Abstract
The structural alteration of chromatin has a key role in regulating gene expression. The alteration of chromatin is mediated by modification of its components. Detailed understanding of the relationship between these modifications, notably, methylation of the full-length CpG island, the association of methyl-CpG binding proteins (MBPs), and the acetylation and methylation of histones in gene silencing is vitally important. Currently, however, the manner in which chromatin components, associated with a specific gene, are modified is poorly understood. Here we provide in vivo evidence in cancer cells of the differential association between CpG methylation, MBPs, and histone modification in the entire CpG island of the human E-cadherin (CDH1) gene. Of the cell lines with CDH1 transcriptional repression, the distribution of methyl-CpGs in the CpG island differed markedly. In a cell line with gene silencing, the promoter region was almost methylation-free. Chromatin immunoprecipitation analysis revealed that the acetylation status of histone H4 differed between cell lines. However, deacetylated histone H3 was associated with the CpG island in all silenced cell lines. Binding of MeCP2 was also detected in all silenced cell lines. Additional binding of MBD1 protein was detected in a cell line in which the promoter region was poorly methylated and only histone H3 was deacetylated. Binding of MBD2 protein was detected in all other silenced cell lines. Histone H3 lysine 9 was methylated in all silenced cells, while histone H3 lysine 4 was methylated in some silenced cell lines. These results demonstrate that chromatin components associated with inactive CDH1 chromatin is heterogeneously modified and suggests the presence of multiple pathways for the formation of inactive chromatin.
INTRODUCTION
CpG islands are associated with the 5′ end of many human genes. A characteristic feature of CpG islands is that they are unmethylated in germ cells and throughout development (with the exception of those on the inactive X chromosome and those associated with some imprinted genes), while the majority of the genome undergoes dramatic changes in methylation (1). In somatic cells, CpG islands are non-methylated irrespective of the expression of the associated genes. CpG islands methylated de novo play important roles in aberrant transcriptional repression and are involved in the inactivation of many known tumor suppressor genes (2). Methylation of the promoter region within CpG islands is considered to be critical for gene silencing given that the promoter region is essential for transcriptional regulation (2). Thus, most analyses of the correlations between CpG island methylation and gene silencing have focused solely on the promoter regions which are in fact only part of the full-length CpG island. As a consequence the methylation status of the entire CpG island required for gene silencing is not clear.
Histone deacetylation is an additional, well-recognized mechanism associated with gene silencing. Non-methylated CpG islands are in a form of active chromatin structure enriched with hyperacetylated histones H3 and H4 (3). In contrast the majority of genomic regions other than CpG islands are in an inactive chromatin structure with deacetylated histones H3 and H4. These two repression mechanisms, DNA methylation and histone deacetylation, are linked by methyl-CpG binding proteins (MBPs) (4,5). MeCP2, MBD1 and MBD2 are known MBPs, which have a conserved methyl-CpG (mCpG) binding domain. Kaiso is a further type of MBP, which has a different binding motif and requires multiple mCpG sites for binding (6). These proteins, and the repressor complexes containing them, repress transcription of methylated genes both in vitro (7,8) and also in cells transfected with methylated DNA fragments (5,8–13). MeCP2 protein can interact with histone deacetylase 1 (HDAC1) and HDAC2 via binding with co-repressor mSin3 (5). MBD2 is a component of the MeCP1 complex (9), interacts with the Sin3 (13) or NuRD (14) complex, and is also associated with HDAC1 and HDAC2. It has been suggested that MBD1 recruits other HDAC activity (10).
Other post-transcriptional histone modifications, such as methylation and phosphorylation, are also reported to be associated with gene activities (15). In particular, histone methylation is attracting increasing research interest (16). Methylation events at lysine 4 of histone H3 (H3-K4) and at lysine 9 of histone H3 (H3-K9) are associated with an active and inactive status of gene expression, respectively (17,18). Although its direct relationship to DNA methylation is unclear, binding of heterochromatin protein to the methylated H3-K9 residue is considered to be an important step for silencing (19,20).
It is critically important to understand how inactive chromatin components associated with a specific gene are modified and what kind of components are involved. Since a CpG island is a distinct methylation-free unit, analysis of the methylation status of the CpG island as a whole and the associated status of modified histones and MBPs in methylated CpG islands in silenced cells will provide clues to understand the molecular mechanism of epigenetic gene silencing. In this study, we have investigated the methylation status of CpG sites of a complete CpG island that is associated with gene silencing mechanisms. We have selected the CpG island of the human E-cadherin (CDH1) gene as a target for full-length CpG island analysis because the CDH1 gene has a CpG island of typical size (∼1.3 kb) containing a well-characterized promoter region close to the 5′ end of the island (21). The gene product of the CDH1 gene is a cell adhesion molecule and inactivation of the CDH1 gene is thought to be associated with metastasis of cancer cells. As a consequence the molecular mechanism of CDH1 gene silencing may be of great importance in understanding the metastatic process. In some human cancer cells a lack of CDH1 gene expression due to methylation has been reported (22,23). Molecular events underlying gene silencing, such as the associated methylation status, histone deacetylation and methylation, and association of MBPs have been further studied.
MATERIALS AND METHODS
Analysis of methylation status of genomic DNA
Methyl-CpG binding domain (MBD) column chromatography was performed as described previously (24,25). Briefly, 50 µg of Tsp509 I (New England BioLabs) digest of genomic DNAs from cancer cell lines were applied to an MBD column (1 ml of resin in a Poly-Prep Column, Bio-Rad 731-1550, 0.8 cm in diameter) and eluted with a stepwise gradient of salt concentrations [0.4–1.0 M NaCl by 40 mM per step, in 20 mM HEPES (pH 7.9), 10% glycerol, 0.1% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and 10 mM β-mercaptoethanol; 4 ml per step]. DNA from 150 µl aliquots of each fraction was subjected to PCR-based detection of fragments containing the CGI as described previously (24,25). Bisulfite modification of Tsp509 I digests of genomic DNA was performed following a published procedure (26) with slight modifications. Approximately 1 µg of bisulfite-treated Tsp509 I-digested genomic DNAs was used for amplification of DNA fragments derived from the predicted CpG island (positions 80 656–81 965, accession no. AC099314, http://www.ebi.ac.uk/cpg/). The first amplification of non-coding strand was ‘hot started’ (3 min at 95°C) with TaqStart antibody (Clontech). PCR was performed with a mixture of Taq DNA polymerase (Gibco BRL) and Taq Extender (Stratagene) following the manufacturer’s recommendation. Twenty-five cycles of amplification (30 s at 95°C, 30 s at 57°C, 8 min at 70°C) were performed. Initial PCR products were re-amplified by heminested PCR with slight modifications (30 cycles for amplification and 3 min for chain elongation). Primers used for the first and second PCR were 5′-GAAGAATTCGTAATTTTAGGTTAGAGGGT-3′ (for both PCR), 5′-GAATCTAGAACCCCTTTCCAAC CCCTCCC-3′ (for the first PCR), 5′-GAATCTAGATT TCCAACCCCTCCCTACTC-3′ (for the second PCR). After EcoRI/XbaI digestion followed by gel purification, the amplified fragments were ligated to EcoRI/XbaI-digested plasmid pMW119 (Nippon gene), and introduced into bacterial strain CJ236. Transformed cells were incubated at 30°C on SOB (2% bacto-tryptone, 0.5% bacto-yeast extract, 0.05% NaCl, 2.5 mM KCl, pH 7.0) plate and cultured in Terrific broth at 30°C overnight. Nucleotide sequencing analysis was performed using thermosequenase fluorescent-labeled primer cycle sequencing kit (Amersham Pharmacia Biotech).
