Abstract
The resistance of Candida albicans to many stresses is dependent on the stress-activated protein kinase (SAPK) Hog1. Hence we have explored the role of Hog1 in the regulation of transcriptional responses to stress. DNA microarrays were used to characterize the global transcriptional responses of HOG1 and hog1 cells to three stress conditions that activate the Hog1 SAPK: osmotic stress, oxidative stress, and heavy metal stress. This revealed both stress-specific transcriptional responses and a core transcriptional response to stress in C. albicans. The core transcriptional response was characterized by a subset of genes that responded in a stereotypical manner to all of the stresses analyzed. Inactivation of HOG1 significantly attenuated transcriptional responses to osmotic and heavy metal stresses, but not to oxidative stress, and this was reflected in the role of Hog1 in the regulation of C. albicans core stress genes. Instead, the Cap1 transcription factor plays a key role in the oxidative stress regulation of C. albicans core stress genes. Our data show that the SAPK network in C. albicans has diverged from corresponding networks in model yeasts and that the C. albicans SAPK pathway functions in parallel with other pathways to regulate the core transcriptional response to stress.
INTRODUCTION
The opportunistic fungal pathogen Candida albicans can colonize numerous niches within its human host (Odds, 1988). This pathogen commonly causes infections of mucosal surfaces in the oral cavity and in the gastrointestinal and urogenital tracts. Furthermore, in immunocompromised patients, C. albicans can enter the bloodstream and disseminate within the host causing life-threatening systemic and deep-seated infections of organs. The success of C. albicans as a pathogen is partly due to its resistance to oxidative stresses and other environmental insults. For example, C. albicans can escape macrophage killing even after phagocytosis (Lo et al., 1997). Also, the inactivation of key stress-protective enzymes or stress-signaling proteins attenuates virulence (Wysong et al., 1998; Alonso-Monge et al., 1999; Hwang et al., 2002). These observations indicate that C. albicans exploits specialized stress responses to protect itself during disease progression in its human host. However, the molecular mechanisms underlying such responses in this fungus are poorly understood.
Genomewide expression profiling has been used to investigate the transcriptional responses of yeast cells to a diverse range of environmental stresses. Such studies in the non-pathogenic model yeasts Saccharomyces cerevisiae (Gasch et al., 2000; Causton et al., 2001) and Schizosaccharomyces pombe (Chen et al., 2003) revealed that these organisms mount a core stress response in which a fraction of the genome responds in a stereotypical manner to diverse stress conditions. In S. pombe, this core stress response is predominantly regulated by the Sty1 stress-activated protein kinase (SAPK). In contrast, in S. cerevisiae, different signaling pathways and transcription factors converge to control a common set of stress genes. Less is known about stress responses in C. albicans. However, recent studies have highlighted clear differences between the stress responses of this fungal pathogen and those of budding and fission yeasts. For example, genome wide expression profiling revealed that C. albicans does not mount a common transcriptional response after exposure to environmental conditions that stimulate core stress responses both in S. cerevisiae and S. pombe (Enjalbert et al., 2003). In addition, homologues of the S. cerevisiae transcription factors Msn2 and Msn4, which play key roles in regulating the core stress response in this budding yeast, do not play equivalent roles in C. albicans (Nicholls et al., 2004).
A number of studies have, however, highlighted the importance of the Hog1 SAPK, the homologue of the S. pombe Sty1 SAPK, in stress responses in C. albicans. The Hog1 SAPK is activated in response to a diverse range of stress conditions, and deletion of HOG1 results in cells with pleiotropic stress phenotypes (San-Jose et al., 1996; Alonso-Monge et al., 2003; Smith et al., 2004). A similar situation is seen in S. pombe, where Sty1 responds to a multitude of environmental insults (reviewed in Toone and Jones, 1998) and regulates the core stress response in this yeast (Chen et al., 2003). However, the activation profiles of the C. albicans and S. pombe SAPKs are not identical. For example, although the S. pombe Sty1 SAPK is activated after temperature upshift, C. albicans Hog1 is not (Smith et al., 2004). In addition, higher levels of oxidative stress are required to activate Hog1 than Sty1 (Smith et al., 2004). Such differences in the activation profiles of Hog1 and Sty1, suggest that specialized Hog1-mediated stress responses have evolved in C. albicans to protect it against host defenses and to promote its survival in the host. Moreover, deletion of HOG1 in C. albicans causes morphological defects and impaired virulence in a mouse model of systemic candidiasis (Alonso-Monge et al., 1999). This emphasizes the link between stress responses and pathogenesis in C. albicans and underlines the importance of understanding the role of the Hog1 SAPK in regulating the cellular responses to stress in this pathogen.
Based on the roles of SAPKs in S. pombe and S. cerevisiae, we predicted that in C. albicans, the Hog1 SAPK would contribute to the regulation of a core transcriptional response to stresses that activate this SAPK. Hence, in this study we have defined the global transcriptional responses in C. albicans to environmental stresses that activate the Hog1 SAPK, as well as the roles of Hog1 in these responses. Our work provides novel insights into the roles of the Hog1 signaling network in the regulation of transcriptional responses to stress in the major systemic fungal pathogen of humans. Furthermore, our data demonstrate that this network has diverged significantly from the corresponding SAPK networks in budding and fission yeasts.
MATERIALS AND METHODS
Strains and Growth Conditions
The strains used in this study are given in Table 1. Strains were grown at 30°C in YPDAU (YPD; Sherman, 1991) containing 0.02% adenine and 0.008% uridine). Cell morphology was analyzed using a Zeiss axioscope (Carl Zeiss, Jena, Germany).
Table 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| RM1000 | ura3:λ imm434/ura3::λimm434,his1::hisG/his1::hisG | Wilson et al. (1999) |
| BWP17 | ura3::λ imm434/ura3::λimm434, his1::hisG/his1::hisG, arg4::hisG/arg4::hisG | Wilson et al. (1999) |
| JC50 | ura3::λ imm434/ura3::λimm434, his1::hisG/his1::hisG, hog1::loxP-ura3-loxP/hog1::loxP-HIS1-loxP CIp20 (URA3, HIS1) | Smith et al. (2004) |
| JC52 | ura3::λ imm434/ura3::λimm434, his1::hisG/his1::hisG, hog1::loxP-ura3-loxP/hog1::loxP-HIS1-loxP CIp20-HOG1(URA3, HIS1) | Smith et al. (2004) |
| JC47 | ura3::λ imm434/ura3::λimm434, his1::hisG/his1::hisG, arg4::hisG/arg4::hisG, hog1::loxP-ARG4-ura3-loxP/hog1::loxP-HIS1-loxP | This work |
| JC118 | ura3::λ imm434/ura3::λimm434, his1::hisG/his1::hisG, arg4::hisG/arg4::hisG, hog1::loxP-ARG4-ura3-loxP/hog1::loxP-HIS1-loxP, cap1::hisG/cap1::hisG-URA3-hisG | This work |
| JC128 | ura3::λ imm434/ura3::λimm434, his1::hisG/his1::hisG, arg4::hisG/arg4::hisG, cap1::hisG/cap1::hisG-URA3-hisG | This work |
| JC45 | ura3::λ imm434/ura3::λimm434, his1::hisG/his1::hisG, hog1::loxP-ura3-loxP/hog1::loxP-HIS1-loxP | This work |
| JC95 | ura3::λ imm434/ura3::λimm434, his1::hisG/his1::hisG, CAP1-GFP-URA3 | This work |
| JC136 | ura3::λ imm434/ura3::λimm434, his1::hisG/his1::hisG, hog1::loxP-ura3 loxP/hog1::loxP-HIS1-loxP, CAP1-GFP-URA3 | This work |
Strain Construction
Oligonucleotide primers used in this study are listed in the Supplementary Data. To analyze the role of Hog1 and Cap1 in the oxidative stress response, hog1, cap1, and hog1 cap1 mutants were created in the same strain background. To delete HOG1, disruption cassettes comprising either the ARG4 or HIS1 gene flanked by loxP sites and 80 base pairs of DNA sequence corresponding to regions of the HOG1 open reading frame (ORF), were generated by PCR using the oligonucleotide primers HOGdelF and HOGdelR (Smith et al., 2004) and the plasmid templates pLAL2 or pLHL2, respectively (Dennison et al., 2005). Disruption cassettes were transformed into C. albicans BWP17 (Wilson et al., 1999) to sequentially disrupt both alleles of HOG1 and generate strain JC47. The CAP1 locus was disrupted by Ura-blasting (Fonzi and Irwin, 1993) in the strains BWP17 (HOG1) and JC47 (hog1), to generate the strains JC128 and JC118, respectively. The HOG1 disruption cassettes deleted codons 45-359 of the 411 codon HOG1 ORF, and the cap1::hisG-URA3-hisG disruption cassette deleted codons 17-500 of the 500-codon CAP1 ORF. Gene disruptions were confirmed by PCR.
To chromosomally tag Cap1 with GFP, CAP1-specific sequences were added to the universal primer sequences described previously (Gerami-Nejad et al., 2001) to generate the oligonucleotide primers CAP1GFP-F and CAP1GFP-R. These primers were used in combination with the template pGFP-URA3 to generate the CAP1-GFP cassettes by PCR (Gerami-Nejad et al., 2001). The CAP1-GFP cassettes were transformed into RM1000 (HOG1) and JC45 (hog1) cells to create C. albicans strains JC95 and JC136, respectively (Table 1). Correct integration at the CAP1 loci was confirmed by PCR and DNA sequencing.
