Abstract
Delta ribozyme possesses several unique features related to the fact that it is the only catalytic RNA known to be naturally active in human cells. This makes it attractive as a therapeutic tool for the inactivation of clinically relevant RNAs. However, several hurdles must be overcome prior to the development of useful gene-inactivation systems based on delta ribozyme. We have developed three procedures for the selection of potential delta ribozyme target sites within the hepatitis B virus (HBV) pregenome: (i) the use of bioinformatic tools coupled to biochemical assays; (ii) RNase H hydrolysis with a pool of oligonucleotides; and (iii) cleavage assays with a pool of ribozymes. The results obtained with delta ribozyme show that these procedures are governed by several rules, some of which are different from those both for other catalytic RNAs and antisense oligonucleotides. Together, these procedures identified 12 sites in the HBV pregenome that can be cleaved by delta ribozymes, although with different efficiencies. Clearly, both target site accessibility and the ability to form an active ribozyme–substrate complex constitute interdependent factors that can best be addressed using a combinatorial library of either oligonucleotides or ribozymes.
INTRODUCTION
The goal of gene therapy is to modulate the expression of a specific gene. The ability of ribozymes (i.e. RNA enzymes) to specifically recognize an RNA substrate, and subsequently catalyze its cleavage, makes them attractive as therapeutic tools for the inactivation of both viral RNAs and the mRNAs associated with various diseases (1–3). Several successful applicable ribozyme models have been tested both in vitro and in a cellular environment (1–3).
Delta ribozyme possesses several unique features that are all related to the fact that it is the only naturally occurring catalytic RNA that has been discovered in humans (4,5). In contrast to other catalytic RNAs, several hurdles remain to be surpassed before we generate a useful gene-inactivation system based on delta ribozyme. The initial step in the development of a ribozyme capable of cleaving a natural RNA molecule is the selection of the cleavage site with the greatest potential for targeting. The specificity of recognition is derived from Watson–Crick base pairs formed between the substrate and the ribozyme. For example, we have engineered a 57 nt delta ribozyme derived from the antigenomic strand of the hepatitis delta virus (HDV) (Fig. 1A) (6,7). According to its double-pseudoknot secondary structure, which is well supported by experimental data, the substrate specificity depends on the formation of the P1 stem (i.e. the sequence binding domain of the ribozyme), which includes one GU wobble base pair followed by six non-specific Watson–Crick base pairs (Fig. 1A) (4). Secondary and tertiary structures of a target that are characterized by significant intramolecular base pairing are important features, which can influence the cleavage activity of ribozymes (8–10). Thus, even though the appropriate target site is present, effective cleavage at that site depends on its accessibility to the ribozyme. Target sequences located in single-stranded regions of an mRNA have a higher potential because they should be more accessible to ribozyme binding than those in double-stranded regions. Within the double-stranded regions the ribozyme might compete unfavorably with intramolecular base pairing when trying to bind its substrate (8–10). The development of several procedures identifying potential target sites in RNA molecules has been reported for various nucleic acid drugs, including other ribozymes (11–22). Based on these reports, we present three procedures developed in order to identify the sites with the greatest potential for cleavage by delta ribozyme in a model RNA. These procedures include the use of: (i) bioinformatic tools coupled to biochemical assays (13,15,16,22); (ii) ribonuclease H (RNase H) mapping in the presence of a library of DNA oligonucleotides (11,20); and (iii) cleavage assays using a library of ribozymes (12,14).
Figure 1.
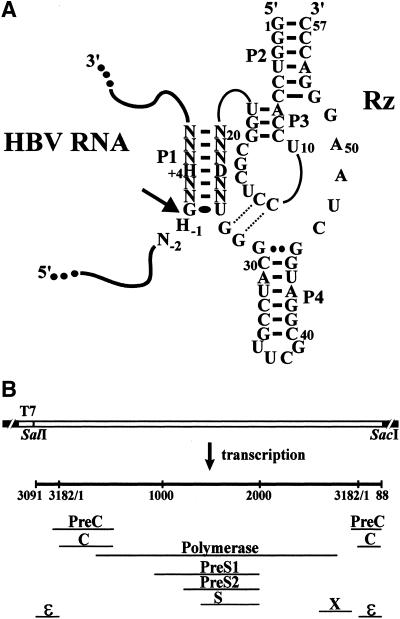
Illustration of delta ribozyme and HBV RNA pregenome. (A) Secondary structure of the engineered trans-acting delta ribozyme bound to the HBV target. The pseudoknot P1.1 is illustrated by the dotted lines. The homopurine base pair at the top of the P4 stem is represented by two large dots, while the wobble base pair is represented by a single large dot. Adjacent and in the P1 stem only the identity of the nucleotides essential for cleavage to occur are shown: H indicates A, C or U; D indicates A, G or U; and N indicates A, C, G, U. The cleavage site is indicated by an arrow. (B) Organization of the pHBVT7 vector. The four overlapping open reading frames are shown (C, P, S and X).
The pregenome RNA from the hepatitis B virus (HBV) was chosen as model target because it offers several advantages. We selected an RNA virus, instead of an mRNA, as our target because the use of an mRNA would have limited our search for potential cleavage sites to those near the 5′-end in order to ensure that cleavage resulted in the production of an inactive protein. HBV has an extremely compact organization as all nucleotides have coding function (Fig. 1B) (23). The 3.2 kb DNA genome replicates via reverse transcription involving an RNA pregenome of ∼3.4 kb whose secondary structure is unknown. Moreover, HBV is a model target that has clinical relevance as 350 million people worldwide are chronic carriers (24,25).
MATERIALS AND METHODS
Bioinformatic analysis of HBV RNA
The sequence of the HBV variant used corresponded to the insert of the plasmid pCHT-9/3091, which was kindly provided by Dr M. Nassal (26). The most stable secondary structures, in terms of energy, of this HBV variant were predicted using the program RNA Structure 3.5 (http://rna. chem.rochester.edu) (27). The resulting file was then analyzed for the probability of binding to complementary RNA oligonucleotides 7 nt long (which corresponds to the size of the P1 stem formed by a substrate and a delta ribozyme) using the software OligoWalk (13).
