Abstract
We have used the duck hepatitis B virus (DHBV) model to study the interference with infection by a myristoylated peptide representing an N-terminal pre-S subdomain of the large viral envelope protein. Although lacking the essential part of the carboxypeptidase D (formerly called gp180) receptor binding site, the peptide binds hepatocytes and subsequently blocks DHBV infection. Since its activity requires an amino acid sequence involved in host discrimination between DHBV and the related heron HBV (T. Ishikawa and D. Ganem, Proc. Natl. Acad. Sci. USA 92:6259-6263, 1995), we suggest that it is related to the postulated host-discriminating cofactor of infection.
The characterization of viral and cellular proteins that take part in attachment and entry of viruses into susceptible host cells is a key objective in molecular virology (11). While a substantial body of knowledge about these early infection events has been achieved for a number of viruses (22), a comparable understanding of hepadnavirus entry into hepatocytes is still lacking. This particularly holds true for human hepatitis B virus (HBV), the causative agent of B-type hepatitis, since in vitro infections are still restricted to the use of primary human hepatocytes (15). Therefore, many studies have been carried out with primary duck hepatocytes (PDHs), which are susceptible to the related duck HBV (DHBV) (18). The recent identification of duck carboxypeptidase D (duCPD) (formerly called gp180) as one receptor component for DHBV (2, 8, 9, 17, 19), the mapping of a pre-S sequence that determines host specificity between DHBV and the related heron HBV (HHBV) (6, 16; T. Ishikawa, personal communication), and the characterization of duCPD interaction with an extended pre-S domain in the large viral surface protein (5, 17, 20, 21) have directed particular attention to this animal model.
The envelope of DHBV encloses the large (L) and the small (S) viral surface proteins. Both proteins are encoded in a single open reading frame such that the L protein contains the complete S part, which serves as a membrane anchor, and an additional N-terminal extension of 161 amino acids called pre-S (14). The L protein of DHBV is myristoylated at glycine 2 of its amino terminus. Replacement of this glycine by an alanine abolishes myristoylation and leads to a significant loss of virus infectivity (10). Since myristoylation was also shown to be important for HBV infection (4), this modification plays an important role in hepadnaviral infection in general.
DHBV infection of duck hepatocytes can be inhibited by an excess of noninfectious subviral particles (SVPs), which correspond to empty virus shells containing only the membrane-embedded L and S proteins (7). Similar effects were observed with Escherichia coli-derived DHBV pre-S polypeptides (Dpre-S). Infection inhibition occurs through direct interference with the duCPD receptor binding (19) when a Dpre-S subdomain (amino acids 30 to 115) is used. However, an approximately 30-fold-higher specific inhibitory activity of SVPs compared to recombinant Dpre-S suggests that an additional inhibitory activity within the L or S protein might be present in SVPs. In this investigation, we confirmed this hypothesis and identified a second pre-S domain, which is probably involved in an additional DHBV entry step distinct from the interaction with duCPD.
Given the major role of N-terminal myristoylation of the pre-S domain in the infection process, we investigated the potential of myristoylated pre-S peptides to interfere with DHBV infection of PDHs. Furthermore, we focused our interest on only the 41 amino-terminal amino acids, whose important role in the infection process has been clearly demonstrated, as they bear a domain determining host discrimination between DHBV and HHBV (6; Ishikawa, personal communication).
