Abstract
Comparisons of expression levels across different cDNA microarray experiments are easier when a common reference is co-hybridized to every microarray. Often this reference consists of one experimental control sample, a pool of cell lines or a mix of all samples to be analyzed. We have developed an alternative common reference consisting of a mix of the products that are spotted on the array. Pooling part of the cDNA PCR products before they are printed and their subsequent amplification towards either sense or antisense cRNA provides an excellent common reference. Our results show that this reference yields a reproducible hybridization signal in 99.5% of the cDNA probes spotted on the array. Accordingly, a ratio can be calculated for every spot, and expression levels across different hybridizations can be compared. In dye-swap experiments this reference shows no significant ratio differences, with 95% of the spots within an interval of ±0.2-fold change. The described method can be used in hybridizations with both amplified and non-amplified targets, is time saving and provides a constant batch of common reference that lasts for thousands of hybridizations.
INTRODUCTION
cDNA microarraying is currently widely used to assess differential gene expression (1). Simultaneous hybridization of two samples labeled with different fluorescent dyes provides an intensity ratio that reflects the relative mRNA levels (2). Though adequate for comparison of two samples, assessment of expression levels across multiple samples, for example in a time series, becomes complicated. For multi-array comparisons, hybridization of a common reference sample simultaneously with each experimental sample is recommended (3,4). Initially one sample, e.g. mRNA originating from one cell line or time point zero, was used as a common reference (5–7). A disadvantage of this approach is that the control sample does not provide a signal in all spots and, since for these no ratio can be calculated, they are usually disregarded in the analysis. Sometimes these gaps are filled in by applying a program that is designed to estimate missing values (8). However, to avoid using an estimation program or other alternatives, the ideal reference should ensure consistent and non-zero values for all probes on the array, guaranteeing that no information is lost when the ratios are calculated (4).
A reference consisting of a labeled PCR product from a part of the vector that all the spotted probes have in common, as has been described for filter hybridizations, meets this criterion (9). However, it will not compete with the target cDNA for hybridization to the specific sequence of the probe. Consequently, the ratios obtained from such a hybridization may not always reflect the amount of RNA present in the experimental sample (e.g. saturated spots). Another described common reference consists of a pool of RNA originating from different cell lines (3,10–12). This approaches the ideal situation, but cell culturing is very time and space consuming. In addition, gene expression in the pooled cell lines may not represent all genes present on the array and it may change over time under even slightly different growth conditions and other variables like passage number. Furthermore, it is difficult to repeatedly quantify and pool large amounts of RNAs from multiple sources in a reliable and reproducible way. Bergstrom et al. used such a common reference and reported a coverage of 90% of the array by the reference (13). An alternative to this method, which does provide signal in all spots that need to be analyzed, is pooling part of the RNA of all the experimental samples (e.g. cell lines or biopsies) which will be used in that particular experiment (4,14). The disadvantage here is that this approach is experiment specific and each time a new experiment is performed, a new reference pool has to be made. Furthermore, if the amount of experimental samples is limiting, it is not possible to use part of it for the common reference and if one wants to study individual samples (e.g. new incoming patients), there is no reference sample present.
The experiments presented here demonstrate the use of a common reference for cDNA microarrays consisting of a mix of all probes spotted on the array. The PCR reference is made by pooling a fraction of all amplified probes before they are printed. Single-stranded products are synthesized in a subsequent in vitro transcription reaction and the product is labeled in parallel with the experimental target. The method can be used in hybridizations with both amplified and non-amplified targets, ensures non-zero values in almost all spots, is time saving, very cheap and provides a consistent batch of common reference that can be used for thousands of hybridizations.
