Abstract
Earlier studies have shown that herpes simplex virus type 1 (HSV-1) activated protein kinase R (PKR) but that the product of the product of the γ134.5 gene binds and redirects the host phosphatase 1 to dephosphorylate the α subunit of eukaryotic translation initiation factor 2 (eIF-2α). In consequence, the γ134.5 gene product averts the threatened shutoff of protein synthesis caused by activated PKR. Serial passages of Δγ134.5 mutants in human cells led to isolation of two classes of second-site, compensatory mutants. The first, reported earlier, resulted from the juxtaposition of the α promoter of the US12 gene to the coding sequence of the US11 gene. The mutant blocks the phosphorylation of eIF-2α but does not restore the virulence phenotype of the wild-type virus. We report another class of second-site, compensatory mutants that do not map to the US10-12 domain of the HSV-1 genome. All mutants in this series exhibit sustained late protein synthesis, higher yields in human cells, and reduced phosphorylation of PKR that appears to be phosphatase dependent. Specific dephosphorylation of eIF-2α was not demonstrable. At least one mutant in this series exhibited a partial restoration of the virulence phenotype characteristic of the wild-type virus phenotype. The results suggest that the second-site mutations reflect activation of fossilized functions designed to block the interferon response pathways in cells infected with the progenitor of present HSV.
Protein kinase R (PKR) is an important component of host responses to infection. The resident enzyme is inactive in unstressed cells but can be induced by double-stranded RNA and interferon. On activation, PKR dimerizes and phosphorylates key proteins, among which is the α subunit of eukaryotic translation initiation factor 2 (eIF-2α) (19). The net effect is a total shutoff of protein synthesis. Activation of PKR is a common mechanism by which eukaryotic cells respond to the presence of and gene expression by infectious agents. The PKR cascade curtails viral replication and thereby spares the organism in the interim between infection and the immune response. Viruses have evolved a variety of mechanism to block or evade the effects of activated PKR (1–3, 10, 22, 23). In the case of herpes simplex virus type 1 (HSV-1), infected cells accumulate a large amount of cRNA, with the consequence that PKR is uniformly induced by wild-type and mutant viruses tested to date (18, 20). However, the product of an HSV-1 gene, γ134.5, recruits and redirects phosphatase 1α to dephosphorylate eIF-2α, enabling uninterrupted viral protein synthesis (16). The efficacy of the host response and, by extension, that of the γ134.5 gene products are evident from the observations that mutants lacking the γ134.5 gene are highly attenuated in experimental animal systems. Although evidence has been presented to the effect that sequences responsible for the virulent phenotype of wild virus only partially overlap with the domain responsible for the dephosphorylation of eIF-2α (14, 17), a Δγ134.5 mutant was highly virulent in mice defective in the interferon pathway (21). These results suggest that both apparent functions of the γ134.5 gene product have evolved to thwart the interferon-mediated host response to infection.
In an early study, Mohr and Gluzman selected by serial passage a Δγ134.5 mutant derived in the background of a laboratory strain [HSV-1(Patton)], a second-site, compensatory revertant capable of sustained protein synthesis (24). This mutant retained the attenuated phenotype of the parent, Δγ134.5 virus (25). Detailed analyses of this second-site, compensatory mutant showed that, as a consequence of a deletion, the promoter domain was juxtaposed to the coding sequence of the US11 open reading frame (ORF). As a consequence, the US11 ORF was expressed early in infection (15). Subsequent studies have shown that US11 protein expressed early in infection is able to block the shutoff of protein synthesis by a mechanism different from that of the γ134.5 protein (4, 5).
The γ134.5 gene is present in only a few related herpesviruses. The carboxyl-terminal domain of the γ134.5 protein is highly homologous to the corresponding domain of the eukaryotic protein GADD34 (8, 30). Since the homologous GADD34 domain can replace the corresponding γ134.5 domain, the data suggest that HSV or its progenitor acquired the sequence from its host. It could be expected therefore that the progenitor of HSV-1 used an alternative method to block the effect of PKR and that the evolved γ134.5 gene replaced the earlier function. The HSV-1 US11 protein expressed early in infection has the earmarks of a mechanism that may have been used to block the activated PKR, except that this gene too, while more widespread than γ134.5, is not a highly conserved gene. Inasmuch as several viruses express more than one function designed to thwart the interferon pathways, we continued to search for additional HSV-1 genes capable of expressing functions antagonistic to activated PKR. In this article we report that serial passage of the Δγ134.5 mutant R3616 in SK-N-SH cells yielded second-site, compensatory mutants that differ from the previously described mutant in two respects. First, the second site maps elsewhere in the HSV genome. Second, at least one of the mutants in the present series exhibits a partial recovery of the virulence associated with HSV-1.
MATERIALS AND METHODS
Cells and viruses.
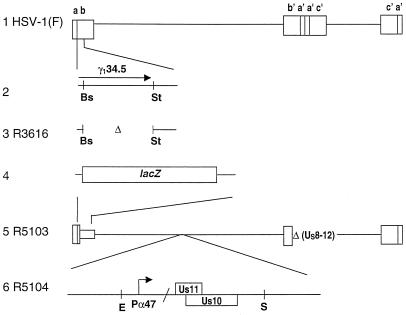
The cell lines used in this study, Vero, HeLa, and SK-N-SH, were obtained from the American Type Culture Collection. The cells were propagated in Dulbecco modified Eagle medium supplemented with 5% newborn calf serum (Vero), 10% (SK-N-SH), or 5% (HeLa) fetal bovine serum. HSV-1(F) is the prototype strain used in the laboratory (12). The recombinant viruses R3616, R5103, and R5104 have been described elsewhere (4, 6). Linear maps of the genomes of these viruses are shown in Fig. 1. R3616 virus lacks 1,000 bp in both copies of the γ134.5 gene. The R5103 virus contains a single copy of the β-galactosidase gene in place of the γ134.5 gene and lacks a portion of the inverted repeats and the ORFs for US8 through US12 (α47) as well as ORF P and ORF O situated antisense to the γ134.5 gene. R5104 contains the native US10 and the α47 promoter-driven US11 ORF inserted into the BglII site of the BamHI Q fragment.
