Abstract
Recent evidence has suggested that plasma membrane sphingolipids and cholesterol spontaneously coalesce into raft-like microdomains and that specific proteins, including CD4 and some other T-cell signaling molecules, sequester into these rafts. In agreement with these results, we found that CD4 and the associated Lck tyrosine kinase of peripheral blood mononuclear cells and H9 leukemic T cells were selectively and highly enriched in a low-density lipid fraction that was resistant at 0°C to the neutral detergent Triton X-100 but was disrupted by extraction of cholesterol with filipin or methyl-β-cyclodextrin. In contrast, the CXCR4 chemokine receptor, a coreceptor for X4 strains of human immunodeficiency virus type 1 (HIV-1), was almost completely excluded from the detergent-resistant raft fraction. Accordingly, as determined by immunofluorescence with confocal microscopy, CD4 and CXCR4 did not coaggregate into antibody-induced cell surface patches or into patches of CXCR4 that formed naturally at the ruffled edges of adherent cells. The CXCR4 fluorescent patches were extracted with cold 1% Triton X-100, whereas the CD4 patches were resistant. In stringent support of these data, CD4 colocalized with patches of cholera toxin bound to the raft-associated sphingoglycolipid GM1, whereas CXCR4 did not. Addition of the CXCR4-activating chemokine SDF-1α did not induce CXCR4 movement into rafts. Moreover, binding of purified monomeric gp120 envelope glycoproteins from strains of HIV-1 that use this coreceptor did not stimulate detectable redistributions of CD4 or CXCR4 between their separate membrane domains. However, adsorption of multivalent gp120-containing HIV-1 virion particles appeared to destabilize the local CD4-containing rafts. Indeed, adsorbed HIV-1 virions were detected by immunofluorescence microscopy and were almost all situated in nonraft regions of the cell surface. We conclude that HIV-1 initially binds to CD4 in a raft domain and that its secondary associations with CXCR4 require shifts of proteins and associated lipids away from their preferred lipid microenvironments. Our evidence suggests that these changes in protein-lipid interactions destabilize the plasma membrane microenvironment underlying the virus by at least several kilocalories per mole, and we propose that this makes an important contribution to fusion of the viral and cellular membranes during infection. Thus, binding of HIV-1 may be favored by the presence of CD4 in rafts, but the rafts may then disperse prior to the membrane fusion reaction.
Recent evidence has suggested that certain membrane lipids, including sphingomyelin, glycolipids, and cholesterol, partition into relatively rigid raft-like condensed microdomains that are resistant to extraction at 0°C with the neutral detergent 1% Triton X-100 and that specific membrane proteins are greatly enriched in these lipid rafts (4, 9, 13, 44). These raft-associated components include phosphatidylinositol glycan-anchored proteins (4, 9), proteoglycans (11, 32), several virally encoded proteins (9, 27, 31, 40), and some proteins involved in T-lymphocyte signaling (13, 44), including CD4 (34) and the CD4-associated cytosolic proteins Lck (13, 34) and Nef (50). A functional role for lipid rafts in T-cell signaling has also been suggested by additional studies (4, 13, 49). Furthermore, stimulation of CD4 endocytosis by activation of protein kinase C is preceded by CD4 emigration from rafts (34). Compounds such as filipin and methyl-β-cyclodextrin that reduce the cholesterol content of viable cells disrupt lipid raft microdomains (10, 12, 42) and inhibit raft-dependent signaling processes (4, 13, 49).
Approximately 15 to 20% of the plasma membrane surface area is believed to consist of rafts (34, 43). In physiological conditions, the raft domains are small and their constituents are believed to be in equilibrium with adjacent nonraft regions of the membrane (4, 9, 13, 16, 43, 44).
Because CD4 is the primary receptor for human immunodeficiency virus type 1 (HIV-1) (23), its presence in lipid rafts has potential implications for HIV-1 infections. For example, exoskeletal and cytoskeletal proteins differentially associate with raft and nonraft microdomains (11, 32), resulting in potential channels or tracks for movements of viruses. HIV-1 and other viruses also bud from the raft region of the cell surface (31, 35). In addition, these results raise questions concerning the degree to which the HIV-1 coreceptors CCR5 and CXCR4 might be associated with rafts. Attachment of HIV-1 to CD4 induces a conformational change in the viral gp120 envelope glycoprotein that exposes a binding site for a coreceptor (19, 48, 51).
Several previous studies suggested that small fractions of CCR5 and CXCR4 might be associated with CD4 even in the absence of HIV-1, as indicated by coimmunoprecipitation from cell extracts made using Triton X-100 or other neutral detergents (52, 53) or by confocal immunofluorescence microscopy (53). However, a degree of apparent association in these studies might occur if a fraction of the CD4 and coreceptors were present together within the lipid raft microdomains that are resistant to extraction with these detergents. Alternatively, such apparent association might be indirectly mediated by detergent-resistant interactions with other substances.
In contrast to these early reports, we and others have been unable to detect significant CD4 and coreceptor colocalization even in cells that contain these proteins in large concentrations, which would be expected to facilitate their association (17, 18, 25). Indeed, a recent study by immunoelectron microscopy indicated that CD4 and coreceptors on the surfaces of T cells were segregated from each other in separate clusters that were nearby but completely nonoverlapping (45). A previous investigation suggested that a small proportion of CCR5 (ca. 11 to 18%) was present in a lipid raft fraction isolated from human adenocarcinoma cells (26). However, CXCR4 was almost entirely absent from a lipid raft fraction isolated by similar methods from human embryonic kidney 293T cells (25). Because lipid rafts are highly dispersed and are usually much smaller than the resolution of confocal microscopy, the latter method may overestimate the apparent colocalization of proteins with rafts (43). This problem can be overcome by causing the rafts to coalesce into microscopically visible patches with antibodies (9, 10).
To address these issues, we adapted a procedure for extraction of cells with a buffer containing 1% Triton X-100 at 0°C (33, 34). The detergent-insoluble lipid microdomains were then isolated by flotation in linear sucrose density gradients. We used the H9 line of human leukemic T cells, which homogeneously coexpress CD4 and CXCR4 and are highly susceptible to laboratory-adapted X4 isolates of HIV-1 (36), as well as stimulated and unstimulated populations of human peripheral blood lymphocytes, which express these proteins heterogeneously (2). We also tested effects of the SDF-1α chemokine and of monomeric and virus-associated multimeric gp120s on raft associations of CD4 and CXCR4. Parallel studies were done by immunofluorescence with confocal microscopy, using coalesced raft patches that were readily detected by this method.
A basic conclusion consistent with electron microscopic evidence (45) is that CD4 and CXCR4 are almost completely segregated into distinct lipid microdomains at 37°C. Although fluxes of these proteins between microdomains must occur in normal cells, the assembly of these proteins into ternary complexes with HIV-1 would be expected to destabilize the lipid bilayer microenvironment, and we propose that this may contribute energetically and kinetically to the membrane fusion step of infection. Consistent with this interpretation, adsorption of HIV-1 to H9 cells appeared to destabilize the CD4-containing rafts, resulting in their enhanced extraction with Triton X-100. In agreement with these results, direct visualization of adsorbed HIV-1 virions established that they were not associated with rafts.
MATERIALS AND METHODS
Cells.
H9 leukemic T cells were obtained from the AIDS Research and Reference Reagent Program (ARRRP, Division of AIDS, National Institutes of Health [NIH], Rockville, Md.; contributed by R. Gallo) and were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). Peripheral blood mononuclear cells (PBMCs) were isolated from freshly drawn blood buffy coat preparations centrifuged through a Histopaque (Sigma Diagnostics, Inc., St. Louis, Mo.) cushion. They were used freshly prepared or stimulated for 2 days with phytohemagglutinin-P (PHA-P) (5 μg/ml) in RPMI 1640 medium with 20% FBS and interleukin-2 (IL-2) (10 U/ml) (Roche Diagnostics Corp., Indianapolis, Ind.). Human astroglioma U87MG cells were from the American Type Culture Collection (Manassas, Va.). Clones of U87MG stably expressing CD4 or both CD4 and CXCR4 were described previously (17). HeLa-CD4 cells (14) were maintained in Dulbecco's modified Eagle's medium with 10% FBS, and human embryonic kidney (HEK) 293T cells were grown in the same medium supplemented with glucose (4.5 g/liter).
Lipid microdomain flotation.
Lipid microdomains were isolated essentially as described (33, 34). Briefly, cells (1 × 108 and 2.5 × 108 cells for H9 and PBMCs, respectively) were washed in ice-cold phosphate-buffered saline (PBS) (Life Technologies, Inc.) and lysed in 1 ml of ice-cold hypotonic buffer A (150 mM NaCl, 20 mM HEPES, 1× complete protease inhibitors cocktail [Roche Diagnostics Corp.] containing 1% Triton X-100). The nuclei were pelleted by centrifugation in a microcentrifuge for 15 s. The postnuclear extract (PNE) supernatant was kept on ice for 30 min, adjusted to 40% (wt/vol) sucrose by the addition of 1 ml of 80% (wt/vol) sucrose in buffer A, and overlaid with a linear 38 to 5% (wt/vol) sucrose gradient in buffer A. Variations of the sucrose overlay conditions are noted in the text. The samples were centrifuged at 100,000 × g in an SW55 rotor for 18 to 20 h at 4°C. A long blunt-tipped needle (inner bore diameter, 0.75 mm) was inserted from the top of the gradient to 5 mm from the bottom of the tube, and fractions (0.5 ml) were collected with the aid of a peristaltic pump. Insoluble proteins pelleted at the bottom were solubilized in buffer A containing 1% sodium dodecyl sulfate (SDS). Collected samples were analyzed by Western immunoblotting (see below).
