Abstract
U4atac snRNA forms a base-paired complex with U6atac snRNA. Both snRNAs are required for the splicing of the minor U12-dependent class of eukaryotic nuclear introns. We have developed a new genetic suppression assay to investigate the in vivo roles of several regions of U4atac snRNA in U12-dependent splicing. We show that both the stem I and stem II regions, which have been proposed to pair with U6atac snRNA, are required for in vivo splicing. Splicing activity also requires U4atac sequences in the 5′ stem–loop element that bind a 15.5 kDa protein that also binds to a similar region of U4 snRNA. In contrast, mutations in the region immediately following the stem I interaction region, as well as a deletion of the distal portion of the 3′ stem–loop element, were active for splicing. Complete deletion of the 3′ stem–loop element abolished in vivo splicing function as did a mutation of the Sm protein binding site. These results show that the in vivo sequence requirements of U4atac snRNA are similar to those described previously for U4 snRNA using in vitro assays and provide experimental support for models of the U4atac/U6atac snRNA interaction.
INTRODUCTION
The recent identification of a minor class of nuclear pre-mRNA introns which are spliced by a distinct alternative spliceosome has provided an unexpected example in which to evaluate the present models of RNA function in splicing (reviewed in 1–4). The snRNPs and their component snRNAs that are involved in splicing this minor (U12-dependent) class of introns in human cells have been shown to be functional analogs of the major (U2-dependent) intron class spliceosomal snRNPs. Thus, U11 snRNA appears to be the functional analog of U1 snRNA, U12 snRNA is the analog of U2 snRNA, U4atac snRNA is the analog of U4 snRNA and U6atac snRNA is the analog of U6 snRNA. U5 snRNA appears to function in both spliceosomes. The functional similarities of the two sets of snRNAs and snRNPs are given added support by the apparent conservation of various RNA–RNA interactions between snRNAs and between snRNAs and the pre-mRNA (1–4).
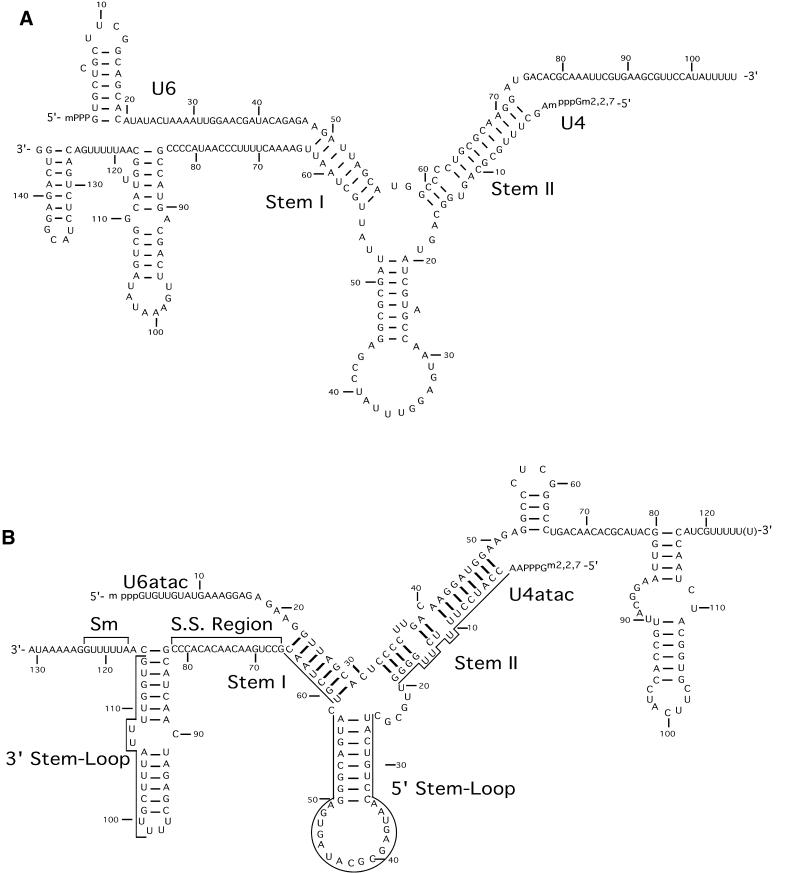
A striking example of this conservation of RNA–RNA interactions is the base pairing between U4atac and U6atac snRNAs, which closely mirrors that of the U4/U6 snRNA interaction (5). These two secondary structure models are compared in Figure 1. In both di-snRNP complexes, the snRNAs are held together by two regions of base pairing called stem I and stem II separated by a stem–loop structure called the 5′ stem–loop.