MATERIALS AND METHODS
Probe preparation
A sub-library of the human sequence-verified 40 K I.M.A.G.E. cDNA clones from Research Genetics (15) was prepared with a re-arraying robot (Genomic Solutions). This sub-library consisted of forty-eight 96-well plates containing muscle-related genes and ESTs. cDNAs were PCR amplified with an M13 forward primer containing an SP6 tail (SP6M13F, Table 1) and an M13 reverse primer containing an extra tail sequence (B1RRV, Table 1). The 100 µl PCR mix contained 50 mM Tris–HCl, pH 9.2 (25°C), 16 mM (NH4)2SO4, 2.25 mM MgCl2, 2.0% (v/v) DMSO, 0.2 µM each primer, 125 µM each dNTP and 5 U Taq polymerase. Bacterial cell cultures were added to the PCR mixture using a 96-pin replicator (Genetix). Samples were incubated for 5 min at 95°C to lyse the bacteria. Amplification was performed by applying 35 PCR cycles consisting of 45 s at 94°C, 1 min at 56°C and 3 min 30 s at 68°C, followed by a final incubation at 68°C for 10 min. Prior to printing, all PCR products were purified using Sephadex G-50 (Sigma) in 96-well format (Millipore) according to the manufacturer’s recommendations. The purified PCR products were concentrated in a vacuum centrifuge (Savant). Dried products were resuspended in 15 µl of Milli-Q water overnight on a shaker at room temperature and 15 µl of 100% DMSO was added to obtain a final concentration of 50% DMSO. The products in the 96-well plates were transferred to twelve 384-well plates.
Table 1. Primer sequences.
| Primer name | Sequence, 5′→3′ |
|---|---|
| Sp6M13F | ATT TAG GTG ACA CTA TAG CTG CAA GGC GAT TAA GTT GGG TAA C |
| B1RRV | ATC ATC ATC ATC GTG AGC GGA TAA CAA TTT CAC ACA GGA AAC AGC |
| GAPDH_3_5 | TAT GAT GAC ATC AAG AAG GTG GTG |
| GAPDH_3_3 | TCT TAC TCC TTG GAG GCC ATG T |
| GAPDH_M_5 | CAG CAA TGC ATC CTG CAC CAC C |
| GAPDH_M_3 | ACA CGG AAG GCC ATG CCA GTG |
| GAPDH_5_5 | GGT GAA GGT CGG TGT GAA CGG A |
| GAPDH_5_3 | CCA TCA CAA ACA TGG GGG CAT C |
| ACTIN_3_5 | ATG AAG TGT GAC GTT GAC ATC CG |
| ACTIN_3_3 | TTG CGG TGC ACG ATG GAG GG |
| ACTIN_M_5 | TCA CCC ACA CTG TGC CCA TC |
| ACTIN_M_3 | CAT AGC TCT TCT CCA GGG AGG A |
| ACTIN_5_5 | GCT GTA TTC CCC TCC ATC GTG G |
| ACTIN_5_3 | GTC TCA AAC ATG ATC TGG GTC AT |
Preparation of glass slides and prehybridization
Cleaning of the glass slides (cut edges, frosted end, 3 × 1 inch; Menzel) and coating with poly-l-lysine (Sigma) was done as described previously (5). Probes were spotted in duplicate with a 417 arrayer (Genetic Microsystems). DNA was crosslinked by UV irradiation at 65 mJ/cm2 (Stratalinker model 1800 UV Illuminator, Stratagene).
To prevent non-specific hybridization, the slides were incubated with 100 µl of prehybridization solution [400 ng/µl yeast tRNA (Roche), 400 ng/µl poly(A) RNA (Sigma), 400 ng/µl herring sperm DNA (Gibco BRL), 100 ng/µl human Cot1 DNA (Gibco BRL), 40 ng/µl vector blocking mix (DNA sequences of the multiple cloning site of all the vectors that have been used in this subset), 5× Denhardt’s solution, 3.2× SSC and 0.4% SDS] at 65°C for 30 min. Prior to hybridization, the slide containing the prehybridization mixture was incubated for 2 min at 80°C to denature the spotted DNA. After prehybridization, the slides were washed twice in 2× SSC for 5 min at room temperature and dehydrated with a series of 70, 90 and 100% ethanol, for 1 min each, respectively.
Common reference
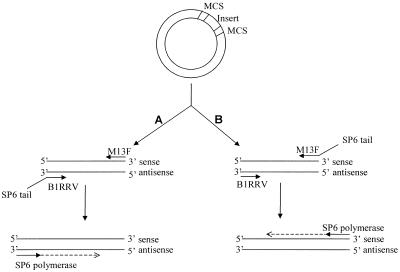
Before purification, 4 µl of each PCR product was pooled to prepare the common reference mix. Purification was performed by phenol extraction in a phase lock heavy 50 ml tube (Eppendorf). Instead of chloroform, the less toxic dichloromethane was used. After extraction, the aqueous layer was transferred to a fresh tube and purified and concentrated by ethanol precipitation. Antisense cRNA transcripts were generated using the Ampliscribe Sp6 High Yield Transcription kit (Epicentre), starting with 1 µg of pooled PCR product (Fig. 1). In addition to the protocol, 1 µl of RNasin (Fermentas) was added and the reaction was incubated at 42°C for 3 h. The generated cRNA was washed three times with 450 µl of diethylpyrocarbonate-treated water using a Microcon-100 column (Millipore). cRNA (750 ng) was reverse transcribed with random hexamers, and labeled through incorporation of Renaissance cyanine 5-dUTP (Cy5) or Renaissance cyanine 3-dUTP (Cy3) (NEN) according to the protocols of Ross et al. (12) with the following modifications: 8 µg of random hexamer primers were used in the reaction and before incubation at 42°C the mixture was incubated at room temperature for 10 min.