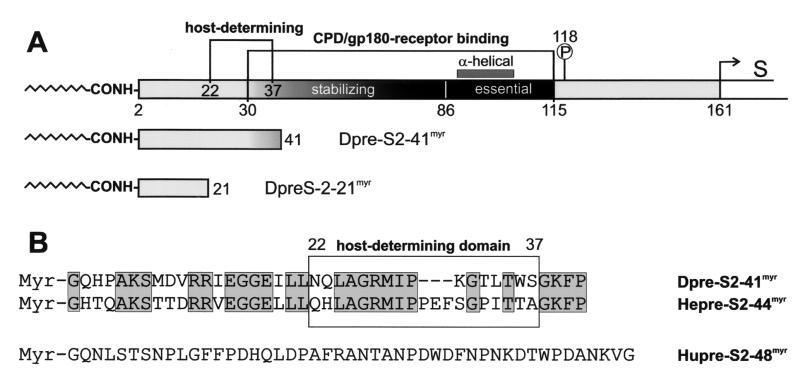
Figure 1 gives an overview of our present knowledge of the organization of the DHBV pre-S domain (Fig. 1A) and depicts the amino acid sequences of the peptides used in infection competition experiments (Fig. 1B). The internal pre-S subdomain, consisting of amino acids 30 to 115, constitutes the duCPD receptor binding site. As shown by surface plasmon resonance and two-dimensional nuclear magnetic resonance analyses, this domain includes an essential, partially α-helical element (amino acids 86 to 115) and a most-randomly structured stabilizing part (amino acids 30 to 85) required for high-affinity complex formation (21). The host-determining pre-S sequence (amino acids 22 to 37) (6; Ishikawa, personal communication) partially overlaps with the stabilizing part of the duCPD binding site but definitely requires amino acids that are N-terminally located. The two synthetic myristoylated DHBV pre-S peptides used in this study (Dpre-S2-41myr and Dpre-S2-21myr) are drawn schematically below the complete pre-S sequence in Fig. 1A. The primary sequences of Dpre-S2-41myr and the two corresponding HHBV and HBV pre-S peptides are shown in Fig. 1B. Both avian peptides exhibit 57% sequence identity, while human pre-S2-48myr (Hupre-S2-48myr) has no sequence similarity.
FIG. 1.
Schematic illustration of the pre-S domain of the DHBV L protein and the two DHBV pre-S-derived myristoylated peptides Dpre-S2-41myr and Dpre-S2-21myr (A) and the primary sequences of myristoylated Dpre-S2-41myr, Hepre-S2-44myr, and Hupre-S2-48myr (B) used in DHBV infection competition experiments. (A) The duCPD (gp180) receptor binding site (amino acids 30 to 115) within the pre-S domain (amino acids 1 to 161) of the DHBV L protein consists of an essential, partially α-helical subdomain (amino acids 86 to 115) and a most-randomly structured element (amino acids 30 to 85) which is needed for the formation of a high-affinity complex. Directly adjacent to the receptor binding site is Ser-118, which becomes phosphorylated (13). Partially overlapping with the stabilizing element but clearly extending it at its N terminus is the sequence that recovers infectivity of HHBV for PDHs (amino acids 22 to 37). (B) Sequence alignment of the two inhibitory peptides Dpre-S2-41myr and the homologous Hepre-S2-44myr. Sequence identity is 57%. Note that within the host-determining region (boxed sequence between amino acids 22 and 37), HHBV pre-S contains the additional insertion 31-PEF-33. The HBV pre-S-derived peptide Hupre-S2-48myr displays no significant homology to its avian counterparts.
