Abstract
The adenovirus (Ad) E1A 243R oncoprotein encodes an N-terminal transcription repression domain that is essential for early viral functions, cell immortalization, and cell transformation. The transcription repression function requires sequences within amino acids 1 to 30 and 48 to 60. To elucidate the roles of the TATA-binding protein (TBP), p300, and the CREB-binding protein (CBP) in the mechanism(s) of E1A repression, we have constructed 29 amino acid substitution mutants and 5 deletion mutants spanning the first 30 amino acids within the E1A 1-80 polypeptide backbone. These mutant E1A polypeptides were characterized with regard to six parameters: the ability to repress transcription in vitro and in vivo, to disrupt TBP-TATA box interaction, and to bind TBP, p300, and CBP. Two regions within E1A residues 1 to 30, amino acids 2 to 6 and amino acid 20, are critical for E1A transcription repression in vitro and in vivo and for the ability to interfere with TBP-TATA interaction. Replacement of 6Cys with Ala in the first region yields the most defective mutant. Replacement of 20Leu with Ala, but not substitutions in flanking residues, yields a substantially defective phenotype. Protein binding assays demonstrate that replacement of 6Cys with Ala yields a mutant completely defective in interaction with TBP, p300, and CBP. Our findings are consistent with a model in which the E1A repression function involves interaction of E1A with p300/CBP and interference with the formation of a TBP-TATA box complex.
Studies with group C human adenoviruses (Ads) have been pivotal to our present understanding of transcriptional regulation and the mechanisms by which cells regulate their growth (for reviews, see references 32 and 35). The first transcription unit expressed during productive infection, early region 1A (E1A), encodes two multifunctional regulatory oncoproteins of 243 and 289 amino acid residues (243R and 289R [Fig. 1]) that interact with key cellular transcriptional regulatory factors. E1A is involved in diverse functions, including transcriptional activation, transcriptional repression, induction of cellular DNA synthesis, cell immortalization, and cell transformation, as well as, paradoxically, the inhibition of tumorigenesis and metastasis. E1A 289R differs from E1A 243R by possessing conserved region 3 (CR3), a 46-amino-acid domain unique to 289R. CR3 is essential (13, 20, 24, 30, 34) and sufficient (15, 25) for activation of viral early genes. Domains common to E1A 243R and E1A 289R are required for the growth-regulatory functions of E1A; these include the nonconserved N terminus (amino acids 1 to 39), CR1 (amino acids 40 to 80), and CR2 (amino acids 120 to 139) (28). Exon 2 possesses autonomous transformation suppression activity (42) localized within a 14-amino-acid region near the C terminus of 243R (3).
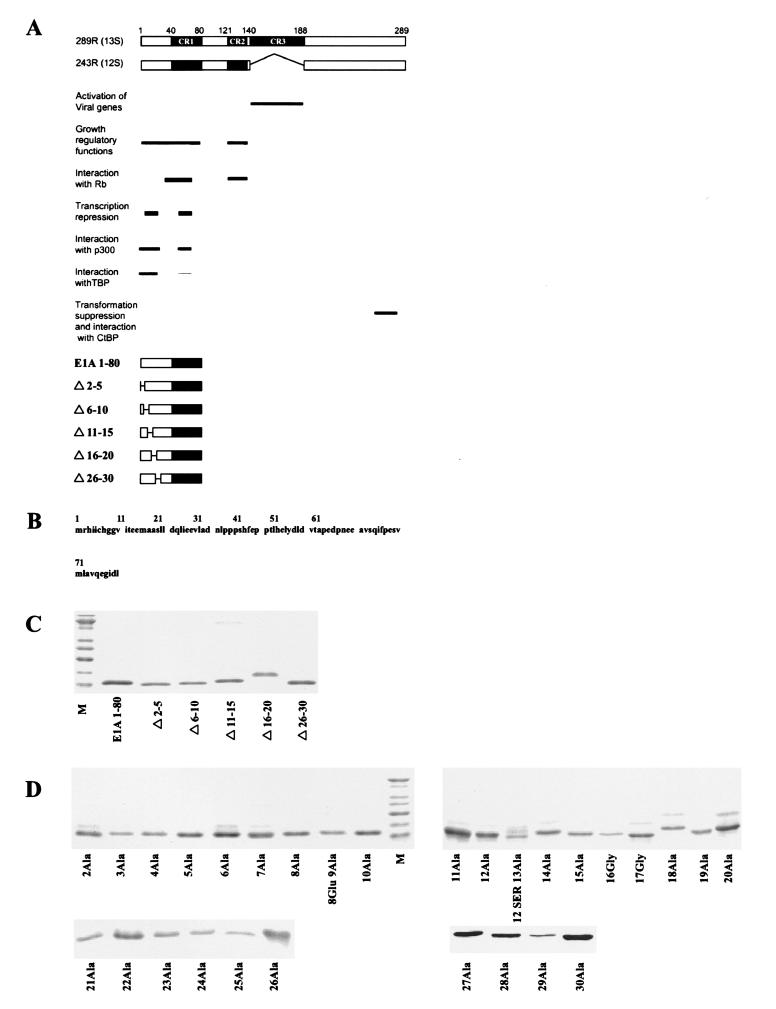
FIG. 1.
(A) Schematic of E1A 289R and 243R proteins, their domain structures, and associated biological activities. Below are schematics of E1A 1-80 and E1A 1-80 deletion polypeptides. (B) Sequence of the first 80 amino acids of E1A (E1A 1-80). (C) Purity of E1A 1-80 deletion mutants. One-microgram amounts of polypeptide were resolved on 15% polyacrylamide gels, stained with SYPRO orange (Molecular Probes), and visualized by blue-green fluorescence on a STORM 840 PhosphorImager. The lowest band of the marker lane (M) is 10 kDa. (D) Purity of E1A 1-80 amino acid substitution mutants. One-microgram amounts of the E1A 1-80 mutants were analyzed as described for panel C above.
E1A 243R can induce progression of quiescent cells to S-phase cellular DNA synthesis by two independent pathways (19, 25). There is good reason to believe that, although independent, these pathways may act synergistically to stimulate progression through the cell cycle (35). The most studied pathway involves sites within E1A CR1 and CR2, which interact with the retinoblastoma tumor suppressor protein pRb. This interaction sequesters pRb and dissociates it from complexes with the E2F family of transcription factors, which are involved in cell cycle regulation (for a review, see reference 9). The second pathway maps within the E1A N-terminal 80 amino acids (37) and is the focus of our studies. The N-terminal pathway is of particular interest because the growth-regulatory functions of E1A require sequences within this region (29). Mutational analysis has identified phenotypic changes, including cellular DNA induction, cell immortalization, and oncogene cooperation, that require the N-terminal pathway. However, the precise sequence requirements, biochemical functions, protein-partner interactions, and cellular genes targeted by the N-terminal pathway are not well understood.
An intriguing biochemical function which maps to the E1A N-terminal pathway is the ability to repress transcriptionally a set of genes involved in cell proliferation and cell differentiation (2, 18, 41, 46, 50; for a review, see reference 35). To understand the mechanism of E1A repression, our laboratory has developed an in vitro transcription system and an in vivo cell microinjection system that faithfully recapitulate repression by E1A 243R (16, 26, 37, 38, 39, 40). A highly purified recombinant protein containing only the N-terminal 80 amino acids (E1A 1-80) strongly represses transcription of E1A-repressible promoters in vitro and in vivo and is used as a “prototype repressor,” which avoids complications introduced by other E1A functional domains. Using these in vitro and in vivo systems, we have previously mapped the E1A repression function to sequences within E1A 1 to 30 and E1A 48 to 60.
Our previous findings reveal several lines of evidence that TATA-binding protein (TBP), the major component of the basal transcription factor TFIID, is a cellular target of E1A repression (37, 38, 39, 40; M. Green and P. Loewenstein, unpublished data). First, E1A repression in vitro is reversed by addition of TFIID or recombinant TBP. Second, addition of TBP restores transcriptional activity to an E1A 1-80 affinity-depleted nuclear extract. Third, overexpression of TBP can partially overcome E1A repression in vivo. Fourth, E1A 243R, and E1A 1-80 can interfere with TBP-TATA interaction in vitro. The ability of E1A to disrupt interaction between TBP and the TATA box is unique among transcriptional repressors that target TBP (40). Of the general transcription factors, TBP is the major sequence-specific DNA binding component of the promoter. Binding of TBP to the TATA box is the initial step in the assembly of a transcription preinitiation complex and is critical for the rate of transcription, and therefore it is an excellent target for a transcription repression function (for a review, see reference 17).