To characterize the lipid raft isolation procedure and to determine the effects of ligand binding on CD4 and CXCR4 distributions, H9 cells were treated with various reagents prior to lysing. To activate protein kinase C, H9 cells were washed in PBS, incubated in warm serum-free RPMI 1640 medium supplemented with 25 mM HEPES and phorbol 12-myristate 13-acetate (PMA) (100 ng/ml) (Sigma Diagnostics, Inc.) at 37°C for 30 min (34). Cell membrane cholesterol was partially extracted by incubating cells in serum-free RPMI 1640 medium supplemented with 25 mM HEPES and filipin III complex (1 μg/ml) (Sigma Diagnostics Inc.) at 37°C for 1 h (34) or with methyl-β-cyclodextrin (10 mM) (Sigma Diagnostics, Inc.) at 37°C, with occasional agitation, for 2 h (33, 41).
H9 cells were also incubated with growth medium supplemented with saturating amounts of human SDF-1 α (100 nM) (PeproTech, Inc., Rocky Hill, N.J.), monomeric X4-tropic gp120IIIB (1 μg/ml) (ImmunoDiagnostics Inc., Woburn, Mass.), or monomeric dual-tropic gp120SF2 (1 μg/ml) (Austral Biologicals, San Ramon, Calif.) at 37°C for 1 h. High-titer replication-defective HIV-gpt virions pseudotyped with LAV/IIIB gp120-gp41 were prepared as previously described (14) and incubated with H9 cells at 37°C for 4 h.
Immunoblot analyses.
Protein samples from the lipid microdomain flotation experiments were adjusted to 1× SDS-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer using 4× stock (1× buffer is 62.5 mM Tris-Cl [pH 6.8], 10% glycerol, 2% SDS, 0.1% bromophenol blue, 2.5% 2-mercaptoethanol). The samples were loaded onto 10% polyacrylamide gels in the presence of 0.1% SDS and subjected to electrophoresis. The proteins were electrotransferred to nitrocellulose membranes and used for immunoblotting. The blots were blocked and sequentially incubated with primary antibodies and horseradish peroxidase (HRP)-conjugated detection proteins in 5% milk in PBS with 0.1% Tween 20. The primary antibodies used were mouse anti-transferrin receptor (Zymed Laboratories, Inc., So. San Francisco, Calif.), rabbit anti-human Lck (Upstate Biotechnology, Lake Placid, N.Y.), rabbit anti-CXCR4 (ProSci, Inc., Poway, Calif.), and sheep anti-CD4 and rabbit anti-CD4 (T4-4) (ARRRP, Division of AIDS, NIAID, NIH; contributed by M. Phelan and R. Sweet, respectively).
Initially, CD4 was detected in the experiments shown in Fig. 1 and 3 using the sheep anti-CD4 but was replaced by the more sensitive rabbit anti-CD4. The HRP-conjugated detection proteins used were goat anti-mouse immunoglobulin G (IgG)-HRP (Southern Biotechnology Associates, Birmingham, Ala.), protein A-HRP (Bio-Rad Laboratories, Hercules, Calif.) for the rabbit primary antibodies, and protein G-HRP (Bio-Rad Laboratories) for sheep anti-CD4. The blots were developed with an enhanced chemiluminescence kit (NEN Life Research Products, Boston, Mass.).
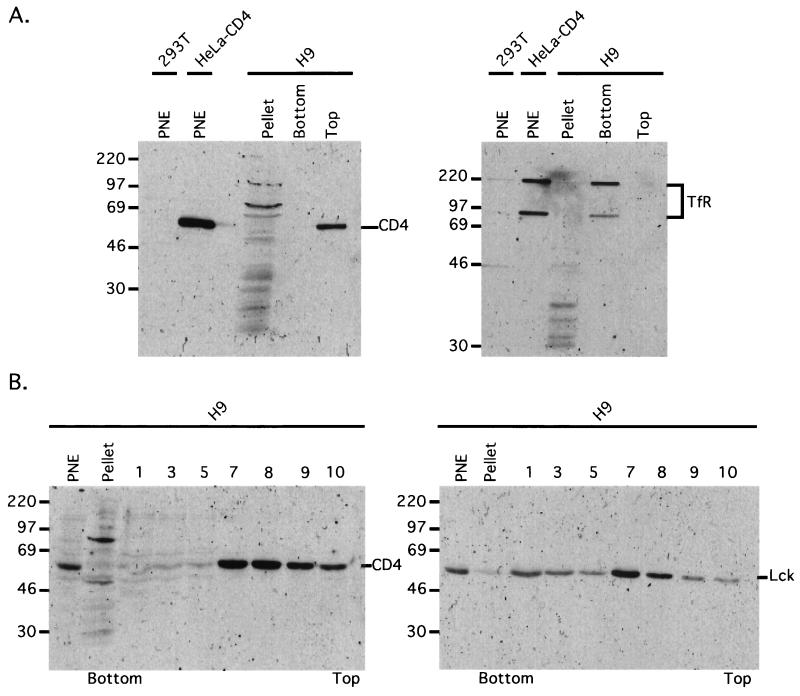
FIG. 1.
CD4 localizes in low-buoyant-density lipid rafts. (A) H9 cells were lysed in a hypotonic buffer containing 1% Triton X-100 at 0°C and gently centrifuged to pellet the nuclei. The PNE was adjusted to 40% sucrose and overlaid with a sucrose step gradient consisting of 2 ml of 30% sucrose and 1 ml of 10% sucrose (see Materials and Methods for details). After ultracentrifugation, samples were collected from the lipid band seen at the top of the 30% sucrose layer (Top), from the 40% sucrose layer (Bottom), and from the insoluble proteins in the pellet, which were dissolved in buffer with 1% SDS (Pellet). These protein samples and PNE samples from control HEK 293T and HeLa-CD4 cells were separated by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with sheep anti-CD4 (left blot) and mouse anti-transferrin receptor (right blot). CD4 localizes in the low-density raft microdomain in the top fraction, whereas transferrin receptor remained in the 40% sucrose bottom layer. Mrs are noted on the left (in thousands). (B) An H9 PNE was adjusted to 40% sucrose (final volume is 2 ml), overlaid with a 3.4 ml of a linear 5 to 38% sucrose buffer gradient, and then ultracentrifuged. Fractions (0.5 ml) were collected from the bottom of the gradient and subjected to immunoblot analyses with CD4 antiserum (left) and Lck antiserum (right). Mrs are noted on the left (in thousands).
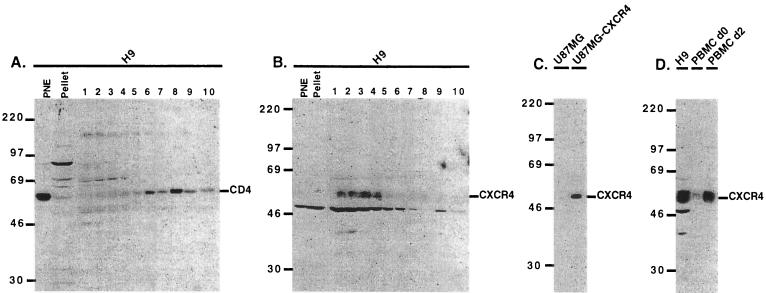
FIG. 3.
CXCR4 is excluded from the low-density lipid rafts. Immunoblot analyses of H9 extracts show CD4 predominantly in the lipid raft top fractions (A), whereas CXCR4 is substantially excluded from these fractions but is strongly concentrated in the high-density fractions (1 to 4) at the bottom of the gradient (B). CXCR4 appears to be absent in the PNE fraction of this H9 sample, although it was detectable in other H9 PNE preparations. We have noticed that CXCR4 was not always stable in sample fractions that were frozen and thawed. Therefore, the sucrose gradient fractions were always electrophoresed immediately after collection without being frozen and thawed. Extracts from unstimulated PBMCs (d0) contain relatively little CXCR4 compared to PBMCs stimulated for 2 days with PHA-P and IL-2 (d2) (D). U87MG and U87MG-CXCR4 extracts were included as negative and positive controls, respectively, for CXCR4 detection (C). Mrs are noted on the left of each blot (in thousands).
Confocal immunofluorescence microscopy.
