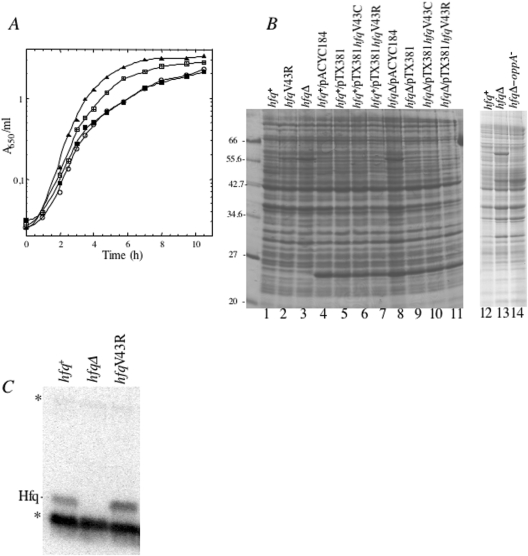
Figure 3.
Effects of overexpression of Hfq variants on cell growth. Growth curves of N3433 (hfq+) and IBPC 953 (hfqΔ) strains transformed with pACYC184 derivative (upper part) and pCL1921 derivative (lower part) plasmids containing hfqwt (giving pCLhfq+ and pTX381), hfqV43C (giving pCLhfqV43C and pTX381hfqV43C) and hfqV43R (giving pCLhfqV43R and pTX381hfqV43R), respectively. Cells were grown in LB media with appropiate antibiotics at 37°C. Turbidity of the cultures was monitored at 650 nm.
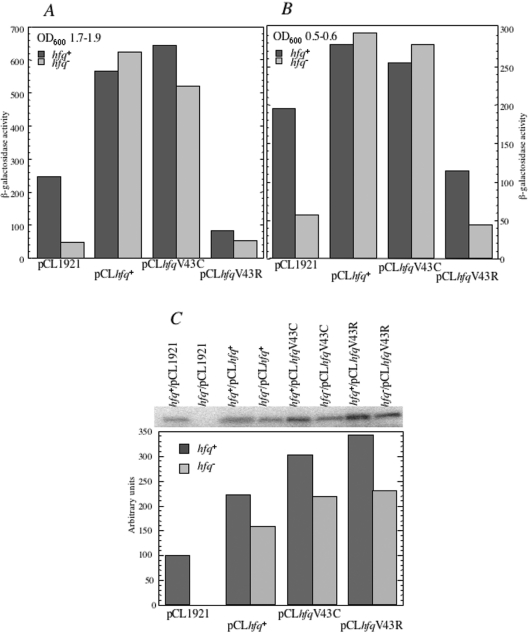
Then, we examined whether Hfq substituted at position 43 regulates the expression of RpoS. This was achieved by following the expression of the rpoS::lacZ translational fusion (46) in hfq-deficient strain containing the hfq10::cat mutation (8) transformed by the pCL1921 series of hfq containing plasmids. β-Galactosidase was measured in cells grown in LB medium to the phase when the growth begins to slow down (OD600 of 1.7–1.9). A 10-fold increase of β-galactosidase synthesis was measured when hfq-deficient cells were transformed with pCLhfq+ and pCLhfqV43C, while plasmid carrying the hfqV43R mutant did not stimulate the expression of the rpoS–lacZ fusion. Interestingly, expression of hfqV43R in the wild-type strain causes repression of the rpoS–lacZ translation to the level observed in the hfq− strain, thus confirming that HfqV43R exerts a severe dominant negative effect on Hfq (Figure 4A).
Figure 4.
