Abstract
Golgi inheritance during mammalian cell division occurs through the disassembly, partitioning, and reassembly of Golgi membranes. The mechanisms responsible for these processes are poorly understood. To address these mechanisms, we have examined the identity and dynamics of Golgi proteins within mitotic membranes using live cell imaging and electron microscopy techniques. Mitotic Golgi fragments, seen in prometaphase and telophase, were found to localize adjacent to endoplasmic reticulum (ER) export domains, and resident Golgi transmembrane proteins cycled rapidly into and out of these fragments. Golgi proteins within mitotic Golgi haze—seen during metaphase—were found to redistribute with ER markers into fragments when the ER was fragmented by ionomycin treatment. The temperature-sensitive misfolding mutant ts045VSVG protein, when localized to the Golgi at the start of mitosis, became trapped in the ER at the end of mitosis in cells shifted to 40°C. Finally, reporters for Arf1 and Sar1 activity revealed that Arf1 and Sar1 undergo sequential inactivation during mitotic Golgi breakdown and sequential reactivation upon Golgi reassembly at the end of mitosis. Together, these findings support a model of mitotic Golgi inheritance that involves inhibition and subsequent reactivation of cellular activities controlling the cycling of Golgi components into and out of the ER.
INTRODUCTION
The Golgi apparatus functions as an essential processing/sorting station at the crossroads of the secretory pathway, receiving newly synthesized and/or recycled proteins from the endoplasmic reticulum (ER) and delivering them to the plasma membrane and other final destinations within cells. Despite knowledge of its roles, the origin and maintenance of the Golgi as a discrete organelle is debated. In one view, the Golgi is an autonomous organelle that is responsible for its biogenesis and maintenance (Pelletier et al., 2002). In a different view, the Golgi is the product of ER export activities and depends on the functioning of ER export domains (Zaal et al., 1999; Miles et al., 2001; Ward et al., 2001; Bevis et al., 2002).
These two views provide very different explanations for the inheritance of the mammalian Golgi during mitosis—a process involving reversible disassembly of the Golgi ribbon into discrete fragments and highly dispersed elements (i.e., mitotic haze). In the first view, known as the autonomous model, mitotic Golgi breakdown/reassembly occurs independently of the ER: the Golgi ribbon is first severed into fragments, the fragments then shed small vesicles, the vesicles later reassemble into fragments, and the fragments recoalesce into a Golgi ribbon (Shorter and Warren, 2002). Throughout this process, Golgi and ER membranes are said to remain distinct and do not communicate. In the second view, called the ER-dependent model, Golgi inheritance occurs through the intermediary of the ER: Golgi fragmentation/dispersal results from changes in the pathways controlling Golgi outgrowth from and absorption into the ER (Zaal et al., 1999; Altan-Bonnet et al., 2004). This is thought to lead to Golgi proteins redistributing into the ER or to ER export domains at different stages of mitosis. Throughout the Golgi disassembly/reassembly process, Golgi and ER membranes are either communicating or exist as a merged compartment.
The autonomous and ER-dependent models of Golgi inheritance are based on different views of Golgi behavior during interphase. Advocates of the autonomous model interpret fragmentation of the interphase Golgi, such as through treatments with ilimaquinone (Takizawa et al., 1993) or okadaic acid (Lucocq et al., 1995), or in cell-free assays from purified Golgi membranes (Misteli and Warren, 1995a,b), as evidence that Golgi membranes have a general capacity to breakdown into smaller elements. This capacity, they argue, is the central causal factor underlying mitotic Golgi disassembly. Advocates of the ER-dependent model instead focus on the fact that conditions that perturb ER-Golgi trafficking pathways in interphase (e.g., treatment with nocodazole or brefeldin A [BFA] or expression of dominant negative Sar1 or Arf1 mutants) causes the Golgi to fragment or to disassemble as a consequence of Golgi membrane proteins continuously entering and leaving Golgi structures as they cycle constitutively through the ER (Lippincott-Schwartz et al., 1989; Cole et al., 1996, 1998; Storrie et al., 1998; Zaal et al., 1999). (For nocodazole, a microtubule-depolymerizing agent, Golgi proteins are unable to cycle back to the centrosomal Golgi structure after cycling through the ER, so these proteins accumulate in new Golgi structures [i.e., fragments] near ER export domains; Cole et al., 1996). Because during mitosis, cytoplasmic microtubules undergo depolymerization (to make tubulin available for forming the spindle apparatus) and ER export is inhibited (Warren et al., 1983; Farmaki et al., 1999; Prescott et al., 2001), proponents of the ER-dependent model argue that these changes are sufficient causal factors for mediating Golgi disassembly during mitosis.
One approach for distinguishing between these two models of Golgi inheritance has been to characterize the mitotic Golgi haze found in metaphase. Whereas mitotic haze in the autonomous model represents free vesicles formed by vesiculation of Golgi stacks, in the ER-dependent model it represents a merged ER-Golgi compartment. Electron microscopy studies have reported the presence of Golgi enzymes in mitotic ER (Thyberg and Moskalewski, 1992; Farmaki et al., 1999; Zaal et al., 1999), but it is unclear whether the enzymes were relocated from the Golgi or were a product of new synthesis, so the findings do not distinguish between the two inheritance strategies. More recent work using filipin to fragment ER in mitotic cells showed that Golgi proteins did not redistribute with ER proteins into ER fragments (Axelsson and Warren, 2004), which favors the autonomous inheritance model. A concern with these experiments, however, is that filipin is a cholesterol-extracting reagent, so it could affect Golgi and ER proteins differentially within ER membranes (Rothberg et al., 1990; Ilangumaran and Hoessli, 1998). Moreover, the Golgi lipid marker BODIPY ceramide was found to redistribute into the ER during mitosis (Axelsson and Warren, 2004), which favors the ER-dependent inheritance model.
A different approach has used rapamycin as an assay to trap Golgi proteins modified with FK506-binding protein (FKBP) with ER proteins tagged with FKBP-rapamycin-associated-protein in the ER during mitosis (Pecot and Malhotra, 2004). Results using this assay revealed that when mitotic cells expressing the modified Golgi and ER proteins were exposed to rapamycin, the Golgi proteins were not trapped in the ER, whereas in cells exposed to BFA treatment and washout the Golgi proteins became trapped in the ER. This suggests that Golgi and ER proteins normally do not mix during mitosis. This interpretation, however, is not without difficulties. Mitotic cells are in the stage of mitosis where Golgi and ER enzymes are proposed to fully mix (i.e., metaphase) for only ∼10 min (Zaal et al., 1999), so FKBP-Golgi proteins may not have been in the ER long enough for them to complex with ER proteins so that rapamycin could rapidly stabilize them. This would explain why ER entrapment of Golgi proteins in rapamycin-treated cells was observed only when cells were treated for 60 min with BFA (which induces complete mixing of Golgi and ER proteins) prior to rapamycin treatment. It further would explain why no ER entrapment of Golgi proteins was observed in cells treated with rapamycin for 90 minutes alone (Pecot and Malhotra, 2004), even though other treatments that block ER export or cause Golgi proteins to misfold in the ER lead to near complete ER entrapment of Golgi proteins within 60 minutes, because of constitutive cycling of these proteins through the ER (Cole et al., 1998; Storrie et al., 1998; Zaal et al., 1999; Ward et al., 2001).
To further address these issues, we have used fluorescence and electron microscopy combined with biophysical techniques. This has allowed us to investigate the identity of the membranes that Golgi proteins reside and circulate through during mitosis and thereby to test key predictions of the autonomous and ER-dependent models of Golgi inheritance. We first examine the kinetic and geographic properties of mitotic fragments seen in prometaphase and telophase, examining whether these structures exchange their content with surrounding nonfragment fluorescence and whether they localize at ER export domains. We next examine the fate of Sar1 and Arf1, two key regulatory GTPases whose activity is essential for maintenance of Golgi structure. Finally, we use a variety of new assays to characterize the mitotic haze seen in metaphase. Our results support the ER-dependent model of Golgi inheritance, in which the membrane absorption and export activities of the ER play an essential role in the disassembly/reassembly cycle of the Golgi during mitosis.
MATERIALS AND METHODS
Plasmids, Cell Synchronization, and Reagents
GalT-CFP, GalT-YFP, Arf1-GFP, Arf1-CFP, Sec13-YFP, and ts045VSVG-YFP have been described previously (Sciaky et al., 1997; Polishchuk et al., 2004; Ward et al., 2001). Signal sequence-RFP-KDEL construct includes the bovine preprolactin signal sequence (first 30 aa) cloned into the Nhe1/Pst1 sites of Clontech (Mountain View, CA) pEGFP-C1 vector (this removes the green fluorescent protein [GFP]). 5′ Sal1 and 3′ BamH1 sites were amplified by PCR onto the monomeric red fluorescent protein (mRFP). The mRFP stop codon was replaced with AAAGATGAGCTCTAAGGATCC to add a KDEL ER retention signal, followed by a stop codon and a BamH1 sequence. The entire construct was inserted into the Sal1 and BamH1 sites of the prolactin signal sequence-containing vector. The signal sequence and mRFP are linked by the sequence TGC AGT CGA CTT (encoding Cys-Ser-Arg-Leu). Anti-GFP antibodies were purchased from Molecular Probes (Eugene, OR). Anti-galactosyltransferase antibodies were a kind gift from E. Berger (University of Zurich, Zurich, Switzerland), anti-mannosidase II (Man II) antibodies were a kind gift from K. W. Moremen (University of Georgia, Athens, GA), anti-sec23 antibodies were purchased from Affinity Bioreagents (Golden, CO), and anti-Sec61β antibodies were a kind gift from Ramanujan Hegde (National Institutes of Health, Bethesda, MD). All secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Nocodazole (Sigma-Aldrich, St. Louis, MO) was used at 5 μg/ml.
Transfection
All transfections were done using FuGENE 6 reagent (Roche Diagnostics, Indianapolis, IN) according to manufacturer's instructions.
Cell Synchronization and Mitotic Stage Identification
Cells were synchronized using 10 μg/ml aphidicolin as described previously (Altan-Bonnet et al., 2003; Pecot and Malhotra, 2004). Cells in interphase were identified as having uncondensed chromosomes and an intact nuclear envelope. Prophase cells were defined as having condensed chromosomes and an intact nuclear envelope. Prometaphase cells were identified as having condensed chromosomes and no nuclear envelope. Metaphase cells were defined as having condensed chromosomes aligned at the metaphase plate and no nuclear envelope. Telophase cells were identified as having condensed chromosomes pulled to opposite spindle poles and having no nuclear envelope. Cells in cytokinesis were identified as having condensed chromosomes at opposite spindle poles, invagination of the cytokinetic furrow, and a nuclear envelope reforming around each chromosomal mass. The extent of chromosome condensation/congression in cells was assessed by phase contrast light microscopy, by coexpression of histone 2B-YFP/CFP, and/or by staining with the DNA dye Hoechst 33342 (Altan-Bonnet et al., 2003). The status of nuclear envelope assembly/disassembly in cells was determined by phase contrast light microscopy.
Confocal Imaging, Temperature Control, Photobleaching Experiments, and Quantification
All images were obtained with a Zeiss LSM 510 confocal microscope. Live cells were held at 37°C on the microscope stage using a Nevtek air blower. High spatial resolution images were collected with a 63×/1.4 numerical aperture (N.A.) Plan Apochromat oil objective (Carl Zeiss, Jena, Germany), ∼1.2 Airy unit pinhole aperture, and 16 line averaging. For quantitative imaging (i.e., measuring fluorescence recovery after photobleaching), a 40×/1.3 N.A oil objective or a 40×/1.2 N.A. water objective was used with the pinhole fully open (∼14 Airy units) to collect fluorescence from the entire depth of the cell. All images were collected on a 12-bit PMT (Carl Zeiss, Jena, Germany). No saturated images were used for quantification.
For ts045VSVG experiments, the temperature of the cells on the microscope stage was kept at 40 or 32°C by a combination of heating the stage and microscope turret from the side with a Nevtek air blower; heating the objective with a heat collar; and heating the cell chamber and stage together with using a cover through which water (40 or 32°C) flowed. Throughout the course of the experiments, thermosensors placed in the media of the cells, and the stage monitored the stability of the temperature.