The multifunctional transcription factor p300 was discovered through its association with E1A (for a review, see reference 29). The E1A regions required for transcription repression (amino acids within 1 to 30 and 48 to 60) overlap those required for binding p300 in vivo (45) and in vitro (Green and Loewenstein, unpublished). p300 and the closely related CREB-binding protein (CBP) (8, 10) belong to a conserved family of coactivators recruited to cellular promoters through interaction with specific transcription factors or as a complex with other cofactors (1, 5, 6, 7, 12, 21, 23, 36, 48, 49). p300 and CBP possess histone acetyltransferase (HAT) activity, and p300 can interact with PCAF, which also possesses HAT activity. Because p300/CBP is involved in the expression of genes necessary for cell differentiation and for maintaining quiescence, genes coactivated by p300/CBP are attractive targets for the E1A repression function. It has been suggested that E1A represses transcription by sequestering p300/CBP and thus preventing p300/CBP from serving as a coactivator. More recently, E1A has been reported to possess several biochemical activities, some of which could be related to the E1A repression function, including inhibition of PCAF-dependent transcription (33, 48) and blocking recruitment of RNA polymerase II on cyclic AMP-dependent promoters (31). These activities of E1A are of great interest, but it is not clear to what extent they relate to the mechanism(s) by which E1A represses transcription or to the function of the E1A N-terminal pathway early during productive infection or in cell transformation. To sort out the proposed E1A N-terminal activities and their in vivo significance, an extensive structure-function analysis of well-defined E1A single-amino-acid substitution mutants is desirable. Similar approaches using radical single-amino-acid substitution mutants and alanine-scanning mutants have been remarkably successful in identifying the precise amino acids on the exposed surface of TBP that interact with the general transcription factors IIA, IIB, IIF, and RNA polymerase II (4, 43). Toward this end, we have constructed, expressed, and purified a panel of 5 small deletion mutants and 29 amino acid substitution mutants within the first 30 amino acids of the E1A 1-80 polypeptide. We probed their abilities to support transcription repression in vitro and in vivo, to interfere with formation of a TBP-TATA complex, and to interact with the cellular partners of the E1A N-terminal repression domain—TBP, p300, and CBP.
MATERIALS AND METHODS
Plasmids.
pSV40 (p1–11), which encodes the simian virus 40 (SV40) T antigen (14), and pEGFPN-1 (Clontech), which expresses the green fluorescent protein, were used in the cell microinjection assay. pHIV-TAR(+) (27) was used in the in vitro transcription repression assay. Glutathione S-transferase (GST)-TBP contains the entire TBP molecule cloned into the pGEX vector. GST-p300-segment B′ and GST-CBP-segment B contain the E1A binding site of p300 and CBP, respectively, cloned into the pGEX vector (48). The pcDNA3-E1A 243R expression vector was constructed by PCR cloning into pcDNA3 (Invitrogen) using 243R cDNA as a template.
Construction of plasmids expressing E1A 1-80 mutants.
Construction of pQE12-E1A 1-80 and pQE12-E1A 1-80Δ4-25 has been described previously (39). We have found that pQE12, although an older expression plasmid, is the most efficient Qiagen vector for the production of biologically active E1A 243R protein and E1A 1-80 polypeptides (37). Five amino acid deletions were constructed within the first 30 amino acids of E1A 1-80 and cloned into pQE12 to express E1A 1-80 polypeptides with six His residues at the C terminus. The deletion mutants were constructed using two PCR cloning strategies. (i) Mutants containing deletion of residues 2 through 5 (Δ2-5) and deletion of residues 6 through 10 (Δ6-10) were constructed by PCR utilizing BamHI-containing upstream deletion primers and a common pQE12 downstream antisense primer containing HindIII. pQE12-E1A 1-80 was used as a template. The PCR products were cloned into an appropriately digested pQE12 vector. (ii) E1A 1-80Δ11-15, -Δ16-20, and -Δ26-30 were constructed using a two-step PCR. Downstream antisense primers with appropriate deletions were used in combination with a wild-type upstream BamHI primer to generate PCR products. These products were gel purified and used as primers for a second round of PCR with pQE12-E1A 1-80 as a template, together with the downstream HindIII primer, to generate full-length deletion mutants which were then cloned into pQE12.
Substitutions of amino acid residues 2 through 30 in an E1A 1-80 backbone were constructed and cloned into pQE12 using four PCR cloning strategies, all with pQE12-E1A 1-80 as a template. (i) Alanine-scanning mutants in amino acids 2 through 9 were constructed with the appropriate alanine substitution built into the upstream primer containing a BamHI site and a common downstream primer containing a BglII site as described in reference 39. PCR products were cloned into the BamHI and BglII sites of pQE12. (ii) Alanine-scanning mutants in residues 10 through 16 were constructed by triple ligation of two PCR products with the pQE12 vector. The first PCR product (used for all clones in this group) was generated using an upstream primer that introduced an NheI restriction enzyme site at nucleotides coding for amino acids 17 and 18 (while conserving the original amino acid sequence) in combination with a downstream BglII primer to generate a wild-type C-terminal fragment containing an NheI site. The second PCR product was constructed with downstream primers producing the NheI site at residues 17 and 18 and introducing the alanine-scanning mutation at the appropriate residue, in combination with an upstream wild-type primer containing the BamHI site. The C-terminal and N-terminal PCR products were digested with NheI (as well as BamHI and BglII) and joined in a triple ligation with the BamHI/BglII-digested pQE12 vector to produce this set of substitution mutants. (iii) Alanine-scanning mutants in residues 19 through 24 were constructed by essentially the reverse of the previous procedure. A wild-type PCR product was generated with a downstream primer to introduce an NheI site at residues 17 and 18 and an upstream, wild-type primer containing a BamHI site. The downstream PCR products were produced with primers containing the NheI site and the appropriate alanine-scanning mutation in combination with the downstream BglII primer. As before, the upstream and downstream PCR products were ligated together with the appropriately digested pQE12 vector. (iv) The alanine-scanning mutants in residues 16, 17, and 25 through 30 were constructed using a different approach. Downstream primers containing the appropriate alanine-scanning mutations were used in combination with a common upstream BamHI primer to generate a PCR product for each mutant. Each PCR product was gel purified and used as a primer in a second round of PCR, in combination with the downstream BglII primer, to generate the full-length E1A 1-80 sequence flanked by BamHI and BglII sites, which were then cloned into pQE12. All E1A 1-80 mutants were sequenced completely to confirm the intended mutation and the accuracy of the nonmutated sequence. Two inadvertent errors were found. E1A 1-80 9Ala also contained a substitution of Glu for 8Gly. E1A 1-80 13Ala also contained a substitution of Ser for 12Thr. Both double mutants were shown to have wild-type transcription repression activity.
Purification of E1A 1-80 and E1A 1-80 mutant polypeptides.
E1A 1-80 polypeptides were prepared by a protocol modified from that suggested by Qiagen for the purification of denatured His6-tagged proteins. This protocol was first developed to produce biologically active E1A 243R protein and E1A 1-80 polypeptide (37). Briefly, cultures of IPTG (isopropyl-β-d-thiogalactopyranoside)-induced bacterial cells (500 ml) containing a pQE12 construct expressing wild-type or mutant E1A 1-80 polypeptide were harvested by centrifugation and frozen at −20°C. The pellets were thawed at room temperature and lysed in 40 ml of Qiagen buffer A (100 mM NaH2PO4, 10 mM Tris-HCl [pH 8.0], and 6 M guanidine-HCl) with gentle mixing for 1 h and clarified by centrifugation. His-tagged E1A 1-80 polypeptides in the clarified lysate were bound overnight to 2 ml of Ni-nitrilotriacetic acid resin (Qiagen). The resin was batch washed five times with 20 ml of buffer A and five times with buffer A at pH 6.3. The resin was then loaded into two 5-ml columns, and each column was washed with 20 ml (10 column volumes) of the same buffer and then with 50 ml of buffer A adjusted to pH 5.9. E1A polypeptides were eluted from the resin with 20 ml of buffer A at pH 4.5, and 2-ml fractions were collected.
To prepare biologically active E1A 1-80 polypeptides, it was necessary to remove guanidine-HCl slowly to facilitate proper folding. Fractions containing eluted protein were pooled and adjusted to 6 ml with elution buffer (buffer A at pH 4.5 containing 6 M guanidine-HCl). The sample was diluted 1:1 with 0.5× buffer D (10 mM HEPES [pH 7.9] at 4°C, 10% glycerol, 50 mM KCl, 0.1 mM phenylmethylsulfonyl fluoride, and 0.25 mM dithiothreitol) (26). The diluted sample (now at 3 M guanidine-HCl) was dialyzed in a 3,000-mw cutoff Slide-A-Lizer dialysis cassette (Pierce) against 0.5× buffer D containing 2 M guanidine-HCl. After 6 to 8 h of dialysis, one-half of the dialysis buffer was removed and replaced with fresh 0.5× buffer D without guanidine-HCl, thereby reducing the guanidine-HCl concentration by half. Dialysis with buffer replacement was continued in this manner until the guanidine-HCl concentration was reduced to 50 to 100 mM. Dialysis was completed against several changes of 0.5× buffer D for 8 h. The E1A polypeptides were then concentrated by size exclusion centrifugation using Centriprep YM-3 followed by Centricon YM-3 (Millipore) to a final concentration of about 1 mg per ml of polypeptide. Using these protocols, multiple preparations of wild-type E1A 1-80 polypeptide and nondefective E1A 1-80 mutant polypeptides were purified with very little variability in biological activity. Further, multiple preparations of biologically defective E1A mutant polypeptides were prepared, minimizing the possibility that variations in folding efficiency account for the lack of biological activity.
