Abstract
Circular dichroism (CD) and UV-melting experiments were conducted with 16 oligodeoxynucleotides modified by the carcinogen 2-aminofluorene, whose sequence around the lesion was varied systematically [d(CTTCTNG[AF]NCCTC), N = G, A, C, T], to gain insight into the factors that determine the equilibrium between base-displaced stacked (S) and external B-type (B) duplex conformers. Differing stabilities among the duplexes can be attributed to different populations of S and B conformers. The AF modification always resulted in sequence-dependent thermal (Tm) and thermodynamic (−ΔG°) destabilization. The population of B-type conformers derived from eight selected duplexes (i.e. -AG*N- and -CG*N-) was inversely proportional to the −ΔG° and Tm values, which highlights the importance of carcinogen/base stacking in duplex stabilization even in the face of disrupted Watson–Crick base pairing in S-conformation. CD studies showed that the extent of the adduct-induced negative ellipticities in the 290–350 nm range is correlated linearly with −ΔG° and Tm, but inversely with the population of B-type conformations. Taken together, these results revealed a unique interplay between the extent of carcinogenic interaction with neighboring base pairs and the thermodynamic properties of the AF-modified duplexes. The sequence-dependent S/B heterogeneities have important implications in understanding how arylamine–DNA adducts are recognized in nucleotide excision repair.
INTRODUCTION
Arylamines and their derivatives are an important class of carcinogens (1). N-acetyl-2-aminofluorene is a strong liver carcinogen that has been studied extensively as a prototype arylamine carcinogen. Upon activation in vivo, it reacts with cellular DNA to produce two C8-substituted acetylaminofluorene (AAF) and aminofluorene (AF) adducts (Figure 1), along with a small amount of N2-substituted adduct (2). The two major adducts are rapidly excised by nucleotide excision repair (NER), whereas the minor dG-N2 adduct persists in mammalian cells (3–5). Translesion synthesis of AF-adducts is achieved with high-fidelity polymerases, whereas replication of AAF-adducts requires specialized bypass polymerases (6). An X-ray study by Hsu et al. (7) has shown that accurate nucleotide incorporation is feasible opposite the AF-lesion in the active site of a large DNA polymerase I Bacillus fragment, but not opposite the AAF-adduct which blocks nucleotide incorporation. Although incorporation opposite AF occurs, the translesion synthesis is slowed due to distortions in the polymerase active site induced by the C:G[AF] base pair. The crystal structures of the AAF and AF-adducts complexed with T7 DNA polymerase yielded similar findings (8). Specifically, they observed weak binding of the AF-adduct to the polymerase and, interestingly, found evidence suggesting that there may be syn–anti glycosidyl conformational heterogeneity within the active site.
Figure 1.
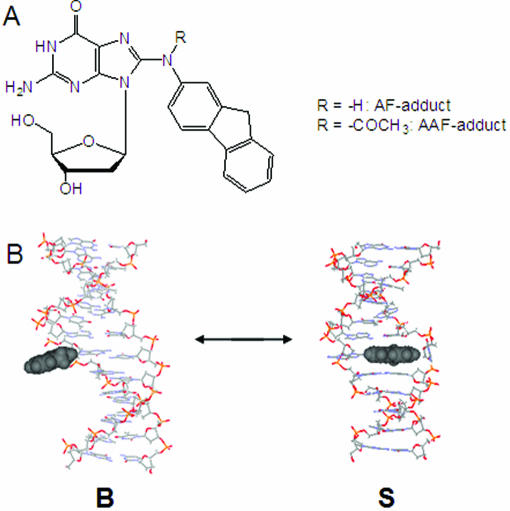
(A) Chemical structures of AF-adduct (N-[deoxyguanosin-8-yl]-2-aminofluorene) and AAF-adduct (N-[deoxyguanosin-8-yl]-2-acetylaminofluorene). (B) Two prototype AF-induced conformations: an external binding B-type (B) conformer and a base-displaced stacked (S) conformer. The hydrophobic AF moiety is shown in CPK. The modified dG in the B-conformer maintains a Watson–Crick base pairing with the AF moiety protruding in the major groove, whereas the S-conformer adopts a base-displacement structure with the AF ring inserted into the helix. The duplex structures of sequence 5 were constructed with the literature protocols (16) and shown here for illustration purpose only.
Distinctly different mutagenic and repair properties of AAF and AF adducts have been observed. In bacteria, AAF yields primarily frameshift mutations, whereas the deacetylated AF produces a mixture of frameshift and point mutations (1). In mammalian cells, however, point mutations are predominant for both adducts, although AAF is associated primarily with deletion mutations in certain sequences (9,10). Zou et al. (11) have shown that the Escherichia coli NER process takes place in a sequential two-step mechanism, i.e. disruption of the Watson–Crick DNA structure is recognized initially, while the type of modification of the nucleotide is important in a subsequent step upon a strand opening. Both the AAF and AF adducts were equally incised when placed in a single strand, indicating they were equally recognized in the second step and discriminated only during the initial recognition. The mutational and repair efficiencies of these adducts depend strongly on the identities of the neighboring bases surrounding the lesion site (9,12). Such sequence effects are likely to be related to adduct-induced conformational heterogeneities in DNA as well as the unique structural features of specific proteins (3,13). Bulky DNA adducts often adopt multiple conformations, which can interconvert mutagenic and non-mutagenic within the active site of replication or repair enzymes (14,15). Duplexes containing AF-adducts exist principally as a mixture of two structurally distinct conformations: an ‘external binding B-type (B)’ conformer and a ‘base-displaced stacked (S)’ conformer (Figure 1) depending on the nature of (anti and syn, respectively) the glycosidyl conformation of the modified dG (15,16). The sequence dependence of the AF-induced S/B-conformational heterogeneity has been well documented for various base sequences that include duplexes, template/primer junction and bulges (15,16).
The structure of duplex DNA influences repair mechanisms. Elucidating the structural and conformation characteristics of carcinogen-modified DNA duplexes is a crucial step towards understanding the mechanisms of DNA repair (3,11,12,17,18). We therefore initiated a systematic study to investigate the sequence-dependent conformational heterogeneity in AF-mutagenesis. In this paper, a complete set of 16 AF-modified 12mer oligodeoxynucleotides (ODNs), in which the lesion was flanked systematically by all possible trimer sequence combinations [d(CTTCTNG*NCCTC), G* = AF adduct, N = G, A, C, T, Table 1], was prepared and then annealed with the appropriate sequences to produce fully complementary heteroduplexes for circular dichroism (CD) and UV-melting experiments. In addition, we obtained 19F NMR spectra of eight selected duplexes (i.e. the -AG*N- and -CG*N- series, sequences 5–12) modified by the fluorine probe 7-fluoro-2-aminofluorene (19,20) in order to determine their S/B population ratios. The systematic sequence variation around the AF-modified guanine enabled us to probe how flanking sequences influence the nature of the base pair interactions at or near the adduct site, and ultimately the S/B-conformational heterogeneities. To the best of our knowledge, this is the first thermodynamic and spectroscopic study on the sequence-dependent conformational heterogeneity of AF-modified oligonucleotides.