Transcript Profiling
Transcript profiling was performed using isogenic C. albicans strains: wild type (JC52: hog1 cells containing HOG1 reintegrated) and hog1 (JC50; Table 1, Smith et al., 2004). The strains were cultured in YPDAU at 30°C, at 180 rpm to OD600 = 0.5. Cells from the same culture were harvested immediately before and 10 min after stress treatment. Three stress conditions were applied to the cells: Oxidative stress: hydrogen peroxide (H2O2) was added to a final concentration of 5 mM; Heavy metal stress: CdSO4 was added to a final concentration of 0.5 mM; and Osmotic stress: prewarmed YPDAU containing 4 M NaCl was added to a final concentration of 0.3 M NaCl. Cells were collected by centrifugation, frozen rapidly in liquid N2, sheared mechanically using a microdismembrator (Braun, Melsungen, Germany) and RNA prepared by extraction with Trizol Reagent (GibcoBRL, Grand Island, NY), as described previously (Hauser et al., 1998). Cy3- and Cy5-labeled cDNAs were prepared from total RNA, and the probes were hybridized with (nearly) whole genome microarrays containing 6000 C. albicans genes (Eurogentec, Seraing, Belgium). Slides were scanned using a ScanArray Lite scanner (PerkinElmer Life Sciences, Beaconsfield, United Kingdom) and quantified using QuantArray software (version 2.0). Data normalization and analysis were performed using GeneSpring (Silicon Genetics, Redwood City, CA), and statistical analysis was performed using SAM (Significance Analysis of Microarrays; Tusher et al., 2001). Expression ratios were calculated by comparing stressed cells with their unstressed control. Data from at least four independent biological replicates were used for each analysis, and the SAM False Discovery Rate was set at 10%. Lists of genes and their expression profiles are available in the Supplementary Data and at the Galar Fungail website (http://www.pasteur.fr/recherche/unites/Galar_Fungail/). C. albicans gene annotations were obtained from CandidaDB (http://genolist.pasteur.fr/CandidaDB; d'Enfert et al., 2005). Functional categories for C. albicans genes were assigned using gene ontology (GO) resources at SGD (www.yeastgenome.org/GOContents.shtml), and on the basis of the MIPS functional assignments for S. cerevisiae homologues (http://mips.gsf.de/proj/yeast/CYGD/db/index.html), as described previously (Yin et al., 2004). Promoter analysis was performed using GeneSpring.
Identification of Hog1-regulated Genes
In the absence of stress, we defined Hog1-regulated genes as those whose basal expression level was at least 50% higher in HOG1 cells than in hog1 cells (HOG1/hog1 ≥ 1.5) or at least 40% lower in HOG1 cells than in hog1 cells (HOG1/hog1 ≤ 0.60). After stress treatment, we defined Hog1-dependent genes as those that 1) were induced at least twofold in wild-type cells in response to stress, and 2) required Hog1 for at least 40% of the induction seen in wild-type cells (hog1 induction/HOG1 induction ≤ 0.6). In addition, we defined genes which displayed increased induction after stress in hog1 cells as those 1) that were induced at least twofold in wild-type cells in response to stress, and 2) displayed at least 40% greater induction in hog1 cells compared with that seen in wild-type cells (hog1 induction/HOG1 induction ≥ 1.4).
Cross-Comparison of Stress Responses and SAPK Function in C. albicans, S. cerevisiae, and S. pombe. Protein sequences from S. cerevisiae, C. albicans, and S. pombe were used to perform an all-against-all BLASTP search (Altschul et al., 1990). S. cerevisiae and S. pombe sequences were downloaded from NCBI (http://www.ncbi.nlm.nih.gov), S. cerevisiae sequences are from the version dated 25 October, 2004, S. pombe sequences are from the version dated 10 September, 2004, except the mitochondrial sequences which are from 9 January, 2004. C. albicans sequences were downloaded from CandidaDB (ftp://ftp.pasteur.fr/pub/GenomeDB/CandidaDB/FlatFiles). Orthologues were selected on the basis of bidirectional blast hits using a cutoff eValue of E-5, and a set of proteins with orthologues in all three genomes was generated. Bidirectional hits were then ranked on the basis of E-value and the best bidirectional hits identified. This yielded an objective list of the orthologues present in C. albicans, S. cerevisiae, and S. pombe. The expression of this set of orthologues was then compared in the three yeasts by combining the transcript profiling dataset for C. albicans (this study) with equivalent sets from S. cerevisiae (Gasch et al., 2000; Fauchon et al., 2002; O'Rourke and Herskowitz, 2004) and S. pombe (Chen et al., 2003; Supplementary Data).
Northern Analysis
RNA preparation and analysis was performed as described previously (Blackwell et al., 2004). Samples of total RNA (10-15 μg) were denatured with glyoxal, separated on 1.2% agarose gels prepared in 15 mM sodium phosphate (pH 6.5), and transferred to GeneScreen membranes (Dupont NEN Research Products, Boston MA). Gene-specific probes were amplified by PCR from genomic DNA by using the oligonucleotide primers listed in the Supplementary Information. All probes were labeled with [α-32P]dCTP with a Prime-a-Gene labeling kit (Promega, Madison WI).
Stress Sensitivity Tests
For stress sensitivity assays, strains were grown at 30°C to midexponential phase. Cells were diluted in YPD and 103 cells, and 10-fold dilutions thereof, were spotted in 5 μl onto YPD agar containing the specific compound at the indicated concentration. Plates were incubated at 30°C for 24 h.
Fluorescence Microscopy
Localization of chromosomally GFP-tagged Cap1 was determined by fluorescence microscopy. Samples of exponentially growing wild-type (JC95) or hog1 (JC136) cells (OD595 = 0.5) expressing chromosomally tagged Cap1-GFP were untreated or treated with a range of H2O2 concentrations for the indicated times. Cells were collected, fixed in 3.7% para-formaldehyde, spread onto poly-l-lysine-coated slides, and coverslips were mounted onto slides using Vectashield mounting medium containing 1.5 mg/ml DAPI (4′-6-diamidino-2-phenylindole; Vector Laboratories, Burlingame, CA). DAPI and GFP fluorescence were captured by exciting cells with 365- and 450-490-nm wavelengths, respectively, using a Zeiss Axioscope, with a 63× oil immersion objective and AxioVision imaging system. Mean GFP fluorescence intensities in the nucleus and cytoplasm of individual cells were quantified with AxioVision LE software, and ratio of intensity in the nucleus to cytoplasm for each cell was calculated.
RESULTS
Genomewide Transcriptional Response of C. albicans to Stresses That Activate the Hog1 SAPK
Based on studies in S. pombe (Chen et al., 2003), we predicted that conditions which activate C. albicans Hog1 would result in the induction of a common set of genes that are regulated by this SAPK (Smith et al., 2004). Hence in this study we compared the global transcriptional responses of wild-type and hog1 C. albicans cells to environmental stresses that are known to activate the Hog1 SAPK. We compared the homozygous hog1/hog1 null mutant (JC50) with the isogenic HOG1 reintegrant (hog1/hog1/HOG1: JC52) because this controlled for any secondary mutations that might have been introduced during the construction of the null mutant. We have shown that this reintegrant is indistinguishable from its parental wild-type strain RM1000 (HOG1/HOG1) with respect to their stress phenotypes (Smith et al., 2004) and their expression of stress genes (Supplementary Data). Three conditions were chosen for transcript profiling: osmotic stress imposed by 0.3 M NaCl, oxidative stress imposed by 5 mM H2O2, and heavy metal stress imposed by 0.5 mM CdSO4. Each of these treatments stimulates the phosphorylation and nuclear accumulation of this Hog1 SAPK within a 10-min time frame (Smith et al., 2004). Furthermore, significant differences in stress regulated gene expression are observed within this time scale (Enjalbert et al., 2003; Smith et al., 2004). Hence, we analyzed the C. albicans transcriptome after a 10-min exposure to each stress condition. Although some stress genes might be missed by analyzing a single time point, most C. albicans stress genes are induced within 10 min (Enjalbert et al., 2003). At least four independent biological replicates were analyzed for each condition.
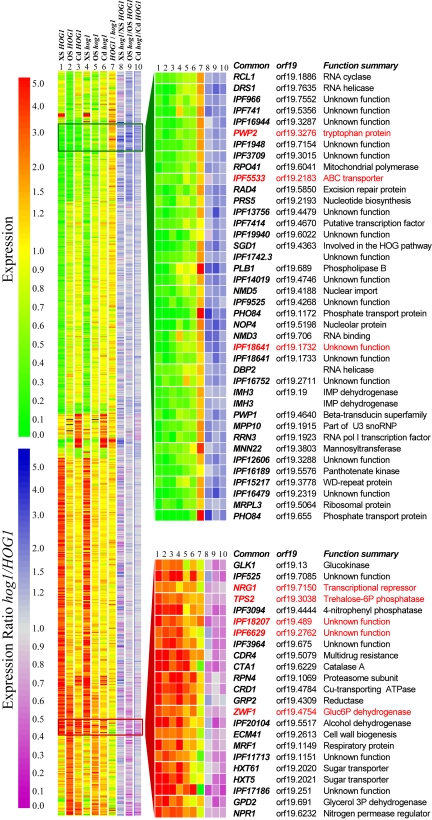
C. albicans genes were clustered on the basis of the degree of similarity in their expression patterns under the conditions examined (Figure 1). Significant differences in both the nature and magnitude of the transcriptional responses of wild-type (HOG1) cells to the three stress conditions were observed. First, more genes responded to the oxidative stress (429 induced, 448 repressed) than to the osmotic stress (178 induced, 190 repressed) and the heavy metal stress (101 induced, 86 repressed). Second, there were clear differences in the subsets of genes that responded to each of the stresses (Figure 1), indicating that C. albicans mounts distinct transcriptional responses to the heavy metal, oxidative, and osmotic stresses examined. However, there was significant overlap between the genes that responded to the osmotic and oxidative stresses (Figure 1, compare lanes 1 and 2). Third, we identified a small subset of genes that responded in a stereotypical manner to all three of the environmental stresses analyzed. These genes represent a core transcriptional stress response in this fungus.
Figure 1.
The global transcriptional response to stress in C. albicans highlights core stress genes. C. albicans genes were clustered on the basis of their expression patterns: left panel, stress-regulated genes; right panel, core stress genes. Expression ratios are indicated by the upper scale bar (red, up-regulation; green, down-regulation): (1) oxidative stress-treated versus untreated HOG1 cells (JC52); (2) osmotic stress-treated versus untreated HOG1 cells; (3) cadmium-treated versus untreated HOG1 cells; (4) oxidative stress-treated versus untreated hog1 cells (JC50); (5) osmotic stress-treated versus untreated hog1 cells; (6) cadmium-treated versus untreated hog1 cells; (7) untreated HOG1 versus untreated hog1 cells. Gene names are provided for core stress genes, with those that did not match the required statistical significance (using SAM) displayed in red. The influence of Hog1 on these expression patterns was estimated by dividing the fold regulation for a particular gene under a particular condition in hog1 cells by the corresponding fold-regulation in HOG1 cells. Calculated hog1/HOG1 ratios are indicated by the lower scale bar (blue, higher expression levels in hog1 cells; purple, lower expression levels in hog1 cells): (8) oxidative stress; (9) osmotic stress; and (10) cadmium stress.