Purification of GST fusion proteins.
One-liter cultures of bacterial cells (DH12S; Life Technologies) containing GST expression plasmids were induced with 1 mM IPTG, grown for 4 h, and harvested by centrifugation. The cells were sonicated, and the GST fusion proteins were immobilized on glutathione (GSH)-agarose and extensively washed with phosphate-buffered saline (PBS)-10% NP-40.
In vitro transcription repression assay.
Preparation of nuclear transcription extracts and in vitro transcription repression assays were performed essentially as described previously (26). The template was prepared from CsCl double-banded pHIV-TAR(+) digested with BamHI followed by phenol-chloroform extraction, chloroform extraction, and ethanol precipitation. Each 25-μl runoff reaction mixture contained 400 ng of template DNA; 20 mM HEPES (pH 7.9; at 4°C); 10% glycerol; 120 mM KCl; 10 mM MgCl2; 4 mM creatine phosphate; 0.5 mM dithiothreitol; 4 U of pancreatic RNase inhibitor (Life Technologies); 0.2 mM EDTA; 0.1 mM phenylmethylsulfonyl fluoride; 500 μM (each) UTP, ATP, and GTP; 25 μM CTP; 1 μl of [32P]CTP (800 to 1,000 Ci/mmol; Pharmacia-Amersham Biotech); and 8 μl of nuclear extract. The repression activity of each E1A 1-80 polypeptide was assayed by addition to the reaction mixture of 62.5 to 500 ng of E1A 1-80 or E1A 1-80 mutant polypeptide. After incubation for 60 min at 30°C, transcripts were isolated and resolved by electrophoresis on 6% polyacrylamide gels containing 6 M urea. The dried gels were exposed to phosphor storage screens, and transcripts were quantitated on a STORM 840 PhosphorImager using ImageQuant software (Molecular Dynamics).
Cell microinjection assay for transcription repression.
Cell microinjection was performed essentially as described previously (16). Briefly, A549 cells were grown on scribed 22-mm2 glass coverslips in 35-mm2 tissue culture dishes containing Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum. To measure transcription repression, the SV40 large T antigen-producing plasmid, pSV40, was injected into the cell nucleus with or without the E1A 243R-producing plasmid pcDNA3-243R, E1A 1-80 polypeptide, or E1A 1-80 mutant polypeptide. In order to identify successfully microinjected cells, the green fluorescent protein-expressing plasmid pEGFPN-1 (Clontech) was coinjected at 12 ng/μl. A concentration of pSV40 (8 ng/μl) was selected which produces readily detectable T antigen in 70 to 80% of successfully injected cells after 4 h. Plasmids and polypeptides were dissolved in PBS prior to microinjection.
Cells were fixed 4 h after injection with 3.7% paraformaldehyde in PBS for 10 min and permeabilized in methanol at −20°C. Following rehydration in PBS and treatment with 0.3% Triton X-100 in PBS for 3 min, the cells were blocked in PBS-0.1% Tween-20-5% bovine serum albumin (BSA) for 15 min. The cells were then incubated with mouse monoclonal anti-SV40 T antigen antibody (Ab-2; Oncogene) in PBS-0.1% Tween-20 for 45 min in a humidified incubator at 37°C. After three washes with PBS-0.1% Tween-20, the cells were blocked again with PBS-0.1% Tween-20-5% goat serum for 15 min followed by incubation with Texas Red-conjugated goat anti-mouse secondary antibody (Jackson Laboratories). After being washed in PBS-0.1% Tween-20, the coverslips were mounted and examined by fluorescence microscopy to identify successfully injected cells (fluorescein isothiocyanate [FITC] filter) and cells expressing SV40 T antigen (rhodamine filter). To help quantitate the results of the microinjection assay, positive cells were counted using a neutral density filter in addition to the rhodamine or FITC filter. In this way, faintly fluorescing cells were eliminated from the count, thus greatly reducing subjective considerations.
Electrophoretic mobility shift analysis (EMSA).
Human TBP (hTBP) was prepared as described previously (43). GST-TBP was obtained from Santa Cruz. A double-stranded oligonucleotide containing −15 to +40 of the Ad major late promoter and a 5′ GG was 3′ end labeled with Klenow fragment and [α-32P]dCTP. In addition, a TFIID consensus oligonucleotide (Santa Cruz) was end labeled with polynucleotide kinase and [γ-32P]ATP. Gel shift reactions were performed as described previously (4, 43). Labeled probe complexed to hTBP (or GST-TBP) was resolved on native 5% polyacrylamide gels, using as the running buffer 12.5 mM Tris-borate-0.05 mM EDTA-5 mM MgCl2. The gels were dried, exposed to phosphor storage screens, and visualized using a STORM 840 PhosphorImager.
Protein binding assay.
Fifty-nanogram amounts of E1A 1-80 or mutant polypeptides were diluted to 500 μl with PBS-0.1% NP-40-0.2% BSA. The protein solution was clarified at 10,000 × g for 15 min in a microcentrifuge to remove aggregates and then preincubated for 60 min at 4°C with GSH-agarose beads (containing ≈1 μg of bound GST) followed by centrifugation to remove any proteins interacting nonspecifically with GST or the agarose beads. The solution was added to 0.5 μg of either GST-TBP, GST-p300-segment B′, or GST-CBP-segment B immobilized on 10 μl of GSH-agarose beads previously equilibrated in PBS-0.1% NP-40-0.2% BSA. The binding reaction was incubated at 4°C overnight. The beads were centrifuged and washed three times with PBS-1% NP-40. Bound E1A 1-80 polypeptide was eluted with sodium dodecyl sulfate sample buffer and resolved by electrophoresis on a 15% polyacrylamide gel. E1A polypeptides were transferred to polyvinylidene difluoride membranes and subjected to Western blot analysis by ECF chemistry (Pharmacia-Amersham Biotech) using an affinity-purified polyclonal antibody raised against an E1A CR1 peptide (E1A 40-80). Aliquots of input polypeptides were analyzed on parallel blots to ensure that equal amounts of E1A polypeptides were present in each binding assay. Input and bound polypeptides were visualized on a STORM 840 PhosphorImager using blue-green fluorescence.
RESULTS
Sequences within the first 30 amino acids of E1A are essential for E1A-mediated transcription repression in vitro and in vivo and contain binding sites for TBP, p300, and CBP. In order to probe structure-function relationships, we expressed and purified a panel of E1A mutants within the N-terminal 30 amino acids. The mutants were constructed within the E1A 1-80 polypeptide backbone, which is a strong transcriptional repressor in vitro and in vivo. First, a series of five-residue deletions was created (Fig. 1A). Then each amino acid in turn from residues 2 through 30 was replaced with alanine (Fig. 1B shows the E1A 1-80 sequence). In those cases where alanine was the native amino acid (16Ala and 17Ala), a glycine residue was substituted. All mutant polypeptides were purified to near homogeneity (Fig. 1C and D).
Amino acid sequence requirement for E1A transcription repression in vitro.