Figure 1.
(A) Secondary structure of human U4 and U6 snRNAs based on the model of Wersig and Bindereif (8). (B) Secondary structure model of human U4atac and U6atac snRNAs. Shown is the structure proposed by Padgett and Shukla (5). The regions of U4atac snRNA that are examined in this study are bracketed.
The regions of the U6 and U6atac snRNAs that pair with U4 and U4atac snRNAs, respectively, to form stem II are also the regions that form an essential RNA stem–loop element in the spliceosome following the dissociation of the U4/U6 or U4atac/U6atac di-snRNP complexes. In the course of examining the function of this intramolecular stem–loop in U6atac snRNA, we found that many U6atac mutants with alterations in this region were inactive in vivo unless they were co-expressed with U4atac snRNAs containing complementary alterations in the stem II pairing region (6,7).
This result showed both that U4atac snRNA was an essential component of the U12-dependent splicing system in vivo and that the stem II interaction was critical to its function. It also gave us a method for determining additional essential elements of U4atac snRNA by combining mutations in other regions of U4atac snRNA with mutations in the stem II region that activate a specific suppressor U6atac snRNA.
Previous investigations of mammalian U4 snRNA were carried out using either in vitro nuclear extracts or microinjected frog oocytes (8–11). The only previous in vivo investigations of U4 snRNA were carried out in budding and fission yeast (12–14). Since U12-dependent splicing has only been detected in metazoan organisms, the functional organization of U4atac snRNA must be investigated in these systems. The in vivo assay for U4atac snRNA function described here represents the first system for investigating the functional organization of a metazoan U4 or U4atac snRNA.
MATERIALS AND METHODS
Construction of U4atac expression plasmids
The starting U4atac snRNA expression plasmid has been described previously (6) and contained mutations in the stem II region to restore complementarity to the U6atac snRNA, which contained the plant-derived sequence in the intramolecular stem–loop region. For the mutations studied here, pairs of mutagenic oligonucleotides were used to prime PCRs using primers at the snRNA 5′ and 3′ ends. The products were then mixed together and reamplified with only the terminal primers. The full length products were then digested with SalI and BglII restriction enzymes and ligated into a U1 expression vector from which the U1 coding region had been excised (15). The sequences of all mutant snRNAs were confirmed by DNA sequencing.
Analysis of in vivo splicing
The P120 minigene plasmid described previously contains exons 5–8 and introns E, F and G of the human nucleolar protein P120 gene (16). The 5′ splice site of the U12-dependent intron F contained the CC5/6GG mutation, which we have shown abolishes U12-dependent splicing of this intron in vivo (17). The U11 snRNA GG6/7CC suppressor mutation was described previously (17). The U6atac snRNA GG14/15CC mutation has also been described (18). These two mutant snRNAs restore U12-dependent splicing to the P120 CC5/6GG 5′ splice site mutant. The suppressor U6atac snRNA was modified further by substituting nucleotides 30–50 of the human sequence with the Arabidopsis thaliana U6atac snRNA sequence of the intramolecular stem–loop element to give the U6atac GG14/15CC + AthSL construct (6). These constructs were combined with the U4atac snRNA suppressor construct described previously (6) or with U4atac snRNA second site mutants as described here.
Transient transfection of the P120 minigene and snRNA expression plasmids into cultured CHO cells was as described (16–18). For these experiments, 0.5 µg of P120 plasmid and 5 µg of each of the snRNA expression plasmids were added to 1 × 106 cells. Where one or more snRNA plasmids were omitted, a corresponding amount of pUC19 plasmid DNA was substituted. Total RNA was isolated from cells 36 h after transfection, reverse transcribed and PCR amplified as described (17,18). The products were analyzed by agarose gel electrophoresis. The DNA bands were visualized using ethidium bromide and photographed using a digital video camera (Kodak). Independent transfections and analyses gave substantially similar results.