FIG. 1.
Schematic representation of DNA sequence arrangement of the wild-type and recombinant viruses use in this study. Line 1, a schematic representation of the HSV-1 genome. The genome consists of two covalently linked components, L and S, each composed of unique sequences (US and UL) flanked by inverted repeat sequences. Line 2 represents the map location of one copy of the γ134.5 gene. In wild-type virus the γ134.5 gene is present in two copies in the ab and b′a′ inverted repeats. Line 3, representation of the genetic modification in the R3616 virus in which the sequence spanning the BstEII to StuI restriction enzyme sites has been deleted in both gene copies of the γ134.5 coding sequences. Line 4, representation of the ab repeat in R5103 in which the γ134.5 gene was replaced by the Escherichia coli β-galactosidase gene. Line 5, representation of the R5103 virus in the Is isoform, with the S component inverted to the prototypical orientation of the L component. Deleted are portions of the inverted repeats and the sequence encoding US8 through US12. Line 6, representation of the R5104 virus in which the UL23 gene in R5103 is disrupted by the insertion of the EcoRI-SalI fragment expressing the US10 gene and the US11 coding sequence under control of the α47 promoter. Abbreviations: Bs, BstEII; E, EcoRI; St, StuI; and S, SalI.
Serial passage of Δγ134.5 viruses R3616 and R5103.
R5103 and R3616 were each independently passaged serially in SK-N-SH cells. The progeny obtained after each passage was titered in Vero cells. When the titer of either virus was reduced below 102 PFU/ml, the virus was enriched by growth on Vero cells to attain a sustainable titer. After six serial passages, 10 plaque isolates were purified from both the R5103- and R3616-infected SK-N-SH cells. High-titer stock of each isolate was generated, and the protein synthesis phenotype was tested. R5103 was serially passaged a total of 10 times in SK-N-SH cells, and the progeny of the last passage was again retested for the protein synthesis phenotype. Viruses R5121 to R5130 all represent plaque-purified progeny from the serially passaged R3616 virus. Viruses R5131 to R5140 represent 10 different plaque-purified viruses derived from R5103 following 10 serial passages in SK-N-SH cells.
Analysis of viral DNA.
Digestion of viral DNAs with BamHI restriction endonuclease, electrophoretic separation in agarose gel, capillary transfer to Zeta Probe membranes, and hybridization by radiolabeled DNA probes were done as previously described (4, 28). The EcoRI-SalI fragment encoding the origin of viral DNA synthesis and the US10-12 of HSV-1 DNA derived from the R5121, R5122, R5124, R5125, R5126, HSV-1(F), or R3616 viral DNA were ligated with the EcoRI-SalI-cleaved pGEM3zF(+). The DNA fragments were then sequenced by automated dye termination technique (Perkin-Elmer) as well as by the T7 Sequenase (Amersham-Pharmacia) chain termination technique according to the protocol recommended by the manufacturer.
In vivo protein synthesis.
Protein labeling experiments were performed as previously described (7). Briefly, SK-N-SH cells grown in 25-cm2 flasks were infected with 10 PFU of virus per cell. At 12.5 h after infection, the medium was replaced with 199V lacking methionine and was incubated for 30 min. At that time the medium was again replaced with 199V lacking methionine but was supplemented with 50 μCi of l-[35S]methionine (>1,000 Ci/mmol; Amersham Pharmacia). After 1 h of labeling, the cells were rinsed twice in ice-cold phosphate-buffered saline lacking Ca and Mg (PBS-A), scraped, rinsed again with PBS-A, resuspended in disruption buffer, boiled, and loaded on a 12% (vol/vol) polyacrylamide gel cross-linked with N,N′-diallyltartardiamide. The proteins were electrophoretically separated, electrically transferred to nitrocellulose membranes, and subjected to autoradiography.
Determination of the status of eIF-2α and of phosphatase activity directed at eIF-2α protein.
The procedures were as previously described (4). Briefly, cytoplasmic S10 fractions were obtained from the HeLa cells harvested at 6 h after infection. The eIF-2α-specific phosphatase activity was measured by reacting radiolabeled eIF-2 [1.0 pM eIF-2(α-32P) and 0.7 pM eIF-2(β-32P) at 0.6 Ci/mmol] with 6 μl of S10 fractions supplemented with 4.5 μl of TKM buffer (10 mM Tris-HCl, [pH 7.5], 20 mM KCl, and 2.25 mM Mg2Cl) and 1.0 mM ATP in a final volume of 12.5 μl at 34°C. Aliquots (6 μl) were removed from the HSV-1(F) reaction at 20 and 60 s, and the reaction was stopped by denaturation in sodium dodecyl sulfate. For the remaining samples, 6-μl aliquots were removed at 60 and 150 s and were treated in a fashion similar to those of the HSV-1(F) samples. All samples were subjected to electrophoresis on a 7% denaturing polyacrylamide gel, stained, dried, and subjected to autoradiography. The eIF-2α band was cut from each gel lane, and its Cerenkov radioactivity was measured.
To measure the phosphorylation of eIF-2α activity present in the infected cell lysates, the equivalent of 0.2 μl of each lysate was incubated with 5 nM purified eIF-2 and 0.04 mM [γ-32P]ATP (25 Ci/mmol) in 10 μl of TKM buffer for 1 min at 34°C. The reactions were terminated and electrophoretically separated on a 7% denaturing polyacrylamide gel (13). The gel was then silver stained, dried, and subjected to autoradiography as described elsewhere (16).
In vitro PKR kinase assays.
