Abstract
We have previously shown that the E protein of the coronavirus infectious bronchitis virus (IBV) is localized to the Golgi complex when expressed exogenously from cDNA. Here, we report that neither the transmembrane domain nor the short lumenal domain of IBV E is required for Golgi targeting. However, an N-terminal truncation containing only the cytoplasmic domain (CTE) was efficiently localized to the Golgi complex, and this domain could retain a reporter protein in the Golgi. Thus, the cytoplasmic tail of the E protein is necessary and sufficient for Golgi targeting. The IBV E protein is palmitoylated on one or two cysteine residues adjacent to its transmembrane domain, but palmitoylation was not required for proper Golgi targeting. Using C-terminal truncations, we determined that the IBV E Golgi targeting information is present between tail amino acids 13 and 63. Upon treatment with brefeldin A, both the E and CTE proteins redistributed to punctate structures that colocalized with the Golgi matrix proteins GM130 and p115 instead of being localized to the endoplasmic reticulum like Golgi glycosylation enzymes. This suggests that IBV E is associated with the Golgi matrix through interactions of its cytoplasmic tail and may have interesting implications for coronavirus assembly in early Golgi compartments.
The Golgi apparatus is a dynamic membrane-bound organelle in which proteins and lipids are modified and sorted to their final destinations. Resident Golgi proteins, such as those involved in the processing and sorting functions of the organelle, must be distinguished from the multitude of proteins that move through the secretory pathway to reach post-Golgi locations. Detailed mechanisms of how this distinction occurs remain unclear, but several types of targeting signals have been found within Golgi resident proteins (27).
Several types of viruses derive their lipid envelopes by budding into the membranes of the Golgi complex (14). While there is still much to learn about the mechanisms and advantages of this interesting process, it is likely that envelope protein targeting plays an important role in selection of the budding site. In addition, the envelope proteins of viruses that assemble in the Golgi complex have served as good models for the study of Golgi protein targeting. The first Golgi targeting signal identified was found in a transmembrane domain of the M protein from the coronavirus infectious bronchitis virus (IBV) (44). The two envelope glycoproteins of rubella virus, E1 and E2, form a heterodimer that is retained in the Golgi complex by targeting information present in the transmembrane domain of E2 (16). A different type of Golgi targeting signal is found in the G1 envelope glycoprotein of Uukuniemi virus virus, a member of the family Bunyaviridae. A 30-amino-acid peptide from the cytoplasmic tail of G1 is sufficient to retain reporter proteins in the Golgi stack (2).
For endogenous integral membrane Golgi proteins, two main types of targeting determinants have been identified. The type II integral membrane glycosyltransferases have Golgi targeting information in their hydrophobic membrane-spanning regions (8). However, a lack of sequence similarity in these transmembrane regions among the different glycosyltransferases obscures a mechanistic understanding of the Golgi retention of these enzymes. By contrast, membrane proteins which are localized to the trans-Golgi network (TGN), such as furin and TGN38, require sequences in their cytoplasmic tails for proper localization (6, 32, 39). These targeting sequences include tyrosine-based internalization motifs similar to those found in the cytoplasmic tails of plasma membrane receptors, and direct the retrieval of TGN proteins that have escaped to the cell surface.
Mechanisms of peripheral Golgi protein targeting have also been studied. Recent work has focused on a class of large cytoplasmic coiled-coil proteins. Many members of this family are called golgins; they are usually autoantigens, and most are peripherally associated with the Golgi complex (4, 11, 29). Well-characterized members of this coiled-coil family include GM130 and p115. GM130 was first identified as part of a detergent-insoluble Golgi matrix (29, 43), and p115 is a factor required for intercisternal Golgi transport (46). GM130 is targeted to the Golgi by virtue of its association with another golgin, GRASP 65, which is N-terminally myristoylated (3, 4). Interestingly, a complex of GM130, p115, giantin, and GRASP 65 has been implicated in cisternal stacking and vesicle transport (3, 41) and it is likely that these proteins play an important role in specifying the structure of the Golgi complex (41). Other peripheral proteins of this class are targeted to the Golgi by a conserved C-terminal region called the GRIP domain (7, 17, 28), although the mechanism by which this occurs is still unknown.
We are investigating mechanisms of coronavirus envelope protein targeting and bud site selection. Coronaviruses are enveloped plus-stranded RNA viruses that obtain their lipid bilayer by budding into the cis-Golgi network (CGN), also known as the endoplasmic reticulum (ER)-Golgi intermediate compartment (20). We are studying the mechanisms by which coronavirus envelope proteins are localized to the budding site, both to fully understand the process of CGN bud site selection and to gain better insight into the ways in which proteins are targeted to the Golgi apparatus. The prototype coronavirus avian IBV contains three envelope proteins. The large spike glycoprotein (S) is transported to the cell surface in both transfected and infected cells, and thus it is likely that localization of S at the budding site depends on interactions with one or both of the other envelope proteins (30). Matrix (M) protein, a triple-spanning membrane glycoprotein, is targeted to the CGN and cis-Golgi complex by information present in its first transmembrane domain (24, 25). We recently determined that the envelope (E) protein is also localized to the Golgi complex when it is expressed in the absence of IBV infection (9).
We studied the Golgi targeting information present in the IBV E protein and found that E is targeted to the Golgi by a signal in its cytoplasmic tail. We show that a truncated form of E consisting only of the tail (CTE) binds to Golgi membranes. An interesting result was obtained with brefeldin A (BFA), a fungal metabolite that induces redistribution of Golgi membrane proteins to the ER and most Golgi coat proteins to the cytoplasm (18). In BFA-treated cells, IBV E and CTE were present in punctate structures throughout the cell that also contained the Golgi matrix proteins GM130 and p115. These Golgi “remnants” are thought to correspond to structural elements of the Golgi that are left behind during BFA treatment and Golgi enzyme redistribution to the ER (40). We therefore propose that the IBV E protein is associated with the Golgi matrix through interactions of its cytoplasmic tail and that this association may have functional implications for coronavirus bud site selection and assembly.
MATERIALS AND METHODS
Cells and viruses.
BHK-21 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 5% fetal calf serum (FCS) and antibiotics, and Vero cells were maintained in DMEM with 10% FCS and antibiotics. The adaptation of IBV (Beaudette strain) to Vero cells has been described (25). Growth and titering of recombinant vaccinia viruses were done as described (47). The recombinant vaccinia virus encoding phage T7 RNA polymerase (vTF7-3) has been described previously (12). The construction of the recombinant vaccinia virus encoding wild-type IBV E (vvIBVE) was reported previously (9). Vaccinia virus recombinants encoding E mutants (vvEG1, vvEG2, vvEG3, and vvCTE) were made using the appropriate plasmids (see below) according to this method.
Plasmids and expression vectors.
The E transmembrane replacement mutants were constructed as follows. Using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) with pBS/IBV E (9) as a template, the first seven amino acids of the E transmembrane domain (Gly 13 through Leu 19) were replaced with the corresponding amino acids of the vesicular stomatitis virus (VSV) G transmembrane domain (Ser 463 through Phe 469) to generate pBS/EG1. Using pBS/EG1 as a template, pBS/EG2 was made by replacing Tyr 20-Leu 26 of IBV E with Phe 470-Ile 476 of VSV G. In pBS/EG2, Ala 27-Gly 32 was replaced with the VSV G sequence Gly 477-Leu 482 to generate pBS/EG3, in which the complete transmembrane domain of IBV E is replaced by that of VSV G.
pBS/CTE was generated from pBS/E by PCR addition of appropriate restriction enzyme sites and a start codon immediately preceding Arg 33 and subcloning this PCR product into Bluescript SK (Stratagene). To construct pBS/GEt, a BamHI restriction site was inserted at the N terminus of the E cytoplasmic tail by QuikChange site-directed mutagenesis. The resulting protein, ETMB (E transmembrane BamH1 site), contained two amino acid changes (Ala 34 Leu 35 to Gly 34 Ile 35), but these changes did not affect normal Golgi targeting of the E protein. The E cytoplasmic tail coding sequence was then cloned into pBS/GTMB (33) BamHI-BamHI in place of the G cytoplasmic tail.