RESULTS AND DISCUSSION
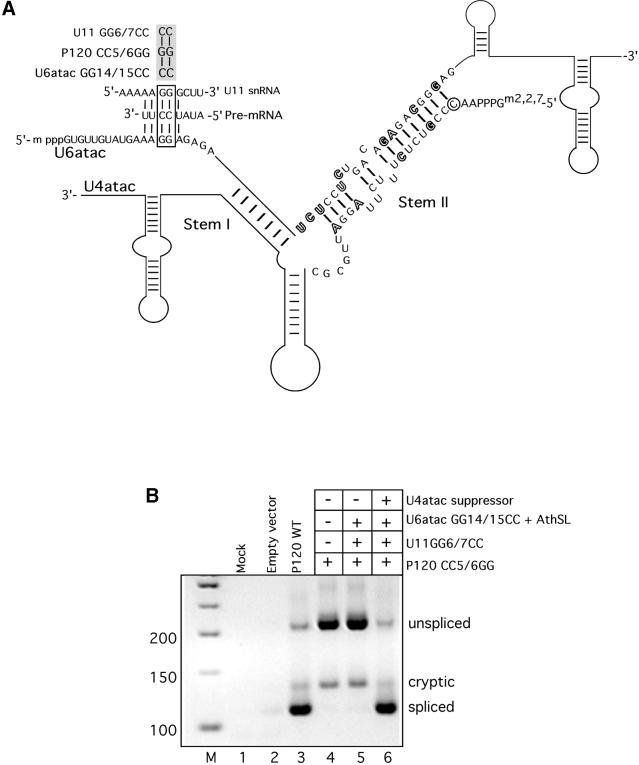
Figure 2 shows the essential features of the in vivo genetic suppression assay for U4atac snRNA. We begin with a previously described minigene derived from the human nucleolar P120 protein gene. This minigene contains three introns. The first and third are U2-dependent introns while the middle intron is a U12-dependent intron (16). The 5′ splice site of this U12-dependent intron has been mutated at two positions (CC5/6GG), which renders it inactive for U12-dependent splicing in vivo (17). Instead, the intron either remains unspliced or splices using two cryptic U2-dependent splice sites. This cryptic splicing event produces spliced exons that are 19 nt longer than the correctly spliced exons (17,18). As we have previously shown, this mutant U12-dependent 5′ splice site can be reactivated for correct splicing by co-transfection of plasmids that express U6atac and U11 snRNAs with compensatory double mutations (18).
Figure 2.
(A) Diagram of the in vivo genetic suppression assay. The sequential base pairing interactions between U11 snRNA and the pre-mRNA 5′ splice site and between U6atac snRNA and the 5′ splice site are shown. The boxed residues are those mutated in the suppression assay as shown at the top of the figure. Both the U6atac GG14/15CC and the U11 GG6/7CC mutations are required to fully suppress the effect of the 5′ splice site CC5/6GG mutation. Also shown are the nine mutations introduced by the AthSL mutant into the stem II region of U6atac snRNA and the compensatory mutations introduced into the U4atac snRNA (outlined residues). This suppressor U4atac snRNA serves as the starting construct for the mutants described in this study. (B) Products of RT–PCR amplification of RNA extracted from CHO cells transfected with the listed constructs. The RT–PCR primers are in P120 exons 6 and 7 and amplify the region of intron F in the P120 minigene. The positions of bands corresponding to unspliced RNA, RNA spliced at the normal U12-dependent splice sites (spliced) and RNA spliced at the cryptic U2-dependent splice sites (cryptic) are indicated. Lane 1 is from mock transfected cells, lane 2 is from cells transfected with the empty expression vector, lane 3 is from cells transfected with the control wild type P120 minigene construct. Lanes 4–6 are from cells transfected with the P120 CC5/6GG 5′ splice site mutant (lane 4), both the U11 and the U6atac suppressor mutants (lane 5), and the combination of the U11, U6atac and U4atac suppressor mutants (lane 6). Lane M is a 50 bp DNA size ladder.
This restoration of correct U12-dependent splicing to the 5′ splice site mutant by a compensatory mutant of U6atac snRNA allowed us to test the effects of other alterations in U6atac snRNA on its function in vivo (6,7). Among the modifications to U6atac snRNA that we tested was the replacement of the intramolecular stem–loop structure in human U6atac snRNA with the analogous region of the U6atac snRNA of the plant A.thaliana (6). The plant sequence of the stem–loop differed from the human sequence in 9 of 21 positions. This chimeric U6atac snRNA mutant (GG14/15CC + AthSL) was inactive in the in vivo suppression assay (Fig. 2B, lane 5) but, as noted above, the changes in the intramolecular stem–loop sequence also weakened the stem II interaction with the endogenous U4atac snRNA. Co-transfecting an appropriately modified U4atac snRNA construct, designed to restore the stem II base pairing, restored suppressor activity to the U6atac snRNA with the plant stem–loop (Fig. 2B, lane 6) (6). Figure 2A shows the changes in the stem II region of these U6atac and U4atac snRNA suppressor mutants. This suppressor U4atac snRNA construct is the starting mutant for the second site mutations described below.