The assays were done as previously described (9, 15). Briefly, S10 fractions were obtained from replicate HeLa cultures harvested 6 h after mock infection or infection as described elsewhere (14). The S10 fractions were reacted with [γ-32P]ATP (100 μCi/sample) for 30 min at 30°C and were then precleared with protein A-agarose and reacted with 1 μg of antibody to PKR (Santa Cruz Biotechnology K-17) in the presence of absence of 50 mM NaF depending upon the assay. The PKR-antibody complexes were precipitated using protein-A agarose and were rinsed with RIPA buffer (PBS containing 0.25% NP-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate) alone or with 50 mM NaF depending on the assay, solubilized in disruption buffer, electrophoretically separated in a 10% denaturing polyacrylamide gel, transferred to a nitrocellulose sheet, and subjected to autoradiography.
Neurovirulence studies.
Female CBA/J mice, 4 to 6 weeks of age, were injected intracerebrally with the aid of a stereotactic frame, in groups of 10 mice with 10 μl of graded doses of HSV R3616 (Δγ134.5; range, 1.3 × 107 to 1.3 × 103 PFU), HSV R5122 (107 to 103 PFU), or R5126 (1.2 × 107 to 1.2 × 103 PFU). As a positive control, three groups of five mice each were injected intracerebrally with 10 μl of graded doses of HSV-1(F) (1,000, 200, and 50 PFU). Survival was followed for 21 days, and the 50% lethal dose (LD50) for each virus was calculated by the Spearman-Karber method (11).
RESULTS
Serial passage of R3616 in SK-N-SH yielded compensatory mutant viruses that exhibit enhanced replication.
The recombinant viruses R3616 and R5103 were each independently serially passaged in SK-N-SH cells. Following six passages, the progeny of R3616 recombinant exhibited enhanced replication when compared with R5103, the progeny of R5103 serial passages, or of R3616. Thus, the R3616 progeny produced plaques on SK-N-SH cells at 100- to 1,000-fold higher dilutions than did equivalent titers of R3616 or R5103. Ten plaques were picked from the serially passaged R3616 virus and grown to high titer on Vero cells, and each was assigned a number from R5121 to R5130. The R5103 progeny population was subjected to an additional four rounds for a total of 10 passages in SK-N-SH cells, and then 10 plaque isolates were purified, grown to high titer on Vero cells, and assigned individual numbers ranging from R5131 to R5140.
Serially passaged viruses R5121 to R5130 maintain viral protein synthesis late in infection, whereas protein synthesis is shut off in cells infected with R3616, R5103, or serially passaged recombinant viruses R5131 to R5140.
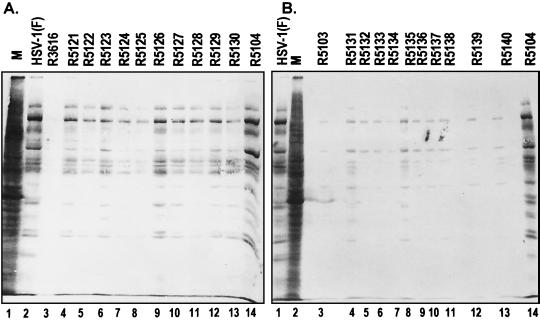
The objective of this series of experiments was to test whether the enhanced replication of serially passaged R3616 virus isolates was the result of improved viral protein synthesis at late times of infection. As described in Materials and Methods, replicate cultures of SK-N-SH cells were mock infected or exposed to 10 PFU per cell of HSV-1(F), R3616, R5121 to R5130, R5103, R5141 to R5150, or R5104. All cultures were incubated in medium containing [35S]methionine at 13 h after infection for 1 h and were then harvested, lysed, denatured, subjected to electrophoresis in denaturing gels, and processed for radiography as described in Materials and Methods. The results (Fig. 2) were as follows:
FIG. 2.
(A) Autoradiographic image of electrophoretically separated [35S]methionine-labeled proteins prepared from mock or virus infected cells. Replicate cultures of SK-N-SH cells were either mock infected (lanes M) or infected with 10 PFU of HSV-1(F), R3616, R5121 to R5130 (10 plaque-purified progeny of serially passaged R3616), or R5104 per cell. At 12 h after infection the mock- or virus-infected cells were incubated in medium that lacked methionine but was supplemented with 50 μCi of [35S]methionine. After 1 h of labeling the cells were rinsed, lysed with disruption buffer, boiled, and electrophoretically separated by denaturing polyacrylamide gel electrophoresis. The proteins were then electrically transferred to nitrocellulose membrane, dried, and subjected to autoradiography. (B) Autoradiographic image of electrophoretically separated [35S]methionine-labeled proteins prepared from mock- or virus-infected cells. The experimental conditions were identical to those described in the legend to panel A except that the SK-N-SH cells were mock infected or infected with HSV-1(F), R5103, R5131 to R5140 (10 plaque-purified progeny of serially passaged R5103), or R5104.
(i) Consistent with previously reported results, at 13 h after infection protein synthesis was totally shut off in cells infected with the R3616 mutant. The synthesis of proteins in cells infected with R5104 was similar to that of the wild-type parent virus HSV-1(F). In contrast protein synthesis in cells infected with the 10 isolates of R3616 progeny passaged serially six times in SK-N-SH cells was better than that observed in R3616 virus-infected cells but was nevertheless highly variable.
(ii) In contrast to the results obtained by serially passaging R3616, the serially passaged R5103 mutant did not acquire compensatory mutations that enabled it to synthesize proteins late in infection. Thus, protein synthesis was uniformly shut off in cells infected with each of the 10 isolates obtained after 10 serial passages of R5103 mutant (Fig. 2B).
These results indicate that six serial passages in SK-N-SH cells enabled R3616 mutant to accumulate compensatory mutations that overcame the absence of the γ134.5 gene. In contrast, 10 serial passages of the R5103 mutant failed to enable the selection of compensatory mutations.
