Abstract
The structure-specific FEN-1 endonuclease has been implicated in various cellular processes, including DNA replication, repair and recombination. In vertebrate cells, however, no in vivo evidence has been provided so far. Here, we knocked out the FEN-1 gene (FEN1) in the chicken DT40 cell line. Surprisingly, homozygous mutant (FEN1–/–) cells were viable, indicating that FEN-1 is not essential for cell proliferation and thus for Okazaki fragment processing during DNA replication. However, compared with wild-type cells, FEN1–/– cells exhibited a slow growth phenotype, probably due to a high rate of cell death. The mutant cells were hypersensitive to methylmethane sulfonate, N-methyl-N′-nitro-N-nitrosoguanidine and H2O2, but not to UV light, X-rays and etoposide, suggesting that FEN-1 functions in base excision repair in vertebrate cells.
INTRODUCTION
FEN-1 is a structure-specific nuclease with both 5′ flap endonuclease and 5′–3′ exonuclease activities (1–3). The enzyme has been implicated in cellular DNA metabolism in many ways. In DNA replication, the enzyme is believed to be responsible for RNA primer removal during Okazaki fragment processing, since it was identified as a factor responsible for the completion of replication in vitro (4–6). Yeast cells lacking the FEN-1 gene (called RAD27) are viable but unable to grow at high temperatures, indicative of a defect in DNA replication (7).
FEN-1 has also been shown to be involved in DNA repair. In higher eukaryotes, apurinic/apyrimidinic sites occur by endogenous and exogenous mechanisms but are efficiently repaired by base excision repair (BER) (reviewed in 8,9). Extensive biochemical studies using cell extracts or purified proteins have established that there are two major BER pathways (10–13). One is a short-patch BER pathway that relies on DNA polymerase β, X-ray repair cross- complementing 1 and DNA ligase I or III, and the other is a long-patch BER pathway that requires DNA polymerase δ/ε, proliferating cell nuclear antigen, FEN-1 and DNA ligase I. In the long-patch pathway, FEN-1 acts to remove 5′ flap nucleotides displaced during gap-filling DNA synthesis. In yeast, the importance of FEN-1 in BER has been well documented (14,15). In addition, with respect to DNA double-strand break (DSB) repair, recent work with yeast rad27 mutants has suggested a role for FEN-1 in the process of non-homologous end-joining (16). However, in higher eukaryotes the biological significance of FEN-1 remains largely unknown because of the lack of its mutant cells. Therefore, in this study, we knocked out the chicken FEN1 gene in the highly recombinogenic DT40 cell line (17). We show in vivo that FEN-1 is not essential for DNA replication, although it is required for repairing lesions caused by methylating agents and H2O2.
MATERIALS AND METHODS
Plasmid construction
A partial chicken FEN1 cDNA was isolated by reverse transcription–PCR (RT–PCR) with primers designed based on the sequences of the human FEN1 (18) and yeast RAD27 (7) genes. The cDNA was used as a probe to screen for a chicken genomic library (Stratagene). An ∼16 kb genomic clone was obtained, subcloned into pTV119N (Takara Shuzo, Kyoto, Japan), yielding pYM1, and partially sequenced (GenBank accession no. AB058602). The 9 kb NcoI–SalI fragment from the subcloned sequence was used to generate FEN-1 targeting constructs by replacing the 0.4 kb ClaI–NotI region with the hygromycin resistance (hygr) or puromycin resistance (purr) gene flanked by the loxP sequences, yielding FEN1-hyg or FEN1-pur, respectively. A chicken FEN-1 expression plasmid (chFEN1) was constructed by ligating the 12 kb BamHI–SalI fragment of pYM1 with a PGKneo cassette.
Cell culture and transfection
Chicken DT40 cells were cultured in a 5% CO2 incubator at 39°C in ES medium (Nissui Seiyaku, Tokyo, Japan) supplemented with 10 µM 2-mercaptoethanol, 10% fetal bovine serum and 1% chicken serum (growth medium) as described previously (19). For colony formation, cells were grown for 7–9 days in ES medium containing 10 µM 2-mercaptoethanol, 20% fetal bovine serum, 2% chicken serum and 0.15% agarose (soft agarose medium). DNA transfection was performed essentially as described previously (19). Briefly, 4 × 106 cells were electroporated with a plasmid vector and, after an 8 h incubation in growth medium, drug-resistant colonies were selected by incubating them in soft agarose medium containing a selective drug. For gene targeting to generate FEN1–/– mutants, 4 µg of a linearized targeting construct were used and colonies resistant to 1.5 mg/ml hygromycin B (Wako Pure Chemical, Osaka, Japan) or 0.5 µg/ml puromycin 2HCl (Wako Pure Chemical) were selected and grown to mass cultures.