U4atac snRNA can be divided into six sub-regions, as shown in Figure 1B, which are arranged in a similar fashion to those in U4 snRNA. From 5′ to 3′ (right to left in Fig. 1) they are: stem II, the 5′ stem–loop, stem I, the internal single stranded (S.S. in Fig. 1) region, the 3′ stem–loop (also called the central stem–loop in U4 snRNA) and the Sm protein binding region. We have tested mutations in U4atac snRNA in each of these regions for their ability to support U12-dependent splicing in vivo. Previous investigations of U4 snRNA in vertebrate and yeast systems have also explored the analogous regions of this snRNA (8,10–12,14).
Stem II
The 5′ proximal region of U4atac snRNA was suggested to base pair to U6atac snRNA on the basis of its sequence complementarity and similar position to the analogous U4 snRNA region (19). For U4 snRNA, the stem II interaction with U6 snRNA is required for splicing activity (8,10,11,14). We have shown previously that base pairing in the stem II region between U4atac and U6atac snRNAs is also essential for U12-dependent splicing in vivo (6). This region of U4atac snRNA can tolerate a large number of simultaneous changes while retaining function as long as complementarity with U6atac is maintained (6,7).
5′ Stem–loop
The region 3′ of stem II in both U4 and U4atac snRNAs is the 5′ stem–loop. U4 snRNA mutants with deletions of the 5′ stem–loop are inactive for splicing both in vivo (10,12) and in vitro (8,9). The loop region of this element in U4 snRNA contains a phylogenetically conserved AAUGAG sequence (residues 29–34) that is also found in U4atac snRNA (residues 34–39). In U4 snRNA, a triple substitution mutation in the AUG of this sequence was defective for splicing (10). Nottrott et al. (20) showed that this region of both U4 and U4atac snRNAs could bind to a 15.5 kDa U4/U6.U5 tri-snRNP protein in vitro and suggested that it might play a functional role in U4 and U4atac snRNA synthesis and/or function. These authors found that several residues in the loop portion of the 5′ stem–loop, including the AAUGAG sequence, were essential for in vitro 15.5 kDa protein binding. In particular, U4 snRNA residues U31, G32 and A33 were found to be the most important for high-affinity binding while resides A29 and A30 were less important (20). These correspond to U4atac residues A34, A35, U36, G37 and A38 (Figs 1 and 3A).
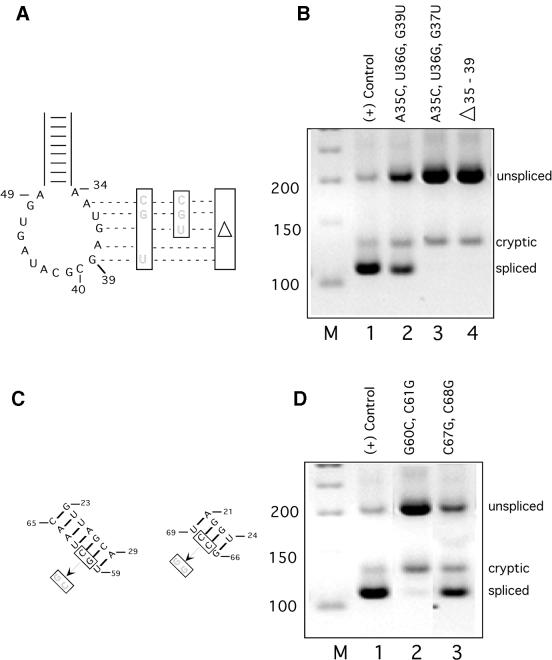
Figure 3.