U87MG-CD4/CXCR4 cells and H9 cells were seeded (24 h and 30 min before the immunofluorescence assay, respectively) on four-chambered Permanox slides (Nalge Nunc International, Naperville, Ill.) that were treated with poly-l-lysine (0.1 mg/ml) in PBS at 37°C for 30 min, to aid the adherence of the cells. All incubations were done in the cells' growth medium at 37°C for 1 h, unless otherwise noted. All fluorescently labeled second antibodies were from Molecular Probes Inc. (Eugene, Oreg.). Cell surface CD4 and CXCR4 were detected on U87MG-CD4/CXCR4 cells by incubating with both rabbit anti-CD4 at 1:100 and mouse anti-CXCR4 (12G5) (PharMingen International, San Diego, Calif.) at 1:100. The cells were washed with growth medium and PBS and fixed with 3% paraformaldehyde in PBS at room temperature for 30 min. Some U87MG-CD4/CXCR4 cells were subjected to cold 1% Triton X-100 in PBS and kept on ice before fixation. Cells were then incubated with goat anti-rabbit IgG conjugated to Alexa 594 at 1:1,000 and goat anti-mouse IgG-Alexa 488 at 1:500. The goat secondary antibodies reacted only with their species-specific primary antibody and showed no cross-reactivity with the opposite primary antibody.
Cholera toxin B subunit (CTx-B) conjugated to rhodamine (CTx-B-Rh;10 μg/ml in cold PBS with 0.1% bovine serum albumin [BSA]) (List Biologicals Laboratories, Inc., Campbell, Calif.) was incubated with the cells on ice for 30 min to label glycosphingolipid GM1 on the cell surface (13). Lipid raft aggregation, or patching, was induced after CTx-B-Rh labeling by incubating the cells with anti-CTx-B (1:250 in PBS with 0.1% BSA) (Calbiochem-Novabiochem Corp., San Diego, Calif.) on ice for 30 min and then moving the cells to 37°C for 5 or 20 min, as noted (13). The cells were washed and fixed as above and subsequently stained with mouse anti-CXCR4 or rabbit anti-CD4, followed by incubations with goat anti-mouse IgG-Alexa 488 or goat anti-rabbit IgG-Alexa 488, respectively.
For visualizing HIV-1 virions (28), H9 cells and HeLa-CD4 cells were incubated with high-titer NL4-3 HIV-1 (propagated as previously described [21]) at 37°C for 4 h prior to lipid raft aggregation at 4°C with CTx-B-Rh, followed by anti-CTx-B (CTx-B-Rh and anti-CTx-B were diluted in cold PBS with 0.1% BSA supplemented with 5 mM HEPES). The cells were moved to 37°C for 20 min, then fixed in 3.7% formaldehyde in PBS with 2% sucrose at room temperature for 30 min, and permeabilized with 0.5% NP-40 in PBS with 10% sucrose at room temperature for 30 min. HIV-1 virions were detected by incubation with HIV-1 p24 hybridoma (183-H12-5C) (from ARRRP, Division of AIDS, NIAID, NIH; contributed by B. Chesebro and K. Wehrly) cell culture supernatant, followed by incubation with goat anti-mouse antibody-Alexa 488. All stained cells were finally treated with ice-cold methanol for 10 min, rinsed with PBS, stored under FluoroGuard antifade mounting reagent (Bio-Rad Laboratories), and viewed by fluorescence confocal microscopy with a Bio-Rad MRC 1024 ES laser confocal imaging system.
RESULTS
Presence of CD4 in low-density lipid rafts.
To initially isolate membrane rafts from H9 leukemic T cells, we lysed the cells at 0°C in the presence of 1% Triton X-100 and gently sedimented the nuclei to obtain a PNE. Two milliliters of the PNE was then adjusted to 40% sucrose and overlaid with 2 ml of 30% sucrose and 1 ml of 10% sucrose for ultracentrifugation. As shown in Fig. 1A, CD4 floated to the top of the 30% sucrose layer, compatible with almost complete localization in low-density membranous rafts, whereas monomeric and dimeric forms of the transferrin receptor remained in the 40% sucrose bottom layer and did not cosediment with large cytoskeletal proteins into the pellet fraction.
Figure 1B shows a more thorough analysis of an H9 extract in which the 2 ml of PNE in 40% sucrose was overlaid with a 3.4-ml linear 5 to 38% sucrose gradient prior to centrifugation and collection of equal volume fractions from the tube. Consistent with the above results, CD4 was present almost exclusively in the lighter-density raft fractions (numbers 7 to 10). Similarly, the tyrosine kinase Lck, which is partially bound to CD4 (13, 34), was also present in the raft fractions, although a portion occurred in the higher-density fractions that contained soluble proteins. In agreement with these interpretations, we also used CTx and found that it adsorbed onto the low-density lipid raft fractions, suggesting that they contain the ganglioside GM1 (results not shown).
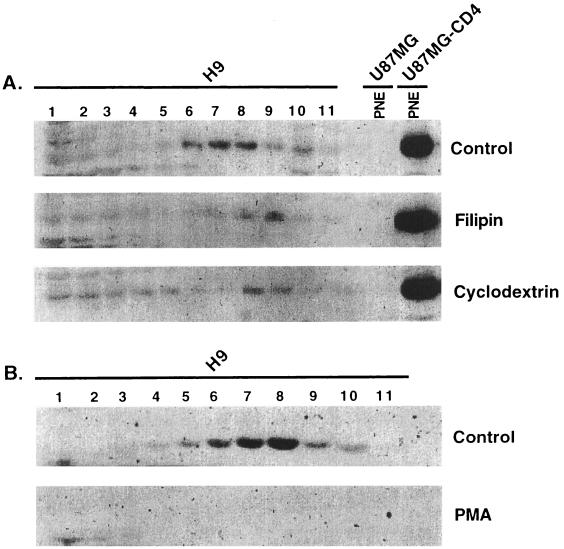
In agreement with the idea that CD4 occurs in the lipid-containing rafts, treatment of H9 cells with the drugs filipin and methyl-β-cyclodextrin, which partially extract cholesterol (10, 12, 42), caused increased efficiencies of CD4 solubilization by cold 1% Triton X-100 and correspondingly reduced quantities of CD4 in the low-density raft fractions (Fig. 2A). Partial extraction of cholesterol also slightly reduced the buoyant densities of the residual CD4-containing rafts. Furthermore, in agreement with previous evidence (34), activation of protein kinase C with PMA stimulated endocytosis and enhanced degradation of CD4 that was preceded by its movement out of the lipid raft fraction (e.g., Fig. 2B and below). Based on many similar analyses and on quantitative densitometric studies, we conclude that approximately 90 to 95% of CD4 is present in the lipid raft fraction of H9 cells. This is a minimum estimate because partial solubilization of raft components sometimes occurs during these experimental procedures (9, 13).
FIG. 2.
Characterization of the CD4-containing lipid rafts. (A) H9 cells were incubated with filipin III complex (1 μg/ml) at 37°C for 1 h or methyl-β-cyclodextrin (10 mM) at 37°C for 2 h prior to cell lysis and isolation of the lipid rafts (see Materials and Methods). These reagents, known to extract cholesterol from the cell membrane, caused CD4 to become more susceptible to solubilization by cold 1% Triton X-100 compared to the untreated H9 control. Fraction 1 is the bottom and 11 is the top. PNEs from U87MG and U87MG-CD4 cells were included as negative and positive controls, respectively, for CD4 detection. (B) Incubation of H9 cells with PMA (100 ng/ml) at 37°C for 30 min resulted in enhanced CD4 endocytosis and degradation. These immunoblots (A and B) were detected using the rabbit anti-CD4 antiserum.
Substantial exclusion of CXCR4 from low-density lipid rafts.
As shown in Fig. 3, CXCR4 was almost completely absent from the CD4-containing raft fractions isolated from H9 cells. The CXCR4 immunoblots from H9 cells contained a cross-reacting unrelated protein with an apparent Mr of approximately 50,000 (Fig. 3B and D) that was absent in the control extracts from U87MG-CD4-CXCR4 cells (panel C) and from PBMCs (panel D). This component is unrelated to CXCR4, since it was also detected by a different rabbit IgG antibody followed by protein A-HRP (results not shown). The data in Fig. 3D also indicate that unstimulated PBMCs contain relatively little CXCR4 compared to cells stimulated for 2 days with IL-2 plus PHA-P.
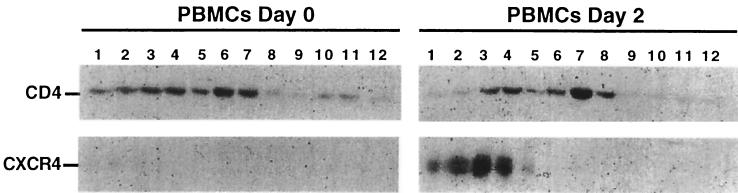
A similar analysis was done using PBMCs. In this experiment, however, the processing was done using 5 to 30% sucrose gradients, which caused the lipid rafts to concentrate more toward the middle in fractions 6 to 9. As shown in Fig. 4 (left panels), only approximately 50% of the CD4 in unstimulated PBMCs was in the lipid rafts, and CXCR4 was barely detectable. After 2 days of stimulation with IL-2 plus PHA-P (right panels), the cells contained dramatically increased amounts of CXCR4, which was completely excluded from the raft fractions. Moreover, a higher proportion of the CD4 (ca. 66%) was present in the low-density raft fractions in these stimulated PBMCs. Results after longer times of PBMC stimulation were similar to the 2-day data. Thus, CXCR4 appeared to be almost completely absent from the CD4-containing membrane raft fractions isolated from both H9 cells and PBMCs.