Chromatin immunoprecipitation analysis of modified histones H3 and H4
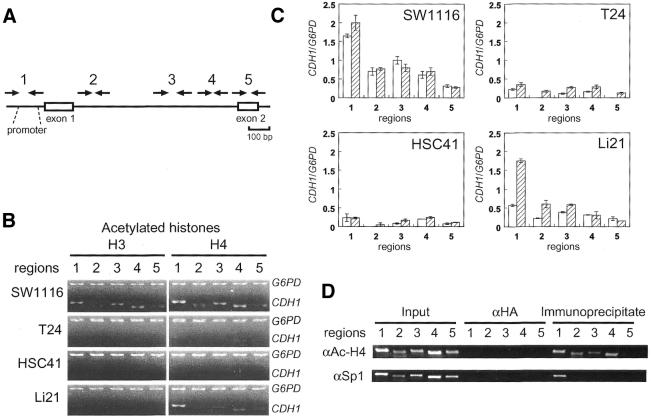
Chromatin immunoprecipitation (ChIP) analysis of modified histones H3 and H4 was performed following the procedure recommended by the supplier of the anti-acetylated H3 (06-599), anti-acetylated H4 (06-866), anti-dimethyl-histone H3 (Lys4) (07-030) and anti-dimethyl-histone H3 (Lys9) (07-212) antibodies (Upstate Biotechnology). Anti-Sp1 antibody (PEP 2) was obtained from Santa Cruz. Briefly, cells treated with 1% formaldehyde were scraped off culture dishes in PBS containing 1 mM PMSF, 1 µg/ml aprotinin and 1 µg/ml pepstatin A). Cells collected by centrifugation were lysed in SDS buffer (1% SDS/10 mM EDTA/50 mM Tris–HCl, pH 8.1) and placed on ice for 10 min. Cross-linked chromatin was sonicated to reduce the size of DNA fragments to <1 kb. After centrifugation, supernatant was diluted 1:10 in dilution buffer (0.01% SDS/1.1% Triton X-100/1.2 mM EDTA/16.7 mM Tris–HCl, pH 8.1/167 mM NaCl). Immune complexes collected by 5 µg of antibodies (4°C, overnight) were eluted (1% SDS/0.1 M NaHCO3, room temperature, 15 min for twice) and cross-links were reversed by heating (65°C, 5 h). DNA fragments were recovered from elute by ethanol precipitation after proteinase K treatment and phenol extraction. Anti-hemagglutinin antibody (αHA) (12CA5) was obtained from Boehringer Mannheim. Duplex PCR-based detection of acetylated histones associated with CpG islands of the CDH1 and the G6PD genes was performed with a mixture of Taq DNA polymerase (Gibco BRL) and Taq Extender (Stratagene) as described above. Thirty cycles of amplification (30 s at 95°C, 20 s at 61°C, 1.5 min at 72°C) were performed with 32P-labeled 5′ primer. Primers used for detection of the CpG island regions were: region 1, 5′-AGAGGGTCACCGCGTCTATG-3′ and 5′-CTCACAGGTGCTTTGCAGTT-3′; region 2, 5′-CAGCCATGGGCCCTTGGAGC-3′ and 5′-CGAACTTTCTTGGAAGAAGG-3′; region 3, 5′-GGTCGTAAATAGGAGTGAGG-3′ and 5′-AGTCACCCCCTCAAGACCTA-3′; region 4, 5′-GGGCTCGGCGGCCGTAGGTT-3′ and 5′-AGATGGAACCGGGTGACTCG-3′; region 5, 5′-CACCCCAGGTCTCCTCTTGG-3′ and 5′-TTTCCAACCCCTCCCTACTC-3′. Primers for the amplification of the G6PD fragment were as described previously (27). PCR products were fractionated on 5% polyacrylamide gels and quantified by BAS 2000 (Fujix) for experiments shown in Figure 3B.
Figure 3.
Differential deacetylation of histones H3 and H4 in the CpG island. (A) Positions of PCR primers used for ChIP analysis. (B) Deacetylation of histones H3 and H4 in the CpG island regions of the CDH1 gene in SW1116, T24, HSC41 and Li21 cells were detected by duplex PCR analysis of immunoprecipitated DNA. The promoter region of the G6PD gene was analyzed for the association with hyperacetylated histones. Input controls are identical to those shown in Figure 4B. (C) Acetylation levels of histone H3 and H4 of the regions 1–5 relative to those of a region within the CpG island of the G6PD gene are shown as open and hatched bars, respectively. Error bars represent the standard deviations of independent duplicate experiments. SW1116, HSC41 and Li21 were established from male patients, and T24 was established from a female patient. The CDH1/G6PD ratio is not compensated for on the basis of the number of X chromosomes in each cell line. (D) Global histone acetylation and localized Sp1 protein binding in the CpG island of SW1116 cell. SW1116 cell lysate was divided into two fractions and each fraction was used for ChIP using anti-acetyl-histone H4 or anti-Sp1 protein antibodies (αAc-H4 or αSp1, respectively).
Western blot analysis
MBPs in nuclear extract (15 µg of protein) from cancer cells were detected by western blotting using ECL system (Amersham Pharmacia Biotech). The antibodies used were: 07-013 (MeCP2, Upstate Biotechnology), S751 [MBD1 (10)], and S923 [MBD2 (9)].
ChIP analysis of MBP binding using cultured cancer cells
ChIP analysis of mCpG binding proteins (MBD1 and MBD2) was performed with 5 µl of polyclonal antiserum essentially according to the procedure for histone analysis (see above). However, additional protease inhibitors (5 µg/ml of leupeptin, antipain and chymostatin) were added and Dynabeads-protein G (Dynal) was used in place of protein A. We also performed ChIP of MeCP2 protein using 5 µg of purified polyclonal IgG of MeCP2 protein and Dynabeads-protein A (Dynal). Antibodies (polyclonal antiserum) against MBD1 [S751 (10)], and MBD2 [S923 or R593 (9)] have been described previously. Antibodies against MBD1 protein (M254) and MBD2 protein (07-199) were obtained from Santa Cruz and Upstate Biotechnology, respectively.