Complementation of the hfq10::cat mutation with Hfq variants assessed by expression of the rpoS-lacZ chromosomal-integrated fusion. MCM11 is a hfq10::cat derivative of RO91 carrying a chromosomal rpoS-lacZ fusion. It was transformed with pCL1921 derived plasmid expressing either wild-type or hfq gene variants (V43C and V43R) giving the pCLhfq+, pCLhfqV43C and pCLhfqV43R plasmids, respectively. The empty vector was used as a control (pCL1921). β-Galactosidase synthesis from the rpoS–lacZ chromosomal-integrated fusion was measured in cells grown until OD600 of 1.6–1.9 (A) or until OD600 of 0.5–0.6. (B) Each value is the mean of at least three independent experiments, with standard deviations not exceeding 15% of magnitude. (C) Expression of the mutated hfq alleles. Soluble protein extracts from strains grown until OD600 of 1.6–1.9 shown on Figure 4A were analysed by western blotting using Hfq antibodies (upper part of the figure). Lower part of the figure is the quantitation of the western blot data coming from four sets of experiments. Hfq antibodies detected monomers in the extracts.
As expected, the β-galactosidase level measured in cells harvested in the middle of the exponential phase (OD600 0.5–0.6) (Figure 4B) was lower than in the prestationary phase. However, the hierarchy of expression levels was the same as in the prestationary phase. This indicates that Hfq controls translation of rpoS both in stationary phase and during exponential growth, when rpoS is expressed at a so-called basal level (47). One can hypothesize that under these conditions in which no characterized sRNA has been hitherto found as a player in RpoS expression, Hfq but not HfqV43R, could directly regulate rpoS translation as earlier reported for Salmonella typhimurium (48).
Quantitative immunoblots were performed to estimate Hfq levels in the different strains entering in stationary phase. Figure 4C shows that the amount of Hfq protein synthesized is higher when both hfq+ and hfq− strains were transformed with pCLhfqV43R compared with pCLhfq+ plasmid, thus suggesting that Hfq expression is affected by the V43R substitution (see below).
These experiments show that the Hfq harbouring the V43R substitution fails to complement Hfq deficiency and has a dominant negative effect on a number of Hfq functions while, in contrast, the V43C substitution does not cause these negative effects, although it is expressed from the plasmid at higher level than Hfqwt (Figure 4C).
Characterization of the hfqV43R mutant
We then examined some phenotypic characteristics of strains containing single copy of the hfqV43R, hfqΔ, hfq1::Ω which were previously attributed to Hfq deficiency in comparison with the wild-type allele (7,49). Interestingly, the hfqV43R mutant exhibits an intermediary phenotype between the hfq1::Ω interrupted mutant and the wild-type strain. Figure 5A shows that hfqΔ growth is diauxic in LB liquid medium containing 0.17 M NaCl as previously reported for hfq1::Ω (7), whereas hfqV43R exhibits only a gradual transition from log growth to stationary phase. Moreover, on LB plates, hfq1::Ω, hfqΔ and hfqV43R colonies were noticeably less opaque than the hfq+ parent, while no marked difference was observed in the size of hfqV43R and wild-type colonies. The same effects were observed in the HfrG6 genetic background (IBhfq95) (data not shown).
Figure 5.
Effect of chromosome encoded hfq+ and hfqV43R alleles on growth, total protein synthesis and relative amounts of Hfq protein. N3433 is the parent strain (filled triangle), hfq1::Ω filled square, hfqV43R (square) and hfqΔ (open circle) mutants correspond to IBPC 929, IBPC 941 and IBPC 953, respectively. (A) Growth curves. Cells were grown in LB medium at 37°C. (B) N3433, IBPC 941 and IBPC 953 strains and N3433 and IBPC 953 transformed with pACYC184 derivative plasmids containing hfqwt (giving pTX381) hfqV43C (giving pTX381hfqV43C) and hfqV43R (giving pTX381hfqV43R) and the empty vector (giving pACYC184) were grown in LB medium with appropriate antibiotics until OD600 of 0.4–0.44. Total cellular proteins were analysed on SDS–PAGE and stained by Coomassie blue. (C) Cellular levels of Hfqwt and HfqV43R in N3433 and IBPC 941, respectively, and IBPC 953 used as a negative control, revealed by immunoblotting. Asterisks indicate two proteins that cross-hybridize with the antibody.