FIG. 4.
CD4 and CXCR4 segregate into different lipid microdomains in stimulated PBMCs. A 5 to 30% sucrose gradient was used in this experiment, which caused the lipid rafts to concentrate in the middle fractions (6 to 9). CXCR4 expression is dramatically increased in PBMCs after 2 days of stimulation with PHA-P and IL-2 and is absent from the raft fractions. Additionally, CD4 is more heavily concentrated in the raft fractions in the stimulated PBMCs compared to unstimulated PBMCs.
Effects of HIV-1 gp120 and the SDF-1α chemokine on raft associations of CD4 and CXCR4.
Since aspects of lymphocyte signaling may require interactions of receptors with lipid rafts (4, 15, 49), we investigated effects of the CXCR4-activating chemokine SDF-1α on this receptor. Moreover, because gp120s derived from HIV-1 strains that use CXCR4 form ternary bridges between CD4 and CXCR4 (19, 29, 48, 51), we also tested the effects of purified gp120s on raft localizations of these membrane proteins. As documented previously, the gp120s that we employed were active in binding to CD4 and CXCR4, and the SDF-1α was active in binding to CXCR4 and in stimulating signal transduction (17, 22). These ligands were adsorbed in saturating concentrations at 37°C, and the cells were then chilled to 0°C prior to extraction of the lipid rafts.
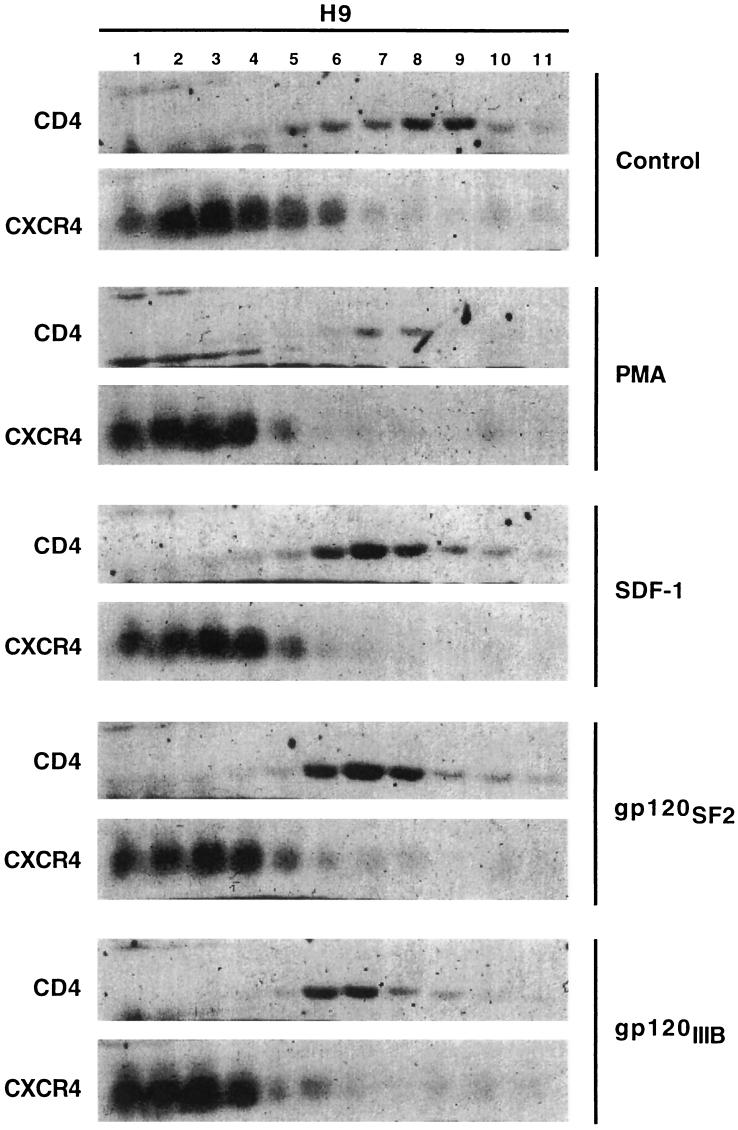
As indicated by the results in Fig. 5, CD4 was localized almost exclusively in the low-density raft fractions (6 to 10) in the untreated control H9 cells, whereas CXCR4 was almost fully excluded from these fractions. Consistent with previous evidence (e.g., see Fig. 2), PMA mobilized CD4 from the rafts and resulted in its enhanced endocytosis and degradation (34). Interestingly, addition of SDF-1α in saturating concentrations did not mobilize CXCR4 into the raft fractions or cause CD4 export from these fractions. Moreover, adsorption of saturating concentrations of purified monomeric gp120s derived from the X4 HIV-1 strain LAV/IIIB or from the dual-tropic strain SF2 did not induce significant CD4 export from rafts or CXCR4 import into rafts.
FIG. 5.
SDF-1α and monomeric HIV-1 gp120 do not significantly shift CD4 or CXCR4 out of their preferred lipid microdomains. CD4 was predominantly localized in the lipid raft fractions, whereas CXCR4 was concentrated in the high-density fractions of control H9 cells. PMA (100 ng/ml) treatment caused the CD4 to leave the raft fractions. H9 cells were incubated with saturating amounts of the chemokine SDF-1α (100 nM), X4-tropic monomeric gp120 IIIβ (1 μg/ml), or dualtropic monomeric gp120 SF2 (1 μg/ml) at 37°C for 1 h prior to lysing the cells. These latter treatments did not noticeably mobilize CXCR4 into the raft fractions or CD4 out of the raft fractions.
These results, which were highly reproducible, suggest that in the conditions of our assays, the bridging of CD4 and CXCR4 by these monomeric gp120s was energetically and/or kinetically insufficient to shift these receptors between their discrete lipid domains or to maintain these shifts throughout our fractionation procedures. However, these experiments do not exclude the possibility that some CXCR4 might occur in the lipid rafts but be susceptible to extraction by the cold 1% Triton X-100 (9, 13).
Previous studies have suggested that multimeric complexes of HIV-1-derived gp120s cause coalescence of cell surface CD4 with attendant activation of the associated tyrosine kinase Lck and with downstream activations of Ras and mitogen-activated protein kinase signaling pathways and of phosphatidylinositol 3-kinase, whereas monomeric gp120s are inactive (3, 38, 47). Moreover, CXCR4 would be expected to bind to multimeric and multivalent virus-CD4n complexes much more strongly than to monomeric gp120-CD4 complexes (29). For these reasons, we determined whether adsorption of intact HIV-1 virions onto H9 cells might mobilize CD4 or CXCR4 out of their segregated membrane sites. For this purpose we used replication-defective HIV-gpt virions, which were pseudotyped with the LAV/IIIB gp120-gp41 envelope glycoproteins as previously described (14, 37).
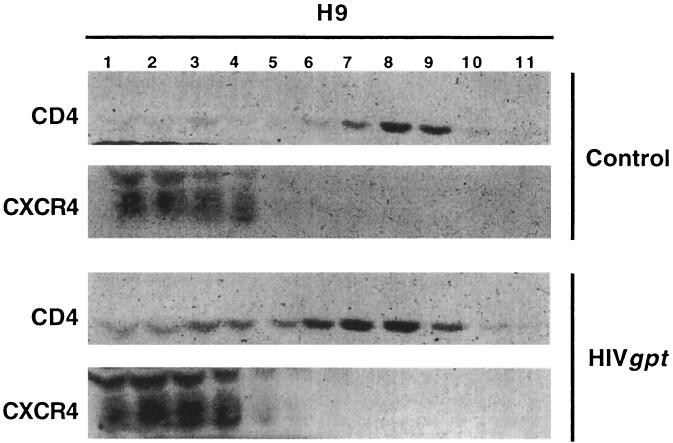
Interestingly, adsorption of these virions onto H9 cells significantly and reproducibly increased the proportion of CD4 that could be extracted from the membrane rafts but had no effect on the extractability of CXCR4 (Fig. 6). As determined by densitometry of appropriately exposed films, approximately 90% of CD4 was in the raft fractions (6 to 10) in the control cells, whereas only approximately 70% was in raft fractions after HIV-gpt adsorption. These results imply that adsorption of HIV-1 virions enhances CD4 emigration from the rafts or that these adsorbed virions destabilize the rafts, resulting in their increased extraction with Triton X-100 (see Discussion).
FIG. 6.
Effect of HIV-gpt virus on CD4 and CXCR4 localizations. H9 cells were incubated with replication-defective HIV-gpt virions at 37°C for 4 h before lysis. CXCR4 localization was unperturbed by this treatment, whereas CD4 was significantly mobilized out of the raft fractions.