Table 1.
The effects of AF modification on the thermal and thermodynamic stability of DNA duplexes 5′-CTTCTNG*NCCTC-3′G* = AF-adduct; N = dG, dA, dT, dC 3′-GAAGANC NGGAG-5′
| Number | Sequencea | −ΔG°b (kcal/mol) | −ΔH°b (kcal/mol) | Tmc (°C) | ΔΔG°d (kcal/mol) | ΔΔH°e (kcal/mol) | ΔTmf (°C) | CD−/+g | % S-conformerh |
|---|---|---|---|---|---|---|---|---|---|
| 1 | -GG*G- | 10.7 (13.7)i | 81.1 (95.4)i | 54.0 (62.3)i | 3.0 | 14.2 | −8.3 | 0.24 | |
| 2 | -GG*A- | 9.4 (13.0) | 74.4 (97.1) | 49.4 (59.1) | 3.6 | 22.8 | −9.7 | 0.33 | |
| 3 | -GG*C- | 9.9 (13.6) | 68.3 (90.9) | 52.8 (63.2) | 3.7 | 22.6 | −10.4 | 0.24 | |
| 4 | -GG*T- | 9.3 (12.0) | 73.2 (86.4) | 49.2 (56.3) | 2.7 | 13.2 | −7.2 | 0.58 | |
| 5 | -AG*G- | 9.9 (12.7) | 84.5 (101.0) | 49.9 (56.2) | 2.8 | 16.5 | −6.3 | 0.24 | 32 |
| 6 | -AG*A- | 8.8 (11.1) | 79.7 (87.8) | 46.1 (54.2) | 2.3 | 8.1 | −8.2 | 0.28 | 39 |
| 7 | -AG*C- | 8.4 (11.0) | 67.8 (74.4) | 45.9 (56.6) | 2.6 | 6.6 | −10.7 | 0.35 | 60 |
| 8 | -AG*T- | 8.1 (10.2) | 66.5 (76.8) | 44.3 (52.8) | 2.1 | 10.3 | −8.5 | 0.44 | 64 |
| 9 | -CG*G- | 10.7 (13.5) | 80.4 (87.8) | 52.6 (63.5) | 2.8 | 7.4 | −10.9 | 0.12 | 36 |
| 10 | -CG*A- | 9.1 (11.7) | 68.0 (77.7) | 49.0 (59.8) | 2.6 | 9.7 | −10.8 | 0.19 | 44 |
| 11 | -CG*C- | 9.6 (12.7) | 70.4 (74.2) | 51.2 (64.4) | 3.1 | 3.8 | −13.2 | 0.23 | 47 |
| 12 | -CG*T- | 8.8 (11.7) | 71.6 (76.0) | 47.5 (59.6) | 2.9 | 4.4 | −12.1 | 0.25 | 63 |
| 13 | -TG*G- | 7.3 (10.4) | 63.7 (75.4) | 41.9 (53.6) | 3.1 | 11.7 | −11.7 | 0.58 | |
| 14 | -TG*A- | 8.8 (11.5) | 70.7 (89.6) | 47.2 (55.8) | 2.7 | 18.9 | −8.6 | 0.41 | |
| 15 | -TG*C- | 8.9 (11.7) | 69.5 (72.2) | 49.0 (59.8) | 2.8 | 2.7 | −10.8 | 0.38 | |
| 16 | -TG*T- | 6.8 (7.9) | 47.0 (61.9) | 39.1 (43.9) | 1.1 | 14.9 | −4.8 | 0.56 |
aThe central trimer portion of the 12mer duplex (G* = AF-adduct, Figure 1).
bThe results of curve fit and Tm − lnCt dependence were within ±15% of each other and therefore these numbers are average of the two methods. The average SDs for −ΔG° and −ΔH° are ±0.22 and ±6.33, respectively.
cTm values at 14 µM taken from the 1/Tm− lnCt/4 Meltwin plots.
dΔΔG = ΔG° (AF-modified duplex) − ΔG° (control duplex).
eΔΔH = ΔH° (AF-modified duplex) − ΔH° (control duplex).
fΔTm= Tm (AF-modified duplex) − Tm (control duplex).
gRatio of the negative over the positive areas in the 250–360 nm region (Figure 3C).
hPercent S-conformer population determined by 19F NMR measurements of duplexes 5–12 modified by the fluorine reporter probe ‘7-fluoro-2-aminfluorene’ (Results).
iData in parentheses are from unmodified control duplexes.
MATERIALS AND METHODS
ODNs were purchased in desalted forms from Sigma-Genosys (The Woodlands, TX). Chemical reagents and enzymes such as DNase I, phosphodiesterase I and alkaline phosphatase were all obtained from Sigma Chemical (Milwaukee, WI). Both 3′- and 5′-phosphodiesterases used in the mass spectrometry digestion experiments were purchased from Worthington Biochemical (Lakewood, NJ). All high-performance liquid chromatography (HPLC) solvents were purchased from Fisher (Pittsburgh, PA).
All AF-modified 12mer ODNs were prepared and characterized according to the literature procedures (21,22). The general base sequence employed in this study (Table 1) was a truncated portion of a mouse ras protoononcogene, which has been used in our earlier 1H (23,24) and 19F NMR (19,20) studies. The synthesis, HPLC purification and enzyme-digest/LC/MS characterization of the modified ODNs are detailed in Supplementary Figures S1–S4.
Optical melting experiments
UV-melting curves were measured using a Beckman DU 800 UV/VIS spectrophotometer equipped with a 6-chamber, 1 cm path length Tm cell. Sample cell temperatures were controlled by a built-in Peltier temperature controller. Duplex solutions with a total concentration in the range of 0.2–14 µM were prepared in solutions containing 0.2 M NaCl, 10 mM sodium phosphate and 0.2 mM EDTA at pH 7.0. The concentration of each ODN sample was estimated based on UV absorbance at a wavelength of 260 nm (25). Melting curves were constructed by varying the temperature of the sample cell (1°C/min) and monitoring the absorbance of the sample at 260 nm. A typical melting experiment consisted of two forward and two reverse scans and was repeated three times.