ChIP analysis of MBP binding using isolated nuclei of cancer cells
ChIP of MBPs using isolated nuclei was performed as described previously (28). Briefly, cells were scraped from four culture dishes in 5 ml of PBS containing protease inhibitors and collected by centrifugation. The pelleted cells were resuspended in 1 ml of ice-cold hypotonic buffer (10 mM Tris–HCl pH 7.4/10 mM NaCl/5 mM MgCl2) containing protease inhibitors. Nuclei were prepared by 10 strokes of homogenization in a glass homogenizer on ice. After centrifugation, nuclei were resuspended in 20 ml of hypotonic buffer containing 0.1% Nonidet P-40 and 0.5% formaldehyde. After incubation at ambient temperature for 15 min, nuclei were collected and sonicated to reduce DNA fragments. Immunoprecipitation of cross-linked chromatin fragments and PCR detection of immunoprecipitated DNA were performed as well as ChIP using cultured cells. Primers for the CDKN2A fragment were as described previously (29).
RESULTS
Differential methylation status of the entire CpG island of the CDH1 gene
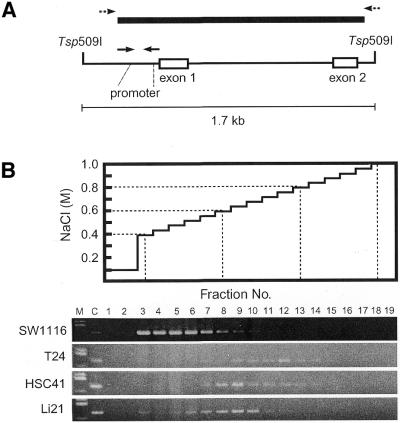
A previous Southern and northern analysis revealed that the CDH1 gene of some human cancer cell lines is silenced by CpG methylation (22). It is not clear whether the methylation status of specific restriction sites represents that of the entire CpG island. To determine the methylation status of the entire CpG island in cell lines, we first performed MBD column chromatography (24,28) which separates DNA fragments primarily on the basis of the number of mCpGs in the DNA fragments. Tsp509 I digests of various cancer cell lines DNA were analyzed by MBD column chromatography. A Tsp509 I fragment containing the entire CpG island (1.3 kb in length) predicted by computation is ∼1.7 kb in size (Fig. 1A) and contains 126 CpGs. The DNA fragment from the non-expressing bladder cancer cell line, T24 (22), had high affinity to the column and was detected by PCR in fractions 9–12 (0.64–0.76 M NaCl, Fig. 1B). Similar results were obtained when the genomic DNAs of Lu65A (lung cancer), Li7NM (liver cancer) and MKN1 (gastric cancer) cells, which are non-expressing (22), were analyzed (data not shown). In the case of non-expressing HSC41 (gastric cancer) and Li21 (liver cancer) cells (22), the DNA fragments showed reduced affinity to the column than that of T24 and were detected in fractions 6–10 (0.52–0.68 M NaCl, Fig. 1B). DNA fragments containing the CpG island of SW1116 (colon cancer) cells which express the CDH1 gene (22) bound only weakly to the column and were detected in fractions 3–7 (0.40–0.56 M NaCl, Fig. 1B), suggesting that most CpG sites (CpGs) are not methylated. Elution profiles of DNA from the expressing MCF7 (breast cancer) cells were similar to that of SW1116 cells and normal human somatic tissue (submandibular gland) DNA (data not shown). These results indicate the variations between cancer cell lines in terms of the methylation status of the entire CpG island associated with transcriptional repression of the CDH1 gene.
Figure 1.
Differences in methylation status of the entire CpG island of the silenced human CDH1 gene. (A) The Tsp509 I fragments associated with the CpG island of the CDH1 gene. Bold line indicates the entire CpG island predicted by computation based on CpG density. Solid arrows indicate primer sites used for PCR detection of elution profiles. Dashed arrows indicate primer sites used for bisulfite genomic sequencing (Fig. 2). PCR products were analyzed by agarose gel electrophoresis. (B) Separation of the DNA fragments by MBD column chromatography. Tsp509 I-digested DNAs were eluted as described (24). M, HaeIII digest of pUC19; C, PCR product from each Tsp509 I-digested DNA. Numbers indicate specific fraction.
Detailed methylation profiling of the entire CpG island
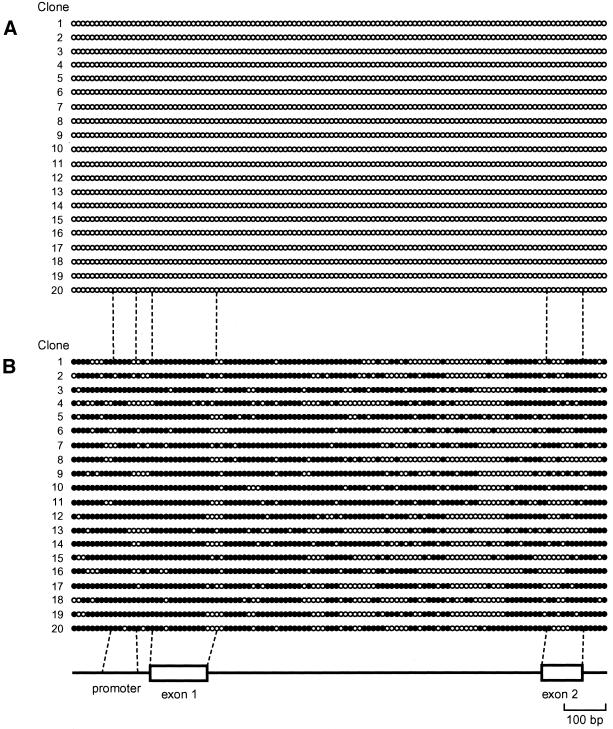
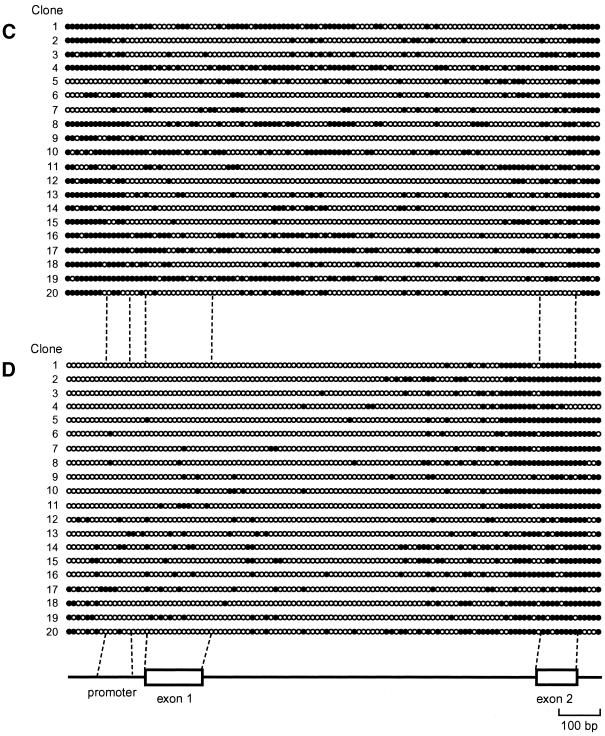
Next we performed detailed methylation profiling of CpGs within the CpG island of the CDH1 gene in cell lines with each cell line representing a different category of elution profile. After bisulfite modification of genomic DNA, a 1.4-kb region encompassing the predicted CpG island (Fig. 1A) was amplified by PCR without fragmentation of the target region (Fig. 1A). PCR products containing the entire CpG island were then cloned and the nucleotide sequences of 20 independent clones from each cell line were determined. Analysis of the entire CpG island clones revealed that all 116 CpGs within the amplified fragments of SW1116 cells were methylation-free (Fig. 2A). Independent experiments revealed that CpGs at both ends of the 1.4-kb PCR fragments and adjacent regions were not methylated in DNA from normal