Abstract
Our interest in the Schizosaccharomyces pombe RecQ helicase, rqh1+, led us to investigate the function of a related putative DNA helicase, srs2+. We identified the srs2+ homolog in S.pombe, and found that srs2+ is not essential for cell viability. A Δsrs2 Δrqh1 double mutant grows extremely slowly with aberrant shaped cells and low viability. This slow growth does not appear to be related to stalled replication, as Δsrs2 Δrqh1 cells showed higher survival rates, compared with Δrqh1, when stalled forks were increased by UV irradiation or hydroxy urea treatment. Consistent with this result, we found that Δsrs2 Δrqh1 cells progress through S-phase with a slight delay, but undergo a checkpoint-dependent arrest presumably at G2/M. Further, we found that Δsrs2 Δrqh1 slow growth is related to recombination, as loss of either the rhp51+ or rhp57+ recombination genes improves cell growth in the double mutant. Δsrs2 is also synthetic lethal with Δrhp54, another homologous recombination gene. This lethality is suppressed in a Δrhp51 background. Together, these results demonstrate a clear genetic interaction between rqh1+, srs2+ and the genes of the homologous recombination pathway.
INTRODUCTION
The faithful replication of DNA is essential for maintaining the integrity of the cell’s genetic information. Many challenges exist for the replication machinery to perform its task. One problem arises when the growing replication fork is halted. Replication fork arrest occurs when the replication complex encounters a site of DNA damage, some secondary DNA structures or nucleotide depletion, as in the case of treatment with hydroxy urea (HU) (1,2). Stalled replication forks have been shown to create sites for recombination, as demonstrated in both bacterial and eukaryotic systems (2–5). The loss of the recQ homolog in various systems leads to hyper-recombination when the cells undergo replication arrest induced by HU or ultraviolet (UV) DNA damage (6–13). These data suggest that the RecQ helicases act to suppress recombination at arrested replication forks, although the RecQ helicases have also been shown to promote certain types of recombination (10,14–16). Different models have been proposed to explain how RecQ helicases might suppress recombination. In Escherichia coli, arrested replication forks are believed to be stabilized by RecQ through the formation of a triple helix (10,17). Studies on the human and Schizo saccharomyces pombe RecQ helicases (Blm and Rqh1, respectively) suggest that they act to remove Holliday junctions formed at stalled replication forks, by reverse branch migration (18,19). In one of these studies, expression of the bacterial Holliday junction resolvase was shown to significantly suppress UV and HU sensitivity in S.pombe Δrqh1 cells (18). These results suggest that cells lacking Rqh1 accumulate Holliday junctions at stalled replication forks.
Recent findings in Saccharomyces cerevisiae demonstrating a synthetic interaction between srs2 and the recQ homolog sgs1 suggest the possibility that Srs2 might also function at stalled replication forks (14,20–22). The S.cerevisiae Srs2 DNA helicase, which is structurally related to the bacterial helicase II (UvrD) (23), was first discovered in a screen for suppressors of trimethoprim sensitivity of rad6 and rad18 mutants (24,25). It was independently characterized as a suppressor of homologous recombination (named HPR5), as mutants showed a hyper-recombination phenotype (24,26,27). SRS2 has since been shown to be associated with the RAD6 post replication repair (PRR) pathway. Specifically, Srs2 functions with the RAD5-dependent error-free branch of PRR, where it has been shown to suppress the UV sensitivities of rad5, ubc13 and mms2 mutants (28). This suppression is lost in cells lacking a functional recombination system, consistent with a role of Srs2 blocking damage from being chanelled into the RAD52-dependent homologous recombination pathway (28,29). The synthetic interaction between srs2 and sgs1 was shown to be suppressed by mutations in the homologous recombination genes rad51, rad55 and rad57 (14). It has also been demonstrated that S.cerevisiae srs2 shows either a lethal phenotype or poor growth when combined with a number of other mutants (rdh54, rad50, mre11, xrs2, top3 and rad54) (21,30,31). The synthetic interactions of these double mutants are furthermore suppressed by mutating the recombination genes rad51, rad52, rad55 and rad57 (14,30–32). In addition, it was shown that srs2 rad54 and srs2 rdh54 lethality can be suppressed by loss of checkpoint function (31). These data are complemented by evidence of genetic interaction between SRS2 and RAD52, based on suppression of rad52 mutations by deletion of SRS2 (33,34). srs2 mutants have also been shown to suppress hyporecombination of some rad51 and rad52 alleles (24,32). Together, these results demonstrate that Srs2 function in S.cerevisiae is linked to homologous recombination.
Both Srs2 and the RecQ helicases are associated with DNA damage response and/or genomic instability. In S.cerevisiae, Sgs1 and Srs2 have been shown to participate in activation of the intra-S checkpoint following exposure to MMS (35,36). In S.pombe, rqh1 mutants show sensitivity to UV and gamma radiation and treatment with HU (7,8,37). rqh1+ was originally identified as rad12+ in a screen for UV sensitive mutants (38). One phenotype associated with rqh1 mutants is that although cells undergo a normal G1–S arrest following DNA damage, they recover poorly. Fluorescence microscopy has revealed that the problem is in mitosis as cells divide with torn chromosomes and uneven distribution of nuclear material (7,8). It has been proposed in S.cerevisiae that Srs2 acts in recombination repair of double strand breaks to stabilize the invading single strand with its homolog, thereby acting against reverse branch migration (39). Srs2 is phosphorylated in response to intra-S DNA damage, and this phosphorylation is dependent on both checkpoint activation and Cdk1 (cyclin-dependent kinase) (36). Following DNA damage in S-phase, srs2 mutants do not fully arrest and Rad53 phosphorylation is reduced, suggesting Srs2 functions in an intra-S checkpoint. In addition, srs2 mutants suppress the lethality of a rad17 checkpoint mutant following intra-S DNA damage (36).
Our interest in rqh1+ has led us to study other genes related to its activity such as top3+ (40). In this context we have sought to identify other potentially interacting proteins. Therefore, we identified a homolog of SRS2 by searching the S.pombe genomic database and created a null mutant, Δsrs2. As reported previously (41), we found that Srs2 is not essential in S.pombe and that the single mutant does not exhibit significant sensitivity to either UV damage or treatment with HU. Furthermore, the Δsrs2 Δrqh1 double mutant shows a severe slow growth defect compared with the single mutants. Our studies found that the Δsrs2 Δrqh1 double mutant progresses through S-phase with a slight delay but arrests in G2/M with many of these cells ultimately undergoing an aberrant mitosis. The G2/M arrest is likely due to a checkpoint dependent DNA damage response, as deletion of the rad9+ checkpoint gene suppresses the accumulation of Δsrs2 Δrqh1 cells in G2/M. This arrest appears to be related to recombination, as deletion of recombination genes also suppresses the slow growth of Δsrs2 Δrqh1 and improves their viability. This result is in contrast to a recently published paper where it was reported that loss of recombination actually worsened the slow growth of Δsrs2 Δrqh1 (41). We will point out why we believe we find these differences. Interestingly, we found that when replication fork stalling is increased by treatment with UV or HU, Δsrs2 Δrqh1 cells show improved survival compared with that of a Δrqh1 single mutant. These data imply that the synthetic phenotype of Δsrs2 Δrqh1 is not a direct result of the double mutant inability to cope with stalled replication. Lastly, in studying the effect of loss of recombination on Δsrs2 Δrqh1 we found that Δsrs2 is synthetically lethal with Δrhp54, and this lethality is suppressed in a Δrhp51 mutant background.
MATERIALS AND METHODS
Yeast strains, molecular techniques and media
The strains with their genotypes used in this study are listed in Table 1. All strains were maintained on YEA media (YEA is 5 g/l yeast extract, 30 g/l of dextrose and 75 mg/l of adenine). Edinburgh minimum media (EMM) is supplied by Bio101 Inc. Double and triple mutants were made by either tetrad analysis or, when appropriate markers were available, by random spore analysis. All newly constructed strains were tested for having the correct genotypes by analytical PCR.