Thermodynamic parameters for bimolecular melting reactions of the duplexes were obtained from melting curve data using the program MELTWIN® version 3.5 (25). Two different methods (25–27) were used to calculate the enthalpy (ΔH°) and entropy (ΔS°) of the helix-coil equilibrium: (i) fits of individual curves and (ii) van't Hoff plots of versus lnCt to fit to = R/ΔH° lnCt/4 + ΔS°/ΔH° (Supplementary Figure S5), in which Tm is a melting transition point in K, Ct is a total duplex concentration, and R is the universal gas constant (1.987 cal/K-mol); ΔG° = ΔH° − TΔS° (25,27). Thermodynamic data derived from van't Hoff plots have been shown to be more reliable than those from fits of curves for non-two-state transitions involving DNA hairpins or slipped duplexes (27). The margins of the parameters derived from the curve fit data and from − lnCt plots in the present study were found to be within ±15%. This close agreement indicates the presence of strictly bimolecular transitions, in which case the data obtained from the two methods are equally reliable (25,28). Table 1 lists the average values of the energetics of thermal transition, enthalpy and free energy change for the duplexes in this study.
Circular dichroism experiments
CD measurements were conducted on a Jasco J-810 spectropolarimeter equipped with a variable Peltier temperature controller. All CD experiments were conducted at 15°C. Typically, 2 ods of a duplex sample were dissolved in 400 µl of a buffer containing 0.2 M NaCl, 10 mM sodium phosphate, 0.2 mM EDTA, pH 7.0, and placed in a 1 mm path length cell. The sample was heated at 85°C for 5 min and then cooled to 15°C over a 10 min period. The spectropolarimeter was scanned from 200 to 400 nm at a rate of 50 nm/min. Data points were acquired every 0.2 nm with a 2 s response time. Spectra were the averages of 10 accumulations and smoothed using a 17 point adaptive smoothing algorithm.
19F NMR experiments
According to the reported procedure (19), preliminary 19F NMR measurements were conducted for eight selected duplexes (sequences 5–12) modified by the ‘fluorine reporter probe’, 7-fluoro-2-aminofluorene. The modified ODNs were purified and characterized similarly as described for the non-fluorinated AF-duplexes (Supplementary Data). Approximately 80–100 ods of pure modified ODNs were annealed with appropriate sequences to form fully complementary heteroduplexes and centrifuged through a Millipore Centricon YM-3 centrifugal filter (Yellow, molecular weight cutoff = 3000). The filtered samples were dissolved in a pH 7.0 buffer (10% D2O/90% H2O) containing 100 mM NaCl, 10 mM sodium phosphate and 100 µM tetrasodium EDTA. Proton-decoupled 19F NMR spectra were obtained using a dedicated 19F/1H dual probe on a Bruker DPX400 Avance spectrometer operating at 376.5 MHz and referenced to CFCl3 by assigning external hexafluorobenzene in C6D6 at −164.90 p.p.m. Spectra exhibited two prominent signals with varying intensities in the −117 to −120 p.p.m. region. Signal assignments have been made by analyses of dynamic NOESY/exchange and H/D solvent isotope data using the protocols described previously (19). The S/B population ratios were obtained by area integration at 20°C and the results are shown in Table 1. Full details of 19F NMR work including dynamic exchange experiments with complete line shape analyses will be published in the near future.
RESULTS
Enzyme-LC/MS characterization
The electrospray-time-of-flight mass spectrometry analysis of each AF-modified ODN was carried out to determine the number and the positions of the AF modifications. Molecular ion spectra did not show any singly charged molecular ions, but rather exhibited the expected multiple charged ions upon electrospray with triply charged species as base peaks. The results of molecular ion spectra of all 16 AF-modified sequences are summarized in Supplementary Table S1.
Each of the ODNs that contained more than one guanine (i.e. sequences 1–5, 9 and 13) was sequenced using the exonuclease strategies similar to those described previously for identifying the position of bases modified by arylamines (29–31). The exonucleases hydrolyze the phosphodiester bonds of the terminal nucleotides sequentially from either the 5′ end (5′–3′ exonuclease) or 3′ end (3′–5′ exonuclease) to give 3′- and 5′-monodeoxynucleotides, respectively. The digestion rate with either exonuclease slowed significantly when a modified guanine base is exposed at the terminus. The details of enzyme-digest/LC/MS characterization are described in Supplementary Data using sequence 13 as an example.
Thermomelting studies
Unmodified control duplexes exhibited melting profiles that were characteristic of a monophasic, sigmoidal, helix-coil transition. Despite the demonstrated conformational heterogeneity, a strong linear association (R2 > 0.974) between and lnCt was observed for all 16 AF-modified duplexes over the 0.2–14 µM concentration range (Supplementary Figure S5). In the present study, the contribution of changes in heat capacity (ΔCp = dΔH/dT) was assumed to be negligible. This assumption is valid because, while ΔCp is considered to be an important factor in analysis of duplex folding, the error associated with neglecting it is important only at lower temperatures (32).
Tm is defined as a midpoint of sigmoidal melting curves. ΔTm is the difference in Tm values between the modified and the control duplex and this shift represents the adduct-induced thermal destabilization. As shown in Table 1, AF modification consistently lowers Tm. Tm in both control and modified sequences increases with increasing % G:C content at the sites flanking the lesion (12). In this study, the Tm values of those sequences that had only A:T flanking base pairs (sequences 6, 8, 14 and 16) ranged from 43.9 to 55.8°C for the control and from 39.1 to 47.2°C for the modified duplexes. An unusually lower Tm value (39.1°C) was obtained for AF-modified sequence 16 (-TG*T-), in which the lesion was flanked by two T:A base pairs. This exceptionally low Tm value differed markedly from the moderately high Tm value of 46.1°C observed for sequence 6 (-AG*A-). The contrast between the Tm values of sequences 16 and 6 illustrates the great impact that base polarity, e.g. A:T versus T:A, can have on thermal stability. The Tm values for sequences with G:C base pairs on both sides of the lesion (sequences 1, 3, 9 and 11) were in the range of 62.3–64.4°C and 51.2–54.0°C for the control and modified duplexes, respectively. The narrow ranges of these values indicate that the base polarity effect for AF-lesions flanked by G:C base pairs is relatively minor. For example, the Tm difference between AF-modified sequences 1 (-GG*G-) and 11 (-CG*C-) was only 2.8°C. The remaining sequences, which contain one A:T and one G:C base pair, exhibited intermediate Tm ranges: 53.3–59.8°C and 41.9–54.0°C, for the control and modified duplexes, respectively.
