Abstract
The human cytomegalovirus UL47 open reading frame encodes a 110-kDa protein that is a component of the virion tegument. We have constructed a cytomegalovirus mutant, ADsubUL47, in which the central portion of the UL47 open reading frame has been replaced by two marker genes. The mutant replicated to titers 100-fold lower than those for wild-type virus after infection at either a high or a low input multiplicity in primary human fibroblasts but was substantially complemented on cells expressing UL47 protein. A revertant virus in which the mutation was repaired, ADrevUL47, replicated with wild-type kinetics. Mutant virions lacked UL47 protein and contained reduced amounts of UL48 protein. The mutant was found to be less infectious than wild-type virus, and a defect very early in the replication cycle was observed. Transcription of the viral immediate-early 1 gene was delayed by 8 to 10 h. However, this delay was not the result of a defect in virus entry or of the inability of virion proteins to transactivate the major immediate-early promoter. We also show that the UL47 protein coprecipitated with the UL48 and UL69 tegument proteins and the UL86-encoded major capsid protein. We propose that a UL47-containing complex is involved in the release of viral DNA from the disassembling virus particle and that the loss of UL47 protein causes this process to be delayed.
Human cytomegalovirus (HCMV) is the prototype betaherpesvirus family member and a ubiquitous human pathogen (18). HCMV infection is normally asymptomatic in healthy children and adults, but infection during pregnancy can result in birth defects, and the virus can cause severe disease in immunocompromised individuals.
HCMV virions have a characteristic herpesvirus structure: an icosahedral capsid containing the viral DNA genome is surrounded by a protein layer termed the tegument and further enveloped by a lipid bilayer containing virus-encoded glycoproteins. The HCMV tegument contains more than 25 virus-encoded proteins (2). Tegument proteins are delivered to the cell at the start of infection and therefore have the opportunity to act at the very start of the infectious process. Several of the HCMV tegument proteins have been characterized, and these proteins play critical roles in immune evasion, activation of viral and cellular promoters, and cell cycle modulation. The UL83-encoded pp65 protein is the most abundant tegument component. Although it is not essential for replication in tissue culture (23), pp65 may aid in viral evasion of immune detection in vivo by causing an immediate-early protein (IE1) to be phosphorylated, rendering it resistant to major histocompatibility complex class I presentation (9). The UL82-encoded pp71 tegument protein has been shown to serve as a transcriptional activator (15) and can increase the infectivity of viral DNA upon transfection into human diploid fibroblasts (human foreskin fibroblasts [HFFs]) (1). A virus lacking pp71, ADsubUL82, displays a multiplicity-dependent growth defect, failing to efficiently activate the accumulation of viral immediate-early mRNAs (4, 5). Another tegument protein, pUL69, blocks progression in the G1 phase of the cell cycle (16), and a UL69-deficient virus, ADsubUL69, is defective for replication in permissive fibroblasts (11). There are additional functions attributed to tegument proteins in other herpesviruses that have not been described for HCMV. For instance, tegument proteins in herpes simplex virus type 1 (HSV-1) function during unpackaging and movement of the capsid to the nuclear pore (3, 14), virion morphogenesis (8, 19), and egress (7).
UL47 is a conserved open reading frame (ORF) encoding a protein of unknown function that is a constituent of the HCMV tegument (2). The UL47 ORF is transcribed as part of a 10-kb transcript that traverses the adjacent UL48 ORF and utilizes the polyadenylation signal located 3′ to UL48 (12). HCMV UL47 is syntenic to HSV-1 UL37, an ORF that encodes a tegument protein whose levels of virion incorporation are tightly controlled (6, 17, 22).
In this report, we describe the construction and characterization of ADsubUL47, a mutant virus that lacks a substantial portion of the UL47 ORF. This mutant virus exhibits a growth defect at low and high multiplicities on HFFs, and this defect is substantially complemented in cells that express pUL47. We demonstrate that the mutant is less infectious than wild-type virus and is defective at a step very early in its replication cycle. Immediate-early 1 (IE1) mRNA accumulation is delayed upon infection with ADsubUL47. However, this delay is not the result of a defect in virus entry into the cell or in the ability of virion proteins to transactivate the major immediate-early promoter (MIEP). In addition, we show that pUL47 coprecipitates with two tegument proteins, pUL48 and pUL69, as well as with the major capsid protein (MCP). pUL47 resides in a protein complex and is involved in an early event in HCMV infection, prior to transcription of IE1 but after translocation of at least some tegument constituents to the nucleus.
MATERIALS AND METHODS
Cells and viruses.
All cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum. Primary HFFs were used between passages 8 and 15.
HCMV strain AD169 stocks were prepared by harvesting the medium from HFFs infected at a multiplicity of 0.1 PFU/cell after cultures displayed complete cytopathic effect. Virus particles were partially purified by sedimentation through a sorbitol cushion as previously described (24) and were resuspended either in DMEM or in lysis buffer. Virus stocks were titered by plaque assay.