Table 1. Strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| SZ132 | h+ ade6-210 leul-32 ura 4-D18 | Laboratory stock |
| SZ242 | h+ srs2::kanR ade6-210 leul-32 ura 4-D18 | This study |
| SZ243 | h+ rqh1:: kanR ade6-210 ura-D18 | This study |
| SZ215 | h+ rhp51:: ura4+ ade6-704 leu1 ura4-D18 | (48) |
| SZ231 | h+ rhp54::ura4+ ade6-704 leu1-32 ura4-D18 | (49) |
| SZ258 | h+ rhp57::ura4+ ade6-216 leul-32 his7-366 ura 4-D18 | (50) |
| SZ76 | h– rad9:: ura4+ ade6-704 leu1-32 ura4-D18 | (51) |
| SZ247 | h– srs2::kanR rqh1:: kanR | This study |
| SZ271 | h+ srs2::kanR rqh1:: kanR rhp51:: ura4+ ade6-210 leul-32 ura 4-D18 | This study |
| SZ277 | h+ srs2::kanR rqh1:: kanR rhp57::ura4+ leu1-32 ura4-D18 | This study |
| SZ279 | h+ srs2::kanR rhp54:: ura4+ rhp51:: ura4+ ade6-210 leul-32 ura4-D18 | This study |
| SZ280 | h+ srs2::kanR rqh1:: kanR rad9:: ura4+ leul-32 ura4-D18 | This study |
| SZ256 | h+ rqh1:: kanR rhp54:: ura4+ ade6-210 leul-32 ura 4-D18 | This study |
| SZ281 | h+ rhp54:: ura4+ rhp51:: ura4+ ade6-704 leu1-32 ura4-D18 | This study |
| SZ613 | h+ cds1::ura4+ srs2::kanR rqh1::kanR leu1-32 ura4-D18 | This study |
| SZ614 | h+ chk1::ura4+ srs2::kanR rqh1::kanR leu1-32 ura4-D18 | This study |
Constructing the rqh1 and srs2 deletion strains
Both the Δrqh1 and Δsrs2 strains were made by deleting the entire gene and replacing it with the kanamycin resistance gene (42,43). The KanMX4 gene was amplified using 95mer primers, Srs2kanA and Srs2kanB (sequences in Table 2), which contain 15 nt complementary to the KanMX4 gene. This disruption cassette was used to directly transform our wild-type haploid strain (SZ132), with cells subsequently plated onto YEA plates containing G418 (200 mg/l).
Table 2. Oligonucleotide sequences used in these studies.
| Name | Sequence |
|---|---|
| SRS2.kanA | ATGGAAACGAAATCATCATACTTGAAGTTTCTGAATGAAGAACAAAGGATTAGTGTTCAGAGTCCTCATAAGTATACTCACAGCTGAAGCTTCGTACG |
| SRS2.kanB | CTATAACATTCGTGAAACTCGTAGCCTTACACCCAATCGTTTTCTCGAACCAGCAGGACGGGCACTCGCGACCTTAGTAAGGCCACTAGTGGATCTG |
Growth kinetics
Analysis of the relative growth rates of strains was carried out by one of two approaches. In the first case, a single colony was picked from a freshly streaked plate and the cells streaked to single colonies on YEA plates. The plates were incubated at 30°C for 3–4 days and photographed. To get a more quantitative value of growth, overnight liquid cultures were started from single colonies and grown to mid log (2 × 105 to 1 × 106 cells/ml) at 30°C overnight. The cells were counted beginning at time 0 and then at every hour for 9 h. The data were plotted as fold increase against time in hours.
UV and HU sensitivity
Overnight liquid cultures were started from single colonies and grown to mid log (1 × 106 to 5 × 106 cells/ml) at 30°C overnight. The cells were counted and an appropriate number of cells for each dose was plated onto YEA plates. Plates were irradiated with 100 J/m2 of 254 nm UV light, incubated for 3–5 days and counted. For HU sensitivity studies, the appropriate dilution of cells was spotted onto YEA plates containing the indicated concentration of HU. Plates were incubated for 4–10 days, counted and photographed. Alternatively, 12 mM HU was added to mid log cultures of cells that were then incubated with shaking at 30°C. Cells were plated onto YEA plates at time 0 and every 3 h after for 9 h. The plates were incubated at 30°C for 3–5 days and counted.
Synchronization and release of cells in G1
To synchronize cells in G1, we used a combination of nitrogen starvation and size fractionation on lactose gradients. Cells were grown to saturation in YEA and then switched to EMM for 24 h. These cells were washed twice with water and then resuspended in 100 ml of EMM without nitrogen and cultured at 30°C for 20 h. The cells were counted and ∼5 × 108 cells in 700 µl were layered onto 13 ml 7–30% lactose gradients made in EMM without nitrogen. The gradients were centrifuged for 4 min at ∼300 g. The tubes were punctured from the bottom and 0.5 ml fractions collected. Fractions were viewed microscopically and fractions containing the smallest cells pooled. These cells were released from the G1 block by switching them to YEA media. At 30 min intervals 1.5 ml aliquots were taken, collected by centrifugation, fixed in 70% ethanol and stored at 4°C.
FACS analysis
Fixed cells were collected by centrifugation and washed five times in 50 mM sodium citrate. These cells were resuspended in 0.5 ml of 50 mM sodium citrate, 100 µg/ml RNase and incubated at 37°C for at least 2 h. The cells were then stained by adding 0.5 ml 50 mM sodium citrate, 1 µM Sytox Green (Molecular Probes) and analyzed within 2 h. The cells were assayed using a Becton Dickson Facs Calibur with Cell Quest 4.0 software.
Cell viability
Fresh plates were struck for each strain, and day cultures started from single colonies, grown in YEA, with shaking at 30°C for 11 h. The cultures were counted and the cells plated on YEA plates. The plates were incubated at 30°C for 4 days and colonies counted.
Fluorescence microscopy
Fluorescence microscopy was carried out as previously described (41). Cells were fixed in 70% ethanol. Following fixation, cells were collected by centrifugation, washed in 50 mM sodium citrate and stained with DAPI.
RESULTS
Loss of srs2+ does not confer UV and HU sensitivity
The srs2+ gene of S.pombe was identified by screening the S.pombe database using the S.cerevisiae SRS2/HPR5 gene sequence as the query sequence with a BLAST search program. A haploid strain of S.pombe, sp30, was used to create Δsrs2 by replacing srs2+ with the KanMX4 disruption cassette which codes for G418/kanamycin resistance (42). Several G418 resistant clones were identified and deletion of srs2+ confirmed by PCR. Thus, we concluded that srs2+ is not essential for viability in S.pombe. The srs2::kanR (Δsrs2) cells were then examined initially for growth properties and morphology. As described previously (41) these clones were found to have normal morphologies and near normal growth rates.
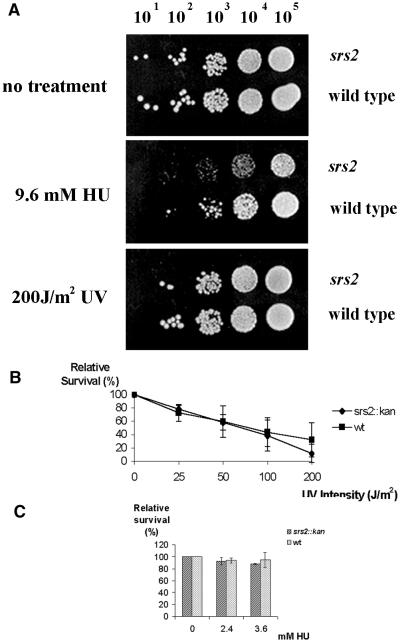
To investigate the potential role of Srs2 in DNA damage repair, we measured sensitivity to UV radiation and HU treatment in our S.pombe Δsrs2 mutant. We conclude that the Srs2 protein plays no significant role in survival following UV irradiation (Fig. 1A and B) or treatment with HU (Fig. 1A and C).
Figure 1.