RT–PCR, northern blot analysis and cell-cycle analysis
RT–PCR was performed as described previously (20). The primers used to amplify the full-length FEN1 cDNA were CFN-F (5′-CCTGGATCCATGGGAATCCACGGCCTGGCCAAG-3′) and CFN-R (5′-CCAGCATGCTTAATGATGATGATGATGATGGTCGACTTTCCCCTTTTTGAACTTGGC TGT-3′). Northern blot analysis was performed as described previously (20). The probe used was a chicken FEN1 cDNA synthesized by PCR with primers CFN-5 (5′-CCGTCTACGTGTTCGATGGCAAACCACCTC-3′) and CFN-6 (5′-CGGCTCTTACTCAGCCTCTTGACCCCATTG-3′). The intensity of mRNA bands was quantified by an NIH Image. Cell-cycle analysis was performed as described previously (21).
Drug sensitivity assays
In order to determine sensitivity to methylmethane sulfonate (MMS), N-methyl-N′-nitro-N-nitrosoguanidine (MNNG; Aldrich Chemical), H2O2 (Mitsubishi Gasu Kagaku, Tokyo, Japan) and VP-16 (Sigma), cells were plated at 102–105 cells per dish into 60 mm bacterial dishes containing 5 ml of soft agarose medium with various concentrations of each drug. X-ray sensitivity of cells was assayed as described previously (22). To determine UV sensitivity, cells were harvested, washed twice with Saline G [138 mM NaCl, 5.4 mM KCl, 0.1 mM CaCl, 0.6 mM MgSO4, 1.1 mM Na2HPO4, 1.1 mM KH2PO4, 1 mM glucose and 5 µg/ml phenol red (23)], resuspended in fresh Saline G and exposed to various doses of germicidal UV light (254 nm, 0.2 J/m2/s). The cells were then diluted appropriately with growth medium and plated at 102–104 cells per dish as above. In all experiments, cells were incubated for 7–9 days, and resulting visible colonies were counted. The percent survival was estimated by comparing the number of colonies generated from untreated cells.
RESULTS AND DISCUSSION
Isolation of FEN1–/– cells
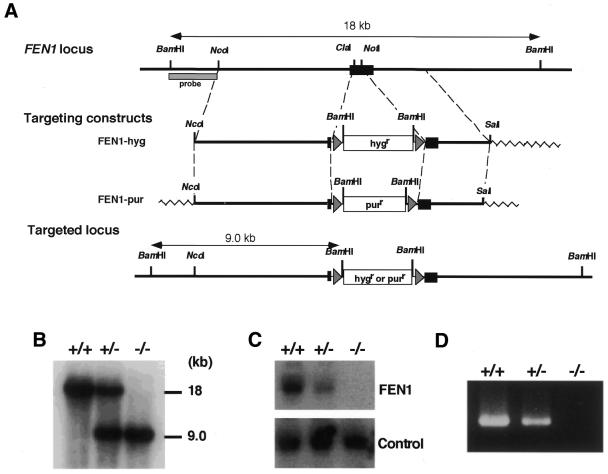
To generate FEN-1 targeting constructs, we isolated a genomic clone containing the chicken FEN1 locus. Inspection of the sequence revealed that the chicken FEN1 gene is encoded by a single exon. An open reading frame in the genomic clone showed ∼84% identity to human FEN-1 protein (18). Southern blot analysis with a chicken FEN1 cDNA probe revealed the existence of a single FEN1 gene in the chicken genome (data not shown). Using this clone, we inserted either the hygr or purr gene in the coding region of the FEN1 locus as shown in Figure 1A. Targeted homologous recombination with these constructs was expected to delete amino acids 34–174, and these targeting events could be detected by the appearance of a 9.0 kb band by Southern blot analysis of BamHI-digested genomic DNA using a 5′ external probe.
Figure 1.