In photobleaching experiments, a region of interest was selected using Zeiss LSM510 software (Carl Zeiss) algorithms. A short, intense laser pulse was delivered to the region of interest to bleach the fluorescence within that region. Images were subsequently collected with lower intensity laser settings.
The number of ER exit sites in a mitotic cell was calculated by counting the number of sec13-yellow fluorescent protein (YFP)-labeled spots whose fluorescence intensity exceeded that of the background cytoplasmic fluorescence as measured by the thresholding algorithm of Zeiss LSM510 software. This was performed from three-dimensional reconstructions of z-sectioned (0.5-μm slice thickness) mitotic cells for every stage of mitosis (n = 20 cells). To quantify the number of Arf1-cyan fluorescent protein (CFP) molecules on the Golgi complex and in the cytoplasm for each stage of mitosis (n = 10 cells), we used a previously reported image processing scheme (Altan-Bonnet et al., 2003) in which cells were cotransfected with both Arf1-CFP and GalT-YFP to identify the Arf1 pool on the Golgi membranes. A simple image processing method involving two regions of interest (ROIs) were used to quantify Golgi and non-Golgi pools of Arf1-CFP and GalT-YFP. One ROI was drawn around GalT-YFP-labeled structures and fragments (representing Golgi labeling), and the other ROI was drawn around the rest of the cell (representing non-Golgi labeling). During each stage of mitosis, the mean fluorescence intensity associated with the Golgi and non-Golgi ROIs were measured for both Arf1-CFP and GalT-YFP. The total Arf1-CFP fluorescence associated with the Golgi was expressed as a fraction of the total cellular fluorescence (i.e., the sum of the Golgi and non-Golgi contributions) and normalized to the initial interphase value.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 10 min at room temperature. After washes in phosphate-buffered saline (PBS) and block in PBS/5% fetal bovine serum (10 min), they were incubated in primary antibody solution in PBS/5% fetal bovine serum/0.2% saponin for 1 h. Postwash in PBS/5% fetal bovine serum, they were incubated with secondary antibody solution in PBS/5% fetal bovine serum/0.2%saponin for 1 h. They were then rinsed in PBS and mounted for microscopy.
Electron Microscopy
Man II-horseradish peroxidase (HRP) expressing mitotic normal rat kidney (NRK) cells were shaken off the dishes and pelleted at 1000 rpm for 5 min (Nizak et al., 2004). They were then were fixed in 4% paraformaldehyde and 0.5% glutaraldehyde for 4 h at room temperature and incubated 35 min with diaminobenzidine tetrahydrochloride (0.25 mg/ml; Sigma-Aldrich) and 0.003% H2O2 (Sigma-Aldrich) in phosphate-buffered solution at pH 7.4. Cells were then incubated in 2.5% glutaraldehyde buffer in sodium cacodylate (0.1 M) at pH 7.4 and placed at 4°C for at least 24 h. Cells were washed by several changes of 0.1 M sodium cacodylate buffer and postfixed 1 h at room temperature in reduced osmium (1:1 mixture of 2% aqueous osmium tetroxide and 3% aqueous potassium ferrocyanide, according to the Karnovsky procedure). After postfixation, cells were pre embedded in agar (2%), dehydrated in ethanol, and processed for Epon embedding. Thin (80-nm) sections were cut and collected on copper grid and stained with lead citrate (2 min).
For conventional transmission electron microscopy, cells were fixed overnight at 4°C in 2.5% glutaraldehyde in sodium cacodylate buffer. Cells were then rinsed and postfixed 1 h at room temperature in reduced osmium (1:1 mixture of 2% aqueous potassium ferrocyanide) as described previously by Karnovsky (1971). After postfixation, the cells were processed as described for Man II-HRP. Sections were then examined with a CM 10 Philips electron microscope at 80 kV.
RESULTS
Characterization of Fluorescent Golgi Marker
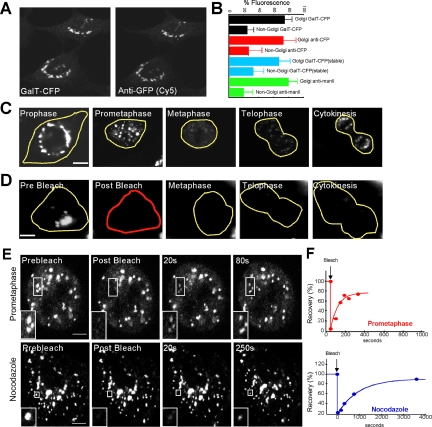
As Golgi marker, we used an Y/CFP-tagged version of the N-linked glycoprotein-processing enzyme galactosyltransferase (GalT) that distributes throughout the Golgi stack (Sciaky et al., 1997). In creating this construct (denoted GalT-Y/CFP), GalT's catalytic domain was substituted with a single Y/CFP, a modification previously shown not to affect the enzyme's distribution within the Golgi (Sciaky et al., 1997; Polishchuk et al., 2004) or its response to pharmacological treatments affecting the Golgi (Ward et al., 2001; Altan-Bonnet et al., 2003; Polishchuk et al., 2004). In cells transiently expressing GalT-CFP, the construct was localized primarily to perinuclear Golgi structures, with identical labeling seen when observed either by CFP fluorescence or by Cy5-labeled anti-GFP immunofluorescence (Figure 1A). Dim, non-Golgi fluorescence was also observed, which probably corresponds to GalT-CFP molecules in the ER, the site where Golgi enzymes are synthesized and through which the enzymes constitutively recycle (Cole et al., 1996, 1998; Storrie et al., 1998; Miles et al., 2001; Ward et al., 2001). When quantified, the non-Golgi fluorescence represented roughly 20% of total cellular fluorescence based on measurements in cells transiently expressing GalT-CFP at a range of expression levels (i.e., from 1 to 10 times the lowest level) (Figure 1B, black bars). Comparable non-Golgi labeling was detected by anti-GFP antibody (Figure 1B, red bars). Non-Golgi fluorescence representing ∼20% of total cellular fluorescence (with Golgi labeling representing 80%) was also observed in a stable cell line expressing GalT-CFP (Figure 1B, blue bars), indicating that the sizes of the Golgi and non-Golgi pools of GalT-CFP were not related to the mode of GalT-CFP expression in cells. An endogenous Golgi enzyme, mannosidase II, when labeled using antibodies and quantified, likewise showed two pools (i.e., Golgi and non-Golgi) with the non-Golgi pool representing ∼20% and the Golgi pool representing ∼80% of total cellular labeling (Figure 1B, green bars). Moreover similar sizes of Golgi and non-Golgi pools of mannosidase II and GalT-CFP were also observed when they were quantified within the same cell (Supplemental Figure S1). This indicated that the distribution of GalT-CFP in Golgi and non-Golgi pools was not unique but was characteristic of endogenous Golgi enzymes.
Figure 1.
Golgi proteins distribute between Golgi and non-Golgi membranes in interphase and rapidly cycle in and out of mitotic fragments during prometaphase. (A) NRK cells transiently expressing GalT-CFP were fixed and immunostained with a primary antibody against GFP and a Cy5-tagged secondary antibody. Left, distribution of CFP fluorescence. Right, Cy5 fluorescence in the same cell. Note the two labeling patterns are identical. (B) Golgi-associated and non-Golgi-associated fluorescence were quantified in individual cells by measuring the mean fluorescence per pixel within each region, subtracting background noise and multiplying by the total area of each region. The plot shows the relative sizes of the two pools for cells transiently expressing GalT-CFP (n = 10; black bars), cells immunostained with antibodies to CFP- and Cy5-labeled secondary antibodies (n = 20; red bars), cells stably expressing GalT-CFP (n = 10; blue bars), and cells that were fixed and immunostained with antibodies against native mannosidase II (n = 20; green bars). (C) Time-lapse images of a single NRK cell transiently expressing GalT-YFP progressing through mitosis. (D) The prebleach image shows an NRK cell transiently expressing GalT-YFP entering mitosis. The entire cell was photobleached in the outlined red area, and cell fluorescence was monitored. Postbleach images at representative stages of mitosis are shown. Fluorescence was monitored with a wide pinhole setting (14 Airy units with a 40×/1.3 N.A. objective) to collect fluorescence from the entire depth of the cell over the course of mitosis. Note no significant fluorescence reappeared as the cell progressed through to cytokinesis, suggesting that newly synthesized GalT-YFP and newly folded YFP did not contribute significant fluorescence to the cells during mitosis. (E) The prebleach image shows an NRK cell expressing GalT-YFP in prometaphase (top) or after treatment with nocodazole for 2 h (bottom). A region of interest containing Golgi fragments (outlined in white) was photobleached. Postbleach images collected at representative time points with a wide pinhole confocal setting (40×/1.3 N.A.; ∼14 Airy units) are shown. The insets show the photobleached region of interest at high magnification. (F) Fluorescent intensity of an individual prometaphase fragment (top) or nocodazole fragment (bottom) was measured at 15-s time intervals in a similar experiment as in G and plotted as a percentage of initial prebleach fluorescence in the fragment (after subtraction of background fluorescence). Recovery of fluorescence into the bleached prometaphase fragment occurred more rapidly than into the bleached nocodazole fragment. Bars, 5 μm.
The Golgi and non-Golgi fluorescent pools of GalT-C/YFP seemed to be maintained through constitutive ER-Golgi cycling pathways because photobleaching Golgi fluorescence, leaving non-Golgi fluorescence intact, in cycloheximide-treated cells resulted in Golgi and non-Golgi fluorescence returning within 2 h to similar percentages of total cellular fluorescence as observed before bleaching (i.e., Golgi 80% and non-Golgi 20%) (our unpublished data) as reported previously (Ward et al., 2001). This suggested that GalT-C/YFP was not retained in the Golgi but could cycle between Golgi and non-Golgi locations.
Only Preexisting Pools of GalT-YFP Contribute to the Observed Changes in Golgi Protein Localization during Mitosis
We imaged single cells expressing GalT-YFP progressing through mitosis (Figure 1C) and found that GalT-YFP-labeled Golgi membranes followed the same sequence of mitotic Golgi disassembly and reassembly as observed using antibodies to the endogenous Golgi enzyme mannosidase II (Supplemental Figure S2), or other Golgi markers in mammalian cells (Lucocq et al., 1987, 1988; Shima et al., 1998; Zaal et al., 1999; Altan-Bonnet et al., 2003). Early during prophase, the Golgi ribbon marked by GalT-YFP redistributed from its original perinuclear position to numerous large fragments surrounding the cell nucleus (Figure 1C, prophase). As prophase progressed into prometaphase, smaller Golgi fragments then occurred throughout the cell (Figure 1C, prometaphase). These fragments were relatively stationary in the cytoplasm (suggesting they are associated with some underlying structure) (Figure 1E), and persisted until metaphase when many of them disappeared and became replaced by mitotic Golgi haze dispersed uniformly across the cell (Figure 1C, metaphase). In telophase, Golgi fragments first reappeared throughout the cell and then coalesced into centralized Golgi structures during cytokinesis (Figure 1C, telophase and cytokinesis).
Given the observed changes in GalT-YFP distribution during mitosis, we asked whether they reflected only the behavior of preexisting pools of GalT-YFP or whether they required newly synthesized and/or folded molecules. This question was important because previous studies have suggested that GFP-tagged Golgi enzymes observed in mitotic Golgi haze and/or fragments might be partially derived from newly synthesized and/or folded pools of these proteins in the ER (Jokitalo et al., 2001; Prescott et al., 2001). To address this issue, we photobleached all GalT-YFP fluorescence within a cell in early prophase and then examined whether any new fluorescence reappeared in the cell as it progressed through mitosis (Figure 1D). Because no significant fluorescence reappeared, we concluded that the changes in GalT-YFP distribution observed during mitosis reflected the movements of preexisting, fluorescent GalT-YFP molecules in the cell.
Golgi Enzymes Rapidly Cycle In and Out of Prometaphase Fragments
We next investigated using a photobleaching approach whether Golgi enzymes within prometaphase fragments were capable of moving in and out of these structures. In the autonomous model, prometaphase fragments function as autonomous structures, so their content of Golgi enzymes should be retained until lost through vesicle shedding. On photobleaching an individual fragment, therefore, no significant fluorescence recovery into the Golgi fragment is predicted. In the ER-dependent model, on the other hand, the contents of Golgi fragments are continually being received from and recycled back to the ER. Hence, upon photobleaching an individual fragment, fluorescence in the fragment is predicted to recover.