Figure 1.
Preparation of the common reference. Pathway A shows the generation of reference sense cRNA using an SP6 promoter sequence attached to the reverse primer to be used in combination with a non- amplified target. Pathway B shows the generation of reference antisense cRNA using an SP6 promoter sequence attached to the forward primer to be used in combination with an amplified target. When using cDNA inserts that are all cloned in a single vector with RNA polymerase sequences flanking the multiple cloning site (MCS), addition of an SP6 tail is not necessary.
Target preparation
Human fibroblast cultures were grown in DMEM without phenol red (Gibco BRL) supplemented with 1% glucose, 2% glutamax, 100 U/ml penicillin, 100 µg/ml streptomycin and 10% heat-inactivated fetal bovine serum (Gibco BRL). Cells were collected at ∼80% confluence and total RNA isolation was performed using the RNeasy Midi kit (Qiagen). During isolation a DNase treatment was performed with the Qiagen RNase-free DNase set. The eluate was precipitated and an ethanol wash was performed. Total RNA (8 µg/reaction) was amplified as described (16), with the following modification: the double-stranded cDNA reaction was stopped and the remaining RNA in the mixture degraded by addition of 7.5 µl of 1 M NaOH, 2 mM EDTA and incubation at 65°C for 10 min. This method was based on the original protocol of Van Gelder et al. (17). Of the obtained cRNA, 1.5 µg was reverse transcribed and labeled as described above.
Purification and hybridization of labeled product
The labeled reference cDNA and labeled target cDNA were pooled, purified and hybridized according to the protocol described (12). Instead of a small volume, the purified product was resuspended in a total volume of 100 µl of hybridization solution that contained an additional 20 ng/µl vector blocking mix (DNA sequences of all the vectors that have been used in this subset). Slides were hybridized in a hybridization station (Genomic Solutions). Slides were preheated for 5 min at 65°C prior to hybridization at 65°C for 13–15 h. After hybridization, each slide was washed in the hybridization station with 2× SSC, 0.1% SDS for five cycles at 30°C, 1× SSC for five cycles at 30°C, 0.2× SSC for three cycles at 25°C, 0.2× SSC for one cycle at 65°C and 0.2× SSC for two cycles at 25°C. Each cycle lasted 1 min and comprised 20 s flow time and 40 s hold. Slides were dehydrated by incubation in 70, 90 and 100% ethanol for 1 min.
Scanning and statistical analysis
Following hybridization, slides were scanned in a Genetic Microsystems 418 scanner to acquire separate images for Cy3 and Cy5. The resulting images were quantified with the software GenePix Pro 3.0.6.81 (Axon Instruments Inc.). Further analysis was performed with the software package SPSS 10.7 (SPSS Inc.). For comparison of different slides, Cy5/Cy3 ratios were normalized to a mean of one using only the spots that gave significant signal (defined as higher than the local median background plus four standard deviations) in each pair (12). To avoid outliers, the standard deviation of the background was calculated per block (32 × 36 spots) by using all median background intensities.
Comparison of the reference intensities in the different hybridizations was performed after variance stabilization and normalization (VSN) of the background-corrected intensities (18).
POPO™-3 DNA staining
POPO™-3 DNA staining was performed to determine the number of spots that contained PCR products (Molecular Probes). The microarray slides were hybridized with 100 µl of POPO™-3 iodide solution (5000× dilution of 1 mM stock in PBS) at room temperature for 30 min in the dark. The slides were washed twice for 5 min in 2× SSC at room temperature, dehydrated in an ethanol series (70, 90 and 100%) and subsequently scanned in the Cy3 channel of the Genetic Microsystems 418 scanner. A spot was regarded as significant if the intensity of the spot was higher than the median local background plus two standard deviations calculated per block.
