Abstract
We report the synthesis of new phosphoramidite building blocks and their use for the modification of oligonucleotides with hydrazides. The reaction of these hydrazide oligonucleotides with active esters and aldehydes is demonstrated for solution conjugation and immobilization. Compared with the established amino modified oligonucleotides, hydrazides show enhanced reactivity at neutral and acidic buffer conditions. One method to introduce hydrazides is using amidites with preformed, protected hydrazides. A completely novel approach is the generation of the hydrazide functionality during the oligonucleotide cleavage and deprotection with hydrazine. Therefore, building blocks for the introduction of esters as hydrazide precursors are described. For the enhanced attachment on surfaces branched modifier amidites, which introduce up to four reactive groups to the oligonucleotide, are applied. The efficiency of branched hydrazide oligonucleotides compared with standard amino modified oligonucleotides for the immobilization of DNA on active electronic Nanogen chips is demonstrated.
INTRODUCTION
The modification of oligonucleotides and analogs with functional groups for reporting and immobilization are common operations in molecular biology and enable the realization of specific assays and applications. Amino modified oligonucleotides, which can be obtained by using amino modifier phosphoramidites in solid phase synthesis are, along with biotinylated and fluorophore labeled oligonucleotides, the most common modifications (1,2).
Amino modified oligonucleotides react with active esters to form stable amide bonds. They also react with aldehydes to produce Schiff’s bases, which, upon reduction, yield stable secondary amines. These two mechanisms are frequently applied for the immobilization of oligonucleotides on substrate surfaces (3,4).
For many specific applications the amino group has the limitation of being pH sensitive in its reaction with active esters. At low to neutral pH amines are protonated, greatly decreasing the reaction rate. At high pH values (8.5) the reaction of amines with active esters is fast, but the decay of the active ester is also accelerated with a half-life being typically in the range of several minutes to a few hours. Amino modified oligonucleotides show low reactivity with active esters under the immobilization conditions used on the Nanogen Nanochip® Molecular Biology Workstation, in which the electronic addressing conditions with positively biased pads generate a localized environment of increased acidity.
In our search for a covalent attachment chemistry as an alternative to the biotin/streptavidine system used on the commercially available Nanogen Nanochip® Molecular Biology Workstation, we developed hydrazide modified oligonucleotides and a series of phosphoramidite building blocks for their convenient preparation. In parallel, we developed a novel permeation layer which contains active esters designed to react rapidly and specifically with hydrazide modified oligonucleotides.
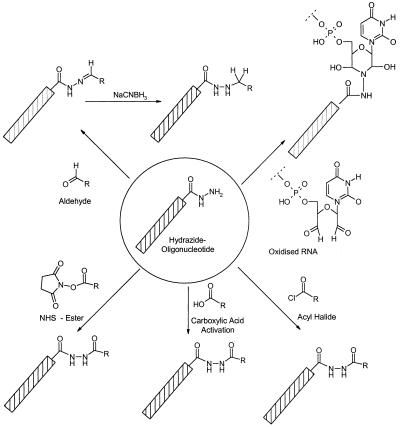
The reactivity of hydrazides to active esters is similar to that of amines (Fig. 1). With their lower pKa of typically 4–5 compared with the pKa of a primary amine of 10–11 they remain reactive at neutral to slightly acidic conditions. Further, hydrazides react with aldehydes to form hydrazones, which are less easily hydrolyzed than Schiff’s bases, thus allowing conjugation reactions without reduction of the products. Furthermore, hydrazone and Schiff’s base formation are ‘traceless’ conjugations, in which no side products or potentially toxic reagents are involved. Therefore, the hydrazone formation is preferred for the in situ conjugation of oligonucleotides (e.g. for those which are to be applied in cell culture experiments). Alternatively, a hydrazide can be used to conjugate a hydrazide modified oligonucleotide to an oligonucleotide whose 3′-terminal ribonucleotide is oxidized to a 1,5-dialdehyde. A stable dihydroxy-morpholino linkage thus is established (5).
Figure 1.
Overview about the reactions of hydrazide modified oligonucleotides.
Hydrazides are reactive functional groups routinely used in protein and carbohydrate chemistry (6). However, there are only a few examples in the literature which utilize hydrazide modified oligonucleotides (7,8). In all examples, the hydrazide is introduced post-synthetically. Only recently a dimethoxytrityl protected hydrazide amidite has been reported, in work carried out in parallel to our development (9). Whereas the utility of hydrazide modified oligonucleotides is evident from the advantages discussed above, the lack of convenient building blocks for their introduction into oligonucleotides has thus far impeded the broad use of this functional modification. Here we present an efficient synthetic approach to produce tailored hydrazide modified oligonucleotides for solution conjugation and surface attachment applications.
MATERIALS AND METHODS
General
Unless otherwise indicated, all reactions were magnetically stirred under a slow stream of dry argon. Reagents were obtained in analytical grade from Sigma-Aldrich Chemical Company (Deisenhofen, Germany) and solvents from Riedel unless otherwise indicated. Column chromatography is accomplished using silica gel 60 (Merck, 230–400 mesh). Melting points are uncorrected. Infrared (IR) spectra are measured on a Perkin Elmer Paragon 1000 FT-IR equipped with a Graseby Specac 10500 ATR unit. 1H-NMR spectra were recorded at 500 MHz; 13C spectra at 125 MHz and 31P at 202 MHz with a Bruker DMX 500 spectrometer. 1H and 13C are reported in units of δ using tetramethyl silane as an internal standard and coupling constants are reported in units of Hz. 31P chemical shifts are reported as δ units and referenced to an external standard of 85% phosphoric acid. ESI mass spectra are recorded on a Finnigan LCQ instrument in positive ionization mode. ‘Trace’ refers to a concentration of 0.1–0.05%.
Synthesis of phosphoramidite building blocks for the introduction of hydrazides into oligonucleotides
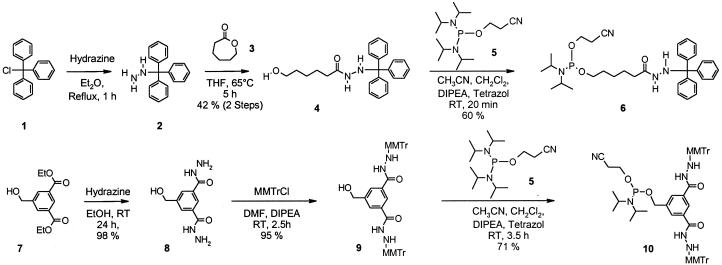
N-Tritylhydrazine hydrochloride (10). To a solution of 84 g (0.3 mol) of trityl chloride in 500 ml ether was slowly added 36 g (0.72 mol; 2.4 eq) of hydrazine hydrate under vigorous stirring. The mixture was heated to reflux for 1 h, cooled to room temperature and washed with 3× 200 ml water. The organic phase was dried over MgCl2, filtered, cooled to –10°C and 200 ml of 1.0 M solution of etheral HCl was added. The resultant precipitate was filtered, washed with 2× 200 ml ether and dried under vacuum to afford 82 g (88%) of N-tritylhydrazine hydrochloride as a colorless solid, which is used without further purification. C19H19ClN2; [310.6].
