Abstract
An important, unresolved issue in mononegavirus biology is whether or not transcription is initiated by the same promoter as RNA replication. In this study, residues important for respiratory syncytial virus (RSV) transcription and RNA replication were identified by subjecting the first 26 nucleotides of genome RNA to saturation mutagenesis. This analysis was performed using a genome analog that allowed transcription and RNA replication to be dissociated from each other and monitored as independent events in an intracellular assay. This analysis showed that nucleotides 3C, 5C, 8U, 9U, 10U, and 11U were important for transcription and RNA replication. Additional nucleotides (1U, 2G, 6U, and 7U) were important for RNA replication, but not transcription. At position 4, G versus C or U augmented transcription and decreased replication, showing that the naturally occurring assignments in the genomic (4G) and antigenomic (4U) promoters are optimal for transcription and RNA replication, respectively. These data show that RSV transcription and RNA replication each involve a cis-acting signal at the very 3" end of the genome. This signal appears to contain a minimum, common element that functions in both transcription and RNA replication, defined by those substitutions that had similar effects on the two processes. Apart from these common nucleotides, other positions were involved in RNA replication but not transcription or had different effects on the two processes. This indicates that the promoters for transcription and replication involve overlapping sets of nucleotides at the very 3" end of the genome and provides evidence that the nucleotide preferences for the two processes are not identical.
Human respiratory syncytial virus (RSV) is the leading viral agent of pediatric respiratory disease worldwide and is a major target for vaccine and antiviral drug development (13). RSV is a member of the subfamily Pneumovirinae of the family Paramyxoviridae of the order Mononegavirales, the nonsegmented negative-strand RNA viruses. Studies involving two prototype mononegaviruses, Sendai virus and vesicular stomatitis virus (VSV) (reviewed in references 31 and 43), showed that transcription by the viral RNA-dependent RNA polymerase initiates at or near the 3" end of the encapsidated RNA genome, in the vicinity of the 3" extragenic leader (Le) region (16), and involves a sequential stop-start mechanism to produce a series of subgenomic mRNAs (2). This process is governed by short cis-acting sequences that mark the boundaries of each gene: the gene start (GS) sequence directs the polymerase to begin the synthesis of the mRNA, and the gene end sequence directs polyadenylation and release of the nascent mRNA (2, 40). Some of the transcribing polymerase remains template bound and crosses the intergenic region to resume synthesis at the next, downstream GS signal, while the remainder dissociates irreversibly, creating a polar transcription gradient (26). During RNA replication, the viral polymerase disregards the GS and gene end signals to produce a complete positive-sense intermediate called the antigenome. The antigenome is encapsidated, like the genome, and contains a promoter at its 3" terminus that directs the synthesis of progeny genome.
The initial events of mononegavirus transcription and RNA replication are not well understood, and it is not clear whether there is regulation between the two processes and, if so, how that occurs. A widely accepted model postulates that the initial stages of both processes are identical, involving a common polymerase that initiates at a single promoter at the 3" (Le) end of the genome. In this model, transcription occurs until there is sufficient soluble intracellular N protein to encapsidate the nascent RNA, which then shifts the polymerase to a readthrough mode to produce encapsidated antigenome. Thus, transcription and RNA replication would be in a dynamic, reciprocal balance controlled by the concentration of soluble N protein (4, 31, 42, 43). Alternatively, transcription and RNA replication could be initiated by separate pathways using distinct promoters and/or different initiation sites. An example of evidence to support this hypothesis comes from work with VSV, where the greater molar abundance of N mRNA over leader RNA synthesized in vitro by the polR1 mutant suggested that the polymerase can initiate transcription directly at the first gene (10). In this scenario, cosynthetic encapsidation remains essential for RNA replication but does not regulate transcription.
RSV follows the general strategy of stop-start transcription and readthrough RNA replication described above (reviewed in reference 13). However, it should be noted that RSV also has a number of differences compared to other mononegaviruses. For example, RSV does not follow the rule of six (38); the cis-acting signals involved in initiation of replication are contained within the Le (11) and so are more circumscribed than those of other paramyxoviruses, which involve sequences beyond the first GS signal; the gene junction regions are highly variable; and RSV encodes several proteins that do not have counterparts in the prototype mononegaviruses, including at least three (M2-1, M2-2, and NS1) that appear to affect transcription or RNA replication (1, 3, 14, 24, 28). It also should be noted that while complementarity between the genomic termini has been suggested to regulate VSV RNA synthesis (44), it does not appear to play a role in either transcription or RNA replication by RSV (19, 36).
The 3" ends of RSV strain A2 genomic and antigenomic RNAs are identical for 10 of the first 11 positions and share 81% identity for the first 24 to 26 positions, after which sequence identity is insignificant (35) (Fig. 1). The 10 residues within the first 3"-terminal 11 positions that are conserved between the RSV A2 genome and its antigenome also are conserved exactly in RSV strain B1, bovine RSV, and avian pneumovirus (5, 29, 37) (Fig. 1). It seemed likely that some or all of these conserved residues constitute a promoter element. However, the boundaries and composition of this putative element were unknown, and nonconserved sequences might also be involved. It would be presumed that this putative promoter element is involved in RNA replication, but this had not been demonstrated directly. Also, it was unclear whether these 3"-terminal residues were important for transcription, since transcription might employ a separate promoter, such as one located at the GS signal of the first gene.
FIG. 1.
Sequence alignment between the 3" terminus of RSV strain A2 genomic RNA (A), the 3" end of its antigenome (B), and the genomic RNAs of subgroup B RSV strain B1 (C), bovine RSV (D), and avian pneumovirus (E). Gaps (dots) were introduced to maximize alignment. Boldface indicates identities between the RSV A2 genome and the sequence in question. Numbering is according to the RSV genome sequence. The Le-proximal GS signal in each genomic RNA sequence is underlined. In the RSV sequence, position 4 is indicated as a G because this assignment was identified in the prototype A2 wt isolate. However, the assignment at this position is a C in certain biologically derived A2 variants. The parental minigenome in the present study contained the 4C assignment because in previous work it increased the efficiency of minigenome RNA synthesis.