A Core Transcriptional Stress Response in C. albicans
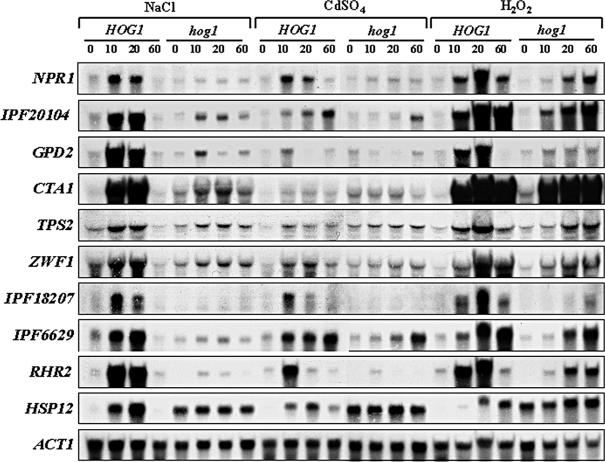
Induced Core Stress Genes. We defined induced core stress genes as those that were induced 1.5-fold or more in response to all three of the stresses examined. The data for 18 of these genes were statistically significant (Figure 1). The induced core stress genes encode proteins involved in redox regulation (CTA1), multidrug resistance (CDR4), carbohydrate metabolism (HXT61, HXT5, GLK1, GPD2, IPF20104), cell wall biogenesis (ECM41), and protein folding and degradation (RPN4, IPF17186). Other core stress genes encode a copper-transporting P-type ATPase (CRD1), an alkaline phosphatase (IPF3094), a mitochondrial respiratory function protein (MRF1), a reductase (GRP2) and a putative protein kinase predicted to regulate the activity of nitrogen source transporters (NPR1). The functions of the remaining induced core stress genes remain obscure (Figure 1). We confirmed the induction of a number of induced core stress genes in response to all three stress conditions by northern blotting (Figure 2). Consistent with the transcript profiling data, the IPF3094, IPF20104, GPD2, and CTA1 mRNAs were shown to be induced by oxidative, osmotic, and heavy metal stresses (Figure 2).
Figure 2.
Northern analysis of core stress genes in C. albicans. RNA was isolated from midlog cultures of HOG1 (JC52) and hog1 (JC50) cells treated with the indicated stresses for 0, 10, 20, and 60 min and analyzed using gene-specific probes. ACT1 was used as a loading control.
According to the transcript profiling data, five additional genes lay just outside the subset of induced core stress genes (Figure 1: genes annotated in red). Changes in transcript level were observed for these five genes under all three stress conditions, but under one of these conditions the change was not statistically significant (according to SAM). However, Northern analyses of these transcripts confirmed that four of these five genes do respond reproducibly to all three of the stress conditions examined (Figure 2). These genes include TPS2 (which is involved in the synthesis of the stress protectant trehalose), ZWF1 (which encodes glucose-6-phosphate dehydrogenase), IPF18207 (which encodes a protein with a high level of sequence similarity to the S. cerevisiae damage response protein, Dap1), and IPF6629 (which encodes a putative thioredoxin peroxidase). The fifth gene, NRG1, was not an induced core stress gene (unpublished data). Northern analysis also confirmed our previous findings that RHR2 (encoding a glycerol phosphatase) and HSP12 (encoding a putative chaperone) are also induced core stress genes (Smith et al., 2004), although these were missed by the transcript profiling analysis.
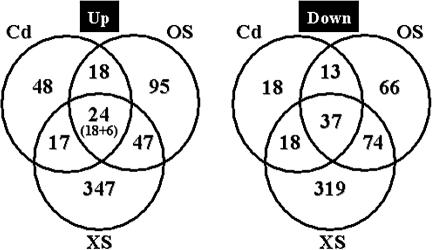
Taken together, our transcript profiling and Northern data have revealed a subset of 24 induced core stress genes in C. albicans. The Venn diagram in Figure 3 illustrates the numbers of genes that are regulated by each of the three stresses analyzed and by the combinations of these stress conditions. This highlights the number of genes that are regulated in a stress-specific manner, the large number of genes that are coregulated in response to osmotic and oxidative stress, and the relatively small subset of core stress genes in C. albicans. The set of induced core stress genes would be reduced to nine genes (ECM41, GLK1, GRP2, HSP12, HXT61, orf19.251, orf19.675, orf19.2762, orf19.7085) if the heat-shock dataset generated by Enjalbert et al., (2003) were included in the analysis (Supplementary Data). Hog1 is not activated in response to heat shock (Smith et al., 2004).
Figure 3.
Venn diagram showing subsets of stress-regulated genes. Genes displaying significant induction of more than 1.5-fold in wild-type cells are included. The numbers of stress regulated genes in each subset are shown: OS, osmotic stress; XS, oxidative stress; Cd, heavy metal stress. The number of induced core stress genes includes those that were revealed by transcript profiling (18) and those that were confirmed or added by Northern analysis (6).
The set of 24 induced core stress genes in C. albicans is smaller than those described in S. cerevisiae and S. pombe (Gasch et al., 2000; Causton et al., 2001, Chen et al., 2003). However, as discussed previously (Enjalbert et al., 2003), these subsets of core stress genes were influenced by the stringency of the criteria used to define them (Gasch et al., 2000; Causton et al., 2001; Chen et al., 2003). In this study, C. albicans core stress genes were defined as those that were induced by all three of the stress conditions examined, whereas Causton et al., (2001) defined S. cerevisiae core stress genes as those that were induced by five of the seven stresses they examined, and Chen et al., (2003) defined S. pombe core stress genes as those that were induced by four of five stresses. Also, the stringency of the statistical analyses has influenced the definition of core stress genes in each organism. In an attempt to parse out these operational differences, we compared roughly equivalent transcript profiling experiments from these three yeasts using equivalent analytical tools. Our C. albicans dataset was reanalyzed alongside S. pombe genes induced after 15 min of exposure to 0.5 mM H2O2, 1 M sorbitol, or 0.5 mM CdSO4 (from the dataset of Chen et al., 2003) and S. cerevisiae genes induced after 10 min of exposure to 0.32 mM H2O2 (Gasch et al., 2000) or 0.5 M KCl (O'Rourke and Herskowitz, 2004), or after 30 min of exposure to 1 mM CdSO4 (Fauchon et al., 2002). For the purposes of this analysis, core stress genes in the three yeasts were defined as being induced in response to all three types of stress (oxidative, osmotic, and heavy metal stress). Using this approach we are likely to have underestimated the proportion of core stress genes in S. cerevisiae. This is because datasets from three different laboratories were required to accumulate the necessary experimental conditions for S. cerevisiae, and hence the level of consistency between these datasets was lower than for the corresponding C. albicans and S. pombe datasets, each of which was generated by a single laboratory. Nevertheless, our comparison revealed that, according to this operational definition, induced core stress genes represented 3.7% of all stress genes in C. albicans, 6.0% of all stress genes in S. cerevisiae, and 22% of all stress genes in S. pombe (Supplementary Data). Therefore, when equivalent transcript profiling datasets are analyzed using equivalent analytical tools, the proportion of core stress genes in C. albicans is smaller than in S. cerevisiae and S. pombe.
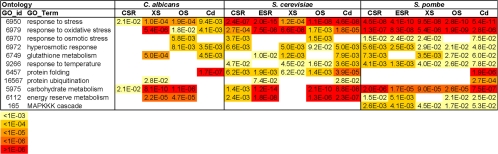
In S. cerevisiae and S. pombe, subgroups of induced core stress genes have been identified with roles in stress responses, metabolism, signaling, and protein folding (Gasch et al., 2000; Causton et al., 2001; Chen et al., 2003). To compare the global roles of core stress genes in the three yeast species, we identified functional categories that are significantly enriched in the sets of core stress genes from C. albicans, S. cerevisiae, and S. pombe. In this analysis, the assignment of genes to specific functional categories was done using the gene ontology (GO) resources at SGD (www.yeastgenome.org/GOContents.shtml). Although fewer induced core stress genes were identified in C. albicans, those that were identified belong to some of the same functional categories as budding and fission yeasts (Figure 4; CSR columns). Response to stress and carbohydrate metabolism genes were significantly overrepresented in the core stress genes from C. albicans, S. cerevisiae, and S. pombe (Figure 4). However, other functional categories that were enriched in the core stress genes of budding and fission yeasts were not overrepresented in C. albicans core stress genes. Although individual oxidative stress, osmotic stress, and energy reserve metabolism genes were part of the C. albicans core stress response, these functional categories were not significantly enriched in the core stress genes. Therefore, there is some overlap, but more functional categories are enriched in the core stress responses of S. cerevisiae and S. pombe than the corresponding response in C. albicans.
Figure 4.
Comparison of GO functional categories that are significantly enriched in subsets of stress genes in C. albicans, S. cerevisiae, and S. pombe. Functional categories that are significantly overrepresented in core, oxidative, osmotic, heavy metal, and heat stress genes sets from C. albicans, S. cerevisiae, and S. pombe were identified using gene ontology (GO) resources at SGD (www.yeastgenome.org/GOContents.shtml). The statistical significance of this enrichment is presented. The list of functional categories was simplified by removing functionally redundant categories and weak probability groups with no clear significance in the response. Core stress genes (CSR) are described in Supplementary Data (Yeast CSRs). Environmental stress response genes (ESR) were as defined by Gasch et al. (2000) and Chen et al. (2003). C. albicans oxidative (XS), osmotic (OS), and heavy metal stress genes (Cd) are defined in this study. S. pombe stress genes were from Chen et al. (2003) (15 min in 0.5 mM H2O2, 1 M sorbitol, 0.5 mM CdSO4,). S. cerevisiae oxidative stress genes were from Gasch et al. (2000) (10 min, 0.32 mM H2O2), osmotic stress genes were from O'Rourke and Herskowitz (2004) (10 min, 0.5 M KCl), and heavy metal stress genes were from Fauchon et al. (2002) (30 min, 1 mM CdSO4).
Repressed Core Stress Genes. We defined repressed core stress genes as being reproducibly repressed 1.5-fold or more under all three of the stress conditions examined. Thirty-four C. albicans genes fell into this category (Figure 1; Supplementary Data), which correlates well with the 38 repressed core stress genes defined previously (Enjalbert et al., 2003). A significant proportion of these genes are involved with protein synthesis and RNA processing (for example, IPF966, IPF3709, NOP4, NMD3, MRPL3, RCL1). Other C. albicans genes that were down-regulated under all three stress conditions are involved in transport (NMD5, PHO84), transcription (RRN3, RPO41, IPF16752), and metabolism (PRS5, IPF7414, IMH3). Again, the subset of repressed core stress genes in C. albicans is relatively small compared with those in S. cerevisiae (Gasch et al., 2000, Causton et al., 2001) and S. pombe (Chen et al., 2003).