Generation of chicken FEN1–/– cells. (A) Schematic representation of targeted disruption of the chicken FEN1 gene. The chicken FEN1 locus, the two targeting constructs and the resulting targeted locus are shown. The black box indicates the open reading frame of the FEN1 gene. The triangles flanking the hygromycin-resistance (hygr) or puromycin-resistance (purr) gene designate loxP sequences. The figure is not drawn to scale. (B) Southern blot analysis. BamHI-digested genomic DNA of wild-type (+/+), heterozygous mutant (+/–) and homozygous mutant (–/–) cells was hybridized with the probe shown in (A). (C) Northern blot analysis. Total RNA of wild-type, FEN1+/– and FEN1–/– cells was hybridized with a chicken FEN1 cDNA probe. As a control, the same filter was rehybridized with a chicken Ku70 cDNA probe. (D) RT–PCR analysis. Total RNA of wild-type, FEN1+/– and FEN1–/– cells was used as a template to amplify the full-length FEN1 cDNA.
The FEN1-hyg construct linearized with NcoI was first transfected into wild-type (FEN1+/+) DT40 cells by electroporation (19), and hygromycin-resistant clones were examined by Southern blot analysis to isolate heterozygous FEN1+/– mutant clones (Fig. 1B). The resulting FEN1+/– cells were phenotypically indistinguishable from wild-type cells (see below), although the FEN1 mRNA level was reduced to ∼50% in a northern blot (Fig. 1C). One of the FEN1+/– clones was subsequently transfected with the FEN1-pur construct linearized with ScaI, and puromycin-resistant clones were subjected to Southern hybridization to select for homozygous FEN1–/– mutant clones. Given the proposed function of FEN-1 for DNA replication in higher eukaryotic cells (4–6), we assumed that the FEN1 gene would be essential for cell viability. Surprisingly, however, homozygous mutant clones were obtained consistently (Fig. 1B). The gene disruption in these clones was further confirmed by northern blotting (Fig. 1C) and RT–PCR (Fig. 1D). Thus, FEN1–/– cells were indeed viable, indicating that the FEN1 gene is dispensable for cell proliferation. In addition, these results indicate that FEN-1 is not essential for Okazaki fragment processing during DNA replication, although the enzyme has been implicated in the completion of in vitro DNA replication (4–6).
The discrepancy between in vitro and in vivo FEN-1 functions in DNA replication is intriguing. Yeast rad27 cells lacking the FEN-1 gene can survive with a temperature-sensitive phenotype, but a double mutant of FEN1 and exonuclease 1 (EXO1) is lethal (24). It was recently reported that human EXO1 also possesses 5′ flap endonuclease activity (25) as well as 5′→3′ exonuclease activity (24). Therefore, a similar nuclease could compensate for the FEN-1 deficiency in FEN1–/– cells. Alternatively, another nuclease activity may contribute to Okazaki fragment processing. In yeast, the DNA2 gene product (Dna2), which can interact with FEN-1 (Rad27), has recently been shown to display not only 5′→3′ DNA helicase activity but also single-strand-specific endonuclease activity (25,26). Bae et al. (27) showed that in conjunction with FEN-1, Dna2 acts sequentially to facilitate the complete removal of RNA primers in Okazaki fragments. A Dna2 homolog has been identified in human cells (28); such a homolog could substitute for at least the function of FEN-1 in DNA replication.
Growth characteristics of FEN1–/– cells
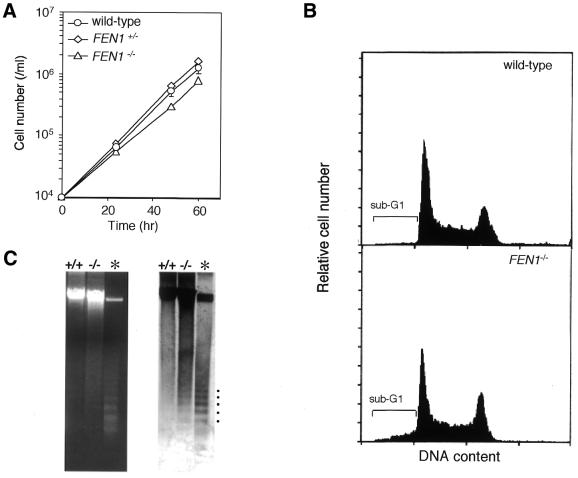
Although FEN1–/– cells were viable, as described above, their growth rate was relatively low compared with wild-type or heterozygous FEN1+/– cells (Fig. 2A). At 39°C, the doubling times of wild-type and FEN1–/– cells were 7.7 and 10.2 h, respectively. This growth defect of FEN1–/– cells was completely restored to normal levels by ectopic expression of the chicken FEN1 gene (data not shown). Remarkably, this growth delay was temperature-independent, contrasting with the temperature-sensitive growth property of yeast rad27 mutants (7): the doubling times of wild-type and FEN1–/– cells were 12.5 and 15.4 h at 35°C, and 7.8 and 10.5 h at 42°C, respectively.
