Abstract
A new type II restriction endonuclease AarI has been isolated from Arthrobacter aurescens SS2-322. AarI recognizes the non-palindromic heptanucleotide sequence 5′-CACCTGC(N)4/8-3′ and makes a staggered cut at the fourth and eighth bases downstream of the target duplex producing a four base 5′-protruding end. AarI activity is stimulated by oligodeoxyribonucleotide duplexes containing an enzyme-specific recognition sequence.
INTRODUCTION
More than 3000 type II restriction endonucleases have been discovered but only 231 are examples of enzymes with different and unique specificities (prototypes) (1). Type II restriction endonucleases recognize short, usually palindromic, sequences of 4–8 bp and, in the presence of Mg2+, cleave DNA within or in close proximity to the target sequence. The main criterion for classifying a restriction endonuclease as a type II enzyme is that it cleaves specifically within or close to its recognition sequence and that it does not require ATP hydrolysis for its nucleolytic activity. Many type II restriction endonucleases do not conform to the standard definition of type II enzymes and, therefore, they were divided into subdivisions, so-called types IIS, IIE, IIF, IIT, IIG, IIB and IIM (2). We report here the isolation and characterization of a new restriction endonuclease AarI from Arthrobacter aurescens SS2-322, which recognizes the novel non-palindromic heptanucleotide sequence 5′-CACCTGC(N)4/8-3′ and cleaves DNA 4 and 8 nt away from the target duplex producing a four base 5′-protruding end. Many AarI sites are not cleaved completely under standard restriction reaction conditions and AarI activity can be stimulated by an oligonucleotide duplex containing the recognition sequence of the enzyme.
MATERIALS AND METHODS
Biological materials
Arthrobacter aurescens strain SS2-322 was from the Fermentas UAB culture collection. The cells were grown aerobically at 37°C in a medium containing 10 g/l peptone, 5 g/l yeast extract, 5 g/l NaCl, 5 g/l glucose and 2.5 g/l MgSO4·7H2O, pH 7.1, up to late logarithmic phase, collected by centrifugation and stored at –20°C until use. Phage λ, plasmid DNAs, the Cycle Reader DNA Sequencing kit, oligonucleotides, all enzymes and DNA size markers were products of Fermentas. Ad2 DNA was purchased from Invitrogen and [α-33P]dATP from Amersham. SssI methylase was purchased from New England Biolabs.
Endonuclease assay
The restriction endonuclease activity was assayed by incubation of 1 µg λ DNA in 50 µl of AarI reaction mixture: 10 mM Bis-Tris propane–HCl pH 6.5, 100 mM KCl, 10 mM MgCl2 and 0.1 mg/ml BSA. Column samples (3 µl) were added to the reaction mixture, incubated for 15 min at 37°C and aliquots from each solution were then electrophoresed on a 0.8% agarose gel. AarI does not cleave λ DNA or any other substrate completely. Therefore, 1 U of AarI was defined as the minimal amount of the enzyme yielding maximal fragmentation (which does not change with a further increase in enzyme concentration) of 1 µg λ DNA in 50 µl of assay buffer for 1 h at 37°C. Various oligonucleotides (see Table 1) were assayed for their ability to stimulate cleavage of the plasmid pASK5metL that contains a single AarI site. An aliquot of 1 µg of the plasmid DNA was incubated with 1–12 U AarI in 50 µl of the AarI buffer for 60 min at 37°C using a 0.05–5.0 µM concentration of oligonucleotides.
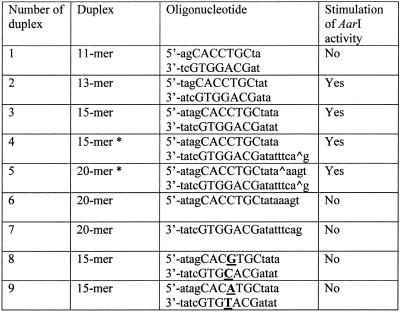
Table 1. The oligonucleotide duplexes used in AarI activity stimulation experiments.
