Abstract
TARGET OF RAPAMYCIN (TOR) kinase controls many cellular functions in eukaryotic cells in response to stress and nutrient availability and was shown to be essential for embryonic development in Arabidopsis thaliana. We demonstrated that Arabidopsis RAPTOR1 (a TOR regulatory protein) interacts with the HEAT repeats of TOR and that RAPTOR1 regulates the activity of S6 kinase (S6K) in response to osmotic stress. RAPTOR1 also interacts in vivo with Arabidopsis S6K1, a putative substrate for TOR. S6K1 fused to green fluorescent protein and immunoprecipitated from tobacco (Nicotiana tabacum) leaves after transient expression was active in phosphorylating the Arabidopsis ribosomal S6 protein. The catalytic domain of S6K1 could be phosphorylated by Arabidopsis 3-phosphoinositide-dependent protein kinase-1 (PDK1), indicating the involvement of PDK1 in the regulation of S6K. The S6K1 activity was sensitive to osmotic stress, while PDK1 activity was not affected. However, S6K1 sensitivity to osmotic stress was relieved by co-overexpression of RAPTOR1. Overall, these observations demonstrated the existence of a functional TOR kinase pathway in plants. However, Arabidopsis seedlings do not respond to normal physiological levels of rapamycin, which appears to be due its inability to bind to the Arabidopsis homolog of FKBP12, a protein that is essential for the binding of rapamycin with TOR. Replacement of the Arabidopsis FKBP12 with the human FKBP12 allowed rapamycin-dependent interaction with TOR. Since homozygous mutation in TOR is lethal, it suggests that this pathway is essential for integrating the stress signals into the growth regulation.
INTRODUCTION
TARGET OF RAPAMYCIN (TOR) is an evolutionarily conserved Ser/Thr kinase that has been shown to control a network of growth signaling pathways in yeast and animal cells in response to environmental cues (Dennis et al., 1999; Thomas, 2000; Rohde et al., 2001; Bjornsti and Houghton, 2004; Abraham, 2005). The regulatory function of TOR kinase requires various interacting partners (Abraham, 2002; Jacinto and Hall, 2003). For instance, it has been shown that the activation of TOR kinase is mediated by interaction with the Regulatory-Associated Protein of TOR (RAPTOR), which is believed to recruit substrates for TOR (Hara et al., 2002; Kim et al., 2002; Nojima et al., 2003). On the other hand, TOR has been known to be inactivated by an antifungal immunosuppressant rapamycin, which binds with TOR and FKBP12 to form a toxic complex that mimics stress signals (Heitman et al., 1991; Koltin et al., 1991; Chen et al., 1995). FKBP12 is a peptidyl-prolyl isomerase that was originally identified as the cytosolic receptor for the immunosuppressive drugs FK506 and rapamycin (Harding et al., 1989). Recent data suggest that rapamycin inhibits the TOR kinase function by causing dissociation of RAPTOR from TOR (Oshiro et al., 2004).
In response to nutrients, hormones, and stress signals, TOR regulates ribosome biogenesis, translation, and transcription of a set of genes involved in the translational machinery (Dennis et al., 1999; Cutler et al., 2001). This integration of energy levels, nutrient availability, and stress information allows TOR to regulate the translational machinery and act as a master regulator that optimizes cellular resources for growth (Abraham, 2002). Maintaining such a balance for energy use is crucial for the cell to ensure its survival under different environmental conditions. One of the well-documented examples of TOR playing a central regulatory role in growth signaling pathways is the control of translation via ribosomal p70 S6 kinase (p70S6K), which phosphorylates ribosomal protein S6 (RPS6) (Chung et al., 1992; Price et al., 1992; Turck et al., 1998; Dufner and Thomas, 1999).
Several lines of evidence suggest that RAPTOR plays a key role in mammalian p70S6K regulation. RAPTOR has been shown to directly bind with p70S6K, and since RAPTOR has been suggested to act as a scaffold to facilitate binding of other proteins to TOR, p70S6K may receive direct signal from TOR (Hara et al., 2002; Kim et al., 2002; Nojima et al., 2003). Another well-established regulatory role of TOR shown in yeast is at the transcriptional level; TOR, through its interaction with the phosphatase PP2A, affects the phosphorylation status of some transcription factors and thereby changes their localization from cytoplasm to the nucleus. Under osmotic stress or nutrient limitation, these transcription factors may shuttle into the nucleus to drive the expression of a set of stress-related genes (Beck and Hall, 1999; Peterson et al., 1999; Rohde et al., 2001).
Despite the importance of the TOR pathway in eukaryotes, little is known about the regulatory mechanism of the plant TOR pathway. Both TOR and RAPTOR homologs have been identified in Arabidopsis thaliana (TOR and RAPTOR1/RAPTOR2, respectively), and their functions have been implicated in embryo development (Menand et al., 2002; Deprost et al., 2005) and meristem-driven cell growth (Anderson et al., 2005). Also, an Arabidopsis homolog of Mei2 (AML1 protein), a meiosis-signaling protein, has been shown to interact with RAPTOR1 by yeast two-hybrid assay, suggesting that the AML1 protein may be a downstream component of the plant TOR signaling pathway (Anderson and Hanson, 2005). Two putative S6K homologs with high sequence homology (87% identity) were identified in Arabidopsis (Mizoguchi et al., 1995). They were originally named ATPK6 and ATPK19 and later referred to as S6K1 and S6K2, respectively (Zhang et al., 1994; Turck et al., 1998). S6K2 was suggested to be an ortholog of p70S6 kinase because it was shown to be able to phosphorylate the ribosomal S6 protein (Zhang et al., 1994; Turck et al., 1998). The activity of plant S6Ks has been shown to increase in response to auxin and cytokinins (Turck et al., 2004). However, no direct link between S6K and TOR has been established yet in plants. Since plants continuously encounter stress during their life cycle, the TOR pathway in plants may be of paramount importance. This pathway may link various stress signals to the growth signal pathways optimizing plant growth under different environmental conditions. Preliminary evidence suggests that a posttranscriptional mechanism may play a role in the regulation of the TOR pathway (Robaglia et al., 2004).
In this study, we cloned the key plant TOR kinase pathway genes known to date from Arabidopsis and attempted to establish the basic TOR kinase regulatory pathway in plants. We found that RAPTOR1 interacts with the HEAT repeats of TOR as well as S6K1 in vivo, and the activity of S6K1 is affected by osmotic stress. Furthermore, we expressed RAPTOR1 along with the S6K1 in tobacco (Nicotiana tabacum) leaves and found that overexpression of RAPTOR1 rendered the S6K1 osmotic stress insensitive. In animals, the activity of p70S6K has also been shown to be controlled by the 3-phosphoinositide-dependent protein kinase-1 (PDK1) pathway (Alessi et al., 1998; Pullen et al., 1998). We demonstrated that the Arabidopsis PDK1 activity was not inhibited by osmotic stress treatment, indicating that osmotic stress inhibition of S6K in plants is likely to be mediated through the TOR kinase pathway.
RESULTS
RAPTOR1 Interacts with the HEAT Repeats of TOR
The Arabidopsis genome contains a single TOR homolog present on chromosome 1, while there are two genes encoding RAPTOR on chromosomes 3 and 5, as previously described (Menand et al., 2002; Anderson et al., 2005; Deprost et al., 2005). We obtained cDNAs of TOR and one of RAPTOR (RAPTOR1) by RT-PCR (Figure 1; see Methods for details). Both cDNA sequences were confirmed to be identical to the GenBank annotation, and the TOR cDNA sequence was also found to be identical to the one described by Menand et al. (2002). TOR and RAPTOR genes were found to be expressed constitutively in various tissues of Arabidopsis at low levels, and the expression levels did not change significantly during different physiological conditions (our unpublished data; see also Menand et al., 2002; Robaglia et al., 2004; Anderson et al., 2005; Deprost et al., 2005), indicating that TOR may be regulated by some posttranscriptional/translational mechanisms (Robaglia et al., 2004).
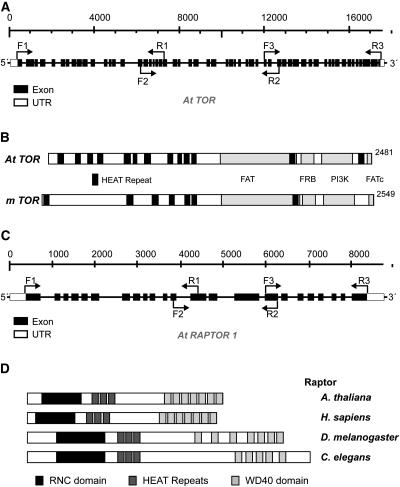
Figure 1.
Cloning of Arabidopsis TOR and RAPTOR1 cDNAs.
(A) Structure of TOR showing exons (black boxes) and intron nucleotide coordinates relative to the start of the gene. Primers used in RT-PCR to clone the TOR cDNA are indicated next to the bent arrows. UTR, untranslated region.
(B) Conserved protein domains found in Arabidopsis TOR, in comparison with mammalian TOR (mTOR). Different domains are indicated with the labeled boxes in black and gray.
(C) Structure of RAPTOR1 showing exons (black boxes) and intron nucleotide coordinates relative to the start of the gene. Primers used in RT-PCR to clone the RAPTOR cDNA are indicated next to the bent arrows.
(D) Conserved protein domains found in different RAPTOR homologs.