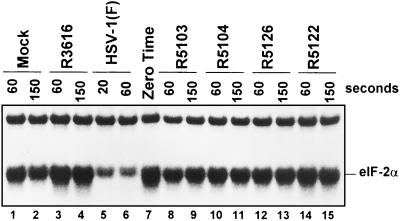
The dephosphorylation of eIF-2α-32P in cells infected with R5122 or R5126 cannot be differentiated from that of cells infected with R3616 recombinant.
Earlier studies demonstrated that ICP34.5, the protein product of the γ134.5 gene, binds to protein phosphatase 1 and diverts it to dephosphorylate eIF-2α, to enable continued protein synthesis in wild-type, infected cells (16). This activity is absent in cells infected with the Δγ134.5 virus. The objective of this series of experiments was to determine whether SK-N-SH cells infected with recombinant virus R5122 or R5126 (i) selectively dephosphorylate eIF-2α and (ii) exhibit the eIF-2α-specific phosphatase activity characteristic of wild-type, virus-infected cells.
As described in Materials and Methods, purified, radiolabeled eIF-2α was incubated with S10 fractions prepared from the lysates of replicate cultures of HeLa cells that were either mock infected or infected with HSV-1(F), R3616, R5103, R5104, R5122, or R5126. The results were consistent with previous studies (4, 16) and demonstrate that, in wild-type virus-infected cells (Fig. 3, compare lanes 5 and 6 with lane 7), a phosphatase rapidly dephosphorylated eIF-2α but not the Mr-39,000 protein that copurifies with eIF-2α. In the mock-infected (Fig. 3, lanes 1, 2, and 7) and Δγ134.5 virus-infected (R3616 [Fig. 3, lanes 3, 4, and 7], R5103 [lanes 7, 8, and 9], R5104 [lanes 7, 10, and 11], R5122 [lanes 7, 12, and 13], and R5126 [lanes 7, 14, and 15]) cell lysates, the specific phosphatase activity was markedly reduced (Fig. 3).
FIG. 3.
Autoradiographic image of purified, in vitro labeled eIF-2α after reaction with mock- or virus-infected cell lysates for the intervals shown. The reactions were terminated by the addition of disruption buffer. The sample marked zero time represents the labeled eIF-2 prior to addition of the cell lysates. The arrow points to the α subunit of eIF-2. The Mr-39,000 protein forms the slower-migrating portion of the band. The slowly migrating protein at the top of the figure is the β subunit of eIF-2.
The results of this experiment indicate that cells infected with representative isolate R5122 or R5126 maintain protein synthesis at late times of infection yet lack the eIF-2α-specific phosphatase activity observed in wild-type virus-infected cells. These results therefore suggest that the secondary mutations in R5122 and R5126 enable continued protein synthesis in infected cells by precluding the activation of PKR or by blocking the phosphorylation of eIF-2α by activated PKR.
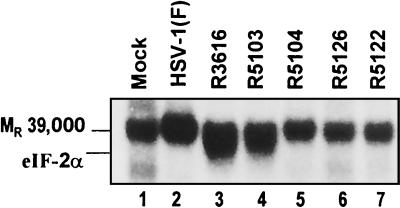
eIF-2α is not phosphorylated in cells infected with R5122 or R5126.
Earlier studies involving a recombinant virus, R5104, capable of wild-type protein synthesis indicated that a virally induced factor precluded the phosphorylation of eIF-2α by activated PKR. To test whether eIF-2α is phosphorylated in cells infected with R5122 or R5126, we measured phosphorylation of eIF-2α by infected cell lysates in the presence of [γ-32P]ATP. The results (Fig. 4) show that the level of eIF-2α phosphorylation correlated with late protein synthesis in the infected cells. As shown in this figure, phosphorylation of the Mr-39,000 protein present in the eIF-2 sample is equivalent (Fig. 4, lanes 1 to 7). The closely migrating eIF-2α band, however, was phosphorylated at high levels only in the presence of S10 fraction from lysates of R3616 mutant-infected cells (Fig. 4, lane 4) or of R5103 mutant-infected cells (Fig. 4, lane 5). The decreased eIF-2α phosphorylation in the wild-type virus-infected cell lysate (Fig. 4, lane 2) is predictable, given the increased phosphatase activity demonstrated in the prior experiment (Fig. 3, lanes 5 to 7). The R5126 and R5122 samples do not contain this eIF-2α-specific phosphatase activity. The reduced phosphorylation of eIF-2α in the presence of R5122-infected (Fig. 4, lane 6) and R5126-infected (Fig. 4, lane 7) cell lysates, therefore, indicates that either PKR is not activated or the phosphorylation of eIF-2α by activated PKR is blocked.
FIG. 4.
Autoradiographic image of electrophoretically separated radiolabeled S10 samples. Lysates prepared from mock- or virus-infected HeLa cells were incubated with [γ-32P]ATP in the presence or absence of added purified eIF-2 as described in Materials and Methods. The arrow indicates the position of eIF-2α relative to the unrelated Mr-39,000 protein present in the eIF-2 preparation.
PKR is activated in cells infected with R5122 or R5126 recombinant virus.
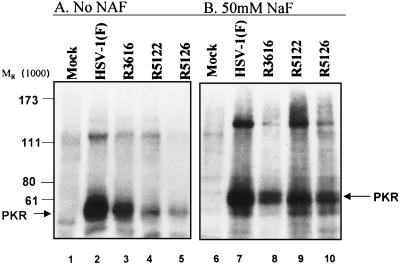
The purpose of this experiment was to determine whether PKR is activated in cells infected with R5122 or R5126. Earlier studies have demonstrated that PKR is activated in HSV-1-infected cells irrespective of the γ134.5 gene expression (4, 9, 15). Earlier studies also indicated that a Δγ134.5 virus with a compensatory mutation that restored late protein synthesis as a consequence of juxtaposition of the promoter of the α47 gene next to the US11 ORF expression also activated PKR (4). To determine whether PKR was activated in cells infected with R5122 or R5126 mutant, we used an assay described earlier in Materials and Methods in which PKR was immune precipitated from S10 fractions and was incubated in the presence of [γ-32P]ATP (150 mCi/ml) (9, 15). The experiment was then repeated, and the immunoprecipitation was performed in the presence of the phosphatase inhibitor NaF. The S10 fractions were radiolabeled with [γ-32P]ATP for 30 min, precleared with protein A, and then incubated with 1 μg of anti-PKR antibody (Santa Cruz Biotechnology K-17) in the presence of 50 mM NaF. The rest of the immunoprecipitation experiment was done in the same fashion, except that 50 mM NaF was present in all of the rinse buffers and protein A immunoprecipitation steps.