6-Hydroxycapronic acid N′-tritylhydrazide 4 (Fig. 2). To a solution of 75.0 g (0.24 mol) of tritylhydrazine hydrochloride in 750 ml of THF was added 26.7 g (0.26 mol, 1.1 eq) of triethylamine. The solution was stirred at room temperature for 15 min, filtered, concentrated, then treated with 27.4 g (0.24 mol, 1 eq) of ε-caprolactone. The mixture was heated to 65°C for 5 h and then allowed to stir overnight at room temperature. After reheating to 65°C ethyl acetate is added until a clear solution is formed. The mixture is allowed to cool to room temperature and then kept at 4°C overnight. The precipitate was filtered and recrystallized from ethyl acetate to afford 39.5 g (45%) of pale yellow solid 4. C25H28N2O2; [388.2]; Mp. 163–165°C; 1H-NMR (CDCl3): 7.49–7.47 (m, 5 H), 7.35–7.10 (m, 10 H), 6.55 (d, J = 7.52, 1 H), 5.55 (d, J = 7.25, 1 H), 3.54 (t, J = 6.45, 2 H), 1.87 (t, J = 7.25, 2 H), 1.62 (bs, 1 H), 1.57–1.34 (m, 4 H), 1.27–1.11 (m, 2 H). IR: 3234(w), 2924(w), 1616(s), 1557(m), 1490(m), 1446(s), 1225(w), 1054(m), 953(w), 909(w), 748(m), 696(s) cm–1; HRMS calculated for C31H44N3O2 [M + TEA]+: 490.34280; found: 490.34269 ± 1 p.p.m.
Figure 2.
Synthesis of phosphoramidites containing hydrazides protected with acid labile trityl and monomethoxytrityl groups.
6-[(2-Cyanethoxy)(diisopropylamino)phosphanyloxy]capronic acid N′-tritylhydrazide 6. To 150 ml acetonitrile, 30.2 ml (176 mmol, 3 eq) diisopropylethyl amine and 132 ml (59 mmol, 1.0 eq) 0.45 M tetrazol solution in anhydrous acetonitrile are added, then 23.2 g (1.47 eq, 87.0 mmol) bis-(diisopropylamino)-(2-cyanoethoxy)-phosphine 5 are added via a syringe over a period of 5 min. The resulting mixture is allowed to stir for 20 min thereby a little white precipitate is formed. The mixture is added dropwise over a period of 10 min to a solution of 23.0 g (59 mmol) 6-hydroxycapronic N′-tritylhydrazide 4 dissolved in 450 ml dry dichloromethane. After 20 min, TLC shows complete consumption of alcohol compound. The reaction mixture is poured onto 400 ml ice cooled phosphate buffer pH 6.5. The organic layer is washed twice with each 400 ml phosphate buffer, dried over anhydrous sodium sulfate, and concentrated in vacuo. The residue is dissolved in a little toluene and purified by column chromatography and eluted with a step gradient of 4.5 l toluene, then toluene/ethyl acetate 4:1, and finally with toluene/ethyl 2:1 acetate (all eluents containing 0.2% triethylamine). Product containing fractions are concentrated and dried in vacuo yielding 24.4 g (60%) of compound 6. C34H45N4O3P; [588.3]; Mp. 91–93°C; 1H-NMR (CDCl3): 7.49–7.46 (m, 5H), 7.34–7.20 (m, 10 H), 6.57 (d, J = 7.2, 1 H), 5.57 (d, J = 7.5, 1H), 3.85–3.74 (m, 2 H), 3.62–3.48 (m, 4 H), 2.62–2.59 (m, 2 H), 1.88–1.84 (m, 2 H), 1.53–1.33 (m, 4 H), 1.27–1.13 (m, 14 H); 31P-NMR (CDCl3): 147.97; IR: 3295(w), 2966(w), 1612(m), 1549(w), 1491(w), 1449(m), 1364(w), 1183(m), 1027(s), 974(s), 877(m), 746(s), 703(s) cm–1; HRMS calculated for C40H61N5O3P [M + TEA]+: 690.45066; found: 690.45073 ± 1 p.p.m.
5-(Hydroxymethyl)isophthalate dihydrazide 8. In a 500 ml round bottom flask, 15 g (59 mmol) diethyl 5-(hydroxymethyl) isophthalate are dissolved in 200 ml dry ethanol. After addition of 57.8 ml (1.12 mol, 19 eq) hydrazine hydrate at room temperature, the mixture is stirred overnight. Precipitated product is collected, washed with EtOH and dried overnight in vacuo. Yield: 13.1 g (98%) of 8 as a colorless solid. C9H12N4O3; [224.1]; Mp. 251°C (decomp); 1H-NMR (DMSO-d6,) 9.75 (bs, 2H), 8.11 (s, 1H), 7.89 (s, 2H), 5.36 (s, 1H), 4.57 (d, J = 3.0 Hz, 2H), 4.51 (bs, 4H); 13C-NMR (DMSO-d6) 165.6, 143.0, 133.4, 127.5, 124.1, 62.4; IR: 3293(m), 1672(sh), 1643(s), 1517(s), 1319(s), 1158(w), 1076(s), 1008(m), 884(m), 736(m), 688(s) cm–1.
5-Hydroxymethylisophthalic acid bis(N′-monomethoxytritylhydrazide) 9. In a 50 ml two-necked round bottom flask 2.0 g (8.9 mmol) 5-(hydroxymethyl)isophthalate dihydrazide 8 are suspended in 20 ml N,N-dimethylformamide (DMF) under argon. Addition of 6.92 g (53.5 mmol, 6 eq) diisopropylethyl amine followed by a portion wise addition of 6.25 g (19.6 mmol, 2.2 eq) 4-methoxytriphenylmethyl chloride and stirring for 1 h results in a clear brown solution. After additional 90 min the solvent is removed in vacuo. The residue is dissolved in 100 ml ethyl acetate and washed two times with water and with saturated sodium bicarbonate solution. The organic layer is dried with sodium sulfate and filtered. After adding of 20 g silica gel the solvent is evaporated. The residue is applied to a chromatography column and the product is eluted by gradient of heptane/ethyl acetate 1/1 to 1/3, containing a few drops of triethylamine. Product containing fractions are collected, concentrated and dried in vacuo. Yield: 6.52 g (95%) of a colorless powder. C49H44N4O5; [768.3] Mp. 127°C (decomp); 1H-NMR: (CDCl3): 7.50 (d, J = 7.7 Hz, 8H), 7.39–7.36 (m, 6H), 7.26–7.23 (m, 10H), 7.20–7.17 (m, 4H), 6.79 (d, J = 8.7 Hz, 4H), 5.79 (d, J = 7.7 Hz, 2H), 4.40 (s, 2H), 3.74 (s, 6H); IR: 3258(w), 1635(m), 1510(s), 1446(m), 1251(s), 1179(m), 1031(s), 750(m), 703(s) cm–1; HRMS calculated for C49H44N4NaO5 [M + Na]+: 791.32044; found: 791.32039 ± 1 p.p.m.