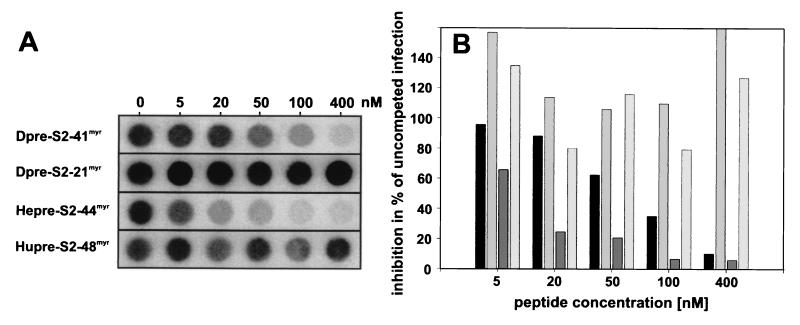
Results of in vitro DHBV infection competition experiments with Dpre-S2-41myr, Dpre-S2-21myr, Hepre-S2-44myr, and Hupre-S2-48myr are summarized in Fig. 2. The Dpre-S2-41myr peptide interferes with DHBV infection of PDHs in a concentration-dependent manner, with 50% inhibition at about 70 nM (Fig. 2A, row 1). In contrast, the shorter peptide Dpre-S2-21myr had no inhibitory effect (Fig. 2A, row 2) even at concentrations of up to 400 nM. The HBV derivative Hupre-S2-48myr did not affect the viral infection level (Fig. 2A, row 4). Notably, when applied at higher concentrations, Dpre-S2-21myr and Hupre-S2-48myr reproducibly enhanced infection slightly, for unknown reasons (Fig. 2 and data not shown). Thus, inhibition of DHBV infection by Dpre-S2-41myr is species specific with respect to mammalian and avian hepadnaviruses and requires amino acids 22 to 41 of the L protein. Since this sequence has been shown to be important in host discrimination between HHBV and DHBV (6; Ishikawa, personal communication), we also tested the respective HHBV peptide for interference with DHBV infection. As shown in Fig. 2A, row 3, Hepre-S2-44myr competed DHBV infection even better than the genuine DHBV homologue (about 10 nM Hepre-S2-44myr was needed for 50% inhibition). Quantification of these results (Fig. 2B) indicates that Dpre-S2-41myr and Hepre-S2-44myr reduced the DHBV infectivity level to less than 10% of that of the control.
FIG. 2.
Competition of DHBV infection by the myristoylated pre-S-derived peptides Dpre-S2-41myr, Dpre-S2-21myr, Hepre-S2-44myr, and Hupre-S2-48myr. PDHs (8 × 105) were infected at a multiplicity of infection of ≈100 in the presence of 5, 20, 50, 100, and 400 nM concentrations of the respective pre-S peptides. Fourteen hours later, peptides and virus were removed. (A) At 7 days postinfection total intracellular DNA was extracted and analyzed by dot blotting as described previously (12). (B) Viral DNA was quantified with a PhosphorImager, and results are presented as percentages of the value for an uncompeted control infection In each group of four bars, results for Dpre-S2-41myr, Dpre-S2-21myr, Hepre-S2-44myr, and Hupre-S2-48myr are shown from left to right, respectively. Three independent experiments were performed, and results from one representative experiment are shown.
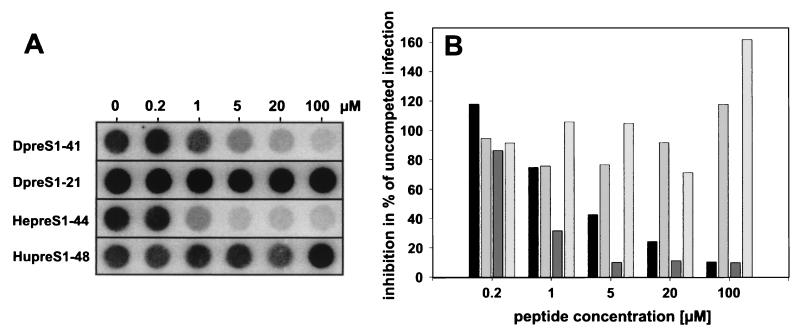
To investigate whether myristoylation is absolutely essential for inhibition, we performed the same set of experiments with the corresponding nonmyristoylated peptides Dpre-S1-41, Dpre-S1-21, Hupre-S1-48, and Hepre-S1-44. Under comparable conditions (concentrations of between 5 and 400 nM), no significant interference with infection was observed (data not shown). However, at higher concentrations, Dpre-S1-41 was able to notably prevent infection. At 100 μM, infection was blocked almost completely (Fig. 3). Like for its myristoylated homologue, activity depends on the presence of amino acids 22 to 41 (Dpre-S1-21 was inactive even at 100 μM) and turns out to be species specific with respect to the HBV analogue Hupre-S1-48. Again, we observed about a sevenfold-higher specific activity for Hepre-S1-44 than for Dpre-S1-41. Taken together, the results indicate that specific inhibition of DHBV infection by pre-S-derived peptides is not absolutely dependent on modification with myristic acid but is drastically improved, by a factor of ≈100.