human somatic tissue (data not shown), demonstrating that these regions are within a CpG island and that the predicted CpG island is bona fide. The majority of CpGs within the CpG island of T24 cells were methylated (Fig. 2B) and the distribution of mCpGs was heterogeneous in the different clones. The CpGs within the CpG island of HSC41 cells were methylated to a lesser extent than those of T24 cells, the distribution of mCpGs was highly heterogeneous (Fig. 2C) and the methylation pattern is not altered after passaging of cell lines (data not shown). In contrast, unexpectedly in the case of Li21 cells, mCpGs were distributed relatively densely in the 3′ region of the CpG island, while the promoter region was poorly methylated (Fig. 2D). Further bisulfite genomic sequencing revealed that this methylation pattern was unchanged after passage (data not shown). We did not find any CpNpG methylation (30).
Figure 2.
(Opposite and above) Heterogeneity of methylation status of the entire CpG island among cell lines or alleles. All 116 CpG sites in the predicted CpG island region in SW1116 (A), T24 (B), HSC41 (C) and Li21 (D) cell lines were analyzed by bisulfite genomic sequencing for methylation status. White and black circles denote unmethylated and methylated CpG sites, respectively (not to scale). Numbers indicate specific plasmid clone.
Differential histone acetylation status in differentially methylated CpG islands
We next performed ChIP analysis of acetylated histones. After cross-linking of DNA-histones by formaldehyde treatment of cells, sonicated chromatin fragments were immunoprecipitated using anti-acetyl-histone H3 or H4 antibodies. Relative acetylation levels of histones H3 and H4 throughout the regions of the CDH1 gene CpG island (regions 1–5, Fig. 3A) were compared with those of a hyperacetylated CpG island region of the G6PD gene. PCR products were not detected in the negative control experiments with anti-HA antibody (data not shown).
In T24 and HSC41 cells, the acetylation levels in regions 1–5 were lower than those of SW1116 cells, particularly at the promoter site (region 1 in Fig. 3B and C). Hyperacetylation of histone H4 was also detected throughout the CpG island, while binding of transcription factor Sp1 was detected at the promoter region by ChIP analysis in the expressing SW1116 cells (Fig. 3D). These results show that deacetylation of histones throughout the CpG island in T24 and HSC41 cells is not an artifact due to insufficient fragmentation of cross-linked chromatin. Therefore, we can conclude that histones H3 and H4 are deacetylated throughout the CpG island in T24 and HSC41 cells regardless of mCpG density (Fig. 2A and B). These results are consistent with the previous findings that acetylation of histones H3 and H4 (27) or H4 (31) around the promoter region is strongly associated with methylation-mediated transcriptional repression. In silenced Li21 cells, the acetylation pattern of histone H4 was similar to that in the expressing SW1116 cells, that is, hyperacetylation of histone H4 was observed within the CpG island (Fig. 3B and C). In contrast, histone H3 within the CpG island was deacetylated in Li21 cells (Fig. 3B and C).
Differential binding of methyl-CpG binding proteins
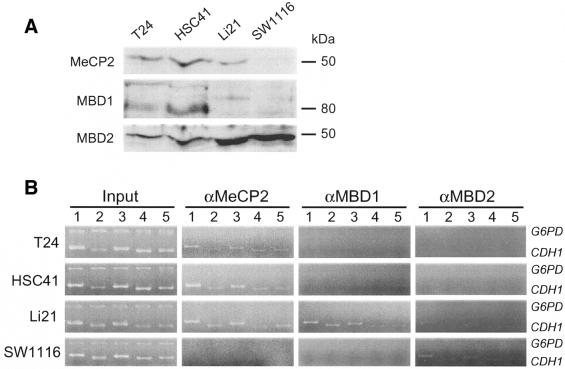
Recent studies show that MBPs are associated with different HDAC activities in vivo (5,9,10). Heterogeneity in the histone modification can be potentially attributed to differential binding of MBPs to the CpG island of silenced cells. In order to understand how MBPs are associated with methylated CpG islands in vivo, we performed a ChIP assay with antibodies specific to each MBP. We first analyzed the presence of endogenous MBPs in cancer cells. Nuclear extracts from each cell were subjected to western blot analysis with antibodies specific to each MBP. We confirmed expression of MeCP2 (52 kDa), MBD1 [multiple bands around 80 kDa, possibly due to splice isoforms (11)] and MBD2 (49 kDa) proteins in all cancer cells although their expression levels differed between cell types (Fig. 4A).
Figure 4.
Differential MBP binding to the CpG island of cancer cells. (A) Presence of MBPs in the nuclei of each cells. Nuclear extract of cells (15 µg of protein) was subjected to western blot analysis using antibodies specific to each MBP. (B) Duplex PCR analysis of DNA fragments derived from the CpG island in input dilution (Input) or chromatin immunoprecipitates of anti-MBP antibodies. Panels depicted are representative of four independent trials. Amplified regions are identical to those used for analysis of acetylated histones. The antibodies that were used are 07-013 (MeCP2), S751 (MBD1) and S923 (MBD2).