5-[(2-Cyanoethyl)(diisopropylamino)phosphanyloxymethyl]-isophthalic acid bis(N′-monomethoxytritylhydrazide) 10 (Fig. 2). A solution of 1.0 g (1.3 mmol) starting alcohol compound 9 in 15 ml dichloromethane, 1.5 ml DMF and a few drops of diisopropylethylamine is prepared. To this solution a mixture containing 0.65 ml (3.9 mmol) diisopropylethylamine, 2.9 ml (1.1 mmol) 0.45 M tetrazol solution in acetonitrile and 0.51 g (1.69 mmol) of bis-(diisopropylamino)- (2-cyanoethoxy)-phosphine 5 in acetonitrile is added. The mixture is stirred for 3.5 h at room temperature. Twenty grams of deacidified silica gel is added followed by solvent removal. Deacification is accomplished by suspending 20 g silica gel in 250 ml ethylacetate containing 1% triethylamine, stirring for 2 h at room temperature, filtration, and drying in vacuo. The residue is chromatographed with gradient elution (1/1 to 1/2 hexane/ethyl acetate with trace triethylamine) yielding 0.89 g (71%) of a colorless foam. C58H61N6O6P; [968.4]; 1H-NMR (CDCl3): 7.53–7.49 (m, 14H), 7.28–7.18 (m, 20H), 6.81 (d, J = 8.7 Hz, 4H), 5.81 (d, J = 8.1 Hz, 2H), 4.65–4.53 (m, 2H), 3.76 (m, 6H), 3.58 (m, 2H), 2.54 (t, J = 6.4 Hz, 2H), 1.18 (d, J = 6.7 Hz, 6H), 1.11 (d, J = 6.7 Hz, 6H), 31P-NMR (CDCl3): 149.6; IR: 2965(w), 1661(m), 1606(m), 1509(s), 1446(s), 1250(s), 1180(s), 1029(s), 977(m), 701(s) cm–1; HRMS calculated for C58H61N6NaO6P [M + Na]+: 991.42824; found: 991.42839 ± 1 p.p.m.
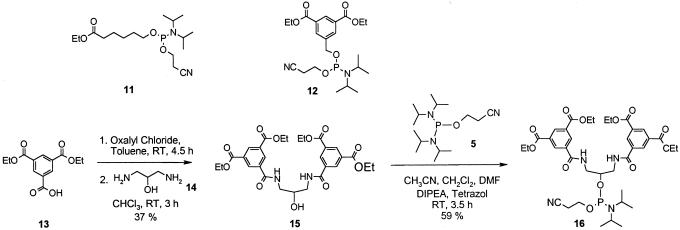
Ethyl 6-[(2-cyanethoxy)(diisporopylamino)phosphanyloxy]hexanoate 11 (Fig. 3). To a solution of 1.65 g (10 mmol) of ethyl 6-hydroxyhexanoate in 30 ml dichloromethane at room temperature are slowly added 5.17 g (40 mmol, 4 eq) of N-ethyldiisopropyl amine and 2.6 g (11 mmol, 1.1 eq) of 2-cyanoethyl N,N-diisopropyl-chloro-phosphoramidite over 15 min. Upon complete addition, the reaction is further stirred for 15 min, concentrated, and chromatographed (ethyl acetate/n-heptane 2/3 with 0.2% triethylamine) to afford 2.47 g (69%) of compound 11 as clear oil: C17H33N2O4P; [360.2]; 1H-NMR (CDCl3): 4.12 (q, J = 7.25, 2 H), 3.90–3.77 (m, 2 H), 3.75–3.55 (m, 4 H), 2.64 (t, J = 6.44, 2 H), 2.30 (t, J = 7.25, 2 H), 1.69–1.59 (m, 4 H), 1.44–1.34 (m, 2 H), 1.25 (t, J = 7.25, 3 H), 1.20–1.12 (m, 12 H); 31P-NMR (CDCl3): 148.01; IR: 2966(m), 2935(sh), 2363(m), 2335(sh), 1733(s), 1463(m), 1364(m), 1183(s), 1157(sh), 1027(s), 975(s), 894(m), 876(sh), 717(s) cm–1; HRMS calculated for C23H49N3O4P [M + TEA]+: 462.34552; found: 462.34505 ± 1 p.p.m.
Figure 3.
Synthesis of phosphoramidites containing esters as hydrazide precursors. 11 and 12 are prepared from the commercially available alcohols.
Diethyl 5-{[(2-cyanethoxy)(diisopropylamino)phosphanyloxy] methyl}isophthalate 12 (Fig. 3). 1.29 g (5 mmol) diethyl 5-(hydroxymethyl)isophthalate are dissolved in 20 ml dry dichloromethane at room temperature and mixed with 2.59 g (40 mmol, 4 eq) N-ethyldiisopropyl amine. 1.3 g (11 mmol, 1.1 eq) 2-cyanoethyl N,N-diisopropyl-chloro-phosphoramidite is added in 15 min with stirring. After 15 min at room temperature TLC ethyl acetate/n-heptane (1:4) shows complete reaction. The mixture is concentrated and mixed with 30 ml ethyl acetate/n-heptane (2:3). Precipitated hydrochloride is filtered; the filtrate is concentrated and directly applied to a chromatography column. Elution with ethyl acetate/n-heptane (1:4) containing a few drops triethylamine affords 1.6 g (70%) diethyl 5-{[(2-cyanethoxy)(diisopropylamino)phosphanyloxy] methyl}isophthalate 12 as a colorless oil. C22H33N2O6P; [453.2]; 1H-NMR (CDCl3): 8.59 (m, 1 H, arom.), 8.21 (m, 2 H, arom.), 4.87–4.75 (m, 2 H, CH2 cyanoethyl), 4.41 (q, J = 6.98 Hz, 4 H, CH2 ethyl), 3.95–3.80 (m, 2 H, 2× CH i-Pr), 3.74–3.61 (m, 2 H, CH2 cyanoethyl), 2.66 [t, J (P,H) = 6.45 Hz, 2 H, O-CH2-arom.], 1.41 (t, J = 6 H, 2× CH3 ethyl), 1.23–1.20 (m, 12 H, CH3, i-Pr); 31P-NMR (CDCl3): 149.94; 13C-NMR (CDCl3): 165.8 (C = O), 140.2 (C-CH2-O-P), 132.1 (2× C arom.), 131.1 (2× C-H arom.), 129.7 (C H arom.), 117.6 (CN), 64.7 (P-O-CH2-arom.), 61.4 (2× CH2 ethyl), 58.6 (O-CH2-CH2-CN), 43.4 (2× C-H i-Pr), 24.7 (4× CH3 i-Pr), 20.5 (O-CH2-CH2-CN), 14.4 (CH3 ethyl); IR: 2968(m), 1722(s), 1607(w), 1454(w), 1366(w), 1324(w), 1238(s), 1195(m), 1026(m), 978(w), 897(w), 754(m) cm–1; HRMS calculated for C28H49N3O6P [M + TEA]+: 554.33535; found: 554.33501 ± 1 p.p.m.