We then tested whether hfq mutations affected the sensitivity of bacterial cells to UV light by streaking liquid cultures onto LB plates and exposing portions of the plates to different doses of UV light. As expected, the hfqΔ and hfq1::Ω mutants were more sensitive than the hfq+ parent (7). In contrast, the hfqV43R mutation did not affect the UV sensitivity (data not shown). We found that the V43R mutation did not reduce the viability of bacteria after a heat-shock at 55°C in contrast to the disruption of the gene that has a dramatic effect on bacterial survival at that temperature (data not shown). The exact function of Hfq underlying these effects remains unknown (49). Together, these results indicate that not all functions of Hfq are affected by a single copy of hfqV43R on the chromosome. However, as shown below, the V43R mutation leads to small (1.9) increase in Hfq levels, which could partly compensate for a reduced activity of the mutant protein. Since HfqV43R fails to stimulate rpoS mRNA translation, it was reasonable to suppose that this mutation may also alter expression of other mRNAs, as shown earlier for the hfq1::Ω and hfq10::cat mutants (5,6,8). Protein patterns from hfq+ and hfqΔ (transformed or not with the pACYC184 derivatives) visualized by Coomassie blue staining show that one band ∼56 kDa is present only in hfqΔ (Figure 5B, lanes 3, 8 and 13), indicating that Hfq negatively regulates synthesis of the corresponding protein. Matrix-assisted laser desorption ionization time-of-flight and LC-MSMS analysis of trypsic peptides generated from the excised band, unambiguously identified these peptides as belonging to the oppA gene product. The 56 kDa OppA protein was also observed in HAT10 (containing hfq10::cat) but not in CSH26 isogenic wild-type parent (data not shown) or in an hfqΔ-oppA::kan strain (Figure 5B, lane 14). OppA is a periplasmic protein component of the oligopeptide transport system (50,51) involved in the active transport of aminoglycoside antibiotics (52). Interestingly, the OppA band is faint (lane 2) or absent (lane 11) in cells containing either hfqV43R or hfqV43C (lane 10), indicating that both variant proteins retained the ability to inhibit the synthesis of this polypeptide. HfqV43R seems, however, to act less efficiently than the wild-type protein. The complete repression of OppA synthesis observed when HfqV43R is overproduced allows us to speculate that a higher level of this variant compensates the lower binding affinity of the protein for the ncRNA, namely GcvB, previously shown to negatively control OppA expression (38).
The HfqV43R mutant fails to autoregulate Hfq expression
Since the above results show that HfqV43R is expressed at a higher level than Hfqwt or HfqV43C (Figure 4C), we speculated that the V43R mutation also affects the negative control that Hfq exerts on its own expression (14). To exclude the possibility that variations in gene copy number may account for this result, we measured the Hfq level originating from a single-copy wild-type or hfqV43R genes on the chromosome. Hfq synthesis in the hfqV43R mutant was found to be 1.9 times higher than in hfq+ cells during exponential growth [Figure 5C and Table 2 column (b)].
Table 2.
Effect of hfqV43R mutation on the expression of hfq-lacZ and the Hfq amounts
| Strain | β-Galactosidase activity (a) | Hfq protein amount (b) | |
|---|---|---|---|
| (c) | (d) | (c) | |
| hfq+ | 4150 ± 50 | 5450 ± 50 | 39884 ± 4280 |
| hfqV43R | 7350 ± 200 (1.7) | 7400 ± 200 (1.3) | 77638 ± 10716 (1.9) |
IBhfq95 and IBhfq95hfqV43R strains carrying a hfq–lacZ translational fusion on the chromosome were grown as described in Figure 3A.
(a) β-Galactosidase levels from the hfq–lacZ fusion. Bacteria were harvested either in exponential phase (c) or upon entry in stationary phase (d). Mean values are calculated from three independent experiments. (b) Relative abundance of Hfq protein in the extracts. Quantification in arbitrary units of the western blot (data not shown), giving the mean values of two independent samples values in parenthesis are the fold increase in β-galactosidase levels or Hfq amount in the hfqV43R mutant compared with the wild-type strain.