In the present study, we conducted mutational analysis of the 3"-terminal 26 nucleotides (nt) of the Le region of a minigenome of RSV strain A2 with the aims of identifying which residues are important for RNA replication and determining if the same nucleotides are also involved in mRNA transcription. The results presented here identified residues within the 3"-terminal 11 positions of Le that are important for both transcription and RNA replication and likely represent a promoter element common to both processes. However, the results also show that the nucleotide preferences are not identical for transcription and replication. Furthermore, there are additional residues that are important for replication but apparently not for transcription. Thus, these results indicate that transcription, like replication, involves recognition of nucleotides at the 3" end of the Le region but that the nucleotides that are important are not identical for the two processes.
MATERIALS AND METHODS
Plasmid mutagenesis.
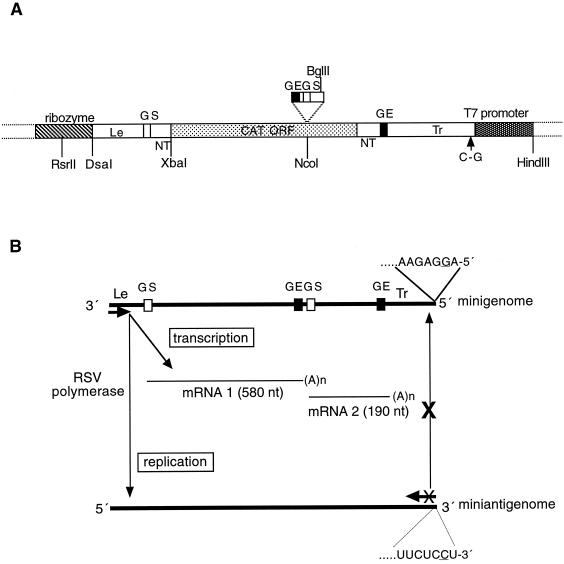
Plasmids encoding minigenomes C75Δ and MP28 were constructed from plasmid C41, which contains the 3"-terminal 75 nt and 5"-terminal 179 nt of RSV genomic RNA flanking a 677-nt negative-sense copy of the chloramphenicol acetyltransferase (CAT) open reading frame (18). The N-P gene junction was inserted into the CAT open reading frame of C41 to generate plasmid MP28. To create plasmid C75Δ, a C-to-G substitution at position 2 relative to the 5" end of the trailer (Tr) was introduced into plasmid MP28 by replacing the BglII-HindIII region containing the Tr with that of plasmid 2G (36). Single-nucleotide substitutions were introduced into the C75Δ Le region by inserting PCR products, generated using mutagenic oligonucleotide primers, into the window created by an XbaI site (at the junction between the NS1 nontranslated region and the CAT open reading frame) and an RsrII site (for mutations at Le positions 1 to 4) or DsaI site (for mutations at Le positions 5 to 26); the RsrII site is located within the ribozyme, and the DsaI site is at the junction between the ribozyme and Le sequence (Fig. 2A). It should be noted that the parental minigenome in this study contained the assignment of C (negative sense) at Le position 4.
Transfections.
Monolayers of HEp-2 cells in six-well dishes were simultaneously infected with 10 PFU (per cell) of vaccinia virus vTF7-3 (provided by Thomas Fuerst and Bernard Moss), which expresses the T7 RNA polymerase (21), and transfected with the following mixture of plasmids per well of a six-well dish: 0.2 μg of minigenome DNA, 0.4 μg of pTM1 N, 0.2 μg of pTM1 P, 0.1 μg of pTM1 M2-1, and 0.1 μg of pTM1 L, as described previously (22). Control reactions lacking L received pTM1 plasmid with no insert so that the amount of transfected DNA was equivalent in each well. Twenty-four hours later, the transfection-infection mixture was replaced with OptiMem containing 2% fetal bovine serum and actinomycin D at 2 μg/ml. The actinomycin D treatment inhibited further T7-mediated RNA synthesis and was done to minimize the possibility that the accumulation of a high level of plasmid-encoded RNA might interfere with the reconstituted RSV system, such as by competing for N protein. The actinomycin D-containing medium was removed after 2 h, replaced with fresh OptiMem containing 2% fetal bovine serum, and incubated for a further 24 h. Because of the large number of mutants to be tested, the experiments were broken into groups that each contained six mutants, a wild-type (wt) positive control, and an L-minus negative control. Also, each transfection reaction was set up in duplicate: RNA was directly extracted from cells from one of the wells, and the cells in the other well were lysed with nonionic detergent and incubated with micrococcal nuclease (MCN) prior to RNA purification to digest unencapsidated RNA, as described previously (17). Cells transfected with minigenome but no support plasmids were used as controls to ensure nuclease activity.
RNA isolation and Northern blot hybridization.
RNA was extracted by dissolving cell pellets or MCN-treated cell lysates in Trizol reagent (Life Technologies) according to the supplier's protocol, except that the RNAs were extracted with phenol-chloroform and ethanol precipitated after the isopropanol precipitation. RNA representing 1/10 of one well of cells was analyzed by electrophoresis in a 1.5% agarose gel containing 0.44 M formaldehyde, transferred to nitrocellulose (Schleicher and Schuell), and fixed by UV cross-linking (Stratagene). Each experimental group consisting of six mutant minigenomes and the wt and L-minus controls was represented on two gels, one containing total RNA and the other containing the MCN-treated counterpart. Negative-sense or positive-sense 32P-labeled CAT-specific probe was synthesized as described previously (22) and hybridized to the Northern blot in a mixture of 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's solution, 0.5% sodium dodecyl sulfate (SDS), and 200 μg of sheared DNA per ml at 65°C for at least 12 h. The blots were washed in 2× SSC-0.1% SDS at room temperature for 30 min and then at 65°C for 2 h and then washed in 0.1× SSC-0.1% SDS at 65°C for 15 to 30 min. Phosphorimager analysis was carried out with a PhosphorImager 445 SI (Molecular Dynamics).
RESULTS
The purpose of this study was to characterize the first 26 nt of RSV strain A2 genomic RNA for their importance in RNA replication and transcription. Saturation mutagenesis was performed on a cDNA-encoded RSV minigenome. Effects on transcription and RNA replication were determined with a previously described system in which RSV RNA synthesis is reconstituted from transfected plasmids expressing the minigenome and RSV nucleocapsid and polymerase proteins (22).