Figure 2.
Growth characteristics of FEN1–/– cells. (A) Growth curves of wild-type, FEN1+/– and FEN1–/– cells. Data shown are the means ± SD of four independent experiments. (B) Cell cycle distribution of wild-type and FEN1–/– cells. Asynchronous cell populations in logarithmic growth phase were subjected to flow cytometric analysis. The brackets indicate sub-G1 fractions. Data shown are from one representative of three independent experiments. (C) Agarose gel electrophoresis of genomic DNA isolated from exponentially growing wild-type and FEN1–/– cells. The gel was stained with ethidium bromide (left panel) and then subjected to Southern blotting using DT40 genomic DNA as probe (right panel). For reference (asterisk), apoptotic ladder bands, which occur in genomic DNA from overgrown DT40 cells, are indicated by dots.
To further compare the proliferative properties of wild-type and FEN1–/– cells, we analyzed the cell-cycle distribution of their asynchronous populations by flow cytometry. As shown in Figure 2B, G1, S and G2/M phase distributions of wild-type and FEN1–/– cells were essentially the same. However, FEN1–/– cells did contain a significantly increased number of sub-G1 cells (8.5% of cells), compared with the wild-type cells (2.5%, usual background levels). Thus, a small fraction of FEN1–/– cells is likely to die by undergoing apoptosis during the cell cycle. To further confirm this death, genomic DNAs were prepared from exponentially growing cells and analyzed by agarose gel electrophoresis. As expected, FEN1–/– DNA consistently exhibited a somewhat smeared pattern, accompanying nucleosomal ladder bands caused by apoptosis (Fig. 2C). These observations suggest that the prolonged doubling time of FEN1–/– cells may be due to a higher rate of spontaneous cell death, rather than an increase in the time length of a single cell cycle. In yeast, rad27 null mutation causes genetic instability at regions of repeated DNA, resulting in DNA strand breaks (29–31). Similarly, in chicken cells, FEN-1 deficiency would cause such genomic instability and strand breaks, thereby leading to apototic cell death.
Increased sensitivity of FEN1–/– cells to methylating agents and H2O2
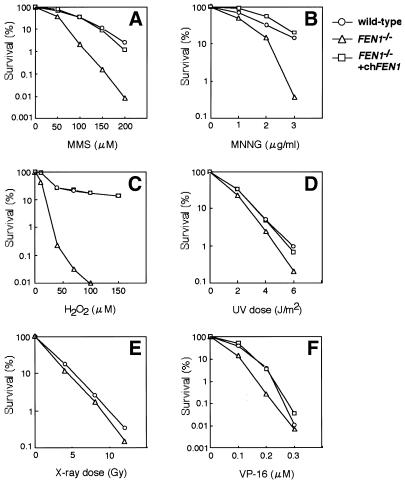
In higher eukaryotic cells, FEN-1 is shown to be involved in long-patch BER by in vitro studies using cell extracts or purified proteins (10–13). However, it is still unclear whether FEN-1 is involved in this repair process in vivo. We therefore investigated the sensitivity to various DNA-damaging agents of FEN1–/– cells. As shown in Figure 3A and B, FEN1–/– cells exhibited a marked sensitivity to alkylating agents MMS and MNNG compared with wild-type cells. These hypersensitivities were almost completely restored to wild-type levels by ectopic expression of the chicken FEN1 gene. Moreover, FEN1–/– cells were extremely sensitive to H2O2; again, this hypersensitivity was completely restored to wild-type levels by ectopic FEN1 expression (Fig. 3C). These results strongly suggest that FEN-1 is involved in BER, in good agreement with in vitro studies (10–13). It has been indicated that MMS-induced lesions can be repaired by both the short- and long-patch BER pathways, whereas H2O2-induced lesions are repaired by the long-patch BER pathway (11,32). Mouse cell lines deficient in DNA polymerase β are more sensitive to MMS than to H2O2 (33), probably reflecting the fact that this polymerase is more active in short-patch BER. In this respect, it is interesting to note that the cytotoxic effect of H2O2 on FEN1–/– DT40 cells was much more pronounced than that of MMS. This may suggest that FEN-1 predominantly participates in the long-patch BER pathway in vivo.
Figure 3.