ADsubUL47 was constructed by recombination within human fibroblasts. Sequences flanking the UL47 region to be deleted were amplified by PCR from cosmid pCM1049 (primers, 5′ TCTCTCTAGAGCGTCCATGATGGCGAGGCG 3′–5′ AGAGATCGATACTAGTCAGACGCGAGATCAGAGCCG 3′ and 5′ TCTCATCGATACTAGTGCACATCGACGCCGCGGTTT 3′–5′ CTCTTCTAGAGTCGCCCTGGTGGCAGCTGG 3′) with Advantage cDNA polymerase (Clontech), digested with XbaI and ClaI, and then ligated together into the XbaI site of the pSP72 vector (Promega). The sequences of all PCR-generated DNAs were determined and found to be wild type. A 2.2-kb DNA XbaI fragment from pGET15 (26) was ligated into the SpeI site of the clone, producing the plasmid construct used for mutagenesis of HCMV. The clone was digested with XbaI, releasing a 4.2-kb fragment that was electroporated along with infectious AD169 DNA and pCGNpp71 into HFFs, as described previously (1). Green fluorescent protein (GFP)-positive plaques were identified, and mutant virus was purified by three additional rounds of plaque purification on complementing cells produced by prior infection with a recombinant retrovirus expressing pUL47. The correct genomic organization was confirmed by Southern blotting.
The recombinant retrovirus was prepared using the system developed by Kinsella and Nolan (13). The UL47 ORF was amplified by PCR from cosmid pCM1049 (primers, 5′ TCTCTCTAGAGCGTCCATGATGGCGAGGCG 3′ and 5′ AGAGGAATTCGGTACCTGGCAGCTGGCCTGTGTGAC 3′) with Advantage cDNA polymerase. Amplified DNA was digested with XbaI and KpnI and ligated into pCGN (25). The hemagglutinin (HA) epitope-tagged UL47 sequence from this construct was amplified by PCR (primers, 5′ CTCTGGATCCGACCATGGCTTCTAGCTATCCTTATG 3′ and 5′ CTCTGGATCCGACCATGGCTTCTAGCTATCCTTATG 3′), digested with BamHI, and inserted into the BamHI site of pRetroEBNA to produce pRetroflu47. Retrovirus was generated by transfection of the plasmid construct into Phoenix packaging cells using calcium phosphate. Supernatants were collected, Retroflu47 was pelleted by centrifugation (for 12 to 16 h at 3,300 × g and 4°C), pelleted virus was resuspended in DMEM supplemented with 10% fetal calf serum, and aliquots were stored at −80°C. Low-passage-number HFFs were infected by incubating subconfluent monolayers with Retroflu47 in the presence of 4 μg of Polybrene/ml.
A revertant of ADsubUL47, ADrevUL47, was constructed by recombination within human fibroblasts. The UL47 gene was amplified from cosmid pCM1049 (primers, 5′ TCTCTCTAGAGCGTCCATGATGGCGAGGCG3 ′–5′ CTCTCGTTGCTCCAGACACGTGCTGT 3′ and 5′ CTCTGCATCGATCCACGTTGATCGGCTCGAAGG 3′–5′ AGAGGAATTCGGTACCTGGCAGCTGGCCTGTGTGAC 3′) with Advantage cDNA polymerase. The boldfaced G in the third primer represents a nucleotide change from the published AD169 sequence (6). The mutation is silent and destroys a StyI site. The two DNA fragments produced in the PCRs were digested with XbaI/ClaI and ClaI/EcoRI, respectively, and then ligated into the XbaI and EcoRI sites of pSP72 (Promega). The 3-kb UL47 fragment was electroporated along with infectious ADsubUL47 viral DNA and pCGNpp71 into HFFs as previously described (1). Clear plaques were selected and purified on HFFs as above, and the correct genomic organization was confirmed by Southern blotting.
To monitor wild-type, mutant, and revertant HCMV growth kinetics, HFFs were infected at 2 (high multiplicity) or 0.001 (low multiplicity) PFU/cell. After 2 h, the inoculum was removed and cells were washed before addition of complete medium. Samples were taken once a day for 6 days (high-multiplicity infection) or once every 3 days for 3 weeks (low-multiplicity infection) and titered by plaque assay on the complementing cells.
The particle-to-PFU ratio was determined for wild-type AD169 and ADsubUL47. Virus stocks were generated from the medium of HFFs infected at a multiplicity of 0.001 PFU/cell. The infectivities of virus stocks were determined by plaque assay on complementing cells. To determine the relative number of viral genomes in the virus preparations, equal volumes of wild-type and ADsubUL47 stocks were mixed, the virus was pelleted by centrifugation through a sorbitol cushion, and the pellet was resuspended in 10.1 TE (10 mM Tris-HCl [pH 8]−0.1 mM EDTA). Viral DNA was extracted and analyzed by Southern blotting as described below using a randomly primed, 32P-labeled UL48 DNA probe (nucleotides 65548 to 66150).
DNA, RNA, and protein analysis.
For Southern blotting, total DNA was isolated from infected cells or virus preparations by incubation in lysis buffer (0.5% sodium dodecyl sulfate [SDS], 25 mM EDTA, 100 mM NaCl, 10 mM Tris [pH 8], 100 μg of proteinase K/ml) for 2 h at 37°C, followed by two extractions with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and ethanol precipitation. After digestion with restriction enzymes, DNA was separated on an 0.8% agarose gel, blotted to nylon membranes (Hybond N+; Amersham), and probed with randomly primed [32P]dCTP-labeled DNA.
For Northern blotting, total RNA was isolated with the TRIZOL reagent (Gibco BRL). RNAs were separated by electrophoresis for 3 h at 100 V in a formaldehyde-containing gel and transferred to a nylon membrane using the Turboblotter system (Schleicher and Schuell). The RNA was cross-linked to the membranes with UV light (Stratalinker) and hybridized with 107 cpm of [32P]dCTP randomly labeled DNA in Quikhyb (Stratagene) or Church buffer (1% bovine serum albumin [BSA], 1 mM EDTA, 0.5 M NaHPO4 [pH 7.2], 7% SDS).