We previously showed that RNA replication mediated by the RSV polymerase in this reconstituted system amplifies the plasmid-supplied encapsidated minigenome up to 50-fold (36). Thus, any introduced mutation affecting the efficiency of RNA replication would change the level of minigenome template and indirectly affect the accumulation of replication and transcription products. Mutations that severely inhibited RNA replication would preclude reliable measurement of transcription. To avoid this complication, these experiments employed a modified minigenome, C75Δ, that contains a point mutation, C to G, at the penultimate nucleotide of the Tr (Fig. 2). As described previously (36), this mutation does not affect the encapsidation of the plasmid-encoded minigenome or its ability to act as a template for the RSV-mediated synthesis of mRNA and encapsidated antigenome. However, the antigenome produced is unable to serve as a template for the synthesis of progeny minigenome, presumably because the mutation at position 2 relative to its 3" end inactivates its promoter (Fig. 2B). Thus, the amount of minigenome template is limited to that supplied directly from the plasmid and is independent of the efficiency of replication. This uncoupled transcription and replication such that effects on one did not affect the other, and the contribution of each nucleotide position to each process could be monitored.
FIG. 2.
Diagram (not to scale) of the C75Δ plasmid (A) and the C75Δ minigenome RNA and its encoded RNA products (B). (A) Structure of the C75Δ plasmid: the minigenome cDNA consists of, in left to right order: the leader (Le) region, NS1 gene-start (GS) signal, nontranslated upstream NS1 gene sequence (NT), CAT open reading frame (ORF; grey stippled), L gene-end signal (GE), nontranslated downstream L gene sequence (NT), and trailer (Tr) region. The minigenome cDNA is flanked by the ribozyme and T7 RNA promoter (hatched and dark stippled boxes, respectively). In addition, as indicated, the CAT gene was divided into two transcriptional units by the introduction of a short cDNA encoding the N-P gene junction. The C to G (negative-sense) mutation that was introduced into the trailer region at position 2 relative to the downstream end is indicated. (B) Diagram (not to scale) of the plasmid-derived, negative-sense C75Δ minigenome and its encoded positive-sense antigenome and mRNAs 1 and 2. The last 7 nt at the 5" end of the minigenome and the complementary sequence at the 3" end of the miniantigenome are shown, and the C-to-G Tr mutation is underlined. Arrowheads indicate direction of RNA synthesis; synthesis of progeny minigenome is prevented by the C-to-G Tr mutation.
Analysis of Le positions 1 to 26.
A series of 78 mutant minigenome plasmids was constructed in which each of the first 26 nt of Le was changed individually to each of the three alternative assignments. Each mutant was assayed for its ability to direct transcription and RNA replication in the reconstituted intracellular system. The levels of mRNA and antigenome produced were measured directly by Northern blot analysis of total intracellular RNA with a negative-sense CAT-specific probe. Duplicate blots were probed with a positive-sense CAT-specific probe to confirm that similar levels of minigenome template were produced by T7 RNA polymerase and that the efficiency of ribozyme cleavage necessary to generate the Le 3" end was unaffected by the mutation in the nearby Le region (not shown).
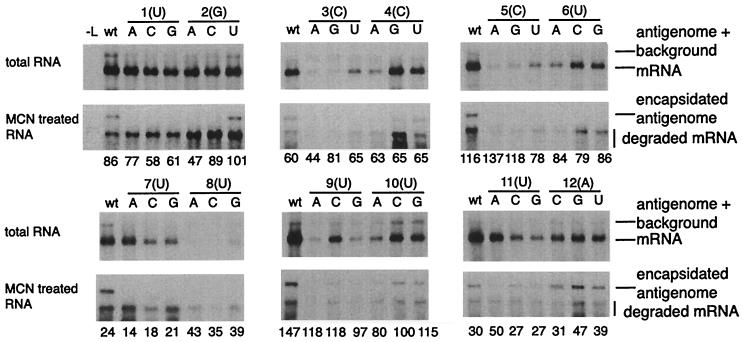
Examples of Northern blots of positive-sense RNAs generated by minigenomes containing substitutions in Le positions 1 to 12 are shown in Fig. 3. Examination of total intracellular RNA showed that, in each transfection experiment, the wt minigenome (C75Δ) generated a large amount of mRNA and a small amount of antigenomic RNA, as is characteristic of this system. The mRNA indicated is the product of the first gene. The smaller mRNA transcribed from the second gene was not visible in these experiments because its small size reduced its efficiency of binding and hybridization in the Northern blot analysis. However, its detection was not necessary since quantitation of the first mRNA was sufficient to measure transcription activity. Quantitation of the accumulated antigenomic RNA was complicated by its low abundance and the presence in the gel lane of a faint background that became evident upon phosphorimager analysis. In order to distinguish the antigenome more clearly, duplicate transfection cell lysates were treated with MCN to digest unencapsidated RNA prior to RNA purification and Northern blot analysis. Each experiment included RNA from a control transfection in which the support plasmids were omitted, and in each case the MCN treatment was sufficient to completely digest the naked minigenomic RNA (not shown). Unexpectedly, a small amount of mRNA was resistant to degradation. Since the conditions of the MCN treatment were sufficient to degrade the unencapsidated control RNA, this suggested that the MCN-resistant mRNA was protected by protein. MCN-resistant mRNA was not observed in experiments in which a replication-competent minigenome was used (19), suggesting that when the minigenome is unable to amplify, the mRNA can be encapsidated nonspecifically due to the resulting excess of N and P proteins. However, the presence of a small amount of undigested mRNA did not interfere with interpretation of the results.
FIG. 3.
Northern blot analysis of antigenomic RNA and mRNA generated from mutant C75Δ-based minigenomes containing single-nucleotide substitutions in the first 12 nt positions of the Le region and complemented by the N, P, M2-1, and L proteins. The six mutant minigenomes and the wt minigenome represented in each blot were transfected and analyzed in parallel. The numbers above each blot indicate the Le position that has been substituted, and the letter in parentheses indicates the wt nucleotide assignment (negative sense). The three alternative mutant assignments at each position are identified. Lane -L, negative control in which L plasmid was omitted. The upper blot for each set of mutants shows total intracellular RNA, and the lower blot shows MCN-treated RNA. All blots were hybridized with a negative-sense, CAT-specific riboprobe. The number beneath each lane represents the level (in arbitrary units) of encapsidated minigenome template that was present in each transfection, assessed by quantitating Northern-blotted MCN-resistant RNA that was detected with a positive-sense, CAT-specific riboprobe. These arbitrary values can be compared within each experimental group contained on a single blot but cannot be compared directly between different gels unless they are first normalized to their respective wt control.