The cysteine to alanine mutants pBS/E-CCAA, pBS/CTE-CCAA, and pBS/GEt-CCAA were all made by QuikChange site-directed mutagenesis of the plasmids described above. The GEt C-terminal truncation mutants were generated by QuikChange insertion of stop codons after IBV E amino acid residues 44, 72, and 95. All mutations were confirmed by dideoxy sequencing.
Antibodies.
The production of the C-terminal antipeptide IBV E rabbit and rat antisera was described previously; affinity-purified rabbit antibody was used for immunofluorescence except in double-label experiments with mannosidase II, in which rat anti-E antibody was used (9). The mouse monoclonal antibodies to GM130 and p115 were purchased from Transduction Laboratories (Lexington, Ky.). The rabbit polyclonal antibody to mannosidase II was a gift from Marilyn Farquhar (University of California-San Diego, La Jolla). The mouse monoclonal anti-VSV G antibody (I1) used in immunofluorescence recognizes the lumenal domain (21). For immunoprecipitation of radiolabeled G and GEt proteins, a rabbit polyclonal antibody to whole VSV was used (48).
Immunofluorescence microscopy.
BHK-21 cells were plated on cover slips in 35-mm dishes 1 day before being infected with recombinant vaccinia virus. vvE, vvEG1, vvEG2, vvEG3, and vvCTE were adsorbed at a multiplicity of infection of 20 in 0.5 ml of serum-free DMEM for 1 h at 37°C, and the inoculum was replaced with regular growth medium. At 3 h postinfection (5 h postinfection for vvCTE), cells were fixed in 3% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature, permeabilized with 0.5% Triton X-100, and stained as previously described (44). BFA (Sigma, St. Louis, Mo.) was added at 5 μg/ml in growth medium 3 h postinfection, and cells were fixed at 5 h postinfection (5 and 7 h postinfection for vvCTE, respectively).
For examination of VSV G surface staining, cells were left unpermeablized and processed for immunofluorescence. For expression of Bluescript plasmids using vTF7-3, the virus was adsorbed at a multiplicity of infection of 10 in Opti-MEM (Life Technologies, Rockville, Md.) for 1 h at 37°C and transfected with 2 μg of DNA using 10 μl of Lipofectin (Life Technologies) as directed by the manufacturer. Cells were fixed, permeabilized, and stained at 2.5 to 3 h postinfection.
Metabolic labeling, immunoprecipitation, and glycosidase digestion.
BHK-21 cells were infected with vTF7-3 and transfected as described above. Vero cells were infected with IBV (passage 12 supernatant) as previously described (25). At 3.5 h postinfection (vTF7-3) or 30 h postinfection (IBV), cells were labeled with 1 mCi of [3H]palmitate (NEN Life Science Products, Inc., Boston, Mass.) per ml in DMEM-10% FCS-50 mM HEPES (pH 7.2)-1× nonessential amino acids or 50 μCi of 35S-Promix (Amersham Pharmacia Biotech, Piscataway, N.J.) per ml in methionine- and cysteine-free medium for 2 h at 37°C. After being rinsed in PBS, cells were lysed in detergent solution (62.5 mM EDTA, 50 mM Tris [pH 8], 0.4% deoxycholate, 1% NP-40) containing protease inhibitors, and the postnuclear supernatants were immunoprecipitated with antibodies to VSV G or IBV E as described (25).
For digestion of samples with endoglycosidase H (endo H), immunoprecipitates were washed and subsequently eluted with 50 mM Tris (pH 6.8)-1% sodium dodecyl sulfate (SDS), and digestion reactions were carried out after addition of an equal volume of 0.15 M sodium citrate, pH 5.5, for 16 h at 37°C, in the absence or presence of 0.4 mU of recombinant endo H (New England Biolabs, Beverly, Mass.). Samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and visualized by fluorography. It was necessary to load five times as many cell equivalents of CTE protein as the other proteins in order to visualize it after a similar exposure time. This is likely due to an increased rate of turnover of CTE relative to the full-length E protein.
Surface immunoprecipitation (31) was used to quantitatively evaluate the surface expression of GEt and its truncation derivatives. BHK-21 cells were infected with vTF7-3 and transfected with plasmids encoding wild-type G, GEt, GEt12, GEt40, or GEt63 using Lipofectin. At 3.5 h postinfection, cells were starved for 15 min in methionine- and cysteine-free medium and pulse-labeled for 10 min with 35S-Promix as described above. Proteins were chased to the cell surface for 1 or 2 h in regular growth medium. Cells were washed twice in cold PBS-1% bovine serum albumin-0.02% NaN3 and incubated for 2 h at 4°C on a platform rocker in 0.5 ml of the same buffer containing 15 μl of anti-VSV antibody. Before lysis in detergent solution with protease inhibitors, cells were washed extensively to remove unbound antibody.
Bound antibody was collected by incubating the postnuclear supernatant with fixed Staphylococcus aureus (Pansorbin; Calbiochem, San Diego, Calif.) in the presence of 0.2% SDS and subsequent centrifugation. The supernatant was then incubated with 5 μl of anti-VSV antibody to immunoprecipitate intracellular proteins, and antibody was again collected with fixed S. aureus. Immunoprecipitates were washed, eluted, and subjected to SDS-10% PAGE as described (25). Surface expression was quantitated using a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Microsome extraction.
To examine membrane association of CTE, BHK-21 cells expressing E, CTE, or CTE-CCAA by infection with vTF7-3 and transfection with Bluescript plasmids were labeled with 35S-Promix, and microsomes were prepared as previously described (9). Samples were treated with 0.1 M NaCl or 0.1 M Na2CO3 (pH 11.5) according to this protocol, and an additional sample was treated with detergent solution under the same conditions. Membranes were pelleted at 100,000 × g for 60 min, and supernatant and pellet fractions were immunoprecipitated with anti-E antibodies. The immunoprecipitates were subjected to SDS-PAGE and fluorography as described (9).
Sequence alignments.
Alignments were done with ClustalW multiple sequence alignment and MacBoxshade 2.15. Accession numbers were as follows: bovine coronavirus E, NP_150081; mouse hepatitis virus E, AAC36597; canine coronavirus E, JQ1723; transmissible gastroenteritis virus E, NP_05826; feline infectious peritonitis virus E, CAA74228; human coronavirus strain 229 E, NP_073554; IBV E, P05139; Uukuniemi virus G1, GNVUUK.
RESULTS
Transmembrane domain of IBV E is not required for proper Golgi localization.
We previously showed that the IBV E protein is localized to Golgi membranes when expressed by itself in transfected cells (9). Since the first transmembrane domain of the IBV M protein is known to contain Golgi targeting information (44), we wondered whether this type of targeting signal also functioned to localize E to the Golgi complex.
To investigate the role of the IBV E transmembrane domain in Golgi targeting, we made chimeric proteins in which part or all of the E membrane-spanning region was replaced with the corresponding region of the plasma membrane-localized VSV glycoprotein (VSV G). These chimeric proteins are diagrammed in Fig. 1. In the mutant protein EG1, the first one-third of the E transmembrane domain was replaced by the first one-third of the VSV G transmembrane domain. Likewise, EG2 contains the first two-thirds of the VSV G membrane span, and in EG3 the entire E transmembrane domain was replaced with that of VSV G. Recombinant vaccinia viruses encoding all three chimeras were generated.