Figure 5 shows results of the PKR immunoprecipitation assays. The autoradiographic image (Fig. 5A) is consistent with data published earlier demonstrating that the antibody precipitates, from lysates of infected cells, a phosphorylated protein with an apparent Mr of 68,000 (Fig. 5A, lanes 2 to 5) that is absent in the mock-infected cell sample (lane 1). The salient feature of the results shown in this figure is a decrease in the relative amount of radioactively labeled PKR from the S10 fraction of cells infected with R5126 or R5122. To determine whether the decreased phosphorylation reflected nonspecific phosphatase activity in the Δγ134.5 virus samples, the experiment was repeated in the presence of the phosphatase inhibitor NaF. The results shown in Fig. 5B indicate that PKR is activated in all of the virus-infected cell samples. The PKR immunoprecipitated (Fig. 5, lane 1) from the mock-infected cells is not phosphorylated, indicating that PKR in mock-infected cells is not phosphorylated; therefore, the phosphorylation of PKR in infected cells is a specific process related to infection. The results lend themselves to the conclusion that a phosphatase activity is enhanced in cell infected with the second-site, compensatory mutants but that this activity is absent from R3616 mutant-infected cells.
FIG. 5.
(A) Autoradiographic image of immunoprecipitated PKR derived from radiolabeled S10 fractions prepared from replicate HeLa cells either mock infected or infected with HSV-1(F), R3616, R5126, or R5122 and harvested at 6 h after infection without added NaF. (B) Autoradiographic image of immunoprecipitated PKR derived from radiolabeled S10 fractions prepared from replicate HeLa cells either mock infected or infected with HSV-1(F), R3616, R5126, or R5122 and harvested at 6 h after infection. After an initial 30-min incubation with [γ-32P]ATP, the samples were subsequently maintained in a 50 mM NaF throughout the remainder of the experiment.
The mutations in R5126 virus partially restore neurovirulence.
Earlier studies on Δγ134.5 viruses with second-site, compensatory mutations demonstrated that early expression of US11 protein enabled protein synthesis at late times after infection of SK-N-SH cells but did not restore the ability of this virus to replicate efficiently in the brains of intracerebrally inoculated mice. These results reinforced the conclusion of earlier studies, indicating that the γ134.5 gene encoded at least two independent functions: the maintenance of viral protein synthesis in infected human cells and enhanced replication in the central nervous systems of infected mice (14, 29).
While the experiments detailed above demonstrated that R5122 and R5126 enable continued viral protein synthesis and enhanced replication in infected SK-N-SH cells, their activity in an in vivo animal model was unknown. The objectives of these experiments were to determine whether the compensatory mutations in R5122 and R5126 enabled the virus to replicate, spread, and cause fatal encephalitis in mice. Specifically, three sets of 50 mice were infected intracerebrally with R3616, R5122, or R5126. Each set of 50 mice was subdivided into five groups, with each group receiving a different logarithmic dilution of virus, ranging from 1.3 × 107 to 1.3 × 103 PFU. In addition, a smaller number of mice were intracerebrally infected with wild-type virus at 50, 200, and 500 PFU as a positive control. The mice were maintained for 21 days. The LD50s were calculated by the Spearman-Karber method (11).
The results of these studies were as follows: as expected, HSV-1(F) yielded an LD50 of 100 PFU, whereas R3616 was avirulent (LD50 > 1.3 × 107 PFU), with only one death among the group of mice receiving the highest dose of virus. R5122 (LD50 = 107 PFU) was not significantly more neurovirulent than R3616, killing three of the mice receiving the highest dose. Of the serially passaged viruses tested, R5126 demonstrated a greater capacity to replicate in SK-N-SH cells and also demonstrated greater neurovirulence, yielding an LD50 of 4.8 × 105 PFU. These data also correlate with the results showing that the level of protein synthesis late in infection was higher in cells infected with R5126 than in those infected with R5122.
The second-site, compensatory mutation of recombinant viruses R5121 to R5130 map outside US10-12 domain of the HSV-1 genome.
The purpose of these studies was to determine whether the mutations underlying the phenotype of mutants R5121 to R5130 map in the same domain of the HSV-1 genome as that of the second-site, compensatory mutant of Mohr and Gluzman. Preliminary experiments have established that the BamHI Z and X fragment of these mutants could not be differentiated with respect to electrophoretic mobility from that of wild-type parent virus and therefore was unlikely to contain large deletions (data not shown). To ascertain whether this domain of the HSV-1 DNA contains small deletions or amino acid substitutions, the EcoRI-SalI fragment encompassing one of the origins of replication and the US10-12 domain of the HSV-1 genome was subcloned from the parent virus (R3616) and five of the serially passaged viruses (R5121, R5122, R5124, R5125, and R5126) and was sequenced using both manual (chain termination method) and automated sequencing (terminal dye termination method). The results, shown in Table 1, were as follows:
TABLE 1.