Stress-specific Responses
A significant number of C. albicans genes were regulated in a stress-specific manner. The key findings are summarized below, and the complete lists of regulated genes are available in the Supplementary Data and can also be viewed at the Galar Fungail II website (http://www.galarfungail.org/data.htm).
Oxidative Stress. We identified 347 genes whose expression was altered specifically in response to 5 mM H2O2. Many of these genes are known to encode factors involved in the detoxification of H2O2 and free radicals, such as superoxide dismutase (SOD2), and enzymes involved in the thioredoxin (TSA1, TRX1, TRR1) and glutaredoxin (GPX1, GSH1) systems. CTA1, which encodes the key antioxidant enzyme, catalase, is one of the induced core stress genes in C. albicans (Figure 1). The transcription factor, Cap1, which has previously been shown to be important for oxidative stress tolerance (Alarco and Raymond, 1999; Zhang et al., 2000), was also strongly induced in response to oxidative stress (5.7-fold), indicating that this factor might play a key role in the transcriptional responses to oxidative stress in C. albicans (see below).
We compared our transcript profile for C. albicans cells exposed to 5 mM H2O2 with the profile obtained previously for cells treated with a mild oxidative stress (0.4 mM H2O2; Enjalbert et al., 2003). This revealed that the global transcriptional responses of C. albicans to H2O2 depend on the level of peroxide stress (Supplementary Data). Genes involved in the detoxification of peroxide stress were induced under both conditions (CAP1, CTA1, GPX1, GST3, TRR1, TRX1). However, other subsets of genes were differentially induced in response to 0.4 or 5 mM H2O2. For example, a large subset of genes involved in carbohydrate metabolism were only induced in response to high levels of peroxide stress (ICL1, GPM2, GSY1, MLS1, NTH1, PCK1), whereas the DNA-damage response appeared to be evoked specifically at low levels of H2O2 (HNT2, IPF4708, IPF4356, RGA2). Furthermore, genes involved in chromatin silencing and epigenetic regulation were specifically repressed in response to high doses of peroxide stress (DOT4, DOT6, IPF9787, ISW2, SAS10). These differences in C. albicans oxidative stress profiles are reminiscent of previous studies in S. pombe in which different transcriptional responses are evoked depending on the level of peroxide stress (Quinn et al., 2002).
Osmotic Stress. Ninety-five genes were induced specifically in response to treatment with 0.3 M NaCl. As described previously (Enjalbert et al., 2003), genes that one might expect to be induced in response to an osmotic/salt stress were up-regulated after exposure to NaCl. These included known and predicted sugar transporters (STL1, IPF4181, IPF12946) and cation transporters (ENA22, ENA21). Genes involved in glycerol accumulation (GPD2 and RHR2), which is a classic response to osmotic stress (Hohmann, 2002), were found to be core stress genes in C. albicans (Figures 1 and 2).
Heavy Metal Stress. Yeast controls the intracellular levels of heavy metal ions primarily through sequestration by glutathione (gamma-l-glutamyl-l-cysteinyl-glycine). Exposure to heavy metals has been suggested to cause oxidative stress either through the depletion of intracellular glutathione levels and/or the displacement of Zn2+ and Fe2+ from proteins, which leads to the generation of highly reactive hydroxyl radicals (OH*; reviewed in Halliwell and Gutteridge, 1984; Stohs and Bagchi, 1995). In C. albicans, we identified 48 genes that were specifically induced in response to the heavy metal Cd2+. These can be broadly split into two main subgroups. The first subgroup includes genes involved in the biosynthesis of sulfur-containing amino acids that are necessary for the synthesis of glutathione. Significantly, most C. albicans genes involved in sulfate assimilation and sulfur-containing amino acid synthesis were induced by Cd2+. Furthermore, genes involved in glutathione synthesis were also induced, either specifically in response to Cd2+ stress (GSH2) or in response to both Cd2+ and oxidative stress (GSH1). Similar responses have been noted upon Cd2+ stress in S. cerevisiae (Momose and Iwahashi, 2001; Vido et al., 2001; Fauchon et al., 2002), whereas in S. pombe scavenging of glutathione from the external environment rather than glutathione synthesis appears to be the primary response of fission yeast to Cd2+ (Chen et al., 2003).
The second subgroup of genes encodes heat shock proteins and chaperones. Such genes are a major component of the core stress response in S. cerevisiae and S. pombe. However, a number of C. albicans genes encoding heat shock proteins (HSP78, SIS1, HSP90, and HSP60) and chaperones (IPF661, CPR6, SBA1, and YDJ1) were specifically induced in response to Cd2+ stress. This suggests that Cd2+ might induce an unfolded protein response in C. albicans. Consistent with this idea, five of the eight classic heat-shock proteins in C. albicans (HSP12, SSA4, HSP90, HSP60, HSP78) were induced both in response to heavy metal stress (Supplementary Data) and to a temperature upshift (Enjalbert et al., 2003).
Comparison with Budding and Fission Yeasts. We compared the global transcriptional responses of C. albicans to oxidative, osmotic, and heavy metal stresses with those of S. cerevisiae and S. pombe. To achieve this, we reexamined the appropriate transcript profiling datasets from S. cerevisiae and S. pombe, described above for the induced core stress genes (Gasch et al., 2000; Fauchon et al., 2002; Chen et al., 2003; O'Rourke and Herskowitz, 2004). Once again, stress-induced genes were assigned to specific functional categories using the gene ontology (GO) resources at SGD (www.yeastgenome.org/GOContents.shtml). This analysis revealed considerable overlap in the global roles of the stress responses in these three yeasts (Figure 4). In C. albicans, S. cerevisiae, and S. pombe, the following functional categories were significantly enriched in their sets of oxidative stress genes: response to oxidative stress, glutathione metabolism, carbohydrate metabolism, and energy reserve metabolism. In all three yeasts, genes involved in the response to osmotic stress, the hyperosmotic response, the response to oxidative stress, carbohydrate metabolism, and energy reserve metabolism were induced by osmotic stress. Also, in all three yeasts, genes involved in the response to oxidative stress, glutathione metabolism and protein folding were induced by heavy metal stress. Therefore, there is a high degree of functional overlap between the global oxidative, osmotic, and heavy metal stress responses in these three yeasts (Figure 4).
We then compared the transcriptional responses of specific stress genes in C. albicans, S. cerevisiae, and S. pombe. First we identified those C. albicans genes that have orthologues in both budding and fission yeasts. This was done in a systematic manner by selecting the most significant bidirectional hit for each C. albicans protein in the other yeasts (Materials and Methods). From this objective list of C. albicans, S. cerevisiae, and S. pombe orthologues, we compared the transcriptional responses of hallmark stress genes in these yeasts (Supplementary Data). Some genes were regulated in a similar manner in all three yeasts. For example, HSP104 and HSP12 were induced in all three yeasts in response to all three of the stresses. However, other genes displayed different regulatory profiles. For example, Cap1/Yap1 was strongly induced in C. albicans and S. cerevisiae in response to the oxidative stress, whereas S. pombe Pap1 was not induced under equivalent conditions. Also, trehalose metabolism genes were induced in S. cerevisiae and S. pombe in response to osmotic and heavy metal stresses, but with the exception of TPS2, this was not observed in C. albicans (Supplementary Data). We conclude that in general, C. albicans, S. cerevisiae, and S. pombe exploit similar global strategies to protect themselves from these stresses, but that the precise mechanisms by which they execute these strategies have diverged.
Regulation of the C. albicans Transcriptome by Hog1 in the Absence of Stress
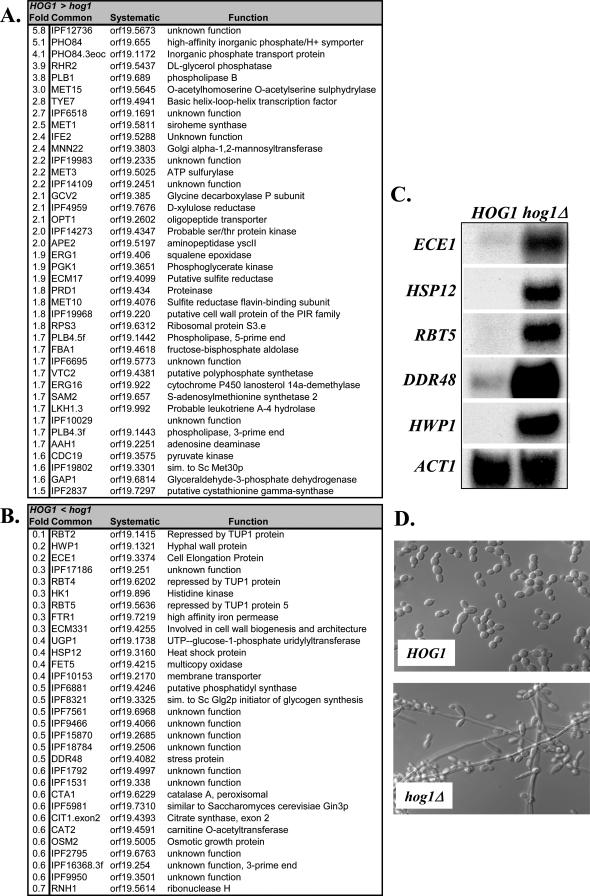
Hog1 plays an important role in the resistance of C. albicans to a wide range of stresses (San-Jose et al., 1996; Alonso-Monge et al., 2003; Smith et al., 2004), indicating that this SAPK is likely to play an important role in transcriptional responses to stress in C. albicans. However, our transcript profiling data revealed that deletion of HOG1 also has significant effects on the C. albicans transcriptome in the absence of stress (Figure 5).
Figure 5.
Deletion of HOG1 has significant effects on the C. albicans transcriptome in the absence of stress. HOG1 (JC52) and hog1 (JC50) cells were grown in YPDUA at 30°C. (A) Transcripts that are elevated in HOG1 cells compared with hog1 cells. (B) Transcripts that are reduced in HOG1 cells compared with hog1 cells. (C) Confirmatory Northern analysis of transcripts in nonstressed HOG1 and hog1 cells. (D) Morphology of nonstressed HOG1 and hog1 cells.
Our genomewide analysis revealed that 40 genes were significantly up-regulated in HOG1 cells compared with the hog1 mutant, suggesting that Hog1 signaling contributes positively to the expression of these genes under basal conditions, either directly or indirectly (Figure 5A). Many of these genes are involved in sulfur amino acid (MET1/3/10/15, ECM17, SAM2, IPF19802/2837), phosphate (PHO84, VTC2), lipid (PLB1/4, ERG1/16), or central carbon metabolism (FBA1, GAP1, PGK1, PYK1). Three of these genes (PHO84, PLB1, MNN22) are repressed core stress genes in C. albicans (Figure 1). There was a corresponding significant overrepresentation of the following functional categories within the subset of genes that were up-regulated in HOG1 compared with hog1 cells: sulfur, methionine, alcohol, and lipid metabolism, and glycolysis.