1,3-Bis-[(3,5-diethyloxycarbonyl)phenylcarbonylamido]-propan-2-ol 15. 9.0 g (34 mmol) diethyl-1,3,5-benzene tricarboxylate 13 are suspended in 100 ml toluene containing two drops of DMF. 5.6 g (3.8 ml, 44 mmol) oxalyl chloride is carefully added (strong gas evolution) over a period of 30 min. The stirred mixture becomes clear after 4 h. After 4.5 h the mixture is washed several times with water until neutral pH of the aqueous phase is achieved. The organic layer is dried over sodium sulfate, filtrated, concentrated and dried in vacuo to yield 9.6 g of crude acid chloride as a pale yellow oil.
1.43 g (15.7 mmol) 1,3-diamino-2-propanol 14 is dissolved in 20 ml chloroform. Five milliliters of DMF and 6.6 ml (47.7 mmol) triethylamine are added. To this mixture a solution of the crude acid chloride in 20 ml chloroform is added dropwise under argon and ice cooling over a period of 45 min. After removal of ice bath, the mixture is allowed to stir at room temperature for 2.5 h and then washed twice with water. The organic layer is dried over sodium sulfate, filtered and concentrated. The residue is mixed with 40 g of silica gel, evaporated to dryness and applied to a silica gel column chromatography. The product is eluted with gradient elution, starting with heptane/ethyl acetate (1/1) and ending with ethyl acetate. Product containing fractions are combined yielding after removal of solvents 4.2 g (37%) of a colorless powder 15: C29H34N2O11; [586.2]; Mp. 189–190°C; 1H-NMR (CDCl3): 8.71 (t, J = 1.6 Hz, 2H), 8.64 (d, J = 1.6 Hz, 4H), 7.78 (m, 2H), 4.58 (m, 1H); 4.39 (q, J = 7.0 Hz, 8H), 4.18 (m, 1H); 3.76–3.65 (m, 4H); 1.40 (t, J = 7.0 Hz, 12H); IR: 3328(w), 2990(w), 1720(s), 1648(s), 1548(sh), 1519(m), 1433(w), 1374(w), 1322(m), 1272(s), 1236(s), 1176(s), 1094(m), 1027(m) cm–1; HRMS calculated for C29H34N2NaO11 [M + Na]+: 609.20578; found: 609.20560 ± 1 p.p.m.
1,3-Bis-[(3′,5′-diethyloxycarbonyl)phenylcarbonylamido]-2-[(2”-cyanoethyloxy)(diisopropylamino)-phosphanyloxy]-propan 16 (Fig. 3). A mixture of 15.2 ml (6.82 mmol) 0.45 M tetrazol solution in acetonitrile, a solution of 2.65 g (3.57 ml, 20.5 mmol) diisopropylethylamine in 5 ml acetonitrile and 2.67 g (8.87) mmol bis-(diisopropylamino)-(2-cyanoethyloxy)phosphan 5 is stirred for 15 min. Within 5 min, this mixture is added to a suspension of 4.0 g (6.28 mmol) alcohol compound 15 in 30 ml dichloromethane and 5 ml DMF. The solution becomes clear within 20 min and is allowed to stir for 3 h. After adding 11 g silica gel the mixture is concentrated to dryness and applied to a silica gel chromatography column. The product is eluted with a gradient of heptane and ethyl acetate, ethyl acetate content ranging from 20 to 66%. Product containing fractions are combined and concentrated to dryness to yield 3.17 g (59%) as a pale yellow foam 16: C38H51N4O12P; [786.3]; 1H-NMR (CDCl3): 8.81 (m, 2H), 8.74 (d, J = 1.6 Hz, 2H), 8.73 (d, J = 1.6 Hz, 2H), 7.58 (m, 2H), 4.43 (q, J = 7.0 Hz, 8H), 4.23 (m, 1H), 3.99 (m, 3H), 3.89 (m, 1H), 3.66 (m, 2H), 3.51 (m, 1H), 3.33 (m, 1H), 2.69 (m, 2H), 1.34 (t, J = 7.0 Hz, 12H), 1.24 (d, J = 6.7 Hz, 6H), 1.21 (d, J = 6.7 Hz, 6H); 31P-NMR (CDCl3): 151.0; IR: 3328(w), 2968(w), 1721(s), 1652(m), 1538(m), 1445(w), 1366(m), 1235(s), 1178(m), 1023(s), 740(s), 715(sh) cm–1; HRMS calculated for C44H67N5O12P [M + TEA]+: 888.45184; found: 888.45149 ± 1 p.p.m.
Synthesis of hydrazide modified oligonucleotides
Oligonucleotides are synthesized using solid phase phosphoramidite chemistry on an automated oligonucleotide synthesizer. The phosphoramidite with the protected hydrazide or hydrazide precursor is applied as 0.1 M solution in acetonitrile and coupled at the desired position in the sequence using standard activation reagents and coupling times. Oligonucle otides containing more than four hydrazides were obtained by coupling branching phosphoramidites (11,12) followed by the coupling of the modifier building block. This approach enables the generation of optimized oligonucleotides for specific applications by choosing the best combination of branching and modifying building blocks. Further, it allows the generation of oligonucleotides with multiple reactive groups for surface immobilization. Depending on the nature of the hydrazide phosphoramidite used, two different deprotection and workup strategies were applied.
Deprotection and isolation of hydrazide oligonucleotides
Protocol I (for phosphoramidites containing protected hydrazides). The controlled pore glass (CPG) bound oligonucleotide (1 μmol) is placed in a 1.5 ml test tube and treated with 1.0 ml conc. aqueous NH4OH. After 2 h at 55°C the ammonia solution is removed and evaporated to dryness under reduced pressure. The residue is dissolved in 1 ml water and filtered through a 0.45 µm syringe filter. The trityl protected hydrazide oligonucleotide is purified by reverse phase HPLC using a Merck LiChrospher® RP 18, 10 µM, column (analytical: 4 × 250 mm, flow = 1.0 ml/min; preparative: 10 × 250, flow = 3.0 ml/min) using 0.1 M triethylammonium acetate (TEAA) pH 7.0 as buffer A and 95% acetonitrile in buffer A as buffer B. A gradient of 0% B to 100% B in 100 min is used for analytical and preparative separations. The fractions containing the trityl-on product were pooled and evaporated to dryness.