(Opposite and above) Splicing phenotypes of U4atac snRNA mutants. (A) Locations of the mutations in the loop region of the 5′ stem–loop element. The first two are three residue substitution mutants and the last mutant is a five residue deletion. (B) RT–PCR products from cells transfected with the indicated U4atac snRNA mutants. Lane 1 in this and subsequent gels is a positive suppression control using the constructs shown in lane 6 of Figure 2B. The experimental lanes in this and subsequent gels are from cells transfected with the P120 CC5/6GG mutant, the GG6/7CC mutant of U11 snRNA, the GG14/15CC + AthSL stem II mutant of U6atac snRNA and the indicated U4atac snRNA mutants. Lanes labeled M are 50 bp DNA size markers. (C) Locations of the mutations in the stem I pairing region of U4atac snRNA. The G60C, C61G mutant would be predicted to disrupt the stem I pairing with U6atac snRNA in the model that we recently proposed (5). The C67G, C68G mutant would be predicted to disrupt the stem I pairing with U6atac snRNA in the model originally proposed by Tarn and Steitz (19). (D) RT–PCR products from cells transfected with the indicated U4atac snRNA mutants. (E) Locations of mutations in the single stranded region of U4atac snRNA. This region is shown base paired to U6atac snRNA according to the model proposed by Jakab et al. (22). (F) RT–PCR products from cells transfected with the indicated U4atac snRNA mutants. (G) Locations of mutations in the 3′ stem–loop element and Sm binding site of U4atac snRNA. Two deletions of the 3′ stem–loop are shown. The first removes the distal portion of the element by deleting residues 92–105 and joining residue 91 to 106 to create a 5 nt terminal loop. The second removes the entire element by deleting residues 84–114 and joining residue 83 to 115. Also shown is the 3 nt mutation of the Sm protein binding site (Sm–). (H) RT–PCR products from cells transfected with the indicated U4atac snRNA mutants.
We made three mutations in this region of U4atac snRNA (Fig. 3A). These were (i) a A35C, U36G, G39U triple substitution mutant, (ii) a A35C, U36G, G37U triple substitution mutant and (iii) a deletion of residues 35–39. When these mutant snRNAs were tested in the in vivo suppression assay, the A35C, U36G, G39U triple substitution mutant showed partial activity (Fig. 3B, lane 2) while the A35C, U36G, G37U triple substitution mutant (lane 3) and the deletion mutant (lane 4) failed to show suppression activity (i.e. the mutant U4atac snRNAs were inactive).
The partial activity of the A35, U36 and G39 triple mutant was unexpected. These mutations were chosen because the analogous U4 triple mutant, A30C, U31G, G34U, was shown by Nottrott et al. (20) to be incapable of binding the 15.5 kDa protein in vitro and to be inactive in a splicing inhibition assay. Possible explanations for the partial activity of the U4atac mutant are that (i) the wild type U4atac sequence has a 5-fold higher affinity for the 15.5 kDa protein than the U4 sequence (20), (ii) only U36 of the most critical UGA sequence was mutated and (iii) the base pairing of G39 in U4atac and G34 in U4 are different and, in fact, the G39U mutation in U4atac was shown to be competent for 15.5 kDa protein binding (20). In addition, the in vitro binding data were obtained using only a fragment of U4 snRNA and the 15.5 kDa protein alone while the in vivo assay used here includes the entire U4atac snRNA, its interacting U6atac snRNA and the full set of snRNP proteins, any or all of which might affect the affinity of the 15.5 kDa protein for the mutant snRNA.
We conclude that the results of the mutations of the U4atac 5′ stem–loop show that the loop residues of this region of U4atac snRNA are required for splicing activity in vivo. They also suggest that the basis of this requirement is the binding of the 15.5 kDa protein and that this protein is required for in vivo splicing of U12-dependent introns.
Stem I
The stem I region of U4atac snRNA has been proposed to base pair to U6atac snRNA. When the original sequence of U4atac snRNA and the proposed U4atac/U6atac base paired structure were published, an error was made in the sequence of U4atac in this region such that the G60 and C61 residues were transposed (19). We have recently shown that the correction of this mistake allows the formation of a different stem I structure that involves more base pairs and is more similar to the stem I structure of U4/U6 (Fig. 1) (5). We have tested this newly proposed model by mutating U4atac residues that are involved in stem I in either the original model (Fig. 3C, right) or our new model (Fig. 3C, left). To test our model, we made the double G60C, C61G mutant, thus creating the originally published sequence. As shown in Figure 3C, left, these mutations would be expected to destabilize the stem I interaction in our new model but not in the original model. On the other hand, as a test of the original U4atac/U6atac stem I pairing model, we made the double C67G, C68G mutant. As shown in Figure 3C, right, these mutations would be expected to destabilize the original model’s stem I but not the stem I of the new model. These constructs were then tested for in vivo suppression activity. As shown in Figure 3D, the G60C, C61G mutant, which disrupted the newly proposed stem I structure, was inactive, while the G67C, G68C mutant, which disrupted the originally proposed stem I structure, was still largely functional. These results provide support for our proposed model of the U4atac/U6atac stem I interaction (5). They also provide genetic evidence in support of the func tional requirement of the U4atac/U6atac stem I interaction in U12-dependent splicing.