When we photobleached an individual mitotic Golgi fragment in a prometaphase cell and monitored fluorescence recovery into the fragment, we observed rapid recovery of fluorescence (Figure 1E). When quantified, the results revealed that within 2 min after photobleaching, the majority of the fragment's prebleach fluorescence was restored (Figure 1F). As other prometaphase fragments maintained their fluorescence intensity over this time period (and did not show a similar increase in fluorescence intensity as observed in the photobleached fragment), the observed recovery into the photobleached fragment represented a replacement of bleached molecules moving out of the fragment by fluorescent molecules moving into it. Hence, Golgi proteins, such as GalT-YFP, do not remain in a prometaphase fragment for any significant time period but are continuously entering and leaving fragments on a rapid time scale.
Enzyme Cycling In and Out of Prometaphase Fragments Compared with That of Nocodazole Fragments
Golgi membrane proteins in interphase cells continuously enter and leave Golgi structures as they cycle constitutively through the ER (Cole et al., 1998; Storrie et al., 1998; Miles et al., 2001; Ward et al., 2001). We aimed, therefore, to compare the rate of this cycling with that of Golgi proteins cycling in and out of prometaphase Golgi fragments in mitotic cells. To accomplish this, we first depolymerized microtubules with nocodazole in cells expressing GalT-YFP, which inhibits transport of pre-Golgi transport intermediates toward the centrosome, causing the existing perinuclear Golgi ribbon to shrink while new Golgi fragments form near ER export domains because of constitutive recycling of Golgi proteins through the ER (Cole et al., 1996; Storrie et al., 1998; Drecktrah and Brown, 1999). We then photobleached a single Golgi fragment in these cells and measured the rate of fluorescence recovery (Figure 1, E and F). Near complete recovery of fluorescence into the nocodazole fragment occurred, but the rate (t1/2 of ∼65 min) was significantly slower than that of a prometaphase fragment (t1/2 of ∼2 min). This indicated, therefore, that enzyme cycling in and out of Golgi structures is accelerated in prometaphase cells.
Prometaphase Fragments Localize Near ER Export Domains as Tubule Clusters
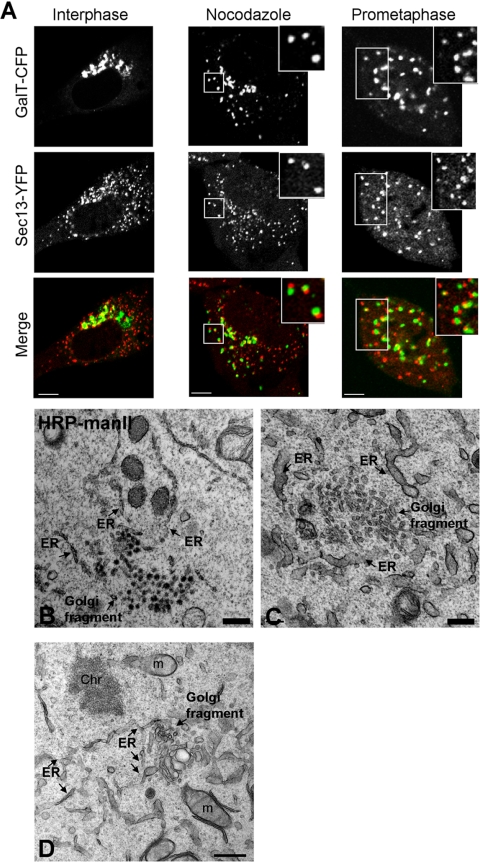
To determine whether Golgi enzyme cycling in and out of prometaphase Golgi fragments took place in areas near ER export domains, as occurs in nocodazole-treated cells (Cole et al., 1996; Hammond and Glick, 2000; Storrie et al., 1998), we compared the spatial arrangement of prometaphase Golgi fragments with that of ER export domains. As reporter for ER export domains, we used the COPII protein Sec13 (Barlowe et al., 1994) tagged with YFP (Sec13-YFP) (Hammond and Glick, 2000; Ward et al., 2001). To confirm that Sec13-YFP correctly targeted to ER export domains, we compared its distribution to that of nocodazole Golgi fragments labeled with GalT-CFP. After a 2-h treatment of interphase cells with nocodazole to generate Golgi fragments, each GalT-CFP-containing fragment was found to be positioned adjacent to an ER export domain marked by Sec13-YFP staining (Figure 2A, nocodazole). This phenotype was also observed when Sec13-YFP-expressing cells were stained with antibodies against native mannosidase II (Supplemental Figure S3). As a further control for Sec13-YFP being an accurate reporter of ER exit sites, we stained cells expressing Sec13-YFP with antibodies to a different COPII component, Sec23p (Fu and Sztul, 2003). Both Sec13-YFP and the anti-Sec23p antibodies labeled the same structures (Supplemental Figure S4), confirming that Sec13-YFP labeled ER export sites. We also compared the overall number and organization of ER exit sites detected with anti-Sec23p antibodies in cells either expressing Sec13-YFP or not (Supplemental Figure S4). No difference in the pattern of ER exit sites was observed, indicating that expression of Sec13-YFP did not perturb ER exit sites.
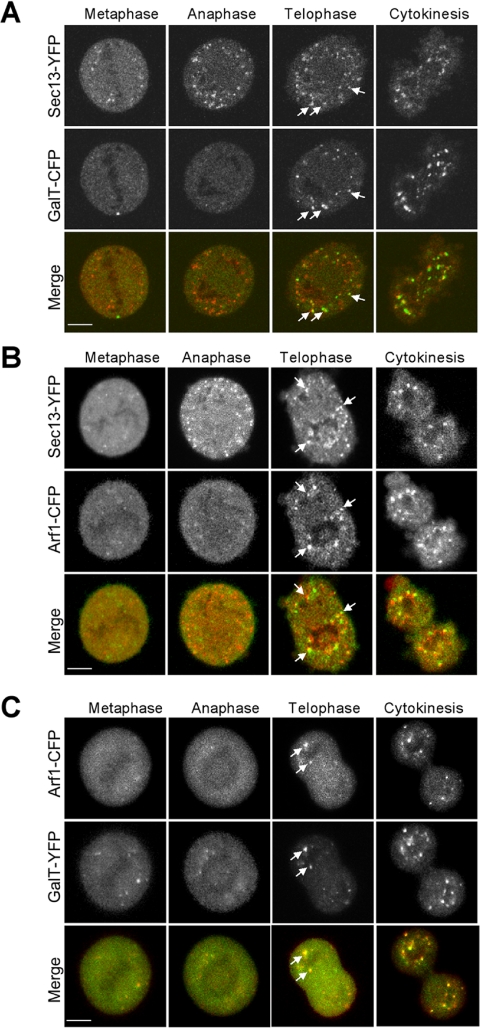
Figure 2.
Prometaphase Golgi fragments localize near ER exit sites. (A) NRK cells stably expressing Sec13-YFP were cotransfected with GalT-CFP and the distribution of the two markers was then imaged using a narrow pinhole confocal setting. Shown are images of a cell in interphase (left), a cell treated with nocodazole (middle), and a cell in prometaphase (right). Note the localization of prometaphase and nocodazole Golgi fragments (labeled with GalT-CFP) near to ER exit sites (labeled with Sec13-YFP). The area in the white box is enlarged in the inset. Bar, 5 μm. (B) NRK cells expressing HRP-mannosidase II in prometaphase were fixed, processed for immunoperoxidase labeling, and prepared for electron microscopy. Images of mitotic Golgi fragments in these cells revealed HRP labeling of vesiculotubular clusters in proximity to ER, with no HRP-positive structures (i.e., vesicles) observed in areas outside the vesiculoubular clusters. (C) NRK cells in prometaphase were fixed and examined by electron microscopy without HRP labeling. Mitotic vesiculoubular clusters adjacent to ER tubules (arrows) are shown. (D) Image of prometaphase cell prepared as in C showing vesiculotubular clusters and their intimate association with ER (arrows). Ch, chromosome; m, mitochondria. Bar, 0.5 μm in electron micrographs.
To determine whether Golgi proteins accumulate next to ER exit sites during mitosis, we examined the distribution of GalT-CFP and Sec13-YFP as cells progressed through mitosis. In prometaphase, a significant coalignment of GalT-CFP-containing prometaphase fragments and Sec13-YFP-labeled structures was observed (Figure 2A), suggesting that prometaphase fragments, like nocodazole fragments, are formed near ER export domains. Quantification of three-dimensional reconstructions through six cells demonstrated that 97% ± 3% of all prometaphase fragments were adjacent to ER export domains. We repeated our study using antibodies to native mannosidase II in NRK cells stably expressing Sec13-YFP, or in NRK cells costained with antibodies to mannosidase II and Sec23p. The Golgi-ER export site alignment was less obvious with the endogenous marker Sec23p than with Sec13-YFP, possibly because of the ER exit sites being more clustered in cells overexpressing Sec13-YFP, which makes it easier to assign colocalization (Supplemental Figure S4). Nevertheless, each Golgi fragment had one or more ER export site closely associated with it.
As a further test for the proximity of prometaphase fragments with ER export domains, we examined prometaphase cells using electron microscopy. Prometaphase fragments were identified using immunoperoxidase staining to label the Golgi enzyme mannosidase II and were found to be tubulovesicular clusters in close association with the ER (Figure 2B). This observation was in agreement with previous results of Lucocq et al. (1987, 1988, 1989), in which mitotic fragments viewed by electron microscopy were also always near smooth ER-resembling ER export domains. In cells whose morphology was better preserved by omitting peroxidase labeling, ER membranes seemed to be giving rise to the prometaphase, tubulovesicular clusters (Figure 2, C and D). Because no small circular membrane profiles positive for immunoperoxidase staining were seen outside a prometaphase cluster, the tubule clusters did not seem to be giving rise to vesicles that moved out into the cytosol (Figure 2B).
Together, these results suggested that prometaphase Golgi fragments were comprised of clusters of membrane tubules and vesicles in close association with ER export domains. Because nocodazole Golgi fragments also reside near ER export domains, the results supported the view that prometaphase fragments share some of the properties of nocodazole-induced Golgi reorganization. One difference between nocodazole Golgi fragments and prometaphase fragments is that the former exist as mini-stacks at ER exit sites (Drecktrah and Brown, 1999), whereas the later exist as tubulovesicular clusters at these sites. This implies that prometaphase cells lack the mechanism(s) responsible for remodeling Golgi membranes into stacks at ER export domains. This could explain the second difference we observed between nocodazole and prometaphase Golgi fragments, which is the difference in cycling rates of Golgi enzymes into and out of these fragments.
Behavior of Arf1 and Sar1 during Prometaphase Fragment Generation
In interphase cells, the GTPase activities of Sar1 and Arf1 have been shown to play important roles in regulating ER export domain morphology and membrane cycling between ER and Golgi membranes (Altan-Bonnet et al., 2004). For example, when Arf1 is inactive and cannot bind to membranes, Golgi membrane proteins redistribute into the ER or to ER export domains and can undergo rapid exchange between these sites (Ward et al., 2001; Puri and Linstedt, 2003). Likewise, when Sar1 is inactive and cannot bind to membranes, ER export domains disappear, and Golgi membrane proteins redistribute completely into the ER (Miles et al., 2001; Ward et al., 2001; Puri and Lindstedt, 2003). The roles of Sar1 and Arf1 in Golgi maintenance in interphase cells raised the possibility that alterations in their activities in mitotic cells might contribute to the morphological changes to the Golgi occurring during mitosis.
To explore this possibility, we visualized Arf1-CFP and Sec13-YFP in cells progressing through mitosis. Because Arf1 exists on Golgi membranes in a GTP-bound active state and in the cytosol as a GDP-bound, inactive form (Goldberg, 1998), any redistribution of Arf1-CFP off membranes during mitosis indicates that Arf1 GTPase activity has decreased. Likewise, because Sec13 requires Sar1-GTPase activity to bind to ER export domains (Barlowe et al., 1994; Miles et al., 2001; Ward et al., 2001; Lee et al., 2004), any redistribution of Sec13-YFP off ER export domains during mitosis suggests that Sar1-GTPase activity has decreased.