Sensitivity of FEN1–/– cells to DNA-damaging agents: MMS (A), MNNG (B), H2O2 (C), UV light (D), X-rays (E) and VP-16 (F). Sensitivity assays were performed as described in Materials and Methods. Data are expressed as mean percentages of survival in three or four independent experiments. chFEN1, an expression plasmid for chicken FEN-1.
Although FEN1–/– cells were hypersensitive to methylating agents and H2O2, they were not hypersensitive to UV light (Fig. 3D), indicating that FEN-1 is not required for nucleotide excision repair, a process that removes lesions caused by UV light. Likewise, FEN1–/– cells were not hypersensitive to X-rays (Fig. 3E), consistent with the observation that yeast rad27 mutants are not sensitive to X-rays (7). Also, FEN1–/– cells were only slightly sensitive to VP-16, a DNA topoisomerase II inhibitor that causes DSBs (Fig. 3F). These results suggest that FEN-1 is dispensable for DSB repair in vertebrate cells.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Shunichi Takeda for the chicken DT40 cell line, DNA constructs and helpful suggestions. We also thank C. Nishigaki for excellent technical assistance. This work was supported in part by grant-in-aids from the Ministry of Health and Welfare and the Ministry of Education, Science, Sports and Culture of Japan.
DDBJ/EMBL/GenBank accession no. AB058602
REFERENCES
- 1.Harrington J.J., and Lieber,M.R. (1994) The characterization of a mammalian DNA structure-specific endonuclease. EMBO J., 13, 1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murante R.S., Huang,L., Turchi,J.J. and Bambara,R.A. (1994) The calf 5′ to 3′ exonuclease is also an endonuclease with both activities dependent on primers annealed upstream of the point of cleavage. J. Biol. Chem., 269, 1191–1196. [PubMed] [Google Scholar]
- 3.Lieber M.R., (1997) The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays, 19, 233–240. [DOI] [PubMed] [Google Scholar]
- 4.Ishimi Y., Claude,A., Bullock,P. and Hurwitz,J. (1988) Complete enzymatic synthesis of DNA containing the SV40 origin of replication. J. Biol. Chem., 263, 19723–19733. [PubMed] [Google Scholar]
- 5.Turchi J.J., and Bambara,R.A. (1993) Completion of mammalian lagging strand DNA replication using purified proteins. J. Biol. Chem., 268, 15136–15141. [PubMed] [Google Scholar]
- 6.Waga S., Bauer,G. and Stillman,B. (1994) Reconstitution of complete SV40 DNA replication with purified replication factors. J. Biol. Chem., 269, 10923–10934. [PubMed] [Google Scholar]
- 7.Reagan M.S., Pittenger,C., Siede,W. and Friedberg,E.C. (1995) Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol., 177, 364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindahl T., Karran,P. and Wood,R.D. (1997) DNA excision repair pathways. Curr. Opin. Genet. Dev., 7, 158–169. [DOI] [PubMed] [Google Scholar]
- 9.McCullough A.K., Dodson,M.L. and Lloyd,R.S. (1999) Initiation of base excision repair: glycosylase mechanisms and structures. Annu. Rev. Biochem., 68, 255–285. [DOI] [PubMed] [Google Scholar]
- 10.Frosina G., Fortini,P., Rossi,O., Carrozzini,F., Raspaglio,G., Cox,L.S., Lane,D.P., Abbondandolo,A. and Dogliotti,E. (1996) Two pathways for base excision repair in mammalian cells. J. Biol. Chem., 271, 9573–9578. [DOI] [PubMed] [Google Scholar]
- 11.Klungland A., and Lindahl,T. (1997) Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J., 16, 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim K., Biade,S. and Matsumoto,Y. (1999) Involvement of flap endonuclease 1 in base excision DNA repair. J. Biol. Chem., 273, 8842–8848. [DOI] [PubMed] [Google Scholar]
- 13.Prasad R., Dianov,G.L., Bohr,V.A. and Wilson,S. (2000) FEN1 stimulation of DNA polymerase β mediates an excision step in mammalian long patch base excision repair. J. Biol. Chem., 275, 4460–4466. [DOI] [PubMed] [Google Scholar]
- 14.Wu X., and Wang,Z. (1999) Relationships between yeast Rad27 and Apn1 in response to apurinic/apyrimidinic (AP) sites in DNA. Nucleic Acids Res., 27, 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen R.J., Friedberg,E.C. and Reagan,M.S. (2000) Sensitivity of a S. cerevisiae RAD27 deletion mutant to DNA-damaging agents and in vivo complementation by the human FEN-1 gene. Mutat. Res., 461, 243–248. [DOI] [PubMed] [Google Scholar]