Previous studies of mutants in the U4 snRNA stem I region have reached mixed conclusions about its requirement in U2-dependent splicing. A deletion of this region in human U4 snRNA was still able to reconstitute splicing in vitro (9) while a substitution mutant was inactive for splicing in microinjected oocytes (10). Yeast U4 snRNA was found to be tolerant of mutations in the stem I region in vivo (14). Nevertheless, the clear conservation of the base pairing potential for this interaction in both splicing systems and in all organisms argues strongly for a functional role in splicing as we found in U4atac snRNA. A formal genetic proof that this region of U4atac snRNA is essential because of base pairing with U6atac snRNA would require the restoration of function with compensatory mutations in U6atac. However, the region of U6atac snRNA that is proposed to be in stem I is both highly conserved and is also proposed to base pair with a region of U12 snRNA at a later stage of spliceosome assembly (19,21).
Single stranded region
The region of U4 snRNA between stem I and the 3′ stem–loop element is tolerant of mutations, suggesting that it is not an essential region of U4 snRNA (11,14). However, Jakab et al. (22) have suggested, based on phylogenetic conservation, that a third interaction between U4 and U6 might involve this region. They also suggested that a similar interaction between human U4atac residues 72–78 and the 5′ end of U6atac could occur (Fig. 3E). To test for the possible functional significance of this interaction as well as a requirement for specific sequences in the single stranded region, we constructed three mutants within this region, as shown in Figure 3E, and tested them in vivo. All three substitution mutants showed nearly wild type suppression activity (Fig. 3F). Each of these mutants would disrupt 3 of the 7 bp between U4atac and U6atac in the model of Jakab et al. (22). In addition, only 4 bp can be modeled between the Drosophila U4atac and U6atac snRNAs described recently (23,24). Therefore, while our mutants in this region showed no impairment of in vivo splicing activity, the possibility remains that only 4 bp are sufficient for this interaction.
Comparison of human and Drosophila U4atac snRNAs shows conservation in this region of the sequence 5′-ACACACCC-3′ (human residues 75–82; Drosophila residues 74–81). However, as shown above, mutation of either human residues 76–79 or 76–80 failed to inactivate U4atac snRNA function in vivo. These results suggest that the pairing proposed by Jakab et al. (22), if it exists at all, is probably not essential for in vivo splicing and that this region of U4atac, similar to the analogous region of U4, is not involved in critical sequence specific interactions.
Recently, Johnson and Abelson (25) described crosslinks between this region of yeast U4 snRNA and the pre-mRNA 5′ splice site that could be detected in an in vitro splicing system. These authors did not investigate the sequence specificity of their crosslinks, leaving open the possibility that these RNAs are in close proximity in the spliceosome but are not involved in a specific interaction. Futhermore, these crosslinks were detected in an artificial trans-splicing in vitro assay rather than a cis-splicing system. Earlier investigations of mutations of yeast U4 snRNA in this region did not affect splicing (14). The yeast U4 and human U4 and U4atac snRNA sequences in this region are too divergent to align unambiguously so it is unclear whether our mutations in this region are informative with respect to this interaction. Nevertheless, the data of Johnson and Abelson (25) raise the possibility that there may be functional interactions involving this region that are not indicated by the mutational data on either U4 or U4atac snRNAs.
3′ Stem–loop
Next, we examined the role of the 3′ stem–loop element of U4atac snRNA. Also called the central stem–loop in U4 snRNA, this element immediately precedes the Sm protein binding site in both U4 and U4atac snRNAs. Complete deletion of this stem–loop element in U4 leads to a loss of in vivo activity, suggesting that it is required for U4 snRNA function (10,12) although it appears to be dispensable for function in vitro (8,9). Furthermore, point mutations were uniformly functional when assayed in vivo (14). We investigated the requirement for this element in U4atac by deleting either the distal portion (residues 92–105) or the entire stem–loop (residues 84–114) (Fig. 3G). Deletion of the distal portion preserved in vivo suppressor activity while deletion of the entire element led to a defective U4atac snRNA molecule (Fig. 3H, lanes 2 and 3). The activity of the distal deletion suggests that this region of the element is not involved in any essential interactions with proteins or RNAs. The defect in the complete deletion of the element could be due to its function as part of the Sm protein binding signal. Sm protein binding sites are usually flanked by either one or two stem–loop elements (26).