RESULTS AND DISCUSSION
Preparation of common reference
The aim of the present study was to develop a common reference that enables comparison of expression levels in multiple microarray hybridizations. An important prerequisite for a common reference is to obtain a signal for each spot. We accomplished this by mixing all arrayed cDNAs and labeling them in one color. We designed the common reference such that it contains the same strand as the test sample: antisense when the sample has been amplified (cRNA) or sense when the sample is directly labeled (mRNA) (Fig. 1). In this way, no binding can take place between the reference and the target in the hybridization reaction, which increases the sensitivity of the experiment. Furthermore, a desired competition is created between sample and reference for binding to the array, resulting in a ratio that reflects the true relative amounts of RNA present in both samples. Since the cDNA inserts we have used were not cloned in a common vector, it was not possible to perform in vitro transcription on the PCR products directly using an internal vector-derived RNA polymerase promoter sequence. We bypassed this problem by adding an SP6 RNA polymerase promoter sequence to either the forward (generating antisense cRNA; see experiments below) or the reverse (generating sense cRNA) primer (Fig. 1). Here the SP6 promoter sequence was selected because the T7 sequence was already present in most vectors, but in both orientations, therefore sometimes generating sense and sometimes generating antisense product. The ability to produce an array-specific common reference depends on the availability of the cDNA clones spotted on the array. To facilitate the use of this approach, array producers could thus make a common reference together with the array or make the clones available to the users.
Quality check for cDNA synthesis and cRNA integrity
To evaluate the common reference, an experiment was performed with six different Cy3-labeled complex cRNA mixtures co-hybridized with Cy5-labeled common reference. The cDNA arrays used contained 4608 genes, spotted in duplicate. These hybridizations were repeated performing a dye-swap labeling of the samples, resulting in a total of 12 hybridizations.
To check the quality of the target, three different parts from the β-actin and GAPDH genes (5′, middle and 3′) were amplified using gene-specific primers (Table 1) and printed in duplicate on the microarray. It appeared that the signal intensities of the 3′ end, middle and 5′ end of both genes were comparable, implying an overall consistency of cDNA synthesis and total RNA integrity. The mean 3′:5′ ratios of the GAPDH and the β-actin genes were, respectively, 1.4 and 1.3. A ratio of <1.5 is indicative of high quality RNA and full-length cDNA synthesis (19).
Quality control of the arrays and common reference
For quality control of the arrays we performed DNA POPO™-3 staining. Of the 9216 spots, there were 1281 spots that did not display a significant signal in the POPO™-3 staining. This is due to growth failure of the bacterial clones, failure of the PCR amplification, failure of the printing or flagging of the spots due to irregularities on the array. To check the performance of the common reference, the number of spots that gave a significant signal was determined in 12 different hybridizations and compared to the positive signals of the DNA staining. Of the properly printed spots 90.3–99.5% were detected with the common reference (Table 2). The variance is the result of a higher background in a few hybridizations, which results in fewer spots passing the thresholds set. Where the common reference is positive, absence of signal from the experimental target indicates that this transcript is not detectably expressed.
Table 2. The number of positive spots in the common reference versus the number of positive spots in the POPO™-3 DNA staining.
| POPO DNA staining positive |
Positive spots in CR (%) | |||
|---|---|---|---|---|
| CR negative | CR positive | |||
| Pair 1 | Normal | 119 | 7816 | 98.5 |
| Dye-swap | 185 | 7750 | 97.7 | |
| Pair 2 | Normal | 54 | 7881 | 99.3 |
| Dye-swap | 36 | 7899 | 99.5 | |
| Pair 3 | Normal | 325 | 7610 | 95.9 |
| Dye-swap | 80 | 7855 | 99.0 | |
| Pair 4 | Normal | 772 | 7163 | 90.3 |
| Dye-swap | 205 | 7730 | 97.4 | |
| Pair 5 | Normal | 432 | 7503 | 94.6 |
| Dye-swap | 128 | 7807 | 98.4 | |
| Pair 6 | Normal | 656 | 7279 | 91.7 |
| Dye-swap | 93 | 7842 | 98.9 | |
CR, common reference; normal, target labeled with Cy3 and CR with Cy5; dye-swap, target labeled with Cy5 and CR with Cy3.