Eleven HBV sequences from various genotypes [DDBJ/EMBL/GenBank accession numbers: A1 (X70185, X80924, X97848, X97849, X97850, X97851 and Z35717), B1 (D00329 and D00330), C1 (X75665) and C2 (L08805)] were retrieved from the database, aligned with the HBV sequence present in plasmid pCHT-9/3091 (26) using the ClustalW package (28), and minor adjustments introduced manually (http://penelope.med.usherb.ca/labojp/pdf/hbv.pdf). This sequence alignment was used to identify potential target sites conserved in most of the genotypes. Other criteria, including substrate specificity for delta ribozyme cleavage, were also used in order to identify the sites with the greatest potential (see Results).
HBV and ribozyme DNA constructs
The plasmid pCHT-9/3091 contains a full-length copy of the HBV pregenome (26). The HBV pregenome insert was subcloned downstream of the T7 RNA promoter within the vector pBlueScript SK (+/–) (Stratagene) using the SalI and SacI restriction sites (Fig. 1B). The resulting plasmid was named pHBVT7, and its identity was confirmed by DNA sequencing.
The construction of all ribozymes was performed as described previously (10). Briefly, pairs of complementary and overlapping DNA oligonucleotides, which corresponded to the P1 stem region, were synthesized (Invitrogen), annealed and ligated to PstI and SphI co-digested pδRzP1.1, yielding a plasmid harboring a delta ribozyme referred to as pδRz-HBVX, where X corresponds to the position of its potential cleavage site within HBV. The sequences of all ribozyme minigenes were confirmed by DNA sequencing.
RNA synthesis
Ribozymes and HBV RNA were transcribed in vitro using SmaI-linearized pδRz-HBVX and pHBVT7, respectively, as templates. Run-off transcriptions were performed in the presence of purified T7 RNA polymerase (10 µg), RNAguard (24 U, Amersham Biosciences), pyrophosphatase (0.01 U, Roche Diagnostics) and linearized plasmid DNA (5 µg) in a buffer containing 80 mM HEPES–KOH pH 7.5, 24 mM MgCl2, 2 mM spermidine, 40 mM DTT, 5 mM of each NTP and with or without 50 µCi [α-32P]GTP (New England Nuclear) in a final volume of 100 µl at 37°C for 4 h. Upon completion, the reaction mixtures were treated with DNase RQ1 (Amersham Biosciences) at 37°C for 20 min, purified by phenol:chloroform extraction and the nucleic acid precipitated with ethanol. The HBV RNA products and ribozymes were respectively fractionated by denaturing 5 and 7.5% polyacrylamide gel electrophoresis (PAGE; 19:1 ratio of acrylamide to bisacrylamide) in buffer containing 45 mM Tris– borate pH 7.5, 7 M urea and 1 mM EDTA. The reaction products were visualized either by UV shadowing or autoradiography. The bands corresponding to the correct sizes for the ribozymes and HBV RNAs were cut out and the transcripts eluted overnight either at room temperature (ribozymes) or at 4°C (HBV RNA) in a solution containing 0.5 M ammonium acetate and 0.1% SDS. The transcripts were desalted on Sephadex G-25 (Amersham Biosciences) spun-columns, and were then precipitated by the addition of 0.1 vol 3 M sodium acetate pH 5.2 and 2.5 vol ethanol, washed, dried, resuspended in ultrapure water and the quantity determined by either absorbance at 260 nm or 32P counting.
Synthesis of a combinatorial library of ribozymes
The library of delta ribozymes with a random P1 stem was constructed using a PCR-based strategy (see inset in Fig. 4). Three DNA oligonucleotides were used: (i) the antisense oligonucleotides (Rz-pool; 5′-GGGTCCCTTAGCCATCCGCGAACGGATGCCCANNNNNNACCGCGAGGAGGTGGACCC-3′, where N = A, C, G or T) served as the primary templates for the library of ribozymes; (ii) the sense primer (T7-5′Rz; 5′-TTAATACGACTCACTATAGGGTCCACCTCCTCGCGGT-3′) permitted the incorporation of the T7 RNA promoter upstream of the minigene; and (iii) the antisense primer (3′Rz; 5′-GGGTCCCTTAGCCATCCGCGAACGG-3′) amplified the template. The PCR mixtures contained 20 mM Tris–HCl pH 8.8, 6 mM MgCl2, 10 mM KCl, 10 mM (NH4)2SO4, 0.1% Triton X-100, 0.2 mM of each dNTP, 2 µM of each primer and 1 U Vent DNA polymerase (New England Biolabs) in a final volume of 100 µl. After an annealing step of 1 min at 94°C, 20 cycles of 1 min at 94°C, 1 min at 50°C and 1 min at 76°C were performed. The PCR products were purified by phenol:chloroform extraction, the nucleic acid precipitated with ethanol, resuspended in water, and then in vitro transcriptions and purifications of the ribozymes were performed as described above.
Figure 4.
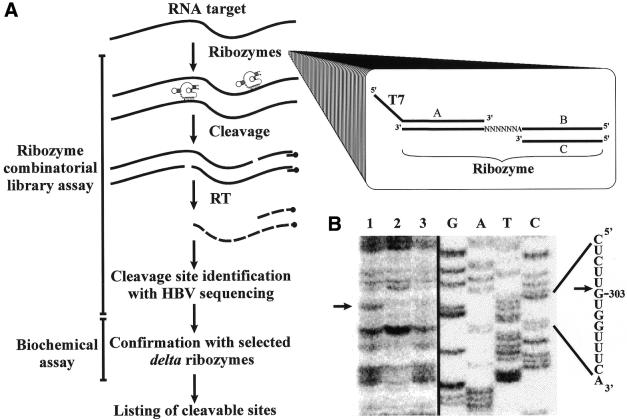
Selection of the sites with the greatest potential for delta ribozyme cleavage sites based on a ribozyme cleavage assay performed in the presence of a ribozyme library. (A) Scheme illustrating the procedure. HBV RNA and cDNA are represented by the full and dashed lines, respectively. RT is reverse transcriptase. The PCR strategy used to synthesize the library is shown in the inset in which oligonucleotides A and C served as sense and antisense primers, respectively, to amplify the randomized sequences of the templates (oligonucleotide B). (B) Autoradiogram of a sequencing gel revealing the cleavage site at position 303. Lane 1 is the reaction performed in the presence of ribozymes and MgCl2, while MgCl2 and the ribozyme were omitted in the reactions in lanes 2 and 3, respectively. Adjacent to the gel is a DNA sequencing reaction, and the corresponding sequence is shown on the right side. The arrow indicates the cleavage site.