We hypothesized that the inability of HfqV43R to control its own synthesis is due to its RNA binding deficiency and that the autocontrol occurs most likely at the translation level. Indeed, long stretches of A and U residues just upstream of the AUG start codon can be regarded as potential Hfq binding sites in the hfq mRNA where Hfq could compete for ribosome binding and repress its translation. To examine this possibility, we measured the yield of β-galactosidase from a hfq::lacZ translational fusion inserted as a single copy in the chromosome of hfq+ and hfqV43R cells. Table 2 column (a) shows that the hfq TIR is 1.7 times more active in the hfqV43R background when compared to wild-type, a repression factor similar to that measured by immunoblot [Figure 5C and Table 2 column (b)].
We then determined the RNA binding affinity of the wild-type and variant proteins to the hfq TIR by mobility shift assay using an RNA fragment extending from the P3hfq promoter (49) to the nucleotides specifying the 12th amino acid of the hfq coding region, hfq(1–101). Table 1 column (b) shows that Hfqwt and HfqV43C bind the hfq(1–101) RNA fragment with the same affinity, while HfqV43R has a Kd value 3.5-fold higher. In conclusion, direct measurements of Hfq cellular levels, expression of the hfq::lacZ translational fusion and Hfq TIR binding experiments gave consistent results, which, in spite of the modesty of the effects, support the idea that Hfq represses its own synthesis about 1.7 times. The results indicate that HfqV43R partly lost the capacity to exert the autocontrol probably because it poorly binds the 5′-part of its own mRNA. Given that HfqV43C appears to be synthesized at higher level than Hfqwt from the pCL plasmid (Figure 4C), it remains to be confirmed that this substitution possibly also affects Hfq autoregulation.
Molecular dynamics
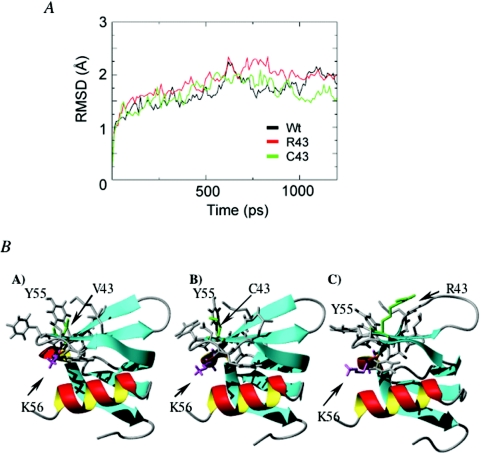
Because affinity and functional defects of the HfqV43R protein may result from folding or oligomerization defaults, we investigated the impact of this change on Hfq structure by performing MD simulations. Figure 6A shows the root-mean square deviations (RMSDs) of the backbone C alpha atoms with respect to the minimized structures as a function of time for the three Hfq proteins (Hfqwt, HfqV43C and HfqV43R). The minimized structures of both variants deviate by 0.2 Å from that of wild-type Hfq. All proteins behave similarly with RMSD versus time and fluctuate around 2 Å. Analysis of the MD-generated secondary structure shows, however, significant differences between the three Hfqs. Supplementary Table 3 reports the average and standard deviation of the helix and the five β-strands for each Hfq protein using the DSSP program (53). The statistics are very similar for the wild-type and HfqV43C proteins, but show a significant decrease in the strands B3 and B4 for V43R, with the mean percentage of B4 being 74% in R43 versus 90, 91% in wild-type and C43. Figure 6B shows a representative MD-generated structure of each Hfq sequence and the destabilizing effect of V43R on the strands B3 and B4. Complementing these analyses, we have followed the percentage of intramolecular/intermolecular side chain contacts in all variants. As expected, V43C does not change the native intramolecular/intermolecular contacts involving residue 43. In contrast, the V43R substitution has no effect on the intermolecular contacts responsible for oligomerization but disrupts the intramolecular interactions between residue 43 (strand B3) and residues Y55 (strand B4) and K56. This disruption destabilizes the strands B3 and B4 and changes the side chain positions of the K56 and the Y55 residues involved in the binding to a AU5G RNA and a 27 nt poly(A) oligomer, respectively (35,54).
Figure 6.
The V43R substitution has a destabilizing effect on the β-strands B3 and B4 and changes the position of the side chain of K56 and Y55. (A) RMSDs of the backbone C alpha atoms with respect to the minimized structures for the three Hfq proteins as a function of time. (B) Representative MD-generated structure of each Hfq sequence.