Confocal immunofluorescence microscopy.
Because certain proteins can be extracted from lipid rafts by 1% Triton X-100 at 0°C (9, 13), we used immunofluorescence procedures with confocal microscopy to obtain additional information about the localizations of CD4 and CXCR4. In order to obtain optimal resolution by confocal microscopy, we initially used U87MG-CD4-CXCR4 cells, which grow as adherent monolayers on the microscope slides. In addition, we used different protocols for immunofluorescent antibody labeling in which the primary antibodies were bound at 37°C either before or after fixation of the cells with paraformaldehyde. Coalescence of these antigens into patches occurs when the primary antibodies are added before fixation, but not when they are added after fixation. These analyses were based on the idea, supported by previous studies of other proteins (9, 10), that antibody-induced coclustering of CD4 and CXCR4 would occur if these proteins were spontaneously associated in the absence of gp120 or if they occurred in a common lipid microdomain in intact cells.
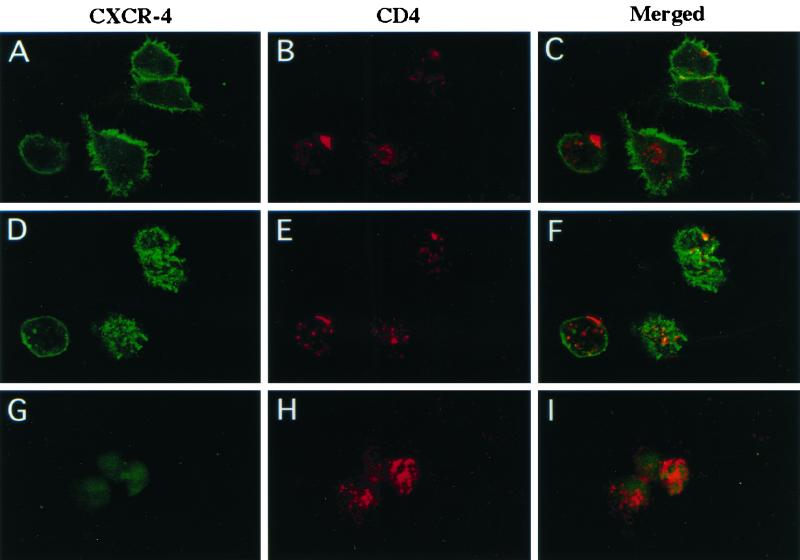
Figure 7 shows a representative analysis in which only the primary antibodies were adsorbed onto the cells at 37°C prior to fixation. The upper six panels (A to F) show the same cellular field, with the confocal sectioning either at the bottom of the cells (panels A to C) or at the top of the cells (panels D to F) in order to view those regions of the plasma membrane. At the bottom of the cells, the CXCR4 (green) was predominantly clustered at the ruffled outer cellular edges or at the leading edges of migrating cells, consistent with the role of CXCR4 in migration (2, 7). On the contrary, CD4 (red) was more randomly distributed in antibody-induced clusters at the bottom of the cells. In contrast, at the upper surface of the cells, the antibody-induced clusters of CD4 and CXCR4 were both more randomly situated. Although the merged image (panel F) shows some apparent colocalization, as indicated by the yellow color, statistical analysis suggests that this is random rather than systematic. Thus, in each cell, the proportion of yellow area was small and was approximately equal to the product of the proportions of the areas that were red and green. Therefore, the large majority of these proteins were not colocalized. Similar conclusions were suggested by our other confocal microscopic analyses.
FIG. 7.
Confocal immunofluorescence microscopy of U87MG-CD4/CXCR4 cells. Primary antibodies, mouse anti-CXCR4 and rabbit anti-CD4, were incubated with U87MG-CD4/CXCR4 cells prior to fixation with 3% paraformaldehyde. CXCR4 was detected with goat anti-mouse IgG-Alexa 488, which fluoresces green, and CD4 was detected with goat anti-rabbit IgG-Alexa 594, which fluoresces red. CXCR4 (green) and CD4 (red) clustered in distinct locations. The same cellular field is shown in panels A to F, with confocal sectioning at the bottom of the cell (A to C) or at the top of the cell (D to F). Cells shown in panels G to I were extracted with cold 1% Triton X-100 for 10 min on ice after primary antibody adsorption but before cell fixation. The CXCR4 (green) was greatly reduced by Triton X-100 extraction, whereas the CD4 (red) fluorescence remained strong.
The cells shown in panels G to I were from the same study except that they were briefly extracted with cold 1% Triton X-100 prior to fixation. Clearly, the green CXCR4 fluorescence was almost completely eliminated from cell surfaces by this extraction, whereas the red CD4 fluorescence was unaffected. This supports our conclusion that cell surface CXCR4 is largely absent from lipid rafts and suggests that its solubilization does not require a prolonged period of extraction and subcellular fractionation.
Additional evidence strongly supporting these conclusions was obtained by using H9 cells to determine whether CD4 and CXCR4 are colocalized with the raft-associated ganglioside GM1. This analysis was based on prior evidence that CTx binds specifically to GM1 and that the rafts then aggregate and coalesce into microscopically visible patches when the viable cells are incubated at 37°C with antibody to CTx but not when the cells are fixed prior to addition of this antibody (9, 10). As shown in Fig. 8 (panels A), CXCR4 and CD4 were both widely dispersed on the cell surfaces when fixation preceded antibody additions, and it was therefore very difficult to determine whether there was any true colocalization of these proteins.
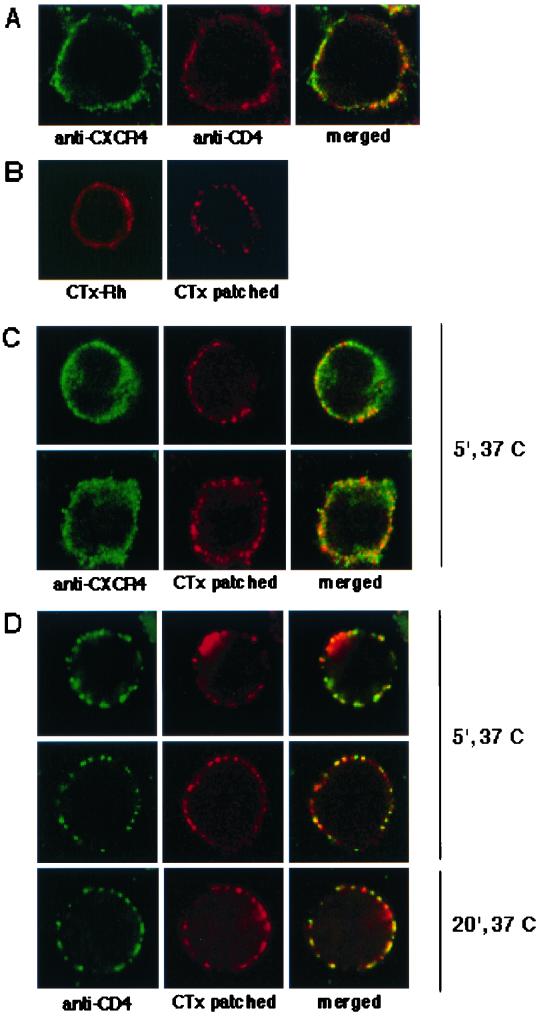
FIG. 8.
CD4 but not CXCR4 is colocalized with CTx patched rafts in H9 cells. (A) H9 cells fixed with 3% paraformaldehyde and then incubated with mouse anti-CXCR4 and rabbit anti-CD4 followed by goat anti-mouse IgG-Alexa 488 (green) and goat anti-rabbit IgG-Alexa 594 (red). The confocal sectioning was through the middle of the cell and shows CXCR4 and CD4 globally distributed throughout the cell surface membrane. (B) Lipid rafts on the cell surface were labeled for 30 min at 0°C with CTx-B-Rh. For patching of GM1 (right frame), anti-CTx was added at 0°C for 30 min. Both slides were then moved to 37°C for 5 min before fixing. H9 cells that were patched with CTx-B-Rh and anti-CTx as above were fixed and then either stained for CXCR4 (panels C, green) or CD4 (panels D, green). CXCR4 staining in the two representative cells in panels C is evenly and globally distributed through the membrane section, whereas the CTx-induced patching shows the aggregation of the rafts (red). In contrast, panels D show that CTx patching resulted in copatching of CD4, as expected for a raft-associated protein.
Because some regions of the cell surface are highly ruffled and therefore contain much more total membrane than nearby regions, two membrane proteins that are randomly situated could appear somewhat colocalized when examined in confocal sections at this magnification (45). As shown in Fig. 8, panels B, the CTx-GM1 complexes were also highly dispersed over the cell surfaces when fixation preceded addition of antibody (left panel), but became patched when the anti-CTx antibody was present at 37°C prior to fixation (right panel). Interestingly, as shown in panels D, the antibody-induced patching of CTx-GM1 complexes resulted in very clear copatching and colocalization of CD4, despite the fact that the antibody to CD4 was added after fixation. In contrast, as shown in panels C, the patching of CTx-GM1 complexes had no effect on the localization of CXCR4, which remained randomly dispersed over the cell surfaces.