RNase H hydrolysis
RNase H reactions with 32P randomly labeled HBV RNA were performed as a biochemical assay coupled with the data obtained from the bioinformatic tools. HBV RNA (∼0.1 µM, ∼20 000 c.p.m.) and DNA oligonucleotides (7 nt, 5 µM) were preincubated for 10 min at 25°C in a final volume of 8 µl containing 20 mM Tris–HCl pH 7.5, 20 mM KCl, 10 mM MgCl2, 0.1 mM EDTA and 0.1 mM DTT. RNase H (0.5 U; United States Biochemicals) was then added (2 µl) and the samples incubated at 37°C for 30 min. The reactions were quenched by adding 3 µl loading buffer (97% formamide, 0.025% xylene cyanol and 0.025% bromophenol blue) and the mixtures fractionated through denaturing 5% PAGE gels (19:1), which were analyzed with a PhosphorImager (Molecular Dynamics).
RNase H reactions were also performed with randomized oligonucleotides (8 nt, 5 µM) and non-radioactive HBV RNA (0.5 µM) under the conditions described above. After the RNase H reaction, water (90 µl) was added and the mixture phenol:chloroform extracted, the nucleic acids precipitated with ethanol, washed, dried and conserved for further analysis by primer extension.
Ribozyme cleavage assays
Cleavage reactions were carried out under single turnover conditions ([Rz] > [S]). Internally 32P-labeled HBV RNA (50 nM) was mixed with ribozyme (1 µM) in a 10 µl mixture containing 50 mM Tris–HCl pH 7.5 and 10 mM MgCl2, and then incubated at 37°C for 3 h. The reactions were stopped by the addition of loading buffer, electrophoresed on denaturing 5% PAGE gels and analyzed with a radioanalytic scanner. The specificity of cleavage of several ribozymes was also verified by primer extension.
The cleavage of full-length HBV RNA (50 nM) was also performed in the presence of the pool of delta ribozymes (10 µM) in a final volume of 10 µl, under the conditions described above. After the reaction, the volume was made up to 50 µl by adding water and the RNA precipitated with ethanol, washed, dried and conserved for further analysis by primer extension.
Primer extension analysis
A collection of DNA oligonucleotides complementary to the HBV RNA was purchased from Invitrogen. The nomenclature adopted referred to their respective complementary sequences on the full-length HBV: HBV199–184 (5′-TCATTAGTTCCCCCC-3′), HBV343–329 (5′-CTGTTTCTCTTCCAA-3′), HBV488–474 (5′-GGCGAGGGAGTTCTT-3′), HBV646– 632 (5′-AGGAAAAGATGGTGT-3′), HBV875–861 (5′-ATA TACCCGCCTTCC-3′), HBV1058–1044 (5′-GTTGGGATTGAAGTC-3′), HBV1245–1231 (5′-GTGGAGACAGCGGGG-3′), HBV1495–1481 (5′-AAAAACCCCGCCTGT-3′), HBV1698–1684 (5′-GCAGGATGAAGAGGA-3′), HBV2040– 2026 (5′-CCCAATACCACATCA-3′), HBV2499–2485 (5′-ACCAAGCCCCAGCCA-3′), HBV2784–2730 (5′-AGACGGAGAAGGGGA-3′), HBV3028–3014 (5′-CCCCCAACTCCTCCC-3′). The oligonucleotides (10 pmol) were 5′-end labeled in a mixture containing 10 µCi [γ-32P]ATP (3000 mCi/mmol; New England Nuclear), 50 mM Tris–HCl pH 7.6, 10 mM MgCl2, 10 mM 2-mercaptoethanol and 12 U of T4 polynucleotide kinase (United States Biochemicals) at 37°C for 30 min. The end-labeled oligonucleotides were purified on denaturing 20% PAGE gels, the relevant bands excised from the gel and eluted overnight at room temperature, passed through G-25 spun-column, ethanol precipitated, washed, dried and resuspended in water (60 µl). 5′-32P-labeled primer (6 µl) and 10× reverse transcription buffer (0.6 µl of 500 mM Tris–HCl pH 8.3, 800 mM KCl and 100 mM MgCl2) were used to dissolve the pellets resulting from either an RNase H hydrolysis or a ribozyme cleavage assay performed with a pool of either oligonucleotides or ribozymes, respectively. The primer annealing step was performed by successively incubating the mixtures at 65°C for 2 min followed by 2 min on ice. The reactions were initiated by adding 0.8 mM of each dNTP, 3.3 mM DTT and Superscript™ II Reverse Transcriptase (100 U; Invitrogen) in a final volume of 12 µl. The samples were incubated at 45°C for 30 min, then ethanol precipitated, washed and analyzed through 5% sequencing PAGE gels. DNA sequencing reactions using the same primer were migrated on the same gels in order to allow for identification of both the primer extension stops and the cleavage sites of the ribozymes. The results were visualized with a PhosphorImager.
RESULTS
Bioinformatic tools coupled to biochemical assays
Initially, an experimental procedure composed of three steps, including the use of bioinformatic tools and biochemical assays, was developed (Fig. 2A). The first step consists of the prediction of the RNA secondary structure and the subsequent identification of the sequences most likely to be in single-stranded regions. Using the RNA folding algorithm in RNA Structure 3.5 (27), eight structures were derived for the HBV RNA pregenome (data not shown). With the exception of minor local differences, the structures were similar and their stabilities, based on the values of relative free Gibbs energy [ΔG determined by free energy minimization (27)], varied by <5%. Regardless of the HBV natural sequence variant in question, the overall organizations of the structures obtained were similar, suggesting that no important differences can be attributed to a given genotype. The eight resulting structures were then analyzed using the OligoWalk program (13). This algorithm predicts the equilibrium affinity of an RNA oligonucleotide for a given sequence, taking into consideration the predicted stability of the oligonucleotide-target helix and the competition with the predicted secondary structures of both the target and the oligonucleotide. In our case the oligonucleotides correspond to the 7 nt composing the P1 binding domain of the delta ribozyme. The HBV sequence is 3358 nt in size and there are N – L + 1 complementary oligonucleotides of length L (where N and L are the lengths of the RNA target and oligonucleotide, respectively, that is 3358 – 7 + 1). Thus, the output consisted of a file of 3352 possible oligonucleotides, for each of which a ΔG value (corresponding to an overall free Gibbs energy value estimated for the interaction between an oligonucleotide and a given target) was calculated. Using an arbitrary cut-off of ΔG < –6.5 kcal/mol, 343 sequences were retained as potential target sites.
Figure 2.