Sm protein binding site
Both U4 and U4atac snRNAs have an Sm protein binding site at the 3′ end of the molecule. Investigations of the requirement for the Sm binding site in U4 snRNA have produced conflicting results. In some cases, there was a clear requirement for this element for function (10,14) while another report suggested that it was not essential for splicing activity in an in vitro reconstitution assay (9).
We tested the role of the U4atac snRNA Sm protein binding site in our in vivo suppression assay. A triple mutation in this element (Sm–) (Fig. 3G) produced a U4atac snRNA that was almost completely inactive (Fig. 3H, lane 4). This indicates that the Sm binding site plays an essential role in the synthesis and/or activity of U4atac snRNA. Since the Sm protein binding site has been shown to be required for the re-importation of U snRNPs to the nucleus (27,28), it is possible that the mutant U4atac snRNA studied here was inactive due to a defect in transport to the nucleus.
CONCLUSIONS
The results presented here show that, although U4 and U4atac snRNAs are highly diverged at the primary sequence level, they are organized into surprisingly similar functional domains. In particular, the stem I and stem II interactions with U6atac snRNA appear to be as predicted on the basis of similarity to the U4/U6 interactions (5,19). In addition, we have shown an in vivo functional requirement for the binding sites of the 15.5 kDa (20) and Sm proteins. In contrast, we did not find a functional requirement for predicted U6atac snRNA base pairing interactions with the single stranded region of U4atac snRNA (22).
This similarity of functional domains has become the dominant theme in comparisons of the minor U12-dependent splicing system with the major U2-dependent system. With few exceptions, all of the known RNA–RNA interactions involved in U2-dependent splicing have identifiable counterparts in the U12-dependent spliceosomal system. Many of these were predicted solely on the basis of comparisons of the RNA sequences of the minor snRNAs (5,19,29) and have gone on to be established experimentally (16–19,21,30,31). Nevertheless, these two splicing systems have been independent since at least the time of the divergence of plants and animals more than one billion years ago (32) and perhaps much longer (33). That these similar RNA–RNA interactions have persisted over this long period suggests that they are part of the fundamental mechanism of spliceosomal splicing.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Meredith Miller for assistance in the construction of some of the mutants described here and Kelly Emmett for assistance with the transfection assays. This work was supported by grants from the National Institutes of Health.
REFERENCES
- 1.Tarn W.Y. and Steitz,J.A. (1997) Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem. Sci., 22, 132–137. [DOI] [PubMed] [Google Scholar]
- 2.Nilsen T.W. (1998) RNA–RNA interactions in nuclear pre-mRNA splicing. In Simons,R.W. and Grunberg-Manago,M. (eds), RNA Structure and Function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 279–307.
- 3.Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosome. In Gestland,R.F., Cech,T. and Atkins,J.F. (eds), The RNA World II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- 4.Wu Q. and Krainer,A.R. (1999) AT-AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol. Cell. Biol., 19, 3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Padgett R.A. and Shukla,G.C. (2002) A revised model for U4atac/U6atac snRNA base pairing. RNA, 8, 125–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shukla G.C. and Padgett,R.A. (1999) Conservation of functional features of U6atac and U12 snRNAs between vertebrates and higher plants. RNA, 5, 525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shukla G.C. and Padgett,R.A. (2001) The intramolecular stem–loop structure of U6 snRNA can functionally replace the U6atac snRNA stem–loop. RNA, 7, 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wersig C. and Bindereif,A. (1990) Conserved domains of human U4 snRNA required for snRNP and spliceosome assembly. Nucleic Acids Res., 18, 6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wersig C. and Bindereif,A. (1992) Reconstitution of functional mammalian U4 small nuclear ribonucleoprotein: Sm protein binding is not essential for splicing in vitro. Mol. Cell. Biol., 12, 1460–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vankan P., McGuigan,C. and Mattaj,I.W. (1990) Domains of U4 and U6 snRNAs required for snRNP assembly and splicing complementation in Xenopus oocytes. EMBO J., 9, 3397–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vankan P., McGuigan,C. and Mattaj,I.W. (1992) Roles of U4 and U6 snRNAs in the assembly of splicing complexes. EMBO J., 11, 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bordonne R., Banroques,J., Abelson,J. and Guthrie,C. (1990) Domains of yeast U4 spliceosomal RNA required for PRP4 protein binding, snRNP–snRNP interactions, and pre-mRNA splicing in vivo. Genes Dev., 4, 1185–1196. [DOI] [PubMed] [Google Scholar]