Characterization of the Δsrs2 mutant. (A) Serial dilutions of Δsrs2 (SZ242) and wild-type (SZ132) cells were plated onto YEA plates (top and bottom panel) or on a YEA plate containing 9.6 mM HU (middle panel). One YEA plate was irradiated with 200 J/m2 of 254 nm UV light. The other YEA plate was left untreated. Following 4 days of incubation at 30°C these plates were photographed. The plate containing HU was incubated for 6 days and photographed. (B) Mid log cultures of Δsrs2 and wild-type cells were plated onto YEA plates and irradiated with the indicated dose of UV light. Plates were incubated at 30°C for 4 days and colonies counted. This represents the average of three experiments. (C) Δsrs2 and wild-type cells were plated onto YEA plates containing the indicated concentrations of HU, grown for 5 days and colonies counted. This is an average of three experiments.
Schizosaccharomyces pombe Δsrs2 shows a synthetic slow growth phenotype with Δrqh1
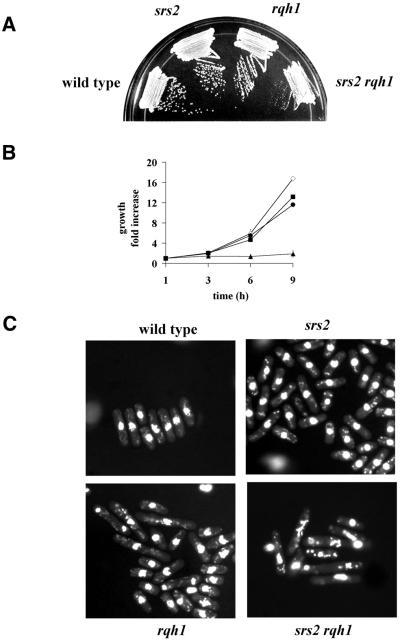
We crossed Δsrs2 with Δrqh1 to create a Δsrs2 Δrqh1 double mutant by random sporulation. Colonies arising from this cross were either normal sized or small. A large screen of over 100 colonies revealed that all normal sized colonies were either single mutants or wild type, while all small colonies were double mutant Δsrs2 Δrqh1. This demonstrates that the Δsrs2 Δrqh1double mutant is viable, unlike in S.cerevisiae where the corresponding double mutant is synthetic lethal (21). However, the double mutant displays a severe slow growth defect (Fig. 2A). Growth kinetic studies demonstrated that the doubling time of the Δsrs2 Δrqh1 mutant is ∼5–6 h (Fig. 2B). Wild-type, Δsrs2, Δrqh1 and Δsrs2 Δrqh1 cells were stained with DAPI and examined microscopically. Figure 2C shows that the double mutant has a phenotype characteristic of fission yeast strains having a growth defect; a mixed population of normal and abnormal appearing cells. The abnormal cells are typically long, often with aberrant cellular morphologies and distorted or fragmented nuclei. The double mutant also shows a very poor plating efficiency (16.7%; see Table 3). We have previously shown that Δrqh1 cells not only grow some 30% slower than wild-type cells but the population also contains a number of elongated and abnormally shaped cells (40). The fact that the Δsrs2 single mutant has normal growth characteristics and nearly normal resistance to both UV and HU further suggests that Srs2 alone plays a relatively minor role in cell viability.
Figure 2.
Δsrs2 shows synthetic slow growth with Δrqh1. The Δsrs2 Δrqh1 double mutant was created from a cross of the single mutants. (A) Wild-type (SZ132), Δsrs2 (SZ242), Δrqh1 (SZ243) single mutants and the Δsrs2 Δrqh1 (SZ247) double mutant were struck onto YEA plates. The plates were incubated for 4 days at 30°C and then photographed. (B) Growth kinetics were carried out on wild-type (diamonds), Δsrs2 (squares) Δrqh1 (circles) and Δsrs2 Δrqh1 (triangles) cells in mid log (1–5 × 105 cells/ml). Cells were collected every hour over a 5 h period and counted in a Coulter Counter. (C) Wild-type, Δsrs2, Δrqh1 and Δsrs2 Δrqh1 cells were isolated from a log growing culture, fixed and then stained with DAPI. A field of cells typical of each strain was photographed.
Table 3. Plating efficiency of mutant strains.
| Strain | Wild type | srs2::kanR | rqh1:: kanR | rhp51::ura4+ | rad9::ura4+ | srs2::kanR rqh1::kanR | srs2:: kanRrqh1::kanRrhp51::ura4+ | srs2:: kanRrqh1::kanRrad9::ura4+ |
|---|---|---|---|---|---|---|---|---|
| Plating efficiency (%) | 95.3 ± 16.7 | 100.2 ± 34.3 | 64.1 ± 8.7 | 63.75 ± 7.1 | 47.0 ± 14.2 | 16.7 ± 0.91 | 38.5 ± 4.2 | 7.5 ± 2.2 |
Δsrs2 Δrqh1 arrests in G2/M and show a mitotic defect
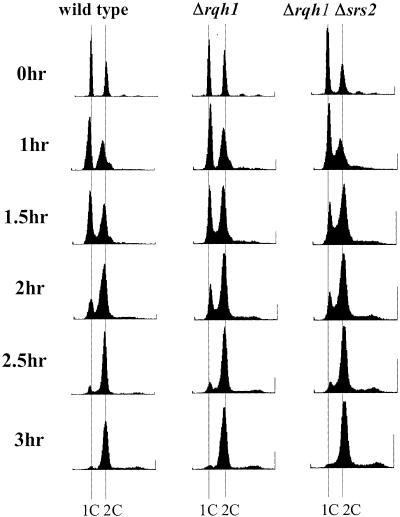
We initially suspected that the slow growth seen in Δsrs2 Δrqh1 cells might be due to a delay in S-phase based on the proposed role of both of these proteins functioning during DNA replication. To investigate this, we carried out FACS analysis on synchronized cells. Overnight cultures of wild type, Δsrs2, Δrqh1 and Δsrs2 Δrqh1 were synchronized in G1 by growth in media lacking nitrogen. Cells were released from the block and samples collected for analysis at time intervals over several hours. In our initial studies, FACS profiles of the Δsrs2 Δrqh1 strain were difficult to interpret, presumably due to the abundance of abnormally large cells present in the population (data not shown). In subsequent experiments, following nitrogen starvation, large cells were eliminated by size selection using 7–30% lactose gradients. These size-selected cells were released from the block by switching to YEA media. FACS analysis of these cells gave much improved results. The results in Figure 3 show that Δsrs2 Δrqh1 cells do exhibit a slightly elongated S-phase. The Δsrs2 Δrqh1 cells exit the G1 block and progress through S-phase somewhat slower than wild-type cells or cells containing either single mutation. This is seen in Δsrs2 Δrqh1 as a greater number of cells in S-phase between 1 and 2 h after release from G1. This slight delay in S-phase would not seem to account for all of the slow growth seen in the double mutant.
Figure 3.
Δsrs2 Δrqh1 cells show a slight delay in S-phase. Wild-type (SZ132), Δrqh1 (SZ243) and Δsrs2 Δrqh1 (SZ247) cells were blocked in G1 by nitrogen starvation and size selected on lactose gradients. The cells were released from the G1 block by shifting to YEA media. Cells were isolated every 30 min after release for 3 h and fixed. Fixed cells were stained with Sytox Green and analyzed by FACS. The profiles are shown here.