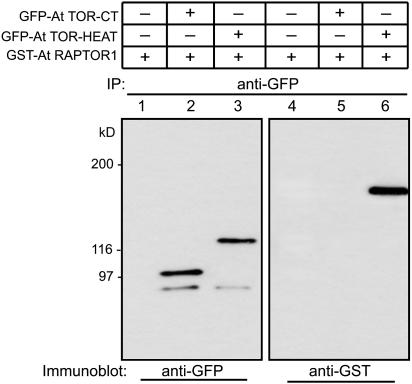
Since TOR and RAPTOR in animals are known to interact with each other (Kim et al., 2002), we examined whether plant RAPTOR1 interacts with the TOR and if this interaction occurs via the HEAT repeat domain of TOR as in the case of animal TOR. To analyze this possible interaction in vivo, we overexpressed glutathione S-transferase (GST)-fused RAPTOR1 (GST-RAPTOR1) and green fluorescent protein (GFP)-fused HEAT repeats of TOR (GFP-TOR-HEAT) constructs in tobacco leaves. The expression was performed in the presence of the tomato bushy stunt virus p19 protein, a system that allows high levels of transient protein expression in plants (Voinnet et al., 2003). The GFP-TOR-HEAT protein expressed in tobacco leaves was immunoprecipitated with GFP antibody. Following SDS-PAGE, the proteins were transferred to membrane, and the presence of GST-RAPTOR1 as a coimmunoprecipitant was observed by protein gel blot analysis using GST antibody. As shown in Figure 2, GST-RAPTOR1 was clearly detected in the immunoprecipitate with GFP-TOR-HEAT protein, suggesting that the interaction between TOR and RAPTOR is also conserved in plants and that the TOR-HEAT repeat motif is involved in the interaction. A construct carrying the FKBP12-rapamycin binding (FRB) and the kinase domains of the TOR (GFP-TOR-CT; see Figure 2 legend) did not show any interaction with RAPTOR1 (Figure 2, lanes 2 and 5), supporting the specificity of the interaction between RAPTOR1 and the HEAT repeats of TOR.
Figure 2.
RAPTOR1 Interacts with TOR-HEAT Repeats in Vivo.
35S:GST-RAPTOR1 and 35S:GFP-TOR-HEAT repeat constructs were expressed in tobacco leaves (Nicotiana benthamiana) by agroinfiltration. The C-terminal 2-kb fragment of TOR that includes the FRB domain and the kinase domain was fused to GFP and expressed as a control (35S:GFP-TOR-CT). The expressed proteins were immunoprecipitated (IP) using GFP antibody, and the immunodetections of the same fractions were made with GFP antibody (lanes 1 to 3) and GST antibody (lanes 4 to 6). Presence of RAPTOR1 as a coimmunoprecipitant of TOR-HEAT repeats is shown in lane 6, while GST-RAPTOR1 expressed alone (lanes 1 and 4) is not pulled down with GFP-antibody.
Differential Localization of the Two Arabidopsis S6Ks
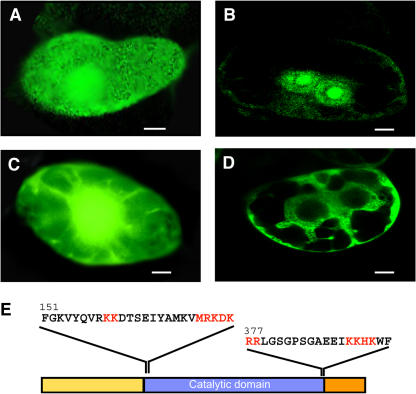
Zhang et al. (1994) and Turck et al. (1998) suggested that S6K2 was the Arabidopsis ortholog of the mammalian p70S6K that has been known to be a downstream component of the mammalian TOR signaling pathway and phosphorylates RPS6. In animals, a closely related isoform, p85S6K, is a product of the same gene using an alternative translational start site and contains an N-terminal nuclear targeting signal (Dufner and Thomas, 1999). This protein is exclusively nuclear and has been suggested to phosphorylate the chromatin-bound nuclear form of RPS6 (Franco and Rosenfeld, 1990), whereas p70S6K, which is mostly cytoplasmic, is involved in the phosphorylation of RPS6 in the cytoplasmic ribosomes. Protein motif analyses of S6K1 and S6K2 sequences by PSORT (http://psort.ims.u-tokyo.ac.jp/) and by PFSCAN (http://myhits.isb-sib.ch/cgi-bin/motif_scan) predicted S6K2 to contain two putative bipartite nuclear targeting signals (Figure 3E). Subcellular localization results with transiently expressed fusion proteins in BY2 cells were consistent with this prediction. As shown in Figure 3, GFP-S6K2 was mainly confined to nucleus/nucleolus (Figure 3B), while the distribution pattern of GFP-S6K2 (Figure 3A) was similar to that of the GFP control (Figure 3C) but with more protein distributed toward cytoplasm, suggesting that the two Arabidopsis S6K homologs may play distinct functional roles in different subcellular locations. The fact that S6K2 is nuclear localized suggests that it may be the functional equivalent of p85S6K rather than p70S6K. To explore the possibility of S6K1 being the cytoplasmic isoform of S6K acting as a functional homolog of the mammalian p70S6K, we tested its interaction with RAPTOR1 and its ability to phosphorylate the bona fide substrate RPS6.
Figure 3.
Differential Localization of the Two Isoforms of S6K.
S6K1 and S6K2 open reading frames were placed downstream of 35S:GFP, and the constructs were transiently expressed in BY2 cells by particle bombardment as described by Gindullis et al. (1999). Bars in (A) to (D) = 10 μm.
(A) 35S:GFP-S6K1.
(B) 35S:GFP-S6K2.
(C) 35S:GFP (control) construct.
(D) 35S:GFP-sucrose synthase as a cytoplasmic localization control.
(E) Location and sequence of the two putative bipartite nuclear localization motifs identified in S6K2. The putative nuclear targeting sequences are indicated in red.
RAPTOR1 Interacts with S6K1 in Vivo
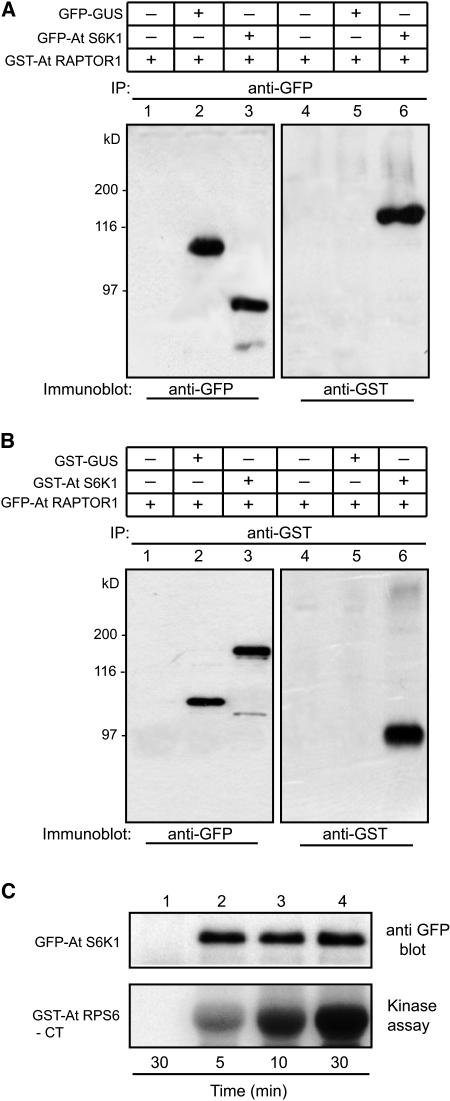
To elucidate the possible link between S6K and TOR signaling pathway in plants, we examined the kinase activity of S6K1 using phosphorylation of RPS6. In addition, we determined a possible link between the S6K1 activity and its interaction with RAPTOR1 by co-overexpression of these proteins. If S6K is under the control of the TOR signaling pathway in plants and is a substrate of TOR kinase, it is likely that S6K would interact with RAPTOR. To determine a direct interaction between S6K1 and RAPTOR1 in vivo, we first isolated Arabidopsis S6K1 and coexpressed transiently the two constructs GFP-S6K1 and GST-RAPTOR1 in tobacco leaves using the p19 system (Voinnet et al., 2003). When the expression levels were optimum, as judged by a GFP control (3 to 5 d after agroinfiltration in general), the leaf samples were analyzed, and S6K1 was immunoprecipitated using specific antibodies. Protein gel blot analysis of the immunoprecipitated fraction using GFP antibodies revealed a distinct band corresponding to the size of GFP-S6K1 chimeric protein (Figure 4A, lane 2). The immunoprecipitate from the same fraction using GST antibody, followed by protein gel blot detection with the GST antibody, revealed the presence of a protein equivalent in mass to GST-RAPTOR (Figure 4A, lane 4). These data suggested that S6K1 interacts with RAPTOR1 in vivo, and this interaction appears to be stable. We repeated the coexpression experiments using alternate constructs (i.e., GST-S6K1 and GFP-RAPTOR1), and the immunoprecipitation was performed using GST antibody. As shown in Figure 4B, using GFP antibody we were able to detect in the immunoprecipitate a band that corresponds to the size of GFP-RAPTOR1. At the same time, GST-S6K1 was also detectable, as shown in lane 4. In both Figures 4A and 4B, the control constructs GFP-GUS (Figure 4A, lanes 2 and 5) and GST-GUS (Figure 4B, lanes 2 and 5) coexpressed with GST-RAPTOR1 or GFP-RAPTOR1 did not show any interaction, suggesting that the interaction between S6K1 and RAPTOR1 demonstrated by transient coexpression in this study is highly specific. These two experiments provided conclusive evidence that S6K1 and RAPTOR do interact in vivo. Finally, we tested the ability of S6K1 immunoprecipitate to phosphorylate its substrate, RPS6. For this experiment, an Escherichia coli–expressed C-terminal fragment of Arabidopsis RPS6 fused to GST (GST-RPS6-CT) was used as a substrate for the kinase reaction. As shown in Figure 4C, the GFP-S6K1 immunoprecipitate using GFP antibody was found to be active and able to phosphorylate RPS6.
Figure 4.
Ectopically Expressed S6K1 Is Active and Interacts in Vivo with RAPTOR1.
(A) 35S:GFP-S6K1 and 35S:GST-RAPTOR constructs were expressed in tobacco (N. benthamiana) leaves by agroinfiltration. Proteins were immunoprecipitated with GFP antibody, and the same fractions were immunodetected by GFP antibody (lanes 1 to 3) and GST antibody (lanes 4 to 6). Lanes 1 and 3, a negative control for immunoprecipitation with GST-RAPTOR1 expressed alone; lanes 2 and 4, control for interaction specificity with GFP-GUS coexpressed with GST-RAPTOR1; note the presence of RAPTOR1 as a coimmunoprecipitant of S6K1 in lane 6.