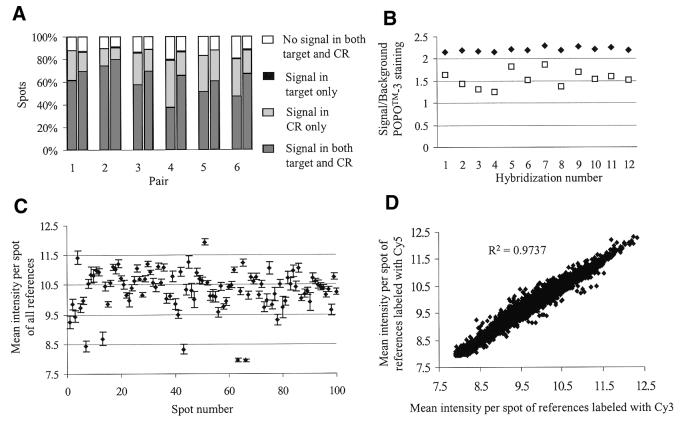
A very low number of spots (0.5% on average) could not be analyzed due to the fact that the reference did not give a significant signal, while the target did (Fig. 2A). This is probably due to the presence of a limited amount of cDNA in these spots. To check this, the signal of these spots in a POPO™-3 staining was evaluated (Fig. 2B). It can be seen that the signal to background ratio of these spots is significantly lower than the ratio in the spots that are positive for target and common reference (P < 0.05). This, in combination with a highly expressed gene in the experimental target, could result in a significant signal from the target and no signal from the reference in the spots. A solution to prevent missing reference signals is to perform a quality control of the PCR products prior to printing on the array, e.g. using agarose gel electrophoresis.
Figure 2.
Data analysis of different hybridizations. (A) Percentage of spots showing significant signal (> median background + 4 SD) in the common reference (CR, light gray), the experimental target (black) or both (dark gray). The left bar in each pair displays the results of the hybridization in which the target is labeled with Cy3 and the CR with Cy5, the right bar the dye-swap hybridization. (B) Signal to background ratio for POPO™-3 staining of the spots that give a signal in the experimental target but not in the reference (open squares) and of the spots that give a signal in both experimental target and reference (closed diamonds). (C) Mean background-corrected intensity per spot (VSN calibrated) of the 12 reference hybridizations of the first 100 spots on the array (closed diamonds). Vertical bars represent standard deviations. (D) Mean background-corrected intensity per spot (VSN calibrated) for the references labeled with Cy3 on the x-axis versus the mean background-corrected intensity per spot (VSN calibrated) for the references labeled with Cy5 on the y-axis.
Reproducibility of the common reference
To test the reproducibility of our method, a dye-swap experiment was carried out. A paired t-test was performed on the natural logarithm of the ratios to determine if there was a significant difference between the ratios of the two hybridizations. No significant differences between dye-swap hybridizations were found in any of the six comparisons (P > 0.989). In addition, 95% of the observed differences fall in the interval of ±0.2-fold change, which can be considered a level of natural variation (Table 3). Whereas the number of genes that gave a significant signal in the reference displayed no significant difference between the two dyes used, the number of genes that gave a significant signal in the target was significantly higher when the target was labeled with Cy5 compared to labeling with Cy3 (5084 versus 6374; Fig. 2A). When using a common reference, the experimental target can always be labeled with the same dye without having different gene–label interactions (20,21) and thus it is not necessary to perform a dye-swap. Therefore, under our experimental conditions, the data suggest labeling the target with Cy5 and the reference with Cy3.
Table 3. Results from the paired Student’s t-test performed on the normal and dye-swap hybridizations.
| Paired differences |
Significance (2-tailed) | |||||
|---|---|---|---|---|---|---|
| Mean | SD | SEM | 95% Confidence interval of the difference |
|||
| Lower | Upper | |||||
| Pair 1 | 8.32E – 05 | 0.4203 | 6.009E – 03 | –1.19E – 02 | 1.170E – 02 | 0.989 |
| Pair 2 | 5.33E – 06 | 0.3266 | 4.061E – 03 | –7.96E – 03 | 7.966E – 03 | 0.999 |
| Pair 3 | –1.88E – 05 | 0.3616 | 5.233E – 03 | –1.03E – 02 | 1.024E – 02 | 0.997 |
| Pair 4 | 5.77E – 05 | 0.4899 | 8.516E – 03 | –1.66E – 02 | 1.675E – 02 | 0.995 |
| Pair 5 | 3.55E – 05 | 0.2957 | 4.561E – 03 | –8.91E – 03 | 8.978E – 03 | 0.994 |
| Pair 6 | 2.13E – 05 | 0.3109 | 4.814E – 03 | –9.42E – 03 | 9.459E – 03 | 0.996 |
Figure 2D shows a comparison between the background-corrected reference intensities labeled with Cy5 and the background-corrected reference intensities labeled with Cy3 after VSN (18). The plot shows a correlation of 0.97, which demonstrates that the reference signal is very similar in every spot, even when it is labeled with the opposite color. To analyze the mean of the background-corrected normalized intensities of the references, we took the first 100 spots present on the array (Fig. 2C). The figure shows that the inter-experimental variation (standard deviation of the mean between the hybridizations) is very low, i.e. the reference gives a constant hybridization signal per spot in different hybridizations. These results are representative for all the spots present on the array (data not shown).