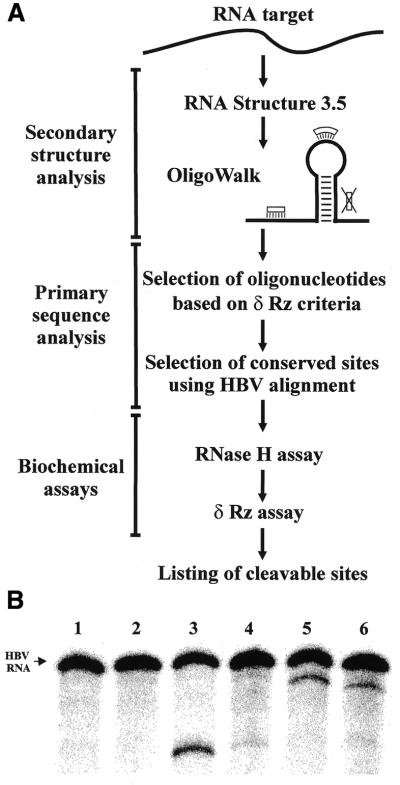
Selection of the sites with the greatest potential for delta ribozyme cleavage based on the use of bioinformatic tools coupled to biochemical assays. (A) Scheme of the steps that compose this procedure. Adjacent to the OligoWalk is an illustration of an example RNA structure that includes two oligonucleotides binding sites located in single-stranded regions and one (indicated by the large X) that is unavailable because it is located in a double-stranded region. (B) Typical results from biochemical assays for two different sites. Lanes 1 and 2 are the negative controls for the RNase H and ribozyme assays, respectively. In each case the HBV RNA was incubated under the reaction conditions, but either the oligonucleotide or the ribozyme were lacking. Lanes 3 and 5 are the RNase H assays in the presence of the oligonucleotide directing the hydrolyses at positions 1154 and 1543, respectively. Lanes 4 and 6 are the ribozyme assays for cleavage at positions 1154 and 1543, respectively.
The second step consists of screening the sites at the nucleotide sequence level based on two criteria. The first of these involved searching for specific sequence features known to be critical for efficient cleavage by a trans-acting antigenomic delta ribozyme. These features include: (i) the first base pair of the P1 stem must be a GU wobble base pair (29,30); (ii) there cannot be a guanosine at the cleavage site (position –1) as this results in an uncleavable substrate (6,7,30); (iii) the presence of two consecutive pyrimidines at position –1 and –2 dramatically reduces the cleavage efficiency of a substrate (30); (iv) the presence of a cytosine at position +4 (i.e. in the middle of the ribozyme strand of the P1 stem) significantly reduces the level of cleavage (31). Searching the 343 sequences for those that possess these four features reduces the list to 15 potential target sites. The second criteria we used was to analyze a sequence alignment of the natural HBV variants in order to identify which of the potential target sites are conserved. The identification of the highly conserved sequences in an RNA species should lead to the development of nucleic acid drugs that have the ability to specifically bind to all, or most, variants of this species. Twelve HBV variants from representative genotypes were aligned (Materials and Methods). This alignment revealed that nine of the 15 potential target sites are relatively highly conserved (i.e. conserved in at least 9 out of the 12 variants). These sequences are listed in Table 1.
Table 1. Compilation of the potential sites identified by the computer method.
| Cleavage position | HBV P1 stem | ORF targeteda | ΔG (kcal/mol) | RNase H assayc | Rz assayc |
|---|---|---|---|---|---|
| 409 | GCCCCUA | C and P genes | –6.6 | n.s. | n.d. |
| 523 | AGAUCUC | P gene | –8.0 | ++ | – |
| 524 | GAUCUCA | P gene | –8.0 | + | – |
| 946 | GCAUGGG | PreS1 gene | –8.5 | + | – |
| 1147 | GCCCUCA | PreS1 gene | –9.7 | – | n.d. |
| 1154 | GGCUCAG | PreS1 gene | –8.5 | +++ | + |
| 1543 | GGACUUC | S genes | –7.1 | +++ | ++ |
| 2250 | GGCCUAU | P gene | –8.3 | + | – |
| 2808 | GCACCUC | X gene | –8.1 | n.s. | n.d. |
| 212b | GGGUGGG | C genes | –0.3 | +++ | + |
| 486b | GCCUCGC | P gene | –0.9 | +++ | – |
| 513b | GCGUCGC | P gene | –3.0 | +++ | +++ |
n.d., not determined; n.s., not specific.
aSee Figure 1B.
bThese sites were suggested not accessible.
cRelative level of cleavage.
In order to give a biochemical dimension to the procedure, the potential sites were then investigated using two assays in solution. First, RNase H hydrolysis using 7 nt long DNA oligonucleotides corresponding to the recognition domain of the ribozyme (i.e. N6C) were performed. RNase H specifically cleaves the RNA of an RNA–DNA duplex and can be used to verify whether or not the binding of an oligonucleotide occurred and is specific to a target sequence (10). Randomly labeled HBV RNA was pre-incubated with unlabeled DNA oligonucleotides, RNase H digested and the mixtures analyzed on denaturing PAGE gels. A typical gel is shown in Figure 2B. The presence of the oligonucleotides corresponding to positions 1154–1160 and 1543–1549 allowed the RNase H to cleave the HBV RNA, although at different levels (lanes 3 and 5). These results are summarized in Table 1 (upper part). Three types of results were obtained: (i) the oligonucleotide corresponding to position 1147 did not result in any detectable cleavage under the conditions used; (ii) those for positions 409 and 2808 gave several non-specific products, suggesting that it bound to the HBV RNA at more than one position, a result that may be explained by the fact that RNase H requires only four consecutive base pairs to be active (32); and (iii) the six others that resulted in specific cleavage, albeit at different levels.
Secondly, the cleavage activity of the ribozymes was tested. Delta ribozymes with appropriate recognition sequences were synthesized by in vitro transcription from a minigene under the control of the T7 RNA promoter cloned into pUC19 for each of the six potential sites that gave specific RNase H hydrolysis. The ability of each ribozyme to cleave the labeled HBV RNA was tested under single-turnover conditions ([Rz] > [S]). After incubation at 37°C, the reactions were quenched and analyzed on PAGE gels. Typical results are shown in Figure 2B. The Rz-1154 cleaved the HBV RNA poorly, while the Rz-1543 appeared to be more efficient. Only two out of the six ribozymes tested exhibited cleavage of the HBV RNA (i.e. Rz-1154 and Rz-1543; see Table 1).