FIG. 3.
Competition of DHBV infection by the nonmyristoylated pre-S-derived peptides Dpre-S1-41, Dpre-S1-21, Hepre-S1-44, and Hupre-S1-48. PDHs (8 × 105) were infected overnight at a multiplicity of infection of ≈100 in the presence of 0.2, 1, 5, 20, and 100 μM concentrations of the respective pre-S peptides. Fourteen hours later, peptides and virus were removed. (A) At 7 days postinfection total intracellular DNA was extracted and analyzed by dot blot analysis as described previously (12). (B) Viral DNA was quantified with a PhosphorImager, and results are presented as percentages of the value for an uncompeted control infection. In each group of four bars, results for Dpre-S1-41, Dpre-S1-21, Hepre-S1-44, and Hupre-S1-48 are shown from left to right, respectively. Two independent experiments were performed.
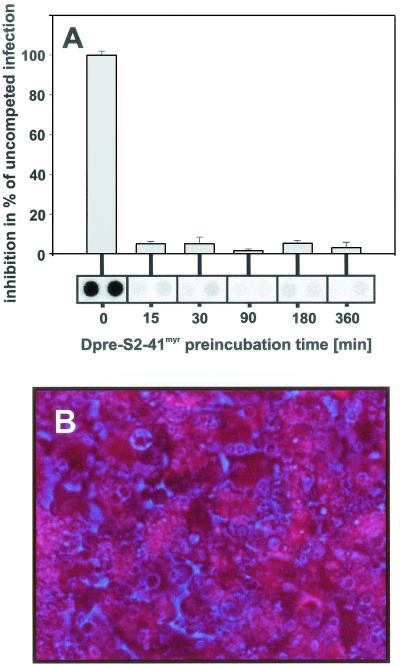
We next asked whether Dpre-S2-41myr can target hepatocytes in the absence of infectious virions. To this end, we preincubated PDHs with Dpre-S2-41myr for different time periods at a concentration of 800 nM, removed the surplus inhibitor, subsequently added infectious DHBV particles, and left them for an additional 12 h. As shown in Fig. 4A,pretreatment of PDHs for only 15 min with Dpre-S2-41myr resulted in a strong reduction of intracellular viral DNA at 7 days postinfection. This indicates that the peptide efficiently targets PDHs and renders them nonsusceptible to subsequent infection. As Dpre-S2-41myr has no significant effect on DHBV replication in already-infected PDHs (data not shown), we assume that it interferes with an early infection event. In addition to this functional indication for hepatocyte binding, we could directly observe that rhodamine-labeled Dpre-S1-41 associates with PDHs after a 3-h incubation period (Fig. 4B). The same was true for rhodamine-labeled myristoylated Dpre-S2-41myr at much lower concentrations (not shown). Taken together, our data support the assumption that the peptide at least acts on hepatocytes and thereby interferes with an early infection event.
FIG. 4.
(A) Competition of DHBV infection following preincubation of PDHs with Dpre-S2-41myr. PDHs were incubated with Dpre-S2-48myr (800 nM) for 15, 30, 90, 180, and 360 min prior to infection with DHBV (14 h; multiplicity of infection, ≈75). Seven days later intracellular viral DNA was extracted, analyzed by dot blotting. and quantified with a PhosphorImager. Values are given as percentages of the value for an uncompeted infection. Error bars indicate standard deviations. (B) Association of Dpre-S1-41 with PDHs. NHS-rhodamine (Pierce) was chemically coupled to Dpre-S1-41 according to the manufacturer's protocol. After purification on a PD10 gel filtration column (Amersham-Pharmacia), the labeled peptide was incubated with PDHs for 3 h. After removal of surplus free peptide, cell-associated Dpre-S1-41 was visualized by fluorescence microscopy (red). The phase-contrast image is shown in blue.