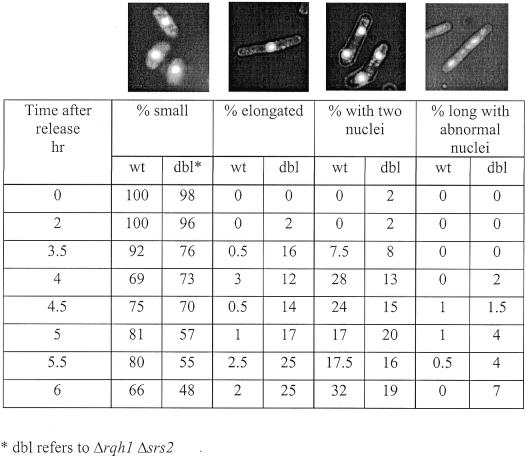
Since Δsrs2 Δrqh1 cells did not show a significant S-phase delay, we went on to determine if there was a more substantial delay in another part of the cell cycle. To address this, we used cells from the same synchronization experiments described above. Cells, fixed at various times after G1 release, were stained with DAPI and examined microscopically. At each time point, random fields of cells were scored as being either small/normal, elongated, having two nuclei or having torn or abnormal nuclei. On average about 250 cells were counted for each time point. Elongated cells were cells judged to be longer than a typical dividing cell. Figure 4 summarizes the results of wild-type and Δsrs2 Δrqh1 cells. Pictures of DAPI stained cells representative of each classification of cells is shown at the top of each column. The percentage of each cell type seen at each time point are listed below each picture. At 0 h, essentially all wild-type and mutant cells were small, consistent with a good G1 arrest. By 2 h we still saw mainly small or normal sized cells in both strains. By 3.5 h post release we began to see cells from both wild-type and Δsrs2 Δrqh1 that contained two nuclei, consistent with cells having completed mitosis. At 4 h, 28% of wild-type cells and 13% of Δsrs2 Δrqh1 cells were found to contain two nuclei. These results demonstrate that a sub-population of Δsrs2 Δrqh1 cells cycle normally. At 4 h many Δsrs2 Δrqh1 were not cycling normally, as indicated by the presence of elongated cells with single nuclei (12% of Δsrs2 Δrqh1 compared with 3% of wild-type cells). This phenotype is indicative of cells delayed in G2/M, suggesting a checkpoint-dependent arrest. These elongated cells appear to accumulate, as their numbers increased to 25% by 5.5 h after release. Only a few percent of elongated cells were ever seen in wild-type cells.
Figure 4.
Δsrs2 Δrqh1 cells appear to arrest at G2/M with many undergoing mitotic catastrophe. Wild-type (SZ132) and Δsrs2 Δrqh1 (SZ247) cells were blocked in G1 by nitrogen starvation and size selected on lactose gradients. Cells were released from the block by growth in YEA. Samples were collected at 30 min intervals, fixed and stained with DAPI. Random fields of cells were then examined microscopically and each cell scored for being in one of the four morphological types; small/normal, elongated (cells without septi which were as long or longer than dividing cells), cells with two nuclei (these are post mitotic cells) and cells with elongated and abnormal nuclei (these are cells that have undergone mitotic catastrophe). Approximately 300 cells were counted for each time point. An example of cells characteristic for each morphological type are shown at the top of each column.
In addition to the elongated cells, at the later time points (5–6 h), there was also an accumulation of cells with torn and abnormally shaped nuclei, indicative of cells that have undergone mitotic catastrophe. This morphology is reminiscent of the phenotype seen in Δrqh1 cells following DNA damage or HU arrest. We also examined synchronized Δsrs2 and Δrqh1 single mutants through this same block and release time course. The Δsrs2 cells were indistinguishable from wild-type cells, while a very small number (<5%) of Δrqh1 cells did elongate with a single nucleus or had torn chromosomes (<2%, data not shown). From these results we conclude that the events that lead to cell growth arrest in Δsrs2 Δrqh1 are stochastic, as some cells fail to progress through the cell cycle while others proceed at what appears to be a normal rate of growth. Also, when cells do arrest it appears to be at G2/M although our FACS data do not rule out the possibility that the arrest is in very late S-phase.
Loss of Srs2 and Rqh1 leads to induction of a checkpoint response
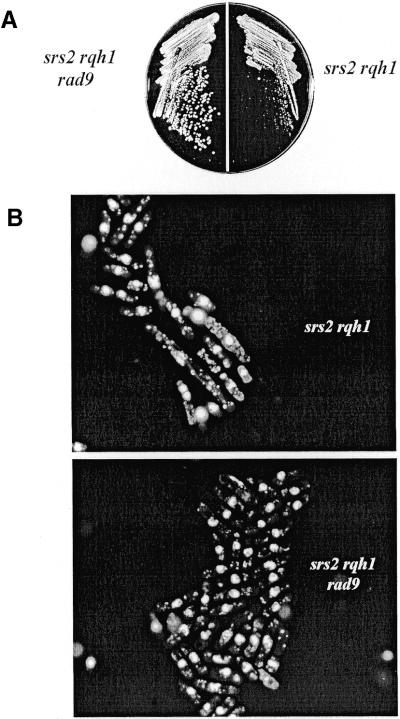
One explanation for the G2/M arrest of Δsrs2 Δrqh1 cells is that their DNA is altered or damaged in such a way that a checkpoint response is elicited. To test this, we created a triple mutant strain in which the rad9+ checkpoint gene was deleted in a Δsrs2 Δrqh1 background. Figure A shows that loss of the rad9+ gene suppresses the slow growth of the Δsrs2 Δrqh1 mutation. Furthermore, when these cells (Δsrs2 Δrqh1 Δrad9) were stained with DAPI and visualized by fluorescence microscopy, fewer elongated cells were detected (Fig. 5B). This result suggests that by deleting rad9+, the Δsrs2 Δrqh1 cells escape a checkpoint-dependent arrest, reducing their elongated cell cycle time and, therefore, partially suppressing their slow growth. We also reasoned that an apparent suppression of slow growth would occur if deletion of rad9+ improved cell survival in Δsrs2 Δrqh1 cells. We suggest this in light of our finding that the Δsrs2 Δrqh1 double mutant shows a high rate of spontaneous cell death, contributing to an appearance of slow growth. Therefore, we measured the plating efficiency of Δsrs2 Δrqh1 and compared it with that of the Δsrs2 Δrqh1 Δrad9 triple mutant. The data shown in Table 3 reveal that the plating efficiency of the triple mutant was actually worse than that of the double mutant, 7.5% for Δsrs2 Δrqh1 Δrad9 versus 17.6% for Δsrs2 Δrqh1. Therefore, the increased growth rate in the triple mutant cannot be explained by improved cell viability. These data support the conclusion that loss of rad9+ in Δsrs2 Δrqh1 improves the growth rate because these cells can no longer induce a checkpoint-dependent arrest.
Figure 5.
Loss of the checkpoint genes suppresses the slow growth and abnormal morphology of Δsrs2 Δrqh1. A Δrad9 Δsrs2 Δrqh1 (SZ280) triple mutant was created to study the loss of the checkpoint response on cell growth and morphology. (A) Strains were struck onto YEA plates and grown at 30°C for 4 days and photographed. The Δrad9 Δsrs2 Δrqh1 triple mutant (SZ280) is seen to have better growth than the Δsrs2 Δrqh1 (SZ247) double mutant. (B) Fluorescence microscopy of DAPI stained cells reveals that the morphology of the triple mutant is much more like wild-type cells than the Δsrs2 Δrqh1 double mutant. See Figure 2C for wild-type cells. (C) Δcds1 Δsrs2 Δrqh1 (SZ613) and Δchk1 Δsrs2 Δrqh1 (SZ614) triple mutants were constructed and analyzed. Fluorescence microscopy of DAPI stained cells shows that while loss of chk1+ suppresses the abnormal morphology of Δsrs2 Δrqh1 cells, loss of cds1+ does not.
We next wanted to ask whether the arrest of Δsrs2 Δrqh1 cells was specific to S-phase or G2. To answer this question we made Δcds1 Δsrs2 Δrqh1 and Δchk1 Δsrs2 Δrqh1 triple mutants and examined the growth and morphology of these cells. Cds1 controls S-phase checkpoint arrest while Chk1 controls G2 checkpoint arrest. While the loss of cds1+ had no effect on cell growth, loss of chk1+ did improve cell growth (data not shown). Consistent with this finding, loss of chk1+ suppressed the appearance of elongated cells normally seen in the Δsrs2 Δrqh1 mutant (Fig. 5C), while loss of cds1+ had no apparent effect on the morphology of Δsrs2 Δrqh1 cells. Δcds1 Δsrs2 Δrqh1 and Δchk1 Δsrs2 Δrqh1 triple mutants had plating efficiencies similar to Δsrs2 Δrqh1, 22 and 14%, respectively. These results are consistent with the Δsrs2 Δrqh1 mutant arresting in G2.