These highly reproducible results strongly suggest that CD4 occurs in the GM1-containing lipid rafts on the surfaces of viable cells at 37°C, but that cell surface CXCR4 is excluded from these rafts. Close inspection of panels D and of other cells suggested that all of the CD4 occurs in the GM1-containing rafts, but that some of the GM1 occurs in the nonraft region of the plasma membrane. Consequently, we believe that CD4 is more quantitatively partitioned into the rafts than GM1.
Direct visualization of adsorbed HIV-1 virions confirms their exclusion from rafts.
We adapted a previously described procedure (28) to directly visualize HIV-1 virions adsorbed onto the cells. As shown in Fig. 9(top panels, green images), HIV-1 virions adsorbed extensively onto the surfaces of HeLa-CD4 cells and were readily detected as spots of fluorescence with a monoclonal antibody to the viral core protein p24. These small green spots were absent from control cells that were not incubated with HIV-1, and their detection required extractions of the viral membranes with detergent, confirming their identity as virion particles. These adsorbed virions were not colocalized with the CTx-induced GM1-raft clusters (red fluorescence) to a statistically significant degree. Indeed, in confocal sections at the tops of the cells, where the raft patches were clearly seen, fewer than 5% of the virions were colocalized with the rafts.
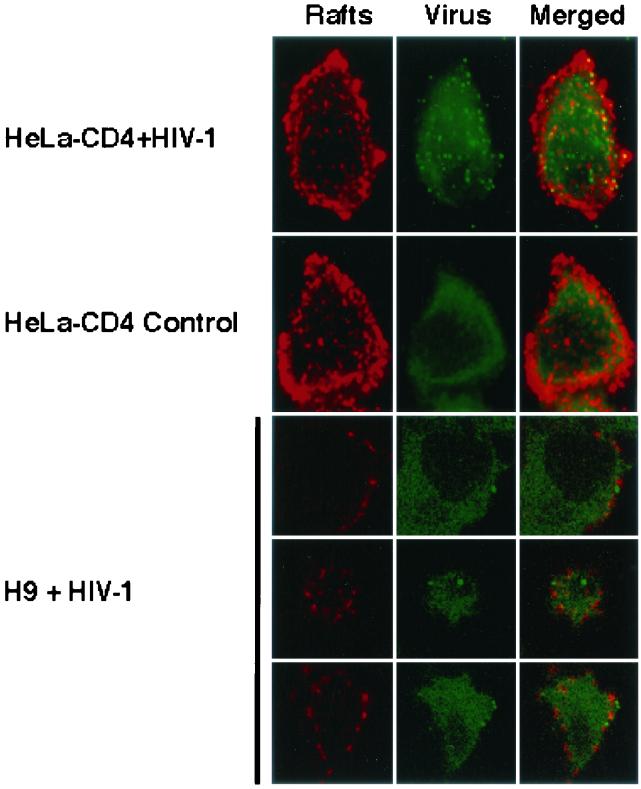
FIG. 9.
HIV-1 virions do not colocalize with CTx-patched rafts in H9 cells. H9 cells and HeLa-CD4 cells were incubated with HIV-1 strain NL4-3 prior to lipid raft aggregation with CTx-B-Rh and anti-CTx as described in Materials and Methods. Cells were fixed, permeabilized, and stained with mouse hybridoma HIV-1 anti-p24 supernatant, followed by goat anti-mouse IgG-Alexa 488 (green). In the top two rows, HeLa-CD4 cells incubated with and without HIV-1 were examined by confocal immunofluorescence microscopy and are shown as a sum projection of all their sections. Numerous HIV-1 virions, appearing as punctate green fluorescence, are shown adsorbed onto the HeLa-CD4 cells that were incubated with HIV-1 but are absent from control cells incubated without HIV-1. The lower panels show confocal slices through three individual H9 cells incubated with HIV-1. The virions (green spots) are distinct from the CTx-patched rafts (red).
Although interesting, we do not emphasize these HeLa-CD4 data because most of these HIV-1 particles are presumably bound to heparan sulfate proteoglycans rather than to CD4 (30). Indeed, in agreement with this idea, we have detected the same number of HIV-1 virions adsorbed onto HeLa cells that lack CD4 (results not shown). Consequently, we use these HeLa-CD4 results principally to illustrate the detection of HIV-1 virions by these methods. In contrast, H9 cells and lymphocytes lack these proteoglycans, and their ability to adsorb HIV-1 virions can be blocked by antibodies to CD4 (30). The lower panels in Fig. 9 are representative confocal sections that show HIV-1 virion particles (green dots) adsorbed onto the surfaces of H9 cells. The HIV-1 particles are clearly bound at sites that are not coincident with the lipid raft patches (red areas). Indeed, in a quantitative survey, only 3 of 99 HIV-1 virion particles on cell surfaces occurred at sites that appeared to overlap the lipid rafts.
DISCUSSION
Apparent segregation of CD4 and CXCR4 into distinct membrane microdomains.
Our results support previous evidence that CD4 and the associated tyrosine kinase Lck are substantially sequestered into a lipid raft fraction of the plasma membrane that is disrupted by extraction of cholesterol with filipin or methyl-β-cyclodextrin but is resistant to extraction with 1% Triton X-100 at 0°C (34). This sequestration occurs in freshly isolated populations of PBMCs, but is even more pronounced in PBMCs that have been activated by exposure to IL-2 plus PHA-P (Fig. 3 and 4) and in H9 leukemic T cells (Fig. 1 to 3), which coexpress CD4 and CXCR4 more homogenously within the cell population (2, 36). Association of CD4 with these raft lipids is reduced by phorbol ester-induced activation of protein kinase C, which results in phosphorylation of CD4 and in its enhanced endocytosis and degradation (34). Some other T-cell signaling components are also preferentially localized in rafts (4, 44, 49).
In contrast, our results suggest that CXCR4 is substantially excluded from CD4-containing lipid raft microdomains. In part, this conclusion is based on the fact that CXCR4 was almost completely absent from rafts extracted from H9 cells or PBMCs with 1% Triton X-100 at 0°C (Fig. 3 to 6) and that CXCR4 was also selectively extracted from the surface membranes of cells that were only briefly incubated with 1% Triton X-100 at 0°C prior to fixation (Fig. 7). By itself, the above evidence would be suggestive but inconclusive, because several proteins have been reported to be extracted from rafts in these conditions (9, 13). In addition, however, we found that antibody-induced patches of CD4 that formed on surfaces of viable cells did not contain CXCR4, and vice versa (Fig. 7, panels A to F).
On the contrary, if these proteins occurred together in lipid rafts, previous evidence implies that they would have coclustered in the conditions of these studies (9). Theoretically, the coalescence of rafts into patches would not be expected to alter the equilibrium partitioning of membrane components into these domains. Moreover, patches of CXCR4 that form naturally at the ruffled edges of adherent or migrating cells also appeared to lack CD4 (Fig. 7, panels A to C), supporting our conclusion that these proteins are largely separated on the surfaces of untreated viable cells at 37°C. A recent study using immunoelectron microscopy also indicated that CD4 and coreceptors occur in segregated nonoverlapping clusters in the surface membranes of untreated human T cells (45).
Further information concerning this matter was obtained by analyzing the sphingoglycolipid ganglioside GM1, which is known to occur preferentially in rafts and to bind CTx (9). When unfixed H9 cells were incubated with CTx and then with antibody to CTx at 37°C, GM1-containing rafts clustered together and coalesced into larger patches that could be easily seen by confocal miscroscopy (Fig. 8). In accordance with our other results, this caused CD4 to coalesce into the same patches (Fig. 8, panels D), but had no effect on CXCR4, which remained randomly dispersed on the cell surfaces (panels C).
Previously, it was reported that approximately 11 to 18% of the chemokine receptor CCR5 in human adenocarcinoma cells was present in isolated lipid rafts, but it was inferred from other evidence that CCR5 might be more extensively present in the rafts in viable cells (26). This evidence did not include studies of antibody-induced patching, but relied instead on apparent colocalization of CCR5 and GM1 at the leading edges of the migrating cells, as seen by confocal microscopy. As described above, however, such evidence can be misleading because the leading edges of cells are highly ruffled and they therefore contain much more total membrane than adjacent regions (45). Moreover, rafts are normally much smaller than the resolution of confocal microscopy (43). Consequently, proteins randomly dispersed in membranes could appear to be concentrated at these sites in the confocal sections. In migrating cells, new-membrane delivery from the Golgi apparatus also occurs at the leading edges, causing these areas to have high concentrations of rapidly cycling components, including chemokine receptors, and a nonrandom composition that is unrelated to rafts.
After this work was completed, we learned of a related investigation that used distinct methods (25). In agreement with our evidence, these other workers found that CD4 and CXCR4 were substantially segregated in PBMCs and HEK 293T cells, but that they became able to interact after formation of multivalent gp120-CD4 complexes. However, several aspects of their methods and conclusions differed from ours. For example, they reported that CXCR4 moves into lipid rafts to form CXCR4-gp120-CD4 ternary complexes at 4°C, a temperature at which protein and lipid movements in membranes should be severely restricted. Their raft flotation studies were done using only 0.2% Triton X-100, which may not fully solubilize nonraft proteins (24). Moreover, their confocal microscopy studies were done using incubations with primary and secondary antisera at 12°C and with fixation following the additions of all antisera. Specificity controls for the secondary antibodies and their species of origins were not described.