- 13.Dandekar T. and Tollervey,D. (1992) Mutational analysis of Schizosaccharomyces pombe U4 snRNA by plasmid exchange. Yeast, 8, 647–653. [DOI] [PubMed] [Google Scholar]
- 14.Hu J., Xu,D., Schappert,K., Xu,Y. and Friesen,J.D. (1995) Mutational analysis of Saccharomyces cerevisiae U4 small nuclear RNA identifies functionally important domains. Mol. Cell. Biol., 15, 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bond U.M., Yario,T.A. and Steitz,J.A. (1991) Multiple processing-defective mutations in a mammalian histone pre-mRNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev., 5, 1709–1722. [DOI] [PubMed] [Google Scholar]
- 16.Hall S.L. and Padgett,R.A. (1996) Requirement of U12 snRNA for the in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science, 271, 1716–1718. [DOI] [PubMed] [Google Scholar]
- 17.Kolossova I. and Padgett,R.A. (1997) U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) introns. RNA, 3, 227–233. [PMC free article] [PubMed] [Google Scholar]
- 18.Incorvaia R. and Padgett,R.A. (1998) Base pairing with U6atac snRNA is required for 5′ splice site activation of U12-dependent introns in vivo. RNA, 4, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarn W.Y. and Steitz,J.A. (1996) Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science, 273, 1824–1832. [DOI] [PubMed] [Google Scholar]
- 20.Nottrott S., Hartmuth,K., Fabrizio,P., Urlaub,H., Vidovic,I., Ficner,R. and Luhrmann,R. (1999) Functional interaction of a novel 15.5 kD [U4/U6.U5] tri-snRNP protein with the 5′ stem–loop of U4 snRNA. EMBO J., 18, 6119–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frilander M.J. and Steitz,J.A. (2001) Dynamic exchanges of RNA interactions leading to catalytic core formation in the U12-dependent spliceosome. Mol. Cell, 7, 217–226. [DOI] [PubMed] [Google Scholar]
- 22.Jakab G., Mougin,A., Kis,M., Pollak,T., Antal,M., Branlant,C. and Solymosy,F. (1997) Chlamydomonas U2, U4 and U6 snRNAs. An evolutionary conserved putative third interaction between U4 and U6 snRNAs which has a counterpart in the U4atac-U6atac snRNA duplex. Biochimie, 79, 387–395. [DOI] [PubMed] [Google Scholar]
- 23.Mount S.M. and Salz,H.K. (2000) Pre-messenger RNA processing factors in the Drosophila genome J. Cell Biol., 150, F37–F44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otake L.R., Scamborova,P., Hashimoto,C. and Steitz,J.A. (2002) The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila. Mol. Cell, 9, 439–446. [DOI] [PubMed] [Google Scholar]
- 25.Johnson T.L. and Abelson,J. (2001) Characterization of U4 and U6 interactions with the 5′ splice site using a Saccharomyces cerevisiaein vitro trans-splicing system. Genes Dev., 15, 1957–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy R. and Busch,H. (1988) Small nuclear RNAs: RNA sequences, structure, and modifications. In Birnstiel,M.L. (ed.), Structure and Function of the Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer-Verlag, Berlin, pp. 1–37.
- 27.Mattaj I.W. (1986) Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell, 46, 905–911. [DOI] [PubMed] [Google Scholar]
- 28.Mattaj I.W. and De Robertis,E.M. (1985) Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell, 40, 111–118. [DOI] [PubMed] [Google Scholar]
- 29.Hall S.L. and Padgett,R.A. (1994) Conserved sequences in a class of rare eukaryotic introns with non-consensus splice sites. J. Mol. Biol., 239, 357–365. [DOI] [PubMed] [Google Scholar]
- 30.Tarn W.Y. and Steitz,J.A. (1996) A novel spliceosome containing U11, U12 and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell, 84, 801–811. [DOI] [PubMed] [Google Scholar]
- 31.Yu Y.T. and Steitz,J.A. (1997) Site-specific crosslinking of mammalian U11 and U6atac to the 5′ splice site of an AT-AC intron. Proc. Natl Acad. Sci. USA, 94, 6030–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H.J., Gaubier-Comella,P., Delseny,M., Grellet,F., Van Montagu,M. and Rouze,R. (1996) Non-canonical introns are at least 109 years old. Nature Genet., 14, 383–384. [DOI] [PubMed] [Google Scholar]
- 33.Burge C.B., Padgett,R.A. and Sharp,P.A. (1998) Evolutionary fates and origins of U12-type introns. Mol. Cell, 2, 773–785. [DOI] [PubMed] [Google Scholar]