The slow growth of Δsrs2 Δrqh1 double mutants is suppressed by loss of the recombination pathway
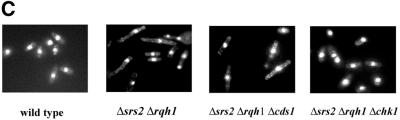
Loss of rqh1+, as well as other members of the recQ family, leads to hyper-recombination, as does loss of srs2+ in S.pombe (8,41,44,45). Thus, the phenotype associated with the simultaneous loss of both srs2+ and rqh1+ might be explained by increased rates of recombination. It follows that eliminating recombination genes might suppress the slow growth seen in this double mutant. To study this, we created two different recombination deficient strains, Δsrs2 Δrqh1 Δrhp51 and Δsrs2 Δrqh1 Δrhp57. One complicating factor with these studies is that in S.pombe recombination mutants themselves are slow growing, although not nearly as severe as Δsrs2 Δrqh1. Despite this, we were able to see clear suppression of the slow growth in Δsrs2 Δrqh1 when either recombination gene was deleted (Fig. 6A and B). We created the triple mutant by random spores and characterized over 100 colonies for each strain. Our Δrhp51 and Δrhp57 strains were disrupted with ura4+ while both Δsrs2 and Δrqh1 were disrupted with kanMX. This allowed us to rapidly screen our colonies. We found that every slow growing colony that we examined was Δsrs2 Δrqh1 while faster growing colonies were always either Δrhp51 or Δrhp57. The figure shows that loss of rhp51+ partially suppresses the slow growth of Δsrs2 Δrqh1 double mutants, to a level similar to that of the Δrhp51 single mutant (Fig. 6A). Δrhp57 mutant was also found to suppress the slow growth of Δsrs2 Δrqh1 (Fig. 6B).
Figure 6.
The slow growth of Δsrs2 Δrqh1 is also suppressed in cells deficient in homologous recombination. (A) Mid log cultures of wild-type (SZ132), Δrhp51 (SZ215), Δsrs2 Δrqh1 (SZ247) and Δsrs2 Δrqh1 Δrhp51 (SZ271) cells were struck onto YEA plates and grown at 30°C for 4 days and photographed. The Δsrs2 Δrqh1 Δrhp51 strain has growth properties similar to the Δrhp51 single mutant. (B) Loss of rhp57+ (SZ258) suppresses as well the slow growth of Δsrs2 Δrqh1 (SZ247) mutant.
Clearly, loss of recombination suppresses the slow growth of Δsrs2 Δrqh1, but does it improve cell survival? To test this we measured the plating efficiency of Δsrs2 Δrqh1 Δrhp51 and compared it with the double mutant as well as the single mutants. In Table 3, we see that the triple mutant showed a plating efficiency of 40% as compared with Δsrs2 Δrqh1, which was only 17.4%. The efficiency was not as good as the single Δrhp51 mutant, which was 65%.
In light of the discrepancy between our data and that of Wang et al. (41) concerning suppression of Δsrs2 Δrqh1 slow growth by loss of recombination genes, we carried out one other experiment to eliminate the possibility that our suppression was the result of some spontaneous suppressor. To determine if a spontaneous suppressor did arise in our strains we crossed Δsrs2 Δrqh1 to Δsrs2 Δrqh1 Δrhp51. The progeny were analyzed by random sporulation. Spores were found to be either large or small at about 50:50. A screen of over 100 colonies from the cross found that all of the large colonies that were tested contained Δrhp51 (data not shown). This demonstrates that the suppression is due to the loss of recombination and not due to a spontaneous suppressor. From these results we show conclusively that the synthetic slow growth of Δsrs2 Δrqh1 is suppressed in a recombination deficient background. These results are consistent with findings reported in S.cerevisiae where it was demonstrated that the synthetic lethality or slow growth of srs2 sgs1 is suppressed by mutations in genes coding for proteins of the recombination machinery (14,30,32).
Loss of srs2+ partially suppresses the HU and UV sensitivity of Δrqh1
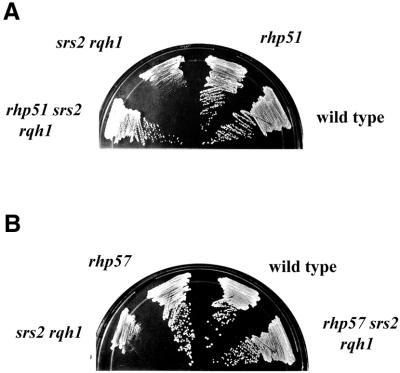
Our FACS analysis of synchronized Δsrs2 Δrqh1 cells indicates that these cells progress through S-phase slightly slower than wild type, which seems to argue against the slow growth being primarily a function of stalled replication. However, it is possible that the impairment seen in Δsrs2 Δrqh1 cells is related to an inability to properly respond to stalled replication forks, even though the major cell cycle arrest occurs outside of S-phase. If stalled forks are the problem, then increasing the number of stalled replication forks in Δsrs2 Δrqh1 cells by DNA damage with UV radiation or depleting the nucleotide pool with HU should have a profound negative effect on cell survival. In Figure 7A, it is seen that rather than being more sensitive to UV the Δsrs2 Δrqh1 double mutant was actually less sensitive to UV than that of the Δrqh1 single mutant. Δsrs2 Δrqh1 double mutants showed an equivalent sensitivity to 12 mM HU in liquid culture when compared with the Δrqh1 single mutant (Fig. 7B). To consider the possibility that the lack of increased sensitivity to HU in the double mutant was a function of its slow growth, we examined the HU sensitivity of the double mutant with continuous HU exposure by plating cells directly onto HU-containing plates. Figure 7C shows plates containing 2.4 mM HU after 8 days of incubation. The Δsrs2 Δrqh1 plate shows not only increased survival relative to the Δrqh1 single mutant but also the presence of larger colonies. Together, these data support the conclusion that loss of srs2+ partially suppresses the HU and UV sensitivity of Δrqh1. More importantly, this result argues against the idea that the synthetic phenotype seen in Δsrs2 Δrqh1 is solely dependent on stalled replication.
Figure 7.
Loss of srs2+ partially suppresses the UV and HU sensitivities of Δrqh1. (A) Mid log cultures of wild-type (SZ132) (circles), Δsrs2 (SZ242) (squares), Δrqh1 (SZ243) (triangles) and Δsrs2 Δrqh1 (SZ247) (diamonds) cells were plated onto YEA plates and irradiated with the indicated doses of UV light. The plates were incubated for 4 days and counted. This is the average of two independent experiments. (B) 12 mM HU was added to mid log cultures of wild-type (circles), Δsrs2 (squares), Δrqh1 (triangles) and Δsrs2 Δrqh1 (diamonds) cells and incubated at 30°C. At the indicated time points, aliquots of cells were plated onto YEA plates, which were incubated for 4 days and colonies counted. This is the average of three independent experiments. (C) Mid log cultures of Δrqh1 and Δsrs2 Δrqh1 cells were plated onto YEA plates containing 2.4 mM HU. Plates were incubated for 8 days and photographed.
Δsrs2 and Δrhp54 show synthetic lethality, which can be suppressed by loss of rhp51+
In our screen for homologous recombination mutants that would suppress the slow growth of Δsrs2 Δrqh1, we found a synthetic lethality between Δsrs2 and Δrhp54. This synthetic lethality also exists in the budding yeast (45). The RAD54 homolog in S.pombe is Rhp54. We crossed srs2::kanR to rhp54:: ura4+ followed by random sporulation onto plates containing G418 to select for srs2::kanR colonies. Two hundred G418 resistant colonies were replica plated on minimum media. No colonies were observed after 4 days of incubation at 30°C indicating that all the colonies were ura– and therefore rhp54+. From this cross we also plated directly onto YEA media and, following this, we identified 100 ura+ colonies. When replica plated onto G418, none of these colonies grew. Thus, we conclude that these mutants are synthetic lethal in S.pombe.