Thirty-one genes displayed significant down-regulation in HOG1 cells compared with the hog1 mutant, suggesting that Hog1 negatively regulates the expression of these genes (Figure 5B). Interestingly, hypha-specific genes were among the most strongly regulated genes in this category (HWP1, ECE1, RBT2/4/5). These data, which were validated by Northern analysis (Figure 5C), are entirely consistent with the observation that hog1 cells form pseudohyphae and hyphae in the absence of a morphogenetic signal (Figure 5D; Alonso-Monge et al., 1999). Interestingly, 61% of these genes are regulated by the Tup1 repressor (Garciá-Sanchez et al., 2005), suggesting that Hog1 signaling might regulate a subset of genes through Tup1. Stress-related functions (CTA1, DDR48, HSP12, OSM2, IPF17186) were also enriched in the subset of genes that were down-regulated in HOG1 cells and some of these (CTA1, HSP12, IPF17186) are induced core stress genes (Figure 1). Indeed the functional category “oxygen and reactive oxygen species metabolism” was significantly overrepresented in this subset of genes that were negatively regulated by Hog1. Hence Hog1 appears to repress the expression of subsets of stress-related and hypha-specific genes under basal conditions. Our analysis of the C. albicans transcriptome in the absence of stress clearly demonstrates both negative and positive roles for Hog1 in gene regulation, as suggested previously (Smith et al., 2004).
Role of Hog1 in the Regulation of the Core Stress Genes in C. albicans
Having established the influence of Hog1 on the C. albicans transcriptome in the absence of stress, we proceeded to examine the role of Hog1 in the regulation of the core stress genes (Figure 1). Originally, we predicted that the Hog1 SAPK would regulate a core transcriptional response to stress under conditions that activate this SAPK. However, when we tested this prediction, it proved to be only partially correct (Figure 1). Inactivation of HOG1 significantly attenuated the transcriptional response of core stress genes to osmotic and heavy metal stress. However, the effect on the activation of core stress genes in response to oxidative stress was less dramatic (Figure 1; compare column 4 with 5 and 6, and column 8 with 9 and 10).
The role of Hog1 in the regulation of core stress genes was further examined by Northern blotting (Figure 2). Congruent with the microarray data, the induction of the majority of these mRNAs in response to osmotic and heavy metal stress was largely Hog1-dependent. Furthermore, with the exception of three genes (GPD2, IPF18207, and RHR2), which did display some dependency on Hog1 for regulation, HOG1 inactivation did not significantly affect the responses of the remaining core stress genes to oxidative stress. Taken together, our microarray and Northern data clearly show that Hog1 plays a key role in the regulation of many core stress genes in response to osmotic and heavy metal stress, but an alternative pathway(s) regulates the majority of these genes in response to oxidative stress.
Role of Hog1 in the Regulation of Specific Stress Responses in C. albicans
Hog1 is activated in response to oxidative stress, and the deletion of HOG1 increases the sensitivity of C. albicans to a range of reactive oxygen species (Alonso-Monge et al., 2003; Smith et al., 2004). Hence, our observation that Hog1 plays a relatively minor role in the induction of core stress genes in response to oxidative stress was unexpected. Nevertheless, a similar picture emerged when we examined the role of Hog1 in the regulation of stress-specific genes in C. albicans. The inactivation of HOG1 significantly attenuated the transcriptional response to osmotic and heavy metal stress (Figure 1, columns 9 and 10), but had a less dramatic effect on the transcriptional response to oxidative stress (Figure 1, column 8). Lists of genes that displayed Hog1 dependency in response to osmotic, oxidative, and heavy metal stress can be found in the Supplementary Data. The main findings are outlined below.
Osmotic Stress. Deletion of HOG1 significantly attenuated the induction of the majority of the genes that were strongly induced in response to osmotic stress (Figure 1). These genes are predicted to encode osmotic stress protective functions, as described above, which is consistent with previous observations that hog1 cells are more sensitive to osmotic stress than wild-type cells (San-Jose et al., 1996; Smith et al., 2004). Interestingly, inactivation of HOG1 also attenuated the osmotic stress-induction of two genes that encode putative substrates for the SAPK. The first is a serine-threonine protein kinase (Rck2), the homologues of which are known SAPK substrates in S. cerevisiae and S. pombe (Bilsland-Marchesan et al., 2000; Teige et al., 2001; Smith et al., 2002). The second is Sko1, a CRE-binding transcriptional repressor that is a well-characterized Hog1-target in S. cerevisiae (Proft et al., 2001). Hence Hog1 might mediate some aspects of the osmotic stress response through these factors.
Heavy Metal Stress. The inactivation of Hog1 attenuated the induction of many core stress genes in response to Cd2+ (CDR1, CDR4, CTA1, GPD2, HXT5, HXT61, IPF525, IPF3094, IPF3964, IPF6629, IPF18207, IPF1718; Figure 1). In addition, the basal level of expression of seven of the top 10 Cd2+-specific-induced genes was significantly reduced in hog1 cells (ECM1, IPF12736, IPF19802, MET1, MET10, MET15, CYS3; Figure 5). Many of these genes have key roles in sulfur amino-acid biosynthesis, a specific response to Cd2+ toxicity (see above). This could account for the increased sensitivity of hog1 cells to heavy metal stress (Smith et al., 2004). In contrast, Hog1 does not play a significant role in the induction of Cd2+-specific genes.
Oxidative Stress. Inactivation of HOG1 had multiple effects on the global transcriptional response of C. albicans to oxidative stress. Of the 246 genes that were induced more than twofold in response to H2O2, 46 displayed Hog1 dependency for their induction (Supplementary Data: gray squares on CSR Up sheet). However, no genes with obvious anti-oxidant functions were identified in this gene set. Further analysis revealed that of these 246 H2O2-induced genes, 62 were induced to a greater extent in cells than in wild-type cells (Supplementary Data: pink squares on CSR Up sheet). Intriguingly, a number of these genes (GSH1, GPX1, TSA1, TRX1) encode proteins with key anti-oxidant functions. The basis for the increased induction of these anti-oxidant genes in hog1 cells is unknown. This might reflect a role for Hog1 in the repression of this gene set. Alternatively, as hog1 cells have impaired tolerance to reactive oxygen species (Alonso-Monge et al., 2003; Smith et al., 2004), increased induction of antioxidant genes may reflect attempts to restore the redox balance in these cells.
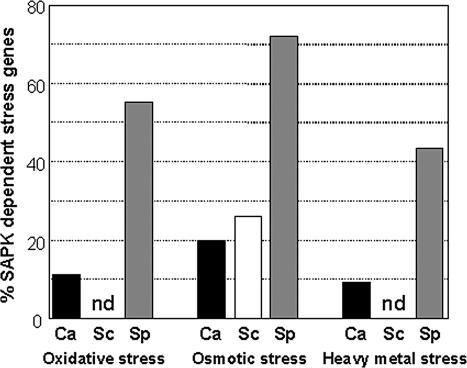
Comparison with Budding and Fission Yeasts. We compared the global roles of the stress activated protein kinases in C. albicans, S. cerevisiae, and S. pombe. To achieve this, the orthologues that displayed transcriptional responses to osmotic, oxidative, or heavy metal stress in at least one of the three yeasts were selected. To assess the impact of the SAPK on this regulation, the transcriptional response of the ortholog in wild-type cells was compared with its response in the corresponding SAPK mutant. Transcript profiling data on S. pombe sty1 mutant cells were from Chen et al. (2003), and data on the transcriptional response of S. cerevisiae hog1 to osmotic stress were from O'Rourke and Herskowitz (2004). However, no data are currently available on the global transcriptional responses of S. cerevisiae hog1 cells to oxidative or heavy metal stresses. Orthologues that displayed any SAPK dependence are listed in Supplementary Data, and the percentage of genes that displayed SAPK dependence under each condition are displayed in Figure 6. This systematic comparison reinforced the view that the Sty1 SAPK plays a central role in the transcriptional responses of S. pombe to osmotic, oxidative, and heavy metal stresses. The Hog1 SAPK also contributes to the corresponding transcriptional responses in C. albicans, and in particular to the response to osmotic stress, as in S. cerevisiae, but to a lesser extent than Sty1 in S. pombe.
Figure 6.
The proportion of gene orthologues that display SAPK-dependent induction in response to oxidative, osmotic, or heavy metal stress in C. albicans, S. cerevisiae, and S. pombe. The set of genes that have orthologues in all three yeasts was identified in a systematic manner (Materials and Methods). From these, those genes that display regulation in response to oxidative, osmotic, or heavy metal stress were selected using the datasets described in Figure 4. The number of genes whose induction was dependent on the SAPK (Hog1 in C. albicans and S. cerevisiae or Sty1 in S. pombe) was then calculated according to the criteria described in Materials and Methods and then expressed as a proportion of the genes that were induced under the same conditions: Ca, C. albicans: Sc, S. cerevisiae; Sp, S. pombe. Note that there are no transcript profiling data that address the role of S. cerevisiae Hog1 during responses to oxidative or heavy metal stresses (nd, no data).
Parallel Stress Pathways in C. albicans
As described above, many core stress genes in C. albicans are dependent on Hog1 for their induction in response to osmotic and heavy metal stress. This suggested that an alternative signaling pathway(s) regulates these genes in response to oxidative stress. One candidate is the Cap1 pathway, given the role of AP-1-related transcription factors in fungal responses to oxidative stress (Moye-Rowley et al., 1989; Toone et al., 1998; Alarco and Raymond, 1999; Zhang et al., 2000; Alonso-Monge et al., 2003) and the observation that CAP1 is significantly induced in response to oxidative stress (see above; Enjalbert et al., 2003).