(B) The reciprocal of the experiment shown in (A). 35S:GST-S6K1 and 35S:GFP-RAPTOR1 were expressed in tobacco leaves by agroinfiltration. Proteins were immunoprecipitated with GST antibody, and the same fractions were immunodetected by GFP antibody (lanes 1 to 3) and GST antibody (lanes 4 to 6). Lanes 1 and 3, a negative control for immunoprecipitation with GFP-RAPTOR1 expressed alone; lanes 2 and 4, control for interaction specificity with GST-GUS coexpressed with GFP-RAPTOR1; note the presence of RAPTOR1 as a coimmunoprecipitant of S6K1 in lane 6.
(C) 35S:GFP-S6K1 expressed in tobacco leaves by agroinfiltration was immunoprecipitated with GFP antibody and subjected to in vitro kinase assay using GST-RPS6-CT as substrate. As a control, 35S:GFP construct expressed and immunoprecipitated with GFP antibody was included in the kinase assay (lane 1). Time in the figure indicates the kinase reaction time at 25°C.
Our data suggests that Arabidopsis S6K1 is more likely to be a functional homolog of animal p70S6K. We do not know if S6K2 also forms a complex with RAPTOR, but the demonstration of the interaction between RAPTOR1 and S6K1, as well as between TOR and RAPTOR1, suggests that S6K is likely to be a downstream component of the TOR signaling pathway in plants.
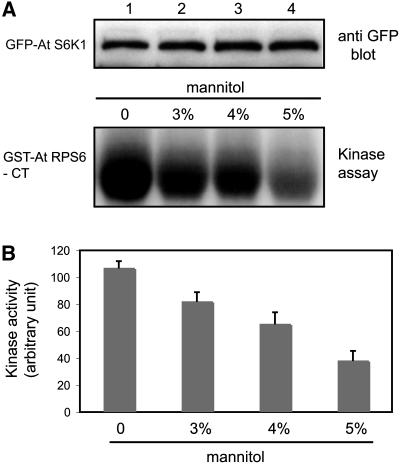
S6K1 Activity Is Affected by Osmotic Stress
To explore the effect of osmotic stress on S6K1 activity, we expressed the GFP-S6K1 fusion protein in tobacco leaves by agroinfiltration and treated the leaves with different concentrations of mannitol. After homogenization of the leaves, GFP-S6K1 was immunoprecipitated with GFP antibody and was subjected to the kinase assay using GST-RPS6-CT peptide as a substrate. With 5% mannitol treatment, the S6K1 activity showed a significant reduction, although at this concentration, leaf tissues did not undergo any plasmolysis. A quantitative analysis of this data showed that >70% reduction in the S6K1 activity occurred after the treatment with 5% mannitol (Figure 5B). This observation suggests that plant S6K may mediate stress response in controlling the phosphorylation state of RPS6, which is known to affect the rate of translation (Jefferies and Thomas, 1994; Thomas and Hall, 1997). The osmotic stress signal may affect the S6K1 activity via the plant TOR kinase pathway. Alternatively, it may come through the PDK1 pathway, which has also been known to regulate S6K in animal cells (Alessi et al., 1998).
Figure 5.
Osmotic Stress Affects S6K1 Kinase Activity.
(A) Leaves of tobacco (N. benthamiana) transiently expressing 35S:GFP-S6K1 construct were treated with different concentration of mannitol. GFP-S6K1 protein was then immunoprecipitated with GFP antibody and subjected to in vitro kinase assay using GST-RPS6-CT as substrate. Top panel: protein gel blot showing equal loading of S6K1 in the kinase reaction. Bottom panel: autoradiograph showing the activity of S6K1 under different concentrations of mannitol treatment.
(B) Quantification of the kinase activity shown in (A) after normalization with the intensity of S6K1 bands in the protein gel blot. The results are represented as the average of four independent experiments ± se (n = 4, P < 0.01 by analysis of variance).
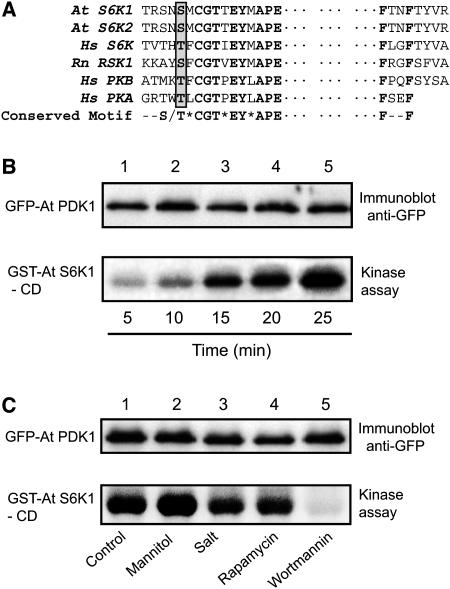
PDK1 Phosphorylates the Activation Loop Domain of S6K1
PDK1 is known to activate the AGC family of protein kinases, to which S6K belongs, and several PDK1 substrates have been identified, including PKB, S6K, RSK, SGK, and PKC (Alessi et al., 1998; Pullen et al., 1998; Biondi et al., 2001). It has been shown that Arabidopsis PDK1 can complement the yeast PDK1 mutation, suggesting the existence of functional conservation of the PDK1 pathway in eukaryotes (Deak et al., 1999). We examined whether S6K is also phosphorylated by PDK1 in plants. Arabidopsis S6K (both S6K1 and S6K2) contains a putative PDK1 phosphorylation motif on the T-activation loop consensus sequences in its catalytic domain (Figure 6A). We expressed a GFP-PDK1 construct in tobacco leaves through agroinfiltration and immunoprecipitated the fusion protein with GFP antibody. The immunoprecipitate was then subjected to a kinase assay using the GST-S6K1 catalytic domain (GST-S6K1-CD; amino acid residues 134 to 389) expressed in E. coli as a substrate to determine the PDK1 activity. The observations from mammalian cells indicated that the phosphorylation of S6K catalytic domain by PDK1 occurs only after a series of consecutive phosphorylations on the C terminus of S6K mediated by TOR. The GFP-PDK1 immunoprecipitate was able to phosphorylate the S6K1 catalytic domain in a time-dependent manner as shown in Figure 6B, indicating that plant PDK1 is also involved in the activation of pS6K. We further determined if plant PDK1 activity is affected by osmotic stress, rapamycin, or wortmannin. Wortmannin is a potent inhibitor of phosphoinositide-3 kinase (PI-3K) and is known to inactivate PDK1 in mammalian cells, an indication that PDK1 is a downstream component of the PI-3K pathway in animal cells (Alessi et al., 1997; Davies et al., 2000). We overexpressed GFP-PDK1 in tobacco leaves and treated a set of leaves with rapamycin (30 μg/mL), mannitol (5%), NaCl (150 mM), or wortmannin (300 nM). As shown in Figure 6C, PDK1 activity was not responsive to osmotic stress, rapamycin, or salt treatments but was very sensitive to wortmannin. These data showed that the downregulation of S6K1 activity upon osmotic stress is not mediated through inactivation of the PDK1 pathway. Thus, it suggests that the osmotic stress signal is most likely to affect the TOR pathway to regulate the S6K activity in plants.
Figure 6.
PDK1 Regulates S6K1 and Its Activity Is Not Affected by Osmotic Stress.
(A) Alignment of the activation loop sequences present in the catalytic domain of different AGC kinases (Rn RSK1, rat p90 ribosomal S6 kinase1; Hs PKB, human protein kinase B; Hs PKA, human protein kinase A). The consensus sequence motif is indicated in bold, and the phosphorylation site on the motif is indicated by a gray box. Dots represent a gap of ∼150 amino acids.
(B) GFP-PDK1 was expressed in tobacco leaves (N. benthamiana) by agroinfiltration and immunoprecipitated with GFP antibody. The immunoprecipitates were then subjected to in vitro kinase assay using GST-S6K1-CD as substrate. Top panel: protein gel blot showing the equal loading of PDK1 in the kinase reactions. Bottom panel: autoradiograph showing the kinase activity over time. Time indicates the kinase reaction time at 25°C.
(C) Tobacco leaves expressing GFP-PDK1 as in (B) were treated with different stresses or inhibitors, as indicated in the figure, before subjecting the protein samples to immunoprecipitation with GFP antibody. The immunoprecipitates were then assayed for the PDK1 activity as in (B).