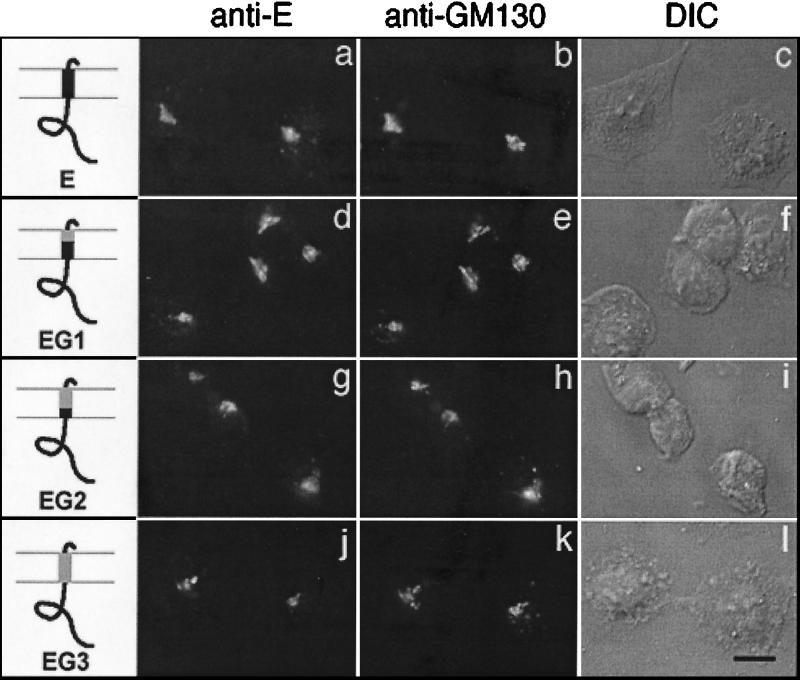
FIG. 1.
Transmembrane domain of IBV E is not required for Golgi targeting. BHK cells infected with vvE (a to c), vvEG1 (d to f), vvEG2 (g to i), or vvEG3 (j to l) were fixed for immunofluorescence at 4 h postinfection, permeabilized, and double labeled with rabbit anti-E antibody (a, d, g, and j) and mouse anti-GM130 antibody (b, e, h, and k). Secondary antibodies were fluorescein-conjugated donkey anti-rabbit immunoglobulin (IgG) and Texas Red-conjugated goat anti-mouse IgG. The third image in each row (c, f, i, and l) is a differential interference contrast (DIC) image of the labeled cells. The diagrams indicate IBV E sequence in black and VSV sequence in gray. Bar, 10 μm.
For localization studies, BHK-21 cells expressing the wild-type E protein or the E transmembrane replacement mutants were analyzed by indirect immunofluorescence using antibodies to the E protein and to the Golgi marker protein GM130. Replacing the E transmembrane domain in part or in entirety with that of VSV G had no effect on IBV E localization (Fig. 1). Therefore, we conclude that the transmembrane domain does not specify Golgi localization of the IBV E protein.
Cytoplasmic tail of IBV E is targeted to the Golgi complex in the absence of the lumenal and transmembrane domains.
To examine the E cytoplasmic tail for Golgi targeting information, we constructed an N-terminal truncation of the E protein by inserting a start codon preceding Arg 33. This truncation mutant, called CTE, consists only of the E cytoplasmic tail. When BHK cells expressing the CTE protein were stained with antibodies to the E protein and to GM130, we observed that CTE was efficiently localized to the Golgi complex (Fig. 2). This indicates that Golgi targeting information is present in the cytoplasmic tail of the IBV E protein.
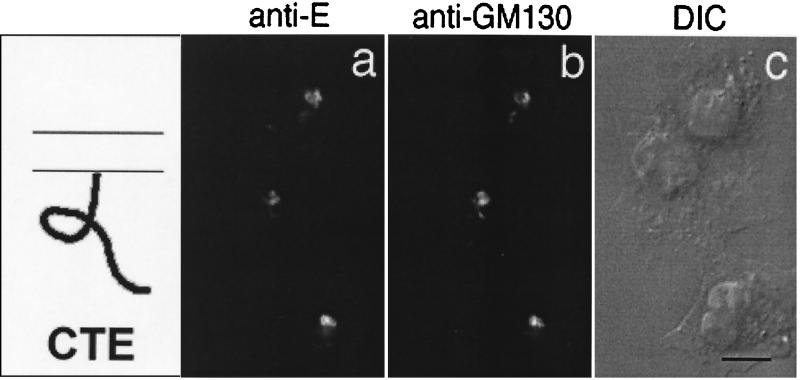
FIG. 2.
Cytoplasmic tail of IBV E is targeted to the Golgi complex. BHK cells infected with vvCTE were fixed for immunofluorescence at 6 h postinfection, permeabilized, and double labeled with rabbit anti-E antibody (a) and mouse anti-GM130 antibody (b). Secondary antibodies were fluorescein-conjugated donkey anti-rabbit IgG and Texas Red-conjugated goat anti-mouse IgG. A DIC image of the labeled cells is shown in panel c. Bar, 10 μm.
To rule out the presence of a redundant targeting signal in the short lumenal domain of E (see Fig. 5A), a chimera consisting of the lumenal domain of E and the transmembrane domain and cytoplasmic tail of VSV G was constructed. This chimera was efficiently transported to the cell surface when transiently expressed in BHK cells (data not shown), indicating that the lumenal N terminus of IBV E does not contain Golgi targeting information. Thus, we conclude that the cytoplasmic tail directs the Golgi localization of the E protein.
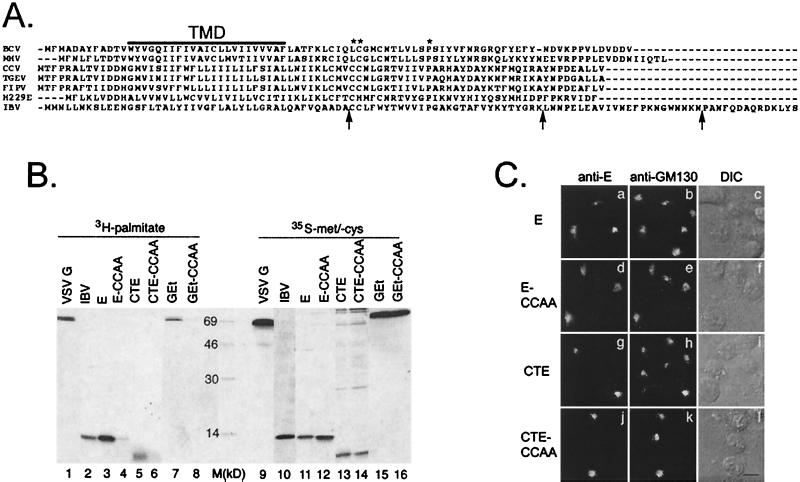
FIG. 5.
Conserved features of coronavirus E tails are dispensable for Golgi targeting. (A) An alignment of coronavirus E proteins from different species. The line labeled TMD shows the location of the transmembrane domain. The conserved cysteine(s) and proline residues that are present in all sequenced coronavirus E proteins are marked with asterisks. Arrows indicate the locations of inserted stop codons in the IBV E tail truncations. BCV, bovine coronavirus; MHV, mouse hepatitis virus; CCV, canine coronavirus; TGEV, transmissible gastroenteritis virus; FIPV, feline infectious peritonitis virus; H229E, human coronavirus strain 229E. (B) IBV-infected Vero cells or transfected BHK cells expressing the indicated proteins were labeled with [3H]palmitic acid or [35S]methionine-cysteine, lysed, and immunoprecipitated with anti-E or anti-VSV G antibodies. The immunoprecipitates were analyzed by SDS-17.5% PAGE and fluorography. Lane M, size markers (in kilodaltons). (C) BHK cells expressing wild-type E (a to c), E-CCAA (d to f), CTE (g to i), or CTE-CCAA (j to l) were fixed for immunofluorescence, permeabilized with Triton X-100, and stained with anti-E and anti-GM130 antibodies. Secondary antibodies were fluorescein-conjugated donkey anti-rabbit IgG and Texas Red-conjugated goat anti-mouse IgG. DIC images of the labeled cells are shown in panels c, f, i, and l. Bar, 10 μm.