This analysis showed that many of the substitutions in the first 26 positions of the Le resulted in reduced levels of mRNA and antigenome (Fig. 3). Quantitation of encapsidated negative-sense RNA confirmed that there was a similar level of minigenome template available in each transfection (Fig. 3). Thus, these data show that this region of the Le contains sequences important for both transcription and RNA replication. The levels of mRNA (in the total RNA fraction) and antigenome (in the MCN-treated RNA fraction) that were produced from each mutant minigenome were quantitated using a phosphorimager and calculated as percentages of the wt levels of mRNA and antigenome in each experiment (Fig. 4). We considered mutations that reduced RNA synthesis to 30% or less of the wt level to be significant. Four categories of mutation were noted, as follows.
FIG. 4.
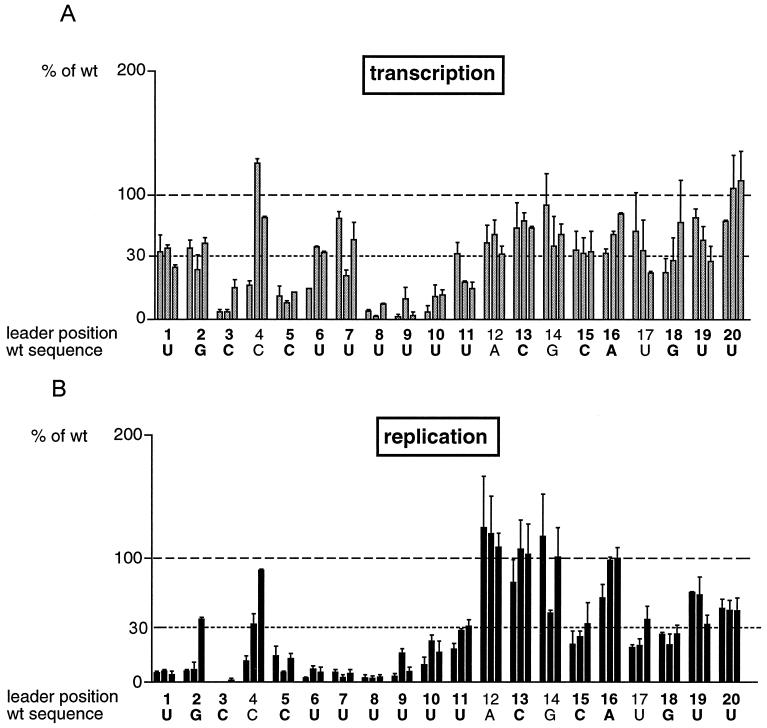
Quantitation of mRNA and antigenomic RNA generated from mutant C75Δ-based minigenomes containing single-nucleotide substitutions in the first 20 nt of the Le region, based on experiments as described for Fig. 3. (A) CAT-specific mRNA 1 measured by Northern blot analysis of total intracellular RNA and phosphorimager analysis. (B) Encapsidated antigenomic RNA bands measured by Northern blot and phosphorimager analysis of RNA purified from MCN-treated cell lysates. The numbers on the x axes indicate the Le positions of the nucleotide substitutions, and the letters indicate the wt nucleotide assignment; assignments that are exactly conserved at the 3" end of antigenomic RNA are in boldface. The three possible substitution mutants for each position are ordered left to right alphabetically by nucleotide assignment (i.e., the wt assignment of position 1 is U, and hence the three substitutions, in order, are A, C, and G). Each RNA value was calculated as a percentage of the wt value (100%, indicated by a dashed line). Each error bar represents the standard error of the mean from two or three independent experiments, depending on the mutation. Mutations that reduced RNA levels to less than 30% of the wt value, indicated by a dotted line, were considered significant.
(i) Substitutions at positions 3, 5, 8, 9, and 10 strongly inhibited both RNA replication and transcription. At each of these positions, none of the three alternative assignments allowed a significant level of either process to occur. Most substitutions at position 11 also inhibited both transcription and replication, although to a lesser extent. Thus, nucleotides 3C, 5C, 8U, 9U, 10U, and 11U are important for both transcription and RNA replication.
(ii) A number of substitutions inhibited replication but not transcription. Substitution with any nucleotide at positions 1, 6, and 7 and substitution with either an A or C at position 2 caused a substantial reduction of antigenome synthesis. In contrast, all substitutions at position 1 or 2 had only modest effects on mRNA synthesis, and mutation of position 6 or 7 had an intermediate effect, with one of the three substitutions inhibiting mRNA synthesis to less than 30% but the other two being well tolerated. Thus, residues 1U, 2G, 6U, and 7U are important for RNA replication but not for transcription.
(iii) One substitution at position 4 had opposing effects on transcription and replication: specifically, replacement of the C residue with G caused an increase in transcription and a decrease in replication. Although these effects were only minor (less than twofold), they were highly reproducible and suggested that the optimal promoter sequences for transcription and RNA replication are not identical. Replacement of the 4C residue with an A reduced both transcription and replication to approximately 25% of the wt level, and replacement with a U (the assignment found at the corresponding position in antigenomic RNA) had little effect on either process.
(iv) Most of the substitutions at positions 12 to 26 had no significant effect on transcription or RNA replication based on the benchmark of 30% (data for positions 12 to 20 are shown in Fig. 4). However, small effects were consistently observed with mutations at certain positions: for example, any substitution at position 15, 17, or 18 reduced RNA replication to approximately 30 to 40% of wt levels and also reduced transcription, although to a lesser extent. It appeared that substitutions within Le nt 12 to 20 had a marginally greater negative effect on RNA replication than on transcription; for example, any substitution at position 20 resulted in a modest decrease in replication, whereas transcription was unaffected. The only exception to this was position 12, at which substitutions had little effect on replication but slightly inhibited transcription. In general, substitutions of nt 21 to 26 had negligible effects on both transcription and RNA replication, with neither process being inhibited to less than 60% of the wt level (data not shown). There were two exceptions; a U-to-A substitution at position 22 reduced replication to 33% and transcription to 43% of wt levels, and a U-to-G substitution at position 23 reduced replication to 38% and transcription to 52% of wt levels. Thus, in general, individual nucleotide substitutions within positions 12 to 26 were well tolerated. However, it should be noted that one drawback to using a minigenome that is limited to a single cycle of replication is that alterations in RNA synthesis that appear to be minor in this assay system could be magnified and become significant during multiple rounds of replication.