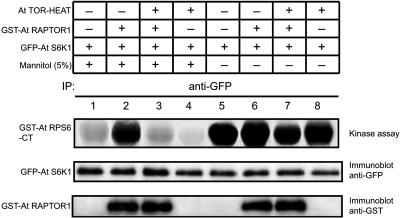
RAPTOR1 Interaction Affects the Osmotic Stress Signaling to S6K
Our data showed that S6K1 interacts with RAPTOR1, and RAPTOR1, in turn, interacts with the HEAT repeats of TOR, suggesting a functional link between these proteins in plants. Our data also suggested that the osmotic stress signal is likely to affect S6K activity through the TOR pathway in plants. Osmotic stress may inhibit TOR kinase activity upstream in the signaling pathway, resulting in the downregulation of S6K1. Alternatively, the osmotic stress may regulate S6K1 activity downstream of the TOR kinase, antagonizing the positive effect of TOR pathway on the S6K1 activity. In order to distinguish these two possibilities, we co-overexpressed GFP-S6K1 and GST-RAPTOR1 in tobacco leaves by agroinfiltration and treated the leaves with 5% mannitol. The leaf tissues were then homogenized and GFP-S6K1 was immunoprecipitated with GFP antibody. The activity of S6K1 was measured by subjecting the immunoprecipitate to the kinase assay. As shown in Figure 7, osmotic stress treatment significantly reduced the activity of S6K1 (lane 1, compare with lane 5). However, the coexpression of RAPTOR alleviated the osmotic stress effect on the activity of S6K1 (Figure 7, lane 2). If osmotic stress regulates the S6K1 activity by inhibition of TOR kinase activity, coexpression of RAPTOR, which is a downstream component of TOR, would have a neutral or additive effect on the S6K1 inhibition. Thus, the data appear to suggest that the osmotic stress affects S6K1 activity downstream of the TOR kinase. The overexpression of RAPTOR probably brings endogenous TOR in closer proximity to its substrate S6K (Hara et al., 2002; Nojima et al., 2003), alleviating the inhibition of S6K under osmotic stress conditions. In order to verify that this effect is indeed RAPTOR specific and requires the function of TOR, we overexpressed TOR-HEAT repeats together with GST-RAPTOR1 and GFP-S6K1 and examined the possible effect of TOR-HEAT repeats. As a nonfunctional fragment that can still interact with RAPTOR1, the TOR-HEAT repeats coexpressed with RAPTOR1 may abrogate the protective effect of RAPTOR on the S6K1 activity under the osmotic stress condition. As shown in Figure 7 (lane 3, compare with lane 2), the co-overexpression of TOR-HEAT repeats restored the sensitivity of S6K1 to osmotic stress in the presence of RAPTOR1. The overexpression of TOR-HEAT repeats along with RAPTOR1 appears to have diluted out the RAPTOR effect on the S6K1 activity. This could have been further verified using the full-length TOR where the kinase domain is present; however, we have not been successful in overexpressing the full-length TOR yet.
Figure 7.
RAPTOR Regulates the Sensitivity of S6K1 to Osmotic Stress.
The 35S:GFP-S6K1, 35S:GST-RAPTOR, and 35S:TOR-HEAT repeat constructs were transiently expressed in tobacco leaves (N. benthamiana) with or without subsequent mannitol treatment as indicated in the figure. Following immunoprecipitation with GFP antibody, the sample fractions were subjected to in vitro kinase assay using GST-RPS6-CT as substrate (top panel), immunodetection with GFP antibody to show the equal loading of S6K1 (middle panel), and another immunodetection with GST antibody to confirm the interaction of RAPTOR with S6K1 (bottom panel).
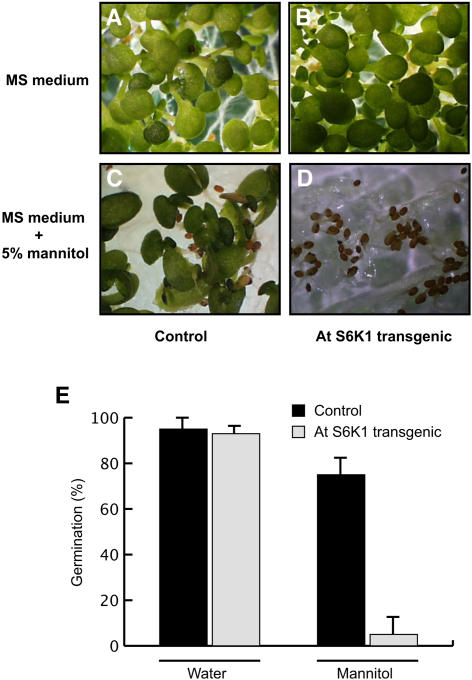
Arabidopsis Plants Overexpressing S6K Are Hypersensitive to Osmotic Stress
We generated transgenic Arabidopsis plants overexpressing S6K1 under the 35S promoter. Overexpression of S6K1 was confirmed by RNA gel blot analysis (data not shown). These plants apparently exhibited no distinct phenotype as compared with the wild type or control transgenic plants expressing the vector alone. However, upon treatment with mannitol, the transgenic seeds of the S6K1-transformed line were found to be overly sensitive to osmotic stress (Figure 8). This does not appear to be due to any germination problem because under normal conditions the germination of the untreated S6K1 transgenic line was similar to the control transgenic plants expressing the vector construct alone (Figure 8E). This observation supports the hypothesis that having S6K activity slowed down under osmotic stress conditions is one of the main mechanisms employed by the plants to sustain harsh environmental conditions.
Figure 8.
Transgenic Arabidopsis Plants Expressing S6K1 Are Hypersensitive to Osmotic Stress.
(A) Arabidopsis plants transformed with the empty vector construct and grown on Murashige and Skoog (MS) plates (Murashige and Skoog, 1962).
(B) Transgenic Arabidopsis plants expressing S6K1 and grown on MS plates.
(C) Arabidopsis plants transformed with the empty vector construct and grown on MS plates containing 5% mannitol.
(D) Transgenic Arabidopsis plants expressing S6K1 and grown on MS plates containing 5% mannitol (note inhibition of seed germination).
(E) Germination of the transgenic seeds under normal conditions and osmotic stress. Three independent lines from both the vector-control transgenic and the S6K1 transgenic plants were measured for the germination level. The average germination rates (in percent) under normal condition and under mannitol treatment are illustrated in the graph with standard error of the mean (n = 3).
Plant Homolog of FKBP12 Does Not Interact with TOR
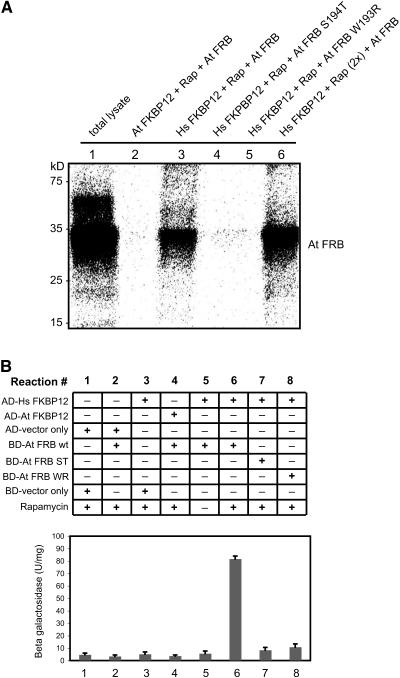
We observed that rapamycin does not inhibit the growth of Arabidopsis seedlings at levels that are effective in yeast and mammalian cells (Heitman et al., 1991; Koltin et al., 1991; Chung et al., 1992). Although we could not rule out the possibility that rapamycin has a lower permeability to the plant cells, that is rather unlikely because rapamycin enters easily in all other eukaryotic cells so far tested. Since the growth inhibitory effect of rapamycin on the cells is known to be due to the formation of an inhibitory complex involving the TOR-FRB domain, rapamycin, and FKBP12 (Van Duyne et al., 1991; Chiu et al., 1994), we tested if plant homolog of FKBP12 associates with the Arabidopsis TOR-FRB domain in a rapamycin-dependent manner, as is the case with mammalian TOR. For this, we performed both a two-hybrid interaction study and a direct binding assay with in vitro–synthesized and labeled TOR-FRB peptide (Figure 9). The labeled peptide was incubated with either GST-At FKBP12 or GST-human FKBP12 (Hs FKBP12). Consistent with a previous observation made with a yeast two-hybrid system (Menand et al., 2002), neither the yeast two-hybrid analysis nor the in vitro translation data could demonstrate a rapamycin-dependent interaction between Arabidopsis FKBP12 and the TOR-FRB domain. Interestingly, in the same experiment, the TOR-FRB domain was able to form a complex with Hs FKBP12. These results suggested that there might be some mutation in the amino acid sequence of the plant FKBP12, rendering it insensitive to rapamycin and thus preventing it from forming an inhibitory complex with the plant TOR.
Figure 9.
Rapamycin-Dependent Interaction between TOR-FRB Domain and Hs FKBP12.
(A) In vitro assay for TOR-FKBP12 interactions. The [35S]Met-labeled TOR-FRB domain and its mutant variants (At FRB S194T and At FRB W193R) were prepared by in vitro transcription/translation system. GST-fusion proteins of At FKBP12 and Hs FKBP12 were expressed in E. coli and purified. All binding assays except for lane 1 (total in vitro translation lysate) included rapamycin (Rap). The amount of rapamycin was doubled in lane 6. The binding assay samples were pulled down with GST-Sepharose and loaded onto SDS-PAGE followed by autoradiography. Hs FKBP12 showed an interaction with At FRB domain in the presence of rapamycin (lanes 3 and 6), while At FKBP12 or the mutant form of Hs FKBP12 failed to form a complex with At FRB (lanes 2, 4, and 5).
(B) Yeast two-hybrid assay showing the TOR-FKBP12 interaction in vivo. GAL-4 (BD) fusion construct of TOR-FRB wild type and mutants W193R (WR) and S194T (ST) were coexpressed with GAL-4 (AD) fusion constructs of At FKBP12 and Hs FKBP12 in the yeast strain SMY4 as indicated in the figure. β-Galactosidase assays were performed in liquid culture using o-nitrophenyl-d-galactopyranoside as a substrate as described in Methods. The results are represented as the average of four independent experiments ± se.
DISCUSSION
Existence of a Functional TOR Kinase Pathway in Plants That Is Insensitive to Rapamycin
In this study, we have provided experimental evidence for the existence of a functional TOR kinase pathway in Arabidopsis. As in other eukaryotes, the pathway appears to be responsible for controlling cell growth through the modulation of S6K activity. Rapamycin, as a specific inhibitor of TOR kinase, has been an instrumental tool in dissecting the TOR signaling pathway in yeast and animals. However, rapamycin does not seem to affect the plant TOR pathway. Our data showed that this was apparently due to some alteration(s) in FKBP12 protein in Arabidopsis that may prevent its interaction with the FRB domain of TOR in a rapamycin-dependent manner. Replacing Arabidopsis FKBP12 with human FKBP12, however, allowed rapamycin-dependent binding with the FRB domain of TOR. A similar observation with yeast FKBP12 and TOR-FRB was made (Menand et al., 2002). Despite the fact that Arabidopsis FKBP12 was unable to bind rapamycin to form an inhibitory complex with TOR, rapamycin was found to have an inhibitory effect on seed germination (our unpublished data) though at a much higher concentration (∼50-fold) than was shown in animal and yeast cells. Therefore, it may be argued that there is a true homolog of FKBP12 as yet identified in plants. However, Robaglia et al. (2004) recently reported that none of the closely related Arabidopsis FKBP12-like proteins were able to bind the TOR-FRB domain, making this possibility rather unlikely. Arabidopsis FKBP12 shares ∼50% identity with human FKBP12 in protein sequences. Since both animal and yeast FKBP12 are sensitive to rapamycin, even though their amino acid sequence identity is only ∼60%, direct comparison of plant and human FKBP protein sequences to identify an area that might be responsible for this difference was not possible. Paradoxically, this observation can lead to construction of a tool in the plant system to study the TOR kinase pathway via chemical genomics approach. Transformation of Arabidopsis with human FKBP12 may produce plants that are sensitive to rapamycin and thus provide a model system enabling functional dissection of the plant TOR kinase pathway.