Sequence comparisons of wild-type and mutant viruses
| Comparison | Nucleotide | Change | Location | Amino acid changeb |
|---|---|---|---|---|
| Difference in nucleotide sequences of HSV-1(F) and HSV-1(17) | 144130 | G in place of A | 3′ end transcripts | |
| 144306 | A in place of G | US10 ORF | ||
| 144516 | G in place of A | US10 ORF | ||
| 144621 | A in place of T | US10 ORF | H in place of Q | |
| 144680 | G in place of T | US10 ORF | L in place of I | |
| 144684 | T in place of G | US10 ORF | ||
| 144702 | C in place of T | US10 ORF | ||
| 144710 | C in place of G | US10 ORF | D in place of H | |
| 144825 | G in place of A | US10 US11 ORF | P in place of S in US11 | |
| No change in US10 | ||||
| 144834–144861a | Δ(CCGGGGAGACCGGGGCTCCCTGGGAGA) | US10, US11 ORF | aa 129–137 in US11c | |
| aa 79–87 in US10 | ||||
| 145258a | Absent G | 5′ transcript (US11) | ||
| 145283 | A in place of G | 5′ transcript (US11) | ||
| 145287 | T in place of C | 5′ transcript (US11) | ||
| 145291 | C in place of T | 5′ transcript (US11) | ||
| 145399 | T in place of C | α47 ORF | K in place of E | |
| 145427 | A in place of G | α47 ORF | ||
| 145455 | T in place of C | α47 ORF | K in place ofR | |
| 145536 | T in place of G | α47 ORF | N in place of T | |
| 145585 | Additional GC | 5′ transcript (α47) | ||
| 145630 | C in place of T | 5′ transcript (α47) | ||
| 145638 | C in place of G | 5′ transcript (α47) | ||
| 145640–145669a | Δ(CGTTCCTCCTGCGGGAAG5′GCACGAACGCGG) | Transcript (α47) | ||
| 145670–145724 | GGAAGCGGTCCACGCAACGGTCGCCGCCGGTCGCCTCGACGAGGACGTTCCTCCT in place of GTGAGCCCCCTCCTCCGCCCCCGCGTCCCCCCTCCTCCGCCCCCGCGTCCCCCCT | 5′ transcript (α47) | ||
| 145725–145759a | Δ(CCTCCGCCCCCGCGTCCCCCCTCCTCCGCCCCCGC) | 5′ transcript (α47) | ||
| 145761–145784 | CGGGAAGGCACGAACGCGGGTGAG in place of TCCCCCCTCCTCCGCCCCCGCGTC | 5′ transcript (α47) | ||
| 145806–145828a | Δ(CCCCCCTCCTCCACCCCCGCGTC) | 5′ transcript (α47) | ||
| 145867 | C in place of A | 5′ transcript (α47) | ||
| 145868a | Absent A | 5′ transcript (α47) | ||
| 145942 | T in place of C | 5′ transcript (α47) | ||
| 145950–145960a | Δ(TCCCCCCCGCG) | 5′ transcript (α47) | ||
| 146046 | G in place of C | 5′ transcript (α47) | ||
| 146152 | T in place of C | |||
| R3616 and progeny versus HSV-1(F) and HSV-1(17) | 144956 | A in place of G | US11, US10 ORF | L in place of P in US11 C in place of R in US10 |
| 145447 | T in place of C | α47 ORF | M in place of V | |
| 145460 | T in place of C | α47 ORF | ||
| 145803 | T in place of C | 5′ transcript (α47) |
Sequence missing in HSV-1(F), R3616, and all derivatives of R3616.
Single-letter code for amino acids.
aa, amino acids.
(i) Four point mutations (nucleotides [nt] 144929 [G to A], nt 145419 [C to T], nt 145432 [C to T], and nt 145712 [C to T]) were identified in the 1.618-kb sequence encompassing the US10-12 coding domains in R5121, R5122, R5124, and R5125 not present in HSV-1(F). These four mutations, however, were also present in passage 0 of the parent virus R3616 and therefore were not responsible for restoring the wild-type protein synthesis phenotype.
(ii) The HSV-1(F) sequence differed from the published HSV-1 (17) sequence, the greatest variability occurring in the c sequence of the inverted repeats. Specifically, five different sets of DNA sequences were absent in HSV-1(F) DNA. These were HSV-1 (17) nt 145630 to 145669, nt 145724 to 145759, nt 145806 to 145,828, nt 145866, and nt 145950 to 145960.
(iii) In addition, a 27-bp region was absent in the HSV-1(F) strain corresponding to the domain coding the carboxyl-terminal region of US11 (HSV-1 [17]) nt 144835 to 144861. These findings are not surprising. Earlier studies have shown that the US11 protein size varies between strains, with intrastrain heterogeneity in the domain encoding the P-R-X amino acid repeats (27).
DISCUSSION
The salient result of the studies described in this report is the identification of a novel second-site, compensatory mutant capable of blocking activated PKR from shutting of protein synthesis late in infection. This mutant differs from that of Mohr and Gluzman in two respects. First, the mutation maps at a site different from that of the Mohr and Gluzman mutant (24). Second, unlike the mutant of Mohr et al. (25), the R5126 mutant described here exhibits a partially restored virulence on intracerebral inoculation of mice. Relevant to these results are the following:
(i) The second-site, compensatory mutant was isolated by serial passage of Δγ134.5 viruses in SK-N-SH cells. Two serial passages were done. The virus passaged in the first series was R3616, which lacked only the γ134.5 genes. The second-site, compensatory mutants emerged and were plaque purified after the sixth passage. A characteristic of these mutants is that they grew to higher titers and blocked the shutoff of protein synthesis caused by activated PKR. The mutants differed in the extent of recovery of neurovirulence associated with wild-type HSV-1. Whereas R5126 was more virulent than the parent virus, R5122 was not. The results suggest that R5126 is the product of additional mutations lacking in the R5122 mutant and that these mutations may or may not map in a single locus. It should be stressed that the selective pressure placed on R3616 was for better replication in cells in culture. We may speculate that, in response to an additional selective pressure, namely, replication in vivo, the mutants that would have emerged would have exhibited still greater neurovirulence.