*, AarI cleavage point.
Purification of restriction endonuclease
Isolation of AarI was carried out at 4°C. Fifty grams of frozen cells were suspended in 150 ml of buffer A (10 mM potassium phosphate pH 7.4, 1 mM EDTA, 7 mM 2-mercaptoethanol) containing 0.15 M NaCl. After sonication, insoluble material was removed from the crude extract by centrifugation at 30 000 g for 1 h. The crude extract was applied to a heparin–Sepharose column (2.5 × 12 cm) and eluted with a 600 ml linear gradient from 0.15 to 0.6 M NaCl. Fractions of 12 ml were collected and assayed for restriction endonuclease activity. Active fractions eluted at 0.45–0.49 M NaCl were dialyzed against buffer A containing 0.2 M NaCl. The sample was applied to a blue-Sepharose column (1.5 × 15 cm) and eluted with a 300 ml linear gradient from 0.2 to 1.4 M NaCl. Fractions of 6 ml were collected. The fractions containing the peak of enzyme activity (0.78–0.90 M NaCl) were dialyzed against buffer A containing 0.2 M NaCl. After dialysis, active fractions were applied to a phosphocellulose P11 column (1.5 × 8 cm) and eluted with a 160 ml linear gradient from 0.2 to 0.8 M NaCl. Fractions containing AarI activity were collected at 0.43–0.48 M NaCl. These fractions were pooled and dialyzed against 10 mM potassium phosphate pH 7.4, 100 mM KCl, 1 mM EDTA, 7 mM 2-mercaptoethanol and 50% glycerol. The final product was stored at –20°C. The yield of the enzyme was ∼100 U/g wet weight of cells.
Determination of the recognition sequence and cleavage site
The recognition sequence of AarI was inferred by restriction mapping of the recognition sites on phage λ DNA. The fragments predicted by cleavage of the inferred recognition sites were compared with the observed fragments from AarI cleavage of different DNAs. λ DNA was used as a template to characterize the cleavage site of AarI. A 20mer oligodeoxyribonucleotide complementary to λ DNA between positions 13860 and 13880 was used in direct sequencing through the AarI recognition site located at position 13941. Four dideoxy sequencing reactions using [α33P]dATP and a Cycle Reader DNA sequencing kit were carried out. The same primer and template were used in an extension reaction, which also included T7 DNA polymerase, dNTP and [α33P]dATP. The extension reaction was heat inactivated, treated with AarI in the presence of 0.5 µM oligonucleotide duplex and the reaction mixture was divided into two. The aliquot was treated with T4 DNA polymerase in the presence of dNTP. All samples were diluted with sequencing dye solution and loaded on a standard sequencing gel along with the dideoxy sequencing reaction.
RESULTS
The recognition sequence and cleavage site of AarI
To determine the substrate specificity of the enzyme six of 12 cleavage sites of AarI on λ DNA (approximate positions 10850, 13900, 16150, 21200, 29900 and 36500) were mapped by double digestion with Bsp120I, Eco105I, EheI, NheI, Pfl23II, XhoI and XbaI (data not shown). A computer-aided search of homologous nucleotide sequences revealed only one common sequence, 5′-CACCTGC-3′, for the mapped AarI sites. The number of DNA fragments generated by AarI cleavage of λ DNA (12 sites), Ad2 DNA (9 sites) and T7 DNA (5 sites) (see Fig. 1) were consistent with this sequence being the AarI recognition sequence. As could be expected, pBR322, pUC19, pTZ19R, M13mp18 and φX174 DNAs, which do not contain 5′-CACCTGC-3′ sequences, were not cleaved by AarI (data not shown).
Figure 1.