Targeting information in the IBV E cytoplasmic tail can retain a reporter protein in the Golgi complex.
To determine whether the E cytoplasmic tail was able to target a reporter protein to the Golgi complex, a chimeric protein was generated that consists of the lumenal head and transmembrane domain of VSV G and the cytoplasmic tail of E. This chimera, called GEt, is efficiently retained in the Golgi complex when expressed in BHK cells (Fig. 3f), as seen by comparison with the wild-type E protein (Fig. 3c) and Golgi marker GM130 (Fig. 3d and g). Surface-localized VSV G (Fig. 3a) is shown for comparison. Thus, the cytoplasmic tail of IBV E is both necessary and sufficient for targeting to the Golgi complex.
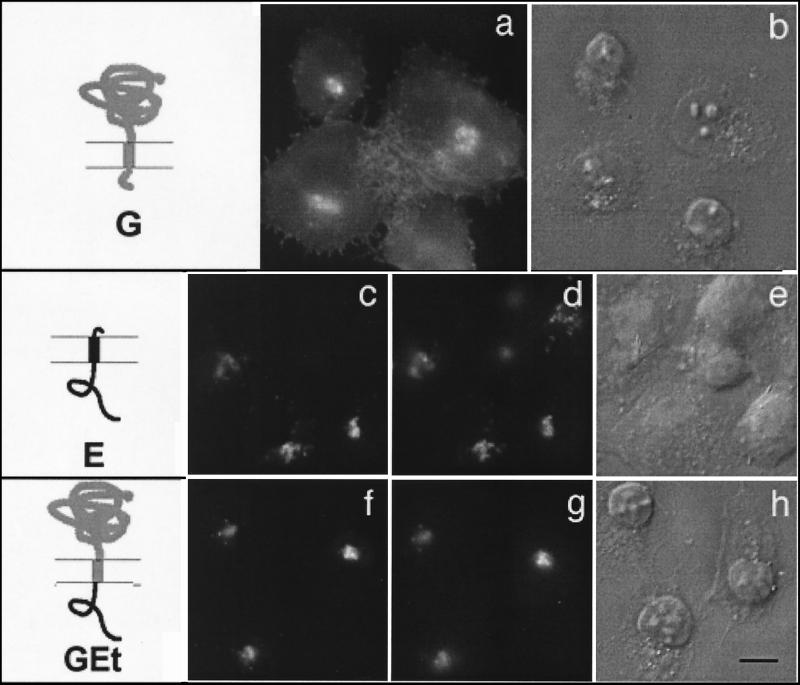
FIG. 3.
Cytoplasmic tail of IBV E is sufficient to retain a reporter protein in the Golgi complex. BHK cells expressing wild-type VSV G (a and b), wild-type IBV E (c to e), or a chimeric protein consisting of the lumenal head and transmembrane domain of VSV G and the cytoplasmic tail of E (GEt; f to h) were fixed for immunofluorescence, permeabilized, and stained with anti-G (a) or double labeled with anti-E (c and f) and anti-GM130 (d and g) antibodies. Secondary antibodies were fluorescein-conjugated donkey anti-rabbit IgG and Texas Red-conjugated goat anti-mouse IgG. DIC images of the labeled cells are shown in panels b, e, and h. The diagrams indicate IBV E sequence in black and VSV sequence in gray. Bar, 10 μm.
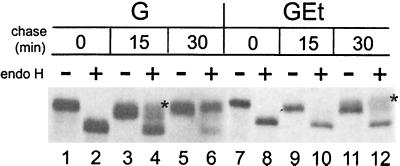
To investigate the Golgi compartment(s) in which the GEt chimera was present, we assessed the processing of the N-linked carbohydrates on the lumenal head domain of VSV G, also present on GEt. Oligosaccharides are modified in the medial Golgi and become resistant to endo H. BHK cells expressing G or GEt were pulse-labeled with [35S]methionine-cysteine and chased for various times in medium containing unlabeled amino acids. G and GEt were immunoprecipitated and treated or mock-treated with endo H (Fig. 4). After 0 min of chase, all of the newly synthesized G and GEt proteins were susceptible to cleavage by endo H (compare lanes 1 and 2 and 7 and 8), indicating that they have not yet reached the medial Golgi. After 30 min of chase, nearly all of the VSV G protein is endo H resistant (lane 6), reflecting its rapid rate of transport to the cell surface (42). A small portion of GEt protein acquired endo H resistance after 30 min of chase, indicating that some GEt has reached the medial Golgi. This small amount of endo H-resistant GEt did not increase after 2 h of chase (data not shown).
FIG. 4.
GEt chimera is partially processed by late Golgi enzymes. BHK cells expressing wild-type G or GEt were pulse-labeled with [35S]methionine-cysteine, chased for 0, 15, or 30 min, lysed, and immunoprecipitated with anti-VSV antibody. The immunoprecipitates were mock treated (lanes −) or treated with endo H (lanes +), and subjected to SDS-10% PAGE and fluorography to determine the extent of endo H resistance and sialylation.
Note the presence of two endo H-resistant bands for both G (lane 4) and GEt (lane 12). The upper bands (marked by asterisks), are sialylated G and GEt (36). Addition of sialic acid to N-linked sugar moieties occurs in the trans-Golgi (37), indicating that a portion of GEt has reached this compartment after 30 min of chase. These results indicate that the GEt chimera is not restricted to the cis-Golgi and are consistent with confocal and immunoelectron microscopy data showing that the wild-type E protein is present in cis-, medial, and trans-Golgi compartments when expressed exogenously (9, 10).
The slow rate of transport of GEt to the medial and trans-Golgi, compared with VSV G, is likely due to inefficient retention in the early Golgi. The lack of a strong ER exit signal, such as the one present in the VSV G cytoplasmic tail (42), could also contribute to the slow oligosaccharide processing of GEt. Thus, the Golgi targeting information present in the cytoplasmic tail of IBV E does not prevent transport to late Golgi compartments. However, this transport is likely to require a transmembrane domain (see Discussion).
Conserved features of coronavirus E protein cytoplasmic tails are dispensable for Golgi localization.
All coronaviruses that have been studied bud into the CGN (19). Since the E protein from several coronaviruses has been implicated in virus assembly and budding (5, 9, 13, 26, 45), all coronavirus E proteins may be localized near the budding site. We examined the cytoplasmic tails of the E proteins from various coronavirus species to look for conserved features that might be involved in Golgi targeting. An alignment of coronavirus E proteins is shown in Fig. 5A. Asterisks indicate the only conserved primary sequence features present in the cytoplasmic tails, which are one or two cysteine residues near the transmembrane domain and a proline residue around amino acid number 55.
Since many viral envelope proteins are palmitoylated on cysteine residues adjacent to their transmembrane domains (34), we asked whether [3H]palmitate could be incorporated into newly synthesized IBV E protein. IBV-infected cells or BHK cells transiently expressing VSV G, wild-type E, or E mutant proteins were labeled with [35S]methionine-cysteine or [3H]palmitate. The VSV G protein is known to be palmitoylated on cysteine residue 489 (35) and thus serves as a positive control for palmitate incorporation. Cells were lysed and proteins were immunoprecipitated and subjected to SDS-PAGE and fluorography (Fig. 5B). The wild-type E protein is labeled with [3H]palmitate in both infected and transfected cells (lanes 2 and 3). This is consistent with a report of increased electrophoretic mobility of the mouse hepatitis virus E protein after treatment with hydroxylamine, which cleaves thioester-linked acyl chains (49). Interestingly, CTE alone is also palmitoylated (lane 5). Changing the two cysteines to alanine (CCAA) eliminated palmitate incorporation in the wild-type E protein, the CTE truncation, and the GEt chimera (lanes 4, 6, and 8, respectively), indicating that the IBV E protein is palmitoylated on one or both of its cysteine residues.