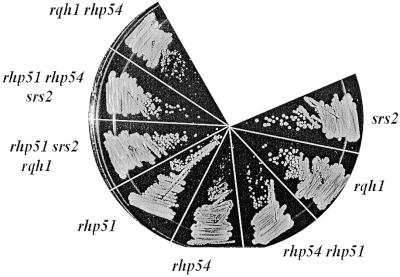
We next looked for a possible suppressor of this lethality. We constructed two double mutant strains Δsrs2 Δrhp51 and Δrhp54 Δrhp51. These strains were crossed and screened for the presence of kanamycin resistance on G418 plates. G418 resistant colonies were isolated and screened for the presence of Δrhp54 by PCR analysis. In this screen several Δsrs2 Δrhp54 Δrhp51 triple mutants were identified. Figure 8 shows one of these strains growing on YEA. Thus, the synthetic lethality of the Δsrs2 Δrhp54 is suppressed by loss of rhp51+.
Figure 8.
The synthetic lethality of Δsrs2 (SZ242) and Δrhp54 (SZ231) is suppressed by loss of rhp51+. Mid log cultures of each indicated strain were grown and struck onto YEA plates. The plates were incubated for 4 days and photographed. The triple mutant, Δrhp51 Δrhp54 Δsrs2 (SZ279) shows a similar growth rate to the Δrhp51 Δsrs2 Δrqh1 (SZ271) triple mutant. Both of these strains grow slower than the Δsrs2 single mutant. The wild-type strain is not shown, however Δsrs2 (SZ242) is pictured, which grows essentially like wild-type cells.
DISCUSSION
A summary of our findings on srs2+ is as follows: (i) Δsrs2 single mutants grow normally and have a normal morphology; (ii) Δsrs2 mutants are not sensitive to UV irradiation and treatment with HU; (iii) simultaneous loss of srs2+ and rqh1+ results in a synthetic phenotype in which the cells show extreme slow growth, aberrant cellular and nuclear morphologies and high spontaneous cell death; (iv) a large portion of Δsrs2 Δrqh1 cells undergo a checkpoint-dependent cell cycle arrest, most likely at G2/M, which is a stochastic event; (v) Δsrs2 Δrqh1 double mutants show high levels of spontaneous mitotic catastrophe; (vi) the slow growth and poor viability of Δsrs2 Δrqh1 are partially suppressed by loss of the rhp51+ and rhp57+ homologous recombination genes; (vii) loss of srs2+ suppresses the UV and HU sensitivity of Δrqh1 mutants; and (viii) Δsrs2 is synthetic lethal with Δrhp54, and this lethality is suppressed by loss of rhp51+. Deletion of the S.pombe srs2+ gene shares many properties in common with deletion of SRS2 in S.cerevisiae although the severity of phenotypes sometimes differs. For example, S.cerevisiae srs2 mutants are more sensitive to UV damage and its synthetic interaction with sgs1 appears more severe than that of Δsrs2 and Δrqh1. In addition we have gone on to show that loss of srs2+ partially suppresses the UV and HU sensitivity of Δrqh1. We also find that Δsrs2 Δrqh1 cells do not show a significant arrest in S-phase but rather arrest at what appears to be G2/M. The spontaneous cell death associated with Δsrs2 Δrqh1 appears to be due to mitotic catastrophe.
Phenotype of Δsrs2
In contrast to Rqh1, Srs2 clearly plays a much less prominent role in cell survival following UV damage or HU treatment. This is demonstrated by the level of sensitivity that each single mutant shows when exposed to these agents (Figs 1 and 7). Consistent with our previous studies we find that the Δrqh1 single mutant has a mild cell growth defect and lower plating efficiency. By comparison, Δsrs2 behaves essentially like wild type for both growth and plating efficiency (Fig. 2B and Table 3).
The synthetic interaction between Δsrs2 and Δrqh1
Schizosaccharomyces pombe Δsrs2 has a synthetic interaction with Δrqh1, manifested as extreme slow growth, poor plating efficiency, high rates of spontaneous cell death and abnormal cellular and nuclear morphologies. One interpretation of a synthetic interaction is that the two genes function in parallel pathways in an essential or very important process. The role of Rqh1 and its homologs is not known, but most evidence points to it playing some role in suppressing recombination arising by stalled replication. The synthetic phenotype of Δsrs2 Δrqh1 would suggest that Srs2 plays a similar or alternative role at stalled forks. However, our data do not support this interpretation. First, if the phenotype of the double mutant results from a major defect in the cells’ ability to cope with spontaneously arising stalled replication forks, then predictably these cells should arrest or slow in S-phase. Our FACS data from synchronized cells show that Δsrs2 Δrqh1 show only a slight delay in S-phase (Fig. 3). Second, if the phenotype of the double mutant results from a defect in its ability to cope with replication fork stalling, then these cells should be extremely sensitive to any increase in the number of stalled forks. Again we find this not to be the case, as the Δsrs2 Δrqh1 double mutant actually shows increased survival when exposed to UV or treated with HU when compared with Δrqh1 (Fig. 7A, B and C). These results argue against the phenotype of the double mutant resulting from stalled replication.
If Δsrs2 Δrqh1 cells are not arresting in S-phase at a significant level, where do they arrest? In an effort to determine this, Δsrs2 Δrqh1, wild-type and single mutant cells were DAPI stained and examined microscopically at times after release from G1 synchronization. Cells from each time point were scored as being either small or normal in size, elongated (assumed to be in G2 arrest), bi-nucleated (cells having completed mitosis) or abnormal (cells with torn or fragmented nuclei). We draw two conclusions from these studies. Our first conclusion, based on the data in Figure 4, is that some double mutant cells appear to cycle normally. Initially both wild-type and Δsrs2 Δrqh1 cells are small with single nuclei. At 3.5 h, which is the earliest time that double nucleated wild-type cells are seen, an equal number of Δsrs2 Δrqh1 are double nucleated. This shows that cells have progressed through mitosis. At later time points we see continued evidence of Δsrs2 Δrqh1 cells cycling normally, as double nucleated cells continue to be present. Our second conclusion is that many Δsrs2 Δrqh1 cells undergo an arrest, most likely at G2/M. This conclusion is based on our observation that large numbers of Δsrs2 Δrqh1 cells accumulate as single nucleated (pre-mitotic) elongated cells. In combination with our FACS data showing no major accumulation of cells in S-phase, these elongated cells with a single nucleus must be in G2, unable to enter M. We cannot rule out the possibility that these cells are in very late S-phase. By 6 h there are essentially no elongated cells found in wild-type, yet 25% of Δsrs2 Δrqh1 cells are elongated. In addition to elongated cells, at 6 h 7% of Δsrs2 Δrqh1 cells have aberrant or torn nuclei. This occurs in elongated cells, suggesting that some arrested cells ultimately undergo mitotic catastrophe. While we did not look at time points beyond 6 h, we assume that the number of cells showing mitotic catastrophe will continue to accumulate with time. We speculate that most, if not all, elongated cells will fail to progress through mitosis properly. This cannot be determined by these experiments, as elongated cells will continue to accumulate. However, if this assumption is correct, then the 25% of elongated cells together with the 7% of cells having already undergone mitotic catastrophe represent one-third of the cell population dying during each round of the cell cycle. This number is in good agreement with a doubling time of ∼6 h for Δsrs2 Δrqh1. A recent study in S.cerevisiae demonstrated that 15% of srs2 sgs1 double mutants arrest in mitosis as large budded cells and that this arrest was checkpoint dependent (20). Cells arrested in mitosis were also found in 7% of srs2 and 2% of sgs1 single mutants, demonstrating that this is not strictly a phenotype of the double mutant. Interestingly, when we examined Δsrs2 in a similar experiment, there was no evidence of G2/M arrested cells or cells undergoing a mitotic catastrophe. We saw evidence for a few elongated Δrqh1 cells suggesting a mild arrest. These data demonstrate that the apparent slow growth of Δsrs2 Δrqh1 cells is not due to a homogeneous slowing of the entire cell population, but that this is a mixed population of normal cycling and arrested cells. This indicates that the cellular events leading to cell cycle arrest are stochastic.