Using an antibody raised against a polypeptide corresponding to the C-terminal tail of the mouse MeCP2 protein (07-013, see Materials and Methods) we detected the binding of MeCP2 protein to the CpG island of all silenced cell lines regardless of methylation status (Fig. 4B). In contrast, using an antibody raised against a recombinant protein corresponding to amino acids 351–556 of the mouse MBD1 protein (S751, see Materials and Methods) we found MBD1 protein binding only to the CpG island of Li21 cells (Fig. 4B). We obtained similar results by using an additional anti-MBD1 antibody raised against recombinant MBD1 protein corresponding to amino acids 383–636 (M254, see Materials and Methods) (data not shown). We did not detect binding of these proteins to non-methylated CpG island of SW1116 cells (Fig. 4B). To confirm the specificity of the ChIP analysis to methylated DNA, we used a non-methylated CpG island region of the G6PD gene, where MeCP2 and MBD1 proteins are not expected to bind. Duplex PCR analysis of both immunoprecipitated DNA fragments and input dilution without immunoprecipitation revealed that the methylated CpG island regions of the CDH1 gene in cells were amplified (Fig. 4B). However, amplification of a region of the G6PD gene was observed by PCR with input dilution alone (Fig. 4B), that is, the CpG island fragment of the CDH1 gene is enriched after immunoprecipitation. These results indicate that the protein bindings in silenced cells are methylation-dependent. We could not detect MBD2 protein binding to the CpG island using antibody S923 (Materials and Methods) (data not shown). In contrast, in the ChIP analysis using an antibody raised against a recombinant protein corresponding to amino acids 152–262 of the mouse MBD2 protein (07-199, see Materials and Methods), we detected protein binding to the CpG island regions in SW1116 cells although binding was not observed in other cell types (Fig. 4B). Similar results were obtained in experiments using antibody R593 (Materials and Methods) (data not shown).
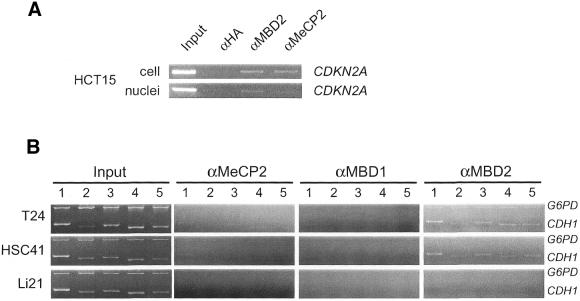
Recently, it has been reported that the MBD2 protein binds to the methylated CDKN2A (p16/INK4A) gene, which is silenced in HCT15 cells (29). However, it has also been reported that the MeCP2 protein bound to the same locus in the identical cell line (32). This difference can potentially be attributed to the experimental conditions that were adopted. In the former study, extracted nuclei were subjected to the ChIP analysis, while in the latter study cultured cells were directly treated with formaldehyde. We repeated these experiments and found that the binding is reproducible although a different antibody against the MBD2 protein was used in our experiments (Fig. 5A). Similar results were obtained at the methylated CDKN2A locus in T24 cell (data not shown). These findings prompted us to investigate MBD protein binding to the CpG island of the CDH1 gene using isolated nuclei as a substrate for the ChIP analysis. We observed MBD2 protein binding to the G6PD locus in T24 cell and the CDH1 CpG island in T24 and HSC41 cells using antibody 07-199 (Fig. 5B). MBD2 protein binding was not observed in the CDH1 CpG island of Li21 cell (Fig. 5B) in which MBD1 protein was found to bind (Fig. 5B). We failed to detect MBD2 protein binding using antibody S923. Binding of MeCP2 and MBD1 proteins to the CDH1 CpG island were not efficiently detected when isolated nuclei was subjected to cross-linking (Fig. 5B).
Figure 5.
Differential MBP bindings to the CpG island revealed by ChIP analysis using isolated nuclei of cancer cells. (A) PCR analysis of DNA fragments derived from the methylated promoter of CDKN2A gene in chromatin immunoprecipitates of anti-MeCP2 or anti-MBD2 antibodies from cell or nuclei. (B) Duplex PCR analysis of DNA fragments derived from the CpG island in input dilution (Input) or chromatin immunoprecipitates of anti-MBP antibodies from nuclei. Panels depicted are representative of duplicate trials. Amplified regions are identical to those used for analysis of cells. Input dilution was prepared from cross-linked chromatin of nuclei. Antibodies that were used are 07-013 (MeCP2), M254 (MBD1) and 07-199 (MBD2). The antibody S751 that was used in the experiments shown in Figure 4 was replaced with the antibody M254 in this experiment due to the lack of the material. We confirmed that similar results were obtained as shown in Figure 4 by using the antibody M254 (data not shown).
Differential methylation of histones
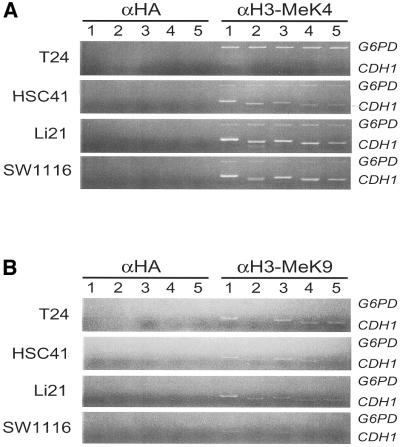
We further investigated how H3-K4 and H3-K9 residues associated with the CDH1 gene are modified in cancer cells. ChIP analysis revealed that the H3-K4 residue associated with the CDH1 gene was methylated in SW1116 cells and weakly methylated in T24 cells (Fig. 6A). Unexpectedly, this residue was methylated in silenced HSC41 and Li21 cells. The H3-K4 residue associated with the X-linked G6PD gene, which is expressed in all cells, was also methylated (Fig. 6A). However, amplification of the G6PD sequence was relatively inefficient in cancer cells.
Figure 6.
Differential methylation of histones H3-K4 (A) and H3-K9 (B) in the CDH1-CpG island. Duplex PCR analysis of DNA fragments derived from the CpG island in chromatin immunoprecipitates of α-HA or anti-methylated histone antibodies (αH3-MeK4 and αH3-MeK9) were performed. Positions of PCR primers used for ChIP analysis are identical to those shown in Figure 3A. Input controls are identical to those shown in Figure 4B.
In contrast, the H3-K9 residue was methylated in all silenced cells and very weakly in expressing SW1116 cells (Fig 6B). T24 cells are derived from a female, and the presence of an inactivated G6PD allele and H3-K9 methylation are expected. However, we could not detect H3-K9 methylation associated with the G6PD gene in T24 cells as well as in other cell lines (Fig. 6B). However, in separate experiments, we detected H3-K9 methylation associated with the X-linked HPRT gene in T24 cells, but not in Li21 cells, which is derived from a male (data not shown).
DISCUSSION
Chromatin structure at CpG islands is closely associated with the regulation of gene expression. CpG islands provide a model for active chromatin (3), and altered chromatin structure is associated with DNA methylation (33). Major chromatin components are DNA, histone and nucleosomal proteins. In this study in cancer cells, we have shown that the CpG island associated with the human CDH1 gene is hetero geneous in terms of DNA methylation, histone modification and MBP binding.