For the removal of the trityl protecting group the oligonucleotide is treated with 80% acetic acid for 30 min at room temperature. The acid is removed in vacuo, and the residue is dissolved in water then extracted twice with ethyl acetate. The aqueous layer is dried again and re-dissolved. Analytical HPLC usually shows a single product (in some cases as double peak), which can be employed for further reactions without purification. Alternatively HPLC purification can be performed using the solvent system described above.
Protocol II (in situ generation of hydrazides). The CPG bound oligonucleotide (1 µmol) is treated with a solution of 50 mg diethylamine in 3.5 ml dichloromethane for cyanoethyl cleavage. After incubation overnight (light exclusion) the supernatant is removed and the support bound oligonucleotide is washed several times with dichloromethane and dried in vacuo.
For the cleavage of the base protecting groups, the conversion of the esters to hydrazides, and the cleavage of the oligonucleotide from the support the CPG with the bound oligonucleotide, is treated with 1 ml 24% hydrazine hydrate. After 18 h under constant agitation at 4°C the reaction is complete. The isolation of the oligonucleotide from the hydrazine solution is achieved by reversed phase extraction. A C18 Sep-Pak® cartridge (0.5 g, Waters) is activated by rinsing with 10 ml acetonitrile and then 10 ml 0.1 M triethylammonium bicarbonate buffer pH 7.0 (TEAB). The hydrazine solution is diluted with the 5-fold volume of TEAB and applied to the cartridge. After binding of the oligonucleotide to the Sep-Pak column the residual hydrazine is washed away with 10 ml TEAB. The oligonucleotide is then eluted from the column with TEAB/acetonitrile (1:2). Oligo nucleotide containing fractions are pooled and evaporated to dryness. For the RP-HPLC characterization and purification of the product the same conditions as described in protocol I are applied.
PCR with hydrazide modified primers
For the amplification, 4 µl of a dNTP mix (2.5 mM; Promega), 2.5 µl of primer 1 (reverse) (10 µM), 2.5 µl of primer 2 (forward) (10 µM), 1 µl of Taq-polymerase (5 U/µl; Promega) and 2 µl of isolated cDNA template are added to 30 µl of DEPC water (Ambion), 5 µl of 10× thermophilic DNA-polybuffer (Promega), 3 µl of MgCl2 (25 mM; Promega). For the PCR, the 50 µl mixture is subjected to the following amplification cycle: pre-incubation at 95°C for 2 min; 35-cycle PCR at 94°C (15 s), 55°C (30 s), 72°C (30 s) and finally 72°C (7 min). After completion of the amplification, the reaction mixture is removed and stored at 4°C. For analysis of the amplicons, a sample (20 µl) is removed from the PCR mixture, mixed with 2 µl of 6× loading buffer (Sigma) and the bands are separated on a 1.7% agarose gel.
Immobilization of oligonucleotides on Nanogen Chips
The use of hydrazide modified oligonucleotides for immobilization on NHS ester containing surfaces will be described in detail elsewhere. Briefly, a microelectronic chip surface with active esters is obtained by the following co-polymerization procedure: to the center of the microarray 1.0 µl of a DMSO (Aldrich Chemical Company, Milwaukee, WI) solution containing N-acryloylsuccinimide (PolySciences, Warrington, PA) acrylamide (Polysciences) and N-methylenebisacrylamide (Polysciences) are added. Darocur™ 4265 (Ciba Speciality Chemicals, Tarrytown, NY) is used as a UV initiator. The chip is placed into a microreaction molding system and the solution is polymerized via UV radiation. The chip surface is rinsed with ethanol, dried and stored under nitrogen. Oligonucleotides (e.g. dT15 controls) labeled with Cy3™ at the 3′ end and bearing different hydrazide structures at the 5′ end (Table 1) are electronically addressed to the pads (100 nM, 2.2 V, 180 s). Fluorescence data is obtained by measuring green Cy3™ fluorescence (570 nm) at the Nanochip™ Molecular Biology Workstation.
Table 1. Sequence listing and analytical data for selected DNA hydrazide oligonucleotides.
| Oligo. no. | Amidite no. | Deprotection protocol | Oligonucleotide sequence (5′–3′) | HPLC retention time (method) | M calculated | M observed |
|---|---|---|---|---|---|---|
| 17 | 6 | I | (A) TTT TTT TTT TTT TTT Cy3 | 16.9 (c) | 5217 | 5215 |
| 18 | 10 | I | (B) TTT TTT TTT TTT TTT Cy3 | 16.8 (d) | 5295 | 5293 |
| 19 | 12 | II | (B) TTT TTT TTT TTT TTT Cy3 | 16.8 (d) | 5295 | 5293 |
| 20 | 12 | II | (C) TTT TTT TTT TTT TTT Cy3 | 16.6 (c) | 5733 | 5733 |
| 21 | 16 | II | (D) TTT TTT TTT TTT TTT Cy3 | 11.1 (d) | 5601 | 5599 |
| 22 | 6 | I | (A) GA TGA GCA GTT CTA CGT GG | 25.1 (a) | 6092 | 6092 |
| 23 | 12 | II | (B) GA TGA GCA GTT CTA CGT GG | 21.2 (a) | 6170 | 6170 |
| 24 | 16 | II | (D) GA TGA GCA GTT CTA CGT GG | – | 6476 | 6475 |
| 25 | 12 | II | (B) ACT ACA GTG ACG TGG ACA TC | – | 6414 | 6411 |
| 26 | 12 | II | (C) TAA TAC GAC TCA CTA TAG GGA GAA AAC CTT GTC ACT AGA TGC AAA GAC | – | – | – |
| 27 | 12 | II | (C) TAC ACC AAC AGA AAA GAT GAG TCC T | – | – | – |
| 28 | 12 | II | (C) ACA ACA ATT TGA AGC TTC TGT AAT TTT G Cy3 | 13.4 (c) | 9809 | 9807 |
| 29 | 16 | II | (D) ACA ACA ATT TGA AGC TTC TGT AAT TTT G Cy3 | 23.5 (b) | 9677 | 9674 |
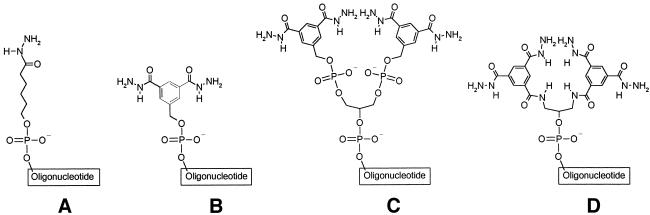
(A), (B), (C) and (D) refer to the hydrazide structure as given in Figure 6. HPLC methods: Merck LiChrospher RP 18, 10 µM, column, analytical: 4 × 250 mm, flow = 1.0 ml/min using 0.1 M TEAA pH 7.0 as buffer A and 95% acetonitrile in buffer A as buffer B. Gradients: a, 0% B to 100% B in 100 min; b, 10% B to 70% B in 60 min; c, 20% B to 100% B in 80 min; d, 25% B to 70% B in 45 min.