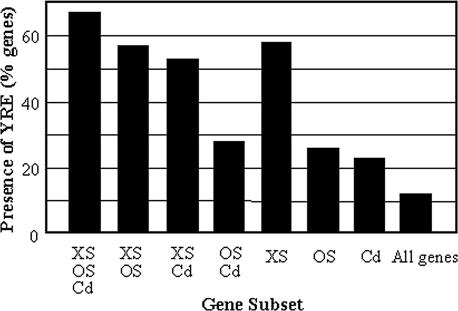
AP-1-like factors activate the transcription of their target genes via the Yap response element (YRE:TKACTAA; Fernandes et al., 1997). In C. albicans, Cap1 has been shown to activate transcription via the YRE (Nicholls et al., 2004), and cap1 cells display increased sensitivity to reactive oxygen species (Alarco and Raymond, 1999; Zhang et al., 2000; Alonso-Monge et al., 2003). Therefore, we performed a systematic promoter analysis to search for putative regulatory elements associated with oxidative-stress induction in C. albicans. This revealed that the YRE is significantly overrepresented in the promoters of genes induced by oxidative stress, compared with osmotic- and heavy metal-induced genes (Figure 7). Hence, those C. albicans genes that are induced in a Hog1-independent manner in response to oxidative stress might be activated by Cap1.
Figure 7.
Proportion of stress-induced genes that contain YRE elements in their promoters. Promoter regions (-800 to -100) were scanned for one or more YRE elements (TKACTAA). Gene subsets: core stress-induced genes (XS, OS, Cd); oxidative and osmotic stress-induced genes (XS, OS); oxidative stress- and cadmium-induced genes (XS, Cd); osmotic stress- and cadmium-induced genes (OS, Cd); top 100 oxidative stress-specific genes (XS); osmotic stress-specific genes (OS); cadmium-specific genes (Cd); all C. albicans genes.
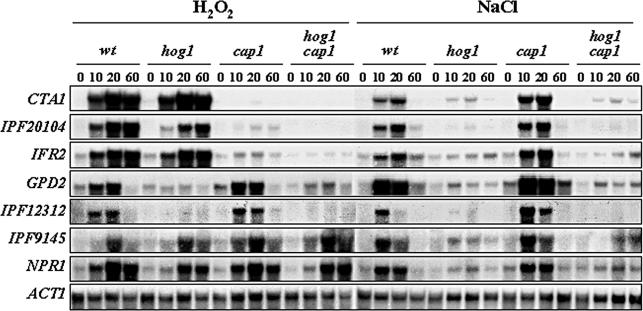
To test this we analyzed the expression of specific C. albicans mRNAs in hog1, cap1, and hog1 cap1 mutants by Northern blotting (Figure 8). Seven mRNAs were chosen for analysis: four core stress genes (CTA1, IPF20104, GPD2, and IPF3094) and three genes that respond strongly to both osmotic and oxidative stresses (IFR2, IPF12312, and IPF9145). As expected, the induction of all seven mRNAs in response to osmotic stress was tightly dependent on Hog1. Furthermore, we found that two of these genes (GPD2 and IPF12312) were also dependent on Hog1 for their expression during oxidative stress. As predicted, the oxidative stress-induction of three genes (CTA1, IPF20104, and IFR2) was significantly attenuated upon inactivation of CAP1 and was independent of Hog1. However, this analysis also uncovered a third oxidative stress signaling pathway in C. albicans: the responses of two genes (IPF3094 and IPF9145) to oxidative stress were independent of both Hog1 and Cap1. Interestingly, all seven genes examined have YREs in their promoters. As only three mRNAs were dependent on Cap1, which suggests either that the sequence context of the YRE within a promoter influences its functionality or that there is redundancy in the oxidative stress induction of some genes.
Figure 8.
Multiple pathways regulate oxidative stress-induced gene expression in C. albicans. Northern blot analysis of RNA isolated from midlog cultures of wild-type (BWP17), hog1 (JC47), cap1 (JC128), and hog1 cap1 (JC118) cells treated with osmotic and oxidative stresses. The indicated gene-specific probes were used and ACT1 was used as a loading control.
In conclusion, this analysis illustrates that Hog1 can act alone or in parallel with distinct signaling pathways in C. albicans to regulate common genes in response to different stress stimuli. The phenomenon of distinct pathways converging on specific promoters to mediate responses to different environmental stimuli is well characterized in S. cerevisiae (Gasch et al., 2000; Causton et al., 2001).
Relationship between Hog1 and Cap1 Signaling in C. albicans
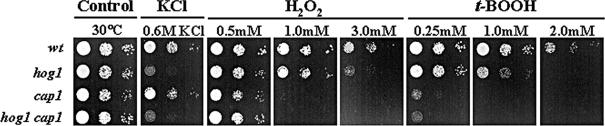
The above analysis indicates that Hog1 and Cap1 execute different roles in the regulation of the transcriptional response to oxidative stress in C. albicans. We predicted, therefore, that this would be reflected in the relative sensitivities of cap1 and hog1 cells to reactive oxygen species. Indeed, cap1 cells were significantly more sensitive than hog1 cells to lower levels of the reactive oxygen species t-BOOH and H2O2 (Figure 9). Also, a hog1 cap1 double mutant was phenotypically identical to the cap1 single mutant with respect to its sensitivity to these reactive oxygen species (Figure 9). In contrast, only hog1 cells were sensitive to osmotic stress. Intriguingly, in a previous study it was reported that hog1 cells are more sensitive than cap1 cells to reactive oxygen species (Alonso-Monge et al., 2003). The basis for this difference is unclear. However, we compared the cap1 mutant constructed in this study with that originally generated by Zhang et al. (2000), and these strains were equally sensitive to low doses of peroxide stress. Furthermore, the phenotypic defects presented in this study were reproducible and were repaired by reintegration of HOG1 or CAP1 into the relevant mutant background (see Supplementary Data). Hence, Cap1 is required for C. albicans to survive both low and high doses of peroxide stress, whereas Hog1 is only necessary for the response to high levels of oxidative stress. This is consistent with our previous observation that Hog1 is activated only in response to high and not low levels of peroxide stress (Smith et al., 2004).
Figure 9.
Phenotypic analysis of C. albicans hog1 and cap1 mutants. Approximately 103 cells, and 10-fold dilutions thereof, of exponentially growing wild-type (BWP17), hog1 (JC47), cap1 (JC128), or hog1cap1 (JC118) cells were spotted onto YPD plates containing the indicated compounds. Plates were incubated at 30°C for 24 h.
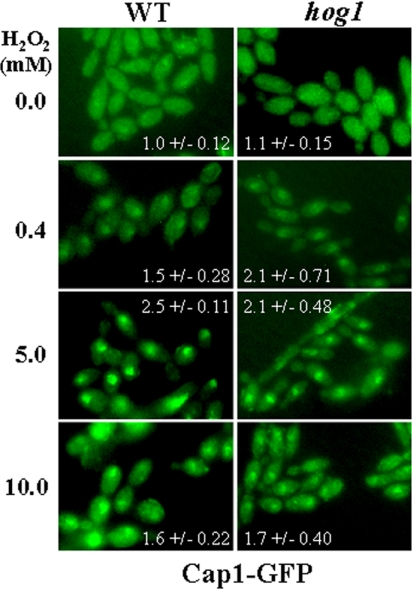
In S. pombe there is well-defined cross-talk between the Sty1 SAPK and the Pap1 pathways (Toone et al., 1998; Quinn et al., 2002). Pap1 accumulates in the nucleus in response to oxidative stress. This nuclear accumulation is dependent on the Sty1 SAPK and is inhibited by increasing peroxide concentrations. To investigate whether Cap1 is regulated in a similar manner in C. albicans, the localization of a Cap1-GFP fusion was examined in HOG1 and hog1 cells after exposure to increasing concentrations of H2O2 (Figure 10). In contrast to the situation in S. pombe, Cap1 rapidly accumulated in the nucleus at both low and high levels of H2O2. Furthermore, no significant differences in the nuclear accumulation of Cap1 were observed between wild-type and hog1 cells, irrespective of the concentration of H2O2. These findings are consistent with the idea that in C. albicans, Cap1 regulates the response to low and high levels of H2O2 independently of Hog1. Furthermore, the observation that Cap1 rapidly accumulates in the nucleus at both low and high levels of H2O2 is consistent with the finding that cap1 cells display sensitivity to both low and high concentrations of oxidative stress (Figure 10).
Figure 10.
The peroxide-induced nuclear accumulation of C. albicans Cap1-GFP occurs at both low and high levels of H2O2 and is independent of Hog1. The localization of Cap1-GFP was determined by fluorescence microscopy in wild-type and hog1 cells treated with both low and high levels of H2O2. Inset numbers represent the ratio of nuclear to cytoplasmic GFP fluorescence in individual cells (mean and SD for more than 30 cells).
DISCUSSION
Previous work showed clearly that C. albicans does not mount a core transcriptional stress response under conditions that activate such responses in S. cerevisiae and S. pombe (Enjalbert et al., 2003). However, the pathogen C. albicans occupies different niches from the benign yeasts, S. cerevisiae and S. pombe. Therefore, it is possible that these yeasts perceive stress differently. Indeed, C. albicans is relatively resistant to certain stress conditions, such as oxidative stress and heat stress, compared with S. cerevisiae and S. pombe (Jamieson et al., 1996; Smith et al., 2004). In addition, although a number of studies have highlighted the importance of the Hog1 SAPK in C. albicans stress responses (San-Jose et al., 1996; Alonso-Monge et al., 2003; Smith et al., 2004), only one of the three stress conditions used in the previous transcript profiling study (Enjalbert et al., 2003) activates the Hog1 SAPK (Smith et al., 2004). Hence in this study we have examined the global transcriptional responses of HOG1 and hog1 C. albicans cells to three stress conditions that activate the Hog1 SAPK: osmotic stress, high level oxidative stress, and heavy metal stress. Several major conclusions can be drawn from our work.
The first main conclusion is that a core transcriptional response to stress does exist in C. albicans. Subsets of core stress genes were identified that were significantly induced or repressed in response to all three stresses (Figures 1-3). Significantly, the core stress genes in C. albicans belong to some of the same functional categories as core stress genes in S. cerevisiae and S. pombe (Figure 4). For example, genes involved in stress responses and carbohydrate metabolism are induced in all three yeasts, suggesting that some of the processes involved in the core stress response are evolutionarily conserved. However, interesting differences were also observed between the core stress responses of C. albicans and those in S. cerevisiae and S. pombe. For example, genes involved in responses to oxidative, osmotic, and temperature stresses are significantly overrepresented in the core stress genes of both S. cerevisiae and S. pombe. In C. albicans, such genes were generally not induced in response to all three stresses and therefore were not defined as being part of the core stress response.