DISCUSSION
The present report shows that a single substitution in the Hfq polypeptide weakens the affinity of the protein for a polyadenylated mRNA and the hfq TIR, decreases the activation of PAP elongation, abolishes stimulation of RpoS expression and repression of its own synthesis while, in contrast, it does not impair repression of oppA expression.
Although both crystallographic data and sequence comparison suggest that D40, F42 and K56 may be involved in oligoribonucleotide binding by analogy with S.aureus Hfq (35) and Lsm–RNA complexes (55,56), none of these residues independently changed to alanine, modified Hfq interaction with polyadenylated RNA (Figure 2A). Our data are consistent with recent experiments showing that these single mutations, which all are supposed to disrupt the RNA binding site surrounding the central channel of the homohexameric ring, have minor effect on binding of a A27 oligomer and of an rpoS mRNA fragment (54). Moreover, a double mutation D40A–F42A still allows Hfq to bind RNA as strongly as the wild-type protein (Figure 2A). As to the K56A substitution, it was shown to affect binding to the small DsrA RNA (54). This fact allowed Mikulecky et al. (54) to propose that Hfq harbours two binding sites, one on the proximal face involving K56 and another on the distal face interacting with poly(A). We suggest that the Hfq interaction with the 3′-rpsO-(A)18 RNA may be realized via the distal face and with another amino acid within the binding site of the proximal face.
While the K56A substitution did not affect Hfq interaction with the polyadenylated RNA, we found that a single valine to arginine substitution at position 43 increased the Kd value 35 times for the same RNA. Moreover, a valine to cysteine substitution at the same position did not alter the RNA affinity. No role in RNA binding was previously assigned to residue 43. Because the bulky residue may affect folding and subunit interactions, we next investigated whether the V43R substitution affects the structure of the protein. MD studies indicate that this replacement destabilizes the strands B3 and B4 and induces reorientation of K56 and Y55 side chains (Figure 6) but does not affect the stability of the protein. The fact that the V43C substitution does not affect the interaction with the polyadenylated RNA indicates that the V43 residue is not directly involved in the binding. Nevertheless, arginine 43 may modify the proximal site containing Y55 and K56 and/or the capability of RNA to interact with the two RNA binding sites. Since our data show that K56 does not interact with 3′-rpsO-(A)18, it is possible that this RNA interacts with Y55. This is consistent with the fact that a Y55A substitution reduces the affinity of Hfq for a A27 oligomer (54). Furthermore, the crystal structure of Hfq associated with AU5G allows to hypothesize that Y55 may interact with an adenine residue of an A-rich oligonucleotide (35). Alternatively, the insertion of the bulky, positively charged arginine may result in modification of the protein surface and, therefore, impedes the simultaneous binding of the polyadenylated RNA to the proximal and distal faces of the protein. It is worth recalling here that the very strong affinity of Hfq for the polyadenylated RNA presumably reflects simultaneous binding to several sites on the surface of Hfq (54).
HfqV43R overproduction affects cell growth and reduces expression of rpoS::lacZ fusion protein in the presence of the wild-type allele. This phenomenon corresponds to the dominant negative effect which was already observed for multimeric proteins acting as repressors or activators (57). Since modelling predicts that the overall structure of the homohexamer is not perturbed by the V43R substitution, we assume that wild-type and HfqV43R subunits could assemble to form heterohexamers, the biological functions of which are impaired. At present, it is the only possible explanation for the dominant negative effect of mutated oligomeric proteins in wild-type cells (57).
Quantitative immunoblots, measurement of binding efficiency and comparative analysis of the hfq–lacZ translation efficiency in hfq+ versus hfqV43R strains argue in favour of modulation of the hfq translation by Hfqwt and the loss of this ability in the case of HfqV43R. However, since Hfq can bind DNA (28) and has been shown to be associated with HN-S and RNA polymerase (29,30) we cannot exclude that Hfq may also control its own expression at the transcriptional level. Tsui and co-workers (14) previously proposed that Hfq modulates its own synthesis. Our data indicate that this modulation, at least partly, takes place at the translational level and that the translational operator region may be restricted to a region present in the transcripts initiated from the hfq promoters functional during exponential growth. It is neither clear, however, whether Hfq binding competes with formation of the translation initiation complex as was proposed recently (58), induces refolding of the mRNA to facilitate degradation or delivers RNase E within the RNase E–Hfq complex (19) nor whether hfq mRNA is targeted by a small RNA. It must be recalled here that Hfq represses very modestly its own synthesis, with a repression factor about 2. This may reflect the fact that Hfq which is implicated in many mechanisms involving various RNAs has a lot of targets in the cell without a strict preference to its own mRNA.