The effects of 5′- and 3′-flanking bases on Tm and −ΔG° were examined and are tabulated in Supplementary Tables S2 and S3. The thermostability of AF-modified duplexes always increased when a 5′ lesion-flanking A was replaced by a 5′ flanking G (by 3.3–6.9°C) and when a 5′ lesion-flanking T was replaced by a 5′ flanking C (by 1.8–10.7°C). The effect of the 5′-T to 5′-C transition was especially pronounced when the lesion was flanked with a G or T (sequence 16→12, sequence 13→9). The 3′-effect was essentially similar except for a transition from -TG*A- (sequence 14) to -TG*G- (sequence 13), which increased Tm significantly (5.3°C). The temperature shift ranges of Tm were 3.6–4.6°C and 1.6–3.7°C for the 3′-A→G and 3′-T→C transitions, respectively. Again, the greatest effect (9.9°C) involved sequence 16 [16→15 (-TG*T- to -TG*C-)]. Note that similar 5′- and 3′-trends were observed for the unmodified duplexes.
As in the case of Tm, −ΔG° of the AF-modified duplexes increased with increasing % G:C content at the sites flanking the adduct (Table 1). The −ΔG° values (6.8–8.8 kcal/mol) observed for AF-modified sequences (6, 8, 14 and 16) with only A:T flanking base pairs were lower than those with G:C flanking base pairs only (sequences 1, 3, 9 and 11; 9.6–10.7 kcal/mol). The effect of base polarity change, however, was significant for those with A:T base pairs. For example, −ΔG° decreased significantly going from -AG*A- (8.8 kcal/mol; sequence 6) to -TG*T- (6.8 kcal/mol; sequence 16). The remaining sequences, which contained one A:T and one G:C base pair, exhibited a borderline −ΔG° (7.3–9.9 kcal/mol). As in the case of Tm, the thermodynamic stabilities for the modified duplexes were always greater with a 5′-G than with a 5′-A (by 0.6–1.5 kcal/mol) and with a 5′-dC than with a 5′-dT (by 0.7–3.4 kcal/mol). Similar differences (0.3–2.1 kcal/mol) were observed when examining the effect of the 3′ flanking base. An exception was again the transition from sequence 14→13 in which a 1.5 kcal/mol −ΔG° increase was found.
The consistent decrease in −ΔG° that arose from AF modification is indicative of a decrease in enthalpy, which implies that there is a loss of base stacking and pairing near the lesion site. The differences in transition free energy (ΔΔG°) for the AF-modified duplexes were significantly smaller than the observed differences in transition enthalpy change (ΔΔH°). This may be due to enthalpy–entropy compensation effects, which have been found to be a commonly occurring phenomenon in modified DNA duplexes (33,34). As expected, an excellent linear correlation (R2 = 0.884) was obtained between −ΔG° and Tm (Supplementary Figure S6); however, the ΔΔG° values did not seem to follow the same trend with Tm (data not shown), suggesting that the identities of the bases flanking the lesion influence ΔΔG° as well (35,36).
Circular dichroism spectra
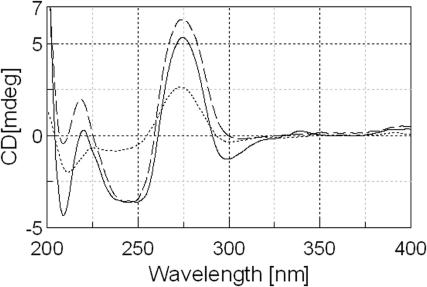
The CD spectra of all the duplexes used in this study were measured under low salt conditions (0.2 M NaCl, Supplementary Figure S7). CD spectra of unmodified and AF-modified sequence 5 (-AG*G-) are shown in Figure 2 as an example. The AF-modified single strand exhibited a positive CD band centered at ∼275 nm and weak negative bands below 250 nm. These CD bands became well organized upon duplex formation and exhibited unique adduct-induced negative ellipticities in the ∼290–365 nm range (see below). The results are compared with that of the unmodified control duplex and indicate that both the control and the AF-modified duplexes adopt an overall B-DNA conformation in solution (33,34,37).
Figure 2.
CD spectral overlays of sequence 5 at 15°C; single-stranded AF-modified sequence (dotted line), unmodified duplex (dashed line), and AF-modified duplex (solid line).
Sequence-dependent CD bands in the 290–365 nm range (CD−/+)
The aforementioned adduct-induced CD bands were present only in the modified duplexes and disappeared as the temperature was raised (Supplementary Figure S8). This phenomenon has been observed previously for AF-modified DNA in limited sequence settings at low to medium NaCl concentrations (22,38–41). The single strand, AF-modified ODNs have broad electronic absorptions in 300–350 nm (Supplementary Figure S1A, inset); therefore, the induced CD must arise from the interaction between the non-chiral AF moiety and the chiral DNA (22).
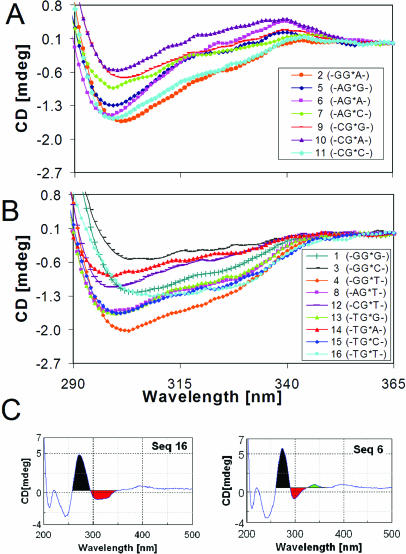
Figure 3 shows overlays of the normalized AF-induced CD bands generated by all of the 16 AF-modified duplexes in the 290–365 nm range. They are uniquely sequence-dependent and can be grouped into two general categories depending on their CD patterns: (i) sequences 2, 5–7, 9–11, which exhibited a negative dip in ∼290–330 nm with varying intensities of positive bump centered around ∼340 nm, forming an ‘S-curved CD’ or (ii) sequences 1,3,4, 8, 12–16, which exhibited broad negative ‘bucket-shaped CD’ in ∼290–350 nm. To assess this interesting phenomenon quantitatively, an arbitrary ratio of the negative area (trough) over the positive area centered ∼275 nm in the range of 260–365 nm (designated herein as CD+/−) was determined to see how this ratio might correlate with the S/B-conformer ratio. This was illustrated in Figure 3C for sequences 16 and 6, which represent typical ‘bucket-shaped’ and ‘S-curved’ CDs, respectively. The negative and the positive area are color-coded as red and black, respectively, for clarity. The plateau at 365 nm was used as a baseline reference point. The small positive bumps around ∼340 nm in the ‘S-curved’ sequences (i.e. green-colored area of sequence 6 in Figure 3C) were ignored in the calculation. The resulting CD−/+ data are tabulated in Table 1. It was noted that the averages CD−/+ value (∼0.41) for ‘bucket-shaped CD’ sequences are much greater than that (∼0.25) from the ‘S-curved’ ones. Also noted was that the series with 5′-T (sequences 13–16) all exhibited ‘bucket-shaped CDs’ (Figure 3B) with consistently higher CD−/+ values compared with those with G, A and C as the 5′-flanking bases.