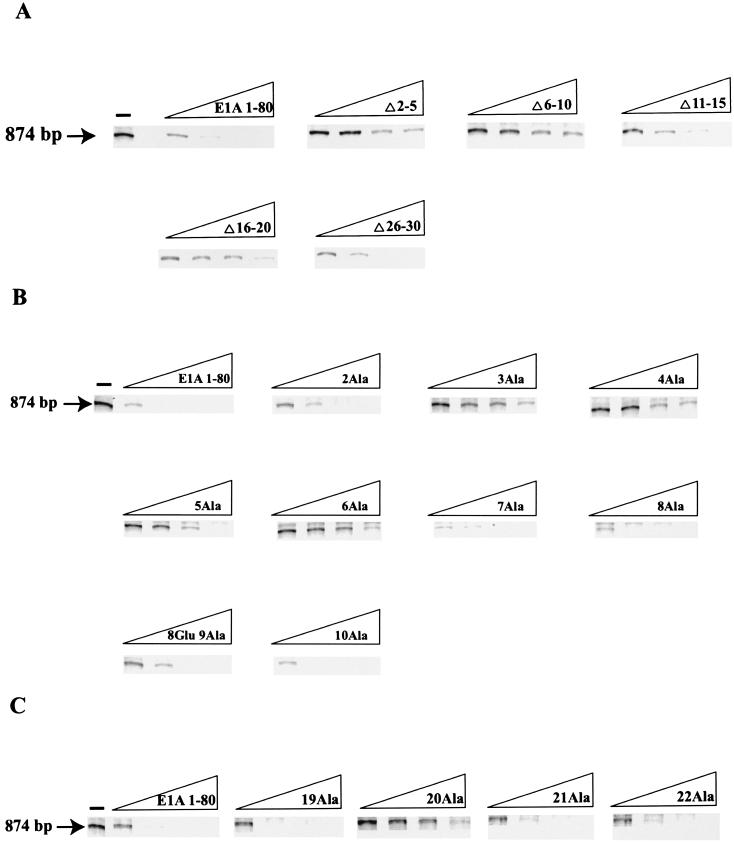
We first examined the ability of the five E1A 1-80 deletion mutants to repress transcription using a sensitive and reliable in vitro transcription repression system (26) with a human immunodeficiency virus long terminal repeat (HIV LTR) promoter template that yields an 874-bp runoff transcript. The HIV LTR promoter is strongly repressed by E1A 243R in vivo (44, 45) and in vitro (38) and by E1A 1-80 in vitro (38). As shown in Fig. 2, in vitro transcription in the absence of E1A polypeptide generates a strong runoff transcript of the expected size, 874 bp. Dose-response measurements with wild-type E1A 1-80 polypeptide show that transcription from the HIV LTR promoter is efficiently repressed at low concentrations (about 60 ng of E1A 1-80 represses transcription over 90%). As shown in Fig. 2A, E1A 1-80Δ2-5, E1A 1-80Δ6-10, and E1A 1-80Δ16-20 are substantially defective in repression activity. E1A 1-80Δ11-15 and E1A 1-80Δ26-30 exhibit marginal loss in repression activity, probably due to alteration in secondary structure as opposed to the deletion of a critical amino acid (see below).
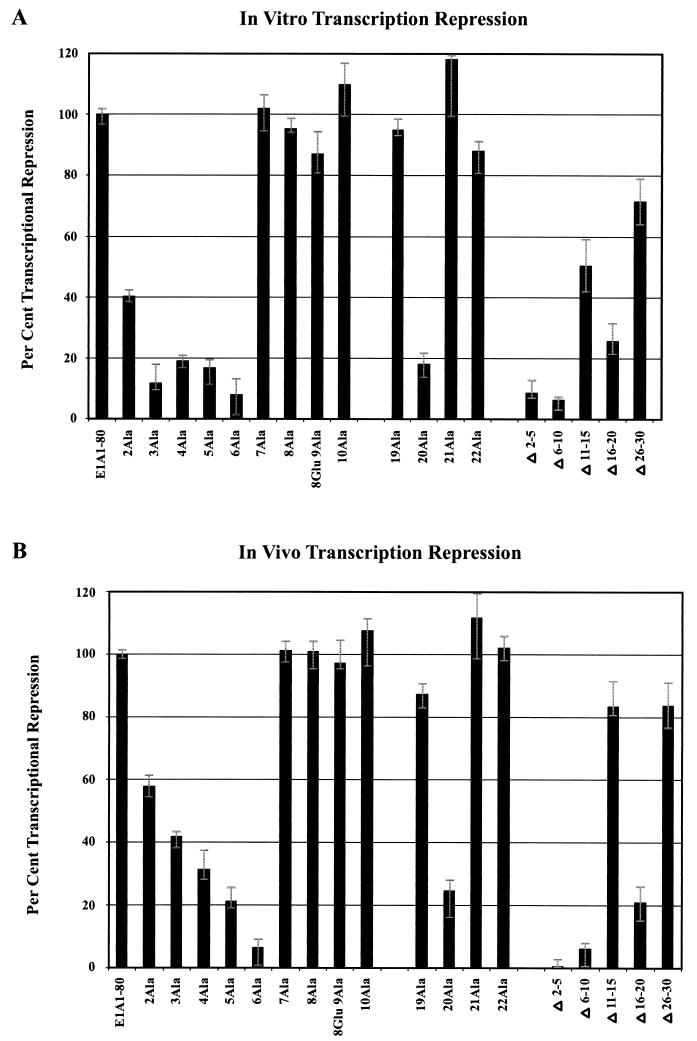
FIG. 2.
Two regions of E1A 1-80, amino acids 2 to 6 and amino acid 20, are critical for E1A repression in vitro. Each E1A polypeptide was assayed for in vitro transcription repression activity at 62.5, 125, 250, and 500 ng per reaction. Each assay was repeated four to six times. At least two independent preparations of each defective mutant were assayed. (A) Representative dose-response measurements of in vitro repression activity of E1A 1-80 and E1A 1-80 deletion polypeptides. (B) Representative dose-response measurements of in vitro repression activity of E1A 1-80 and E1A 1-80 with Ala substituted for residues 2 to 10 (the polypeptide with Ala substituted for 9Gly also contains a Glu substituted for 8Gly). (C) Representative dose-response measurements of in vitro repression activity of E1A 1-80 and E1A 1-80 19Ala, E1A 1-80 20Ala, E1A 1-80 21Ala, and E1A 1-80 22 Ala.
Because small deletions cannot identify critical single amino acids required for repression activity and because deletions may alter secondary structure, leading to erroneous conclusions, we examined each of the 29 amino acid substitution mutants for the ability to repress transcription (Fig. 2B and C). E1A 1-80 with Ala substituted for 2Arg (E1A 1-80 2Ala) was found to be partially defective (about 40% of its activity was retained at 60 ng over the course of multiple experiments). E1A 1-80 3Ala, E1A 1-80 4Ala, E1A 1-80 5Ala, and E1A 1-80 6Ala were found to be substantially defective (strong transcription was evident in reactions with as much as 500 ng of E1A mutant polypeptide). Dose-response measurements with E1A 1-80 containing Ala substituted for residues 7 through 18 showed no significant defect in repression function (results with 7Ala to 10Ala are shown in Fig. 2B, whereas those with 11Ala to 18Ala are not shown). Substitution of Ala for 20Leu (E1A 1-80 20Ala) resulted in a defective mutant, whereas its flanking substitutions (19Ala, 21Ala, and 22Ala) were found to be essentially wild type in activity (Fig. 2C). Dose-response measurements with E1A 1-80 containing Ala substituted for residues 23 through 30 showed wild-type repression activity (data not shown). We conclude that there are two regions within the first 30 amino acids that are critical for the E1A repression function in vitro, residues 2 to 6 and residue 20. Our results analyzing the single-amino-acid substitution mutants indicate that the partial loss of activity of E1A 1-80Δ11-15 and E1A 1-80Δ26-30 are not due to deletion of critical amino acids.
Amino acid sequence requirement for E1A transcription repression in vivo.
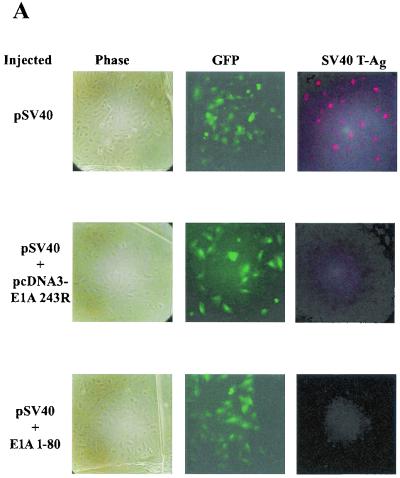
To determine whether the amino acid residues critical for repression in vitro are also important for repression in vivo, we analyzed the same panel of E1A 1-80 mutant polypeptides by an efficient and reproducible cell microinjection assay (16, 37). This assay has the benefit of comparing the biological effects of E1A 1-80 polypeptides within cells using a known amount of protein delivered at a precise time. The reporter in the cell microinjection assay, pSV40, expresses the SV40 T antigen, which is visualized by indirect fluorescence microscopy. Successfully microinjected cells are identified by direct fluorescence microscopy of the green fluorescent protein expressed from coinjected plasmid pEGFPN-1 (Clontech). Figure 3A illustrates this assay. Each row represents the same field of cells visualized under different conditions. The first column is visualized under phase microscopy, the second column is visualized under conditions to identify the green fluorescent protein (successfully injected cells), and the third column is visualized under conditions to identify SV40 T antigen-expressing cells. Figure 3A (top row) shows that T antigen is expressed in the majority of successfully microinjected cells in the absence of E1A. When coinjected with either the E1A 243R-expressing plasmid pcDNA3-E1A 243R (Fig. 3A, middle row) or wild-type E1A 1-80 polypeptide (Fig. 3A, bottom row), SV40 T antigen synthesis is severely repressed.