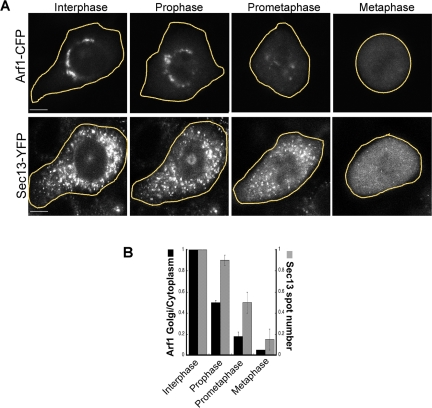
In cells expressing Arf1-CFP or Sec13-YFP, Arf1-CFP's pool on the Golgi decreased and pool in the cytoplasm increased in prophase before any significant change in Sec13-YFP labeling of ER exit sites occurred (Figure 3, A and B, prophase). By prometaphase, Golgi-associated Arf1-CFP (i.e., on prometaphase fragments) decreased even further, to 20% of interphase levels. Because Sec13-YFP-positive structures had only declined to 50% at this stage of mitosis, the data suggested that before complete ER exit site disassembly in mitosis (Farmaki et al., 1999; Prescott et al., 2001; Stephens, 2003), Arf1 had largely redistributed into the cytoplasm and become inactive. At metaphase, Arf1-CFP and Sec13-YFP were both almost entirely cytoplasmic, as reported previously using techniques that included fluorescence correlation spectroscopy (Altan-Bonnet et al., 2003) and biochemical fractionation and light microscopy (Hammond and Glick, 2000; Prescott et al., 2001; Stephens, 2003). This suggested that during metaphase, Arf1 is inactive and ER exit sites are disassembled.
Figure 3.
Arf1 and Sec13 sequentially redistribute into the cytosol as cells progress from prophase to metaphase. (A) NRK cells expressing Arf1-CFP and Sec13-YFP were imaged as they progressed from interphase through mitosis. A wide-open pinhole confocal setting (14 Airy units with a 40×/1.3 N.A. objective) was used to collect fluorescence from the entire cell depth. Selected images during interphase, prophase, prometaphase, and metaphase are shown. Note that at prometaphase very little Arf1-CFP was associated with subcellular structures, whereas a large fraction of Sec13-YFP was associated with ER exit sites. This indicated that during mitosis Arf1-CFP redistributed into the cytosol sooner than Sec13-YFP. Bars, 5 μm. (B) Time-lapse images of individual NRK cells coexpressing Arf1-CFP and GalT-YFP (GalT-YFP was used to identify the Golgi membrane) (n = 10 cells) or expressing Sec13-YFP alone (n = 20) were collected as the cells progressed through mitosis. The fluorescence from Arf1-CFP (black bars) and Sec13-YFP (gray bars) at different stages of mitosis were quantified (described in Materials and Methods) and plotted in a histogram. The data showed that Arf1-CFP was lost from Golgi membranes before ER export domains labeled with Sec13-YFP disappeared.
These data were consistent with the possibility that decreased Arf1 activity on Golgi membranes contributes to the kinetic properties of prometaphase fragments (i.e., their more rapid exchange of membrane content, and tubule cluster rather than stacked morphology). The following speculative scenario, therefore, could be responsible for prometaphase fragment generation. As cells progress from prophase to prometaphase, decreased Arf1 activity (and its downstream effects) would disrupt Golgi stack formation and lead to accelerated retrograde traffic of Golgi proteins; microtubule depolymerization during this period combined with continued Sar1 activity would then lead to cycling Golgi proteins accumulating near ER export domains as tubulovesicular clusters.
Characteristics of Telophase Golgi Fragments
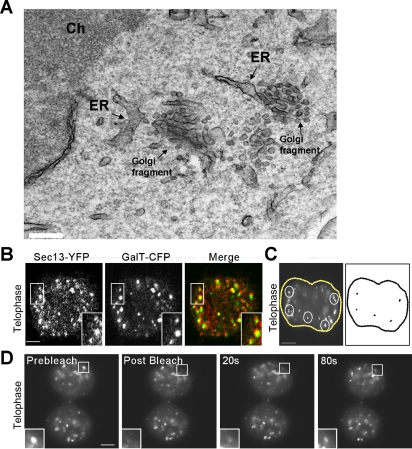
We next studied the properties of telophase Golgi fragments, which have been shown to emerge from the mitotic Golgi haze of metaphase (Shima et al., 1998; Zaal et al., 1999; Altan-Bonnet et al., 2003; Axelsson and Warren, 2004; Pecot and Malhotra, 2004). Electron microscopy of cells in telophase revealed that the telophase fragments closely resembled prometaphase fragments in that they occurred as clusters of tubules located adjacent to and in some cases occurring as emerging from the ER (Figure 4A). The close association with ER exit sites was confirmed at the light microscopy level, in which virtually all GalT-CFP-containing fragments were positioned close to sites labeled with Sec13-YFP (Figure 4B). Time-lapse movies tracking the movement of these fragments revealed that before microtubule repolymerization in late telophase, the fragments were relatively immobile within cells (Figure 4C), as found for ER exit sites (Hammond and Glick, 2000; Stephens, 2003). Furthermore, upon photobleaching an individual fragment containing GalT-YFP, there was rapid recovery of GalT-YFP fluorescence back into the fragment (Figure 4D). This indicated that Golgi enzymes reside in telophase fragments only briefly before cycling out of these structures to surrounding membranes (such as ER). Hence, telophase Golgi fragments exhibited morphological and kinetic properties that are similar to prometaphase Golgi fragments.
Figure 4.
Characterization of mitotic Golgi fragments seen in telophase. (A) Electron microscopy of mitotic Golgi fragments observed during telophase. Note the proximity of mitotic vesiculotubular clusters to ER. Ch, chromosome. Bar, 0.2 μm. (B) Mitotic NRK cells coexpressing GalT-CFP and Sec13-YFP were imaged during telophase using a narrow pinhole confocal setting. The area in the white box is enlarged in the inset. Note the coalignment of telophase Golgi fragments labeled with GalT-CFP and ER export domains labeled with Sec13-YFP. Bar, 5 μm. (C) An NRK cell expressing GalT-CFP in telophase was imaged every 1.2 s over 3 min using a wide pinhole confocal setting, and the images were merged into a single file (left). The trajectories of five fragments (outlined in white) were analyzed using particle track algorithm of NIH Image software and are plotted in black (right). Bar, 5 μm. (D) A region of interest containing a fragment (outlined in white) was selectively photobleached in a telophase cell expressing GalT-YFP. Images were collected every 20 s with a wide pinhole confocal settings. Note the rapid recovery of fluorescence (within 80 s) into the photobleached region of interest. Bar, 5 μm.
To determine the sequence of telophase Golgi fragment formation relative to ER exit site biogenesis at the end of mitosis, we performed dual time-lapse imaging of cells coexpressing GalT-CFP and Sec13-YFP as they progressed from metaphase through cytokinesis (Figure 5A and Supplemental Figure S5 movie). As early as late metaphase, Sec13-YFP was seen reassociating with punctate structures in the cytoplasm. The reassociation continued through telophase, when the pool of membrane-bound Sec13-YFP reached the steady-state level observed in interphase. There was minimal GalT-CFP labeling of punctate structures until telophase, and the labeled structures all were localized adjacent to puncta containing Sec13-YFP (Figure 5A and Supplemental Figure S5 movie). Dual time-lapse imaging of cells expressing Arf1-CFP and Sec13-YFP (Figure 5B) revealed that Arf1-CFP also reassociated with membranes after Sec13-YFP. Notably, Arf1-CFP membrane reassociation coincided with telophase Golgi fragment generation (containing GalT-YFP) near ER exit sites (Figure 5, B and C), suggesting the membrane reassociation of Arf1 was related to GalT-CFP emergence from ER. By cytokinesis, Sec13-YFP, Arf1-CFP, and GalT-YFP all were located in punctate structures and many of these had an overlapping pattern of distribution (Figure 5, A and C).
Figure 5.
Dynamics of GalT-CFP, Sec13-YFP, and Arf1-CFP during exit from mitosis. (A) A single NRK cell stably expressing Sec13-YFP and cotransfected with GalT-CFP progressing through mitosis was imaged with a wide pinhole confocal setting. Representative images of the cell in metaphase, anaphase, telophase, and cytokinesis are shown. Arrows point to the telophase fragments containing both Sec13-YFP and GalT-CFP labeling. Note that Sec13-YFP labeling preceded GalT-CFP labeling of the fragments (Supplemental Figure S5 movie). (B) Representative confocal images of metaphase, anaphase, telophase, and cytokinesis from a single NRK cell stably expressing Sec13-YFP and cotransfected with Arf1-CFP progressing through mitosis are shown. Arrows point to telophase fragments containing both Sec13-YFP and Arf1-CFP. Note that Sec13-YFP reassociated with the punctate fragments before Arf1-CFP. (C) Representative confocal images of metaphase, anaphase, telophase, and cytokinesis from a single NRK cell cotransfected with Arf1-CFP and GalT-YFP progressing through mitosis are shown. Arrows point to telophase fragments containing both Arf1-CFP and GalT-YFP. Note that both Arf1-CFP and GalT-YFP reassociated with fragments only during telophase. Bars, 5 μm.
Together, these findings suggested that telophase Golgi fragments grow out from ER export domains at the end of mitosis. This process occurs after ER exit sites are reassembled and coincides with Arf1 recruitment onto the newly forming structures and their import of Golgi enzymes.
Analysis of Mitotic Golgi Haze
Our findings suggesting that Arf1 and Sar1 undergo sequential inactivation during prometaphase fragment formation and sequential reactivation during telophase fragment formation led us to ask whether in metaphase—when Arf1 and Sar1 both seem to be inactive—Golgi proteins reside in ER membranes. This question is relevant because when Arf1 and Sar1 are inactive during interphase, Golgi proteins redistribute into the ER (Storrie et al., 1998; Miles et al., 2001; Ward et al., 2001; Puri and Linstedt, 2003). Although previous work has provided evidence consistent with Golgi proteins redistributing into the ER during metaphase (Thyberg and Moskalewski, 1992; Zaal et al., 1999), more recent pharmacological and morphological studies (Axelsson and Warren, 2004; Pecot and Malhotra, 2004) have argued the contrary based on findings that 1) Golgi proteins do not colocalize with Golgi markers before or after mitotic ER is fragmented with filipin (Axelsson and Warren, 2004), and 2) Golgi proteins capable of forming a complex within the ER in the presence of rapamycin are not trapped in the ER upon addition of rapamycin to mitotic cells (Pecot and Malhotra, 2004). To determine whether this contrary data can be reconciled with the view that Golgi inheritance is mediated through ER export activities, we performed three types of experiments designed to discern whether mitotic haze has properties of ER.
In our first experiment, we examined mitotic Golgi haze by high-resolution confocal microscopy in cells expressing GalT-YFP. We started this experiment by photobleaching all non-Golgi fluorescence before the cell's entry into mitosis (Figure 6A). This ensured that fluorescence within the mitotic haze came only from GalT-YFP fluorescence associated with the Golgi ribbon. Fluorescence from newly synthesized GalT-YFP fluorescence and/or YFP folding was not a factor in these experiments because when an entire cell entering mitosis was photobleached, no fluorescence reappeared over the time period of the experiment (Figure 1D).
Figure 6.
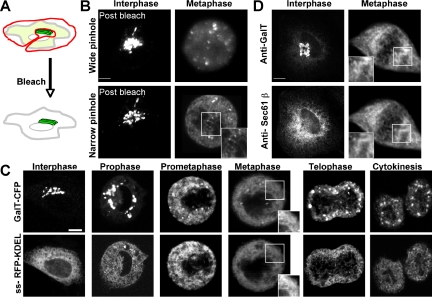
Distributions of Golgi enzymes and ER markers during mitosis. (A) Diagram of bleaching protocol in which the area surrounding the Golgi is selectively photobleached to remove all non-Golgi fluorescence in the cell. The non-Golgi area was photobleached rapidly (∼15 s) with a high-intensity laser beam. (B) A single NRK cell transiently expressing GalT-YFP imaged in interphase and metaphase after the photobleaching protocol described in A. The images were obtained using either a wide pinhole (∼8 Airy units with 63×/1.4 N.A. objective) (top) or narrow pinhole (1.2 Airy units with 63×/1.4 N.A. objective) (bottom). The area in the white box is enlarged to show the gain in resolution of subcellular structures when the pinhole is narrowed. (C) GalT-CFP and ss-RFP-KDEL distributions in an NRK cell undergoing mitosis. Images were obtained using a narrow pinhole (1.2 Airy units with 63×/1.4 N.A. objective). (D) Nonsynchronized HeLa cells were fixed and immunostained with antibodies against native Sec61β and galactosyltransferase. Interphase and metaphase stage cells were selected and imaged on a confocal microscope with a narrow pinhole (1.2 Airy units with 63×/1.4 N.A. objective). Representative images from 10 cells each of interphase and metaphase stage are shown. Bars, 5 μm.