Thus, from the nine potential sites identified in the bioinformatic approach, only two were positive for in vitro delta ribozyme cleavage. Clearly, the biochemical assays are essential in order to validate the predictions derived from the bioinformatic analyses. The robustness of the initial computer predictions were further tested by analyzing seven additional sites with ΔG > –6.5 kcal/mol (data not shown). The sequences of these sites respected the nucleotide requirements considered previously. Among these sites, three appear to be accessible based on the RNase H assay (Table 1, lower part). The ribozymes corresponding to these three sites were synthesized and their cleavage activities accessed. Rz-486 did not detectably cleave the HBV RNA, but both Rz-212 and Rz-513 did, although at different levels. Clearly, this approach appears to have important limitations, at least in the case of the delta ribozyme.
Biochemical approaches based on the use of libraries
Target site accessibility and the ability to form an active ribozyme–substrate complex are two interdependent factors whose relationship is complex. Such a relationship can be better addressed by using a combinatorial method. Conse quently, we developed two procedures with a delta ribozyme based on previous reports for both the hammerhead and hairpin ribozymes (12).
A library of oligonucleotides. Figure 3A is a schematic representation of the procedure using a library of oligonucleotides. In theory, all accessible sites within an RNA molecule (i.e. those in single-stranded regions) would be specifically bound by an oligonucleotide and the resulting RNA–DNA heteroduplex subsequently hydrolyzed by RNase H (examples in 11,20). The cleavage sites can then be identified by electrophoresis of primer extension reactions using 5′-end-labeled primers and the RNase H reaction products. From the sequence of the substrate (HBV RNA), the sequence at the cleavage site may be determined and, thus, that of the complementary oligonucleotide (i.e. that acting as the P1 domain in the ribozyme). Finally, the corresponding ribozymes are synthesized and the cleavage activity tested.
Figure 3.
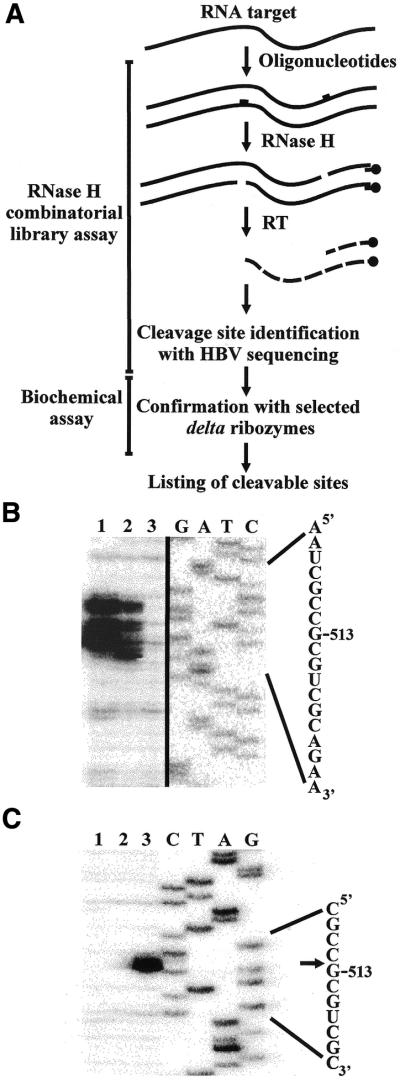
Selection of the sites with the greatest potential for delta ribozyme cleavage based on the use of an RNase H assay in the presence of a library of oligonucleotides. (A) Scheme illustrating the procedure. HBV RNA and cDNA are represented by the full and dashed lines, respectively. RT is reverse transcriptase. (B) Autoradiogram of a sequencing gel revealing the single-stranded region that includes position 513. Lanes 1 and 2 correspond to the reactions performed in the presence of the 5′-SSNDMSCD-3′ and 5′-N6CD-3′ libraries, respectively (where N = A, C, G or U; D = A, G or U; M = A or C; and S = C or G). Lane 3 is the reaction performed in the absence of any oligonucleotide. Adjacent to the gel is a DNA sequencing reaction, and the corresponding sequence is shown on the right side. (C) Autoradiogram of a sequencing gel of the cleavage assay for Rz-513. Lanes 1 and 2 are reactions performed in the absence of either ribozyme or MgCl2, respectively. Lane 3 is the complete reaction. After the cleavage reaction, primer extension was performed and migrated beside a sequencing reaction. The sequence is shown adjacent to the gel. The arrow indicates the cleavage site.
The library used was composed of oligonucleotides 8 nt in length corresponding to one residue before the cleavage site, which has to be single-stranded (i.e. position –1), and the seven residues of the recognition domain of the ribozyme (i.e. the P1 stem). The libraries were designed taking into account the sequence specificities described earlier for delta ribozyme cleavage. More specifically, the nucleotide at position –1 relative to the cleavage site cannot be a guanosine (30), therefore the 3′-end nucleotide of the oligonucleotide cannot be a C. The first base pair of the P1 stem has to be a GU wobble pair (29,30), therefore the seventh nucleotide of all oligonucleotides was a cytosine (i.e. allowing the formation of a GC base pair). The larger library of oligonucleotides corresponds to the sequence 5′-N6CD-3′ (i.e. 12 288 different sequences), while the smaller one corresponds to the sequence 5′-SSNDMSCD-3′ (i.e. 576 different sequences) (where N indicates A, C, G or U; D indicates A, G or U; S indicates C or G; and, M indicates A or C). The latter library was designed to include more of the sequence preferences of trans-acting delta ribozymes, and to favor stronger binding of the substrate to the ribozyme due to the presence of a higher percentage of GC residues.
A typical example of the identification of a potential site is illustrated in Figure 3B. In this case the primer extension was performed using an oligonucleotide complementary to positions 632–646 of the HBV RNA. In the absence of any oligonucleotide no band was observed in the specific region as compared with lanes 1 and 2 (lane 3). In the presence of the oligonucleotides from the 5′-N6CD-3′ library, several bands were detected (lane 2). This indicates the presence of a single-stranded region accessible to several oligonucleotides binding on different copies of the RNA target, and results in cleavage at various contiguous positions. In the presence of oligonucleotides from the 5′-SSNDMSCD-3′ library (lane 1), fewer bands were observed. This reflects the smaller expansion of the sequences composing the library. However, since each sequence is present in a higher concentration (i.e. for the same absolute amount of oligonucleotides) the concentration of cleaved RNA, and consequently cDNA, is larger. This assay suggested that a ribozyme may be engineered for cleavage at position 513 (i.e. Rz-513). Rz-513 specifically and efficiently cleaved the HBV RNA as shown by the primer extension assay in Figure 3C.