For Western blotting, protein samples were separated by electrophoresis in an SDS-containing 7.5 to 15% polyacrylamide gel and transferred to nitrocellulose membranes (Opti-tran; Schleicher and Schuell). Blots were probed with the indicated antibodies and then developed using the ECL reagent (Amersham). Antibodies against pUL32, pUL69, pUL86, pUL99, pIRS1, and pTRS1 have been described previously (11, 20, 21, 27). The HA-specific antibody was purchased from Babco; a UL48-specific monoclonal antibody was generated using a glutathione S-transferase (GST) fusion protein containing the first 259 amino acids of the UL48 protein; UL47-specific polyclonal sera were produced in a mouse injected with a GST fusion protein containing amino acids 880 to 930 of the UL47 protein.
Immunoprecipitations were performed as described previously (10). Cells were starved in DMEM lacking cysteine and methionine with 5% dialyzed fetal calf serum for 30 min and were then incubated with [35S]cysteine and [35S]methionine for 5 h prior to harvesting and immunoprecipitation. Polyacrylamide gels containing immunoprecipitated proteins that were labeled with [35S]cysteine and [35S]methionine were treated with Enhance (Dupont) and analyzed in a phosphorimager.
For immunofluorescent analysis, HFFs were plated onto 12-mm coverslips and 12 h later were either mock infected or infected with either AD169 (20 PFU/cell) or ADsubUL47 (5 PFU/cell). Coverslips were removed 4 h later, rinsed in phosphate-buffered saline (PBS), and fixed in 0.4% paraformaldehyde. Cells were blocked in 0.5% BSA and 5% goat serum in PBST (0.5% Tween-20, 1% Triton X-100 in PBS). Primary (anti-UL82, anti-UL83, anti-UL69) and secondary (anti-mouse Alexa 568; Molecular Probes) antibodies were diluted in 0.5% BSA and 5% goat serum in PBST. Cells were incubated with the primary antibody for 1 h and with the secondary antibody for 30 min and were washed with PBST three times after each incubation. Cells were then washed twice with distilled water, incubated with Hoescht 33342 DNA stain, washed twice in distilled water, and mounted onto slides.
Assay for transactivation of the MIEP.
HFFs immortalized by telomerase and containing an integrated copy of the MIEP controlling expression of the tetracycline activator protein were either mock infected or infected with either AD169 or ADsubUL47 at a multiplicity of 2 PFU/cell for 4 h. Cells were then washed twice with PBS, and RNA was isolated using the TRIZOL reagent and analyzed by Northern blotting using a 32P-labeled probe for the tetracycline activator sequence.
RESULTS
Construction and propagation of a mutant virus lacking the UL47 coding sequence.
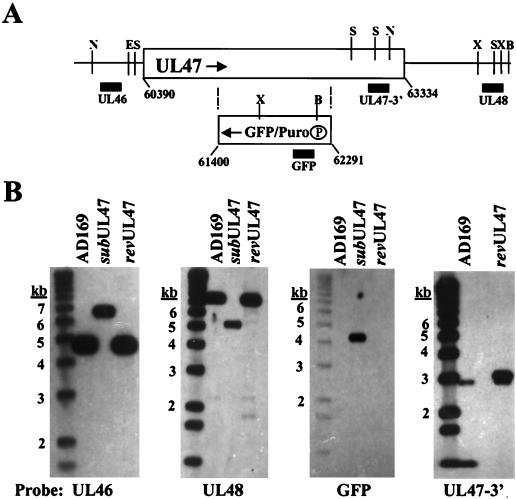
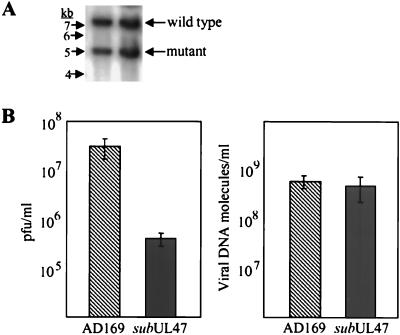
To explore the role of pUL47 during HCMV infection, we generated a substitution mutant carrying marker genes encoding GFP and puromycin-N-acetyltransferase in place of a portion of the UL47 ORF (Fig. 1A). The mutant, ADsubUL47, was produced by homologous recombination in HFFs. It was purified on complementing cells (R47 HFFs) created by infecting HFFs with a recombinant retrovirus (Retroflu47) that constitutively expresses an HA epitope-tagged UL47 fusion protein from the retroviral promoter. R47 HFFs did not fully complement the growth of the mutant virus; the yield of ADsubUL47 was reduced by a factor of 10 from that of wild-type virus (data not shown). The purity and genomic organization of ADsubUL47 were confirmed by Southern blot assay, probing for regions on both sides of the mutation and for the GFP insert (Fig. 1B). The mutation was repaired by homologous recombination in HFFs with a UL47 coding sequence containing a single missense mutation onto the ADsubUL47 viral genome. Southern blotting of ADrevUL47 DNA demonstrated that it was pure and contained the missense mutation, as shown by loss of a StyI restriction site (Fig. 1B).
FIG. 1.