After photobleaching non-Golgi fluorescence in the above-mentioned manner, we performed time-lapse confocal imaging to follow the fate of the fluorescent Golgi pool during mitosis (Figure 6B, postbleach). By metaphase, the fluorescence had redistributed into small punctate structures in addition to a diffuse haze-like pattern when observed using a wide-open confocal aperture (Figure 6B, wide pinhole). To resolve the haze, we imaged the same cell by closing down the confocal aperture to ∼1.2 Airy units (Figure 6B, narrow pinhole). This minimized out-of-focus light and maximized in-focus light collection (Pawley, 1995). Under these microscope settings, the metaphase haze resolved into a fine reticulum that extended throughout the cytoplasm, including the area between the two spindle poles (Figure 6B, inset).
To determine whether the fine reticulum corresponded to ER, we used the optimized imaging settings to compare the distribution of GalT-CFP with ss-RFP-KDEL, (an exclusively lumenal ER marker with no Golgi pool; Supplemental Figure S6) in an NRK cell progressing through mitosis that coexpressed these markers (Figure 6C). In prometaphase as well as metaphase, GalT-CFP was readily seen in reticular elements that significantly overlapped with those labeled by the ER marker. Because the images for GalT-CFP and ss-RFP-KDEL were collected sequentially rather than simultaneously (with each image requiring ∼3-5 s to collect), and membranes within the cell were moving, perfect overlap between the reticular patterns was impossible. Nevertheless, the patterns were very similar (Figure 6C, inset, metaphase). During telophase, the two markers still seemed to be within the same tubular network with parts of the network invading the spindle/chromatin area. The observed codistribution of both markers in tubule elements projecting into the spindle region (Figure 6C, telophase) contrasted with previous light microscopy studies reporting only Golgi proteins localized to this area (Jesch et al., 2001; Axelsson and Warren, 2004). Given the EM and light-level studies demonstrating the presence of ER in the spindle region (Chaldakov and Vankov, 1985; Waterman-Storer et al., 1993; Terasaki, 2000), one possible explanation for why the previous reports (Jesch et al., 2001; Axelsson and Warren, 2004) did not see significant pools of ER markers in the spindle region by light microscopy is that transmembrane proteins were used as markers for the ER rather than a soluble, lumenal ER marker (which was used in our study). If some transmembrane ER markers were confined to the rough ER during mitosis and the rough ER was less enriched in the spindle region, then this might explain why the transmembrane ER markers appeared depleted from the spindle region.
Immunofluorescence experiments using antibodies against native galactosyltransferase and Sec61β, an ER integral protein, gave similar overall results to those found using GalT-CFP and ss-RFP-KDEL. However, the labeling intensity for Sec61β was slightly weaker than that of GalT in the tubular projections within the spindle region (Figure 6D). This is consistent with the previous reports for a differential distribution of rough ER and Golgi transmembrane markers in the spindle region, and suggests that rough ER membrane markers are differentially distributed within the ER of mitotic cells— unlike lumenal ER markers that freely diffuse (such as ss-RFP-KDEL). Whether this arises because of a differential lipid environment in the spindle ER region or because of physical restrictions (i.e., the abundance of microtubules) remains to be investigated.
In our second experiment, we used treatments that perturb ER structure to determine whether they similarly affected the distribution of ER and Golgi markers. To accomplish this, we fragmented mitotic ER by altering calcium levels in cells using the calcium ionophore ionomycin, which has been widely used to fragment the ER and to produce disconnected ER elements (Subramanian and Meyer, 1997; Park et al., 2000; Olah et al., 2001). We then looked to see whether Golgi and ER markers codistributed in the ER fragments and whether these markers became immobilized as a consequence.
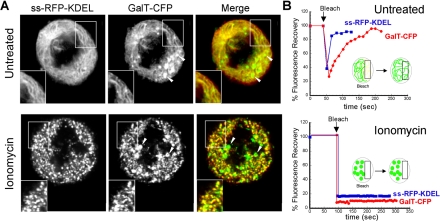
Cells were cotransfected with GalT-CFP and ss-RFP-KDEL to visualize both Golgi and ER markers. Before the cell's entry into mitosis, GalT-CFP fluorescence outside the Golgi region-of-interest was removed by photobleaching. This ensured that only Golgi-derived GalT-CFP was being visualized in these experiments. Cells were then allowed to proceed undisturbed through mitosis until late prometaphase, whereupon ionomycin was added for 10 min to the medium. After treatment, high-resolution images of both GalT-CFP and ss-RFP-KDEL were acquired using a narrow pinhole setting (1.2 Airy units) on the confocal microscope. As expected, the ER became completely fragmented in the ionomycin-treated cells, with ss-RFP-KDEL localized in small structures scattered throughout the cytoplasm (Figure 7A, ionomycin). Furthermore, no recovery of ss-RFP-KDEL fluorescence into a photobleached strip occurred in these cells (Figure 7B, ionomycin) unlike in untreated cells where recovery was rapid (Figure 7B, untreated). This indicated that ionomycin treatment caused ER continuity to be lost. Notably, in virtually all puncta containing ss-RFP-KDEL, a significant amount of GalT-CFP was also detected (>85%, n = 3 cells), and no diffusely distributed GalT-CFP (i.e., mitotic Golgi haze) remained (Figure 7A). GalT-CFP fluorescence did not recover into puncta when they were photobleached (Figure, 7B, ionomycin), in contrast to full recovery of GalT-CFP in untreated cells at this stage of mitosis (Figure 7B, untreated). A proportion of GalT-CFP was also found in fragments distinct from those containing ss-RFP-KDEL (Figure 7A, ionomycin, arrowheads). These fragments likely correspond to Golgi fragments seen in untreated prometaphase cells (Figure 7A, untreated, arrowheads). Given that mitotic Golgi haze disappears and its fluorescence redistributes into fragments containing ER proteins upon ER fragmentation by ionomycin treatment, the data supported the view that mitotic Golgi haze represents ER rather than dispersed Golgi vesicles.
Figure 7.
Golgi and ER markers codistribute when the ER is fragmented with ionomycin. (A) HeLa cells coexpressing GalT-CFP and ss-RFP-KDEL entering mitosis were identified and the non-Golgi pool of GalT-CFP fluorescence was photobleached (as shown in Figure 6A). Once the cells had entered late prometaphase/metaphase, they were incubated with or without 10 μg/ml ionomycin for ∼10 min. The distribution of GalT-CFP and ss-RFP-KDEL was then imaged using a narrow pinhole confocal setting (1.2 Airy units with 63×/1.4 N.A. objective). The area in the white box is enlarged to show the coalignment of fragments containing GalT-CFP and ss-RFP-KDEL. Arrowheads point to mitotic fragments that contain GalT-CFP but not ss-RFP-KDEL (which are seen in both treated and untreated cells). (B) Cells coexpressing GalT-YFP and ss-RFP-KDEL were treated with or without ionomycin as described above. A region of interest was photobleached (see diagram), and recovery into the photobleached area was monitored over time with a wide pinhole confocal setting (∼14 Airy units with 40×/1.3 N.A. objective). The recovery rates are plotted.
In our third experiment aimed at identifying mitotic Golgi haze, we used the well-characterized, thermoreversible folding mutant of vesicular stomatitis virus known as ts045VSVG as a tool for determining whether molecules localized in the Golgi at the time of mitotic Golgi breakdown are retrieved to the folding/misfolding environment of the ER. At 40°C, ts045VSVG misfolds, forms noncovalently associated aggregates and is retained within the ER (Doms et al., 1987). When the temperature is decreased to 32°C, the aggregates disassemble, and ts045VSVG exits the ER normally (de Silva et al., 1990). Significantly, ts045VSVG that has exited the ER is not thermosensitive, whereas ts045VSVG returned to the ER can reaggregate at 40°C and become trapped in the ER (Cole et al., 1998; Mezzacasa and Helenius, 2002).
NRK cells were cotransfected with GalT-CFP and ts045VSVG-YFP and were kept at 40°C during synchronization to the G1/S boundary by aphidicolin treatment (Altan-Bonnet et al., 2003; Pecot and Malhotra, 2004). Aphidicolin was washed out while cells were kept at 40°C (see scheme in Figure 8A). At 7.5 h after washout, when cells soon would enter mitosis, the temperature was shifted down to 32°C. This allowed ts045VSVG-YFP to properly fold and be released into the secretory pathway. Once cells had begun to enter mitosis and a significant pool of ts045VSVG-YFP had accumulated in the Golgi (∼15 min after temperature shift), all non-Golgi fluorescence was removed by photobleaching. This ensured only Golgi-derived ts045VSVG-YFP was followed during the experiment. The temperature was then either maintained at 32°C or shifted to 40°C, and time-lapse images of both GalT-CFP and ts045VSVG-YFP were acquired as the cells progressed through mitosis (Figure 8, B and C). Our working assumption was that if Golgi membranes mix with ER at any time during mitosis, then in cells shifted to and maintained at 40°C, ts045VSVG-YFP will become trapped in the ER and fail to reassociate with Golgi membranes at the end of mitosis. On the other hand, if Golgi membranes remain distinct from ER membranes during mitosis, then at the end of mitosis in cells incubated at 40°C, vesicles containing ts045VSVG-YFP should fuse back into Golgi structures and ts045VSVG then should be transported to the plasma membrane.
Figure 8.
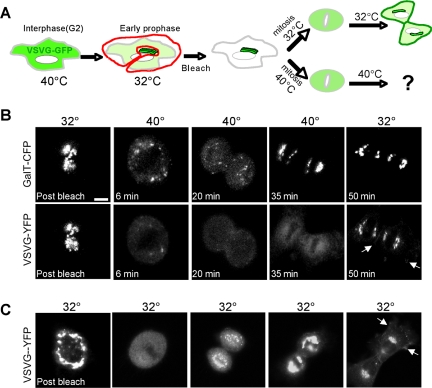
Retrieval of ts045VSVG-YFP to the ER during mitosis. (A) Scheme of experiment. (B and C) NRK cells cotransfected with ts045VSVG-YFP and GalT-CFP were synchronized with aphidicolin (Altan-Bonnet et al., 2003) and grown overnight at 40°C. Then, 7.5 h after aphidicolin washout, before entry into mitosis, cells were transferred to 32°C for 10-15 min to redistribute ts045VSVG-YFP into the Golgi. The fluorescence of ts045VSVG-YFP and GalT-CFP outside the Golgi region of interest was then photobleached in early prophase cells (B and C, postbleach). The cells were then incubated either at 32°C (C) or at 40°C and imaged while going through mitosis (B). Images at representative time points postbleach are shown. At the end of mitosis (corresponding to times after the 35-min time point), the cells at 40°C were shifted back to 32°C. Arrows at the 50-min time point in B and C point to the pool of ts045VSVG-YFP at the plasma membrane. All images were collected with a 40×/1.3 N.A. objective and wide pinhole confocal settings (∼14 Airy units) to collect fluorescence from entire depth of the cell. Bars, 5 μm.
In cells maintained at 32°C or shifted to 40°C, Golgi-localized pools of ts045VSVG-YFP and GalT-CFP redistributed into a widely dispersed pattern (i.e., mitotic haze) as the cells progressed into metaphase (Figure 8, B and C). At the end of mitosis, ts045VSVG-YFP in cells at 32°C redistributed into newly forming Golgi structures before moving to the plasma membrane (Figure 8C). In cells at 40°C, by contrast, ts045VSVG-YFP failed to reassociate with newly forming Golgi elements (labeled with GalT-CFP) and remained diffusely localized (Figure 8B). This result is explained if ts045VSVG-YFP molecules had redistributed into the ER during mitosis and upon shift to 40°C had undergone misfolding and retention in this compartment. Consistent with this hypothesis, if the temperature was shifted from 40°C back to 32°C in cells undergoing cytokinesis, ts045VSVG-YFP molecules rapidly redistributed from their diffuse distribution into Golgi structures (Figure 8B). Control experiments showed that 1) newly synthesized and/or newly folded ts045VSVG-YFP molecules played no role in the observed changes in ts045VSVG-YFP distribution because photobleaching all ts045VSVG-YFP within cells resulted in no fluorescence recovery during the experimental time period (Supplemental Figure S7), and 2) Golgi-localized ts045VSVG molecules in interphase cells shifted to 40°C did not return to the ER but were efficiently delivered to the plasma membrane (Cole et al., 1998; Mezzacasa and Helenius, 2002). Based on these results, we concluded that Golgi proteins can return to the ER in mitosis, resulting in the observed behavior of ts045VSVG in cells at 40°C.