Plant S6K Activity Appears to Be Regulated by the TOR Pathway
RAPTOR has been shown to positively and negatively regulate the TOR kinase pathway in mammalian cells (Hara et al., 2002; Kim et al., 2002). By coimmunoprecipitation analyses, we showed that RAPTOR1 associated in vivo with TOR via its HEAT repeat domain. We also demonstrated in vivo interaction between RAPTOR1 and S6K1. We showed that S6K1 can phosphorylate RPS6 and interact with RAPTOR1. In addition, our localization data showed that S6K2 is localized in the nucleus, while S6K1 is primarily cytoplasmic. Our finding of two functional S6Ks in Arabidopsis, one of which preferentially localized in the nucleus, is consistent with the animal systems where two isoforms of S6K (p70S6K and p85S6K) function exclusively in the cytoplasm and in nucleus, respectively (Koh et al., 1999; Valovka et al., 2003). Our interaction and phosphorylation data, taken together with the localization data, suggest that S6K1 is a functional ortholog of p70S6K and is likely to be regulated by the plant TOR pathway. The primary function of S6K2 may in fact be in regulating the phosphorylation of RPS6 in the nucleolus (Franco and Rosenfeld, 1990). We recently observed that Arabidopsis RPS6 interacts with the nucleolus-localized histone deacetylase (HD2B; our unpublished data). The activity of nucleolus-localized S6K2 may thus be involved in the regulation of the interaction of HD2B with RPS6, which may have a functional implication in the ribosome biogenesis.
Mammalian S6Ks contain a conserved TOR signaling motif (TOS) that is critical in receiving the TOR/RAPTOR-mediated regulatory signal (Schalm and Blenis, 2002; Nojima et al., 2003). Although both TOR and RAPTOR protein sequences are highly homologous to their mammalian counterparts, the primary structure of the two S6Ks (other than the catalytic domain) is quite different from that of mammalian S6K (Turck et al., 1998, 2004), and it does not appear to carry a conserved TOS motif. Further characterization of the N- and C- terminal regulatory domains of the two S6Ks would be necessary for a better understanding of the protein motifs important in the regulation of plant S6K activity by RAPTOR/TOR.
S6K1 Activity Is Regulated by Osmotic Stress and This Signaling Is PDK1 Independent
S6K has been identified as the key downstream effector of the TOR kinase pathway through which the translational regulation of cell growth and proliferation is mediated in animal cells (Price et al., 1989; Pearson et al., 1995). The TOR pathway in animal and yeast cells has been found to be affected by various cellular stresses, including osmotic stress and starvation. We showed that the activity of S6K1 was affected by osmotic stress (mannitol treatment). This observation indicates that TOR may be involved in the regulation of the metabolic adjustment of plant cells to osmotic stress. This is consistent with the current hypothesis that TOR kinase is an energy sensor in eukaryotic cells (Dennis et al., 2001). The sensitivity of S6K1 to osmotic stress may slow down the protein synthesis and ribosome biogenesis under stress conditions. Therefore, plants overexpressing S6K may be more sensitive to osmotic stress treatment. This was found indeed to be the case, as the transgenic Arabidopsis seeds overexpressing S6K1 were overly sensitive to the mannitol treatment. This result suggests that slowing down the S6K activity under unfavorable growth conditions may be one of the main mechanisms employed by plants to sustain harsh environmental conditions. However, it was reported that the effect of osmotic stress on the phosphorylation status of ribosomal S6 protein in maize (Zea mays) root tips was inconsistent (Williams et al., 2003). This observation may suggest that different physiological and/or developmental status of the plant plays a role in modulating the stress response. In Arabidopsis culture cells, osmotic stress has almost no effect on the actively growing cells, whereas the S6K2 activity in the stationary-phase cells or starvation-induced cells is highly sensitive to osmotic stress (our unpublished data).
Activation of p70S6K in animal cells is achieved through a series of consecutive phosphorylation events mediated by different kinases, including TOR, mitogen-activated protein kinase, and PDK1 (Pullen and Thomas, 1997). As a member of the AGC kinase family (Hanks and Hunter, 1995), the ultimate activation of S6K requires phosphorylation of its activation loop located in the catalytic domain, which is mediated by PDK1 (Alessi et al., 1998; Pullen et al., 1998). Phosphorylation of p70S6K by mammalian TOR is considered to be a prerequisite for PDK1 to be able to act on the activation loop of the p70S6K catalytic domain (Burnett et al., 1998; Dennis et al., 1998). Although the overall amino acid sequence of the plant S6Ks is significantly different from their mammalian counterparts, the catalytic domain is highly conserved with that of other AGC kinases. We showed that the S6K1 domain is a substrate for PDK1 when subjected to in vitro kinase reaction. Taken together with the in vivo interaction data of S6K1 and RAPTOR1, this result suggests that the activity of plant S6K is regulated by TOR kinase as well as PDK1, similar to what was found in animal cells (Long et al., 2004).
Activation of animal PDK1 requires a second messenger phosphatidylinositol-3,4,5-phosphate, which is generated by the catalytic action of class-I PI-3K (Chan et al., 1999). No evidence for the existence of phosphatidylinositol-3,4,5-phosphate or class-I PI-3K is available in plants. However, our result showed that the PDK1 activity was inhibited by wortmannin, an inhibitor of class-I PI-3K (Brunn et al., 1996). This result was also consistent with the recent observation made by Turck et al. (2004). Plant PDK1 may be activated by a different type of phosphatidylinositol phosphate, as suggested by Deak et al. (1999), produced by a yet-to-be-identified lipid kinase.
The observation that the PDK1 activity was not sensitive to osmotic stress suggests that the osmotic stress signal is likely to regulate S6K1 activity through the TOR pathway. It is possible that the TOR kinase activity itself is downregulated by osmotic stress signal, leading to the inhibition of S6K1 activity. Alternatively, osmotic stress signal may act downstream of the TOR (see below), antagonizing the positive regulatory effect of TOR on the S6K1 activity.
The Sensitivity of S6K1 to Osmotic Stress Is Reduced by RAPTOR1 and the Effect Requires the Functional TOR
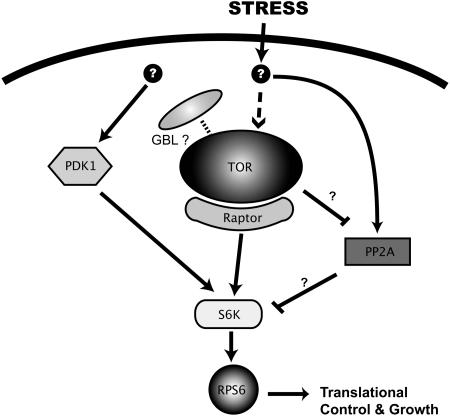
In order to distinguish the two possible modes of osmotic stress signaling that may affect the activity of S6K1 (i.e., via an inhibition of the TOR kinase or through another effector acting downstream of the TOR pathway), we designed an experiment in which both S6K1 and RAPTOR1 were overexpressed in tobacco leaves and subjected to osmotic stress treatment. The results showed that the sensitivity of S6K1 to osmotic stress was significantly reduced by coexpression of RAPTOR1, indicating that osmotic stress signal may be conveyed through an effector downstream of the TOR kinase pathway. This is consistent with the observation that the effect of RAPTOR1 is diluted out by coexpression of the TOR-HEAT repeats, demonstrating that the effect is RAPTOR specific and dependent on the function of TOR. Although these data should be interpreted with caution, one possible candidate for such an antagonist of TOR is the phosphatase PP2A. As PP2A has been implicated in the osmotic stress response in mammalian culture cells (Parrott and Templeton, 1999), TOR, with the help of RAPTOR, may be competing with PP2A for S6K as a substrate in plants (Figure 10). In yeast system, the inactivation of PP2A by TOR has been well documented (Rohde et al., 2001; Düvel et al., 2003). Whether a similar regulation occurs in plants in response to various environmental cues is yet to be determined.
Figure 10.
TOR Kinase Signaling Pathway in Plants.
Data from this study support the existence of the plant TOR pathway modulating plant cell growth in response to stress. The basic skeleton of the pathway, as illustrated above, is conserved as in other eukaryotes. It remains to be answered whether the activity of plant TOR also requires the G-β-like (GBL) protein.
TOR Controls Plant Growth and Development
Two recent studies have indicated that RAPTOR may be involved in the control of embryo development and/or meristem-driven growth of plants (Anderson et al., 2005; Deprost et al., 2005). Taken together with the fact that the homozygous mutant of TOR is lethal (Menand et al., 2002), it suggests that the TOR pathway plays a key role in the regulation of plant growth and development, particularly under stress. Dissecting the role and the regulation of each component of the TOR pathway in plants will greatly contribute to a better understanding of plant growth and development. Development of a transgenic line expressing human FKBP12 followed by isolation of a suppressor(s) of this pathway may provide a functional genomic approach to analyze this key regulatory pathway essential for plant growth under diverse environmental conditions.