Figure 3.
CD spectral overlays of AF-modified duplexes in the 290–365 nm region at 15°C for (A) ‘S-curved’ and (B) ‘bucket-shaped’ sequences. (C) Calculation of CD−/+ (Results).
19F NMR-based S/B ratios for the -AG*N- and the -CG*N- sequences
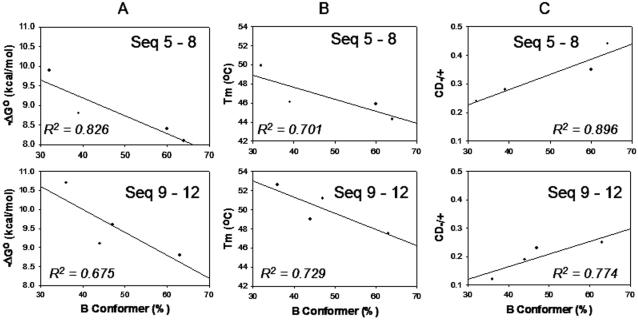
The S/B population ratios of the four different 3′flanking bases have been studied with either A (sequences 5–8) or C (sequences 9–12) as the 5′-flanking bases. The results were plotted against −ΔG° (Figure 4A) and Tm (Figure 4B). In each case, the % B-conformation exhibited inverse correlations. We observed linear correlations between % B-conformation and the CD−/+ values (R2 = 0.896 and 0.774 for sequences 5–8 and 9–12, respectively, Figure 4C). It was noted that both the % B and CD−/+ values increased in the order of 3′-T > C > A > G regardless of the nature of the 5′-flanking bases (A or C).
Figure 4.
Correlations of % B-conformation with (A) −ΔG°, (B) Tm and (C) CD−/+ for eight selected AF-modified duplexes (sequences 5–12).
DISCUSSION
The precise mechanisms by which carcinogen–DNA motifs are recognized by repair or replication proteins are of immense interest from the standpoint of the structure–function relationships. A key outstanding question is to define how base sequence context governs equilibria between different conformations in carcinogen–DNA adducts. Is a given sequence context better repaired than another? Is one sequence context more prone to miscode during replication than another? Are there unique conformers that are more or less repair susceptible, and/or more likely to produce mutation on replication? Answers to these questions are directly related to our understanding of mutational hotspots for carcinogen damage. The AF-system is uniquely well placed to address these questions because of its known sequence-dependent conformational heterogeneity (15,16). In the present study, we conducted a systematic thermomelting and CD investigation to probe the sequence effects on the AF-induced conformational heterogeneity.
Thermomelting data
In accordance with observations made employing other bulky lesion-modified DNA duplexes including mismatches and bulges (12,16–18,25), we found that AF modification consistently lowered thermal- (Tm) and thermodynamic (−ΔG°) stabilities. DNA destabilization arises from decreased enthalpy, which can be produced by a loss of base stacking and pairing near the lesion site (12,18). The differences in the thermal (ΔTm) and thermodynamic (ΔΔG°, ΔΔH°) stabilities represent the direct effect of AF modification on the corresponding unmodified duplex. The stabilities of both the control and the modified duplexes were found to be sensitive to the specific nature of the flanking bases. In general, duplex stability was enhanced by purine bases (G, A) flanking the lesion site. Specifically, Tm and −ΔG° of both the unmodified and the modified duplexes were lower with A and T adjacent to the lesion site, than with G and C, in both 5′- and 3′-directions (Supplementary Tables S2 and S3). The effect of base polarity was pronounced, especially for those flanked with A:T base pairs. For example, −ΔG° increased by 2 kcal/mol when T:A base pairs in sequence 16 were replaced by A:T base pairs in sequence 6. A smaller increase (1.1 kcal/mol) was observed when G:C base pairs were replaced by C:G base pairs (i.e. sequence 11 to 1). An excellent correlation (R2 = 0.884) between −ΔG° and Tm was obtained for all the unmodified and modified duplexes indicates that the AF-modified duplexes follow a typical pattern of bulky adduct-induced destabilization of the secondary structure of DNA at or near the adduct site due to the loss of base stacking and base pairing.
An important finding from current study is the inverse correlative relationship observed between Tm/−ΔG° and % B-conformer population of sequences 5–8 (Figure 4A and B). The Tm and −ΔG° values observed are the consequence of relative contributions from the B- and S-conformer components of the AF-duplex populations. While adduct formation is generally known to induce thermal and thermodynamic destabilization (18), it is not clear which conformer, S or B, actually contributes more to destabilization. This question is critical as DNA conformation is a major determining factor for translesion synthesis and repair susceptibility (3,15–17). The lack of Watson–Crick base pairing at the lesion site in the S-conformer suggests that it may be the S-conformer. However, it has been shown that deletion duplexes containing bulky carcinogens (AAF and BP-N2-dG adducts) predominantly adopt the S-type conformation, yet exhibit unusually higher Tm values (16,17). Clearly, the stacking interaction between the carcinogenic moiety and flanking DNA bases is a driving force for such stabilization. Although the modified dG of B-conformer is hydrogen-bonded to the dC at the modification site, it is possible that the highly hydrophobic AF moiety in the major groove may have an unfavorable solvation, destabilizing the local DNA structure thereby weakening Watson–Crick pairing. Although the available S/B population data are limited (Table 1), the results shown in Figure 4 suggest strongly that the B-conformer is responsible for the AF-induced thermal and thermodynamic destabilization of the duplex.