DISCUSSION
Recently, it has been proposed that Golgi and ER membranes in mammalian cells remain distinct during mitosis and that the ER's absorptive/export activities play no role in mitotic Golgi breakdown and reassembly (Axelsson and Warren, 2004; Pecot and Malhotra, 2004). In testing this hypothesis—through dynamic cell imaging of Golgi and ER markers, electron microscopy, ER fragmentation with ionomycin, and ER entrapment through misfolding—we have reached the opposite conclusion: the Golgi is, in fact, inherited through the ER in mitosis. The basis for this conclusion comes from two major overall findings. First, that mitotic Golgi haze, seen in metaphase, represents recycled Golgi proteins trapped in the ER, a consequence that is likely related to the mitosis-specific disassembly of ER exit sites (Farmaki et al., 1999; Prescott et al., 2001; Stephens, 2003) and inactivation of Arf1 (Altan-Bonnet et al., 2003). Second, that mitotic Golgi fragments, seen in prometaphase and telophase, are not isolated breakdown products of the Golgi; rather, they are structures undergoing continuous exchange of their components through the ER and dispersed ER exit sites. Together, these findings lead us to propose a model for mitotic Golgi disassembly/reassembly involving the inhibition and subsequent reactivation of cellular activities that control recycling of Golgi components into and out of the ER (see model in Figure 9).
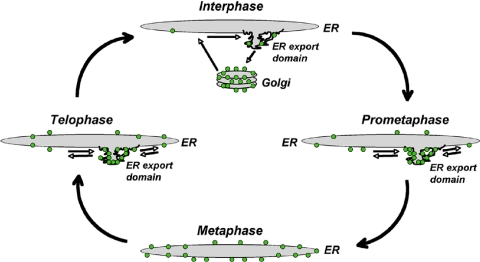
Figure 9.
ER-dependent model for Golgi breakdown and reassembly in mitosis. During interphase, Golgi enzymes (green dots) undergo constitutive cycling through the Golgi apparatus, ER, and ER export domains as part of a general mechanism for maintaining the identity of Golgi membranes (Altan-Bonnet et al., 2004). At steady state, roughly 80% of Golgi enzymes reside in the Golgi and 20% in the ER or ER export domains. Between prophase and prometaphase, Golgi enzymes undergo enhanced retrograde transport back to the ER because of reduced Arf1 activity on membranes. Because cytoplasmic microtubules are depolymerized at this stage of mitosis, the Golgi enzymes cannot return to a centrosomal location and instead accumulate in membrane adjacent to ER export domains (cycling in and out of these domains). During metaphase, both Sar1 and Arf1 become largely inactivated. This causes ER export domains to disappear and Golgi enzymes to relocate to the ER. By telophase, Sar1 is reactivated and ER export domains are reformed. Once Arf1 is reactivated as well, Golgi enzymes are exported from the ER to ER export domains (cycling in and out of them). By cytokinesis, both Sar1 and Arf1 are fully active and cytoplasmic microtubules have repolymerized. ER export domains can now differentiate into pre-Golgi structures that bud off the ER and track along microtubules into the perinuclear region where they fuse into a Golgi stack. Golgi enzymes now undergo normal constitutive cycling between Golgi, ER, and ER export domains.
Ruling Out the Autonomous Golgi Model
Our primary finding supporting an ER-mediated inheritance strategy was that Golgi haze in metaphase represents Golgi proteins within the ER. This was supported by three lines of evidence: 1) mitotic haze can be resolved into ER by high-resolution confocal microscopy, 2) it redistributes with ER into fragments upon ionomycin treatment, and 3) it displays quality control features characteristic of ER such as misfolding and retention of proteins.
To establish the first type of evidence—that mitotic Golgi haze can be resolved into ER—we developed a confocal imaging protocol aimed at determining both whether Golgi and ER markers could be observed in mitotic ER and whether Golgi proteins in mitotic ER derive from preexisting Golgi. Answers to these questions have not been obtained in previous light microscopy studies for several reasons. First, microscope settings in such studies are typically set up for imaging bright mitotic Golgi fragments rather than mitotic haze, meaning thick sections of mitotic cells are examined under relatively low brightness settings (Shima et al., 1998; Jokitalo et al., 2001; Seemann et al., 2002; Pecot and Malhotra, 2004). Because mitotic Golgi haze is much dimmer than mitotic Golgi fragments, such settings usually result in mitotic Golgi haze being too dim and/or out of focus to be resolved. Second, studies using deconvolution microscopy to compare ER and Golgi markers in mitotic cells run into the problem that deconvolution often causes dim structures to disappear. This could explain the discrepancy between ER and Golgi levels in the spindle region reported in studies using deconvolution techniques (Jesch et al., 2001; Pecot and Malhotra, 2004). And third, when an ER pattern within mitotic Golgi haze is observed, it is difficult to establish whether this is because of 1) retrieval of Golgi proteins back to the ER, 2) newly synthesized Golgi enzymes being trapped in the ER (Farmaki et al., 1999; Prescott et al., 2001), and/or 3) refolding of misfolded GFP pools in the ER (Jokitalo et al., 2001; Shorter and Warren, 2002).
To overcome these problems, we used a confocal imaging protocol in which mitotic Golgi haze was imaged with a narrow pinhole setting and with enough laser power so that mitotic haze was bright and could be focused. To make sure that Golgi protein fluorescence within mitotic haze arose only from GalT-YFP proteins preexisting in the Golgi, we photobleached all cellular fluorescence outside the Golgi once cells entered mitosis. To demonstrate that mitotic haze did not come from newly synthesized and/or folded pools of GalT-YFP proteins in the ER, we showed that no new fluorescence occurred during mitosis after all cellular fluorescence was first photobleached. To define ER, we expressed the soluble ER marker ss-RFP-KDEL (which labels ER uniformly because of its free diffusion within the ER lumen) in mitotic cells coexpressing GalT-CFP. On imaging in this manner, we found that the pattern of labeling of ER and Golgi markers significantly overlapped during metaphase, the stage of mitosis in which there is predominantly Golgi haze. The labeling pattern occurred as a tubular network characteristic of mitotic ER (Ellenberg et al., 1997), with the network extending into the spindle region as tubule strands containing both GalT-CFP and ss-RFP-KDEL. Our finding that native Golgi and ER markers also colocalize in a reticular pattern in metaphase cells further supported our observations with GalT-CFP and ss-RFP-KDEL markers. These results suggested that mitotic Golgi haze consists of proteins derived from the Golgi, and, when resolved, looks like ER.
Our second line of evidence linking mitotic Golgi haze and ER—that mitotic Golgi haze acts like ER— used an assay in which ER and Golgi markers were examined in mitotic cells before and after the ER was fragmented with ionomycin, a well-characterized ER fragmenting reagent (Subramanian and Meyer, 1997; Park et al., 2000; Olah et al., 2001). If mitotic Golgi behaves as ER, then Golgi and ER proteins should both be found in ER fragments in this experiment. Consistent with this, we found that during ionomycin-treatment Golgi proteins in mitotic haze redistributed with ER proteins into ER fragments, where both types of proteins became immobile. Interestingly, use of a similar assay with filipin (a cholesterol-extracting agent) (Rothberg et al., 1990; Ilangumaran and Hoessli, 1998) to fragment mitotic ER showed that Golgi proteins did not redistribute into filipin-induced mitotic ER fragments (Axelsson and Warren, 2004). A possible explanation is that filipin may differentially affect the membrane stability of Golgi proteins, which typically favor a cholesterol/sphingolipid-rich microenvironment (Munro, 1998) and thus may get extracted from the ER during the filipin treatment. Consistent with this, Warren and colleagues found that Golgi proteins diffused ∼50% faster in filipin-treated mitotic cells (Axelsson and Warren, 2004).
The third type of evidence relating mitotic Golgi haze to ER—that mitotic Golgi haze shares the ER's capacity to retain misfolded proteins—was acquired using an ER trapping assay. The impetus for this assay came from the recent work of Malhotra and colleagues (Pecot and Malhotra, 2004) who used a different ER trapping assay, involving rapamycin, to address the nature of mitotic Golgi haze. These authors found that FKBP-protein fused to a Golgi enzyme (FKBP-Golgi protein) became stably bound to FKBP-rapamycin-associated protein (FRAP) attached to an ER resident protein (FRAP-ER protein) within 2.5 min of rapamycin treatment in BFA-treated but not in mitotic cells, a result they interpreted as evidence that Golgi and ER do not mix during mitosis. Interestingly, the FKBP-Golgi protein in this system did not get trapped in the ER during interphase after a 90-min treatment with rapamycin, which is predicted given normal Golgi protein cycling rates through the ER (Cole et al., 1998; Zaal et al., 1999; Miles et al., 2001; Ward et al., 2001). This implies that trapping is very sensitive to 1) the amount of FKBP-Golgi proteins in the ER and 2) the length of time the FKBP-Golgi proteins have been in the ER before rapamycin treatment. In the Pecot and Malhotra (2004) experiments, cells were treated with BFA (which induces complete mixing of Golgi and ER proteins) for 60 min before rapamycin treatment. By contrast, mitotic cells are in the stage of mitosis where Golgi and ER enzymes are proposed to fully mix (i.e., metaphase) for only ∼10 min (Zaal et al., 1999). The different lengths of time FKBP-Golgi proteins were in the ER in the two situations (i.e., BFA versus mitosis) therefore may be the reason why there was ER entrapment of Golgi proteins in BFA but not mitotic cells. Under this interpretation, FKBP-Golgi proteins in mitotic cells would not have been in the ER at a high enough concentration for a long enough time to permit them to develop low-affinity interactions with FRAP-ER proteins, which may be necessary for a stable ternary complex of these proteins and rapamycin to rapidly form (i.e., within 2.5 min) upon addition of the drug. Given these concerns, we searched for a different strategy for trapping Golgi proteins in the ER in mitotic cells.
Previous work has shown that the ER has the capacity to misfold and trap proteins undergoing constitutive cycling through the ER in interphase cells (Cole et al., 1998; Mezzacasa and Helenius, 2002). For example, ts045VSVG protein with a Golgi targeting signal in interphase cells will redistribute from the Golgi into the ER and be trapped there upon shift from 32 to 40°C (Cole et al., 1998). This is because of the chimera undergoing misfolding in the ER's unique quality control environment as it constitutively cycles through this compartment at 40°C. Given this property of the ER, we reasoned that if Golgi membranes mix with the ER at any time during mitosis, then in cells shifted to and maintained at 40°C, ts045VSVG pools in the Golgi should become trapped in the ER and fail to reassociate with Golgi membranes at the end of mitosis. This is precisely what we observed upon performing this experiment; moreover, upon shift back to 32°C, ts045VSVG redistributed from the ER into Golgi structures.
In assessing these findings, account must be taken of those studies that have failed to detect Golgi proteins in the ER in mitosis. This failure, however, could be because of the methodologies that were used and may not, in fact, be inconsistent with our findings. The period of mitosis during which Golgi proteins reside in highly dispersed membranes is short relative to the total time that cells are in mitosis (Zaal et al., 1999). It is, thus, easy to miss this stage when observing populations of mitotic cells, unless correlative light and electron microscopy (Polishchuk et al., 2000) is used to identify mitotic cells with a dispersed Golgi pattern. During the period in which Golgi proteins are highly dispersed, these proteins may not be mixed uniformly with ER resident proteins within the ER and hence may not be capable of forming complexes with resident ER proteins as is necessary for detection in the rapamycin assay. Nocodazole is often used to synchronize mitotic cells for ultrastructural and biochemical analysis, but this treatment causes Golgi fragments to persist (Jesch et al., 2001) so the dispersed mitotic Golgi phenotype is absent in these cells. In subcellular fractionation experiments, large quantities of mitotic cells are harvested so cells will necessarily be at different states of mitotic Golgi breakdown, which could explain why Golgi proteins from mitotic cells have not been shown to cofractionate with ER markers on density gradients (Jesch et al., 2001).