Construction of ADsubUL47. (A) Schematic representation of the UL47 locus and the ADsubUL47 substitution mutation. The rectangular box represents the UL47 coding sequence with the relevant nucleotide numbers (6) shown. The GFP/Puro box represents the DNA recombined into the UL47 ORF, with the boundaries of the UL47 nucleotides replaced given below the box. The circled P represents the promoter for the GFP/Puro cassette. Arrows indicate the opposite directions of transcription for UL47 and the GFP/Puro cassette. Vertical lines represent restriction enzyme recognition sites (B, BamHI; E, EcoRI; N, EcoNI; S, StyI; X, XmnI). Solid boxes indicate locations of probes used in Southern blotting. (B) Southern blots of AD169, ADsubUL47, and RUL47 viral DNA digested with EcoNI (leftmost panel), BamHI and EcoRI (center panels), and StyI (rightmost panel). Probes are shown below the blots: UL46 probe (nucleotides 60388 to 60562), UL48 probe (nucleotides 65548 to 66150), GFP probe, and UL47 3′ probe (nucleotides 62352 to 62853). Sizes of DNA markers are shown to the left of each blot.
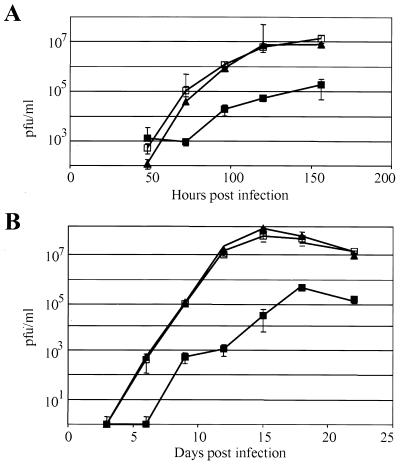
We compared the kinetics of replication for the wild-type AD169, ADsubUL47, and ADrevUL47 viruses at two input multiplicities. At a relatively high multiplicity of 2 PFU/cell (Fig. 2A), ADsubUL47 replicated to titers 100-fold lower than those for the wild-type or revertant viruses. ADsubUL47 also displayed a delay in the production of infectious particles compared to the wild-type and revertant viruses, accumulating virus beginning at 96 h postinfection (hpi). Wild-type AD169 and ADrevUL47 exhibited identical growth kinetics, indicating that the mutation was successfully repaired and that the phenotype of ADsubUL47 was due to the substitution mutation and not to another mutation in the genome. At a multiplicity of 0.001 PFU/cell (Fig. 2B), ADsubUL47 again produced yields 100-fold lower than those of the wild-type or revertant viruses at 22 days postinfection. Again, ADsubUL47 exhibited a delay in the production of infectious virus, and consequently, at earlier points in the time course the mutant yield was reduced as much as 10,000-fold from those of wild-type viruses. Wild-type AD169 and ADrevUL47 exhibited the same growth kinetics and accumulated identical yields, confirming that the mutation was repaired in the revertant.
FIG. 2.
Growth of wild-type AD169, ADsubUL47, and ADrevUL47. HFFs were infected at a multiplicity of 2 (A) or 0.001 (B) PFU/cell with AD169 (▴), ADsubUL47 (▪), or ADrevUL47 (squlo). Samples were removed at the indicated times after infection and titered by plaque assay on R47 HFFs.
Characterization of ADsubUL47 virions.
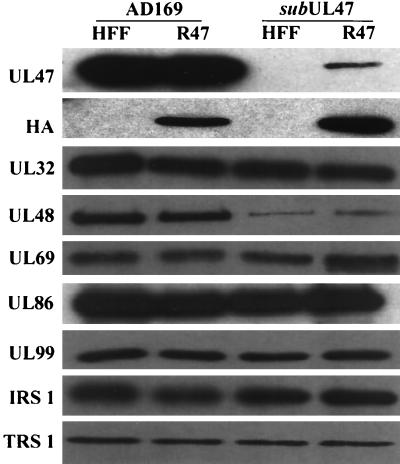
We examined the tegument composition of ADsubUL47 virions in order to determine the efficiency with which pUL47 is incorporated into ADsubUL47 virions when the mutant is propagated on R47 HFFs and to ascertain whether the loss of pUL47 affected the tegument incorporation of other HCMV proteins. Virions were isolated from HFFs or R47 HFFs infected with wild-type AD169 or ADsubUL47 and were subjected to Western blot analysis using a polyclonal serum against pUL47 (Fig. 3, UL47). pUL47 was absent from ADsubUL47 virions propagated on HFFs and was present, but at substantially reduced levels in comparison to those of the wild-type virus, when propagated on the complementing cells. This suggests that the incomplete complementation of the mutant virus on these cells may be due to an insufficient level of pUL47 incorporated into virions. Virions probed with the anti-HA antibody confirmed that the epitope-tagged pUL47 was incorporated into both wild-type and mutant virions propagated on R47 HFFs (Fig. 3, HA).
FIG. 3.
Composition of ADsubUL47 virions. HFFs or R47 HFFs were infected with wild-type AD169 or ADsubUL47 at a multiplicity of 2 PFU/cell; virions were purified from the culture medium and lysed; and equal amounts of proteins were subjected to Western blot analysis using antibodies to the indicated viral proteins or to the influenza virus HA epitope tag.