Time saving
To prepare the common reference, 4 µl of each PCR product was pooled. This yielded 2.2 mg of double-stranded PCR products. Starting with 1 µg of PCR product (ideal amount) in the in vitro transcription, the final yield will be 40–50 µg of cRNA. If more is needed it is better to pool cRNAs obtained from individual in vitro transcriptions to ensure that one constant batch of common reference is made. With the 2.2 mg PCR product, 2200 in vitro transcriptions can be performed, yielding 88 mg of cRNA. In our experiments we use 750 ng of common reference per hybridization as this amount results in a similar hybridization signal to 1.5 µg of the experimental target. Therefore, 2.2 mg of PCR product is sufficient for 117 000 hybridizations. The time necessary for preparation of the described common reference will not be more than 3 days, which is considerably less than the weeks of culturing and RNA isolation required when making a common reference from cultured cells (11).
CONCLUSION
In this report we have described a fast and cheap method for the preparation of a common reference that can be used in cDNA microarray hybridization experiments. By pooling a small part of all the probes before they are spotted on the array and performing subsequent amplification, a batch of common reference is produced that shows a reproducible hybridization signal. Our results demonstrate that, in a good quality hybridization, up to 99.5% of the properly printed spots are detected, providing a ratio when compared to the experimental target. Using this reference in the analysis of cDNA microarray data enables reliable computations of expression levels, which is especially useful when comparing more than two hybridizations. When this manuscript was in preparation, a similar approach for oligo arrays was described by Dudley et al. (22). They suggest using a common reference that consists of the reverse complement of the spotted oligos. Although, due to cost, this is not a feasible approach for many laboratories, it does provide a very good common reference. An alternative approach would be to add a common tag to all oligos spotted and then use the reverse complement as a common reference. This approach would increase product cost of the arrays by ∼20%, but production and use of the reference would be very cheap. However, as already mentioned in the Introduction, the aspect of competition is not taken into account when a tag sequence is used. Another possibility would be to produce a reference based on a mixture of cDNAs, one for each oligo spotted. This reference could then be prepared as shown in this paper, yielding a signal in every spot and, hence, facilitating the analysis of the absolute expression levels.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Peter-Bram ’t Hoen and Stefan White for critical reading of the manuscript and valuable suggestions. This work was supported by grants of the Nederlands Wetenschappelijk Onderzoek (NWO), Muscular Dystrophy Campaign (MDC) and the Center for Biomedical Genetics (CBG).