Further analysis of Le position 4 mutations.
Position 4 has been shown to be variable among biologically derived isolates and derivatives of RSV. wt strain A2 contains the assignment of G at this position (35). Certain attenuated derivatives have the assignment of C, although this did not appear to contribute significantly to the attenuation phenotype (20). The parental C75Δ minigenome used in the present work contained the 4C assignment because in previous studies it was associated with more efficient minigenome RNA synthesis (12, 22). In the present study, the assignment of C at Le position 4 was associated with increased replication and decreased transcription, whereas the assignment of G had the converse effect, as described above.
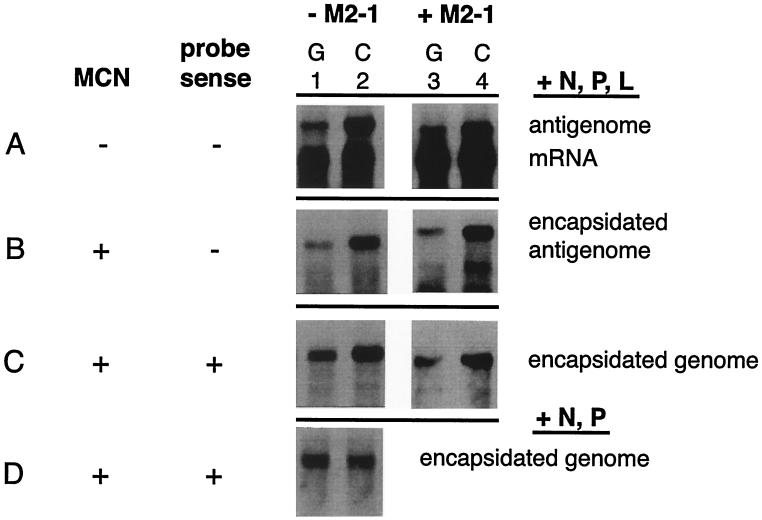
The C versus G assignment at position 4 also was examined in the MP28 minigenome backbone (see Materials and Methods), which contained a wt Tr and thus was competent for amplification by the reconstituted RSV polymerase. This provided the opportunity to examine each mutation under conditions where its effect should be magnified by multiple rounds of replication; however, as noted above, analysis of transcription per se would be complicated by the differences in the accumulation of minigenome template. The experiment was performed in the presence and absence of the M2-1 transcription elongation factor. Although previous studies indicated that M2-1 affects transcription antitermination but not initiation and does not affect RNA replication (14, 18), we wanted to confirm that the differential effect of the position 4 mutation was not dependent on the presence of M2-1.
Analysis of positive-sense RNA (Fig. 5A and B) showed that the 4C minigenome template generated significantly more antigenome than did the 4G minigenome template. This was evident from analysis of total intracellular RNA (Fig. 5A) as well as from analysis of lysates that had been treated with MCN (Fig. 5B). The results were the same in the absence of M2-1 (Fig. 5A and B, lanes 1 and 2) or in its presence (lanes 3 and 4). Analysis of negative-sense, encapsidated (i.e., resistant to digestion with MCN) RNA showed that the 4C minigenome also accumulated to a higher level than did the 4G minigenome (Fig. 5C), as would be expected from enhanced antigenome synthesis under conditions permissive to genome amplification. As a control, we monitored the accumulation of encapsidated 4G and 4C minigenome RNAs from transfections in which the L plasmid had been omitted, which would preclude RSV-mediated RNA replication and would restrict the minigenome to that derived directly from the transfected plasmid. Under these conditions, the accumulation of the 4G and 4C minigenomes was reduced approximately 50-fold, reflecting the lack of amplification, and the two minigenomes accumulated to comparable levels (Fig. 5D). This confirmed that the higher level of 4C minigenome observed in the presence of complete polymerase was the result of more efficient RNA replication. In contrast to the pattern observed with the C75Δ minigenome backbone, the 4C mutant in the MP28 minigenome yielded more mRNA than its 4G counterpart (Fig. 5A). However, this can be attributed to the increased level of minigenome template resulting from its more efficient replication. Thus, the assignment at position 4 affects both RNA replication and transcription in a manner that varies depending on the residue. The assignment of C versus G had a reciprocal effect on the two processes that was magnified under conditions permissive for RNA replication.
FIG. 5.
Northern blot analysis of mRNA and antigenomic RNA generated from minigenomes containing a wt Tr region (minigenome MP28) and either the G (lanes 1 and 3) or C (lanes 2 and 4) assignment at Le position 4. Each minigenome plasmid was transfected together with plasmids N, P, and L (panels A, B, and C, lanes 1 and 2); N, P, L, and M2-1 (panels A, B, and C, lanes 3 and 4); or N and P only (panel D). (A) Total intracellular RNA hybridized with negative-sense probe, showing antigenomic RNA and mRNA 1. (B) RNA from MCN-treated lysates hybridized with negative-sense probe, showing encapsidated antigenome and residual mRNA. (C and D) RNA from MCN-treated lysates hybridized with positive-sense probe, showing encapsidated minigenome RNA. The conditions for panel C were permissive for reconstituted RSV-mediated amplification, whereas the transfections for panel D were in the absence of L and thus the only source of minigenomic RNA was the transfected plasmid. The Northern blot indicated in panel D was exposed to the detector screen approximately 50-fold longer than that in panel C.
DISCUSSION
In this study, the 3"-terminal 26 nt of RSV genomic RNA were analyzed by saturation mutagenesis. Importantly, this analysis was performed with a minigenome that contained a mutation in the Tr region that prevented its amplification, so effects on replication and resulting changes in template concentration would not affect transcription. This analysis showed that the 3" end of the Le region contains sequence important for both transcription and RNA replication. The most important residues were contained within the highly conserved first 11 positions; somewhat surprisingly, other well-conserved residues were relatively unimportant. The sets of residues important for transcription versus RNA replication overlapped but were not identical.