It is important to note that the subset of core stress genes identified in C. albicans is smaller than those in S. cerevisiae and S. pombe (Gasch et al., 2000; Causton et al., 2001, Chen et al., 2003). Although Hog1 is activated under each of the stress conditions examined (Smith et al., 2004), it is possible that the small number of core stress genes in C. albicans might result, not from a small regulon, but from the imposition of relatively mild stress. Also, the number of core stress genes defined by a transcript profiling study is dependent on experimental design and the criteria used to define such genes, as discussed previously (Enjalbert et al., 2003). For example, Causton et al., (2001) defined core stress genes in S. cerevisiae as those that were induced by five of the seven stresses they examined, whereas Chen et al., (2003) defined core stress genes in S. pombe as those that were induced by four of five stresses. In this study, C. albicans core stress genes were defined as those that were induced by all three of the stress conditions examined. Therefore, to compare the core stress responses in C. albicans, S. cerevisiae, and S. pombe in a more direct manner, we analyzed transcript profiling datasets that were generated from these yeasts under roughly equivalent experimental conditions (Supplementary Data). This approach did require S. cerevisiae data from three different laboratories to be used, and the resultant variation did lead to the size of S. cerevisiae core stress response being underestimated relative to the other yeasts. Nevertheless, this systematic analysis reinforced the view that the set of induced core stress genes is genuinely smaller in C. albicans than in budding and fission yeasts (Supplementary Data).
The second main conclusion is that C. albicans induces different subsets of genes in response to osmotic, oxidative, and heavy metal stress. Although the genomewide response to osmotic stress has been published previously (Enjalbert et al., 2003), this is the first report of the transcriptional response to heavy metal stress and high level peroxide stress in C. albicans. Analyses of stress-specific responses in this study revealed three important findings. First, our osmotic stress dataset is consistent with that previously generated by Enjalbert et al. (2003). Second, C. albicans responds to heavy metal stress by inducing genes required for glutathione biosynthesis. Third, exposure of C. albicans to high levels of H2O2 results in a profile that is significantly different from that obtained after treatment of cells with low levels of H2O2 (Enjalbert et al., 2003). Hence, as noted in S. pombe (Quinn et al., 2002) and S. cerevisiae (O'Rourke and Herskowitz, 2004), the intensity of a particular stress plays a crucial role in determining the cellular response to the stress in C. albicans.
The global transcriptional responses of C. albicans to oxidative, osmotic, or heavy metal stress display significant overlap with those in S. cerevisiae and S. pombe (Figure 4). For example, genes involved in the response to oxidative stress and glutathione metabolism were significantly overrepresented among the sets of genes induced by oxidative stress in all three yeasts. Genes involved in the hyperosmotic response and carbohydrate and energy reserve metabolism were enriched in osmotic stress-induced genes in the three yeasts. Also, genes involved in oxidative and hyperosmotic stress responses, glutathione metabolism and protein folding were significantly overrepresented in the sets of Cd2+-induced genes in C. albicans, S. cerevisiae, and S. pombe (Figure 4). Therefore in general, these diverse species appear to exploit similar global strategies to protect themselves from oxidative, osmotic, and heavy metal stress.
However, this similarity does not extend to heat shock. In both S. cerevisiae and S. pombe, heat shock induces a wide range of protective functions (Gasch et al., 2000; Causton et al., 2001, Chen et al., 2003), and a comparison of the functional categories that are significantly overrepresented in these gene sets reveals a high degree of overlap between these yeasts (Supplementary Data). These functional categories include responses to osmotic stress, oxidative stress, and temperature, as well as protein folding and carbohydrate and energy reserve metabolism. In contrast, the C. albicans response to heat shock appears to be more specialized than in budding and fission yeasts, involving the induction of genes involved in the response to stress and carbohydrate metabolism (Supplementary Data).
The third main conclusion is that the role of the Hog1 SAPK in C. albicans gene regulation, according to the Hog1 transcriptome defined in this study, is entirely consistent with the majority of observations relating to Hog1 function in C. albicans (Figure 11). This role is complex, with Hog1 affecting basal levels of gene expression in the absence of stress and gene regulation after exposure to stress. In the absence of stress, hypha-specific genes were significantly up-regulated in hog1 cells. This correlates with the observation that hog1 cells form pseudohyphae and hyphae in the absence of a morphogenetic signal (Figure 5; Alonso-Monge et al., 1999). A number of other genes, many with stress-related functions, were also affected in hog1 cells in the absence of stress (Figure 5). We compared our data from hog1 cells in the absence of stress, with those obtained from cells carrying a deletion in the Ssk1 response regulator protein, which relays stress signals to the Hog1 SAPK module (Chauhan et al., 2003). Consistent with Ssk1 regulating Hog1 function, some genes are deregulated both in ssk1 and hog1 cells (HSP12, CHK1, GPH1, and APE2).
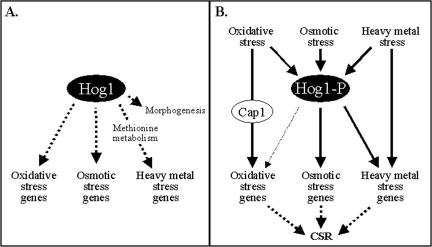
Figure 11.
Model depicting the role of the Hog1 SAPK in the regulation of gene expression in C. albicans. Deletion of HOG1 has effects on both basal and stress-induced gene expression. Notably, hyphal-specific genes and stress-related genes are deregulated in hog1 cells under basal conditions (A). In response to stress, Hog1 plays a central role in the regulation of osmotic- and heavy metal stress-induced gene expression, but a less significant role in the regulation of oxidative stress genes (B). Instead, other pathways, such as the Cap1 pathway, play key roles in the regulation of oxidative stress genes (see text). Hence, the Hog1 SAPK pathway functions in parallel with other pathways, to regulate the core stress response (CSR) in C. albicans.
Our analyses also revealed that Hog1 plays a key role in the global transcriptional response to osmotic and heavy metal stress. In particular, C. albicans genes involved in processes required for heavy metal tolerance (such as sulfur amino-acid biosynthesis) and osmotic stress tolerance (such as glycerol accumulation and sugar and cation transport) were deregulated in hog1 cells (Figure 1). This is consistent with previous findings that Hog1 is activated and translocates to the nucleus in response to osmotic and heavy metal stress and that hog1 cells are sensitive to such stresses (San-Jose et al., 1996; Smith et al., 2004). In addition, our transcript profiling data can account for the previously observed stress-cross protection in C. albicans. Pretreatment of C. albicans cells with an osmotic stress confers significant cross-protection against oxidative stress (Smith et al., 2004). This is consistent with the large number of genes that were induced by both of these conditions (Figures 1-3). Furthermore, we noted that Hog1 regulates the expression of genes encoding key antioxidant enzymes (CTA1, IPF6629) after osmotic stress, which can account for the Hog1 dependency for this stress cross-protection.
In addition, our current work has confirmed that Hog1 contributes to the regulation of the core transcriptional response to stress in C. albicans, although this contribution is smaller than we initially envisaged. The inactivation of HOG1 exerts a less dramatic effect on the transcriptional response to oxidative stress than the responses to osmotic and heavy metal stress, and this is reflected in the role of Hog1 in the regulation of core stress genes in C. albicans. Only three core stress genes displayed some dependency on Hog1 for their activation in response to all stress conditions tested. Instead the majority of core stress genes displayed Hog1 dependency for osmotic and heavy metal stress induction, but not for oxidative stress activation. Our subsequent analyses revealed that Cap1 regulates such core stress genes in response to oxidative stress, in combination with at least one other unidentified pathway. Hence, the Hog1 SAPK pathway functions alone, or in parallel with other pathways, to regulate the core transcriptional response to stress in C. albicans (Figure 11).
Our fourth main conclusion is that the role of the C. albicans SAPK pathway in the regulation of gene expression differs from analogous pathways in S. pombe and S. cerevisiae. In S. pombe, the induction of core stress genes by all stresses, including oxidative stress, is predominantly regulated by the Sty1 SAPK (Chen et al., 2003). The central role of Sty1 was supported by our systematic analysis of orthologues that exist in S. pombe, S. cerevisiae, and C. albicans (Supplementary Data). The induction of large proportion of S. pombe stress genes is dependent on Sty1, whether cells are exposed to an osmotic, oxidative, or heavy metal stress (Figure 6). Hog1 plays a less central role in C. albicans. In this pathogen, the Hog1 SAPK is robustly phosphorylated in response to oxidative stress, and deletion of HOG1 results in C. albicans cells with impaired tolerance to a range of oxidative stress-inducing agents (Alonso-Monge et al., 2003; Smith et al., 2004). It was unexpected, therefore, that Hog1 does not play a major role in the induction of core stress genes in response to oxidative stress in C. albicans. Nonetheless, Hog1 does play a key role in the regulation of core stress genes in response to osmotic and heavy metal stresses in C. albicans (Figures 1 and 2) and contributes to the regulation of some oxidative, osmotic, and heavy metal stress genes (Figure 6; Supplementary Data). This contrasts with the situation in S. cerevisiae, where the maximal induction of core stress genes in response to osmotic stress requires both Hog1 and the general-stress transcription factors Msn2 and Msn4 (O'Rourke and Herskowitz, 2004). Interestingly, the functions of Msn2- and Msn4-related proteins also appear to have been reassigned in C. albicans (Nicholls et al., 2004).
The differing relationships between the SAPK and AP-1 signaling pathways in S. pombe and C. albicans might account for the different roles of the Sty1 and Hog1 SAPKs in oxidative stress signaling in these fungi. The AP-1 transcription factors, Pap1 and Cap1, play central roles in the activation of the transcriptional responses to oxidative stress in S. pombe and C. albicans. However, in S. pombe, the activation of Pap1 is dependent on the Sty1 SAPK (Toone et al., 1998; Quinn et al., 2002; Vivancos et al., 2004; Bozonet et al., 2005), whereas in C. albicans, activation of Cap1 occurs independently of the Hog1 SAPK (Figure 10). This might explain why Sty1 plays a major role in oxidative stress responses in S. pombe, whereas Hog1 plays a less central role in the global transcriptional response of C. albicans to oxidative stress. Nevertheless, the Hog1 SAPK is important for the physiological response to oxidative stress in C. albicans (Figure 9; Alonso-Monge et al., 2003; Smith et al., 2004), possibly because it does regulate some genes in response to oxidative stress (Figures 1, 2, and 6). However, Hog1 might also regulate nontranscriptional responses to oxidative stress in C. albicans. Significantly, a number of SAPK substrates, other than transcription factors, have been identified in S. cerevisiae and S. pombe, including various protein kinases (Bilsland-Marchesan et al., 2000; Teige et al., 2002; Sanchez-Piris et al., 2002; Smith et al., 2002) and cell cycle regulators (Escote et al., 2004).