Since each primer produced a readable sequence of 200–300 bases, several primers were required in order to map the full HBV RNA. An average of two potential sites were detected for each primer extension reaction (i.e. per ∼200–300 bases) based on the detection of either one, or stretches of, band(s). Most of the time, either one or two consecutive bands of weak intensity were detected. Preliminary experiments have shown that such bands do not indicate productive sites. When we considered the detection of three or more consecutive bands in primer extension, we retrieved only six potential sites (Table 2). In each case, the region was analyzed to determine the most appropriate sequence in terms of cleavage by a delta ribozyme (i.e. position –1 to the cleavage site must be A, C or U). With the exception of Rz-887, five of the six ribozymes exhibited a relatively efficient cleavage of the HBV RNA under the conditions used (Table 2). Of these five active ribozymes, four were conserved (only Rz-574 was not) with respect to the sequence alignment of the HBV variants (i.e. present in at least 9 out of the 12 variants).
Table 2. Compilation of the potential sites identified by the RNase H method.
| Cleavage position | HBV P1 stem | ORF targeteda | RNase H assay (N)b | Rz assayc |
|---|---|---|---|---|
| 167 | GGCAAGC | C genes | + (3) | +++ |
| 279 | GGGCCUA | C genes | + (4) | +++ |
| 461 | GGUCCCC | P gene | + (4) | ++ |
| 513 | GCGUCGC | P gene | + (9) | +++ |
| 574 | GGGGAAC | P gene | + (3) | + |
| 887 | GAAACAA | P gene | + (7) | – |
A combinatorial library of ribozymes. Finally, we tried an approach based on a library of delta ribozymes (Fig. 4A). The most important advantage of this approach should be that the detection of a cDNA band is directly related to the cleavage of a ribozyme, whereas the bands produced by the previous RNase H-based assay were an indirect indication. Conse quently, this method is theoretically better because the selection is based not only on a mimic of the binding site, but rather on both binding and cleavage by the ribozyme.
A pool of ribozymes was synthesized using a minigene under the control of a T7 RNA polymerase promoter (see inset, Fig. 4A). The minigene was made from a pool of antisense oligonucleotides in which the positions corresponding to the P1 stem were randomized, and the second strand synthesized using a sense primer that formed the T7 RNA promoter. The library was based on the randomization of the six positions of the P1 stem (i.e. 5′-N6U-3′, giving 4096 ribozymes). This library took into account the requirement for a U for the formation of the GU wobble base pair adjacent to the cleavage site (29,30). The ribozymes (i.e. 2.4 nM of each individual ribozyme) were incubated with HBV RNA (50 nM), and the cleavage sites identified by electrophoresis of primer extension reactions as was done for the RNase H approach described above. A typical example of the identification of a site is illustrated in Figure 4B. In this case the primer extension was performed using an oligonucleotide complementary to positions 329–343 of the HBV RNA. The presence of the pool of ribozymes resulted in the detection of an additional band corresponding to cleavage at position 303 (lane 1) as compared with the results obtained in the absence of either MgCl2 (lane 2) or the ribozyme (lane 3). Scanning the entire length of the HBV RNA revealed six potential sites (Table 3). Subsequently, specific delta ribozymes were made from the minigenes and their cleavage activities assessed. Five out of the six cleaved the HBV RNA, with the Rz-303, Rz-1543, Rz-2833 and Rz-2861 recognizing conserved sequences of the HBV variants (i.e. present in at least in 9 out of 12 variants).
Table 3. Compilation of the potential sites identified by the library of delta ribozymes.
| Cleavage position | HBV P1 stem | ORF targeteda | Rz pool assayb | Rz assayb |
|---|---|---|---|---|
| 303 | GUGGUUU | C genes | ++ | +++ |
| 1365 | GUAAACC | Pre S2 | + | – |
| 1543 | GGACUUC | S genes | ++ | ++ |
| 2624 | GUUGUCC | P gene | + | ++ |
| 2833 | GUCUGUG | X gene | + | +++ |
| 2861 | GUGCACU | X gene | + | +++ |
aSee Figure 1B.
bRelative level of cleavage.
DISCUSSION
Procedures for the selection of sites
This work presents the development of three procedures for the identification of delta ribozyme cleavage sites within an RNA target, a step essential to the development of a gene therapy. The first approach consists of an experimental scheme composed of three steps, including the use of bioinformatic tools coupled to biochemical assays (Fig. 2). At its heart, this procedure is based on computer prediction of the RNA secondary structure, a procedure that is the most popular method of determining the structures of all kinds of RNA (13,16) and elucidating the regions that are most likely to be single-stranded. Of the nine potential sites obtained by the computer-assisted analysis, six were both specific and effective for the RNase H hydrolysis (Table 1). However, only two members of this subset were cleaved by the appropriate ribozymes. Although the two first steps were performed rapidly, as the coupling of the biochemical approach is essential to providing relevant results, this procedure did not result in any significant gains in terms of time. We concluded that this bioinformatic approach has severe limitations, at least in the case of delta ribozyme. Many reasons can be put forth to explain the discrepancy between the computer predictions and sites effectively cleaved by the ribozymes, including the fact that the tertiary structures of both the ribozyme and the RNA target were not taken into account by the software. However, the most important reason is the fact that, even today, the prediction of RNA structure remains a complicated task for long RNA molecules (8–10).
Based on our results, a combinatorial method is the best means of overcoming the complexed interdependent nature of target site accessibility and the ability to form an active ribozyme–substrate complex. In the RNase H procedure, the presence of combinatorial libraries of oligonucleotides mimics the binding of the ribozyme. Taking into account some of the substrate specificity features of the delta ribozyme cleavage allowed us to both limit the expansion of the library and to ‘personalize’ it to this catalytic RNA. Preliminary results showed that only the presence of a stretch of bands, corresponding to consecutive stops of the primer extension, was indicative of a productive site. The binding domain of the substrate is restricted to seven bases, but a larger single-stranded domain is required in order to both satisfy the tertiary structure of the ribozyme and permit its molecular mechanism (which involves a conformational transition of the ribozyme– substrate complex) (7). The procedure based on a combinatorial library of delta ribozymes randomized for their recognition sequences offers the advantage of selecting sites that are bound by a ribozyme and subsequently cleaved. In that case only single bands were seen. Both combinatorial methods allowed us to identify six potential sites, five of which were cleaved in vitro by the appropriate ribozymes in each case. However, none of the selected sites were identified by both the oligonucleotide and ribozyme libraries. This observation indicates that none of the methods permits the identification of all potential sites, which is clearly a limiting factor.