The use of a plasmid-based minigenome system in which amplification of the minigenome was blocked was an important feature of this study. We arrived at this strategy after extensive preliminary experimentation beginning in 1991 (unpublished data). We began with a previously described system in which the minigenomic RNA is synthesized by T7 polymerase in vitro and transfected into RSV-infected cells, with the trans-acting viral proteins provided by the infecting standard virus (11, 30, 38). We previously noted that this system has the advantage of taking place in the context of a natural infection and in the absence of vaccinia virus or any other heterologous biological agent (30, 38). However, we found that this RSV-complemented system requires a very high level of amplification of the input minigenome in order to detect products of RNA replication and transcription, and mutations that reduced the level of RNA replication were difficult to monitor for either process. We also explored the use of a plasmid-based system in which the minigenome was competent for amplification by RSV-mediated RNA replication (as with minigenome MP28 in Fig. 5) (22). This system was more reliable and robust, but it was difficult to monitor transcription in cases where RNA replication was reduced substantially. We believe that this precludes reliable analysis of transcription in situations where RNA replication is significantly inhibited. We therefore adopted the present strategy of use of a plasmid-supplied, nonamplifiable minigenome template, whereby we could (i) monitor the intracellular synthesis and encapsidation of the plasmid-supplied minigenome to determine the amount of template and (ii) dissociate RNA replication and transcription such that effects on one do not alter the other. One drawback of a minigenome system is that one cannot extrapolate reliably to predict the magnitude of the effect of individual mutations on RNA synthesis by infectious virus. On the other hand, use of a minigenome system provides assurance that the effects of each mutation can be monitored, in contrast to infectious virus, where many of the mutations likely would not be recoverable and where the more complex nature of the system would complicate analysis of viable mutations. The next step for future work will be to place selected mutations in complete recombinant virus.
This study showed that positions 1 to 3 and 5 to 11 are important for efficient RNA replication. Since antigenome synthesis is presumed to initiate at the first nucleotide of the genome, it was not surprising that the adjacent sequence would be involved in replication initiation. Similarly, it was recently shown that the first 12 nt of the genome of human parainfluenza virus type 3 (PIV3) are important for RNA replication (25), and several studies indicate that the 3" terminus of the VSV Le is necessary for RNA replication (32, 39, 44). In the present study, substitutions at positions 12 to 26 had relatively little effect on the efficiency of replication, which was somewhat surprising given that many of these positions are conserved between genomic and antigenomic RNAs (Fig. 1). However, residues in this region appeared to have a role in replication if they were mutated in aggregate, based on the observation that replacement of Le nt 16 to 44 with an equivalent length of heterologous sequence inhibited replication, whereas replacement of Le nt 26 to 44 had little effect (R. Fearns and P. L. Collins, unpublished data). It should also be noted that effects of single-nucleotide substitutions that seem small in a nonamplifying minigenome might become significant when amplified through many cycles of replication. This will be evaluated in complete recombinant virus in future work.
We also observed that the 3" terminus of genomic RNA contains residues important for transcription, specifically, nucleotides at positions 3, 5, 8, 9, 10, and 11. Single-nucleotide substitutions further downstream within Le, involving positions 12 to 26, had no effect or little effect on transcription. A minigenome in which Le residues 16 to 44 were replaced with an equivalent length of heterologous sequence was able to direct transcription, supporting the idea that downstream Le nucleotides were not important for transcription. The finding that 3"-terminal residues are important for transcription was not necessarily anticipated, because it is not known whether transcription by RSV (or any mononegavirus) initiates at the first nucleotide of the genome or, alternatively, at the Le-proximal GS signal (genome position 45 in the case of RSV). In the latter case, the transcription promoter need not necessarily involve the very 3" end at all. The present findings do not identify the site of initiation of RSV transcription (since the polymerase might contact nucleotides at the 3" end but initiate at the GS signal), but our results do show that at least part of the transcription promoter lies at the very 3" end of the genome. Previously, in vitro reconstitution studies with VSV suggested that all RNA synthesis initiates solely at the 3" end of the VSV genome (16, 39), although this has been questioned (10). Studies with a VSV minigenome concluded that the 3"-terminal 24 nt are part of the transcription promoter (32), although that study involved 6-nt deletions rather than fine mapping. Thus, sequence at the very 3" end of the genome is required for transcription of both RSV and VSV, but that of RSV appears to be more circumscribed. A previous study with a PIV3 minigenome examined the role of residues within the first 13 nt at the 3" terminus of Le in transcription (25). Mutations at a number of positions resulted in significantly reduced mRNA synthesis. However, in that study, experiments were carried out using a minigenome that was competent for amplification, and the same mutations severely inhibited RNA replication. Thus, it is not possible to conclude whether the observed inhibition of transcription was direct or a consequence of the effects on replication and template abundance. Therefore, the role of these nucleotides in PIV3 transcription remains undefined. For RSV, the present findings provide direct evidence that at least part of the transcription promoter lies at the very 3" end of the genome, irrespective of the location of the site of transcription initiation, and overlaps with the replication promoter.
Among the first 11 positions there was flexibility at position 4: the assignment of C, G, or U at this position was compatible with a high level of RNA synthesis, but the optimal assignment was different for transcription versus replication. A G residue was associated with lower level of replication and a higher level of transcription than either a U or a C residue. The four wt human or bovine RSV genome sequences that have been determined to date, i.e., human RSV strains A2, S2, and B1 and bovine RSV strain A51908, each have G at genome position 4 (Fig. 1) (5, 29, 35, 37, 41) and U at antigenome position 4 (each relative to the 3" end). Thus, this single nucleotide difference would be predicted to bias the genomic RNA promoter for transcription and the antigenomic promoter for RNA replication. When the 4C mutation was introduced into a wt backbone, the resulting 4C virus produced smaller plaques and exhibited slightly reduced growth in vitro (27; K.-C. Tran and P. L. Collins, unpublished observations).