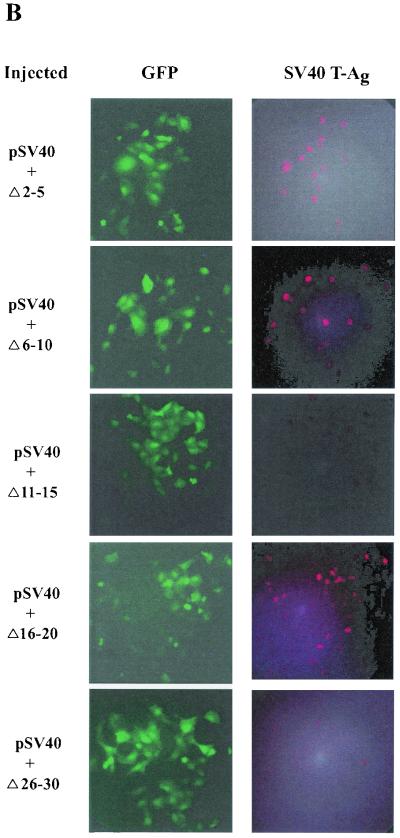
FIG. 3.
Two regions of E1A 1-80, amino acids 2 to 6 and amino acid 20, are critical for E1A repression in vivo. Each individual experiment involved the microinjection of about 100 cells, but only a portion of each microinjected area is seen in the representative photographic fields in these figures. (A) Representative cell microinjection assays demonstrating the transcriptional-repression function of E1A 243R and E1A 1-80. The photomicrographs in each row are of the same field of cells visualized under different conditions. The left column (Phase) is visualized under phase microscopy. The middle column (GFP) is visualized under fluorescence microscopy through an FITC transparent filter and shows cells expressing green fluorescent protein (GFP), a marker for successfully injected cells. The right column (SV40 T-Ag) is visualized under fluorescence microscopy through a rhodamine transparent filter and shows cells expressing SV40 T antigen. Cells in the first row were coinjected with 8 ng of pSV40/μl and 10 ng of pEGFPN-1/μl. About 75% of successfully injected cells were positive for SV40 T antigen. Cells in the second row were coinjected with pSV40, pEGFPN-1, and 10 ng of the E1A 243R-expressing plasmid pcDNA3-E1A 243R/μl. Cells in the third row were coinjected with pSV40, pEGFPN-1, and 25 ng of E1A 1-80 polypeptide/μl. (B) Representative microinjection assays demonstrating that E1A 1-80Δ2-5, E1A 1-80Δ6-10, and E1A 1-80Δ16-20 are defective in transcription repression function in vivo. Cells were coinjected with 8 ng of pSV40/μl, 10 ng of pEGFPN-1/μl, and 25 ng of the indicated E1A 1-80 deletion mutant/μl. (C) Representative assays demonstrating that E1A 1-80 2Ala, 3Ala, 4Ala, 5Ala, and 6Ala are defective to various degrees in transcription repression function in vivo. Cells were injected as described for panel B above except that 25 ng of the indicated Ala substitution mutant/μl was injected. (D) Representative assays demonstrating that E1A 1-80 20Ala is defective in transcription repression function in vivo. Cells were injected as described for panel B above. It should be noted that for E1A 1-80 20Ala, the field shown has fewer cells than fields coinjected with other mutants. E1A 1-80 20 Ala is nonetheless clearly defective: 17 cells were positive for T antigen of 21 cells injected, representing 80% of successfully injected cells visualized in this field.
We first determined whether the E1A 1-80 deletion mutants defective in transcription repression in vitro are also defective in vivo. E1A 1-80Δ2-5, E1A 1-80Δ6-10, and E1A 1-80Δ16-20 are clearly defective in E1A repression function; a large proportion of coinjected cells expressed T antigen (Fig. 3B). On the other hand, E1A 1-80Δ11-15 and E1A 1-80Δ26-30 had essentially wild-type repression activity in that few coinjected cells expressed T antigen (Fig. 3B).
The results described above suggest that some amino acid residues within positions 2 to 5 and 6 to 10 are critical for E1A repression in vivo. We therefore examined the single-amino-acid substitution mutants from residues 2 to 10. Figure 3C shows the results of a typical experiment. E1A 1-80 2Ala is partially defective, whereas E1A 1-80 3Ala, E1A 1-80 4Ala, E1A 1-80 5Ala, and E1A 1-80 6Ala are substantially defective in the ability to repress expression from the SV40 early promoter. E1A 1-80 7Ala, E1A 1-80 8Ala, E1A 1-80 9Ala, and E1A 1-80 10Ala retain a wild-type phenotype.
Because E1A 1-80Δ16-20 was defective for in vivo repression (Fig. 3B) and E1A 1-80 20Ala was defective for in vitro repression (Fig. 2C), we analyzed by microinjection the amino acid substitutions around amino acid 20. Figure 3D demonstrates that E1A 1-80 20Ala is substantially defective in E1A repression ability whereas E1A 1-80 19Ala, E1A 1-80 21Ala, and E1A 1-80 22Ala retain wild-type activity.
Quantitative comparison of the E1A amino acid requirements for transcription repression in vitro and in vivo.
The averages of four to six independent in vitro transcription repression analyses are presented in Fig. 4A, and the averages of two to three cell microinjection analyses are presented in Fig. 4B. Both in vitro and in vivo, substitution of Ala for 6Cys leads to virtually complete loss of transcription repression activity. Substitution of Ala for 2Arg leads to only partial defectiveness, whereas substitution for residues 3His, 4Ile, and 5Ile leads to a substantially more defective phenotype. Finally, substitution of Ala for 20Leu results in a strongly defective phenotype. Thus, the patterns of E1A N-terminal sequence requirements for in vitro and in vivo activity are remarkably similar.
FIG. 4.
Quantitative comparison of in vitro and in vivo transcription repression activity of mutant E1A 1-80 polypeptides. (A) Quantitative PhosphorImage results of four to six independent in vitro experiments at 62.5 ng of polypeptide per reaction were averaged and normalized to the amount of repression exhibited by wild-type E1A 1-80 polypeptide. (B) Quantitative results of two to three independent cell microinjection experiments were averaged and normalized to repression exhibited by wild-type E1A 1-80 polypeptide. The error bars indicate the high and low values of the averaged data.
E1A amino acid sequence requirements for disruption of TBP-TATA interaction.
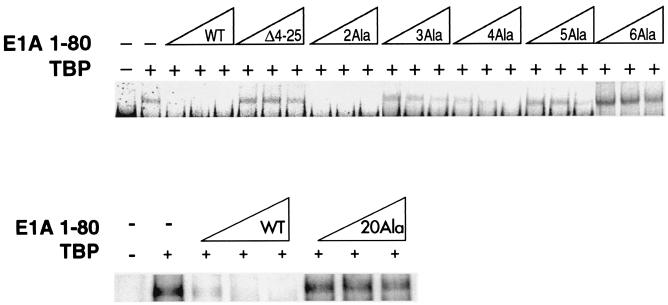
We have previously reported that the E1A 243R protein and the E1A 1-80 polypeptide can dissociate the interaction between TBP and TATA DNA, as shown by both DNase footprint analysis and EMSA (40). This ability implies an in vivo function for the E1A repression domain. It was therefore important to examine the panel of E1A 1-80 mutant polypeptides for the ability to interfere with TBP-TATA interaction. An end-labeled oligonucleotide probe containing the TATA element was incubated with recombinant TBP in the absence or presence of wild-type E1A 1-80 or mutant E1A 1-80 polypeptide. The TBP-TATA complex was resolved by gel electrophoresis and visualized by PhosphorImage analysis. As shown in Fig. 5, when TBP is present in the binding mixture, a band containing the TBP-TATA complex is formed (compare top and bottom, lanes 1 and 2). Wild-type E1A 1-80 polypeptide completely prevents the formation of the TBP-TATA complex (top and bottom, lanes 3 to 5). However, E1A 1-80Δ4-25, a mutant lacking most of the first 30 amino acids and defective in vitro and in vivo for repression ability, has no effect on TBP-TATA complex formation (top, lanes 6 to 8).
FIG. 5.
Ability of E1A 1-80 polypeptide mutants to interfere with complex formation between TBP and TATA DNA. EMSA was performed using as the probe a 32P-labeled oligonucleotide containing a TATA element. hTBP (TBP) was added (+) as indicated (GST-TBP was used in the lower lanes). E1A 1-80 or mutant E1A 1-80 polypeptide was added as indicated at three concentrations, 50, 100, and 200 ng. The reaction products were analyzed by native polyacrylamide gel electrophoresis and visualized by PhosphorImage analysis as described in Materials and Methods. These analyses were repeated two to three times. Two independent preparations of E1A 1-80 polypeptides exhibiting a mutant phenotype were analyzed with essentially the same results.