OppA is a periplasmic component involved in the transport of nutritional peptides and in the sensitivity to aminoglycoside antibiotics (52,59). It was previously reported that GcvB ncRNA co-immunoprecipitates with Hfq and represses oppA expression by sequestering the ribosome binding site (16,38). As numerous data show that multiple functions of Hfq as a regulator of gene expression are realized via its interaction with small RNAs, it is reasonable to expect that GcvB exerts repression of OppA expression in concert with Hfq. Consistent with this idea, we show here that negative regulation of OppA synthesis takes place only in the presence of Hfq. Interestingly, the V43R mutation does not abrogate this function. Secondary structure predictions suggest that GcvB possesses U-rich single-stranded sequences which may be equally well bound by Hfqwt and HfqV43R. It remains to be determined whether Hfq acts by promoting pairing between GcvB and the oppA mRNA or by stabilizing GcvB. Yet, we cannot completely exclude that Hfq facilitates degradation of the oppA mRNA by delivering RNase E (19).
Hfq is an important regulator of gene expression through its RNA chaperon activity, the mechanism of which is still poorly understood. In particular, it is not clear how the protein interacts with its targets and facilitates RNA–RNA annealing. The HfqV43R protein is affected in RNA binding and in some, but not all Hfq functions. Among them, stimulation of RpoS expression and repression of OppA synthesis were shown to involve ncRNAs but only RpoS stimulation was affected by the V43R substitution. We concluded that the V43R substitution affects RpoS expression, probably by impairing the interaction of Hfq with an ncRNA, while it only marginally modifies the GcvB mediated regulation of OppA. Interestingly, this suggests that the GcvB and DsrA (57) RNAs may bind Hfq differently. We thus hypothesize that there are several ways for ncRNAs to accommodate the different binding sites of the Hfq protein.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors are grateful to J. Plumbridge for careful critical reading of the manuscript. The authors thank M. Wachi, M. E. Winkler, R. Hengge-Aronis, M. Springer, T. Elliot, K. Igarashi and the Church lab for kindly supplying strains and plasmids and A. Ishihama for Hfq antibodies used in early stage of experiments. Mass spectroscopy analysis was carried out by V. Labas and J. Vinh at Ecole Supérieure de Physique et Chimie Industrielles, Paris. I.V.B. was supported by Université Paris 7, Denis Diderot as Invited Professor and by the RFBR grant 03-04-49131. Funding to pay the Open Access publication charges for this article was provided by CNRS Departement des sciences de la vie.
Conflict of interest statement. None declared.