We believe that these differences may be important because cross-reacting secondary antibodies could potentially cause separated antigens to coalesce and because structures and compositions of microdomains are likely to differ at 4 to 12°C compared to our confocal microscopy studies at 37°C. Although additional work will be needed to address these matters, we believe that this other investigation supports our conclusion that CXCR4 and CD4 are separated on cell surfaces and that their interaction can be enhanced by formation of multivalent gp120-CD4 complexes. Our conclusion that monomeric gp120-CD4 complexes are substantially segregated from CXCR4 on cell surfaces is also consistent with a binding study by Mondor et al. (29).
Implications for HIV-1 infections.
In apparent conflict with our evidence that CD4 and CXCR4 are largely segregated into distinct lipid microdomains on cell surfaces, it is clear that infections by X4 strains of HIV-1 require coalescence of CD4 and CXCR4 around the adsorbed virions (19, 29, 48, 51). From our results, we believe that this coalescence could occur by several nonexclusive mechanisms. One possibility compatible with our data in Fig. 6 is that formation of HIV-1-CD4n complexes might stimulate emigration of these complexes out of the rafts and into the adjacent CXCR4-containing regions of the plasma membranes. Because these virus-CD4n complexes contain multivalent attachment sites for CXCR4, they would be expected to bind CXCR4 much more avidly than monomeric gp120-CD4 complexes. According to this idea, the adsorbed virions might partition out of the rafts as a consequence of their strong associations with multiple CXCR4 molecules that are anchored in the nonraft region. Formation of multivalent virus-CD4n complexes also causes activation of Lck (3, 38, 47), and it is conceivable that this might contribute to the emigration or dissolution of the rafts.
Although additional studies will be required to evaluate these possibilities, the latter mechanism could not be necessary for HIV-1 infections because cells lacking Lck can be infected and because CD4 mutants that cannot interact with Lck can also mediate infections (1, 47). Alternatively, the data in Fig. 6 and 9 are consistent with the hypothesis that HIV-1 adsorption onto the cells decreases the stability of the local lipid raft microenvironment, so that the CD4 is more efficiently extracted with Triton X-100. According to this interpretation, CXCR4 would gradually partition into the rafts due to its high affinity for the tightly anchored multivalent virus-CD4n complexes. This immigration of CXCR4 would destabilize the local raft microenvironment by the mechanism described below, thereby enhancing its solubilization by Triton X-100.
Based on our evidence that CD4 and CXCR4 occur almost exclusively in distinct lipid domains of the membrane, it seems likely that their coalescence around the adsorbed HIV-1 virion would cause enhanced mixing of lipids at this site and/or movement of one of these receptor proteins into a lipid domain distinct from its most stable location. Inevitably, such changes would destabilize the membrane microenvironment and would lower the activation energy required for membrane fusion. Although the extent of this destabilization can be estimated only roughly, it is relevant that cell surface CD4 is often 95% localized in rafts, that CXCR4 appears to be almost completely absent from rafts, and that rafts are believed to comprise approximately 10 to 20% of plasma membrane surfaces (34, 43). These considerations suggest that the concentration of CD4 may be approximately 100 times higher in the rafts than in the nonraft regions, whereas our microscopy and cell fractionation results imply that CXCR4 is probably at least 10 times more concentrated in the nonraft area of the plasma membrane at both 0 and 37°C. Consequently, the free energy for partition of these proteins into their preferred lipid environments is likely to be in the range of −1 to −2 kcal/mol, consistent with other evidence that the lipid domain associations of proteins are unstable and are rapidly fluctuating in physiological conditions (4, 9, 16). Nevertheless, even small reductions in activation energies can dramatically increase the rates and efficiencies of reactions (46). Moreover, infections with HIV-1 are believed to require coalescence of approximately four to six CD4 and coreceptor molecules (18, 20), which would be expected to destabilize the local membrane microenvironment in an approximately additive manner.
We propose that this destabilization of the host membrane could be important for the membrane fusion reaction. Moreover, since many or perhaps even all enveloped viruses bind to multiple cell surface proteins or onto sugar residues that occur on diverse glycoproteins, proteoglycans, and glycosphingolipids (5), it is possible that destabilization caused by mixing of raft and nonraft components might be a widespread or even general mechanism that contributes to infections of membrane-enveloped viruses. For example, in addition to CD4 and coreceptors, HIV-1 also binds to other membrane components, including heparan sulfate proteoglycans and glycosphingolipids (8, 39). Presumably, this lipid destabilization mechanism would function synergistically with structural changes in the viral envelope glycoproteins (6) to induce the membrane fusion reaction. Thus, we propose that HIV-1 may enter cells not in a raft microdomain but rather in a destabilized transition state that contains a mixture of raft and nonraft components.
Finally, we note that extraction of cellular cholesterol with filipin or methyl-β-cyclodextrin can strongly inhibit infections with HIV-1 (25). Although this is compatible with the hypothesis that lipid rafts contribute to HIV-1 infections, in agreement with the membrane destabilization model described above, removal of cholesterol is toxic and could have myriad effects on cellular functions. Moreover, cholesterol and other lipids occur in both raft and nonraft regions of plasma membranes, and they are likely to be important in both locales (16). Consequently, such infectivity data may be unrelated to lipid rafts. Possibly, the rafts may facilitate HIV-1 adsorption onto CD4 and then disperse prior to the ultimate membrane fusion reaction. In contrast, release of virions from cells may occur specifically from rafts (31, 35). Additional studies will be needed to test these ideas and to understand the roles of membrane lipid domains in infections with HIV-1 and other viruses.
Acknowledgments
We are grateful to Aurelie Snyder at the MMI Research Core Facility (OHSU) for expert technical assistance with the confocal microscopy and to Mariana Marin, Emily Platt, and Chetankumar Tailor for help and critical advice.
This research was supported by NIH grant CA67358 (to D.K.) and by the Agence National de Recherche contre le SIDA (ANRS) (to J.M.H.). The project was initiated during a sabbatical visit of J.M.H. to Oregon and was subsequently continued at both institutions.
REFERENCES
- 1.Bedinger, P., A. Moriarty, R. C. D. von Borstel, N. J. Donovan, K. S. Steimer, and D. R. Littman. 1988. Internalization of the human immunodeficiency virus does not require the cytoplasmic domain of CD4. Nature 334:162-165. [DOI] [PubMed] [Google Scholar]
- 2.Bleul, C. C., L. Wu, J. A. Hoxie, T. A. Springer, and C. R. Mackay. 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. USA 94:1925-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briand, G., B. Barbeau, and M. Tremblay. 1997. Binding of HIV-1 to its receptor induces tyrosine phosphorylation of several CD4-associated proteins, including the phosphatidylinositol 3-kinase. Virology 228:171-179. [DOI] [PubMed] [Google Scholar]
- 4.Brown, D. A., and E. London. 2000. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275:17221-17224. [DOI] [PubMed] [Google Scholar]
- 5.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka. 2000. Virus attachment to host cells, p. 101-131. In Principles of virology: molecular biology, pathogenesis, and control. ASM Press, Washington, D.C.
- 6.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka. 2000. Virus entry into cells, p. 133-161. In Principles of virology: molecular biology, pathogenesis, and control. ASM Press, Washington, D.C.