Virions were probed for a variety of tegument proteins, only one of which, pUL48, was found to be incorporated at reduced levels in the mutant virions; the level of incorporation was independent of complementation (Fig. 3, UL48). Thus, the ADsubUL47 virions are deficient in pUL47 and pUL48.
pUL48 protein levels are reduced in ADsubUL47-infected cells.
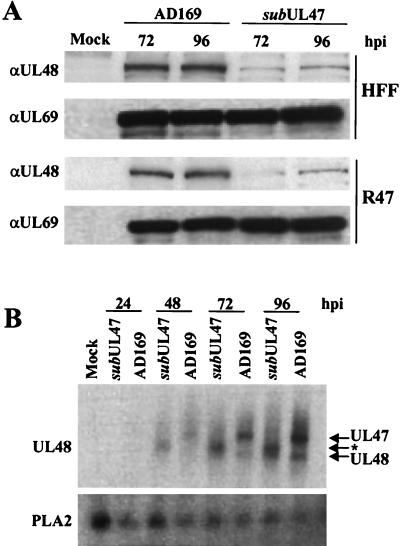
The lower levels of pUL48 present in ADsubUL47 virions led us to ask whether the decrease was due to a defect in pUL48 tegument incorporation or to a decrease in pUL48 levels within infected cells. To determine the pUL48 levels in wild-type AD169- and ADsubUL47-infected cells, lysates were prepared and subjected to Western blot analysis (Fig. 4A). Probing with a pUL48-specific monoclonal antibody revealed that pUL48 levels were significantly reduced in ADsubUL47-infected cells compared to AD169-infected cells. Levels of pUL48 were not restored by the presence of the epitope-tagged pUL47 produced in the R47 HFFs. We conclude that the lower level of pUL48 in ADsubUL47 virions is due to the accumulation of reduced pUL48 levels in mutant virus-infected cells.
FIG. 4.
Synthesis of UL48 mRNA and protein. (A) HFFs and R47 HFFs were either mock infected or infected with either wild-type AD169 or ADsubUL47 at a multiplicity of 2 PFU/cell, and protein extracts were prepared at the indicated times. Forty micrograms of protein was analyzed by Western blotting using pUL48- and pUL69-specific monoclonal antibodies. (B) RNA was isolated at various times postinfection from mock-infected cells or from cells infected at a multiplicity of 2 PFU/cell with AD169 or ADsubUL47 and was analyzed by Northern blotting using a probe to the UL48 gene. The cellular PLA2 mRNA was used as a loading control. Arrows indicate the migration of the UL47-, UL48-, and ADsubUL47-specific (*) transcripts.
We next asked if the substitution mutation had affected transcription of the UL48 gene. RNA was isolated at various times after mock infection or infection with wild-type or mutant viruses and was analyzed by Northern blotting (Fig. 4B). The UL48 probe hybridized to several bands in wild-type virus-infected cells. The largest transcript, a 10-kb RNA observed only in AD169 lanes, represents the UL47 mRNA, which traverses the UL48 ORF. ADsubUL47-infected cells do not contain this species. However, the UL48 probe also hybridizes to an RNA unique to the mutant virus, which migrates more rapidly than the UL47 mRNA. The mutant virus-specific RNA contains sequences corresponding to the 3′ domain of the UL47 mRNA (data not shown), and its size indicates that it initiates at the simian virus 40 promoter within the GFP/Puro marker cassette. The smallest transcript, a 7-kb RNA, is the UL48 mRNA. At 72 and 96 hpi, the 7-kb UL48 transcript was clearly detected in wild-type and mutant virus-infected cells, and the level of this RNA was the same for the two viruses as shown by phosphorimager quantitation (data not shown). These data demonstrate that the steady-state level of UL48 RNA was not affected by the substitution mutation. Although we have not directly demonstrated a posttranscriptional defect, it is likely that the level of pUL48 is reduced at the translational or posttranslational level.
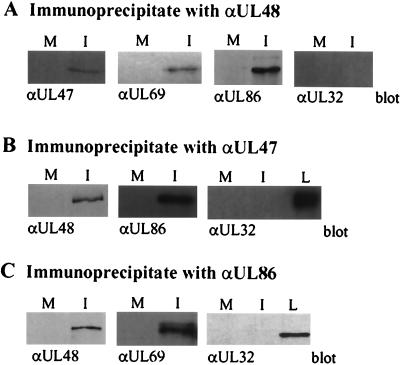
pUL47 coimmunoprecipitates with pUL48, pUL69, and pUL86 (MCP).
Given the marked reduction in the intracellular level of pUL48 in ADsubUL47-infected cells, we tested the possibility that pUL47 and pUL48 interact. We employed R47 HFFs, which express epitope-tagged pUL47, to search for interactions so that pUL47 could be detected with an HA-specific antibody. Mock- or wild-type AD169-infected cells were pulse-labeled with [35S]cysteine plus [35S]methionine, and proteins in lysates were immunoprecipitated with antibodies specific for pUL47 or pUL48. Electrophoretic analysis revealed that several infected-cell proteins coprecipitated with pUL47 (∼105, 150, and 250 kDa) and pUL48 (105 and 150 kDa) (data not shown). We hypothesized that these were virion proteins and tested candidates of the appropriate size by Western blot assays of proteins in the immunoprecipitates. pUL47 (110 kDa), pUL69 (105 kDa), and pUL86 (150 kDa), but not pUL32 (150 kDa), coimmunoprecipitated with pUL48 (Fig. 5A). The experiment was repeated by immunoprecipitating with antibodies specific for pUL47 or pUL86 (Fig. 5B and C), and pUL48 (250 kDa) was coprecipitated in each case, confirming the interactions. The inability of antibodies to pUL47 or pUL48 to coprecipitate pUL32, a very abundant tegument protein, provided evidence for the specificity of the interactions. We interpret these results to indicate that pUL47, pUL48, pUL69, pUL86, and perhaps other proteins exist in a complex late after infection. We do not yet know which specific protein-protein contacts are made within the complex. It is interesting that the level of pUL48, but not pUL69 or pUL86, is reduced in the absence of pUL47 (Fig. 3), even though the complete set of proteins can be coimmunoprecipitated (Fig. 5).