We first examined the effect of each E1A 1-80 deletion polypeptide on the formation of the TBP-TATA complex. E1A 1-80Δ2-5, E1A 1-80Δ6-10, and E1A 1-80Δ16-20 are defective in the ability to interfere with TBP-TATA complex formation, whereas E1A 1-80Δ11-15 and E1A 1-80Δ26-30 retained near wild-type activity (data not shown). Thus, the same deletion mutants that are defective for the ability to support transcription repression in vitro and in vivo (Fig. 4) are defective for the ability to interfere with TBP-TATA interaction.
We next analyzed the ability of the E1A 1-80 substitution mutants to interfere with TBP-TATA interaction. The pattern of defectiveness observed for disruption of a TBP-TATA complex is strikingly similar to the pattern of defectiveness observed for repression of transcription in vitro and in vivo. As shown in Fig. 5 (top), E1A 1-80 4Ala is partially defective, E1A 1-80 3Ala and E1A 1-80 5Ala are substantially defective, and E1A 1-80 6Ala is completely defective. It is of interest to note that E1A 1-80 6Ala appears to induce the formation of a greater quantity of TBP-TATA complex than is observed in the absence of E1A. The significance of this reproducible phenomenon is not clear. Nuclear magnetic resonance structural studies may provide insight. E1A 1-80 2Ala in Fig. 5 (top) does not appear to be appreciably defective, but in other analyses with different preparations, a 20 to 30% reduced ability to interfere with TBP-TATA complex formation was observed. Substitutions in amino acids 7 through 19 had no effect on the ability of the E1A polypeptide to interfere with TBP-TATA interaction (data not shown). Significantly, E1A 1-80 20Ala, which is defective in repression activity in vitro and in vivo, is also strongly defective in the ability to interfere with TBP-TATA complex formation (Fig. 5, bottom lanes). Mutants with substitutions in amino acids 21 through 30 exhibit wild-type phenotypes (data not shown).
In vitro binding of mutant E1A 1-80 polypeptides to TBP, p300, and CBP.
TBP, p300, and CBP have been implicated as cellular targets or partners in E1A repression, although molecular mechanisms are yet to be defined. To further understand the interactions between the E1A N-terminal 30-amino-acid sequence and these cellular proteins, in vitro protein binding assays with the E1A 1-80 deletion and substitution polypeptides were performed. Fusion proteins between GST and (i) full-length TBP (GST-TBP), (ii) a p300 fragment containing the E1A binding site (GST-p300-segment B′), and (iii) a CBP fragment containing the E1A binding site (GST-CBP-segment B) were immobilized on GSH-agarose and incubated with approximately equimolar amounts of wild-type and mutant E1A 1-80 polypeptides. The input, in all binding experiments, was monitored to ensure that approximately equal levels of E1A polypeptides were available for binding (data not shown). E1A 1-80 polypeptides bound to the GST fusion proteins were resolved on sodium dodecyl sulfate-polyacrylamide gels and analyzed by Western blot analysis using an affinity-purified polyclonal antibody raised against the E1A CR1 domain.
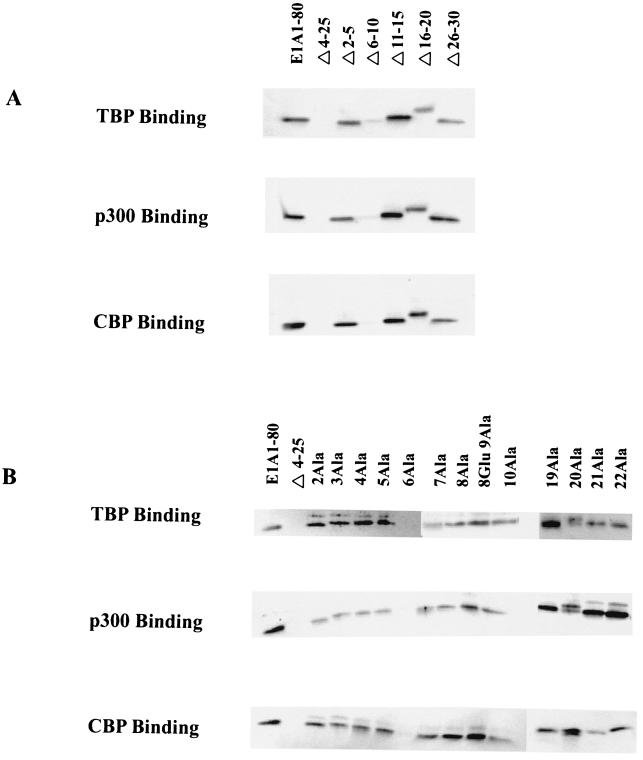
Wild-type E1A 1-80 binds strongly to GST-TBP, GST-p300, and GST-CBP (Fig. 6A and Fig. 6B). This interaction is specific, because an E1A 1-80 deletion mutant which lacks most of the first 30 amino acids (E1A 1-80Δ4-25) does not bind to any of the GST fusion proteins (Fig. 6). This E1A 1-80 deletion mutant was previously shown to lack E1A repression activity and to lack the ability to bind TBP and p300 (40) (Green and Loewenstein, unpublished). Analysis of the E1A 1-80 deletion mutants shows clearly that E1A 1-80Δ6-10 is defective in the ability to interact with TBP, p300, and CBP (Fig. 6A). Of interest, E1A 1-80Δ2-5 and E1A 1-80Δ16-20, which are defective in repression function in vitro and in vivo, bind to GST-TBP, GST-p300, and GST-CBP (Fig. 6A).
FIG. 6.
In vitro binding of E1A 1-80 mutant polypeptides to GST-TBP, GST-p300-segment B, and GST-CBP-segment B. E1A 1-80 or mutant E1A 1-80 polypeptide was incubated with an equimolar amount of GSH-agarose immobilized ligand (TBP, p300, or CBP). Bound E1A polypeptide was analyzed by Western blotting and visualized by blue-green fluorescence on a STORM 840 PhosphorImager as described in Materials and Methods. (A) Analysis of E1A 1-80 deletion polypeptides. (B) Analysis of E1A 1-80 substitution polypeptides.
The ability of the E1A substitution mutants to interact with TBP, p300, and CBP was next assayed. E1A 1-80 6Ala is completely deficient in binding to TBP, p300, and CBP (Fig. 6B). Of interest, E1A 1-80 6Ala is the substitution mutant most defective in the ability to repress transcription in vitro and in vivo (Fig. 4). Consistent with the ability of E1A 1-80Δ2-5 to bind TBP, p300, and CBP, E1A 1-80 2Ala, 3Ala, 4Ala, and 5Ala exhibited wild-type ability to bind these proteins. Likewise, when the substitution mutants around residue 20 were assayed, there appeared to be no defect in the ability of E1A 1-80 19Ala, 20Ala, 21Ala, and 22Ala to bind TBP, p300, and CBP (Fig. 6B).
DISCUSSION
The E1A N-terminal amino acid requirements for interference with TBP-TATA box complex formation are the same as those required for repression of transcription in vitro and in vivo.
Data showing a strong correlation between the ability of a mutant to repress transcription in vitro and in vivo and to interfere with TBP-TATA complex formation are summarized in Table 1. Two regions were identified, amino acids 2 to 6 and amino acid 20, that are required for all three functions. Substitution of Ala for 6Cys produced a mutant particularly deficient in these functions. Since disulfide bridges are often essential elements of secondary structure, it is an attractive possibility that E1A 1-80 6Ala is defective due to structural alteration. However, 6Cys is the only cysteine residue in the E1A repression domain and therefore is probably not simply a structural element. Although 6Cys is not conserved among all Ad serotypes, it is conserved among group C Ads and could play an essential role for these Ad serotypes, for example, in homodimer formation. Replacement of amino acids N terminal to 6Cys results in defective phenotypes in all three functions. Replacement of 3His or 5Ile leads to mutants substantially defective in all three functions. Replacement of 4Ile yields a mutant partially defective in the ability to block TBP-TATA interaction but strongly defective in repression function in vitro and in vivo. Substitution of Ala for 2Arg yields a mutant only partially defective in the ability to block TBP-TATA formation and to support repression in vitro and in vivo.
TABLE 1.