- 16.Wu X., Wilson,T.E. and Lieber,M.R. (1999) A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc. Natl Acad. Sci. USA, 96, 1303–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buerstedde J.-M., and Takeda,S. (1991) Increased ratio of trageted to random integration after transfection of chicken B cell lines. Cell, 67, 179–188. [DOI] [PubMed] [Google Scholar]
- 18.Hiraoka L.R., Harrington,J.J., Gerhard,D.S., Lieber,M.R. and Hsieh,C.-L. (1994) Sequence of human FEN-1, a structure-specific endonuclease, and chromosomal localization of the gene (FEN1) in mouse and human. Genomics, 25, 220–225. [DOI] [PubMed] [Google Scholar]
- 19.Adachi N., Ishino,T., Ishii,Y., Takeda,S. and Koyama,H. (2001) DNA ligase IV-deficient cells are more resistant to ionizing radiation in the absence of Ku70: implications for DNA double-strand break repair. Proc. Natl Acad. Sci. USA, 98, 12109–12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi M., Adachi,N. and Koyama,H. (1998) Characterization of the 3′ untranslated region of mouse DNA topoisomerase IIα mRNA. Gene, 215, 329–337. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama C., Adachi,N. and Koyama,H. (1998) Bleomycin enhances random integration of transfected DNA into a human genome. Mutat. Res., 409, 1–10. [DOI] [PubMed] [Google Scholar]
- 22.Sado K., Ayusawa,D., Enomoto,A., Suganuma,T., Oshimura,M., Sato,K. and Koyama,H. (2001) Identification of a mutated DNA ligase IV gene in the X-ray hypersensitive mutant SX10 of mouse FM3A cells. J. Biol. Chem., 276, 9742–9748. [DOI] [PubMed] [Google Scholar]
- 23.Puck T.T., Ciecura,S.J. and Robinson,A. (1958) Genetics of somatic mammalian cells: III. Long-term cultivation of euploid cells from human and animal subjects. J. Exp. Med., 108, 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tishkoff D.X., Boerger,A.L., Bertrand,P., Filosi,N., Gaida,G.M., Kane,M.F. and Kolodner,R.D. (1997) Identification and characterization of Saccharomyces cerevisiaeEXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl Acad. Sci. USA, 94, 7487–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee B.-I., and Wilson,D.M. (1999) The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J. Biol. Chem., 274, 37763–37769. [DOI] [PubMed] [Google Scholar]
- 26.Bae S.-H., and Seo,Y.-S. (2000) Characterization of the enzymatic properties of the yeast Dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem., 275, 38022–38031. [DOI] [PubMed] [Google Scholar]
- 27.Bae S.-H., Bae,K.-H., Kim,J.-A. and Seo,Y.-S. (2001) RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature, 412, 456–461. [DOI] [PubMed] [Google Scholar]
- 28.Eki T., Okumura,K., Shiratori,A., Abe,M., Nogami,M., Taguchi,H., Shibata,T., Murakami,Y. and Hanaoka,F. (1996) Assignment of the closest human homologue (DNA2L:KIAA0083) of the yeast Dna2 helicase gene to chromosome band 10q21.3–q22.1. Genomics, 37, 408–410. [DOI] [PubMed] [Google Scholar]
- 29.Johnson R.E., Kovvail,G.K., Prakash,L. and Prakash,S. (1995) Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science, 269, 238–240. [DOI] [PubMed] [Google Scholar]
- 30.Tishkoff D.X., Filosi,N., Gaida,G.M. and Kolodner,R.D. (1997) A novel mutation avoidance mechanism dependent on S. cerevisiaeRAD27 is distinct from DNA mismatch repair. Cell, 88, 253–263. [DOI] [PubMed] [Google Scholar]
- 31.Freudenreich C.H., Kantrow,S.M. and Zakian,V.A. (1998) Expansion and length-dependent fragility of CTG repeats in yeast. Science, 279, 853–856. [DOI] [PubMed] [Google Scholar]
- 32.Gary R., Kim,K., Cornelius,H.L., Park,M.S. and Matsumoto,Y. (1999) Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem., 274, 4354–4363. [DOI] [PubMed] [Google Scholar]
- 33.Fortini P., Pascucci,B., Belisario,F. and Dogliotti,E. (2000) DNA polymerase β is required for efficient DNA strand break repair induced by methyl methanesulfonate but not by hydrogen peroxide. Nucleic Acids Res., 28, 3040–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]