FIG. 5.
Interaction of pUL47 with pUL48, pUL69, and pUL86. R47 HFFs were mock infected or infected with wild-type AD169 at a multiplicity of 2 PFU/cell; after 72 h, protein lysates were prepared and then immunoprecipitated with antibodies against pUL48 (A), pUL47 (HA epitope) (B), or pUL86 (C), separated by SDS-polyacrylamide gel electrophoresis, and blotted to nitrocellulose membranes. Blots were probed with antibodies recognizing pUL47 (HA epitope), pUL48, pUL69, pUL82, and pUL32. M, mock-infected cells; I, HCMV-infected cells; L, lysate.
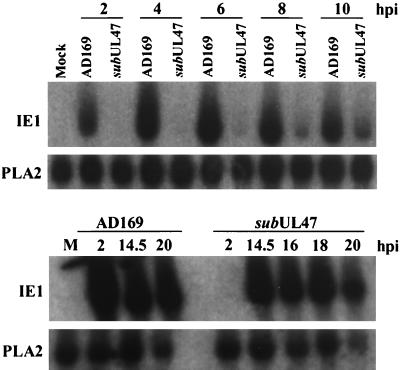
ADsubUL47 exhibits a delay in IE1 transcription.
The delayed release of virus particles from ADsubUL47-infected cells indicated that there is a defect in the mutant’s replication cycle. We examined the accumulation of an immediate-early mRNA to determine if there was a delay at the start of ADsubUL47 infection. RNA was isolated from mock-infected, wild-type AD169-infected, and ADsubUL47-infected cells at various times postinfection and subjected to Northern blot analysis. There was a significant amount of IE1 RNA present in AD169-infected cells at 2 hpi (Fig. 6, upper panel), and levels of the RNA remained high through 20 hpi (Fig. 6, lower panel). IE1 transcript was not detected in ADsubUL47-infected cells until 8 hpi (Fig. 6, upper panel), and it had accumulated to significant levels by 14.5 hpi (Fig. 6, lower panel). The ADsubUL47 genome was competent for IE1 RNA expression, but its accumulation was delayed by at least 8 h.
FIG. 6.
IE1 mRNA expression in ADsubUL47-infected cells. Cells were either mock infected or infected at a multiplicity of 2 PFU/cell with either wild-type AD169 or ADsubUL47; and RNA was harvested at various times and analyzed by Northern blotting. Blots were probed with 32P-labeled IE1 or PLA2 (loading control) cDNAs. M, mock-infected cells.
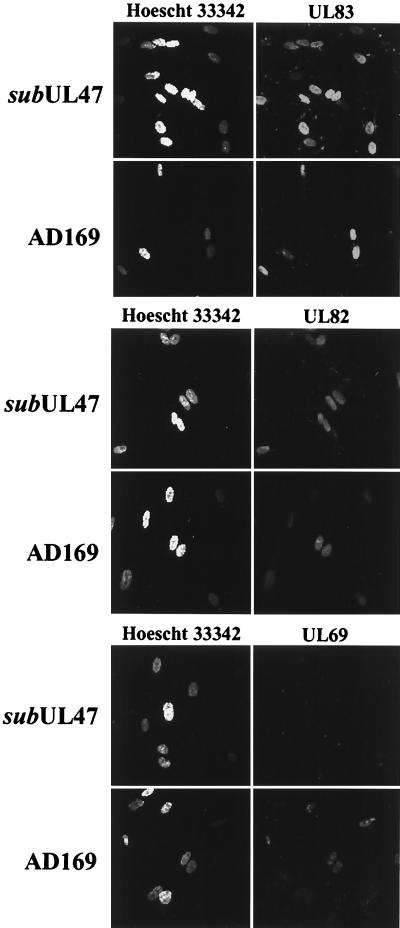
ADsubUL47 exhibits a defect subsequent to virion binding and membrane fusion.
The delay in IE1 mRNA accumulation suggested that there was a defect at a step in ADsubUL47 infection prior to expression of immediate-early genes. We examined the ability of ADsubUL47 to enter cells by monitoring the release of three virion tegument proteins into infected cells (Fig. 7). Immunofluorescence assays of AD169-infected cells demonstrated that pUL69, pUL82, and pUL83 proteins localized to the nucleus of the infected cell by 4 hpi. In ADsubUL47-infected cells, pUL82 and pUL83 localized to the nucleus within 4 hpi, but pUL69 was not detected. Consistent with the immunofluorescence data, pUL69 was detected by Western blotting in lysates from AD169-infected but not from ADsubUL47-infected cells at 4 hpi (data not shown). pUL69 is present at wild-type levels in ADsubUL47 virions (Fig. 3), and it coprecipitated with pUL47-interacting proteins (pUL48 and MCP) (Fig. 5). We suspect that pUL69 was destabilized by the loss of pUL47 and/or the reduced levels of pUL48 in ADsubUL47 particles, rendering it undetectable by Western blot analysis and immunofluorescence. Nevertheless, the abilities of pUL82 and pUL83 to rapidly localize to the nucleus after infection with ADsubUL47 argue that the particle enters the cell and is at least partially disassembled before the block imposed by the lack of pUL47.