REFERENCES
- 1.Valentin-Hansen P., Eriksen M., Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 2.Vassilieva I.M., Garber M.B. The regulatory role of the HFq protein in bacterial cells. Mol. Biol. 2002;36:785–791. [PubMed] [Google Scholar]
- 3.Nogueira T., Springer M. Post-transcriptional control by global regulators of gene expression in bacteria. Curr. Opin. Microbiol. 2000;3:154–158. doi: 10.1016/s1369-5274(00)00068-0. [DOI] [PubMed] [Google Scholar]
- 4.Storz G., Opdyke J.A., Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Muffler A., Traulsen D.D., Fischer D., Lange R., Hengge-Aronis R. The RNA-binding protein HF-1 plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 1997;179:297–300. doi: 10.1128/jb.179.1.297-300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takada A., Wachi M., Nagai K. Negative regulatory role of the Escherichia coli hfq gene in cell division. Biochem. Biophys. Res. Com. 1999;266:579–583. doi: 10.1006/bbrc.1999.1863. [DOI] [PubMed] [Google Scholar]
- 7.Tsui H.-C.T., Leung H.-C.E., Winkler M.E. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 8.Wachi M., Takada A., Nagai K. Overproduction of the outer-membrane proteins FepA and FhuE responsible for iron transport in Escherichia coli hfq::cat mutant. Biochem. Biophys. Res. Com. 1999;264:525–529. doi: 10.1006/bbrc.1999.1537. [DOI] [PubMed] [Google Scholar]
- 9.Mohanty B.K., Maples V.F., Kushner S.R. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 10.Le Derout J., Folichon M., Briani F., Dehò G., Régnier P., Hajnsdorf E. Hfq affects the length and the frequency of short oligo(A) tails at the 3′ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res. 2003;31:4017–4023. doi: 10.1093/nar/gkg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajnsdorf E., Régnier P. Host factor HFq of Escherichia coli stimulates elongation of poly(A) tails by poly(A)polymerase I. Proc. Natl Acad. Sci. USA. 2000;97:1501–1505. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vytvytska O., Jakobsen J.S., Balcunate G., Andersen J.S., Baccarini M., von Gabain A. Host-factor I, Hfq, binds to Escherichia coli ompA mRNA in a growth rate-dependent fashion and regulates its stability. Proc. Natl Acad. Sci. USA. 1998;95:14118–14123. doi: 10.1073/pnas.95.24.14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massé E., Escorcia F.E., Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsui H-C.T., Feng G., Winkler M.E. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 1997;179:7476–7487. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sledjeski D.D., Whitman C., Zhang A. HFq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang A., Wassarman K.M., Rosenow C., Tjaden B.C., Storz G., Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 17.Folichon M., Arluison V., Pellegrini O., Huntzinger E., Regnier P., Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moll I., Afonyushkin T., Vytvytska O., Kaberdin V.R., Blasi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morita T., Maki K., Aiba H. RNase E based-ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geissmann T.A., Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. Embo J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vytvytska O., Moll I., Kaberdin V.R., von Gabain A., Bläsi U. HFq (HFI) stimulates ompA mRNA decay by interfering with ribosomes binding. Genes Dev. 2000;14:1109–1118. [PMC free article] [PubMed] [Google Scholar]
- 22.Wassarman K.M., Repoila F., Rosenow C., Storz G., Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang A., Wassarman K.M., Ortega J., Steven A.C., Storz G. The Sm-like HFq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 24.Moller T., Franch T., Hojrup P., Keene D., Bächinger H.P., Brennan R.G., Valentin-Hansen P. HFq: a bacterial Sm-like protein that mediates RNA–RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 25.Massé E., Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Udekwu K.I., Darfeuille F., Vogel J., Reimegard J., Holmqvist E., Wagner E.G.H. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanderpool C.K., Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 28.Azam T.A., Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. J. Biol. Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 29.Kajitani M., Ishihama A. Identification and sequence determination of the host factor gene for bacteriophage Qβ. Nucleic Acids Res. 1991;9:1063–1066. doi: 10.1093/nar/19.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sukhodolets M.V., Garges S. Interaction of Escherichia coli RNA Polymerase with the Ribosomal Protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42:8022–8034. doi: 10.1021/bi020638i. [DOI] [PubMed] [Google Scholar]
- 31.Senear A.W., Steitz J.A. Site-specific interaction of Qβ host factor and ribosomal protein S1 with Qβ and R17 bacteriophage RNAs. J. Biol. Chem. 1976;251:1902–1912. [PubMed] [Google Scholar]