Circular dichroism data
The CD of DNA is sensitive to a variety of structural disturbances such as the distortion of Watson–Crick base pairs, adduct formation, and the presence of bulges and mismatches (12,25,42–48). All of the AF-modified duplexes studied here exhibited a characteristic CD pattern in the 210–300 nm range with the positive band at ∼275 nm being the most intense, which is typical for a global B-form DNA (Figure 3 and Supplementary Figure S7) (37).
AF-modified duplexes exhibited sequence-dependent adduct-induced negative CD bands in the 290–365 nm range (defined as CD−/+) which arise from the interaction between AF and the chiral DNA. The extent (CD−/+) of the induced ellipticities was found to be directly correlated with % B-conformation, at least for duplexes 5–12 (Figure 4C). The physical and spectroscopic basis underlying the significance of CD−/+ is not clear. The tall positive ∼275 nm CD is likely to contain contribution from the DNA itself and the excitonic AF-C8-dG interactions, whereas the induced negative CD reflects some asymmetric aspects of the micro-environment of the AF-chromophore. The two CD bands are probably interactive because of their close proximity. The B-type conformers would maintain a non-random arrangement with a certain degree of mobilization of the AF-chromophore in the major groove. In contrast, the S-conformers are likely to favor a strong interaction between the immobilized AF and the neighboring bases. The situation is somewhat similar to the induced CD observed at around 350 nm for BP-N2-dG-modified adducts. Pradhan et al. (49,50) have shown that the intensity of the induced CD of the pyrene chromophore at 350 nm is modulated by the nature of the adduct structure and the sequence context around the lesion site, ultimately leading to the BP-N2-dG-induced conformational heterogeneity.
The available correlations (Table 1) indicate that the CD−/+ ratio and the S/B ratios, although influenced by the 3′-flanking bases, seem to be less dependent on the 5′-flanking bases except for the -AG*C- and -CG*C- sequences, where the % S-conformation is 60 and 47%, respectively. It should also be noted that the % S-conformer increases in the order of 3′-dG > dA > dC > dT, indicating the importance of the 3′-purine flanking bases for stacked conformation. Taken together, these results suggest a unique interplay between the extent of the AF-interaction with neighboring base pairs and S/B-conformational heterogeneity, i.e. the greater the negative 290–350 nm ellipticity, the greater the % B population, a major contributor to the destabilization of AF-modified duplexes. It should be emphasized, however, that the available NMR-based S/B ratios are limited only to the -AG*N- and CG*N- sequences. The S/B effect of the remaining 5′-flanking bases (T and G) must be obtained in order to establish the general potential of CD−/+ in assessing the AF-induced conformational heterogeneity in other unknown sequences.
Biological implications
Most bulky carcinogenic lesions in duplex DNA are eliminated by NER (3,12,17). The AF-lesion excision efficiency by the E.coli UvrABC system is known to depend on the extent of base pair destabilization and the sequence context around the lesion site (3,11,12). Mekhovich et al. (51) have shown that the rate of incision of AF adducts is significantly faster when it is positioned in the mutation hot spot NarI sequence (5′-GGCG*CC-3′), than when located in a non-NarI sequence (5′-GATG*ATA-3′). According to previous high resolution 1H NMR studies (16), AF-modified duplexes with -CG* C- and -TG*A- sequence contexts have been shown to adopt 30–50% and 60–70% B-conformation, respectively. This is in good agreement with our CD results, which showed that the CD−/+ value for the -TG*A- sequence context was nearly twice that for the -CG*C- context (Table 1). The base composition of the central trimer of these sequences is identical to those of sequence 11 (47% B) in the present work and of sequence 14 (55%) in our previous study (19). While the overall sequence contexts and chain lengths of these sequences differ, the results seem to suggest that the S-conformer has greater repair susceptibility than the B-type conformer. However, Zou et al. (11) have shown that AF-adducts in a -TG*T- sequence context are incised more efficiently than those in a -CG*C- sequence context. The presence of a T:A base pair in the flanking positions relative to the modified dG* may allow for greater local flexibility and conformational heterogeneity than in a -CG*C- sequence. This is consistent with our findings which showed lower Tm (39°C) and −ΔG° (6.8 kcal/mol) values for sequence 16 (-TG*T-, Table 1). NMR data are not yet available for the -TG*T- context; however, its relatively large CD−/+ value (0.56) implies that the sequence may exist primarily in the B-type conformation. These results illustrate the complexity of how multi-conformeric adducts interact with repair proteins, information that is crucial to the first step in damage recognition in NER (17,18,52).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We are grateful to the NIH (R01CA098296) for their financial support for this work. This research was made possible in part by the use of Research Core Facility supported by the NCRR/NIH (P20 RR016457). Funding to pay the Open Access publication charges for this article was provided by the NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Heflich R.H., Neft R.E. Genetic toxicity of 2-acetylaminofluorene, 2-aminofluorene and some of their metabolites and model metabolites. Mutat. Res. 1994;318:73–114. doi: 10.1016/0165-1110(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 2.Beland F.A., Kadlubar F.F. Metabolic activation and DNA adducts of aromatic amines and nitroaromatic hydrocarbons. In: Cooper C.S., Grover P.L., editors. Handbook of Experimental Pharmacology. Heidelberg: Springer-Verlag; 1990. pp. 267–325. [Google Scholar]