- 7.Ganju, R. K., S. A. Brubaker, R. D. Chernock, S. Avraham, and J. E. Groopman. 2000. β-Chemokine receptor CCR5 signals through SHP1, SHP2, and Syk. J. Biol. Chem. 275:17263-17268. [DOI] [PubMed] [Google Scholar]
- 8.Hammache, D., N. Yahi, M. Maresca, G. Pieroni, and J. Fantini. 1999. Human erythrocyte glycosphingolipids as alternative cofactors for human immunodeficiency virus type 1 (HIV-1) entry: evidence for CD4-induced interactions between HIV-1 gp120 and reconstituted membrane microdomains of glycosphingolipids (Gb3 and GM3). J. Virol. 73:5244-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harder, T., P. Scheiffele, P. Verkade, and K. Simons. 1998. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 141:929-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huby, R. D., R. J. Dearman, and I. Kimber. 1999. Intracellular phosphotyrosine induction by major histocompatibility complex class II requires coaggregation with membrane rafts. J. Biol. Chem. 274:22591-22596. [DOI] [PubMed] [Google Scholar]
- 11.Ilangumaran, S., B. Borisch, and D. C. Hoessli. 1999. Signal transduction via CD44: role of plasma membrane microdomains. Leuk. Lymphoma 35:455-469. [DOI] [PubMed] [Google Scholar]
- 12.Ilangumaran, S., and D. C. Hoessli. 1998. Effects of cholesterol depletion by cyclodextrin on the sphingolipid microdomains of the plasma membrane. Biochem. J. 335:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janes, P. W., S. C. Ley, and A. I. Magee. 1999. Aggregation of lipid rafts accompanies signaling via the T cell antigen receptor. J. Cell Biol. 147:447-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabat, D., S. L. Kozak, K. Wehrly, and B. Chesebro. 1994. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J. Virol. 68:2570-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasahara, K., and Y. Sanai. 1999. Possible roles of glycosphingolipids in lipid rafts. Biophys. Chem. 82:121-127. [DOI] [PubMed] [Google Scholar]
- 16.Kenworthy, A. K., N. Petranova, and M. Edidin. 2000. High-resolution FRET microscopy of cholera toxin B-subunit and GPI-anchored proteins in cell plasma membranes. Mol. Biol. Cell 11:1645-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozak, S. L., S. E. Kuhmann, E. J. Platt, and D. Kabat. 1999. Roles of CD4 and coreceptors in binding, endocytosis, and proteolysis of gp120 envelope glycoproteins derived from human immunodeficiency virus type 1. J. Biol. Chem. 274:23499-23507. [DOI] [PubMed] [Google Scholar]
- 18.Kuhmann, S. E., E. J. Platt, S. L. Kozak, and D. Kabat. 2000. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J. Virol. 74:7005-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Layne, S. P., M. J. Merges, M. Dembo, J. L. Spouge, and P. L. Nara. 1990. HIV requires multiple gp120 molecules for CD4-mediated infection. Nature 346:277-279. [DOI] [PubMed] [Google Scholar]
- 21.Madani, N., and D. Kabat. 2000. Cellular and viral specificities of human immunodeficiency virus type 1 Vif protein. J. Virol. 74:5982-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madani, N., S. L. Kozak, M. P. Kavanaugh, and D. Kabat. 1998. gp120 envelope glycoproteins of human immunodeficiency viruses competitively antagonize signaling by coreceptors CXCR4 and CCR5. Proc. Natl. Acad. Sci. USA 95:8005-8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maddon, P. J., A. G. Dalgleish, J. S. McDougal, P. R. Clapham, R. A. Weiss, and R. Axel. 1986. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47:333-348. [DOI] [PubMed] [Google Scholar]
- 24.Madore, N., K. L. Smith, C. H. Graham, A. Jen, K. Brady, S. Hall, and R. Morris. 1999. Functionally different GPI proteins are organized in different domains on the neuronal surface. EMBO J. 18:6917-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mañes, S., G. del Real, R. A. Lacalle, P. Lucas, C. Gómez-Moutón, S. Sánchez-Palomino, R. Delgado, J. Alcamí, E. Mira, and A. C. Martínez. 2000. Membrane raft microdomains mediate lateral assemblies required for HIV-1 infection. EMBO Rep. 1:190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manes, S., E. Mira, C. Gomez-Mouton, R. A. Lacalle, P. Keller, J. P. Labrador, and A. C. Martinez. 1999. Membrane raft microdomains mediate front-rear polarity in migrating cells. EMBO J. 18:6211-6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manie, S. N., S. Debreyne, S. Vincent, and D. Gerlier. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74:305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marechal, V., F. Clavel, J. M. Heard, and O. Schwartz. 1998. Cytosolic Gag p24 as an index of productive entry of human immunodeficiency virus type 1. J. Virol. 72:2208-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mondor, I., M. Moulard, S. Ugolini, P. J. Klasse, J. Hoxie, A. Amara, T. Delaunay, R. Wyatt, J. Sodroski, and Q. J. Sattentau. 1998. Interactions among HIV gp120, CD4, and CXCR4: dependence on CD4 expression level, gp120 viral origin, conservation of the gp120 COOH- and NH2-termini and V1/V2 and V3 loops, and sensitivity to neutralizing antibodies. Virology 248:394-405. [DOI] [PubMed] [Google Scholar]
- 30.Mondor, I., S. Ugolini, and Q. J. Sattentau. 1998. Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J. Virol. 72:3623-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliferenko, S., K. Paiha, T. Harder, V. Gerke, C. Schwarzler, H. Schwarz, H. Beug, U. Gunthert, and L. A. Huber. 1999. Analysis of CD44-containing lipid rafts: recruitment of annexin II and stabilization by the actin cytoskeleton. J. Cell Biol. 146:843-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orlandi, P. A., and P. H. Fishman. 1998. Filipin-dependent inhibition of cholera toxin: evidence for toxin internalization and activation through caveolae-like domains. J. Cell Biol. 141:905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parolini, I., S. Topa, M. Sorice, A. Pace, P. Ceddia, E. Montesoro, A. Pavan, M. P. Lisanti, C. Peschle, and M. Sargiacomo. 1999. Phorbol ester-induced disruption of the CD4-Lck complex occurs within a detergent-resistant microdomain of the plasma membrane. Involvement of the translocation of activated protein kinase C isoforms. J. Biol. Chem. 274:14176-14187. [DOI] [PubMed] [Google Scholar]
- 35.Pickl, W. F., F. X. Pimentel-Muinos, and B. Seed. 2001. Lipid rafts and pseudotyping. J. Virol. 75:7175-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platt, E. J., S. L. Kozak, and D. Kabat. 2000. Critical role of enhanced CD4 affinity in laboratory adaptation of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 16:871-882. [DOI] [PubMed] [Google Scholar]
- 37.Platt, E. J., N. Madani, S. L. Kozak, and D. Kabat. 1997. Infectious properties of human immunodeficiency virus type 1 mutants with distinct affinities for the CD4 receptor. J. Virol. 71:883-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popik, W., J. E. Hesselgesser, and P. M. Pitha. 1998. Binding of human immunodeficiency virus type 1 to CD4 and CXCR4 receptors differentially regulates expression of inflammatory genes and activates the MEK/ERK signaling pathway. J. Virol. 72:6406-6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puri, A., P. Hug, K. Jernigan, J. Barchi, H. Y. Kim, J. Hamilton, J. Wiels, G. J. Murray, R. O. Brady, and R. Blumenthal. 1998. The neutral glycosphingolipid globotriaosylceramide promotes fusion mediated by a CD4-dependent CXCR4-utilizing HIV type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 95:14435-14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheiffele, P., A. Rietveld, T. Wilk, and K. Simons. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038-2044. [DOI] [PubMed] [Google Scholar]
- 41.Scheiffele, P., M. G. Roth, and K. Simons. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 16:5501-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnitzer, J. E., P. Oh, E. Pinney, and J. Allard. 1994. Filipin-sensitive caveolae-mediated transport in endothelium: reduced transcytosis, scavenger endocytosis, and capillary permeability of select macromolecules. J. Cell Biol. 127:1217-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schutz, G. J., G. Kada, V. P. Pastushenko, and H. Schindler. 2000. Properties of lipid microdomains in a muscle cell membrane visualized by single molecule microscopy. EMBO J. 19:892-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 45.Singer, I. I., S. Scott, D. W. Kawka, J. Chin, B. L. Daugherty, J. A. DeMartino, J. DiSalvo, S. L. Gould, J. E. Lineberger, L. Malkowitz, M. D. Miller, L. Mitnaul, S. J. Siciliano, M. J. Staruch, H. R. Williams, H. J. Zweerink, and M. S. Springer. 2001. CCR5, CXCR4, and CD4 are clustered and closely apposed on microvilli of human macrophages and T cells. J. Virol. 75:3779-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tinoco, I., K. Sauer, and J. C. Wang. 1978. Transition-state theory, p. 294-298. In Physical chemistry: principles and applications in biological sciences. Prentice-Hall, Inc., Englewood Cliffs, N.J.
- 47.Tremblay, M., S. Meloche, S. Gratton, M. A. Wainberg, and R. P. Sekaly. 1994. Association of p56lck with the cytoplasmic domain of CD4 modulates HIV-1 expression. EMBO J. 13:774-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 49.Viola, A., S. Schroeder, Y. Sakakibara, and A. Lanzavecchia. 1999. T lymphocyte costimulation mediated by reorganization of membrane microdomains. Science 283:680-682. [DOI] [PubMed] [Google Scholar]
- 50.Wang, J. K., E. Kiyokawa, E. Verdin, and D. Trono. 2000. The Nef protein of HIV-1 associates with rafts and primes T cells for activation. Proc. Natl. Acad. Sci. USA 97:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 52.Xiao, X., A. Kinter, C. C. Broder, and D. S. Dimitrov. 2000. Interactions of CCR5 and CXCR4 with CD4 and gp120 in human blood monocyte-derived dendritic cells. Exp. Mol. Pathol. 68:133-138. [DOI] [PubMed] [Google Scholar]
- 53.Xiao, X., L. Wu, T. S. Stantchev, Y. R. Feng, S. Ugolini, H. Chen, Z. Shen, J. L. Riley, C. C. Broder, Q. J. Sattentau, and D. S. Dimitrov. 1999. Constitutive cell surface association between CD4 and CCR5. Proc. Natl. Acad. Sci. USA 96:7496-7501. [DOI] [PMC free article] [PubMed] [Google Scholar]