Thus, both RNA replication and transcription involve a common core element, namely, genome positions 3, 5, 8, 9, 10, and 11. Consistent with the idea of a common element, nucleotide substitutions within these positions affected transcription and replication similarly; for example, a G substitution at position 5 resulted in lower levels of transcription and replication than either an A or a U substitution, and likewise a C substitution at position 9 was tolerated slightly better than either an A or a G substitution. As mutations in this region affect transcription and replication in a similar way, this suggests that this element plays the same role in both transcription and RNA replication. RNA transcription and replication each involve additional cis-acting sequences specific to each process. We previously showed that efficient mRNA synthesis also requires the first GS signal, spaced correctly relative to the 3" end of the genome (19). In the present study, nucleotides at positions 1, 2, 6, and 7 were found to be important for RNA replication but were relatively unimportant for transcription. It is not known whether these replication-specific nucleotides are involved in polymerase contact and entry, the initiation of antigenome synthesis, stability of the encoded antigenome, or encapsidation. It is tempting to speculate that these residues encode an encapsidation signal in the nascent antigenome. There is evidence that the VSV and measles virus Le regions encode an encapsidation signal (4, 6); however, in previous experiments we showed that the RSV Le-CAT readthrough mRNA is not encapsidated, even though sufficient N protein was apparently available (19), and we and others did not find evidence for specific encapsidation of RNA containing RSV sequence (34, 36).
The data in the present paper can be interpreted into two alternative models, which employ some general features of models described previously for Sendai virus and VSV and which also take into account our previous data involving the role of the Le-proximal GS signal in transcription (19).
Model 1.
In the first model, transcription and RNA replication are initiated from a single common promoter that directs RNA synthesis to initiate at the 3" terminus of the Le. According to this hypothesis, the promoter consists of the residues 3C, 5C, 8U, 9U, 10U, and 11U. The additional residues identified as being important for RNA replication, namely, 1U, 2G, 6U, and 7U, would be postulated to encode an encapsidation signal in the antigenome, and the GS signal would be required for transcription reinitiation following Le RNA synthesis. Like others (15, 42), we suggest that the initiation complex is unstable and requires further signals (either cis or trans acting) to transition into a stable elongation mode. Thus, the newly initiated RNA polymerase can enter three alternative pathways, as follows.
(i) The unstable initiation complex terminates RNA synthesis at the end of the Le but reinitiates at the GS signal and begins transcription. This termination step might involve a specific cis-acting signal but alternatively might occur over a range of positions near the end of Le and might simply reflect the limit of elongation by the unstable complex. Once the polymerase has initiated mRNA synthesis at the NS1 GS signal, its association with the M2-1 antitermination factor would allow transcription elongation.
(ii) The unstable initiation complex associates with N protein and switches into a stable replication complex that synthesizes progeny antigenome, as previously suggested for Sendai virus (42).
(iii) The unstable initiation complex neither reinitiates at the GS signal nor associates with N and instead dissociates from the template near the end of the Le region. By analogy to bacterial and eukaryotic RNA polymerases, this abortive initiation might be the most frequent outcome.
According to this model, transcription and RNA replication might be in reciprocal balance; however, this need not be the case if a large percentage of initiation events are abortive. Thus, this model is not inconsistent with our previous finding that the N protein concentration does not alter the balance between transcription and replication (17).
Model 2.
In the second model, transcription and RNA replication are distinct processes that have overlapping but distinct promoters. The two promoters share common elements (nucleotides 3C, 5C, 8U, 9U, 10U, and 11U) but also have specific, different elements, namely, nucleotides 1U, 2G, 6U, and 7U in the case of RNA replication (which in this scenario might be a direct part of the promoter rather than encoding an encapsidation signal) and the Le-proximal GS signal in the case of transcription. In this model the GS site might represent an initiation domain of the transcription promoter, analogous to the initiation domain of the T7 promoter (7, 8, 9, 23, 33). Transcription and replication would thus be independently controlled events that involve overlapping but distinct promoters and, presumably, functionally distinct forms of the polymerase complex. Based on the finding that the GS signal must be spaced appropriately relative to the 3" end, the results from our RSV minigenome studies seem more consistent with model 2, but it is not yet possible to conclusively identify the correct model.
Acknowledgments
We thank Thomas Fuerst and Bernard Moss for vTF7-3, Myron Hill and Kim-Chi Tran for technical assistance, and Brian Murphy and Robert Chanock for critical reviews of the manuscript.
REFERENCES
- 1.Atreya, P. L., M. E. Peeples, and P. L. Collins. 1998. The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication. J. Virol. 72:1452-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barr, J. N., S. P. Whelan, and G. W. Wertz. 1997. cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J. Virol. 71:8718-8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bermingham, A., and P. L. Collins. 1999. The M2-2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc. Natl. Acad. Sci. USA 96:11259-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumberg, B. M., C. Giorgi, and D. Kolakofsky. 1983. N protein of vesicular stomatitis virus selectively encapsidates leader RNA in vitro. Cell 32:559-567. [DOI] [PubMed] [Google Scholar]
- 5.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castaneda, S. J., and T. C. Wong. 1990. Leader sequence distinguishes between translatable and encapsidated measles virus RNAs. J. Virol. 64:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, K. A., and R. R. Burgess. 1987. Construction of bacteriophage T7 late promoters with point mutations and characterization by in vitro transcription properties. Nucleic Acids Res. 15:5413-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheetham, G. M. T., D. Jeruzalmi, and T. A. Steitz. 1999. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature 399:80-83. [DOI] [PubMed] [Google Scholar]
- 9.Cheetham, G. M. T., and T. A. Steitz. 2000. Insights into transcription: structure and function of single-subunit DNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 10:117-123. [DOI] [PubMed] [Google Scholar]