- 3.Gillet L.C., Scharer O.D. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006 doi: 10.1021/cr040483f. DOI: 10.1021/cr040483f, http://dx.doi.org/10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 4.Gillet L.C., Alzeer J., Scharer O.D. Site-specific incorporation of N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-AAF) into oligonucleotides using modified ‘ultra-mild’ DNA synthesis. Nucleic Acids Res. 2005;33:1961–1969. doi: 10.1093/nar/gki335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui X.S., Eriksson L.C., Moller L. Formation and persistence of DNA adducts during and after a long-term administration of 2-nitrofluorene. Mutat Res. 1999;442:9–18. doi: 10.1016/s1383-5718(99)00053-4. [DOI] [PubMed] [Google Scholar]
- 6.Pages V., Fuchs R.P. How DNA lesions are turned into mutations within cells? Oncogene. 2002;21:8957–8966. doi: 10.1038/sj.onc.1206006. [DOI] [PubMed] [Google Scholar]
- 7.Hsu G.W., Kiefer J.R., Burnouf D., Becherel O.J., Fuchs R.P., Beese L.S. Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase. J. Biol. Chem. 2004;279:50280–50285. doi: 10.1074/jbc.M409224200. [DOI] [PubMed] [Google Scholar]
- 8.Dutta S., Li Y., Johnson D., Dzantiev L., Richardson C.C., Romano L.J., Ellenberger T. Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis. Proc. Natl Acad. Sci. USA. 2004;101:16186–16191. doi: 10.1073/pnas.0406516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibutani S., Suzuki N., Tan X., Johnson F., Grollman A.P. Influence of flanking sequence context on the mutagenicity of acetylaminofluorene-derived DNA adducts in mammalian cells. Biochemistry. 2001;40:3717–22. doi: 10.1021/bi0027581. [DOI] [PubMed] [Google Scholar]
- 10.Shibutani S., Suzuki N., Grollman A.P. Mutagenic specificity of (acetyl amino) fluorine-derived DNA adducts in mammalian cells. Biochemistry. 1998;37:12034–12041. doi: 10.1021/bi981059+. [DOI] [PubMed] [Google Scholar]
- 11.Zou Y., Shell S.M., Utzat C.D., Luo C., Yang Z., Geacintov N.E., Basu A.K. Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease. Biochemistry. 2003;42:12654–12661. doi: 10.1021/bi034446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singer B., Hang B. Nucleic acid sequence and repair: role of adduct, neighbor bases and enzyme specificity. Carcinogenesis. 2000;21:1071–1078. [PubMed] [Google Scholar]
- 13.Seo K.Y., Jelinsky S.A., Loechler E.L. Factors that influence the mutagenic patterns of DNA adducts from chemical carcinogens. Mutat. Res. 2000;463:215–246. doi: 10.1016/s1383-5742(00)00047-8. [DOI] [PubMed] [Google Scholar]
- 14.Seo K.Y., Nagalingam A., Tiffany M., Loechler E.L. Mutagenesis studies with four stereoisomeric N2-dG benzo[a]pyrene adducts in the identical 5′-CGC sequence used in NMR studies: G→T mutations dominate in each case. Mutagenesis. 2005;20:441–448. doi: 10.1093/mutage/gei061. [DOI] [PubMed] [Google Scholar]
- 15.Cho B.P. Dynamic conformational heterogeneities of carcinogen–DNA adducts and their mutagenic relevance. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2004;22:57–90. doi: 10.1081/LESC-200038217. [DOI] [PubMed] [Google Scholar]
- 16.Patel D.J., Mao B., Gu Z., Hingerty B.E., Gorin A., Basu A.K., Broyde S. Nuclear magnetic resonance solution structures of covalent aromatic amine–DNA adducts and their mutagenic relevance. Chem. Res. Toxicol. 1998;11:391–407. doi: 10.1021/tx9702143. [DOI] [PubMed] [Google Scholar]
- 17.Isaacs R.J., Spielmann H.P. A model for initial DNA lesion recognition by NER and MMR based on local conformational flexibility. DNA Repair. 2004;4:455–464. doi: 10.1016/j.dnarep.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Geacintov N.E., Broyde S., Buterin T., Naegeli H., Wu M., Yan S., Patel D.J. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 19.Zhou L., Rajabjadeh M., Traficante D.D., Cho B.P. Conformational heterogeneity of aryl amine-modified DNA: 19F NMR Evidence. J. Am. Chem. Soc. 1997;119:5384–5389. [Google Scholar]
- 20.Cho B.P., Zhou L. Probing the conformational heterogeneity of the acetylaminofluorene-modified 2′-deoxyguanosine and DNA by 19F NMR spectroscopy. Biochemistry. 1999;38:7572–7583. doi: 10.1021/bi990182d. [DOI] [PubMed] [Google Scholar]
- 21.Lee M.S., King C.M. New syntheses of N-(guanosin-8-yl)-4-aminobiphenyl and its 5′-monophosphate. Chem. Biol. Interact. 1981;34:239–248. doi: 10.1016/0009-2797(81)90135-6. [DOI] [PubMed] [Google Scholar]
- 22.Marques M.M., Beland F.A. Synthesis, characterization and solution properties of ras sequences modified by arylamine carcinogens at the first base of codon 61. Chem. Res. Toxicol. 1990;3:559–565. doi: 10.1021/tx00018a011. [DOI] [PubMed] [Google Scholar]
- 23.Cho B.P., Beland F.A., Marques M.M. NMR structural studies of a 15-mer DNA duplex from a ras proto-oncogene modified with the carcinogen 2-aminofluorene: conformational heterogeneity. Biochemistry. 1992;31:9587–9602. doi: 10.1021/bi00172a013. [DOI] [PubMed] [Google Scholar]
- 24.Cho B.P., Beland F.A., Marques M.M. NMR structural studies of a 15-mer DNA duplex from a ras protooncogene modified with the carcinogen 2-aminofluorene: conformational heterogeneity. Biochemistry. 1994;33:1373–1384. doi: 10.1021/bi00172a013. [DOI] [PubMed] [Google Scholar]
- 25.Arghavani M.B., SantaLucia J., Romano L.J. Effect of mismatched complementary strands and 5′-change in sequence context on the thermodynamics and structure of benzo[a]pyrene-modified oligonucleotides. Biochemistry. 1998;37:8575–8583. doi: 10.1021/bi980237w. [DOI] [PubMed] [Google Scholar]
- 26.Marky L.A., Breslauer K.J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka F., Kameda A., Yamamoto M., Ohuchi A. Thermodynamic parameters based on a nearest-Neighbor model for DNA sequences with a single-bulge loop. Biochemistry. 2004;43:7143–7150. doi: 10.1021/bi036188r. [DOI] [PubMed] [Google Scholar]
- 28.Allawi H.T., SantaLucia J. Thermodynamics of internal C.T mismatches in DNA. Nucleic Acid Res. 1998;26:2694–2701. doi: 10.1093/nar/26.11.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown K., Harvey C.A., Turteltaub K.W., Shields S.J. Structural characterization of carcinogen-modified oligodeoxynucleotide adducts using matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 2003;38:68–79. doi: 10.1002/jms.401. [DOI] [PubMed] [Google Scholar]