Having established that the IBV E protein is palmitoylated, we asked whether this modification played a role in Golgi targeting. The nonpalmitoylated E-CCAA and GEt-CCAA proteins were transiently expressed in BHK cells by infection with vTF7-3 and transfection with Bluescript plasmids. Indirect immunofluorescence microscopy using anti-E and anti-GM130 antibodies (Fig. 5C) demonstrated that the localization of the CCAA mutants was indistinguishable from that of their palmitoylated counterparts. Evaluation of the effect of palmitoylation on the Golgi targeting of the CTE protein was made difficult by the saturable binding of the CTE protein to the Golgi when expressed at high levels with vTF7-3. Under these expression conditions, both CTE and CTE-CCAA localized to other membranes as well as to the Golgi complex (predominantly mitochondria; data not shown), whereas infection with a recombinant vaccinia virus expressing CTE (vvCTE) resulted in a lower and more homogeneous level of CTE expression and exclusive Golgi localization of CTE. Potential saturable binding of the CTE protein to the Golgi is consistent with its association with a component of the Golgi matrix (see Discussion).
We also mutated proline 56 to alanine to examine its role in the Golgi targeting of the IBV E protein. We found that this mutation had no effect on proper Golgi localization of E (data not shown). We conclude that the obvious conserved features of coronavirus E protein cytoplasmic tails, which are cysteines that serve as sites of palmitate addition, and proline 56 do not play a role in targeting the IBV E protein to the Golgi complex.
Golgi targeting information in the cytoplasmic tail of IBV E is contained between amino acids 44 and 95.
To define the region(s) within the E tail that was required for Golgi localization, we generated C-terminal truncations of the GEt chimera protein by inserting stop codons after amino acids 44, 72, and 95 of IBV E (corresponding to residues 12, 40, and 63, respectively, of the tail; see Fig. 5A). We analyzed the GEt chimeric protein because the lack of a strong antibody to the N terminus of E made evaluation of C-terminally truncated wild-type E protein difficult.
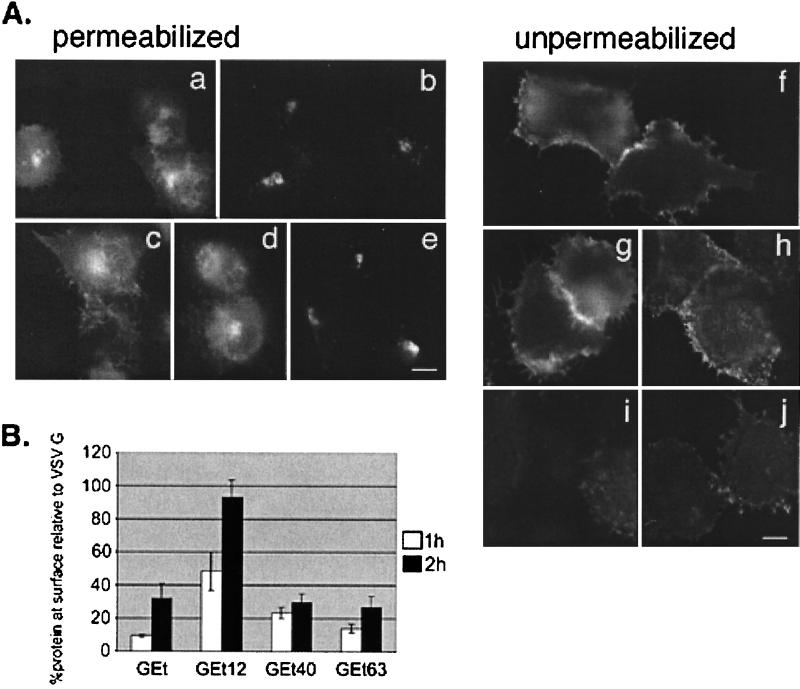
We examined BHK cells transiently expressing wild-type G protein, the GEt chimera, and its truncation derivatives by indirect immunofluorescence using a monoclonal anti-VSV G antibody (Fig. 6A). In permeabilized cells, we observed that truncating the E cytoplasmic tail to 12 amino acids (GEt12) resulted in cell surface staining (panel c). By contrast, GEt63, which contains 63 of the 77 amino acids of the E cytoplasmic tail, was efficiently localized to the Golgi (panel e). An intermediate truncation, GEt40, was found on the cell surface (panel d), although at a lower level than GEt12.
FIG. 6.
Golgi targeting information in the IBV E protein is between residues 12 and 63 of the cytoplasmic tail. (A) BHK cells expressing VSV G (a and f), GEt (b and i), GEt12 (c and g), GEt40 (d and h), or GEt63 (e and j) were fixed for immunofluorescence and permeabilized (a to e) or fixed and left unpermeabilized (f to j) and stained with an antibody to the ectodomain of VSV G. Secondary antibody was Texas Red-conjugated goat anti-mouse IgG. (B) BHK cells expressing GEt, GEt12, GEt40, or GEt63 were pulse-labeled with [35S]methionine-cysteine and chased for 1 h (white bars) or 2 h (black bars). Intact cells were incubated with polyclonal anti-VSV antibody for 2 h at 4°C. After cell lysis, antibody complexes were collected, and supernatants were incubated with anti-VSV to immunoprecipitate intracellular proteins. Samples were run on SDS-PAGE, and quantitation of surface and internal proteins was performed. A ratio of the amount of surface to internal protein was calculated for GEt and truncation derivatives. Shown on the graph is this ratio taken as a percentage of the surface/internal ratio of wild-type VSV G protein at the corresponding time point. Each value represents an average of three experiments ± SEM.
Unpermeabilized cells were examined to more easily compare the levels of surface staining of the different truncations, and the same trend was observed. GEt12 exhibited as much surface staining as the wild-type G protein (panels g and f, respectively), and both GEt and GEt63 showed little or negligible surface staining (panels i and j, respectively). The amount of GEt40 surface staining (panel h) was intermediate between that of GEt12 and GEt63.
To quantitatively evaluate the surface expression of the GEt truncations in comparison with that of wild-type G and the parent GEt chimera, we performed surface immunoprecipitations of BHK cells expressing these constructs. Cells were pulse-labeled with [35S]methionine-cysteine and chased in medium containing unlabeled amino acids for 1 or 2 h, and intact cells were incubated with polyclonal anti-VSV antibody for 2 h at 4°C. After cell lysis, antibody-antigen complexes (representing surface protein) were collected, and supernatants were subjected to another round of immunoprecipitation to collect intracellular proteins. Samples were run on SDS-PAGE, and quantitation of surface and internal proteins was performed on a PhosphorImager (Fig. 6B). A ratio of the amount of surface to internal protein was calculated for GEt and truncation derivatives. Shown on the graph is this ratio taken as a percentage of the surface/internal ratio of wild-type VSV G protein at the corresponding time point.
GEt12 reached the plasma membrane to the same extent as VSV G protein, only more slowly, indicating that it is not retained in the Golgi complex. The low levels of surface GEt and GEt63 detected at 2 h of chase is consistent with the small amount of GEt carbohydrate processing shown in Fig. 4. This could reflect a low level of escape from the Golgi complex. The level of GEt40 at the surface after 1 h of chase was greater than that of either GEt or GEt63 (and did not increase after 2 h of chase), indicating that GEt40 lacks at least some of the Golgi targeting information present in GEt and GEt63. However, the bulk of targeting information appears to reside between E tail residues 12 and 40.