FIG. 7.
Tegument protein release in ADsubUL47-infected cells. HFFs were plated onto coverslips and infected with wild-type AD169 at a multiplicity of 20 PFU/cell or with ADsubUL47 at a multiplicity of 5 PFU/cell. Coverslips were fixed and stained for pUL69, pUL82, or pUL83 at 4 hpi. Nuclei were visualized by staining with Hoescht 33342.
Although the ADsubUL47 virus entered permissive fibroblasts, pUL69 rapidly disappeared. pUL69 transcriptionally activates various promoters, including the MIEP (1). We reasoned that the incoming virion proteins, including pUL69, might fail to transactivate the MIEP or might transactivate it to lower levels and that this could explain the delay in accumulation of IE1 mRNA. We addressed this question by examining the level of transactivation induced by wild-type and mutant viruses on a MIEP integrated into the cellular genome of human fibroblasts that were life-extended by constitutive expression of telomerase (4). The MIEP controls the expression of the tetracycline transcriptional activator protein. Transcriptional activation was measured by quantifying the levels of tetracycline activator RNA in mock-, AD169-, and ADsubUL47-infected cells in the presence of cycloheximide. The protein synthesis inhibitor prevents the synthesis of IE1 and IE2, viral proteins that dramatically activate the MIEP. ADsubUL47 virion proteins activated the promoter by a factor of 2.6 ± 0.4, while wild-type virion proteins activated it by a factor of 2.0 ± 0.4. This experiment must be interpreted with caution given the low level of transactivation observed. Nevertheless, the ability of the mutant to activate the MIEP to the same level as wild-type virus suggests that the delay in immediate-early transcription is probably not the result of a transactivation defect of the virion proteins. This implies that early in infection, the ADsubUL47 genome is unavailable for transactivation by viral and cellular proteins.
Relative infectivity of ADsubUL47 particles.
Finally, we asked if the loss of pUL47 affected the infectivity of the mutant virions. We had noticed that more tegument proteins were delivered to cells infected with ADsubUL47 than to cells infected with wild-type AD169 at the same input multiplicity. Thus, we determined the particle-to-PFU ratio for the two viruses. Aliquots of mutant and wild-type virus stocks were removed for titering by plaque assay. Then equal volumes of the remainder of the stocks were mixed, virus particles were pelleted, and DNA was isolated and analyzed by Southern blotting. Phosphorimager analysis of these blots (Fig. 8A) was performed to quantify the relative amounts of bands diagnostic for the two viral DNAs, and thus virions, in the stock. Comparison of the amount of infectious virus present in the stocks (Fig. 8B, left panel) to the amount of viral DNA present (Fig. 8B, right panel) demonstrated that, although there was a difference of almost 100-fold in titers for the two viruses, there was only a 1.2-fold difference in the amount of viral DNA. We interpret this to mean that the ADsubUL47 mutant virus is less infectious than the wild-type virus; there is a ∼60-fold difference in their particle-to-PFU ratios.
FIG. 8.
Relative particle-to-PFU ratios. Wild-type AD169 and ADsubUL47 virus stocks were prepared, and small aliquots were titered to determine the concentration of infectious virus. Then equal volumes of the two stocks were mixed, virions were partially purified, and their DNA was extracted. (A) Viral DNA was digested with BamHI and EcoRI and subjected to a Southern blot assay using a 32P-labeled UL48-specific probe. (B) Comparison of the virus titers for AD169 and ADsubUL47 to the relative amounts of the two DNAs, determined by phosphorimager quantification of the bands visualized in panel A.
DISCUSSION
To analyze the function of pUL47, we generated a mutant virus containing a substitution mutation in the UL47 ORF (Fig. 1). Characterization of this mutant virus, ADsubUL47, demonstrated that it replicated poorly in cultured fibroblasts (Fig. 2) and displayed a substantially increased particle-to-PFU ratio (Fig. 8). The mutant virus exhibited a defect early after infection. IE1 mRNA accumulation is delayed by at least 8 h in ADsubUL47-infected HFFs (Fig. 6). The ability of ADsubUL47 to eventually produce IE1 mRNA suggests that its genome is not defective for the production of mRNA but that there is a delay in the onset of viral gene expression. The delay in IE1 expression is not due to a defect in membrane fusion or entry of the newly infecting ADsubUL47 particle, since tegument proteins reach the nucleus rapidly after infection (Fig. 7). Thus, it appears the defect lies somewhere between viral entry and the onset of immediate-early mRNA accumulation.
pUL47 coimmunoprecipitates with other tegument proteins, pUL48 and pUL69, as well as pUL86, the MCP. These coprecipitations suggest the formation of a complex late after infection in the cell and presumably in the tegument of virus particles. pUL48, a constituent of the complex, is present at significantly reduced levels in pUL47-deficient ADsubUL47 virus particles (Fig. 3) and ADsubUL47-infected cells (Fig. 4). Consequently, the tegument of ADsubUL47 particles lacks two proteins, pUL47 and pUL48, and the very early defect that we observe could be due to the lack of either or both proteins. A temperature-sensitive mutation, tsB7, in the HSV UL36 homolog of HCMV UL48 exhibits a defect in the release of viral DNA from capsids (3, 14). At the nonpermissive temperature, DNA-containing capsids accumulate at the nuclear membrane. Thus, it is possible that the function of the HCMV pUL47-containing complex is the efficient release of viral DNA from the capsid. If this is the case, pUL47, likely together with pUL48, might influence a step in the disassembly of the tegument from the capsid: it could influence proper localization of the disassembling viral particle within the cell or its movement to the nuclear pore, or it could directly influence release of viral DNA from the capsid. Work is in progress to distinguish among these possibilities.