Conjugation of hydrazide oligonucleotides
Conjugation of a hydrazide oligonucleotide with a peptide aldehyde. (By hydrazone formation and subsequent reduction): 4.4 nmol monohydrazide oligonucleotide 22 is dissolved in 60 µl 10 mM ammonium acetate buffer (pH 4.0). Forty-four nanomoles (10 eq) antipain hydrochloride [C27H44N10O6·2 HCl; (677.6304); Calbio No. F 178220] in 15 µl buffer are added and agitated for 3 h at room temperature. The intermediate product is reduced with NaBH3CN (100 eq) for 1 h at room temperature. The product is isolated by HPLC (Column: Merck LiChrospher RP 18, 10 µM, 4 × 250 mm; buffer A = 0.1 M TEAA pH 7.0, buffer B = 75% acetonitrile in buffer A; flow = 1.0 ml/min; gradient: 10% B to 85% B in 60 min). The retention time of the product (oligonucleotide peptide conjugate) is 16.5 min, oligonucleotide 22 elutes at 13.9 min. MS (ESI): calculated: 6680.6; found: 6679.6).
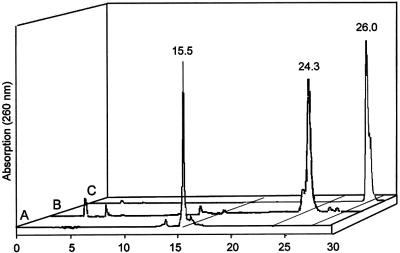
Conjugation of a hydrazide oligonucleotide with an aldehyde oligonucleotide. (Obtained by periodate oxidation of a 3′-terminal ribo U): the 3′-modified DNA oligonucleotide, 5′-dT15-rU-3′ is obtained from a commercial DNA oligo supplier (BioSpring, Frankfurt am Main, Germany; standard DNA synthesis on a ribo-U CPG support). An aliquot of this nucleic acid is dissolved in deionized water to obtain a 5 mM solution. Twenty microliters (100 nmol) of the solution are introduced into a 1.5 ml Eppendorf microreaction vessel. One microliter of a 0.1 M aqueous sodium periodate solution is added. The vessel is left at room temperature in the dark for 1 h. Excess periodate is removed by adding 1 µl of a 0.5 M sodium sulfite solution. After 15 min, 10 µl of a 1 M sodium phosphate buffer pH 7.4 are added and the resulting solution is transferred to a 1.5 ml Eppendorf microreaction vessel in which 8.4 nmol of a Mono-hydrazide modified Cy3™-labeled oligonucleotide, Hy-dT15-Cy3 17, had been evaporated to dryness beforehand. After mixing, the solution is left standing in the dark for 1 h. The product is purified by RP-HPLC (conditions protocol I, gradient 10% B to 60% B in 50 min). Figure 4 shows HPLC traces of the starting oligonucleotides and the isolated conjugate. Retention time: 5′-dT15-rU-3′: 15.5 min; hydrazide Hy-dT15-Cy3: 26.0 min; conjugate: 24.3 min. The conjugate is characterized by ESI MS: M calculated: 10001; M found: 10001.
Figure 4.
HPLC traces of a conjugation reaction. The oligonucleotide with a 3′-ribo uridine (A; 5′-dT15-rU-3′) is oxidized with periodate to a 1,5-bisaldehyde and reacted with a 5′-monohydrazide oligonucleotide (C; Hy-dT15-Cy3). The isolated conjugate is displayed in trace B.
RESULTS
Starting from commercial available building blocks the synthesis of phosphoramidites for the introduction of hydrazide functional groups was efficient and provided the modifiers in gram scale. For the conversion of the alcohols into phosphoramidites we used either bis-(diisopropylamino)-(2-cyanoethoxy)-phosphine 5 or the more reactive N,N-diisopropyl-chloro-phosphoramidite as phosphitylating reagent. Both reagents were applicable in all cases. We preferred the bis-(diisopropylamino) reagent for trityl or MMTr protected or complex structures 6, 10 and 16 and used the chloro-reagent only for the preparation of phosphoramidites 11 and 12.
Phosphoramidites 6 and 11 provide two straightforward modifiers for the preparation of monohydrazide oligonucleotides. These amidites, and the resultant monohydrazide oligonucleotides, are a direct substitute for the amino modifier C6 and amino modified oligonucleotides in many applications (1,2 and references therein).
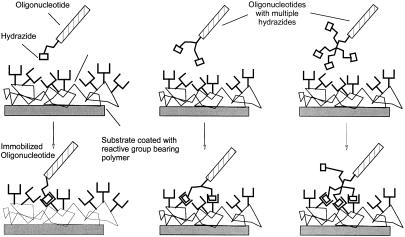
To synthesize oligonucleotides with multiple hydrazides (Fig. 5), we initially used commercially available branching phosphoramidites (11,12) followed by the coupling of phosphoramidite 6. This approach yielded good results in oligonucleotide synthesis (structure C in Fig. 6). As our assay and application development proved the utility of such constructs (data not shown), we decided to synthesize phosphoramidites for the direct introduction of several hydrazides, thus avoiding the use of the expensive branching amidites and enable introduction of multiple hydrazides in a single coupling step.
Figure 5.
Attachment of oligonucleotides with multiple hydrazides on chips surfaces: immobilization efficiency increases with the number of functional groups within the oligonucleotide.
Figure 6.
Structures of hydrazide oligonucleotides. Monohydrazide A is obtained from phosphoramidite 6 (deprotection protocol I) and 11 (deprotection protocol II); dihydrazide B is generated from phosphoramidite 10 (deprotection protocol I) and 12 (deprotection protocol II); bis-(dihydrazide) C from 12 in conjunction with branching amidite (12) (deprotection protocol II); tetrahydrazide D is obtained from amidite 16 (deprotection protocol II).
Mono- and di-hydrazide modified oligonucleotides (Fig. 6A and B) were easily obtained from the amidites 6 and 10 with trityl-on mode of standard solid phase synthesis and deprotection methods. RP-HPLC purification of the trityl-on products followed by acidic cleavage of the trityl group provided the desired oligonucleotide in high purity. Minor amounts of side products were observed during deprotection with aqueous ammonia, a result of the nucleophilic attack of the ammonia at the carbonyl of the hydrazide to produce an amide rather than a protected hydrazide. This side product formation was dependent on the time of the ammonia deprotection and never exceeded 15% under the conditions of protocol I. Oligonucleotide yields were typically in the experimentally acceptable range of 50%, therefore optimization to suppress side product formation was not undertaken. For the mono-hydrazide phosphoramidite 6 we used the inexpensive trityl protecting group instead of the DMTr group used by Kurz et al. (9) or the MMTr used for the commercially available amino modifier C6. For the synthesis of the intermediate tritylhydrazine and its purification as hydrochloride we could adapt the procedure published by Wygand and Steglich (10). The cleavage of this group with acetic acid was complete after 30 min at room temperature, so that we were fully satisfied with the performance of this protecting group for the synthesis of monohydrazides. For the dihydrazide building block 10 we compared the performance of the trityl, the MMTr and the DMTr protecting group in initial experiments. The stability towards acidic cleavage increased, as expected, from DMTr to MMTr to trityl. Conversely, the amide formation due to nucleophilic attack of ammonia at the hydrazide carbonyl increased from Trityl over MMTr to DMTr. We found MMtr as optimal protecting group for this specific phosphoramidite structure.