Correlations among E1A N-terminal amino acid requirements for transcription repression in vitro and in vivo and for interference with TBP-TATA complex formationa
| Mutant polypeptide | In vitro repression | In vivo repression | TBP-TATA interference |
|---|---|---|---|
| E1A 1-80 wild type | ++++ | ++++ | ++++ |
| E1A 1-80Δ2-5 | − | − | − |
| E1A 1-80Δ6-10 | − | − | − |
| E1A 1-80Δ11-15 | ++ | +++ | ++++ |
| E1A 1-80Δ16-20 | − | − | − |
| E1A 1-80Δ26-30 | +++ | +++ | +++ |
| E1A 1-80 2A1a | ++ | ++ | ++ |
| E1A 1-80 3A1a | − | + | − |
| E1A 1-80 4A1a | − | ± | ± |
| E1A 1-80 5A1a | − | − | − |
| E1A 1-80 6A1a | − | − | − |
| E1A 1-80 7A1a | ++++ | ++++ | ++++ |
| E1A 1-80 8A1a | ++++ | ++++ | ++++ |
| E1A 1-80 9A1a | ++++ | ++++ | ++++ |
| E1A 1-80 10A1a | ++++ | ++++ | ++++ |
| E1A 1-80 19A1a | ++++ | ++++ | ++++ |
| E1A 1-80 20A1a | − | − | − |
| E1A 1-80 21A1a | ++++ | ++++ | ++++ |
| E1A 1-80 22A1a | ++++ | ++++ | ++++ |
All of the single E1A 1-80 mutants were assayed for the three parameters. Mutants not shown in the Table (E1A 1-80 11 A1a to 18 A1a and 23 A1a to 30 A1a) were wild type in their ability to repress transcription in vitro and in vivo and to interfere with TBP-TATA complex formation. ++++ indicates wild-type activity (80 to 100%); − is defined as a completely mutant phenotype (greater-than-five-fold reduction in activity). Intermediate values are indicated by ±, +, ++, and +++. The percentages of wild-type activity for repression in vitro and in vivo are presented in Fig. 4.
The second critical site within the first 30 amino acids in E1A 1-80 is 20Leu, which is required for transcription repression in vitro and in vivo as well as for interference with the formation of a TBP-TATA complex. That 20Leu is a critical amino acid is supported by the finding that a mutant lacking residues 16 to 20 (E1A 1-80Δ16-20) is defective in these functions. It is of interest that 20Leu is one of the few strongly conserved amino acids in the nonconserved N-terminal domain among different Ad serotypes (11, 22).
Replacement of amino acid 6Cys abrogates the ability of E1A 1-80 to interact with its cellular partners.
Protein binding assays at approximately equimolar concentrations of E1A 1-80 mutant polypeptide and TBP, p300, or CBP revealed that only amino acid 6Cys is unequivocally required for interaction. Under these binding conditions, E1A 1-80 2Ala, 3Ala, 4Ala, 5Ala, and 20Ala were found to bind to their ligands, TBP, p300, and CBP, as well as does wild-type E1A 1-80. Inasmuch as these mutants are functionally defective in transcription repression in vitro and in vivo (Fig. 4), as well as for disruption of a TBP-TATA complex (Fig. 5), it is possible that some or all of these amino acids have critical functions other than that of interacting with TBP, p300, or CBP. If this were so, the in vitro binding assay would not detect the necessity of these amino acids for these other in vivo activities. It is also possible that these E1A residues are required for interacting with cellular partners under native conditions where E1A may function as part of an intracellular complex containing TBP, p300/CBP, and PCAF, as well as other factors (7, 12, 21, 23, 48, 49). In any case, it is clear that amino acid 6 is pivotal for repression function and for TBP-TATA disruption, as well as for binding to TBP, p300, and CBP.
Replacement of E1A 2Arg with Gly in Ad has been reported to result in a defective ability to bind p300 by coimmunoprecipitation of infected cell lysates and to transform cells (47). Wang et al. (45) analyzed Ad with a substitution in E1A residue 2, 3, 20, or 21. Substitution in residue 2 substantially reduced the ability to coimmunoprecipitate p300, whereas substitution in residue 3 or 20 partially reduced the ability to coimmunoprecipitate p300. However, substitution in residue 2 or 3, but not 20 or 21, resulted in a loss of repression activity as determined by transfection analysis with E1A 12S mutant plasmids. These data did not allow for a clear correlation between p300 binding and repression activity. The continued ability of E1A 12S plasmid with a substitution in E1A residue 20 to support the E1A repression function (45) differs from our findings that E1A 1-80 20Ala is defective in repression function in vitro and in vivo. Possibly this difference reflects the expression of other E1A functional domains from the E1A 12S plasmid in the study by Wang et al. (45); this possibility is avoided by the use of mutant E1A 1-80 polypeptides.
Molecular mechanism of E1A transcription repression.
We have identified two regions within the N-terminal 30 amino acids that are important for transcription repression. 6Cys is required not only for transcription repression function but also for binding TBP and p300/CBP. p300 is able to interact with another sequence at the E1A N terminus within residues 48 to 60. Although sequences within residues 48 to 60 are required for p300 binding, they are not required for TBP binding because deletion of residues 48 to 60 from E1A 1-80 (E1A 1-80Δ48-60) reduces binding to TBP only marginally (20 to 30%) (40). Because p300/CBP is likely involved in E1A repression and because p300/CBP functions as a coactivator for promoters that regulate the cell cycle and cell differentiation, it is an attractive possibility that promoter-bound p300/CBP serves as a “molecular scaffold” that E1A utilizes to gain access to specific promoters involved in growth regulation. We therefore suggest a two-step working model: (i) E1A uses p300/CBP as a high-affinity molecular scaffold to access specific promoters where (ii) E1A can inhibit the general transcription machinery by interfering with the formation of a TBP-TATA complex. It is also possible that TBP and p300/CBP represent independent targets of the E1A repression domain on different cellular promoters.
One can speculate that binding sites within E1A residues 48 to 60 initially strongly interact with promoter-bound p300, which could then permit the E1A 6Cys region to be released and bind to TBP. This interaction could alter the conformation of TBP, thus melting it from the TATA box. The role of 20Leu in E1A repression is not known. It could be involved in the interaction with TBP or the melting of TBP from the TATA box. Alternatively, it could be required for an essential conformation of the E1A repression domain. To help distinguish among these alternatives, nuclear magnetic resonance studies of wild-type and mutant E1A 1-80 polypeptides are being performed.
To summarize, the study described here provides strong evidence that TBP is a functional target of the E1A N-terminal repression domain. Further, a molecular mechanism of E1A repression is suggested by the fact that interference with TBP-TATA formation maps to E1A sequences that are required for repression in vivo and in vitro. The promoter specificity of E1A repression most likely involves interaction between E1A and p300/CBP. This will be the subject of future studies.
Acknowledgments
We thank Y. Nakatane for generous gifts of plasmids.
This work was supported by Research Career Award AI-04739 and Public Health Service grant CA29561 to M.G. from the National Institutes of Health.