Finally, it is noteworthy that an HSV-1 null mutant lacking its UL36 gene exhibited a defect in virion assembly (8). Capsids accumulated in the cytoplasm of infected cells, but they were not packaged into envelopes. This mutant did not display the disassembly defect observed for the temperature-sensitive UL36 allele, possibly because mutant virions contained pUL36 derived from the complementing cells, which was available to perform the very early function. Since HCMV UL48 is homologous to HSV-1 UL36, and since pUL48 accumulates to reduced levels in ADsubUL47-infected cells (Fig. 4), it is possible that part of the growth deficiency we have observed for ADsubUL47 results from a late block to assembly in addition to the early block to disassembly that we have documented.
Acknowledgments
We thank W. Gibson for the polyclonal UL86 antibody and for sharing his unpublished observation that pUL47 and pUL48 interact. We thank R. Kalejta for helpful discussions and critical reading of the manuscript.
This work was supported by a grant from the National Cancer Institute (CA82396).
REFERENCES
- 1.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (UL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batterson, W., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in the release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., G. E. Hultman, and T. Shenk. 2000. Replication of wild-type and mutant cytomegalovirus in life-extended diploid fibroblasts. J. Virol. 74:10816–10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bresnahan, W. A., and T. E. Shenk. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 97:14506–14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chee, M. S., et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125–169. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham, C., A. J. Davidson, A. R. MacLean, N. S. Taus, and J. D. Baines. 2000. Herpes simplex virus type 1 gene UL14: phenotype of a null mutant and identification of the encoded protein. J. Virol. 74:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai, P. 2000. A null mutation in UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608–11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert, M. J., S. R. Riddell, B. Plachter, and P. D. Greenberg. 1996. Cytomegalovirus selectively blocks antigen processing and presentation of its immediate-early gene product. Nature 383:720–722. [DOI] [PubMed] [Google Scholar]
- 10.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 11.Hayashi, M. L., C. Blankenship, and T. Shenk. 2000. Human cytomegalovirus UL69 protein is required for the efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. USA 97:2692–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyun, J. J., H. S. Park, K. H. Kim, and H. J. Kim. 1996. Analysis of transcripts expressed from the UL47 gene of human cytomegalovirus. Arch. Pharm. Res. 22:542–549. [DOI] [PubMed] [Google Scholar]
- 13.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405–1413. [DOI] [PubMed] [Google Scholar]
- 14.Knipe, D. M., W. Batterson, C. Nosal, B. Roizman, and A. Buchanan. 1981. Molecular genetics of herpes simplex virus. VI. Characterization of a temperature-sensitive mutant defective in the expression of all early viral gene products. J. Virol. 38:539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, B., and M. F. Stinski. 1992. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATP and AP-1 cis-acting elements. J. Virol. 66:4434–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu, M., and T. Shenk. 1999. Human cytomegalovirus UL69 protein induces cells to accumulate in the G1 phase of the cell cycle. J. Virol. 73:676–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLauchlan, J. 1997. The abundance of the herpes simplex virus type 1 UL37 tegument protein in virus particles is closely controlled. J. Gen. Virol. 78:189–194. [DOI] [PubMed] [Google Scholar]
- 18.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p.2629–2673. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 19.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowak, B., C. Sullivan, P. Sarnow, R. Thomas, F. Bricout, J. C. Nicolas, B. Fleckenstein, and A. J. Levine. 1984. Characterization of monoclonal antibodies and polyclonal immune sera directed against human cytomegalovirus virion proteins. Virology 132:325–338. [DOI] [PubMed] [Google Scholar]
- 21.Romanowski, M. J., and T. Shenk. 1997. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. J. Virol. 71:1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitz, J. B., A. G. Albright, P. R. Kinchington, and F. J. Jenkins. 1995. The UL37 protein of herpes simplex virus type 1 is associated with the tegument of purified virions. Virology 206:1055–1065. [DOI] [PubMed] [Google Scholar]
- 23.Schmolke, S., H. F. Kern, P. Drescher, G. Jahn, and B. Plachter. 1995. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J. Virol. 69:5959–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stinski, M. F. 1977. Sequence of protein synthesis in cells infected by human cytomegalovirus: early and late virus-induced polypeptides. J. Virol. 23:751–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka, M., and W. Herr. 1990. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell 60:375–386. [DOI] [PubMed] [Google Scholar]
- 26.Tullis, G. E., and T. Shenk. 2000. Efficient replication of adeno-associated virus type 2 vectors: a cis-acting element outside of the terminal repeats and a minimal size. J. Virol. 74:11511–11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood, L. J., M. K. Baxter, S. M. Plafker, and W. Gibson. 1997. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. J. Virol. 71:179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]