- 30.McLuckey S.A., Habibigoudarzi S. Decompositions of multiply charged oligonucleotide anions. J. Am. Chem. Soc. 1993;115:12085–12095. [Google Scholar]
- 31.Wu H., Aboleneen H. Sequencing oligonucleotides with blocked termini using exonuclease digestion and electrospray mass spectrometry. Anal. Biochem. 2000;287:126–135. doi: 10.1006/abio.2000.4827. [DOI] [PubMed] [Google Scholar]
- 32.Mikulecky P.J., Feig A.L. Heat capacity changes in RNA folding: application of perturbation theory to hammerhead ribozyme cold denaturation. Nucleic Acids. Res. 2004;32:3967–3976. doi: 10.1093/nar/gkh723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fasman G.D. Circular Dichroism and the Conformational Analysis of Biomolecules. NY: Plenum Press; 1996. [Google Scholar]
- 34.Nakanishi K., Berova N., Woody R. Circular Dichroism: Principle and Applications. New York, NY: VCH; 1994. [Google Scholar]
- 35.Sagi J., Hang B., Singer B. Sequence-dependent repair of synthetic AP sites in 15-mer and 35-mer oligonucleotides: role of thermodynamic stability imposed by neighbor bases. Chem. Res. Toxicol. 1999;12:917–923. doi: 10.1021/tx990088y. [DOI] [PubMed] [Google Scholar]
- 36.Sagi J., Guliaev A.B., Singer B. 15-mer DNA duplexes containing an abasic site are thermodynamically more stable with adjacent purines than with pyrimidines. Biochemistry. 2001;40:3859–3868. doi: 10.1021/bi0024409. [DOI] [PubMed] [Google Scholar]
- 37.Baase W.A., Johnson W.C. Circular dichroism and DNA secondary structure. Nucleic Acids Res. 1979;6:797–814. doi: 10.1093/nar/6.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spodheim-Maurizot M., Saint-Ruf G., Leng M. Conformational changes induced in DNA by in vitro reaction with N-hydroxy-N-2-aminofluorene. Nucleic Acids Res. 1979;6:1683–1694. doi: 10.1093/nar/6.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Houte L.P., Westra J.G., Retel J., Van Grondelle R. A spectroscopic study of the conformation of poly d(G-C).poly d(G-C) modified with the carcinogenic 2-aminofluorene. Carcinogenesis. 1988;9:1017–10127. doi: 10.1093/carcin/9.6.1017. [DOI] [PubMed] [Google Scholar]
- 40.Van Houte L.P.A., Bokma J.T., Lutgerink J.T., Westra J.G., Retel J., Van Grondelle R., Blok J. An optical study of the conformation of the aminofluorene–DNA complex. Carcinogenesis. 1987;8:759–766. doi: 10.1093/carcin/8.6.759. [DOI] [PubMed] [Google Scholar]
- 41.Van Houte L.P.A., Westra J.G., Retel J., Van Grondelle R. A circular dichroism study on the conformation of d(CGT) modified with N-acetyl-2-aminofluorene or 2-aminofluorene. J. Biomol. Struct. Dyn. 1991;9:45–59. doi: 10.1080/07391102.1991.10507892. [DOI] [PubMed] [Google Scholar]
- 42.Marky L.A., Rentzeperis D., Luneva N.P., Cosman M., Geacintov N.E., Kupke D.W. Differential hydration thermodynamics of stereo isomeric DNA-benzo[a]pyrene adducts derived from diol epoxide enantiomers with different tumorigenic potentials. J. Am. Chem. Soc. 1996;118:3804–3810. [Google Scholar]
- 43.Sagi J., Chenna A., Hang B., Singer B. A single cyclic p-benzoquinone adduct can destabilize a DNA oligonucleotide duplex. Chem. Res. Toxicol. 1998;11:329–334. doi: 10.1021/tx9701992. [DOI] [PubMed] [Google Scholar]
- 44.Gelfand C.A., Plum G.E., Grollman A.P., Johnson F., Breslauer K.J. The impact of an exocyclic cytosine adduct on DNA duplex properties: significant thermodynamic consequences despite modest lesion-induced structural alterations. Biochemistry. 1998;37:12507–12512. doi: 10.1021/bi981090b. [DOI] [PubMed] [Google Scholar]
- 45.Hang B., Sagi J., Singer B. Correlation between sequence-dependent glycosylase repair and the thermal stability of oligonucleotide duplexes containing 1, N6-ethenoadenine. J. Biol. Chem. 1998;273:33406–33413. doi: 10.1074/jbc.273.50.33406. [DOI] [PubMed] [Google Scholar]
- 46.Sagi J., Perry A., Hang B., Singer B. Differential destabilization of the DNA oligonucleotide double helix by a T·G mismatch, 3,N4-ethenocytosine, 3,N4-ethanocytosine, or an 8-(hydroxymethyl)-3,N4-ethenocytosine adduct incorporated into the same sequence contexts. Chem. Res. Toxicol. 2000;13:839–845. doi: 10.1021/tx000040g. [DOI] [PubMed] [Google Scholar]
- 47.Gelfand C.A., Plum G.E., Grollman A.P., Johnson F., Breslauer K.J. Thermodynamic consequences of an abasic lesion in duplex DNA are strongly dependent on base sequence. Biochemistry. 1998;37:7321–7327. doi: 10.1021/bi9803372. [DOI] [PubMed] [Google Scholar]
- 48.Venkatarangan L., Sivaprasad A., Johnson F. Site-specifically located 8-amino-2′-deoxyguanosine: thermodynamic stability and mutagenic properties in Escherichia coli. Nucleic Acids Res. 2001;29:1458–1463. doi: 10.1093/nar/29.7.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pradhan P., Jernstrom B., Seidel A., Norden B., Graslund A. Induced circular dichroism of benzo[a]pyrene-7,8-dihydrodiol 9,10-epoxide stereoisomers covalently bound to deoxyribooligonucleotides used to probe equilibrium distribution between groove binding and intercalative adduct conformations. Biochemistry. 1998;37:4664–4673. doi: 10.1021/bi972783f. [DOI] [PubMed] [Google Scholar]
- 50.Pradhan P., Tirumala S., Liu X., Sayer J.M., Jerina D.M., Yeh H.J. Solution structure of a trans-opened (10S)-dA adduct of (+)-(7S,8R,9S,10R)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a fully complementary DNA duplex: evidence for a major syn conformation. Biochemistry. 2001;40:5870–5881. doi: 10.1021/bi002896q. [DOI] [PubMed] [Google Scholar]
- 51.Mekhovich O., Tang M., Romano L.J. Rate of incision of N-acetyl-2-aminofluorene and N-2-aminofluorene adducts by UvrABC nuclease is adduct- and sequence-specific: comparison of the rates of UvrABC nuclease incision and protein–DNA complex formation. Biochemistry. 1998;37:571–579. doi: 10.1021/bi971544p. [DOI] [PubMed] [Google Scholar]
- 52.Buterin T., Meyer C., Giese B., Naegeli H. DNA quality control by conformational readout on the undamaged strand of the double helix. Chem. Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.