REFERENCES
- 1.Brown P.O. and Botstein,D. (1999) Exploring the new world of the genome with DNA microarrays. Nature Genet., 21, 33–37. [DOI] [PubMed] [Google Scholar]
- 2.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 3.Alizadeh A., Eisen,M., Davis,R.E., Ma,C., Sabet,H., Tran,T., Powell,J.I., Yang,L., Marti,G.E., Moore,D.T. et al. (1999) The lymphochip: a specialized cDNA microarray for the genomic-scale analysis of gene expression in normal and malignant lymphocytes. Cold Spring Harbor Symp. Quant. Biol., 64, 71–78. [DOI] [PubMed] [Google Scholar]
- 4.Eisen M.B. and Brown,P.O. (1999) DNA arrays for analysis of gene expression. Methods Enzymol., 303, 179–205. [DOI] [PubMed] [Google Scholar]
- 5.DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 6.Khan J., Simon,R., Bittner,M., Chen,Y., Leighton,S.B., Pohida,T., Smith,P.D., Jiang,Y., Gooden,G.C., Trent,J.M. et al. (1998) Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res., 58, 5009–5013. [PubMed] [Google Scholar]
- 7.Perou C.M., Jeffrey,S.S., van de Rijn,M., Rees,C.A., Eisen,M.B., Ross,D.T., Pergamenschikov,A., Williams,C.F., Zhu,S.X., Lee,J.C. et al. (1999) Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc. Natl Acad. Sci. USA, 96, 9212–9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Troyanskaya O., Cantor,M., Sherlock,G., Brown,P., Hastie,T., Tibshirani,R., Botstein,D. and Altman,R.B. (2001) Missing value estimation methods for DNA microarrays. Bioinformatics, 17, 520–525. [DOI] [PubMed] [Google Scholar]
- 9.Zehetner G., Pack,M. and Schafer,K. (2001) Preparation and screening of high-density cDNA arrays with genomic clones. Methods Mol. Biol., 175, 169–188. [DOI] [PubMed] [Google Scholar]
- 10.Alizadeh A.A., Eisen,M.B., Davis,R.E., Ma,C., Lossos,I.S., Rosenwald,A., Boldrick,J.C., Sabet,H., Tran,T., Yu,X. et al. (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature, 403, 503–511. [DOI] [PubMed] [Google Scholar]
- 11.Scherf U., Ross,D.T., Waltham,M., Smith,L.H., Lee,J.K., Tanabe,L., Kohn,K.W., Reinhold,W.C., Myers,T.G., Andrews,D.T. et al. (2000) A gene expression database for the molecular pharmacology of cancer. Nature Genet., 24, 236–244. [DOI] [PubMed] [Google Scholar]
- 12.Ross D.T., Scherf,U., Eisen,M.B., Perou,C.M., Rees,C., Spellman,P., Iyer,V., Jeffrey,S.S., van de Rijn,M., Waltham,M. et al. (2000) Systematic variation in gene expression patterns in human cancer cell lines. Nature Genet., 24, 227–235. [DOI] [PubMed] [Google Scholar]
- 13.Bergstrom D.A., Penn,B.H., Strand,A., Perry,R.L., Rudnicki,M.A. and Tapscott,S.J. (2002) Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell, 9, 587–600. [DOI] [PubMed] [Google Scholar]
- 14.van’t Veer L.J., Dai,H., van de Vijver,M.J., He,Y.D., Hart,A.A., Mao,M., Peterse,H.L., van der Kooy,K., Marton,M.J., Witteveen,A.T. et al. (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature, 415, 530–536. [DOI] [PubMed] [Google Scholar]
- 15.Lennon G., Auffray,C., Polymeropoulos,M. and Soares,M.B. (1996) The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics, 33, 151–152. [DOI] [PubMed] [Google Scholar]
- 16.Luo L., Salunga,R.C., Guo,H., Bittner,A., Joy,K.C., Galindo,J.E., Xiao,H., Rogers,K.E., Wan,J.S., Jackson,M.R. et al. (1999) Gene expression profiles of laser-captured adjacent neuronal subtypes. Nature Med., 5, 117–122. [DOI] [PubMed] [Google Scholar]
- 17.Van Gelder R.N., von Zastrow,M.E., Yool,A., Dement,W.C., Barchas,J.D. and Eberwine,J.H. (1990) Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl Acad. Sci. USA, 87, 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber W., Von Heydebreck,A., Sultmann,H., Poustka,A. and Vingron,M. (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics, 18 (Suppl 1), S96–S104. [DOI] [PubMed] [Google Scholar]
- 19.Graf L., Heimfeld,S. and Torok-Storb,B. (2001) Comparison of gene expression in CD34+ cells from bone marrow and G-CSF-mobilized peripheral blood by high-density oligonucleotide array analysis. Biol. Blood Marrow Transplant., 7, 486–494. [DOI] [PubMed] [Google Scholar]
- 20.Yue H., Eastman,P.S., Wang,B.B., Minor,J., Doctolero,M.H., Nuttall,R.L., Stack,R., Becker,J.W., Montgomery,J.R., Vainer,M. et al. (2001) An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res., 29, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tseng G.C., Oh,M.K., Rohlin,L., Liao,J.C. and Wong,W.H. (2001) Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res., 29, 2549–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dudley A.M., Aach,J., Steffen,M.A. and Church,G.M. (2002) Measuring absolute expression with microarrays with a calibrated reference sample and an extended signal intensity range. Proc. Natl Acad. Sci. USA, 99, 7554–7559. [DOI] [PMC free article] [PubMed] [Google Scholar]