METHODS
RNA Isolation and RT-PCR Reactions
Total RNA was isolated from leaves of 2-week-old Arabidopsis thaliana seedlings (ecotype Columbia) using Trizol reagent (Invitrogen) following the manufacturer's instructions. Approximately 2 μg of total RNA prepared by the above method was used in each RT-PCR reaction using the Thermoscript RT-PCR system (Invitrogen) following the standard amplification conditions.
Cloning of Arabidopsis TOR cDNA
The full-length cDNA of the TOR gene on chromosome 1 (At1g50030) was cloned by RT-PCR using the Thermoscript RT-PCR system following the manufacturer's protocol. Since this cDNA is very long, 7.67 kb, we attempted to amplify it in three overlapping fragments that were fused together upon cloning using specific restriction enzymes (BspEI and EcoICRI) to generate the full-length clone (Figure 1A). The following three primer pairs were used to generate the three overlapping PCR fragments: TOR F1, 5′-GCCCTGCAGCGATGTCTACCTCGTCGC-3′; TOR R1, 5′-GCATCAGCTCCGGAATATATTGTCTC-3′; TOR F2, 5′-CAAGAGTGGGTGGCTTGGCAATGAGAC-3′; TOR R2, 5′-ACCTCCTCTAGTTCTGACAGCTGCTG-3′; TOR F3, 5′-TCAGCAGCTGTCAGAACTAGAGGAGG-3′; TOR R3, 5′-TCACCAGAAAGGGCACCACCCAACATAG-3′.
The initial PCR cloning and the subsequent fragment fusion were performed in pCR2.1 vector (Invitrogen). The entire region of the obtained full-length TOR cDNA was sequenced and confirmed to be identical to the GenBank database (NM_103891) and the one described by Menand et al. (2002). The TOR cDNA consists of 56 exons and predicts a protein product of 279 kD, which contains all the conserved domains found in the mammalian TOR, including HEAT repeats, FAT domain, FRB domain, kinase domain, and the FATc domain (Figure 1B).
For downstream applications, including protein expression and mutagenesis, the full-length TOR cDNA in pCR2.1 was subcloned into the Gateway entry vector pENTR1A (Invitrogen) and subsequently moved in to appropriate destination vectors.
Cloning of RAPTOR1 cDNA
Cloning of Arabidopsis RAPTOR homolog (RAPTOR1) was achieved by RT-PCR amplification of At3g08850 cDNA. As with the TOR cDNA cloning, its cDNA was amplified as three overlapping fragments to be joined together by unique restriction enzyme sites (StuI and XbaI). Using the Thermoscript RT-PCR kit, Arabidopsis leaf RNA was amplified with the following primer sets corresponding to the sequences surrounding the unique restriction enzyme sites in the deduced RAPTOR1 (At3g08850) exons (Figure 1C): RapF1, 5′-GAATTCATGGCATTAGGAGACTTAATGGT-3′; RapF2, 5′-GAATTCGCTCTTGTACTTCTTGGAAGATT-3′; RapF3, 5′-GAATTCAACTACCTGAGTGGATTGAGGA-3′; RapR1, 5′-CTCGTCGATGTCCATCCACAATGACAGCCAGAAC-3′; RapR2, 5′-GGCGGTACCATTTAAATTTGACTAGCATCCGCACCA-3′; RapR3, 5′-CCCGGGAAATGCACCCGAGCTATCTAGA-3′. The products of these three independent reactions were cloned in pCR2.1 vector and subsequently joined together in the Gateway entry vector pENTR1A (Invitrogen). The identity of the full-length clone was confirmed by sequencing. The full-length RAPTOR cDNA was then moved in to various Gateway downstream vectors for protein expression in Escherichia coli as well as in tobacco (Nicotiana tabacum) leaves.
Cloning and Expression of Arabidopsis S6K
Arabidopsis ribosomal S6K1 (ATPK6) and S6K2 (ATPK19) cDNAs (Mizoguchi et al., 1995) were cloned by PCR amplification from an Arabidopsis leaf cDNA library. The PCR primers were designed to cover 5′- and 3′-untranslated regions of the corresponding mRNAs to ensure specificity of the PCR amplification because their coding regions are highly homologous to each other. The amplified PCR products were directly cloned into the Gateway entry vector pENTR1A (Invitrogen), and the sequences were verified by DNA sequencing. Various fusion protein constructs of these clones were made either by transferring them to appropriate Gateway destination vectors through LR recombination reaction (Invitrogen) or by restriction digestion–based subcloning into the target vectors.
Cloning and Expression of Arabidopsis FKBP12, RPS6, S6K-CD, and Human FKBP12
The GST-human FKBP12 fusion construct was a kind gift from Jie Chen (University of Illinois, Urbana-Champlain, IL). Full-length cDNA clones of Arabidopsis FKBP12 homolog (FKBP12; At5g64350) and Arabidopsis 40S ribosomal protein S6 (RPS6B; At5g10360) were obtained by PCR amplification of the corresponding cDNA libraries. The PCR products were directly cloned into pGEX-KG vector (Guan and Dixon, 1991) to generate GST-fusion proteins. The GST-fusion protein clone of the RPS6 C-terminal end (GST-RPS6-CT) was generated by PCR cloning of the amino acid residues 150 to 249 of GST-RPS6. The GST-S6K1 catalytic domain fusion construct (GST-S6K1-CD) was generated by PCR amplification and subcloning of the S6K1 catalytic domain (amino acid residues 134 to 389) into pGEX-KG vector. E. coli cells containing the GST-fusion protein clones were induced with 1 mM isopropylthio-β-galactoside, and the protein was purified essentially as described by Lam et al. (1995).
Bacteria producing the recombinant protein were pelleted and resuspended in 5 mM phosphate buffer, pH 6.8, containing 1 mM EDTA and 5 mM β-mercaptoethanol. The pellet was resuspended in STE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 150 mM NaCl). Lysozyme was added, the suspension was incubated on ice for 15 min, and 1 mM DTT was added just before sonication. Sonication was done several times, 1 min each, with incubation on ice after each sonication. The lysate was centrifuged at 14,000 rpm on a swing bucket centrifuge, and the supernatant was taken and incubated with agarose beads for binding at 4°C for 30 min. After rigorous washes with STE buffer containing Triton X-100 (0.2%), β-mercaptoethanol (2 mM), and PMSF (10 mM), small aliquots of agarose beads containing the bound proteins were checked for the purity of purification on SDS-PAGE. GST-RPS6 protein was used in the kinase assay as bound to the beads without further purification. Both GST-At FKBP12 and GST-Hs FKBP12 were subjected to further purification by elution with 50 mM glutathione and dialysis before use in the in vitro binding assays.
Rapamycin-Dependent in Vitro Binding Assay for FKBP12 and TOR-FRB Domain
The TOR-FRB domain (amino acids 1980 to 2280) was subcloned by PCR into pAGA3 vector (Qin et al., 1997). Using the in vitro TNT quick coupled transcription/translation system (Promega), [35S]Met-labeled Arabidopsis FRB domain was prepared according to the manufacturer's instructions. Briefly, 1 μg of pAGA3 plasmid containing the Arabidopsis FRB cDNA was incubated with TNT reticulocyte lysate and 20 μCi of [35S]Met in a total volume of 50 μL for 90 min at 30°C. The labeling efficiency and the level of protein expression were evaluated by SDS-PAGE and autoradiography.
Mutations in the FRB domain of TOR were made using the Quick Change kit (Stratagene) according to the manufacture's instructions. The following two pairs of primers were used to generate two mutations in two sites, W193F and S194T: TOR-WF-F, 5′-GCCATACTTTGGCATGAAATGTTTCATGAAGCACTAGAAGAAGC-3′; TOR-WF-R, 5′-GCTTCTTCTAGTGCTTCATGAAACATTTCATGCCAAAGTATGGC-3′; TOR-ST-F, 5′-GAAGCACTAGAAGAAGCTACTCGCTTGTATTTTGGTGAAC-3′; TOR-ST-R, 5′-GTTCACCAAAATACAAGCGAGTAGCTTCTTCTAGTGCTTC-3′.
GST-FKBP12 expressed and purified from E. coli was premixed with rapamycin in 1 mL of binding buffer (PBS with 1 mM DTT and 0.3% Triton X-100) at a concentration of 1 μM each, followed by addition of 1 μM FRB protein and brief incubation on ice. Glutathione-Sepharose (20 μL) was then added to the mixture and rocked at 4°C for 1 h. The matrix was subsequently washed three times in 1 mL of binding buffer, boiled in SDS sample buffer, and loaded on a 15% SDS-PAGE gel.
Rapamycin-Dependent Binding by Yeast Two-Hybrid Assay
The two-hybrid strain SMY4 (TOR1-3 fpr1::ADE2) is an isogenic derivative of strain Y190 (MATatrp1-901 his3 leu2-3112 ura3-52 ade2 gal4 gal80 URA3::GAL-lacZ LYS2::GAL-HIS3; Connell-Crowley et al., 1993) in which the endogenous TOR1 and FKBP12 genes were disabled (Cardenas et al., 1994). Gal4(AD)-fusion constructs of Arabidopsis FKBP12 and human FKBP12, as well as Gal4(BD)-fusion constructs of TOR-FRB wild type and mutants, were prepared by PCR subcloning. Yeast strain SMY4 was grown in yeast extract/peptone/dextrose or supplemented synthetic dextrose medium. Transformation of yeast was performed using the lithium acetate method with carrier DNA (Gietz et al., 1992). Yeast colonies transformed with the fusion vectors were selected on synthetic medium lacking uracil, Trp, and/or Leu.
For β-galactosidase assays, yeast transformants were grown at 30°C in liquid synthetic medium (SD) with selection (Trp-Leu). Rapamycin was added at a concentration of 1.0 μg/mL when necessary. β-Galactosidase activity was measured with the substrate o-nitrophenyl-d-galactopyranoside following the protocol described by Miller (1972). Each independent transformant was assayed in quadruplets.