REFERENCES
- 1.Arany, Z., D. Newsome, E. Oldread, D. M. Livingston, and R. Eckner. 1995. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature 374:81–84. [DOI] [PubMed] [Google Scholar]
- 2.Boulukos, K. E., and E. B. Ziff. 1993. Adenovirus 5 E1A proteins disrupt the neuronal phenotype and growth factor responsiveness of PC12 cells by a conserved region 1-dependent mechanism. Oncogene 8:237–248. [PubMed] [Google Scholar]
- 3.Boyd, J. M., T. Subramanian, U. Schaeper, M. LaRegina, and G. Chinnadurai. 1993. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24 ras mediated transformation, tumorigenesis and metastasis. EMBO J. 12:469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant, G. O., L. S. Martel, S. K. Burley, and A. J. Berk. 1996. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev. 10:2491–2504. [DOI] [PubMed] [Google Scholar]
- 5.Chakravarti, D., V. LaMorte, M. Nelson, T. Nelson, T. Nakahima, I. Schulman, H. Juguilon, M. Montminy, and R. Evans. 1996. Role of CBP/P300 in nuclear receptor signalling. Nature 365:99–103. [DOI] [PubMed] [Google Scholar]
- 6.Chakravarti, D., V. Ogryzko, H.-Y. Kao, A. Nash, H. Chen, Y. Nakatani, and R. M. Evans. 1999. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96:393–403. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor co-activator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569–580. [DOI] [PubMed] [Google Scholar]
- 8.Chrivia, J. C., R. P. S. Kwok, M. Lamb, M. Nagiwara, M. R. Montminy, and R. H. Goodman. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365:855–859. [DOI] [PubMed] [Google Scholar]
- 9.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245–2262. [DOI] [PubMed] [Google Scholar]
- 10.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1a-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869–884. [DOI] [PubMed] [Google Scholar]
- 11.Gedrich, R. W., S. T. Bayley, and D. A. Engel. 1992. Induction of AP-1 DNA-binding activity and c-fos mRNA by adenovirus 243R E1A protein and cyclic AMP requires domains necessary for transformation. J. Virol. 66:5849–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass, C. K., D. W. Rose, and M. G. Rosenfeld. 1997. Nuclear receptor coactivators. Curr. Opin. Cell Biol. 9:222–232. [DOI] [PubMed] [Google Scholar]
- 13.Glenn, G. M., and R. P. Ricciardi. 1987. An adenovirus type 5 E1A protein with a single amino acid substitution blocks wild type E1A transactivation. Mol. Cell. Biol. 7:1004–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gluzman, Y., J. F. Sambrook, and R. J. Frisque. 1980. Expression of early genes of origin-defective mutants of simian virus 40. Proc. Natl. Acad. Sci. USA 7:3898–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, M., P. M. Loewenstein, R. Pusztai, and J. S. Symington. 1988. An adenovirus E1A protein domain activates transcription in vivo and in vitro in the absence of protein synthesis. Cell 53:921–926. [DOI] [PubMed] [Google Scholar]
- 16.Green, M., A. Thorburn, and P. M. Loewenstein. 1998. Cell microinjection: in vivo analysis of the functional domains of viral regulatory proteins. Methods Mol. Med. 21:169–193. [DOI] [PubMed] [Google Scholar]
- 17.Green, M. R. 2000. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25:59–63. [DOI] [PubMed] [Google Scholar]
- 18.Hen, R., E. Borrelli, and P. Chambon. 1985. Repression of the immunoglobulin heavy chain enhancer by the adenovirus-2 E1A products. Science 230:1391–1394. [DOI] [PubMed] [Google Scholar]
- 19.Howe, J. A., J. S. Mymryk, C. Egan, P. E. Branton, and S. T. Bayley. 1990. Retinoblastoma growth suppressor and a 300-kDa protein appear to regulate cellular DNA synthesis. Proc. Natl. Acad. Sci. USA 87:5883–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jelsma, T. N., J. Howe, J. S. Mymryk, C. M. Evelegh, N. F. A. Cunniff, and S. T. Bayley. 1989. Sequences in E1A proteins of human adenovirus 5 required for cell transformation, repression of a transcriptional enhancer, and induction of proliferating cell nuclear antigen. Virology 171:120–130. [DOI] [PubMed] [Google Scholar]
- 21.Kamei, Y., L. Xu, T. Heinzel, J. Torchia, R. Purokawa, B. Gloss, S. Lin, R. Heyman, D. Rose, C. Glass, and M. G. Rosenfeld. 1996. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85:403–414. [DOI] [PubMed] [Google Scholar]
- 22.Kimelman, D., J. S. Miller, D. Porter, and B. E. Roberts. 1985. E1a regions of the human adenovirus and of the high oncogenic simian adenovirus 7 are closely related. J. Virol. 53:399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurokawa, R., D. Kalafus, M.-H. Ogliastro, C. Kioussi, L. Xu, J. Torchia, M. G. Rosenfeld, and C. K. Glass. 1998. Differential use of CREB binding protein-coactivator complexes. Science 279:700–703. [DOI] [PubMed] [Google Scholar]
- 24.Lillie, J. W., M. Green, and M. R. Green. 1986. An adenovirus E1a protein region required for transformation and transcriptional repression. Cell 46:1043–1051. [DOI] [PubMed] [Google Scholar]
- 25.Lillie, J. W., P. M. Loewenstein, M. R. Green, and M. Green. 1987. Functional domains of adenovirus type 5 E1a proteins. Cell 50:1091–1100. [DOI] [PubMed] [Google Scholar]
- 26.Loewenstein, P. M., C.-Z. Song, and M. Green. 1998. In vitro transcription: probing the molecular functions of adenovirus regulatory protein. Methods Mol. Med. 21:157–168. [Google Scholar]
- 27.Marciniak, R. A., and P. A. Sharp. 1991. HIV-1 Tat protein promotes formation of more processive elongation complexes. EMBO J. 10:4189–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moran, E., and M. B. Mathews. 1987. Multiple functional domains in the adenovirus E1A gene. Cell 48:177–178. [DOI] [PubMed] [Google Scholar]
- 29.Moran, E. 1993. DNA tumor virus transforming proteins and the cell cycle. Curr. Opin. Genet. Dev. 3:63–70. [DOI] [PubMed] [Google Scholar]
- 30.Moran, E., B. Zerler, T. M. Harrison, and M. B. Mathews. 1986. Identification of separate domains in the adenovirus E1A gene for immortalization activity and the activation of virus early genes. Mol. Cell. Biol. 6:3470–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakajima, T., C. Uchida, S. F. Andreon, J. D. Parvin, and M. Montminy. 1997. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 11:738–747. [DOI] [PubMed] [Google Scholar]
- 32.Philipson, L. 1995. Adenovirus—an eternal archetype. Curr. Top. Microbiol. Immunol. 199:1–24. [DOI] [PubMed] [Google Scholar]
- 33.Puri, P. L., V. Sartorelli, X. J. Yang, Y. Hamamori, V. V. Ogryzko, B. H. Howard, L. Kedes, J. Y. Wang, A. Graessmann, Y. Nakatani, and M. Levrero. 1997. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell 1:35–45. [DOI] [PubMed] [Google Scholar]
- 34.Schneider, J. F., F. Fisher, C. R. Goding, and N. C. Jones. 1987. Mutational analysis of the adenovirus E1a gene: the role of transcriptional regulation in transformation. EMBO J. 6:2053–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shenk, T. 1996. Adenoviridae: the viruses and their replication, p.2111–2148. In B. Fields et al. (ed.), Virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 36.Smith, C. L., S. A. Onate, M.-J. Tsai, and B. W. O’Malley. 1996. CREB binding protein acts synergistically with steroid receptor coactivator 1 to enhance steroid receptor-dependent transcription. Proc. Natl. Acad. Sci. USA 93:8884–8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song, C.-Z., C. J. Tierney, P. M. Loewenstein, R. Pusztai, J. S. Symington, Q. Tang, K. Toth, S. T. Bayley, and M. Green. 1995. Transcriptional repression by human adenovirus E1A N-terminus/conserved domain 1 polypeptides in vivo and in vitro in the absence of protein synthesis. J. Biol. Chem. 40:23263–23267. [DOI] [PubMed] [Google Scholar]
- 38.Song, C.-Z., P. M. Loewenstein, and M. Green. 1995. Repression in vitro, by human adenovirus E1A protein domains, of basal or Tat-activated transcription of the human immunodeficiency virus type 1 long terminal repeat. J. Virol. 69:2907–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song, C.-Z., P. M. Loewenstein, K. Toth, and M. Green. 1995. TFIID is a direct functional target of the adenovirus E1A transcription-repression domain. Proc. Natl. Acad. Sci. USA 92:10330–10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song, C.-Z., P. M. Loewenstein, K. Toth, Q. Tang, A. Nishikawa, and M. Green. 1997. The adenovirus E1A repression domain disrupts the interaction between the TATA binding protein and the TATA box in a manner reversible by TFIIB. Mol. Cell. Biol. 17:2186–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stein, R. W., and E. B. Ziff. 1987. Repression of insulin gene expression by adenovirus type E1A proteins. Mol. Cell. Biol. 7:1164–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian, T., M. LaRegina, and G. Chinnadurai. 1989. Enhanced ras oncogene mediated cell transformation and tumorigenesis by adenovirus 2 mutants lacking the C-terminal region of E1a protein. Oncogene 4:415–420. [PubMed] [Google Scholar]
- 43.Tang, H., X. Sun., D. Reinberg, and R. H. Ebright. 1996. Protein-protein interactions in eukaryotic transcription initiation: structure of the preinitiation complex. Proc. Natl. Acad. Sci. USA 93:1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ventura, A. M., M. Q. Arens, A. Srinivasan, and G. Chinnadurai. 1990. Silencing of human immunodeficiency virus long terminal repeat expression by an adenovirus E1a mutant. Proc. Natl. Acad. Sci. USA 87:1310–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, H.-G. H., Y. Rikitake, M. C. Carter, P. Yaciuk, S. E. Abraham, B. Zerler, and E. Moran. 1993. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J. Virol. 67:476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webster, K. A., G. E. O. Muscat, and L. Kedes. 1988. Adenovirus E1A products suppress myogenic differentiation and inhibit transcription from muscle-specific promoters. Nature 332:553–557. [DOI] [PubMed] [Google Scholar]
- 47.Whyte, P., N. M. Williamson, and E. Harlow. 1989. Cellular targets for transformation by the E1A proteins. Cell 56:67–75. [DOI] [PubMed] [Google Scholar]
- 48.Yang, X.-J., V. V. Ogrysko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with adenoviral oncoprotein E1A. Nature 382:319–324. [DOI] [PubMed] [Google Scholar]
- 49.Yao, T.-P., G. Ku, N. Zhou, R. Scully, and D. M. Livingston. 1996. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl. Acad. Sci. USA 93:10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu, D., T.-C. Suen, D.-H. Yan, L.-S. Chang, and M.-C. Hung. 1990. Transcriptional repression of the neu protooncogene by the adenovirus 5 E1A gene products. Proc. Natl. Acad. Sci. USA 87:4499–4503. [DOI] [PMC free article] [PubMed] [Google Scholar]