Defining Mitotic Golgi Fragments
Our second major finding was that mitotic Golgi fragments represent ER-derived structures through which Golgi proteins cycle rapidly (e.g., from ER exit sites to a fragment and then back again into the ER). A variety of different methodologies were used to establish this, including fluorescence double-labeling, immunoelectron microscopy, and photo-bleaching recovery experiments.
Fluorescence imaging using markers for mitotic Golgi fragments and ER exit sites showed that ER exit site markers surround and coalign with mitotic Golgi fragments. Because a previous light microscopy study examining the distribution of ER exit sites and Golgi fragments in mitosis interpreted the lack of direct overlap between Golgi fragments and ER exit sites as evidence that these two structures were not functionally linked (Hammond and Glick, 2000), we relied on other methods to establish whether there was a dynamic association between Golgi fragments and ER exit sites in mitotic cells. Live cell imaging of a single cell coexpressing Sec13-YFP and GalT-CFP revealed that mitotic Golgi fragments grow out from ER export domains at the end of mitosis, remain near these sites for a short period, and then undergo clustering into a Golgi ribbon. Immunoperoxidase electron microscopy of cells in prometaphase and telophase further showed that mitotic Golgi fragments were clusters of tubules/vesicles localized adjacent to ER export sites, and in some cases, were in direct continuity with ER export domains. Finally, when a mitotic Golgi fragment was photobleached in cells expressing GalT-YFP, fragment fluorescence rapidly recovered >80% of its original intensity within 2 min, indicating Golgi proteins continuously move in and out of mitotic fragments while maintaining steady-state pools in these fragments.
These findings suggest that mitotic Golgi fragments share key properties of nocodazole Golgi fragments, which are known to 1) localize near ER exit sites, 2) form by outgrowth from ER rather than by direct fragmentation of Golgi, and 3) undergo continuous cycling of their membrane components in and out of fragments. Nocodazole-induced and mitotic Golgi fragments nevertheless have other properties that are distinct. Mitotic fragments exist as clusters of branching tubules and vesicles, whereas nocodazole fragments exist as mini-Golgi stacks at the ultrastructural level. Moreover, the cycling rate of Golgi components in and out of mitotic Golgi fragments is much faster than that for nocodozole fragments. These differences provide clues to how each type of Golgi fragment is formed and maintained. For example, the inability of mitotic Golgi fragments to assemble into stacks at ER exit sites is likely related to the inability of these membranes to efficiently recruit sorting and transport machinery (e.g., Arf1 and its effectors) necessary for such conversion: this, in turn, may underlie the increased speed that Golgi components cycle into and out of these structures.
Toward a Model of Golgi Inheritance from ER
Given our data suggesting that mitotic Golgi haze represents ER and that mitotic Golgi fragments (both in prometaphase and telophase) are dynamic membrane outgrowths from the ER, we envision a multistep process for Golgi inheritance from the ER (Figure 9). In this model, the formation of prometaphase Golgi fragments near ER exit sites results from 1) accelerated retrograde traffic into the ER and 2) the inability of newly emerging membrane at ER exit sites to recruit machinery necessary for conversion into Golgi. The next step—appearance of mitotic Golgi haze— occurs when ER export domains undergo complete disassembly, leading to entrapment of Golgi components in the ER. In cells in which ER exit sites do not all disassemble, some fragments would remain within the mitotic haze, explaining why in some cell types Golgi fragments persist during mitosis (Schroeter et al., 1985; Jokitalo et al., 2001). The final step— reappearance of mitotic Golgi fragments— occurs when ER export domains reassemble and the exported membrane becomes capable of recruiting necessary machinery for transforming this membrane into Golgi. Golgi components would exit the ER at ER exit sites and assemble into membrane tubular clusters near these sites. The clusters later would transform into Golgi cisternae/stacks, and then coalesce into a juxtanuclear site.
An alternative model for explaining how new Golgi elements coalesce near ER exit sites during telophase is that the ER exit site exports selected components at the end of mitosis, which then provide a “platform” for the capture of Golgi membrane components that are present in small vesicle carriers in the cytosol (Shorter and Warren, 2002; He et al., 2004; Soderholm et al., 2004). Although this “platform-capture” model can readily explain why new Golgi elements arise near ER exit sites during telophase, it has difficulty explaining other key observations from our study. First, the platform-capture model's view that Golgi enzymes in vesicles are selectively “captured” at an ER exit site is at odds with the continuous, rapid cycling of Golgi enzymes into and out of ER exit sites in mitotic cells demonstrated in our photobleaching experiments. Second, the putative mitotic vesicles carrying Golgi enzymes in the platform-capture model are predicted to diffuse freely throughout the cytoplasm, even during fragmentation of ER membrane by ionomycin treatment. Yet, ionomycin treatment of mitotic cells led to Golgi enzymes becoming completely immobilized as well as colocalized with ER markers in large fragments, consistent with an ER rather than vesicle localization of the Golgi enzymes in mitotic cells. Third, the platform-capture model cannot explain why incubation at the nonpermissive temperature of 40°C led to ER entrapment of ts045VSVG molecules at the end of mitosis when these molecules were localized in the Golgi before mitosis (readily explained by Golgi inheritance from the ER). Because the model of Golgi inheritance from the ER, by contrast, can account for all of these data, it would seem to be the more comprehensive representation of the events occurring during mitotic Golgi breakdown and reassembly.
The molecular mechanism for Golgi inheritance from the ER is only just beginning to be understood, but our data suggest some possibilities. By monitoring the behavior of CFP-tagged Arf1 and ER exit site markers during prometaphase fragment formation in mitosis, we found that Arf1 levels on prometaphase Golgi membranes were depleted relative to interphase Golgi membranes. Because this occurred before ER exit sites (detected by Sec13-YFP labeling) had fully disassembled, the data suggested Arf1 inactivation precedes ER exit site disassembly during mitosis. This finding is significant because Arf1 inactivation in interphase cells (produced by expression of Arf1T31N or BFA treatment) accelerates Golgi retrograde traffic and leads to the appearance of tubulovesicular membranes enriched in Golgi proteins at ER exit sites (Miles et al., 2001; Ward et al., 2001; Puri and Linstedt, 2003). Because such structures resemble prometaphase fragments, the data suggest that the formation of prometaphase Golgi fragments is related to Arf1 inactivation.
Consistent with this possibility, previous work has shown that when Arf1 inactivation in mitosis is prevented by expression of a constitutively active Arf1 mutant or by treatment with H89 (Altan-Bonnet et al., 2003), prometaphase Golgi fragments do not form and the Golgi remains localized to centrosomal elements. Thus, enhanced recycling, potentially mediated by Arf1 inactivation, might play an important role in the early disassembly of cisternal Golgi stacks in mitosis. ER export arrest, which occurs after Arf1 inactivation, would subsequently lead to trapping of rapidly cycling Golgi residents within the ER, causing prometaphase fragments to disappear and mitotic haze to occur.
The mechanism for Arf1 inactivation and ER export arrest in mitosis is not known. However, because our data suggest these two processes occur sequentially rather than simultaneously, there would seem to be distinct mitotic mechanisms affecting each of these blocks. Consistent with this, when Arf1 remains active in mitotic cells, Golgi membranes do not disassemble yet ER export domains still disperse (Altan-Bonnet et al., 2003). Furthermore, at the end of mitosis, ER exit site reassembly occurs in anaphase, whereas rebinding of Arf1 to membranes only occurs in telophase. Because Arf1 rebinding to membranes in telophase coincides with emergence of Golgi enzymes from the ER, the data suggest these processes are linked.
This role of Arf1 and ER exit site machinery in mitotic Golgi assembly/disassembly seems to parallel the role of Arf1 and Sar1 in Golgi breakdown/biogenesis during interphase (Altan-Bonnet et al., 2004). There, Sar1 activity induces recruitment of peripheral proteins to ER export domains, including COPII, Golgi matrix proteins, Rab proteins, SNAREs, and matrix proteins (Altan-Bonnet et al., 2004; Lee et al., 2004), which differentiate from surrounding ER. This leads to the recruitment of Arf1 and its numerous effectors, including phospholipid-modifying enzymes, COPI, spectrin, ankyrin, and signaling molecules (Altan-Bonnet et al., 2003, 2004), causing the domains to further differentiate into Golgi structures. Our data on the behavior in mitotic cells of Arf1, Sec13, and Golgi enzymes are consistent with a similar sequential process. Thus, there seems to be a fundamental commonality in Golgi breakdown/biogenesis throughout the cell cycle. Because Golgi inheritance in Saccharomyces cerevisiae is also linked to ER inheritance (Reinke et al., 2004), the mechanisms underlying Golgi inheritance from yeast and vertebrate cells may be conserved.
What triggers the alterations in ER/Golgi membrane flux during mitosis is an important area of future work. Cellular kinases such as cdc2 and mitogen-activated protein kinase kinase (MEK) 1 have been reported to be important for the disassembly of Golgi membranes in mitosis (Acharya et al., 1998; Lowe et al., 1998). A possible activity of these kinases could be to inactivate proteins that play a role the ER-to-Golgi trafficking of membranes along microtubules. Indeed, a recent report showing that MEK kinases regulate the trafficking of melanosomes on microtubules is consistent with this role (Deacon et al., 2005). Moreover, GRASP65 has been recently reported to play a role in the organization of the mitotic spindle (Sutterlin et al., 2005). The fact that the Golgi matrix proteins GM130 and GRASP65 are potential targets of MEK and cdc2 kinases, raises the possibility that in addition to their previous proposed roles (Lowe et al., 2000), these matrix proteins facilitate Golgi membrane association with the cytoskeleton.