The virus passaged in the second series was R5103 (Fig. 1). This virus lacks, in addition to the γ134.5 genes, also the sequences encoding the genes US8 through US12. No second-site, compensatory mutations were isolated after 10 passages of this virus under conditions identical to those used in the first series. It should be noted that the sequence of the genes missing from this series, US8 through US12, of R5126 DNA does not differ from that of the parent, R3616 mutant (Table 1 and data not shown), and that therefore the second-site mutation does not map in this domain of the HSV-1 genome. One hypothesis that could explain our failure to isolate second-site mutations in this passage series is that the product of the gene encoding the second-site mutation requires the function of one or more genes absent from this series of mutants (US8 through US12).
(ii) The mechanism by which the mutants of the series R5121 to R5130 overcome the effects of activated PKR is not known. As reported earlier, PKR is activated in cells infected with wild-type or mutant virus but not in mock-infected cells. The results obtained in the studies described in this report (Fig. 5A) show decreased activation of PKR recovered from cells infected with mutant viruses (as determined by autophosphorylation) from that recovered from wild-type virus- or R3616 mutant-infected cells. This difference disappeared on addition of NaF to block the action of phosphatases. Since we were unable to demonstrate a phosphatase activity specific for eIF-2α recruited in wild-type virus-infected cells, the data suggest that the mutated gene product(s) induces a phosphatase activity that specifically or nonspecifically dephosphorylates PKR.
(iii). In order to investigate the mechanisms underlying the mutation which confers on the mutant the phenotype described in this report, it is necessary to map it. In principle, there are three ways in which this can be done. The first involves analysis of the mutant genome for the presence of deletions or genome rearrangements that would hint at the site of the mutation. The deletion of a small DNA sequence was in fact the hint that led to the mapping of the mutation in the serially passaged virus of Mohr and Gluzman (24). Comparisons of the parent and mutant genome failed to yield evidence of deletions or rearrangements in the mutant genome (unpublished studies). The second method involves marker transfer, that is, cotransfection of random fragments of mutant genome with intact DNA of parent virus followed by selection of a virus with the mutant phenotype. In this instance the key experimental objective is to differentiate between spontaneous mutations which gave rise to the mutant being mapped and the expected mutant arising from marker transfer. Marker transfer yields unambiguous results when the discriminator is all or none, as in, for example, the discrimination between thymidine kinase-positive and -negative mutants. The problems becomes impossible if the readout can be partial, as in, for example, slight increases in protein synthesis and or neurovirulence. Once there is a hint of potential site of the mutation, the problem becomes much simpler. For example, Mohr and Gluzman mapped their mutant by marker transfer in cells that restrict the growth of the parent virus (24). To unambiguously map the mutation, it was necessary to reconstruct the mutation in a cell line which did not discriminate between the parent and mutant viruses (5). The third, brute-force approach is to sequence both parent and mutant viruses and reconstruct in a nonselective cell line each amino acid substitution, deletion, or rearrangement of the mutant virus in the genome of the parent virus. While theoretically possible, this approach is far beyond the scope of the project at the present time. An additional problem noted earlier in the text is that the mutant phenotype may reflect mutations in several genes.
It seems relevant to note that Mulvey and colleagues reported that PKR is not activated in wild-type virus-infected cells (26). PKR resident in uninduced cells is present in small amounts. The experimental design published by Mohr and colleagues indicates that the specific activity of 32P is at least an order of magnitude lower that that used in our present and earlier studies. Given the paucity of PKR and the low specific activity of 32P in the labeling medium, it is not surprising that they could not detect labeled PKR.
From studies on cells infected with HSV-1 and many other viruses, it is becoming clear that activation of interferon pathways is the major cell defense to infection. This conclusion is amply supported by the fact that deletion of the γ134.5 gene totally abrogates the capacity of the virus to spread and cause disease (6, 29). The evidence that some viruses have evolved more than one mechanism to abrogate this host response, and, finally, the observation that the Δγ134.5 virus is fully virulent in mice from which a vital component of the interferon pathway was knocked out attest to the importance of the PKR activation pathway as a host response to infection. Inasmuch as the loci of the sites of the two second-site mutations are not active in abrogating the interferon pathway in wild-type HSV-1 but become activated only after they had been mutated suggests that they are fossils of earlier functions evolved to abrogate the interferon pathway. The evolution of these genes could reflect simply better and better ways to block the host response or a viral response to the evolution of stronger host defenses. As noted earlier, γ134.5, the presently active anti-PKR gene evolved in part by the acquisition of a portion of the GADD34 gene. To date, this gene has been detected in the closely related HSV-1, HSV-2, and simian B virus genes and therefore is likely to have been acquired shortly before the evolution of these three viruses from a common progenitor. A clue to the order of the evolution of viral genes designed to abrogate the interferon pathway may be deduced from the extent of conservation of the fossils. The US11 gene expressed early in infection does block the interferon pathway, but this gene, albeit more highly conserved than the γ134.5 gene, is nevertheless not highly conserved among the members of the herpesvirus family. It remains to be seen whether the gene carrying the second-site mutations of the R5122 to R5130 series of mutants is more widespread and therefore may represent the fossilized version of a gene active in a more distant HSV-1 progenitor.
Acknowledgments
We thank Giang Tong and Li Na for their technical assistance.
These studies were initiated at the University of Chicago with the aid of U.S. Public Health Service grants from the National Cancer Institute (CA87761, CA83939, CA71933, and CA78766). The studies were completed at the University of Alabama with the aid of a grant from the National Institutes of Health (AI01680).