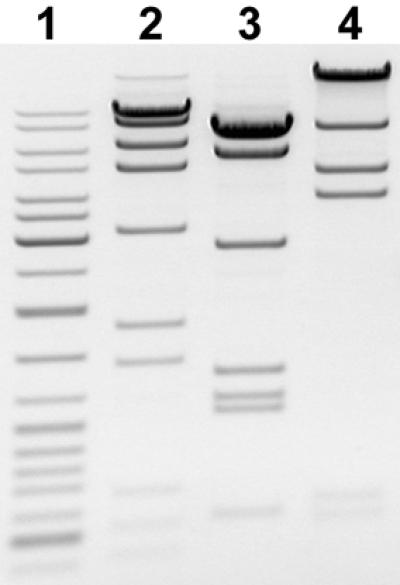
AarI digests of λ (lane 2), Ad2 (lane 3) and T7 (lane 4) DNAs. The molecular weight marker is GeneRuler™ DNA Ladder Mix (lane 1).
The cleavage position of AarI was determined by comparison of the dideoxy sequence ladders with fragments generated by AarI cleavage and T4 DNA polymerase action on the digestion product. The results of the determination of the AarI cleavage site are shown in Figure 2. The fragment generated by AarI digestion co-migrates with the T band of the sequence ladder (lane 2), indicating that the cutting point is 4 nt away from the recognition sequence 5′-CACCTGC. The single band obtained after treatment with T4 polymerase co-migrates with the C band (lane 1) of the sequence ladder, confirming that the cleavage point on the complementary DNA strand 3′-GTGGAGC is 8 nt away from the recognition sequence. Summarizing, we conclude that AarI cleaves double-stranded DNA 4 and 8 bases downstream of its recognition sequence generating fragments with 5′-protruding ends. AarI is a new prototype of the type II restriction endonucleases whose recognition sequence and cleavage specificity is
Figure 2.
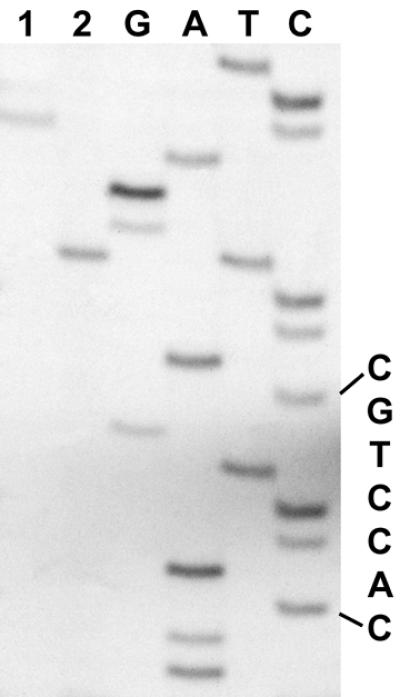
Determination of the AarI cleavage site. Lanes G, A, T and C, the sequence ladders through an AarI site; lane 2, the product of the primed synthesis reaction cleaved with AarI; lane 1, T4 DNA polymerase action on the AarI digest.
5′-CACCTGCNNNN
3′-GTGGACGNNNNNNNN
Enzymatic properties
We have studied the influence of different factors (metal ions, pH, ionic strength and cofactors) on the activity of the AarI restriction endonuclease. The enzyme shows an absolute requirement for Mg2+. The pH optimum for AarI is 6.5–7.0 and the optimal salt concentration 50–100 mM. The optimal temperature for cleavage was found to be 37°C. ATP at a concentration up to 2 mM or AdoMet at a concentration up to 0.3 mM did not show perceptible influence on the endonucleolytic activity of AarI. Since the AarI recognition sequence may overlap a CG dinucleotide due to the 3′-external cytosine nucleotide (5′-CACCTGm5CG), the enzyme sensitivity to CG methylation was investigated. Two of nine AarI sites on Ad2 DNA overlap CG. Their premethylation with SssI methylase significantly decreased cleavage rate of the methylated sites. Incubation of 1 µg of the premethylated DNA with 10 U AarI for 1 h did not reveal complete cleavage of the methylated target sites. More prolonged incubation (for 4 h) was required to achieve complete cleavage.