Cytoplasmic tail of IBV E is peripherally associated with Golgi membranes.
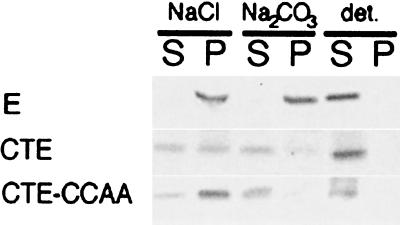
We previously showed that the IBV E protein remains associated with cell membranes upon treatment with alkaline carbonate, which is typical behavior of an integral membrane protein (9). We examined the membrane association of the CTE protein by preparing microsomes from radiolabeled cells transiently expressing E, CTE, or CTE-CCAA. Samples were treated with 0.1 M NaCl, 0.1 M Na2CO3 (pH 11.5), or detergent, membranes were centrifuged, and supernatants and pellets were immunoprecipitated with anti-E antibodies (Fig. 7). As expected, the wild-type E protein remained in the pellet upon treatment with alkaline carbonate but was released into the supernatant when the membranes were treated with detergent. CTE and CTE-CCAA partially pelleted with membranes. CTE was partially extracted from the membranes by 0.1 M NaCl and almost completely extracted by alkaline carbonate, indicating that it has a peripheral association with membranes. This is not surprising, since it lacks a transmembrane domain. CTE-CCAA behaved in a similar way, although it seemed to be totally extracted from membranes upon treatment with alkaline carbonate, which may reflect its lack of palmitoylation (see Fig. 5B).
FIG. 7.
CTE protein is peripherally associated with Golgi membranes. BHK cells expressing E, CTE, or CTE-CCAA were labeled with [35S]methionine-cysteine, and microsomes were prepared from homogenized cells. The microsomes were extracted with 0.1 M NaCl, 0.1 M Na2CO3 (pH 11.5), or detergent as indicated, and membranes were pelleted. Pellets (P) and supernatants (S) were immunoprecipitated with anti-É antibodies in the presence of detergent, and immunoprecipitates were analyzed by SDS-17.5% PAGE and fluorography.
E and CTE localize with Golgi remnants in BFA-treated cells.
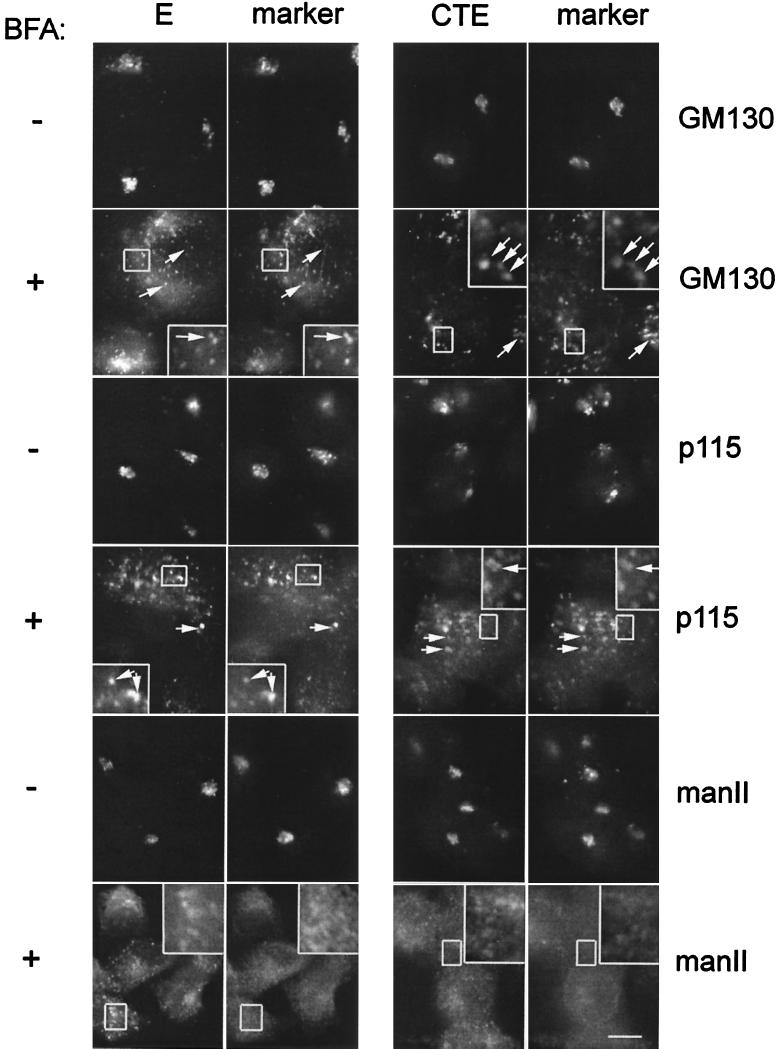
Since CTE was associated with Golgi membranes, we were interested to know where it would be localized after treatment with BFA, which inhibits secretion and induces redistribution of Golgi membrane proteins, such as mannosidase II, to the ER (18). BHK cells expressing IBV E or CTE were treated with BFA or mock treated and stained with antibodies to E and GM130, p115, or mannosidase II as indicated (Fig. 8). We observed redistribution of both the E and CTE proteins in treated cells (+BFA panels). Instead of being localized to the ER, however, E and CTE were found to colocalize with GM130 and p115 in punctate structures that are distributed throughout the cell. These BFA-induced structures have been observed previously by others (15, 38, 40) and are postulated to represent a Golgi structural matrix that remains after Golgi membranes and glycosylation enzymes are redistributed to the ER by treatment with BFA (40). Other Golgi matrix proteins that are found in Golgi remnants are p115, giantin, GRASP 65, and GRASP 55 (40). The punctate structures containing E and CTE are quite distinct from the ER, since they do not colocalize with the glycosylation enzyme mannosidase II, as was previously shown for the other Golgi remnant proteins (40). This is especially evident in the enlarged insets of each micrograph in Fig. 8.
FIG. 8.
IBV E and CTE are localized with Golgi remnants after BFA treatment. BHK cells expressing E or CTE by infection with recombinant vaccinia viruses were treated with BFA for 2 h at 3 and 5 h postinfection, respectively, fixed for immunofluorescence, permeabilized, and double labeled with rabbit anti-E antibody and anti-GM130 antibody or mouse anti-p115 antibody, or rat anti-E antibody and rabbit anti-mannosidase II (manII) antibody as indicated. Secondary antibodies were fluorescein-conjugated donkey anti-rabbit IgG and Texas Red-conjugated goat anti-mouse IgG, or fluorescein-conjugated goat anti-rat IgG and Texas Red-conjugated goat anti-mouse IgG. Boxed regions are enlarged in the insets of the BFA-treated panels. Bar, 10 μm.
DISCUSSION
We are interested in understanding how coronavirus envelope protein targeting to the Golgi complex is involved in bud site selection and assembly in this compartment. In the work described here, we studied the Golgi targeting information present in the IBV E protein. We report that the cytoplasmic tail of IBV E contains a Golgi targeting signal, and here discuss possible mechanisms by which this signal functions and its relevance to virus assembly in the Golgi complex.
Golgi targeting of the IBV E cytoplasmic tail.
We found that the cytoplasmic tail of the IBV E protein (CTE) is efficiently targeted to the Golgi complex when expressed in the absence of the rest of the protein. Under expression conditions which result in the accumulation of very high levels of the CTE protein, however, we observed that CTE was found in other membranes besides the Golgi, including mitochondria (see Results). This suggests that, unlike the wild-type E protein, the binding of CTE to the Golgi complex may be saturable. This is consistent with the possibility that CTE is targeted by associating with a protein or lipid constituent of the cytoplasmic face of the Golgi. Although this association is also likely to occur in the context of the entire protein, we have noticed qualitative differences in the Golgi localization of the wild-type E and CTE proteins.