REFERENCES
- 1.Black, T. L., G. N. Barber, and M. G. Katze. 1993. Degradation of the interferon-induced 68,000 Mr protein kinase by poliovirus requires RNA. J. Virol. 67:791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brand, S. R., R. Kobayashi, and M. B. Mathews. 1997. The tat protein in human immunodeficiency virus type 1 is a substrate and inhibitor of the interferon-induced, virally activated protein kinase, PKR. J. Biol. Chem. 272:8388–8395. [DOI] [PubMed] [Google Scholar]
- 3.Carroll, K., O. Elroy-Stein, B. Moss, and R. Jagus. 1993. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2α-specific protein kinase. J. Biol. Chem. 268:12837–12842. [PubMed] [Google Scholar]
- 4.Cassady, K. A., M. Gross, and B. Roizman. 1998. The second-site mutation in the herpes simplex virus recombinants lacking the γ134.5 gene precludes shutoff of protein synthesis by blocking phosphorylation of eIF-2α. J. Virol. 72:7005–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassady, K. A., M. Gross, and B. Roizman. 1998. The herpes simplex virus US11 protein effectively compensates for the γ134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the α subunit of eukaryotic translation initiation factor 2. J. Virol. 72:8620–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou, J., E. R. Kern, R. J. Whitley, and B. Roizman. 1990. Mapping of herpes simplex virus-1 neurovirulence to γ134.5, a gene nonessential for growth in cell culture. Science 250:1262–1266. [DOI] [PubMed] [Google Scholar]
- 7.Chou, J., and B. Roizman. 1992. The γ134.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programed cell death in neuronal cells. Proc. Natl. Acad. Sci. USA 89:3266–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou, J., and B. Roizman. 1994. Herpes simplex virus 1 γ134.5 gene function, which blocks the host response to infection, maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Proc. Natl. Acad. Sci. USA 91:5247–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou, J., J.-J. Chen, M. Gross, and B. Roizman. 1995. Association of a Mr 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation intiation factor eIF-2α and premature shutoff of protein synthesis after infection with γ134.5− mutants of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 92:10516–10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, M. V., H.-W. Chang, B. L. Jacobs, and R. J. Kaufman. 1993. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J. Virol. 67:1688–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dougherty, R. M. 1964. Animal virus titration techniques, p.169–223. In R. J. C. Harris (ed.), Techniques in experimental virology. Academic Press, New York, N.Y.
- 12.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effect on social behavior of cells. J. Gen. Virol. 2:357–364. [DOI] [PubMed] [Google Scholar]
- 13.Gross, M., and D. A. Kaplansky. 1983. Differential effect of Mn2+ on the hemin-controlled translational repressor and the double-stranded RNA-activated inhibitor. Biochim. Biophys. Acta 740:255–263. [DOI] [PubMed] [Google Scholar]
- 14.He, B., J. Chou, D. A. Liebermann, B. Hoffman, and B. Roizman. 1996. The carboxyl terminus of the murine MyD116 gene substitutes for the corresponding domain of the γ134.5 gene of herpes simplex virus to preclude the premature shutoff of total protein synthesis in infected human cells. J. Virol. 70:84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He, B., J. Chou, R. Brandimarti, I. Mohr, Y. Gluzman, and B. Roizman. 1997. Suppression of the phenotype of γ134.5− herpes simplex virus 1: failure of activated RNA-dependent protein kinase to shut off protein synthesis is associated with a deletion in the domain of the α47 gene. J. Virol. 71:6049–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, B., M. Gross, and M. Roizman. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, B., M. Gross, and B. Roizman. 1998. The γ134.5 protein of herpes simplex virus 1 has the structural and functional attributes of a protein phosphatase 1 regulatory subunit and is present in a high molecular weight complex with the enzyme in infected cells. J. Biol. Chem. 273:20737–20743. [DOI] [PubMed] [Google Scholar]
- 18.Jacquemont, B., and B. Roizman. 1975. RNA synthesis in cells infected with herpes simplex virus. X. Properties of viral symmetric transcripts and of double-stranded RNA prepared from them. J. Virol. 15:707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katze, M. 1995. Regulation of interferon-induced PKR: can viruses cope? Trends Microbiol. 3:75–78. [DOI] [PubMed] [Google Scholar]
- 20.Kozak, M., and B. Roizman. 1975. RNA synthesis in cells infected with herpes simplex virus. IX. Evidence for accumulation of abundant symmetric transcripts in nuclei. J. Virol. 15:36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leib, D. A., M. A. Machalek, B. A. Williams, R. H. Silverman, and H. W. Virgin. 2000. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc. Natl. Acad. Sci. USA 97:6097–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology 214:222–228. [DOI] [PubMed] [Google Scholar]
- 23.Mathews, M. B., and T. Schenk. 1991. Adenovirus virus-associated RNA and translation control. J. Virol. 65:5657–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohr, I., and Y. Gluzman. 1996. A herpesvirus genetic element which affects translation in the absence of viral GADD34 function. EMBO J. 15:4759–4766. [PMC free article] [PubMed] [Google Scholar]
- 25.Mohr, I., D. Sternberg, S. Ward, D. Leib, M. Mulvey, and Y. Gluzman. 2001. A herpes simplex virus γ34.5 mutant that exhibits enhanced growth in cultured glioblastoma cells is severely attenuated in animals. J. Virol. 75:5189–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulvey, M., J. Poppers, A. Ladd, and I. Mohr. 1999. A herpesvirus ribosome-associated RNA-binding protein confers a growth advantage upon mutants deficient in a GADD34-related function. J. Virol. 73:3375–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rixon, F. J., and D. J. McGeoch. 1984. A 3′ co-terminal family of mRNAs from the herpes simplex virus type 1 short region: two overlapping reading frames encode unrelated polypeptides one of which has highly reiterated amino acid sequence. Nucleic Acids Res. 12:2473–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503–517. [DOI] [PubMed] [Google Scholar]
- 29.Whitley, R. J., E. Kern, S. Chatterjee, J. Chou, and B. Roizman. 1993. Replication, establishment of latency, and induced reactivation of herpes simplex virus γ134.5 deletion mutants in rodent models. J. Clin. Investig. 91:2837–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhan, Q., K. A. Lord, I. Alamo, Jr., M. C. Hollander, F. Carrier, D. Ron, K. W. Kohn, B. Hoffman, D. A. Liebermann, and A. J. Fornace, Jr. 1994. The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress cell growth. Mol. Cell. Biol. 14:2361–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]