We also found that high-density methylation of the entire CpG island or promoter methylation is not a prerequisite for transcriptional repression. In Li21 cells the promoter region was sparsely methylated (Fig. 2C). The number of mCpGs in the CpG island of these cells is consistent with the relative order of elution in MBD column chromatography experiments (Figs 1 and 2). These results indicate that the methylation status (as revealed by bisulfite genomic sequencing) of representative clones, particularly the unexpected methylation status observed in Li21 cells, is not an artifact of bisulfite modification and/or PCR. In addition, the methylation status of the CpG island in Li21 cells is not tissue specific because the DNA fragment containing the CpG island of an additional liver cancer cell line (Li7NM) had high affinity to the MBD column as in the case of hypermethylated CpG island in T24 cells (data not shown). It is reported that the CDH1 gene becomes silenced by binding of the repressor protein Snail (34,35). However, as binding factors were not detected by in vivo footprint experiments (data not shown), repressor binding cannot be the cause of the observed silencing.
A common feature of the CpG islands in T24, HSC41 and Li21 cells, is that they are always highly methylated at one or both ends of the CpG island (Fig. 2). Our results raise the possibility that high-density methylation occurring at, at least, one end of the CpG island is a general feature of methylated CpG islands associated with stably repressed genes. This observation is consistent with the finding that de novo methylation of the CDH1 CpG island initially involves sequences at both ends (36). However, it is not clear whether this spread of methylation is an event associated with silencing because in this experiment fibroblasts (in which the CDH1 gene is not intrinsically expressed) were used. On the other hand, it has been reported that methylation of the downstream CpG island does not block upstream promoter activities of endothelin receptor B (EDNRB) (37) gene in cancer. This phenomenon is in contrast to the CDH1 gene silencing associated with non-methylated promoter in Li21 cell. These results suggest that methylation at the 3′ region of the CpG island alone is not sufficient for silencing of the corresponding gene.
Analysis of histone acetylation status demonstrated that histones H3 and H4 associated with the CDH1 gene in T24 and HSC41 cells are not acetylated (Fig. 3). In contrast, only histone H4 associated with the same gene in Li21 cells were acetylated (Fig. 3). A recent study demonstrated that treatment of cells from fragile X-syndrome patients with trichostatin A, a potent inhibitor of HDAC(s), failed to reacetylate histone H3 in a methylated CGG repeat tract at the 5′ end of the FMR1 gene while histone H4 became acetylated (27). It is also reported that histone H3 deacetylation, but not H4, is associated with CpG methylation and MeCP2 recruitment (38,39). In our investigations we observed a similar differential deacetylation. Acetylation of histone H3 and that of histone H4 may not be equivalent in terms of methylation-mediated gene silencing.
We detected binding of MeCP2 protein to the CpG island of all silenced cells. Binding of MBD1 protein was only detected in the CpG island of Li21 cells (Fig. 4B), while that of MBD2 was detected in T24 and HSC41 cells (Fig. 5B). MBPs bind differentially to the methylated CpG islands. MBP bindings in Li21 cells to the promoter region of the CpG island was evident (Fig. 4B) although most of CpGs in the region were sparsely methylated. A limited number of mCpG would be sufficient for the recruitment of MBPs in vivo.
It is not clear how MBPs are differentially employed in physiological settings. It is expected that there are both general and specific in vivo roles for each MBP. MeCP2 protein is essential in mice for maintenance of normal neurological function (40,41). Furthermore, a defect of this gene results in the neurological disorder, Rett syndrome (42). These observations suggest that the specific role of a particular MBP cannot always be functionally compensated by other MBPs. It is reported that the MeCP2 protein binds to the inactive, maternal allele of the imprinted U2af1-rs1 gene in mice (38). However, the recruitment of other MBPs to the same CpG island is not clear. We found MBD2 protein binding at the methylated CpG island of the X-linked G6PD gene (Fig. 5B) in T24 cells, which is derived from a female patient. However, binding of other MBPs was not detected (Figs 4B and 5B). These results suggest that specific MBPs are recruited to methylated CpG islands in the physiological setting. Our observation of the binding of multiple MBPs to the methylated CpG island of the CDH1 gene suggests that the mechanism of the recruitment of MBPs to methylated CpG islands varies between the physiological setting and in disease-associated de novo methylation.
ChIP analysis of MBPs gave different results depending on the cross-linking conditions. We detected MBD2 protein binding to the CpG island only when isolated nuclei were used for ChIP, while other MBPs could not be detected (Fig. 5B). The binding is not artificially caused during nuclei isolation steps because we could not detect MBD2 protein binding in Li21 cells (Fig. 5B) although the protein is present in nuclear extract of all cancer cells (Fig. 4A). Furthermore, it is unlikely that loss of other MBPs during the isolation step was compensated by the binding of MBD2 since we detected MeCP2 and MBD1 in nuclei (Fig. 4A). These results show that both MeCP2 and MBD2 proteins bind to the CpG island in T24 and HSC41 cells in vivo. Differences in methylation status of the CpG island may be required for targeting and binding of either MBD1 or MBD2 proteins among cell lines. Some factor(s) specifically expressed in each cell type and bound at the CpG island may attract selective protein bindings. Our data also suggest that the binding of complexes containing MBPs is not necessarily stable during nuclei isolation steps and that differences in stability can occur. It is reported that in vivo footprints of transcription factors at the human PGK1 locus disappeared when isolated nuclei rather than whole cells were used (43). This could be due to detachment of transcription factors from promoter regions by the alteration of chromatin structure caused during nuclei isolation (43). Our ChIP analysis using nuclei of SW1116 cell failed to detect Sp1 protein binding at the promoter region of the CDH1 gene (data not shown) although the binding was detected during cross-linking experiments when cells were used (Fig. 3D). Therefore, failure in the detection of MeCP2 and MBD1 protein bindings at the CDH1 locus in nuclei (Fig. 5B) may be due to the detachment from DNA. In this case, MBD2 protein is stably associated with chromatin in the methylated CpG island of the CDH1 gene even when the nuclei are isolated. Therefore, failure in detection of MBD2 protein binding by ChIP using cells (Fig. 4B) may be due to epitope masking by other components involved in the repressor complex. In any event ChIP analysis using both cells and nuclei will lead to a greater understanding of protein bindings in vivo.
Our studies demonstrated the unexpected observation that MBD2 binding was detected in SW1116 cells, although the CpG island was methylation-free (Fig. 2A). This binding can be attributed to the presence of an Alu sequence that resides in close proximity to the promoter region. It is highly probable that the majority of sheared DNA fragments that contain the promoter region of the CDH1 gene also contain an adjacent Alu sequence. As most Alu sequences are methylated, binding of MBDs to the Alu sequence is expected. Precipitation of Alu-containing fragments can be detected by CDH1 primers.