Our last main conclusion is that C. albicans utilizes different strategies, compared with S. cerevisiae and S. pombe, to regulate the core transcriptional response to stress. This conclusion is supported by several observations. First, as described above, the role of the SAPK pathways in the regulation of core stress genes differs in these fungi. Second, there are significant differences in the relationship between the SAPK and AP-1 signaling pathways in C. albicans and S. pombe. Third, although Msn2 and Msn4 play an important role in the controlling the core transcriptional response to stress in S. cerevisiae (Gasch et al., 2000; Causton et al., 2001), their roles have diverged in C. albicans (Nicholls et al., 2004). Hence, our data reinforce the view that there has been significant divergence between the stress responses in this pathogenic fungus and those in the benign budding and fission yeasts (Enjalbert et al., 2003; Nicholls et al., 2004; Smith et al., 2004). As stress responses are intimately linked with C. albicans virulence, it appears that this fungal pathogen has evolved specialized stress response mechanisms to allow it to adapt to numerous environmental niches within the human host.
Supplementary Material
Acknowledgments
We thank Jill Cheetham, Brian Morgan, Simon Whitehall, and Elizabeth Veal for discussions and comments on the manuscript; Malcolm Whiteway and Andre Nantel for their help and advice; Cheryl Gale and Judith Berman for their kind gift of GFP cassettes; and Dominique Sanglard for the cap1 strain. J.Q. was funded by the Medical Research Council (Career Development Award G120/581) and the Biotechnology and Biological Sciences Research Council (BBSRC) (BB/C510383/1). A.B. was funded by the European Commission (MRTN-CT-2003-504148), the British Council (CRP004), and the BBSRC (BB/C510391/1).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-06-0501) on December 7, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Alarco, A. M., and Raymond, M. (1999). The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181, 700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Monge, R., Navarro-Garcia, F., Molero, G., Diez-Orejas, R., Gustin, M., Pla, J., Sanchez, M., and Nombela, C. (1999). Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 181, 3058-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Monge, R., Navarro-Garcia, F., Roman, E., Negredo, A .I., Eisman, B., Nombela, C., and Pla, J. (2003). The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2, 351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- Bilsland-Marchesan, E., Arino, J., Saito, H., Sunnerhagen, P., and Posas, F. (2000). Rck2 kinase is a substrate for the osmotic stress-activated mitogen-activated protein kinase Hog1. Mol. Cell. Biol. 20, 3887-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell, C., Martin, K. A., Greenall, A., Pidoux, A., Allshire, R. C., and Whitehall, S. K. (2004). The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol. Cell. Biol. 24, 4309-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozonet, S. M., Findlay, V. J., Day, A. M., Cameron, J., Veal, E. A., and Morgan, B. A. (2005). Oxidation of a eukaryotic 2-Cys peroxiredoxin is a molecular switch controlling the transcriptional response to increasing levels of hydrogen peroxide. J. Biol. Chem. Apr 11, [Epub ahead of print]. [DOI] [PubMed]
- Causton, H. C., Ren, B., Koh, S. S., Harbison, C. T., Kanin, E., Jennings, E. G., Lee, T. I., True, H. L., Lander, E. S., and Young, R. A. (2001). Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12, 323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, N., Inglis, D., Roman, E., Pla, J., Li, D., Calera, J. A., and Calderone, R. (2003). Candida albicans response regulator gene SSK1 regulates a subset of genes whose functions are associated with cell wall biosynthesis and adaptation to oxidative stress. Eukaryot. Cell 2, 1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., Toone, W. M., Mata, J., Lyne, R., Burns, G., Kivinen, K., Brazma, A., Jones, N., and Bahler, J. (2003). Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14, 214-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Enfert, C. et al. (2005). CandidaDB: a genome database for Candida albicans pathogenomics. Nucleic Acids Res. 33, D353-D357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison, P.M.J., Ramsdale, M., Manson, C. L., and Brown, A.J.P. (2005). Gene disruption in Candida albicans using a synthetic, codon-optimised Cre-loxP system. Fungal Genet. Biol. 42, 737-748. [DOI] [PubMed] [Google Scholar]
- Enjalbert, B., Nantel, A., and Whiteway, M. (2003). Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14, 1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escote, X., Zapater, M., Clotet, J., and Posas, F. (2004). Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat. Cell Biol. 6, 997-1002. [DOI] [PubMed] [Google Scholar]
- Fauchon, M., Lagniel, G., Aude, J.-C., Lombardia, L., Soularue, P., Petat, C., Marguerie, G., Sentenac, A., Werner, M., and Labarre, J. (2002). Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell 9, 713-723. [DOI] [PubMed] [Google Scholar]
- Fernandes, L., Rodrigues-Pousada, C., and Struhl, K. (1997). Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 17, 6982-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi, W. A., and Irwin, M. Y. (1993). Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sánchez, S., Mavor, A., Russell, C. L. Argimon, S., Dennison, P., Enjalbert, B. and Brown, A.J.P. (2005). Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol. Biol. Cell 10.1091/mbc.E05-01-0071. [DOI] [PMC free article] [PubMed]
- Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D., and Brown, P. O. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerami-Nejad, M., Berman, J., and Gale, C. A. (2001). Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18, 859-864. [DOI] [PubMed] [Google Scholar]
- Halliwell, B., and Gutteridge, J. M. (1984). Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219, 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser, N. C., Vingron, M., Scheideler, M., Krems, B., Hellmuth, K., Entian, K. D., and Hoheisel, J. D. (1998). Transcriptional profiling on all open reading frames of Saccharomyces cerevisiae. Yeast 14, 1209-1221. [DOI] [PubMed] [Google Scholar]
- Hohmann, S. (2002). Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. 66, 300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, C. S., Rhie, G. E., Oh, J. H., Huh, W. K., Yim, H. S., and Kang, S. O. (2002). Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology 148, 3705-3713. [DOI] [PubMed] [Google Scholar]
- Jamieson, D. J., Stephen, D. W., and Terriere, E. C. (1996). Analysis of the adaptive oxidative stress response of Candida albicans. FEMS Microbiol. Lett. 138, 83-88. [DOI] [PubMed] [Google Scholar]
- Lo, H. J., Kohler, J. R., DiDomenico, B., Loebenberg, D., Cacciapuoti, A., and Fink, G. R. (1997). Nonfilamentous C. albicans mutants are avirulent. Cell 90, 939-949. [DOI] [PubMed] [Google Scholar]
- Momose, Y., and Iwahashi, H. (2001). Bioassay of cadmium using a DNA microarray: genome-wide expression patterns of Saccharomyces cerevisiae response to cadmium. Env. Toxicol. Chem. 20, 2353-2360. [DOI] [PubMed] [Google Scholar]
- Moye-Rowley, W. S., Harshman, K. D., and Parker, C. S. (1989). Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 3, 283-292. [DOI] [PubMed] [Google Scholar]
- Nicholls, S., Straffon, M., Enjalbert, B., Nantel, A., Macaskill, S., Whiteway, M., and Brown, A.J.P. (2004). Msn2/4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen, Candida albicans. Eukaryot. Cell 5, 1111-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds, F. C. (1988). Candida and Candidosis, 2nd ed., Bailliere Tindall, London.
- O'Rourke, S. M., and Herskowitz, I. (2004). Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell 15, 532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft, M., Pascual-Ahuir, A., de Nadal, E., Arino, J., Serrano, R., and Posas, F. (2001). Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 20, 1123-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn, J., Findlay, V. J., Dawson, K., Millar, J. B., Jones, N., Morgan, B. A., and Toone, W. M. (2002). Distinct regulatory proteins control the graded transcriptional response to increasing H2O2 levels in fission yeast Schizosaccharomyces pombe. Mol. Biol. Cell 13, 805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Piris, M., Posas, F., Alemany, V., Winge, I., Hidalgo, E., Bachs, O., and Aligue, R. (2002). The serine/threonine kinase Cmk2 is required for oxidative stress response in fission yeast. J. Biol. Chem. 277, 17722-17727. [DOI] [PubMed] [Google Scholar]
- San-Jose, C., Monge, R. A., Perez-Diaz, R., Pla, J., and Nombela, C. (1996). The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J. Bacteriol. 178, 5850-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3-21. [DOI] [PubMed] [Google Scholar]
- Smith, D. A., Toone, W. M., Chen, D., Bahler, J., Jones, N., Morgan, B. A., and Quinn, J. (2002). The Srk1 protein kinase is a target for the Sty1 stress-activated MAPK in fission yeast. J. Biol. Chem. 277, 33411-33421. [DOI] [PubMed] [Google Scholar]
- Smith, D. A., Nicholls, S., Morgan, B. A., Brown, A. J., and Quinn, J. (2004). A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell 15, 4179-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohs, S. J., and Bagchi, D. (1995). Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 18, 321-336. [DOI] [PubMed] [Google Scholar]
- Teige, M., Scheikl, E., Reiser, V., Ruis, H., and Ammerer, G. (2001). Rck2, a member of the calmodulin-protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc. Natl. Acad. Sci. USA 98, 5625-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toone, W. M., and Jones, N. (1998). Stress-activated signalling pathways in yeast. Genes Cells 3, 485-498. [DOI] [PubMed] [Google Scholar]
- Toone, W. M., Kuge, S., Samuels, M., Morgan, B. A., Toda, T., and Jones, N. (1998). Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 12, 1453-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher, V. G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vido, K., Spector, D., Lagniel, G., Lopez, S., Toledano, M. B., and Labarre, J. A. (2001). Proteome analysis of the cadmium response in Saccharomyces cerevisiae. J. Biol. Chem. 276, 8469-8474. [DOI] [PubMed] [Google Scholar]
- Vivancos, A. P., Castillo, E. A., Jones, N., Ayte, J., and Hidalgo, E. (2004). Activation of the redox sensor Pap1 by hydrogen peroxide requires modulation of the intracellular oxidant concentration. Mol. Microbiol. 52, 1427-1435. [DOI] [PubMed] [Google Scholar]
- Wilson, R. B., Davis, D., and Mitchell, A. P. (1999). Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181, 1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysong, D. R., Christin, L., Sugar, A. M., Robbins, P. W., and Diamond, R. D. (1998). Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect. Immun. 66, 1953-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Z., Stead, D., Selway, L., Walker, J., Riba-Garcia, I., McLnerney, T., Gaskell, S., Oliver, S. G., Cash, P., and Brown, A. J. (2004). Proteomic response to amino acid starvation in Candida albicans and Saccharomyces cerevisiae. Proteomics 8, 2425-2436. [DOI] [PubMed] [Google Scholar]
- Zhang, X., De Micheli, M., Coleman, S. T., Sanglard, D., and Moye-Rowley, W. S. (2000). Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol. Microbiol. 36, 618-629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.