DNA Constructs for Agroinfiltration
The 35S::p19 and 35S::mGFP5 constructs were described previously (Voinnet et al., 2003). The 35S::RAPTOR-GFP fusion construct was made by mobilization of the full-length RAPTOR cDNA from pENTR1A into pB2WG7 vector (Karimi et al., 2002) through LR recombination reaction following the manufacturer's instructions (Invitrogen). The 35S::GFP-S6K1 was constructed by PCR amplification of S6K1 cloned in pENTR1A and subsequent subcloning into pEGAD vector (Cutler et al., 2000) via BamHI and SmaI restriction digestion and ligation. The 35S::GSTS6K2 was constructed by first cloning S6K2 into pGEX-KG to generate GST-S6K2 fusion protein and then by subcloning the GST-S6K2 open reading frame into pENTR1A and LR recombination reaction with pH2WG7 vector (Karimi et al., 2002).
Agroinfiltration and Transient Protein Expression
Agroinfiltration procedure for the transient protein expression in tobacco leaves was basically performed according to the method described by Voinnet et al. (2003). Briefly, Agrobacterium tumefaciens strains ABI, C58C1, and GV3101 carrying binary constructs were grown at 29°C in Luria-Bertani medium with antibiotic selection and acetosyringone to stationary phase. Bacterial cells were collected by centrifugation at 6000g for 15 min at room temperature and resuspended in 10 mM MgCl2 and 150 μg/mL of acetosyringone. For cotransfections, Agrobacterium cultures carrying different constructs were mixed at equal ratio and left for 6 h at room temperature before infiltration. Leaves of 2-week-old Nicotiana benthamiana plants were infiltrated with the bacterial cultures through abaxial air spaces. In all experiments, Agrobacterium C58C1 carrying the 35S:p19 construct (Voinnet et al., 2003) was cotransfected to achieve maximum level of protein expression.
The treatments of osmotic stress and inhibitors in Figures 5 to 7 were achieved by a similar manner as described by Lu et al. (2002). Briefly, 3 to 4 d after the agroinfiltration of the DNA constructs for transient protein expression, solutions containing a desired concentration of mannitol (up to 5%), salt (150 mM NaCl), rapamycin (30 μg/mL), wortmannin (300 nM), or control (water) were infiltrated to the same leaves, and protein extraction was performed after 30 min. Rapamycin and wortmannin were initially dissolved in DMSO and then diluted with water to make the desired concentration right before the treatment.
Growth and Transformation of Arabidopsis Plants
Arabidopsis plants (ecotype Columbia) were grown at 21°C with 16 h of light (100 μmol photons m−2 s−1) per day and 60% humidity. Plant transformation was performed by the floral dip technique (Chung et al., 2000) using Agrobacterium strain GV3101 containing pB2WG7 vector with Arabidopsis S6K1 insert. Transformants were recovered by sowing seeds into soil. Ten-day-old seedlings were sprayed by Basta at a concentration of 20 mg/L or 1:1500 dilution. For each transformation, 20 to 24 transformants were screened for expression of the transgene. One transformant from each transgenic line was used for detailed characterization. Plants were grown for 4 to 7 d on plates containing sterile solidified MS medium supplemented with 3% sucrose and 5% mannitol. Three independent transgenic lines were used in the osmotic stress treatment (mannitol) experiments illustrated in Figure 8. The germination rate of these transgenic lines under normal conditions was >95%, which was comparable to the control transgenic line transformed with vector alone (Figure 8E).
Immunoprecipitation and Kinase Assay
Agroinfiltrated tobacco leaves were collected generally 5 d after infiltration. The leaves were ground in liquid nitrogen, and the powder was resuspended in 200 μL of IP buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1 mM DTT, and freshly added protease/phosphatase inhibitors: 10 μg/mL of pepstatin, antipain, leupeptin, 1 mM PMSF, 1 mM NaVO3, and 5 mM NaF) on ice for 3 min and vortexed briefly. Antibody was added (GST antibody from Santa Cruz Biotechnology [catalog number SC-459] and GFP antibody from Molecular Probes [catalog number A-11122]), and the samples were incubated at 4°C for 1 to 2 h with gentle rocking. Five microliters of protein-A agarose beads (Amersham) was added to each sample and rocked for another 1 h at 4°C. The beads were washed with 200 μL of the IP buffer with 0.1% SDS five times. The beads were then resuspended in 30 μL of SDS sample buffer and heated at 95°C for 3 min before loading onto 10 to 15% SDS-PAGE for protein gel blot analysis. After transferring the protein samples to nitrocellulose membrane (Millipore), immunodetection using the GST antibody or GFP antibody was performed as described previously (Hong et al., 2001).
For in vitro kinase assay, the final immunoprecipitates were resuspended in kinase buffer (20 mM HEPES, pH 7.5, 125 mM NaCl, 1 mM DTT, 10 mM MgCl2, and 5 mM MnCl2), and the reaction was performed at 25°C by adding 100 μM cold ATP and 0.1 μCi [γ-32P]ATP (Perkin-Elmer) in the presence of GST-fusion substrate protein. The reaction was stopped by adding 5× SDS sample buffer and heating at 95°C for 5 min. Samples were then run on SDS-PAGE followed by autoradiography. Aliquots of the samples were also subjected to protein gel blotting to ensure equal loading of the samples and normalization for quantitative analysis of the result.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At1g50030 (TOR), At3g08850 (RAPTOR1), At3g08730 (S6K1), At3g08720 (S6K2), At5g64350 (FKBP12), and At5g10360 (RPS6B).
Acknowledgments
We thank David Baulcombe for providing vectors for the p19 expression system, Jie Chen for the human FKBP12 clone, Mike Hall for providing the yeast strain SMY4, and Zonglie Hong for the GFP-SUSY1 construct. We also thank Iris Meier and J.C. Jang for their comments on the manuscript. This work was supported by a USDA grant (2002-35100-12185).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Desh Pal S. Verma (verma.1@osu.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035931.
References
- Abraham, R.T. (2002). Identification of TOR signaling complexes: More TORC for the cell growth engine. Cell 111 9–12. [DOI] [PubMed] [Google Scholar]
- Abraham, R.T. (2005). TOR signaling: An odyssey from cellular stress to the cell growth machinery. Curr. Biol. 15 R139–R141. [DOI] [PubMed] [Google Scholar]
- Alessi, D.R., James, S.R., Downes, C.P., Holmes, A.B., Gaffney, P.R., Reese, C.B., and Cohen, P. (1997). Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase B alpha. Curr. Biol. 7 261–269. [DOI] [PubMed] [Google Scholar]
- Alessi, D.R., Kozlowski, M.T., Weng, Q.P., Morrice, N., and Avruch, J. (1998). 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr. Biol. 8 69–81. [DOI] [PubMed] [Google Scholar]
- Anderson, G.H., and Hanson, M.R. (2005). The Arabidopsis Mei2 homologue AML1 binds AtRaptor1B, the plant homologue of a major regulator of eukaryotic cell growth. BMC Plant Biol. 5 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, G.H., Veit, B., and Hanson, M.R. (2005). The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 3 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, T., and Hall, M.N. (1999). The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402 689–692. [DOI] [PubMed] [Google Scholar]
- Biondi, R.M., Kieloch, A., Currie, R.A., Deak, M., and Alessi, D.R. (2001). The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J. 20 4380–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsti, M.A., and Houghton, P.J. (2004). The TOR pathway: A target for cancer therapy. Nat. Rev. Cancer 4 335–348. [DOI] [PubMed] [Google Scholar]
- Brunn, G.J., Williams, J., Sabers, C., Wiederrecht, G., Lawrence, J.C., Jr., and Abraham, R.T. (1996). Direct inhibition of the signaling functions of the mammalian target of rapamycin by the phosphoinositide 3-kinase inhibitors, wortmannin and LY294002. EMBO J. 15 5256–5267. [PMC free article] [PubMed] [Google Scholar]
- Burnett, P.E., Barrow, R.K., Cohen, N.A., Snyder, S.H., and Sabatini, D.M. (1998). RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E–BP1. Proc. Natl. Acad. Sci. USA 95 1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas, M.E., Hemenway, C., Muir, R.S., Ye, R., Fiorentino, D., and Heitman, J. (1994). Immunophilins interact with calcineurin in the absence of exogenous immunosuppressive ligands. EMBO J. 13 5944–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, T.O., Rittenhouse, S.E., and Tsichlis, P.N. (1999). AKT/PKB and other D3 phosphoinositide-regulated kinases: Kinase activation by phosphoinositide-dependent phosphorylation. Annu. Rev. Biochem. 68 965–1014. [DOI] [PubMed] [Google Scholar]
- Chen, J., Zheng, X.F., Brown, E.J., and Schreiber, S.L. (1995). Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. USA 92 4947–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, M.I., Katz, H., and Berlin, V. (1994). RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. USA 91 12574–12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, J., Kuo, C.J., Crabtree, G.R., and Blenis, J. (1992). Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 69 1227–1236. [DOI] [PubMed] [Google Scholar]
- Chung, M.H., Chen, M.K., and Pan, S.M. (2000). Floral spray transformation can efficiently generate Arabidopsis transgenic plants. Transgenic Res. 9 471–476. [DOI] [PubMed] [Google Scholar]
- Connell-Crowley, L., Solomon, M.J., Wei, N., and Harper, J.W. (1993). Phosphorylation independent activation of human cyclin-dependent kinase 2 by cyclin A in vitro. Mol. Biol. Cell 4 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, N.S., Pan, X., Heitman, J., and Cardenas, M.E. (2001). The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol. Biol. Cell 12 4103–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Griffitts, J.S., and Somerville, C.R. (2000). Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, S.P., Reddy, H., Caivano, M., and Cohen, P. (2000). Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak, M., Casamayor, A., Currie, R.A., Downes, C.P., and Alessi, D.R. (1999). Characterisation of a plant 3-phosphoinositide-dependent protein kinase-1 homologue which contains a pleckstrin homology domain. FEBS Lett. 451 220–226. [DOI] [PubMed] [Google Scholar]