Substrate specificity for delta ribozyme cleavage
Using small model substrates, the specificity for delta ribozyme cleavage has been determined in vitro (29–31,33). Briefly, the P1 stem is composed of the 7 nt that base pair with the ribozyme; hence, these nucleotides can be defined as internal determinants of substrate specificity. Moreover, it has been demonstrated that the nucleotide composition at positions –1 to –4 from the scissile bond influences the efficiency of cleavage of small substrates (31,33). For example, the presence of two, single-stranded, consecutive pyrimidines at positions –1 and –2 decreases the levels of cleavage to varying degrees depending on the identity of the residues in question. The nucleotides in positions –1 to –4 do not directly contribute to substrate recognition (e.g. substrate binding), but the identity of these residues is essential for efficient cleavage (10,30). Therefore, they contribute to substrate specificity, likely as external determinants. In this study, the nucleotides at positions –1 to –4 had various compositions. Most of the cleavage sites retained do not possess consecutive pyrimidines at positions –1 and –2, or if they did they were inefficiently cleaved, supporting the idea that the rules established with small substrates are consistently respected in a long RNA target. However, we also observed exceptions to these rules: for example, the Rz-513 exhibited an efficient cleavage even though two cytosines are located at positions –1 and –2. This result shows that the situation of a small substrate is slightly different to that of a long molecule such as HBV RNA. In fact, there seems to be a complex situation in which several factors, including the secondary structure motifs in close proximity to a potential site, as well as long interactions, modulate the efficiency of the cleavage.
Any contribution to the substrate specificity for delta ribozyme cleavage is important for further development in vivo. As any given 7mer that corresponds to the length of the P1 stem (the binding domain) might be found at a frequency of one in 47 nt (i.e. 16 384), duplicate targets might be present in the population of cellular mRNA, and cleavage of these RNAs could prove toxic to the cell. Therefore, the modulation of cleavage efficiency caused by the sequences in positions –1 to –4 of the cleavage site might be of primary importance in minimizing this problem. The net effect of this is to extend from 7 to 11 contiguous nucleotides the domain of the substrate that contributes to determining the ability of an RNA molecule to be cleaved by delta ribozyme. Furthermore, it is important to remember that the complexity of an RNA structure drastically reduces the number of potential sequences that can be considered as potential sites to target. In this study, the use of a combinatorial library including thousands of different ribozymes did not allow us to retrieve too many active ribozymes even though the sequence complementary to the recognition sequence was present within the HBV RNA. Clearly, the secondary and tertiary structures of the RNA molecules, as well as their complexation with other cellular components (e.g. with proteins forming RNPs) also make important contributions to minimizing the problem of lack of specificity.
Targeting HBV
Together, the three procedures used in this work yield a collection of 12 delta ribozymes with the potential to target throughout the HBV RNA pregenome. Comparing the sites identified here, within the HBV RNA, with those of other reports, based on the use of other catalytic RNAs like the hammerhead and the hairpin ribozyme (14,24,25,34–37), shows no similarities. Moreover, when we consider the sites targeted in other HBV works it appears impossible to engineer delta ribozyme cleaving at these positions, primarily because the delta ribozyme has different characteristics than other catalytic RNAs. On the other hand, the extension of this work would be to show whether or not some, or all, of the delta ribozymes of the collection would be effective in controlling the propagation of HBV in vivo. However, previous reports have demonstrated the existence of relatively good correlations between in vitro and in vivo for both oligonucleotide and ribozyme effects (19,20,22,38).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Michael Nassal for providing the HBV vector. This work was supported by grants from both the Canadian Institutes of Health Research (CIHR) and Health Canada to J.-P.P. The RNA group is supported by grants from both the CIHR and Fonds FCAR (Québec). L.J.B. was a recipient of a pre-doctoral fellowship from FCAR/FRSQ and the Georges Phénix Fondation. J.-P.P. is an investigator from the CIHR.
REFERENCES
- 1.Lewin A.S. and Hauswirth,W.W. (2001) Ribozyme gene therapy: application for molecular medicine. Trends Mol. Med., 7, 221–228. [DOI] [PubMed] [Google Scholar]
- 2.Michienzi A. and Rossi,J.J. (2001) Intracellular applications of ribozymes. Methods Enzymol., 341, 581–596. [DOI] [PubMed] [Google Scholar]
- 3.Sun L.Q., Cairns,M.J., Saravolac,E.G., Baker,A. and Gerlach,W.L. (2000) Catalytic nucleic acids: from lab to application. Pharmacol. Rev., 52, 325–347. [PubMed] [Google Scholar]
- 4.Shih I.H. and Been,M.D. (2002) Catalytic strategies of the hepatitis delta virus ribozymes. Annu. Rev. Biochem., 71, 887–917. [DOI] [PubMed] [Google Scholar]
- 5.Lévesque D., Choufani,S. and Perreault,J.P. (2002) Delta ribozyme benefits from a good stability in vitro that becomes outstanding in vivo. RNA, 8, 464–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ananvoranich S. and Perreault,J.P. (1998) Substrate specificity of δ ribozyme cleavage. J. Biol. Chem., 273, 13182–13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercure S., Lafontaine,D., Ananvoranich,S. and Perreault,J.P. (1998) Kinetic analysis of δ ribozyme cleavage. Biochemistry, 37, 16975–16982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell T.B. and Cech,T.R. (1995) Identification of ribozymes within a ribozyme library that efficiently cleave a long substrate RNA. RNA, 1, 598–609. [PMC free article] [PubMed] [Google Scholar]
- 9.Birikh K.R., Berlin,Y.A., Soreq,H. and Eckstein,F. (1997) Probing accessible sites for ribozymes on human acetylcholinesterase RNA. RNA, 3, 429–437. [PMC free article] [PubMed] [Google Scholar]