For amidites containing protected hydrazides the number of hydrazides introduced to a single oligonucleotide is limited. Building blocks containing four trityl protected hydrazides were prepared (data not shown). Unfortunately, neither the oligonucleotides obtained by these amidites nor oligonucleotides prepared by using branching amidites together with 10 could be isolated by RP-HPLC presumably due to the high number of nonpolar trityl groups.
To further develop and introduce multiple hydrazide moieties into a single oligonucleotide, we have developed phosphoramidites containing esters as hydrazide precursors (Fig. 3). Upon completion of the oligonucleotide synthesis in which ester phosphoramidites are employed, the product is treated with hydrazine. The use of hydrazine as a deprotecting agent has been described previously (13); additionally it converts the esters into hydrazides. Hydrazine deprotection is also a standard procedure for the syntheses of pyranosyl RNA in our laboratories (14). This in situ generation of hydrazides during the deprotection of oligonucleotides with hydrazine is an approach of specific utility for oligonucleotides with multiple hydrazides. Highly modified oligonucleotides are obtained by using branching amidites in conjunction with amidites 12 or 16. For practical considerations we here limit our samples to structures containing four hydrazides, although samples with more hydrazides were obtained (data not shown). RP-HPLC purification is hampered due to the lack of a hydrophobic trityl protecting group in the product. We observed only minor amounts (<5% determined by mass spectroscopy) of transamination with cytosine, which did not interfere with the applicability of the oligonucleotides in our assays. Mono-, di-, bis-(di)- and tetra-hydrazide oligonucleotides were obtained from amidites 11, 12 and 16 in typical overall synthesis yields ranging from 30 to 50%. Purity and structure were confirmed by ESI-MS and in comparison with the oligonucleotides obtained from protected hydrazide building blocks.
Both approaches yielded oligonucleotides modified with one to four hydrazides in good quality and amounts adequate to investigate the application in conjugation and immobilization reactions.
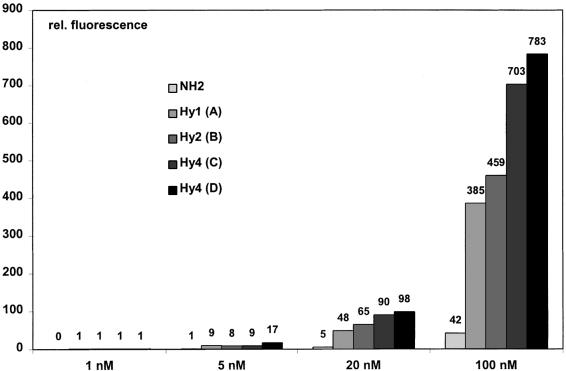
For the immobilization of oligonucleotides on active electronic chips we used 3′ Cy3™ fluorescently labeled oligonucleotides with hydrazide modification at the 5′-end. Mono-, di,- bis-(di)- and tetra-hydrazides (Fig. 6) were compared with an oligonucleotide with a 5′-amino modifier C6. In Figure 7 the fluorescence determined after immobilization of the samples to the surface is plotted for four different concentrations as a measure for the amount of immobilized oligonucleotide. The results are uncorrected for the gradual decomposition of the active ester in the aqueous buffer, which starts with the application of an aqueous solution to the chips surface. At 100 nM the first immobilized amino modified oligonucleotide, where the active ester concentration is the highest, shows the lowest immobilization efficiency being only 11% of the mono-hydrazide oligonucleotide signal as direct counterpart. Even without compensation for the effect of the active ester decomposition, the di-, bis-(di)- and tetra-hydrazides give the highest surface immobilization. Repeated immobilization experiments at different oligonucleotide concentrations between 1 and 100 nM confirm the relative immobilization efficiency observed at 100 nM with continuously decreasing absolute fluorescence.
Figure 7.
Immobilization of amino modified and hydrazide oligonucleotides on Nanogen chips. Cy3™ labeled hydrazide oligonucleotides were addressed at four different concentrations (1, 5, 20 and 100 nM). The fluorescence measured after washing is plotted as a measure for the amount of immobilized oligonucleotide. The amount of immobilized oligonucleotide increases with increasing oligonucleotide concentration. At 100 and 20 nM, monohydrazides (A; see Fig. 6) show nine times more efficient immobilization than amino modified (NH2) oligonucleotides. The quantity of immobilized oligonucleotide can be increased further by introducing two (B) and four (C and D) hydrazide moieties to the oligonucleotide.
The stability of the immobilization by the hydrazide/NHS attachment method was assayed by submitting the covalently attached labeled oligonucleotide 20 to a series of conditions. Fluorescence was measured before and after the treatment. A 48 h wash with high salt buffer (50 mM sodium phosphate at pH 7 with 500 mM NaCl), heating to 60°C for 5 min, and applying a negative bias of 1.2 V for 2 min resulted in each case only in a 2% loss of the fluorescence signal. Finally, the chip containing the bound Cy3™ labeled oligonucleotide was washed with 0.2 M NaOH for 15 min. Under these conditions, a 10% loss of fluorescence signal was measured. Further analysis under solution phase conditions indicated that the Cy3™ labeled oligonucleotide experiences a 10% loss of signal when exposed to 0.2 M NaOH for 10 min then neutralized. The 10% loss of fluorescence observed on the chip can therefore be attributed to the instability of the dye and not to the loss of the oligomer from the surface.
To ensure the function of hydrazide oligonucleotides as a substitute for biotinylated and amino modified oligonucleotides, we tested their performance as PCR primers. No difference in amplicon formation was observed in comparison with unmodified primers (see Supplementary Material).
The practical applicability of hydrazides as covalent attachment system for the immobilization of amplicons on the Nanogen Nanochip® Molecular Biology Workstation was demonstrated with the factor V Leiden SNP assay (data to be published elsewhere).
Another attractive application of hydrazide oligonucleotides is their use for conjugation reactions. As an example we tested the reaction of a hydrazide modified oligonucleotide with the peptide aldehyde antipain, with subsequent reduction of the product with NaBH3CN. We could detect the conjugation product by HPLC (Supplementary Material) and characterize the isolated conjugate by mass spectroscopy. Equally a 1,5-bisaldehydo oligonucleotide, obtained by the oxidation of a terminal ribonucleotide with periodate, was conjugated with the monohydrazide oligonucleotide 17. As shown in Figure 4 a conjugate is readily formed, and subsequently isolated and characterized by HPLC and mass spectroscopy.