There are several different histone methyltransferases. It is not clear which enzyme mediates histone modification at a specific chromosomal position. It is reported that H3-K4 and H3-K9 methylation mediated by histone methyltransferases, SET7 and SUV39H1, respectively, inhibit each other (44). However, H3-K9 methylation by a different histone methyltransferase, SETDB1, is not affected by H3-K4 methylation (45). We found both H3-K4 and H3-K9 methylation in HSC41 and Li21 cells, but only H3-K4 or H3-K9 methylation was detected in SW1116 or T24 cells, respectively (Fig. 6). These results suggest the involvement of different histone methyltransferases in the modification of the CDH1-CpG island. These results also suggest that H3-K4 methylation is not simply associated with gene expression. Recently, it has been reported that methylation at both H3-K4 and H3-K9 disrupted the binding of the NuRD repressor complex (46). This phenomenon is in contrast to our finding that both H3-K4 and H3-K9 were methylated in some silenced cell lines, although binding of MBD2 protein, which forms a repressor complex with the NuRD complex, was not detected in Li21 cells.
DNA methylation is known to cause gene silencing and recent studies have shown that histone modification regulates DNA methylation. Mammalian DNA methyltransferase DNMT1 is associated with HDAC activity and DNA methylation is dependent on histone deacetylation (47–49). In Neurospora crassa and Arabidopsis thaliana, it is also reported that DNA methylation is controlled by histone methylation (50,51). These results suggest that altered chromatin structure regulates DNA methylation. Methylation status of cell clones differed even within the same cell line. The simple deficiency of maintenance of methylation may not be sufficient to explain this heterogeneity. Furthermore, the global methylation status within the CpG island is maintained. The most probable explanation for this intercellular heterogeneity is the difference in the propagation of inactive chromatin structure associated with histone modification and differences between individual cell clones.
The involvement of different MBPs, and therefore different repressor complexes, in the same CpG islands raises several problems. Do MBD1 or MBD2 contribute (and if so, equally) to silencing? What kinds of factors regulate their specific binding? Is binding responsible for silencing or a consequence of chromatin alteration? In any event the chromatin components associated with CpG islands of inactive genes are heterogeneous in nature and the answers to the above questions will require the further molecular dissection of molecules associated with these complexes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank A. Bird and T. Sugimura for critical review of the manuscript; M. Munakata, J. Yasuda, T. Ohta and M. Saito for technical advice and assistance. Some antibodies (antiserum S923, R593 and S751) were kindly provided by A. Bird. This work was supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports and Science, a Grant-in-Aid for Cancer Research from the Ministry of Health and Welfare of Japan (to M.S.), a Grant-in-Aid from the Ministry of Health and Welfare of Japan for the Second-Term Comprehensive 10-Year Strategy for Cancer Control (to M.S. and T.S.) and a Research Grant on Human Genome and Gene Therapy from the Ministry of Health and Welfare, Japan (to T.S.). S.K. was a recipient of Research Resident Fellowship from the Foundation for Promotion of Cancer Research, Japan.
REFERENCES
- 1.Yoder J.A., Walsh,C.P. and Bestor,T.H. (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet., 13, 335–340. [DOI] [PubMed] [Google Scholar]
- 2.Herman J.G. and Baylin,S.B. (2000) Promoter-region hypermethylation and gene silencing in human cancer. Curr. Top. Microbiol. Immunol., 249, 35–54. [DOI] [PubMed] [Google Scholar]
- 3.Tazi J. and Bird,A. (1990) Alternative chromatin structure at CpG islands. Cell, 60, 909–920. [DOI] [PubMed] [Google Scholar]
- 4.Jones P.L., Veenstra,G.J.C., Wade,P.A., Vermaak,D., Kass,S.U., Landsberger,N., Strouboulis,J. and Wolffe,A.P. (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet., 19, 187–191. [DOI] [PubMed] [Google Scholar]
- 5.Nan X., Ng,H.-H., Johnson,C.A., Laherty,C.D., Turner,B.M., Eisenman,R.N. and Bird,A. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- 6.Prokhortchouk A., Hendrich,B., Jørgensen,H., Ruzov,A., Wilm,M., Georgiev,G., Bird,A. and Prokhortchouk,E. (2001) The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev., 15, 1613–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaludov N.K. and Wolffe,A.P. (2000) MeCP2 driven transcriptional repression in vitro: selectivity for methylated DNA, action at a distance and contacts with the basal transcription machinery. Nucleic Acids Res., 28, 1921–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nan X., Campoy,F.J. and Bird,A. (1997) MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell, 88, 471–481. [DOI] [PubMed] [Google Scholar]
- 9.Ng H.-H., Zhang,Y., Hendrich,B., Johnson,C.A., Turner,B.M., Erdjument-Bromage,H., Tempst,P., Reinberg,D. and Bird,A. (1999) MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nature Genet., 23, 58–61. [DOI] [PubMed] [Google Scholar]
- 10.Ng H.-H., Jeppesen,P. and Bird,A. (2000) Active repression of methylated genes by the chromosomal protein MBD1. Mol. Cell. Biol., 20, 1394–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita N., Shimotake,N., Ohki,I., Chiba,T., Saya,H., Shirakawa,M. and Nakao,M. (2000) Mechanism of transcriptional regulation by methyl-CpG binding protein MBD1. Mol. Cell. Biol., 20, 5107–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu F., Thiesen,J. and Strätling,W.H. (2000) Histone deacetylase-independent transcriptional repression by methyl-CpG-binding protein 2. Nucleic Acids Res., 28, 2201–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boeke J., Ammerpohl,O., Kegel,S., Moehren,U. and Renkawitz,R. (2000) The minimal repression domain of MBD2b overlaps with the methyl-CpG-binding domain and binds directly to Sin3A. J. Biol. Chem., 275, 34963–34967. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y., Ng,H.-H., Erdjument-Bromage,H., Tempst,P., Bird,A. and Reinberg,D. (1999) Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev., 13, 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 16.Kouzarides T. (2002) Histone methylation in transcriptional control. Curr. Opin. Genet. Dev., 12, 198–209. [DOI] [PubMed] [Google Scholar]
- 17.Noma K.-I., Allis,C.D. and Grewal,S.I. S. (2001) Transitions in distinct histone H3 methylation patterns are the heterochromatin domain boundaries. Science, 293, 1150–1155. [DOI] [PubMed] [Google Scholar]