In previous work we examined the localization of the IBV E protein by confocal microscopy and showed that it colocalized with markers of the cis-, medial, and trans-Golgi. We concluded that E is distributed throughout the Golgi complex when expressed in the absence of a virus infection (9). This result was supported by immunoelectron microscopy (10). In preliminary confocal microscopy experiments, we observed that the CTE protein showed a greater degree of colocalization with the cis-Golgi marker GM130 and less colocalization with markers of the medial and trans-Golgi than the wild-type E protein. This suggests that CTE is restricted to the cis-Golgi, possibly because it lacks a transmembrane domain and thus cannot be moved by vesicular transport to other Golgi compartments. This idea is supported by the observation that the oligosaccharides of GEt are partially processed (Fig. 4), demonstrating that GEt can reach late Golgi compartments. Thus, transport of the CTE protein to the medial and trans-Golgi can be accomplished by the addition of lumenal and transmembrane domains from either wild-type E or an unrelated protein (VSV G). This could explain why Golgi targeting of CTE but not wild-type E or the GEt chimera can be saturated at high expression levels.
The Golgi targeting information that we have identified within the IBV E protein is quite different from that found in the IBV M protein. IBV M has been shown to contain Golgi targeting information within its first transmembrane domain (25, 44). Specifically, four amino acid residues that make up a polar face within a predicted α-helix are critical for retention in the Golgi complex. Golgi localization of chimeric proteins containing this IBV M transmembrane domain is lost if the polar face is mutated (23). Thus, two distinct types of Golgi localization signals seem to function in IBV envelope protein targeting to the Golgi complex. These two targeting signals result in somewhat different subcompartment distributions of the two proteins when they are expressed exogenously.
The localization of the IBV M protein is restricted to the cis-Golgi network and cis-Golgi complex (24), whereas IBV E is localized throughout the Golgi stack (9). The mouse hepatitis virus E protein and IBV E can induce formation of virus-like particles, into which the M protein is efficiently incorporated if present (9, 26). It seems likely that interaction between the IBV E and M proteins results in their precise localization at the cis-Golgi network budding site (10, 22) and subsequent assembly into virus particles. We are currently defining regions of IBV E and M that are involved in their direct interaction and incorporation into virus-like particles.
Golgi matrix and IBV E protein.
When we treated cells expressing E or CTE with BFA, which causes many Golgi membrane proteins to redistribute to the ER (18), we found that both E and CTE precisely colocalized with GM130 and p115 in punctate Golgi remnants (Fig. 8). GM130- and p115-containing puncta are thought to represent a cytoplasmic structural component or matrix of the Golgi complex that is separable from most Golgi membranes and glycosylation enzymes found in the ER after BFA treatment (40). The exact nature of the Golgi matrix remains elusive, but it contains the integral membrane protein giantin and GRASP 65 and GRASP 55, as well as GM130 and p115 (40), and has been proposed to mediate vesicle tethering during intra-Golgi transport (3, 41).
Our results suggest that IBV E associates with the Golgi matrix by interactions involving its cytoplasmic tail. A direct interaction of IBV E with any of the known components of the Golgi matrix could mediate its targeting. Interaction with the matrix could also modulate vesicular traffic, which might be advantageous to a virus that must collect its envelope proteins for assembly in a specific Golgi compartment. If association with the Golgi matrix is important in specifying the virus budding site, then it is possible that IBV E recruits the other envelope proteins to this site through direct interactions. The IBV M protein does not appear to be associated with the Golgi matrix in the same way as IBV E, since it redistributes to the ER during BFA treatment. We are currently addressing the possibility that IBV E mediates the localization of IBV M with the Golgi matrix.
Are cytoplasmic Golgi targeting signals and association with the Golgi matrix common features among envelope proteins of viruses that assemble in the Golgi complex?
A comparison of the Golgi targeting mechanisms of IBV E and the Uukuniemi virus envelope glycoprotein G1 reveals some remarkable similarities. Uukuniemi virus is a member of the family Bunyaviridae, a group of viruses that also derive their envelope by budding into Golgi membranes. Uukuniemi virus G1 was shown to be targeted to the Golgi complex by a 30-amino-acid region in its cytoplasmic tail (2). Although Uukuniemi virus G1 is a large protein, the tail is very similar in length to the tail of IBV E (about 80 amino acids), and an alignment of the two tails reveals a substantial number of conserved residues, both within and outside of the region defined as being important for Golgi targeting (Fig. 9).
FIG. 9.
Conserved residues in the IBV E and Uukuniemi virus G1 cytoplasmic tails. Cytoplasmic tail sequences were aligned using ClustalW. Identical residues are shaded in black, and similar residues are shaded in gray. The brackets indicate the regions of each protein that contain Golgi targeting information.
Like the IBV E protein, Uukuniemi virus G1 contains two cysteine residues adjacent to its transmembrane domain, and these serve as sites for the addition of palmitic acid (1). As we have shown here for IBV E, palmitoylation is not necessary for proper Golgi targeting of Uukuniemi virus G1, since mutation of the cysteines has no effect on targeting (1). The similarities between IBV E and Uukuniemi virus G1 suggest a common mechanism of Golgi targeting, with potential relevance to the assembly process of enveloped viruses that bud into the Golgi complex.
IBV E is more similar in primary tail sequence to Uukuniemi virus G1 than it is to the E proteins of other species of coronavirus (Fig. 5A). This suggests that the Golgi targeting mechanisms of coronavirus E proteins may not be conserved or that a three-dimensional structure not revealed by primary sequence is important. Even though all coronaviruses studied bud into the cis-Golgi network (19), it is possible that they have different mechanisms of accumulating their envelope proteins at the budding site. Quite interestingly, Uukuniemi virus G1 has also been found to be associated with GM130 (R. Rönnholm, personal communication), which supports the possibility that Golgi matrix proteins are involved in the protein targeting and/or assembly process of enveloped viruses that bud into Golgi membranes. Future experiments will directly address this possibility.
Acknowledgments
This work was supported by National Institutes of Health grant GM42522.
We thank Ragna Rönnholm for sharing unpublished results and the members of the Machamer lab for critical reading of the manuscript.