- 10.Roy G., Ananvoranich,S. and Perreault,J.P. (1999) Delta ribozyme has the ability to cleave in trans an mRNA. Nucleic Acids Res., 27, 942–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lima W.F., Brown-Driver,V., Fox,M., Hanecak,R. and Bruice,T.W. (1997) Combinatorial screening and rational optimization for hybridization to folded hepatitis C virus RNA of oligonucleotides with biological antisense activity. J. Biol. Chem., 272, 626–638. [PubMed] [Google Scholar]
- 12.Yu Q., Pecchia,D.B., Kingsley,S.L., Heckman,J.E. and Burke,J.M. (1998) Cleavage of highly structured viral RNA molecules by combinatorial libraries of hairpin ribozymes. J. Biol. Chem., 273, 23524–23533. [DOI] [PubMed] [Google Scholar]
- 13.Mathews D.H., Burkard,M.E., Freier,S.M., Wyatt,J.R. and Turner,D.H. (1999) Predicting oligonucleotide affinity to nucleic targets. RNA, 5, 1458–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.zu Putlitz J., Yu,Q., Burke,J.M. and Wands,J.R. (1999) Combinatorial screening and intracellular antiviral activity of hairpin ribozymes directed against hepatitis B virus. J. Virol., 73, 5381–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scherr M., Rossi,J.J., Sczakiel,G. and Patzel,V. (2000) RNA accessibility prediction: a theoritical approach is consistent with experimental studies in cell extracts. Nucleic Acids Res., 28, 2455–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amarzguioui M., Brede,G., Babaie,E., Grotli,M., Sproat,B. and Prydz,H. (2000) Secondary structure prediction and in vitro accessibility of mRNA as tools in the selection of target sites for ribozymes. Nucleic Acids Res., 28, 4113–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scherr M., LeBon,J., Castanotto,D., Cunliffe,H.E., Meltzer,P.S., Ganser,A., Riggs,A.D. and Rossi,J.J. (2001) Detection of antisense and ribozyme accessible sites on native mRNAs: application to NCOA3 mRNA. Mol. Ther., 4, 454–460. [DOI] [PubMed] [Google Scholar]
- 18.Barroso-delJesus A. and Berzal-Herranz,A. (2001) Selection of targets and the most efficient hairpin ribozymes for inactivation of mRNAs using a self-cleaving RNA library. EMBO Rep., 21, 1112–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krüger M., Beger,C., Welch,P.J., Berber,J.R. and Wong-Staal,F. (2001) C-SPACE (cleavage-specific amplification of cDNA ends): a novel method of ribozyme-mediated gene identification. Nucleic Acids Res., 29, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sohail M., Hochegger,H., Klotzbücher,A., Le Guellec,R., Hunt,T. and Southern,E.M. (2001) Antisense oligonucleotides selected by hybridization to scanning arrays are effective reagents in vivo. Nucleic Acids Res., 29, 2041–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan W.H., Devlin,H.F., Kelley,C., Isom,H.C. and Clawson,G.A. (2001) A selection system for identifying accessible sites in target RNAs. RNA, 7, 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding Y. and Lawrence,C.E. (2001) Statistical prediction of single-stranded regions in RNA secondary structure and application to predicting effective antisense target sites and beyond. Nucleic Acids Res., 29, 1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong C.Y.W. (2000) Genetic variations of hepatitis B virus. Curr. Opin. Infect. Dis., 13, 481–487. [DOI] [PubMed] [Google Scholar]
- 24.Welch P.J., Tritz,R., Yei,S., Barber,J. and Yu,M. (1997) Intracellular application of hairpin ribozyme genes against hepatitis B virus. Gene Ther., 4, 736–743. [DOI] [PubMed] [Google Scholar]
- 25.Weinberg M., Passman,M., Kew,M. and Arbuthnot,P. (2000) Hammerhead ribozyme-mediated inhibition of hepatitis B virus X gene expression in cultured cells. J. Hepatol., 33, 142–151. [DOI] [PubMed] [Google Scholar]
- 26.Nassal M. (1992) The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and production viral positive-strand DNA synthesis but not for virus assembly. J. Virol., 66, 4107–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters provides improved prediction of RNA secondary structures. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Clustal W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishikawa F., Fauzi,H. and Nishikawa,S. (1997) Detailed analysis of base preferences at the cleavage site of a trans-acting HDV ribozyme: a mutation that changes cleavage site specificity. Nucleic Acids Res., 27, 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deschênes P., Lafontaine,D.A., Charland,S. and Perreault,J.P. (2000) Nucleotides –1 to –4 of hepatitis delta ribozyme substrate increase the specificity of ribozyme cleavage. Antisense Nucleic Acid Drug Dev., 10, 53–61. [DOI] [PubMed] [Google Scholar]
- 31.Ananvoranich S., Lafontaine,D.A. and Perreault,J.P. (1999) Mutational analysis of the antigenomic trans-acting delta ribozyme: the alterations of the middle nucleotides located on the P1 stem. Nucleic Acids Res., 17, 1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hogrefe H.H., Hogrefe,R.I., Walder,R.Y. and Walder,J.A. (1990) Kinetic analysis of Escherichia coli RNase H using DNA-RNA-DNA/DNA substrates. J. Biol. Chem., 265, 5561–5566. [PubMed] [Google Scholar]
- 33.Shih I.H. and Been,M.D. (2001) Energetic contribution of non-essential 5′ sequence to catalysis in a hepatitis delta virus ribozyme. EMBO J., 20, 4884–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Y., Kong,Y.Y., Wang,Y. and Qi,G. (2001) Inhibition of hepatitis B virus by hammerhead ribozyme targeted to the poly(A) signal sequence in cultured cells. Biol. Chem., 382, 655–660. [DOI] [PubMed] [Google Scholar]
- 35.Beck J. and Nassal,M. (1995) Efficient hammerhead ribozyme-mediated cleavage of the structured hepatitis B virus encapsidation signal in vitro and in cell extracts, but not in intact cells. Nucleic Acids Res., 23, 4954–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim Y.K., Junn,E., Park,I., Lee,Y., Kang,C. and Ahn,J.K. (1999) Repression of hepatitis B virus X gene expression by hammerhead ribozymes. Biochem. Biophys. Res. Commun., 257, 759–765. [DOI] [PubMed] [Google Scholar]
- 37.Passman M., Weinberg,M., Kew,M. and Arbuthnot,P. (2000) In situ demonstration of inhibitory effects of hammerhead ribozymes that are targeted to the hepatitis Bx sequence in cultured cells. Biochem. Biophys. Res. Commun., 268, 728–733. [DOI] [PubMed] [Google Scholar]
- 38.Kato Y., Kuwabara,T., Warashina,M., Toda,H. and Taira,K. (2001) Relationships between the activities in vitro and in vivo of various kinds of ribozyme and their intracellular localization in mammalian cells. J. Biol. Chem., 276, 15378–15385. [DOI] [PubMed] [Google Scholar]