- 32.Arluison V., Folichon M., Marco S., Derreumaux P., Pellegrini O., Seguin J., Hajnsdorf E., Regnier P. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur. J. Biochem. 2004;271:1258–1265. doi: 10.1111/j.1432-1033.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 33.Sonnleitner E., Moll I., Bläsi U. Functional replacement of the Escherichia coli hfq gene by the homologue of Pseudomonas aeruginosa. Microbiology. 2002;148:883–891. doi: 10.1099/00221287-148-3-883. [DOI] [PubMed] [Google Scholar]
- 34.Arluison V., Derreumaux P., Allemand F., Folichon M., Hajnsdorf E., Régnier P. Structural modelling of the Sm-like protein Hfq from Escherichia coli. J. Mol. Biol. 2002;320:705–712. doi: 10.1016/s0022-2836(02)00548-x. [DOI] [PubMed] [Google Scholar]
- 35.Schumacher M.A., Rearson R.F., Moller T., Valentin-Hansen P., Brennan R.G. Structures of the pleiotropic translational regulator HFq and an HFq-RNA complex: a bacterial Sm-like protein. Embo J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauter C., Basquin J., Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaminski P.A., Desnoues N., Elmerich C. The expression of nifA in Azorhizobium caulinodens requires a gene product homologous to Escherichia coli HF-1, an RNA-binding protein involved in the replication of the phage Qβ RNA. Proc. Natl Acad. Sci. USA. 1994;91:4663–4667. doi: 10.1073/pnas.91.11.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urbanowski M.L., Stauffer L.T., Stauffer G.V. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 39.Lerner C.G., Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Link A.J., Philipps D., Church G.M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame charaterization. J. Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boni I.V., Artamonova V.S., Dreyfus M. The last RNA-binding repeat of the Escherichia coli ribosomal protein S1 is specifically involved in autogenous control. J. Bacteriol. 2000;182:5872–5879. doi: 10.1128/jb.182.20.5872-5879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dreyfus M. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J. Mol. Biol. 1988;204:79–94. doi: 10.1016/0022-2836(88)90601-8. [DOI] [PubMed] [Google Scholar]
- 43.Folichon M., Allemand F., Regnier P., Hajnsdorf E. Stimulation of poly(A) synthesis by E. coli poly(A)polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J. 2005;272:454–463. doi: 10.1111/j.1742-4658.2004.04485.x. [DOI] [PubMed] [Google Scholar]
- 44.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 45.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 46.Lange R., Hengge-Aronis R. The cellular concentration of the σS subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 47.Muffler A., Fischer D., Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 48.Brown L., Elliott T. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurum. J. Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsui H.C.T., Feng G., Winkler M.E. Transription of the mutL repair, miaA tRNA modification, hfq pleiotropic regulator, and hflA region protease genes of Escherichia coli K12 from clustered Eσ32-specific promoters during heat-shock. J. Bacteriol. 1996;178:5719–5731. doi: 10.1128/jb.178.19.5719-5731.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hogarth B.G., Higgins C.F. Genetic organization of the oligopeptide permease (opp) locus of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 1983;153:1548–1551. doi: 10.1128/jb.153.3.1548-1551.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrews J.C., Short S.A. Genetic analysis of Escherichia coli oligopeptide transport mutants. J. Bacteriol. 1985;161:484–492. doi: 10.1128/jb.161.2.484-492.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kashiwagi K., Tsuhako M.H., Sakata K., Saisho T., Igarashi A., da Costa S.O., Igarashi K. Relationship between spontaneous aminoglycoside resistance in Escherichia coli and a decrease in oligopeptide binding protein. J. Bacteriol. 1998;180:5484–5488. doi: 10.1128/jb.180.20.5484-5488.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 54.Mikulecky P.J., Kaw M.K., Brescia C.C., Takach J.C., Sledjeski D.D., Feig A.L. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nature Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Törö I., Thore S., Mayer C., Basquin J., Séraphin B., Suck D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archael Sm protein complex. Embo J. 2001;20:2293–2303. doi: 10.1093/emboj/20.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thore S., Mayer C., Sauter C., Weeks S., Suck D. Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA: common features of RNA-binding in Archae and Eukarya. J. Biol. Chem. 2003;278:1239–1247. doi: 10.1074/jbc.M207685200. [DOI] [PubMed] [Google Scholar]
- 57.Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 58.Vecerek B., Moll I., Blasi U. Translational autocontrol of the Escherichia coli hfq RNA chaperone gene. RNA. 2005;11:976–984. doi: 10.1261/rna.2360205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Higgins C.F., Hardie M.M. Periplasmic protein associated with the oligopeptide permeases of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 1983;155:1434–1438. doi: 10.1128/jb.155.3.1434-1438.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.