REFERENCES
- 1.Andersson, A. M., L. Melin, A. Bean, and R. F. Pettersson. 1997. A retention signal necessary and sufficient for Golgi localization maps to the cytoplasmic tail of a Bunyaviridae (Uukuniemi virus virus) membrane glycoprotein. J. Virol. 71:4717–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, A. M., and R. F. Pettersson. 1998. Targeting of a short peptide derived from the cytoplasmic tail of the G1 membrane glycoprotein of Uukuniemi virus virus (Bunyaviridae) to the Golgi complex. J. Virol. 72:9585–9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr, F. A., N. Nakamura, and G. Warren. 1998. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 17:3258–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr, F. A., M. Puype, J. Vandekerckhove, and G. Warren. 1997. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell 91:253–262. [DOI] [PubMed] [Google Scholar]
- 5.Baudoux, P., C. Carrat, L. Besnardeau, B. Charley, and H. Laude. 1998. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 72:8636–8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos, K., C. Wraight, and K. K. Stanley. 1993. TGN38 is maintained in the trans-Golgi network by a tyrosine-containing motif in the cytoplasmic domain. EMBO J. 12:2219–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, D. L., K. Heimann, J. Lock, L. Kjer-Nielsen, C. Van Vliet, J. L. Stow, and P. A. Gleeson. 2001. The GRIP domain is a specific targeting sequence for a population of trans-Golgi network derived tubulo-vesicular carriers. Traffic 2:336–344. [DOI] [PubMed] [Google Scholar]
- 8.Colley, K. J. 1997. Golgi localization of glycosyltransferases: more questions than answers. Glycobiology 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corse, E., and C. E. Machamer. 2000. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 74:4319–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corse, E., and C. E. Machamer. 2001. Infectious bronchitis virus envelope protein targeting: implications for virus assembly. Adv. Exp. Med. Biol., in press. [DOI] [PubMed]
- 11.Fritzler, M. J., J. C. Hamel, R. L. Ochs, and E. K. Chan. 1993. Molecular characterization of two human autoantigens: unique cDNAs encoding 95- and 160-kD proteins of a putative family in the Golgi complex. J. Exp. Med. 178:49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuerst, T. R., E. G. Giles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godeke, G. J., C. A. de Haan, J. W. Rossen, H. Vennema, and P. J. Rottier. 2000. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J. Virol. 74:1566–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths, G., and P. Rottier. 1992. Cell biology of viruses that assemble along the biosynthetic pathway. Semin. Cell Biol. 3:367–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendricks, L. C., S. L. McClanahan, M. McCaffery, G. E. Palade, and M. G. Farquhar. 1992. Golgi proteins persist in the tubulovesicular remnants found in brefeldin A-treated pancreatic acinar cells. Eur. J. Cell Biol. 58:202–213. [PubMed] [Google Scholar]
- 16.Hobman, T. C., L. Woodward, and M. G. Farquhar. 1995. Targeting of a heterodimeric membrane protein complex to the Golgi: rubella virus E2 glycoprotein contains a transmembrane Golgi retention signal. Mol. Biol. Cell 6:7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kjer-Nielsen, L., R. D. Teasdale, C. van Vliet, and P. A. Gleeson. 1999. A novel Golgi-localisation domain shared by a class of coiled-coil peripheral membrane proteins. Curr. Biol. 9:385–388. [DOI] [PubMed] [Google Scholar]
- 18.Klausner, R. D., J. G. Donaldson, and J. Lippincott-Schwartz. 1992. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116:1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klumperman, J., J. K. Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krijnse-Locker, J., M. Ericsson, P. J. Rottier, and G. Griffiths. 1994. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 124:55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lefrancois, L., and D. S. Lyles. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. Virology 121:168–174. [DOI] [PubMed] [Google Scholar]
- 22.Lim, K. P., and D. X. Liu. 2001. The missing link in coronavirus assembly: retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 276:17515–17523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machamer, C. E., M. G. Grim, A. Esquela, S. W. Chung, M. Rolls, K. Ryan, and A. M. Swift. 1993. Retention of a cis Golgi protein requires polar residues on one face of a predicted alpha-helix in the transmembrane domain. Mol. Biol. Cell 4:695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machamer, C. E., S. A. Mentone, J. K. Rose, and M. G. Farquhar. 1990. The E1 glycoprotein of an avian coronavirus is targeted to the cis Golgi complex. Proc. Natl. Acad. Sci. USA 87:6944–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machamer, C. E., and J. K. Rose. 1987. A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. J. Cell Biol. 105:1205–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda, J., A. Maeda, and S. Makino. 1999. Release of coronavirus E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology 263:265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munro, S. 1998. Localization of proteins to the Golgi apparatus. Trends Cell Biol. 8:11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munro, S., and B. J. Nichols. 1999. The GRIP domain - a novel Golgi-targeting domain found in several coiled-coil proteins. Curr. Biol. 9:377–380. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura, N., C. Rabouille, R. Watson, T. Nilsson, N. Hui, P. Slusarewicz, T. E. Kreis, and G. Warren. 1995. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131:1715–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Opstelten, D. J., M. J. Raamsman, K. Wolfs, M. C. Horzinek, and P. J. Rottier. 1995. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 131:339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitta, A. M., J. K. Rose, and C. E. Machamer. 1989. A single-amino-acid substitution eliminates the stringent carbohydrate requirement for intracellular transport of a viral glycoprotein. J. Virol. 63:3801–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ponnambalam, S., C. Rabouille, J. P. Luzio, T. Nilsson, and G. Warren. 1994. The TGN38 glycoprotein contains two nonoverlapping signals that mediate localization to the trans-Golgi network. J. Cell Biol. 125:253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puddington, L., C. E. Machamer, and J. K. Rose. 1986. Cytoplasmic domains of cellular and viral integral membrane proteins substitue for the cytoplasmic domain of the vesicular stomatitis virus glycoprotein in transport to the plasma membrane. J. Cell Biol. 102:2147–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Resh, M. D. 1999. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451:1–16. [DOI] [PubMed] [Google Scholar]
- 35.Rose, J. K., G. A. Adams, and C. J. Gallione. 1984. The presence of cysteine in the cytoplasmic domain of the vesicular stomatitis virus glycoprotein is required for palmitate addition. Proc. Natl. Acad. Sci. USA 81:2050–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenwald, A. G., C. E. Machamer, and R. E. Pagano. 1992. Effects of a sphingolipid synthesis inhibitor on membrane transport through the secretory pathway. Biochemistry 31:3581–3590. [DOI] [PubMed] [Google Scholar]
- 37.Roth, J. 1991. Localization of glycosylation sites in the Golgi apparatus using immunolabeling and cytochemistry. J. Electron Microsc. Tech. 17:121–131. [DOI] [PubMed] [Google Scholar]
- 38.Sandvig, K., K. Prydz, S. H. Hansen, and B. van Deurs. 1991. Ricin transport in brefeldin A-treated cells: correlation between Golgi structure and toxic effect. J. Cell Biol. 115:971–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schafer, W., A. Stroh, S. Berghofer, J. Seiler, M. Vey, M. L. Kruse, H. F. Kern, H. D. Klenk, and W. Garten. 1995. Two independent targeting signals in the cytoplasmic domain determine trans-Golgi network localization and endosomal trafficking of the proprotein convertase furin. EMBO J. 14:2424–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seemann, J., E. Jokitalo, M. Pypaert, and G. Warren. 2000. Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature 407:1022–1026. [DOI] [PubMed] [Google Scholar]
- 41.Seemann, J., E. J. Jokitalo, and G. Warren. 2000. The role of the tethering proteins p115 and GM130 in transport through the Golgi apparatus in vivo. Mol. Biol. Cell 11:635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sevier, C. S., O. A. Weisz, M. Davis, and C. E. Machamer. 2000. Efficient export of the vesicular stomatitis virus G protein from the endoplasmic reticulum requires a signal in the cytoplasmic tail that includes both tyrosine-based and di-acidic motifs. Mol. Biol. Cell 11:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slusarewicz, P., T. Nilsson, N. Hui, R. Watson, and G. Warren. 1994. Isolation of a matrix that binds medial Golgi enzymes. J. Cell Biol. 124:405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swift, A. M., and C. E. Machamer. 1991. A Golgi retention signal in a membrane-spanning domain of coronavirus E1 protein. J. Cell Biol. 115:19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co expression of viral envelope protein genes. EMBO J. 15:2020–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waters, M. G., D. O. Clary, and J. E. Rothman. 1992. A novel 115-kD peripheral membrane protein is required for intercisternal transport in the Golgi stack. J. Cell Biol. 118:1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weisz, O. A., and C. E. Machamer. 1994. Use of recombinant vaccinia virus vectors for cell biology. Methods Cell Biol. 43:137–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weisz, O. A., A. M. Swift, and C. E. Machamer. 1993. Oligomerization of a membrane protein correlates with its retention in the Golgi complex. J. Cell Biol. 122:1185–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, X., W. Bi, S. R. Weiss, and J. L. Leibowitz. 1994. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology 202:1018–1023. [DOI] [PubMed] [Google Scholar]