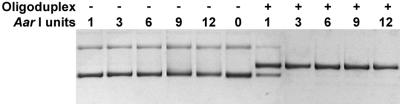
Cleavage of DNA by AarI is never complete even on non-modified substrates. Complete digestion of a single AarI site-containing plasmid pASKmet5L DNA was observed only after the addition of an oligonucleotide duplex harboring the AarI recognition sequence (see Fig. 3). Of all oligonucleotides tested (see Table 1) only duplexes that contain an AarI recognition sequence and are no shorter than a 13mer enhanced activity of the enzyme. Single-stranded oligonucleotides (constituents of the stimulating duplex) and duplexes in which the middle C in the recognition sequence 5′-CACCTGC was substituted by A or G (5′-CACGTGC or 5′-CACATGC) did not show any effect on AarI activity. A concentration of duplexes containing the enzyme recognition sequence ranging between 0.5 and 2.5 µM (100–500-fold molar excess over substrate) was found to be optimal for the activation of AarI. Higher oligonucleotide duplex concentrations inhibited AarI activity.
Figure 3.
Digestion of the single AarI site-containing plasmid DNA (pASKmet5L) using different amounts of the enzyme with (+) or without (–) the addition of 1 µM 15mer oligonucleotide duplex no. 3 (see Table 1).
DISCUSSION
AarI is a novel restriction endonuclease that recognizes the asymmetric nucleotide sequence 5′-CACCTGC(N)4/8-3′ and makes a staggered cut at a distance of four bases from the recognition sequence on the upper strand and at the eighth base on the complementary strand, producing four base 5′-protruding ends. AarI is a new prototype of type IIS restriction endonucleases and is the second after SapI among them that recognizes a heptanucleotide sequence (REBASE version 207). On the other hand, stimulation of the endonuclease activity by an oligonucleotide duplex containing the cognate recognition site indicates that AarI is similar to type IIE or type IIF enzymes that need to interact with two copies of the target site for effective cleavage (3). In the case of type IIE enzymes one of the target sequences serves as an allosteric effector for the effective cleavage of the other recognition site. Type IIF endonucleases cleave both recognition sequences in a concerted reaction (4–6). The exact subtype to which AarI belongs has yet to be determined.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Egle Cesnaviciene for help in preparation of the manuscript. Our special thanks to Tomas Tarvainis for figure preparation and technical help.
REFERENCES
- 1.Roberts R.J. and Macelis,D. (2001) REBASE—restriction enzymes and methylases. Nucleic Acids Res., 29, 268–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pingoud A. and Jeltsch,A. (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res., 29, 3705–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bath A.J., Milsom,S.E., Gormley,N.A. and Halford,S.E. (2002) Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem., 277, 4024–4033. [DOI] [PubMed] [Google Scholar]
- 4.Grazulis S., Deibert,M., Rimseliene,R., Skirgaila,R., Sasnauskas,G., Lagunavicius,A., Repin,V., Urbanke,C., Huber,R. and Siksnys,V. (2002) Crystal structure of the Bse634I restriction endonuclease: comparison of two enzymes recognizing the same DNA sequence. Nucleic Acids Res., 30, 876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deibert M., Grazulis,S., Sasnauskas,G., Siksnys,V. and Huber,R. (2000) Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nature Struct. Biol., 7, 792–799. [DOI] [PubMed] [Google Scholar]
- 6.Siksnys V., Skirgaila,R., Sasnauskas,G., Urbanke,C., Cherny,D., Grazulis,S. and Huber,R. (1999) The Cfr10I restriction enzyme is functional as a tetramer. J. Mol. Biol., 291, 1105–1118. [DOI] [PubMed] [Google Scholar]