The G2/M arrest seen in Δsrs2 Δrqh1 cells is likely due to the accumulation of some type of DNA damage or structure that induces a checkpoint response. This is further corroborated by our finding that deletion of the rad9+ checkpoint gene suppresses the accumulation of the elongated, G2/M arrested cells in the Δsrs2 Δrqh1 mutant (Fig. 5B). While we cannot be certain what DNA structure is recognized as damage, it is tempting to speculate that it is recombination intermediates. This is based on the fact that Δsrs2 mutants have been reported to have a hyper-recombination phenotype and our findings that loss of recombination genes suppresses the slow growth of Δsrs2 Δrqh1 cells and improves their viability (Fig. 6A and B, and Table 3). It is known that Cds1 regulates the S-phase arrest while Chk1 regulates the G2 arrest. Our data, showing that loss of chk1+ but not cds1+ suppresses the elongated morphology seen in Δsrs2 Δrqh1, further support the conclusion that these cells are arresting in G2.
Deletion of recombination genes suppress the Δsrs2 Δrqh1 phenotype
Previous studies demonstrated that Δrqh1 mutants have no increase or only slightly elevated recombination rates (8,18). However, loss of Srs2 is associated with a 12-fold increase in spontaneous recombination rates (41,45). The elevated recombination rates in Δsrs2 do not appear to have any obvious deleterious effect on cells, as they grow normally and have normal morphologies. We wondered if the severe phenotype seen in Δsrs2 Δrqh1 was due to the inability of these cells to deal effectively with recombination. This would appear to be the case as we found that deletion of either rhp51+ or rhp57+ markedly suppresses the slow growth and low viability of Δsrs2 Δrqh1 cells. Loss of recombination also returns Δsrs2 Δrqh1 cells to a more normal cellular phenotype. This normalization suggests that recombination is at least a partial cause of the aberrant phenotype seen in the double mutant. One interpretation of these results is that potential problems caused by hyper-recombination in Δsrs2 cells are suppressed by Rqh1 function, and so the problems are only seen upon simultaneous loss of both genes.
Our findings of slow growth suppression in Δsrs2 Δrqh1 cells by loss of recombination are in contrast to the results of Wang et al. (41). They reported that Δrhp51 failed to suppress the slow growth of Δsrs2 Δrqh1 and, in fact, the triple mutant grew worse. We found clear suppression by not only loss of rhp51+, but also by loss of rhp57+. Additionally, they reported that their Δsrs2 Δrhp51 double mutant looked like a Δsrs2 Δrqh1 mutant and were more sensitive to UV damage than the Δrhp51 single mutant. Our experiments showed neither a synthetic interaction between Δsrs2 and Δrhp51 nor increased UV sensitivity in the double mutant compared with the Δrhp51 single mutant (data not shown). Other results with Δsrs2 were the same between our laboratories. This suggests that the discrepancies may arise from the use of different Δrhp51 mutants which could contain uncharacterized strain differences. We used the Δrhp51 reported previously (46), while Wang et al. used a Δrhp51 created for their studies. However, we are confident in our conclusion that loss of recombination suppresses the Δsrs2 Δrqh1 phenotype based on the fact that we observed suppression with two different recombination mutants and that comparable results have been established for the budding yeast S.cerevisiae. We also addressed the possibility that our suppression was due to a spontaneous suppressor. Our results argue against the presence of a spontaneous suppressor of the Δsrs2 Δrqh1 phenotype.
Δsrs2 is synthetic lethal with Δrhp54
While searching for recombination mutants that would suppress the slow growth of Δsrs2 Δrqh1, we found synthetic lethality between Δsrs2 and Δrhp54. This lethality was suppressed by deletion of rhp51+. Two possible explanations could account for this result. One is that Srs2 functions in recombination in a process in parallel with Rhp54 and that one of these two proteins is necessary to complete recombination initiated by Rhp51. This would explain why deletion of rhp51+ suppresses this synthetic lethality. An alternative explanation comes from studies in S.cerevisiae where Srs2 is known to function in the PRR pathway (24,29,47). Loss of srs2 in a rad6 mutant suppresses the UV sensitivity of these strains presumably by allowing repair of DNA damage to occur by homologous recombination. While we do not know if Srs2 plays the same role in PRR in S.pombe, the same synthetic lethality between Δsrs2 and Δrad54 exists in S.cerevisiae (31). We also know that loss of srs2+ in S.pombe leads to a hyper-recombination phenotype (41). From this, we can speculate that in the absence of srs2, damaged or incompletely replicated DNA is funneled into the Rhp51-dependent homologous recombination pathway. Presumably the process initiated by Rhp51 in a Δsrs2 background requires Rhp54 to be completed, and the initiation of this process in the absence of Rhp54 leads to lethality. In this scenario, Srs2 plays an indirect role in recombination.
Why does loss of srs2+ suppress rqh1 sensitivity to UV and HU?
It is difficult to explain how deletion of srs2+ in a Δrqh1 background can have a negative effect on cell growth and viability, yet have a protective effect on the ability of these cells to cope with UV and HU. We have already given a reasonable possible explanation for the synthetic interaction between Δsrs2 and Δrqh1 as it relates to normal growth. It is not necessary to link these two phenotypes since one happens in damaged or replication arrested cells and the other is a spontaneous response. One possibility is as follows: upon replication arrest, caused by DNA damage or HU treatment, recombination takes place at a high rate in the absence of Rqh1 (8,18). It is the process of recombination that leads to the decrease in viability seen in these cells. We base this latter statement on the finding that expression of a recombination resolvase partially suppresses the HU and UV sensitivity of Δrqh1 cells. In addition, unpublished data from our laboratory show that loss of either rhp51+ or rhp57+ completely or nearly completely suppresses the HU or UV sensitivity of Δrqh1. One proposed function of SRS2 in S.cerevisiae is to act downstream of RAD51 to stabilize recombination intermediates (39). By enhancing recombination, Srs2 might increase the HU and UV sensitivity of Δrqh1 cells. This proposed function might explain why the loss of Srs2 improves the survival of Δrqh1 cells when challenged with UV or HU. Obviously, further investigation will be necessary to answer this question with certainty.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to the following for providing us with strains: V. I. Bashkirov, W.-D. Heyer, H. Iwasaki, S. D. Park and A. Carr. This work was funded by NIEHS grant ES07940.