DISCUSSION
For the introduction of hydrazides two synthetic routes have been exploited. First, phosphoramidites of mono- and di-hydrazides (6 and 10) protected with an acid labile protecting group were prepared. Figure 2 summarizes the synthesis of these building blocks. They can be employed in the standard oligonucleotide synthesis process and yield, upon ammonia deprotection, oligonucleotides which can be purified by reversed phase HPLC. The protecting group on the hydrazide is then cleaved with dilute acetic acid giving a hydrazide oligonucleotide. Because of the application of trityl-on mode of standard oligonucleotide deprotection conditions, this is the easiest route to access hydrazide modified oligonucleotides.
In a second approach the reactive hydrazide is generated in situ during the oligonucleotide deprotection with hydrazine. To our knowledge this is the first time that a desired functionalization of an oligonucleotide is realized by the deprotecting reagent itself during the deprotection and cleavage of the oligonucleotide from the support, whereas the post-synthetic functionalization of H-phosphonate oligonucleotides has been achieved previously with the oxidation reagent as described by Tang and Agrawal (15,16). As a precursor for the hydrazide we used esters. The phosphoramidites 11, 12 and 16 are available by very short synthesis routes from commercially available hydroxyl containing esters. Using this approach it is possible to introduce four, and potentially even more, hydrazides into an oligonucleotide by a single coupling of a modifier phosphoramidite.
In summary, an efficient synthetic path to hydrazide modified oligonucleotides has been established which enables the supply of these materials for our internal and external applications.
The advantages of hydrazide oligonucleotides in various conjugation reactions, in comparison with established amino modification, have to be further assessed. As an example, we demonstrated here the conjugation of a hydrazide oligonucleotide with a second oligonucleotide and a peptide. Both reactions succeeded at the first attempt, indicating the robustness of the chemistry involved.
Our focus was the use of hydrazide oligonucleotides for the attachment of capture sequences and amplified DNA on active electronic Nanochips®. For the immobilization of oligonucleotides on substrate surfaces an efficient and fast reaction is desired. We can significantly increase the immobilization efficiency by attaching more than one hydrazide reactive group to an oligonucleotide (Fig. 5). Initially we used the branching phosphoramidites developed by Horn et al. (11) and Shchepinov et al. (12) followed by the reaction of hydrazide phosphoramidites to generate oligonucleotides with more than one reactive group. For a convenient modification of the oligonucleotide, a single coupling step of a modifier building block is preferred. For that purpose we synthesized and tested synthons capable of introducing two and four hydrazides at once. These are compared with monohydrazides for immobilization on active Nanochips®. The tetrahydrazides obtained by amidite 16 (Fig. 6, structure D) were even more efficient than the bis-(dihydrazides) generated with branching amidites (structure C). On the chip even the monohydrazides showed a 9-fold higher immobilization efficiency than the amino modified reference oligonucleotide.
The set of hydrazide phosphoramidites presented here provides a robust method of obtaining hydrazide modified oligonucleotides. It allows a tuning of the attachment chemistry for the specific conditions of the assay by choosing the hydrazide structure with the optimal immobilization efficacy. This flexibility allows integration with the tests and application developments on the existing Nanochips® Molecular Biology Workstation.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
Norbert Windhab and Christoph Brücher are gratefully acknowledged for their support and fruitful discussions. Marc Pignot is acknowledged for performing PCR with hydrazide primers.
REFERENCES
- 1.Beaucage S.L. and Iyer,R.P. (1993) The functionalization of oligonucleotides via phosphoramidite derivatives. Tetrahedron, 49, 1925–1963. [Google Scholar]
- 2.Eckstein F. (1991) Oligonucleotides and Analogues. A Practical Approach. IRL Press, Oxford.
- 3.Schena M. (2000) Microarray Biochip Technology. Eaton Publishing, Natick, MA.
- 4.Beier M. and Hoheisel,J.D. (1999) Versatile derivatisation of solid support media for covalent bonding on DNA-microchips. Nucleic Acids Res., 27, 1970–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timofeev E.N., Kochetkova,S.V., Mirzabekov,A.D. and Florentiev,V.L. (1996) Regioselective immobilization of short oligonucleotides to acrylic copolymer gels. Nucleic Acids Res., 24, 3142–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermanson G.T. (1996) Bioconjugate Techniques. Academic Press, San Diego, CA.
- 7.Grimm G.N., Boutorine,A.S. and Hélène,C. (2000) Rapid routes of synthesis of oligonucleotide conjugates from non-protected oligonucleotides and ligands possessing different nucleophilic or electrophilic functional groups. Nucleosides Nucleotides Nucleic Acids, 19, 1943–1965. [DOI] [PubMed] [Google Scholar]
- 8.Ghosh S.S., Kao,P.M. and Kwoh,D.Y. (1989) Synthesis of 5′ oligonucleotide hydrazide derivatives and their use in preparation of enzyme-nucleic acid hybridization probes. Anal. Biochem., 178, 43–51. [DOI] [PubMed] [Google Scholar]
- 9.Kurz M., Lohse,P. and Wagner,R. (2001) Peptide acceptor ligation methods. Patent Application WO 0107657.
- 10.Wygand F. and Steglich,W. (1959) Peptidsynthesen mit N-TFA-Aminosäure- bzw. N-TFA-Peptid-tritylhydraziden. Chem. Ber., 92, 313–319. [Google Scholar]
- 11.Horn T., Warner,B.D., Running,J.A., Downing,K., Clyne,J. and Urdea,M.S. (1989) The synthesis of branched oligonucleotides as signal amplification multimers for use in nucleic acid assays. Nucleosides Nucleotides, 8, 875–877. [Google Scholar]
- 12.Shchepinov M.S., Udalova,I.A., Bridgman,A.J. and Southern,E.M. (1997) Oligonucleotide dendrimers: synthesis and use as polylabelled DNA probes. Nucleic Acids Res., 25, 4447–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polushin N.N., Morocho.A.M., Chen,B.-C. and Cohen,J.S. (1994) On the rapid deprotection of synthetic oligonucleotides and analogs. Nucleic Acids Res., 22, 639–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamon C., Brandstetter,T. and Windhab,N. (1999) Pyranosyl-RNA supramolecules containing non-hydrogen-bonding base-pairs. Synlett, 940–944. [Google Scholar]
- 15.Tang J.Y. and Agrawal,S. (1990) Incorporation of multiple reporter groups on synthetic oligonucleotides. Nucleic Acids Res., 18, 6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agrawal S. and Tang,J.-Y. (1990) Site specific functionalization of oligodeoxynucleotides for non-radioactive labelling. Tetra. Lett., 31, 1543–1546. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.