- 10.Chuang, J. L., and J. Perrault. 1997. Initiation of vesicular stomatitis virus mutant polR1 transcription internally at the N gene in vitro. J. Virol. 71:1466-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, P. L., M. A. Mink, and D. S. Stec. 1991. Rescue of synthetic analogs of respiratory syncytial virus genomic RNA and effect of truncations and mutations on the expression of a foreign reporter gene. Proc. Natl. Acad. Sci. USA 88:9663-9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, P. L., M. A. Mink, M. G. Hill, E. Camargo, H. Grosfeld, and D. S. Stec. 1993. Rescue of a 7502-nucleotide (49.3% of full-length) synthetic analog of respiratory syncytial virus genomic RNA. Virology 195:252-256. [DOI] [PubMed] [Google Scholar]
- 13.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, p. 1433-1485. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 14.Collins, P. L., M. G. Hill, J. Cristina, and H. Grosfeld. 1996. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc. Natl. Acad. Sci. USA 93:81-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupuy, L. C., S. Dobson, V. Bitko, and S. Barik. 1999. Casein kinase 2-mediated phosphorylation of respiratory syncytial virus phosphoprotein P is essential for the transcription elongation activity of the virus polymerase: phosphorylation by casein kinase 1 occurs mainly at Ser215 and is without effect. J. Virol. 73:8384-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emerson, S. U. 1982. Reconstitution studies detect a single RNA polymerase entry site on the vesicular stomatitis virus genome. Cell 31:635-642. [DOI] [PubMed] [Google Scholar]
- 17.Fearns, R., M. E. Peeples, and P. L. Collins. 1997. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology 236:188-201. [DOI] [PubMed] [Google Scholar]
- 18.Fearns, R., and P. L. Collins. 1999. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J. Virol. 73:5852-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fearns, R., M. E. Peeples, and P. L. Collins. 2000. Functional analysis of the genomic and antigenome promoters of human respiratory syncytial virus. J. Virol. 74:6006-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firestone, C. Y., S. S. Whitehead, P. L. Collins, B. R. Murphy, and J. E. Crowe, Jr. 1996. Nucleotide sequence analysis of the respiratory syncytial virus subgroup A cold-passaged (cp) temperature sensitive (ts) cpts-248/404 live-attenuated virus vaccine candidate. Virology 225:419-422. [DOI] [PubMed] [Google Scholar]
- 21.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grosfeld, H., M. G. Hill, and P. L. Collins. 1995. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins: transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J. Virol. 69:5677-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guajardo, R., P. Lopez, M. Dreyfus, and R. Sousa. 1998. NTP concentration effects on initial transcription by T7 RNAP indicate that translocation occurs through passive sliding and reveal that divergent promoters have distinct NTP concentration requirements for productive initiation. J. Mol. Biol. 281:777-792. [DOI] [PubMed] [Google Scholar]
- 24.Hardy, R. W., and G. W. Wertz. 1998. The product of the respiratory syncytial virus M2 gene ORF 1 enhanced readthrough of intergenic regions during viral transcription. J. Virol. 72:520-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman, M. A., and A. K. Banerjee. 2000. Precise mapping of the replication and transcription promoters of human parainfluenza virus type 3. Virology 269:201-211. [DOI] [PubMed] [Google Scholar]
- 26.Iverson, L. E., and J. K. Rose. 1981. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell 23:477-484. [DOI] [PubMed] [Google Scholar]
- 27.Jin, H., D. Clarke, H. Z. Zhou, X. Cheng, K. Coelingh, M. Bryant, and S. Li. 1998. Recombinant human respiratory syncytial virus (RSV) from cloned cDNA and construction of subgroup A and B chimeric RSV. Virology 251:206-214. [DOI] [PubMed] [Google Scholar]
- 28.Jin, H., X. Cheng, H. Z. Zhou, S. Li, and A. Seddiqui. 2000. Respiratory syncytial virus that lacks open reading frame 2 of the M2 gene (M2-2) has altered growth characteristics and is attenuated in rodents. J. Virol. 74:74-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karron, R. A., D. A. Buonagurio, A. F. Georgiu, S. S. Whitehead, J. E. Adamus, M. L. Clements-Mann, D. O. Harris, V. B. Randolph, S. A. Udem, B. R. Murphy, and M. S. Sidhu. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA 94:13961-13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo, L., H. Grosfeld, J. Cristina, M. G. Hill, and P. L. Collins. 1996. Effect of mutations in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J. Virol. 70:6892-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 32.Li, T., and A. K. Pattnaik. 1999. Overlapping signals for transcription and replication at the 3" terminus of the vesicular stomatitis virus genome. J. Virol. 73:444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez, P. J., J. Guillerez, R. Sousa, and M. Dreyfus. 1997. The low processivity of T7 RNA polymerase over the initially transcribed sequence can limit productive initiation in vivo. J. Mol. Biol. 269:41-51. [DOI] [PubMed] [Google Scholar]
- 34.Marriott, A. C., J. M. Smith, and A. J. Easton. 2001. Fidelity of leader and trailer sequence usage by the respiratory syncytial virus and avian pneumovirus replication complexes. J. Virol. 75:6265-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mink, M. A., D. S. Stec, and P. L. Collins. 1991. Nucleotide sequence of the 3" leader and 5" trailer regions of human respiratory syncytial virus genomic RNA. Virology 185:615-624. [DOI] [PubMed] [Google Scholar]
- 36.Peeples, M. E., and P. L. Collins. 2000. Mutations in the 5" trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step. J. Virol. 74:146-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Randhawa, J. S., A. C. Marriott, C. R. Pringle, and A. J. Easton. 1997. Rescue of synthetic minireplicons establishes the absence of the NS1 and NS2 genes from avian pneumovirus. J. Virol. 71:9849-9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samal, S. K., and P. L. Collins. 1996. RNA replication by a respiratory syncytial virus RNA analog does not obey the rule of six and retains a nonviral trinucleotide extension at the leader end. J. Virol. 70:5075-5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smallwood, S., and S. A. Moyer. 1993. Promoter analysis of the vesicular stomatitis virus RNA polymerase. Virology 192:254-263. [DOI] [PubMed] [Google Scholar]
- 40.Stillman, E. A., and M. A. Whitt. 1997. Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J. Virol. 71:2127-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tolley, K. P., Marriott, A. C., A. Simpson, D. A. Plows, D. A. Matthews, S. J. Longhurst, J. E. Evans, J. L. Johnson, P. A. Cane, V. B. Randolph, A. J. Easton, and C. R. Pringle. 1996. Identification of mutations contributing to the reduced virulence of a modified strain of respiratory syncytial virus. Vaccine 14:1637-1646. [DOI] [PubMed] [Google Scholar]
- 42.Vidal, S., and D. Kolakofsky. 1989. Modified model for the switch from Sendai virus transcription to replication. J. Virol. 63:1951-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner, R. R., and J. K. Rose. 1996. Rhabdoviridae: the viruses and their replication, p. 1121-1135. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Raven Press, New York, N.Y.
- 44.Whelan, S. P., and G. W. Wertz. 1999. Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: identification of distinct sequences essential for transcription but not replication. J. Virol. 73:297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]