REFERENCES
- 1.Lowndes N.F. and Murguia,J.R. (2000) Sensing and responding to DNA damage. Curr. Opin. Genet. Dev., 10, 17–25. [DOI] [PubMed] [Google Scholar]
- 2.Hyrien O. (2000) Mechanisms and consequences of replication fork arrest. Biochimie, 82, 5–17 [DOI] [PubMed] [Google Scholar]
- 3.Seigneur M., Bidnenko,V., Ehrlich,S.D. and Michel,B. (1998) RuvAB acts at arrested replication forks. Cell, 95, 419–430. [DOI] [PubMed] [Google Scholar]
- 4.van Brabant A.J., Ye,T., Sanz,M., German,J.L.,III, Ellis,N.A. and Holloman,W.K. (2000) Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry, 39, 14617–14625. [DOI] [PubMed] [Google Scholar]
- 5.Bierne H., Seigneur,M., Ehrlich,S.D. and Michel,B. (1997) uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol. Microbiol., 26, 557–567. [DOI] [PubMed] [Google Scholar]
- 6.Prince P.R., Emond,M.J. and Monnat,R.J. (2001) Loss of Werner syndrome protein function promotes aberrant mitotic recombination. Genes Dev., 15, 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davey S., Han,C.S., Ramer,S.A., Klassen,J.C., Jacobson,A., Eisenberger,A., Hopkins,K.M., Lieberman,H.B. and Freyer,G.A. (1998) Fission yeast rad12+ regulates cell cycle checkpoint control and is homologous to the Bloom’s syndrome disease gene. Mol. Cell. Biol., 18, 2721–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart E., Chapman,C.R., Al-Khodairy,F., Carr,A.M. and Enoch,T. (1997) rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J., 16, 2682–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heo S.J., Tatebayashi,K., Ohsugi,I., Shimamoto,A., Furuichi,Y. and Ikeda,H. (1999) Bloom’s syndrome gene suppresses premature ageing caused by Sgs1 deficiency in yeast. Genes Cells, 4, 619–625. [DOI] [PubMed] [Google Scholar]
- 10.Karow J.K., Wu,L. and Hickson,I.D. (2000) RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev., 10, 32–38. [DOI] [PubMed] [Google Scholar]
- 11.Luo G., Santoro,I.M., McDaniel,L.D., Nishijima,I., Mills,M., Youssoufian,H., Vogel,H., Schultz,R.A. and Bradley,A. (2000) Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nature Genet., 26, 424–429. [DOI] [PubMed] [Google Scholar]
- 12.Myung K., Datta,A., Chen,C. and Kolodner,R.D. (2001) SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nature Genet., 27, 113–116. [DOI] [PubMed] [Google Scholar]
- 13.Watt P.M., Hickson,I.D., Borts,R.H. and Louis,E.J. (1996) SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics, 144, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gangloff S., Soustelle,C. and Fabre,F. (2000) Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nature Genet., 25, 192–194. [DOI] [PubMed] [Google Scholar]
- 15.Harmon F.G. and Kowalczykowski,S.C. (1998) RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev., 12, 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onoda F., Seki,M., Miyajima,A. and Enomoto,T. (2001) Involvement of SGS1 in DNA damage-induced heteroallelic recombination that requires RAD52 in Saccharomyces cerevisiae. Mol. Gen. Genet., 264, 702–708. [DOI] [PubMed] [Google Scholar]
- 17.Courcelle J. and Hanawalt,P.C. (1999) RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet., 262, 543–551. [DOI] [PubMed] [Google Scholar]
- 18.Doe C.L., Dixon,J., Osman,F. and Whitby,M.C. (2000) Partial suppression of the fission yeast rqh1(-) phenotype by expression of a bacterial Holliday junction resolvase. EMBO J., 19, 2751–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karow J.K., Constantinou,A., Li,J.L., West,S.C. and Hickson,I.D. (2000) The Bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc. Natl Acad. Sci. USA, 97, 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McVey M., Kaeberlein,M., Tissenbaum,H.A. and Guarente,L. (2001) The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is composite of normal aging processes and mitotic arrest due to defective recombination. Genetics, 157, 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S.K., Johnson,R.E., Yu,S.L., Prakash,L. and Prakash,S. (1999) Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science, 286, 2339–2342. [DOI] [PubMed] [Google Scholar]
- 22.Mullen J.R., Kaliraman,V. and Brill,S.J. (2000) Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics, 154, 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aboussekhra A., Chanet,R., Zgaga,Z., Cassier-Chauvat,C., Heude,M. and Fabre,F. (1989) RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res., 17, 7211–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rong L., Palladino,F., Aguilera,A. and Klein,H.L. (1991) The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics, 127, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence C.W. and Christensen,R.B. (1979) Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol., 139, 866–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguilera A. and Klein,H.L. (1988) Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics, 119, 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguilera A. and Klein,H.L. (1989) Genetic and molecular analysis of recombination events in Saccharomyces cerevisiae occurring in the presence of the hyper-recombination mutation hpr1. Genetics, 122, 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ulrich H.D. (2001) The srs2 suppressor of UV sensitivity acts specifically on the RAD5- and MMS2-dependent branch of the RAD6 pathway. Nucleic Acids Res., 29, 3487–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schiestl R.H., Prakash,S. and Prakash,L. (1990) The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics, 124, 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein H.L. (1997) RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics, 147, 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein H. (2001) Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2delta with other DNA repair genes in Saccharomyces cerevisiae. Genetics, 157, 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schild D. (1995) Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics, 140, 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaytor M.D., Nguyen,M. and Livingston,D.M. (1995) The complexity of the interaction between RAD52 and SRS2. Genetics, 140, 1441–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milne G.T., Ho,T. and Weaver,D.T. (1995) Modulation of Saccharomyces cerevisiae DNA double-strand break repair by SRS2 and RAD51. Genetics, 139, 1189–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frei C. and Gasser,S.M. (2000) The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev., 14, 81–96. [PMC free article] [PubMed] [Google Scholar]
- 36.Liberi G., Chiolo,I., Pellicioli,A., Lopes,M., Plevani,P., Muzi-Falconi,M. and Foiani,M. (2000) Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J., 19, 5027–5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freyer G.A., Davey,S., Ferrer,J.V., Martin,A.M., Beach,D. and Doetsch,P.W. (1995) An alternative eukaryotic DNA excision repair pathway. Mol. Cell. Biol., 15, 4572–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phipps J., Nasim,A. and Miller,D.R. (1985) Recovery, repair, and mutagenesis in Schizosaccharomyces pombe. Adv. Genet., 23, 1–72. [DOI] [PubMed] [Google Scholar]
- 39.Paques F. and Haber,J.E. (1997) Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 6765–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maftahi M., Han,C.S., Langston,L.D., Hope,J.C., Zigouras,N. and Freyer,G.A. (1999) The top3(+) gene is essential in Schizosaccharomyces pombe and the lethality associated with its loss is caused by Rad12 helicase activity. Nucleic Acids Res., 27, 4715–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S.W., Goodwin,A., Hickson,I.D. and Norbury,C.J. (2001) Involvement of Schizosaccharomyces pombe Srs2 in cellular responses to DNA damage. Nucleic Acids Res., 29, 2963–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bahler J., Wu,J.Q., Longtine,M.S., Shah,N.G., McKenzie,A.,III, Steever,A.B., Wach,A., Philippsen,P. and Pringle,J.R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast, 14, 943–951. [DOI] [PubMed] [Google Scholar]
- 43.Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- 44.Gangloff S., McDonald,J.P., Bendixen,C., Arthur,L. and Rothstein,R. (1994) The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol., 14, 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palladino F. and Klein,H.L. (1992) Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics, 132, 23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muris D.F., Vreeken,K., Carr,A.M., Broughton,B.C., Lehmann,A.R., Lohman,P.H. and Pastink,A. (1993) Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res., 21, 4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osman F., Tsaneva,I.R., Whitby,M.C. and Doe,C.L. (2002) UV irradiation causes the loss of viable mitotic recombinants in Schizosaccharomyces pombe cells lacking the G(2)/M DNA damage checkpoint. Genetics, 160, 891–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jang Y.K., Jin,Y.H., Shim,Y.S., Kim,M.J., Yoo,E.J., Seong,R.H., Hong,S.H. and Park,S.D. (1995) Evidences for possible involvement of Rhp51 protein in mitotic events including chromosome segregation. Biochem. Mol. Biol. Int., 37, 329–337. [PubMed] [Google Scholar]
- 49.Muris D.F., Vreeken,K., Carr,A.M., Murray,J.M., Smit,C., Lohman,P.H. and Pastink,A. (1996) Isolation of the Schizosaccharomyces pombe RAD54 homologue, rhp54+, a gene involved in the repair of radiation damage and replication fidelity. J. Cell Sci., 109 (Pt 1), 73–81. [DOI] [PubMed] [Google Scholar]
- 50.Tsutsui Y., Morishita,T., Iwasaki,H., Toh,H. and Shinagawa,H. (2000) A recombination repair gene of Schizosaccharomyces pombe, rhp57, is a functional homolog of the Saccharomyces cerevisiae RAD57 gene and is phylogenetically related to the human XRCC3 gene. Genetics, 154, 1451–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lieberman H.B., Hopkins,K.M., Laverty,M. and Chu,H.M. (1992) Molecular cloning and analysis of Schizosaccharomyces pombe rad9, a gene involved in DNA repair and mutagenesis. Mol. Gen. Genet., 232, 367–376. [DOI] [PubMed] [Google Scholar]