Supplementary Material
Acknowledgments
We thank Graham Warren and George Patterson for helpful discussions. We thank Ramanujan Hegde and Grégoire Altan-Bonnet for critical reading of the manuscript. This research was supported in part by the Intramural Research Program of the National Institute of Child Health and Human Development, National Institutes of Health.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-02-0155) on November 23, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Acharya, U., Mallabiabarrena, A., Acharya, J. K., and Malhotra, V. (1998). Signaling via mitogen-activated protein kinase kinase (MEK1) is required for Golgi fragmentation during mitosis. Cell 92, 183-192. [DOI] [PubMed] [Google Scholar]
- Altan-Bonnet, N., Polishchuk, R., Phair, R. D., Weigert, R., and Lippincott-Schwartz, J. (2003). A role for Arf1 in mitotic Golgi disassembly, chromosome segregation and cytokinesis. Proc. Natl. Acad. Sci. USA 100, 13314-13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altan-Bonnet, N., Sougrat, R., and Lippincott-Schwartz, J. (2004). Molecular basis for Golgi maintenance and biogenesis. Curr. Opin. Cell Biol. 16, 364-372. [DOI] [PubMed] [Google Scholar]
- Axelsson, M. A., and Warren, G. (2004). Rapid, endoplasmic reticulum-independent diffusion of the mitotic Golgi haze. Mol. Biol. Cell 15,1 843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe, C., Orci, L., Yeung, T., Hosobuchi, M., Hamamoto, S., Salama, N., Rexach, M. F., Ravazzola, M., Amherdt, M., and Schekman, R. (1994). COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77, 895-907 [DOI] [PubMed] [Google Scholar]
- Bevis, B. J., Hammond, A. T., Reinke, C. A., and Glick, B. S. (2002). De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat. Cell Biol. 4, 750-756. [DOI] [PubMed] [Google Scholar]
- Chaldakov, G. N., and Vankov, V. N. (1985). Association of smooth membranes with spindle microtubules in the arterial smooth muscle cell. Acta Biol. Hung. 36, 179-183. [PubMed] [Google Scholar]
- Cole, N. B., Ellenberg, J., Song, J., DiEuliis, D., and Lippincott-Schwartz, J. (1998). Retrograde transport of Golgi-localized proteins to the ER. J. Cell Biol. 140, 1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, N. B., Sciaky, N., Marotta, A., Song, J., and Lippincott-Schwartz, J. (1996). Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol. Biol. Cell 7, 631-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon, S. W., Nascimento, A., Serpinskaya, A. S., and Gelfand, V. I. (2005). Regulation of bidirectional melanosome transport by organelle bound MAP kinase. Curr. Biol. 8, 459-463. [DOI] [PubMed] [Google Scholar]
- de Silva, A. M., Balch, W. E., and Helenius, A. (1990). Quality control in the endoplasmic reticulum: folding and misfolding of vesicular stomatitis virus G protein in cells and in vitro. J. Cell Biol. 111, 857-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms, R., Keller, D. S., Helenius, A., and Balch, W. E. (1987). Role for adenosine triphosphate in regulating the assembly and transport of vesicular stomatitis virus G protein trimers. J. Cell Biol. 105, 1957-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drecktrah, D., and Brown, W. J. (1999). Phospholipase A(2) antagonists inhibit nocodazole-induced Golgi ministack formation: evidence of an ER intermediate and constitutive cycling. Mol. Biol. Cell 10, 4021-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg, J., Siggia, E. D., Moreira, J. E., Smith, C. L., Presley, J. F., Worman, H. J., and Lippincott-Schwartz, J. (1997). Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138, 1193-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmaki, T., Ponnambalam, S., Prescott, A. R., Clausen, H., Tang, B. L., Hong, W., and Lucocq, J. M. (1999). Forward and retrograde trafficking in mitotic animal cells. ER-Golgi transport arrest restricts protein export from the ER into COPII-coated structures. J. Cell Sci. 112, 589-600. [DOI] [PubMed] [Google Scholar]
- Fu, L., and Sztul, E. (2003). Traffic-independent function of the Sar1p/COPII machinery in proteasomal sorting of the cystic fibrosis transmembrane conductance regulator. J. Cell Biol. 160, 157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, J. (1998). Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell 95, 237-248. [DOI] [PubMed] [Google Scholar]
- Hammond, A. T., and Glick, B. S. (2000). Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol. Biol. Cell 11, 3013-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, C. Y., Ho, H. H., Malsam, J., Chalouni, C., West, C. M., Ullu, E., Toomre, D., and Warren, G. (2004). Golgi duplication in Trypanosoma brucei. J. Cell Biol. 165, 313-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilangumaran, S., and Hoessli, D. C. (1998). Effects of cholesterol depletion by cyclodextrin on the sphingolipid microdomains of the plasma membrane. Biochem. J. 335, 433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesch, S. A., Mehta, A. J., Velliste, M., Murphy, R. F., and Linstedt, A. D. (2001). Mitotic Golgi is in a dynamic equilibrium between clustered and free vesicles independent of the ER. Traffic 2, 873-884. [DOI] [PubMed] [Google Scholar]
- Jokitalo, E., Cabrera-Poch, N., Warren, G., and Shima, D. T. (2001). Golgi clusters and vesicles mediate mitotic inheritance independently of the endoplasmic reticulum. J. Cell Biol. 154, 317-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky, M. J. (1971). Use of ferrocyanide-reduced osmium tetroxide in electron microscopy. In: Proceedings of the 11th Meeting of the American Society of Cell Biology, New Orleans, LA. Abstract 284.
- Lee, M. C., Miller, E. A., Goldberg, J., Orci, L., and Schekman, R. (2004). Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 20, 87-123. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., Yuan, L. C., Bonifacino, J. S., and Klausner, R. D. (1989). Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56, 801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, M., Gonatas, N. K., and Warren, G. (2000). The mitotic phosphorylation cycle of the cis-Golgi matrix protein GM130. J. Cell Biol. 149, 341-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, M., Rabouille, C., Nakamura, N., Watson, R., Jackman, M., Jamsa, E., Rahman, D., Pappin, D. J., and Warren, G. (1998). Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell 94, 783-793. [DOI] [PubMed] [Google Scholar]
- Lucocq, J., Berger, E., and Hug, C. (1995). The pathway of Golgi cluster formation in okadaic acid-treated cells. J. Struct. Biol. 115, 318-330. [DOI] [PubMed] [Google Scholar]
- Lucocq, J. M., Berger, E. G., and Warreng, G. (1989). Mitotic Golgi fragments in HeLa cells and their role in the reassembly pathway. J. Cell Biol. 109, 463-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucocq, J. M., Pryde, J. G., Berger, E. G., and Warren, G. (1987). A mitotic form of the Golgi apparatus in HeLa cells. J. Cell Biol. 104, 865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucocq, J. M., Pryde, J. G., Berger, E. G., and Warren, G. (1988). A mitotic form of the Golgi apparatus identified using immuno-electronmicroscopy. Prog. Clin. Biol. Res. 270, 431-440. [PubMed] [Google Scholar]
- Nizak, C., Sougrat, R., Jollivet, F., Rambourg, A., Goud, B., and Perez, F. (2004). Golgi inheritance under a block of anterograde and retrograde traffic. Traffic 5, 284-299. [DOI] [PubMed] [Google Scholar]
- Mezzacasa, A., and Helenius, A. (2002). The transitional ER defines a boundary for quality control in the secretion of tsO45 VSV glycoprotein. Traffic 3, 833-849. [DOI] [PubMed] [Google Scholar]
- Miles, S., McManus, H., Forsten, K. E., and Storrie, B. (2001). Evidence that the entire Golgi apparatus cycles in interphase HeLa cells: sensitivity of Golgi matrix proteins to an ER exit block. J. Cell Biol. 155, 543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T., and Warren, G. (1995a). A role for tubular networks and a COP I-independent pathway in the mitotic fragmentation of Golgi stacks in a cell-free system. J. Cell Biol. 130, 1027-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T., and Warren, G. (1995b). Mitotic disassembly of the Golgi apparatus in vivo. J. Cell Sci. 108, 2715-2727. [DOI] [PubMed] [Google Scholar]
- Munro, S. (1998). Localization of proteins to the Golgi apparatus. Trends Cell Biol. 8, 11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olah, Z., Szabo, T., Karai, L., Hough, C., Fields, R. D., Caudle, R. M., Blumberg, P. M., and Iadarola, M. J. (2001). Ligand-induced dynamic membrane changes and cell deletion conferred by vanilloid receptor 1. J. Biol. Chem. 276, 11021-11030. [DOI] [PubMed] [Google Scholar]
- Park, M. K., Petersen, O. H., and Tepikin, A. V. (2000). The endoplasmic reticulum as one continuous Ca(2+) pool: visualization of rapid Ca(2+) movements and equilibration. EMBO J. 19, 5729-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawley, J. B. (1995). Handbook of Biological Confocal Microscopy, 2nd ed., Dordrecht, Netherlands: Kluwer Academic Publishers (Springer).
- Pecot, M. Y., and Malhotra, V. (2004). Golgi membranes remain segregated from the endoplasmic reticulum during mitosis in mammalian cells. Cell 116, 99-107. [DOI] [PubMed] [Google Scholar]
- Pelletier, L., et al. (2002). Golgi biogenesis in Toxoplasma gondii. Nature 418, 548-552. [DOI] [PubMed] [Google Scholar]
- Polishchuk, R., Di Pentima, A., and Lippincott-Schwartz, J. (2004). Delivery of raft-associated, GPI-anchored proteins to the apical surface of polarized MDCK cells by a transcytotic pathway. Nat. Cell Biol. 6, 297-307. [DOI] [PubMed] [Google Scholar]
- Polishchuk, R. S., Polishchuk, E. V., Marra, P., Alberti, S., Buccione, R., Luini, A., and Mironov, A. A. (2000). Correlative light-electron microscopy reveals the tubular-saccular ultrastructure of carriers operating between Golgi apparatus and plasma membrane. J. Cell Biol. 148, 45-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott, A. R., Farmaki, T., Thomson, C., James, J., Paccaud, J. P., Tang, B. L., Hong, W., Quinn, M., Ponnambalam, S., and Lucocq, J. (2001). Evidence for prebudding arrest of ER export in animal cell mitosis and its role in generating Golgi partitioning intermediates. Traffic 2, 321-335. [DOI] [PubMed] [Google Scholar]
- Puri, S., and Linstedt, A. D. (2003). Capacity of the Golgi apparatus for biogenesis from the endoplasmic reticulum. Mol. Biol. Cell 14, 5011-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke, C. A., Kozik, P., and Glick, B. S. (2004). Golgi inheritance in small buds of Saccharomyces cerevisiae is linked to endoplasmic reticulum inheritance. Proc. Natl. Acad. Sci. USA 101, 18018-18023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg, K. G., Ying, Y. S., Kamen, B. A., and Anderson, R.G.W. (1990). Cholesterol controls the clustering of the glycophospholipid-anchored membrane receptor for 5-methyltetrahydrofolate. J. Cell Biol. 111, 2931-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciaky, N., Presley, J., Smith, C., Zaal, K. J., Cole, N., Moreira, J. E., Terasaki, M., Siggia, E., and Lippincott-Schwartz, J. (1997). Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J. Cell Biol. 139, 1137-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter, D., Ehemann, V., and Paweletz, N. (1985). Cellular compartments in mitotic cells: ultrahistochemical identification of Golgi elements in PtK-1 cells. Biol. Cell 53, 155-163. [DOI] [PubMed] [Google Scholar]
- Seemann, J., Pypaert, M., Taguchi, T., Malsam, J., and Warren, G. (2002). Partitioning of the matrix fraction of the Golgi apparatus during mitosis in animal cells. Science 295, 848-851. [DOI] [PubMed] [Google Scholar]
- Sharff, M. D., and Robinson, E. (1966). Polyribosome disaggregation during metaphase. Science 151, 992-995. [DOI] [PubMed] [Google Scholar]
- Shima, D. T., Cabrera-Poch, N., Pepperkok, R., and Warren, G. (1998). An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J. Cell Biol. 141, 955-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter, J., and Warren, G. (2002). Golgi architecture and inheritance. Annu. Rev. Cell Dev. Biol. 18, 379-420. [DOI] [PubMed] [Google Scholar]
- Soderholm, J., Bhattacharyya, D., Strongin, D., Markovitz, V., Connerly, P. L., Reinke, C. A., and Glick, B. S. (2004). The transitional ER localization mechanism of Pichia pastoris Sec12. Dev. Cell 6, 649-659. [DOI] [PubMed] [Google Scholar]
- Stephens, D. J. (2003). De novo formation, fusion and fission of mammalian COPII-coated endoplasmic reticulum exit sites. EMBO Rep. 4, 210-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie, B., White, J., Rottger, S., Stelzer, E. H., Suganuma, T., and Nilsson, T. (1998). Recycling of Golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 143, 1505-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, K., and Meyer, T. (1997). Calcium-induced restructuring of nuclear envelope and endoplasmic reticulum calcium stores. Cell 89, 963-971. [DOI] [PubMed] [Google Scholar]
- Sutterlin, C., Polishchuk, R., Pecot, M., and Malhotra, V. (2005). The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol. Biol. Cell 16, 3211-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, P. A., Yucel, J. K., Veit, B., Faulkner, D. J., Deerinck, T., Soto, G., Ellisman, M., and Malhotra, V. (1993). Complete vesiculation of Golgi membranes and inhibition of protein transport by a novel sea sponge metabolite, ilimaquinone. Cell 73, 1079-1090. [DOI] [PubMed] [Google Scholar]
- Terasaki, M. (2000). Dynamics of the endoplasmic reticulum and Golgi apparatus during early sea urchin development. Mol. Biol. Cell 11, 897-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyberg, J., and Moskalewski, S. (1992). Reorganization of the Golgi complex in association with mitosis: redistribution of mannosidase II to the endoplasmic reticulum and effects of brefeldin A. J. Submicrosc. Cytol. Pathol. 24, 495-508. [PubMed] [Google Scholar]
- Ward, T. H., Polishchuk, R. S., Caplan, S., Hirschberg, K., and Lippincott-Schwartz, J. (2001). Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 155, 557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, G., Featherstone, C., Griffiths, G., and Burke, B. (1983). Newly synthesized G protein of vesicular stomatitis virus is not transported to the cell surface during mitosis. J. Cell Biol. 97, 1623-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer, C. M., Sanger, J. W., and Sanger, J. M. (1993). Dynamics of organelles in the mitotic spindles of living cells: membrane and microtubule interactions. Cell Motil. Cytoskeleton 26, 19-39. [DOI] [PubMed] [Google Scholar]
- Zaal, K. J., et al. (1999). Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell 99, 589-601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.