- 18.Litt M.D., Simpson,M., Gaszner,M., Allis,C.D. and Feisenfelt,G. (2001) Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science, 293, 2453–2455. [DOI] [PubMed] [Google Scholar]
- 19.Lachner M., O’Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- 20.Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromodomain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- 21.Hennig G., Behrens,J., Truss,M., Frisch,S., Reichmann,E. and Birchmeier,W. (1995) Progression of carcinoma cells is associated with alterations in chromatin structure and factor binding at the E-cadherin promoter in vivo. Oncogene, 11, 475–484. [PubMed] [Google Scholar]
- 22.Yoshiura K., Kanai,Y., Ochiai,A., Shimoyama,Y., Sugimura,T. and Hirohashi,S. (1995) Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc. Natl Acad. Sci. USA, 92, 7416–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graff J.R., Gabrielson,E., Fujii,H., Baylin,S.B. and Herman,J.G. (2000) Methylation patterns of the E-cadherin 5′ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J. Biol. Chem., 275, 2727–2732. [DOI] [PubMed] [Google Scholar]
- 24.Shiraishi M., Chuu,Y.H. and Sekiya,T. (1999) Isolation of DNA fragments associated with methylated CpG islands in human adenocarcinomas of the lung using a methylated DNA binding column and denaturing gradient gel electrophoresis. Proc. Natl Acad. Sci. USA, 96, 2913–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiraishi M., Sekiguchi,A., Chuu,Y.H. and Sekiya,T. (1999) Tight interaction between densely methylated DNA fragments and the methyl-CpG binding domain of the rat MeCP2 protein attached to a solid support. Biol. Chem., 380, 1127–1131. [DOI] [PubMed] [Google Scholar]
- 26.Frommer M., McDonald,L.E., Millar,D.S., Collis,C.M., Watt,F., Grigg,G.W., Molloy,P.L. and Paul,C.L. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA, 89, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coffee B., Zhang,F., Warren,S.T. and Reines,D. (1999) Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nature Genet., 22, 98–101. [DOI] [PubMed] [Google Scholar]
- 28.Cross S.H., Charlton,J.A., Nan,X. and Bird,A.P. (1994) Purification of CpG islands using a methylated DNA binding column. Nature Genet., 6, 236–244. [DOI] [PubMed] [Google Scholar]
- 29.Magdinier F. and Wolffe,A.P. (2001) Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc. Natl Acad. Sci. USA, 98, 4990–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark S.J., Harrison,J. and Frommer,M. (1995) CpNpG methylation in mammalian cells. Nature Genet., 10, 20–27. [DOI] [PubMed] [Google Scholar]
- 31.Gilbert S.L. and Sharp,P.A. (1999) Promoter-specific hypoacetylation of X-inactivated genes. Proc. Natl Acad. Sci. USA, 96, 13825–13830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen C.T., Gonzales,F.A. and Jones,P.A. (2001) Altered chromatin structure associated with methylation-induced gene silencing in cancer cells: correlation of accessibility, methylation, MeCP2 binding and acetylation. Nucleic Acids Res., 29, 4598–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antequera F., Boyes,J. and Bird,A. (1990) High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell, 62, 503–514. [DOI] [PubMed] [Google Scholar]
- 34.Cano A., Pérez-Moreno,M.A., Rodrigo,I., Locascio,A., Bianco,M.J., del Barrio,M.G., Portillo,F. and Nieto,M.A. (2000) The transcription factor Snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nature Cell Biol., 2, 76–83. [DOI] [PubMed] [Google Scholar]
- 35.Batlle E., Sancho,E., Franci,C., Dominguez,D., Monfar,M., Baulida,J. and de Herreros,A.G. (2000) The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nature Cell Biol., 2, 84–89. [DOI] [PubMed] [Google Scholar]
- 36.Graff J.R., Herman,J.G., Myöhänen,S., Baylin,S.B. and Vertino,P.M. (1997) Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J. Biol. Chem., 272, 22322–22329. [DOI] [PubMed] [Google Scholar]
- 37.Pao M.M., Tsutsumi,M., Liang,G., Uzvolgyi,E., Gonzales,F.A. and Jones,P.A. (2001) The endothelin receptor B (EDNRB) promoter displays heterogeneous, site specific methylation patterns in normal and tumor cells. Hum. Mol. Genet., 10, 903–910. [DOI] [PubMed] [Google Scholar]
- 38.Gregory R.I., Randall,T.E., Johnson,C.A., Khosla,S., Hatada,I., O’Neil,L.P., Turner,B.M. and Feil,R. (2001) DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs2. Mol. Cell. Biol., 21, 5426–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lorincz M., Schübeler,D. and Groudine,M. (2001) Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol. Cell. Biol., 21, 7913–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen R.Z., Akbarian,S., Tudor,M. and Jaenisch,R. (2001) Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nature Genet., 27, 327–331. [DOI] [PubMed] [Google Scholar]
- 41.Guy J., Hendrich,B., Holmes,M., Martin,J.E. and Bird,A. (2001) A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nature Genet., 27, 322–326. [DOI] [PubMed] [Google Scholar]
- 42.Amir R.E., Van den Veyver,I.B., Wan,M., Tran,C.Q., Francke,U. and Zoghbi,H.Y. (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genet., 23, 185–188. [DOI] [PubMed] [Google Scholar]
- 43.Pfeifer G.P. and Riggs,A.D. (1991) Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells using DNase I and ligation-mediated PCR. Genes Dev., 5, 1102–1113. [DOI] [PubMed] [Google Scholar]
- 44.Wang T., Cao,R., Xia,L., Erdjument-Bromage,H., Borchers,C., Tempst,P. and Zhang,Y. (2001) Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell, 8, 1207–1217. [DOI] [PubMed] [Google Scholar]
- 45.Schultz D.C., Ayyanathan,K., Negorev,D., Maul,G.G. and Rauscher,F.J.,III (2002) SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev., 16, 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zegerman P., Canas,B., Pappin,D. and Kouzarides,T. (2002) Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) complex. J. Biol. Chem., 277, 11621–11624. [DOI] [PubMed] [Google Scholar]
- 47.Fuks F., Burgers,W.A., Brehm,A., Hughes-Davies,L. and Kouzarides,T. (2000) DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nature Genet., 24, 88–91. [DOI] [PubMed] [Google Scholar]
- 48.Rounter M.R., Bachman,K.E. and Baylin,S.B. (2000) DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nature Genet., 25, 269–277. [DOI] [PubMed] [Google Scholar]
- 49.Robertson K.D., Ait-Si-Ali,S., Yokochi,T., Wade,P., Jones,P.L. and Wolffe,A. (2000) DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2-responsive promoters. Nature Genet., 25, 338–341. [DOI] [PubMed] [Google Scholar]
- 50.Tamaru H. and Selker,E.U. (2001) A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature, 414, 277–283. [DOI] [PubMed] [Google Scholar]
- 51.Jackson J.P., Lindroth,A.M., Cao,X. and Jacobsen,S.E. (2002) Control of CpNpG methylation by the KRYPTONITE histone H3 methyltransferase. Nature, 416, 556–560. [DOI] [PubMed] [Google Scholar]