- Dennis, P.B., Fumagalli, S., and Thomas, G. (1999). Target of rapamycin (TOR): Balancing the opposing forces of protein synthesis and degradation. Curr. Opin. Genet. Dev. 9 49–54. [DOI] [PubMed] [Google Scholar]
- Dennis, P.B., Jaeschke, A., Saitoh, M., Fowler, B., Kozma, S.C., and Thomas, G. (2001). Mammalian TOR: A homeostatic ATP sensor. Science 294 1102–1105. [DOI] [PubMed] [Google Scholar]
- Dennis, P.B., Pullen, N., Pearson, R.B., Kozma, S.C., and Thomas, G. (1998). Phosphorylation sites in the autoinhibitory domain participate in p70(s6k) activation loop phosphorylation. J. Biol. Chem. 273 14845–14852. [DOI] [PubMed] [Google Scholar]
- Deprost, D., Truong, H.N., Robaglia, C., and Meyer, C. (2005). An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem. Biophys. Res. Commun. 326 844–850. [DOI] [PubMed] [Google Scholar]
- Dufner, A., and Thomas, G. (1999). Ribosomal S6 kinase signaling and the control of translation. Exp. Cell Res. 253 100–109. [DOI] [PubMed] [Google Scholar]
- Düvel, K., Santhanam, A., Garrett, S., Schneper, L., and Broach, J.R. (2003). Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 11 1467–1478. [DOI] [PubMed] [Google Scholar]
- Franco, R., and Rosenfeld, M.G. (1990). Hormonally inducible phosphorylation of a nuclear pool of ribosomal protein S6. J. Biol. Chem. 265 4321–4325. [PubMed] [Google Scholar]
- Gietz, D., St Jean, A., Woods, R.A., and Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindullis, F., Peffer, N.J., and Meier, I. (1999). MAF1, a novel plant protein interacting with matrix attachment region binding protein MFP1, is located at the nuclear envelope. Plant Cell 11 1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, K.L., and Dixon, J.E. (1991). Eukaryotic proteins expressed in Escherichia coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192 262–267. [DOI] [PubMed] [Google Scholar]
- Hanks, S.K., and Hunter, T. (1995). Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 9 576–596. [PubMed] [Google Scholar]
- Hara, K., Maruki, Y., Long, X., Yoshino, K., Oshiro, N., Hidayat, S., Tokunaga, C., Avruch, J., and Yonezawa, K. (2002). RAPTOR, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110 177–189. [DOI] [PubMed] [Google Scholar]
- Harding, M.W., Galat, A., Uehling, D.E., and Schreiber, S.L. (1989). A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature 341 758–760. [DOI] [PubMed] [Google Scholar]
- Heitman, J., Movva, N.R., and Hall, M.N. (1991). Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253 905–909. [DOI] [PubMed] [Google Scholar]
- Hong, Z., Delauney, A.J., and Verma, D.P. (2001). A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell 13 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto, E., and Hall, M.N. (2003). Tor signalling in bugs, brain and brawn. Nat. Rev. Mol. Cell Biol. 4 117–126. [DOI] [PubMed] [Google Scholar]
- Jefferies, H.B., and Thomas, G. (1994). Elongation factor-1 alpha mRNA is selectively translated following mitogenic stimulation. J. Biol. Chem. 269 4367–4372. [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kim, D.H., Sarbassov, D.D., Ali, S.M., King, J.E., Latek, R.R., Erdjument-Bromage, H., Tempst, P., and Sabatini, D.M. (2002). mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110 163–175. [DOI] [PubMed] [Google Scholar]
- Koh, H., Jee, K., Lee, B., Kim, J., Kim, D., Yun, Y.H., Kim, J.W., Choi, H.S., and Chung, J. (1999). Cloning and characterization of a nuclear S6 kinase, S6 kinase-related kinase (SRK); a novel nuclear target of Akt. Oncogene 18 5115–5119. [DOI] [PubMed] [Google Scholar]
- Koltin, Y., Faucette, L., Bergsma, D.J., Levy, M.A., Cafferkey, R., Koser, P.L., Johnson, R.K., and Livi, G.P. (1991). Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol. Cell. Biol. 11 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E., Martin, M.M., Timerman, A.P., Sabers, C., Fleischer, S., Lukas, T., Abraham, R.T., O'Keefe, S.J., O'Neill, E.A., and Wiederrecht, G.J. (1995). A novel FK506 binding protein can mediate the immunosuppressive effects of FK506 and is associated with the cardiac ryanodine receptor. J. Biol. Chem. 270 26511–26522. [DOI] [PubMed] [Google Scholar]
- Long, X., Muller, F., and Avruch, J. (2004). TOR action in mammalian cells and in Caenorhabditis elegans. Curr. Top. Microbiol. Immunol. 279 115–138. [DOI] [PubMed] [Google Scholar]
- Lu, C., Han, M.H., Guevara-Garcia, A., and Fedoroff, N.V. (2002). Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc. Natl. Acad. Sci. USA 99 15812–15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand, B., Desnos, T., Nussaume, L., Berger, F., Bouchez, D., Meyer, C., and Robaglia, C. (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA 99 6422–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.H. (1972). Experiments in Molecular Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Mizoguchi, T., Hayashida, N., Yamaguchi-Shinozaki, K., Kamada, H., and Shinozaki, K. (1995). Two genes that encode ribosomal-protein S6 kinase homologs are induced by cold or salinity stress in Arabidopsis thaliana. FEBS Lett. 358 199–204. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Nojima, H., Tokunaga, C., Eguchi, S., Oshiro, N., Hidayat, S., Yoshino, K., Hara, K., Tanaka, N., Avruch, J., and Yonezawa, K. (2003). The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E–BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278 15461–15464. [DOI] [PubMed] [Google Scholar]
- Oshiro, N., Yoshino, K., Hidayat, S., Tokunaga, C., Hara, K., Eguchi, S., Avruch, J., and Yonezawa, K. (2004). Dissociation of raptor from mTOR is a mechanism of rapamycin-induced inhibition of mTOR function. Genes Cells 9 359–366. [DOI] [PubMed] [Google Scholar]
- Parrott, L.A., and Templeton, D.J. (1999). Osmotic stress inhibits p70/85 S6 kinase through activation of a protein phosphatase. J. Biol. Chem. 274 24731–24736. [DOI] [PubMed] [Google Scholar]
- Pearson, R.B., Dennis, P.B., Han, J.W., Williamson, N.A., Kozma, S.C., Wettenhall, R.E., and Thomas, G. (1995). The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J. 14 5279–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, R.T., Desai, B.N., Hardwick, J.S., and Schreiber, S.L. (1999). Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycin associated protein. Proc. Natl. Acad. Sci. USA 96 4438–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, D.J., Grove, J.R., Calvo, V., Avruch, J., and Bierer, B.E. (1992). Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science 257 973–977. [DOI] [PubMed] [Google Scholar]
- Price, D.J., Nemenoff, R.A., and Avruch, J. (1989). Purification of a hepatic S6 kinase from cycloheximide-treated rats. J. Biol. Chem. 264 13825–13833. [PubMed] [Google Scholar]
- Pullen, N., Dennis, P.B., Andjelkovic, M., Dufner, A., Kozma, S.C., Hemmings, B.A., and Thomas, G. (1998). Phosphorylation and activation of p70s6k by PDK1. Science 279 707–710. [DOI] [PubMed] [Google Scholar]
- Pullen, N., and Thomas, G. (1997). The modular phosphorylation and activation of p70s6k. FEBS Lett. 410 78–82. [DOI] [PubMed] [Google Scholar]
- Qin, N., Platano, D., Olcese, R., Stefani, E., and Birnbaumer, L. (1997). Direct interaction of gbetagamma with a C-terminal gbetagamma-binding domain of the Ca2+ channel alpha1 subunit is responsible for channel inhibition by G protein-coupled receptors. Proc. Natl. Acad. Sci. USA 94 8866–8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia, C., Menand, B., Lei, Y., Sormani, R., Nicolai, M., Gery, C., Teoule, E., Deprost, D., and Meyer, C. (2004). Plant growth: The translational connection. Biochem. Soc. Trans. 32 581–584. [DOI] [PubMed] [Google Scholar]
- Rohde, J., Heitman, J., and Cardenas, M.E. (2001). The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 276 9583–9586. [DOI] [PubMed] [Google Scholar]
- Schalm, S.S., and Blenis, J. (2002). Identification of a conserved motif required for mTOR signaling. Curr. Biol. 12 632–639. [DOI] [PubMed] [Google Scholar]
- Thomas, G. (2000). An encore for ribosome biogenesis in the control of cell proliferation. Nat. Cell Biol. 2 E71–E72. [DOI] [PubMed] [Google Scholar]
- Thomas, G., and Hall, M.N. (1997). TOR signalling and control of cell growth. Curr. Opin. Cell Biol. 9 782–787. [DOI] [PubMed] [Google Scholar]
- Turck, F., Kozma, S.C., Thomas, G., and Nagy, F. (1998). A heat-sensitive Arabidopsis thaliana kinase substitutes for human p70s6k function in vivo. Mol. Cell. Biol. 18 2038–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck, F., Zilbermann, F., Kozma, S.C., Thomas, G., and Nagy, F. (2004). Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis. Plant Physiol. 134 1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valovka, T., et al. (2003). Protein kinase C phosphorylates ribosomal protein S6 kinase betaII and regulates its subcellular localization. Mol. Cell. Biol. 23 852–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne, G.D., Standaert, R.F., Karplus, P.A., Schreiber, S.L., and Clardy, J. (1991). Atomic structure of FKBP-FK506, an immunophilin-immunosuppressant complex. Science 252 839–842. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33 949–956. [DOI] [PubMed] [Google Scholar]
- Williams, A.J., Werner-Fraczek, J., Chang, I.F., and Bailey-Serres, J. (2003). Regulated phosphorylation of 40S ribosomal protein S6 in root tips of maize. Plant Physiol. 132 2086–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S.H., Broome, M.A., Lawton, M.A., Hunter, T., and Lamb, C.J. (1994). atpk1, a novel ribosomal protein kinase gene from Arabidopsis. II. Functional and biochemical analysis of the encoded protein. J. Biol. Chem. 269 17593–17599. [PubMed] [Google Scholar]