With respect to the finding that the sequence required for restoration of HHBV infectivity in PDHs and the sequence essential for duCDP-independent infection competition map to the same location within Dpre-S, we assume that it is the host-discriminating entry step which is blocked by the pre-S peptide inhibitors described here. The ability to directly target hepatocytes supports this assumption. The fact that both myristoylated and nonmyristoylated inhibitory peptides are able to associate with hepatocytes suggests that they affect an identical molecule and that myristic acid only favors this interaction, possibly by increasing the local peptide concentration at membranes. Interestingly, a recent reinvestigation of the infectivity of a DHBV Lmyr-negative mutant showed that when applied at high titers for longer time periods, the mutated virus exhibited residual infectivity (1). With the hypothesis of an efficient interaction of duck and heron pre-S with a cellular host-determining factor, the poor infectivity of HHBV for PDHs can be explained by the lack of a subsequent and specific processing of Hepre-S, which could, for example, consist of a proteolytic cleavage.
We cannot, however, exclude the possibility that these inhibitory peptides are also able to interact with the viral particles. Human immunodeficiency virus envelope protein-derived peptides have similarly been shown to be capable of efficiently inhibiting the entry of this virus into its host cell. These peptides, by interacting specifically with a defined domain of the envelope proteins, interfere with the formation of a transient fusion-active state (3). If this also holds true for hepadnaviruses, we would predict that a peptide corresponding to the target sequence of Dpre-S1-41 within the L protein will also exhibit inhibition activity. Identification of such an element together with the cofactor addressed by the N-terminal peptide would allow further elucidation of the hepadnaviral fusion mechanism.
Acknowledgments
This work was performed in the laboratory of Heinz Schaller, ZMBH Heidelberg, whom we thank for continuous excellent support. We are grateful to Margrit Ellis for the synthesis and purification of synthetic peptides, Hans Ulrich Schairer for generous financial help, Bärbel Glass and Uta Klöcker for the preparation of PDHs, Stephanie Held for technical assistance, and Elizabeth Grgacic and Jacques Le Seyec for comments and critical reading of the manuscript.
This work was made possible by an H. and C. Schaller Stiftung fellowship to S.U. and by a grant from the Deutsche Forschungsgemeinschaft (UR 72/4-1) to S.U.
REFERENCES
- 1.Borst, E. M. 1997. Untersuchungen zur Interaktion des Enten Hepatitis B Virus L-Proteins mit primären Entenhepatozyten. Ph.D. thesis. University of Ulm, Ulm, Germany.
- 2.Breiner, K. M., S. Urban, and H. Schaller. 1998. Carboxypeptidase D (gp180), a Golgi-resident protein, functions in the attachment and entry of avian hepatitis B viruses. J. Virol. 72:8098-8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 4.Gripon, P., J. Le Seyec, S. Rumin, and C. Guguen-Guillouzo. 1995. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology 213:292-299. [DOI] [PubMed] [Google Scholar]
- 5.Ishikawa, T., K. Kuroki, R. Lenhoff, J. Summers, and D. Ganem. 1994. Analysis of the binding of a host cell surface glycoprotein to the pre-S protein of duck hepatitis B virus. Virology 202:1061-1064. [DOI] [PubMed] [Google Scholar]
- 6.Ishikawa, T., and D. Ganem. 1995. The pre-S domain of the large viral envelope protein determines host range in avian hepatitis B viruses. Proc. Natl. Acad. Sci. USA 92:6259-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klingmüller, U., and H. Schaller. 1993. Hepadnavirus infection requires interaction between the viral pre-S domain and a specific hepatocellular receptor. J. Virol. 67:7414-7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuroki, K., R. Cheung, P. L. Marion, and D. Ganem. 1994. A cell surface protein that binds avian hepatitis B virus particles. J. Virol. 68:2091-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuroki, K., F. Eng, T. Ishikawa, C. Turck, F. Harada, and D. Ganem. 1995. gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J. Biol. Chem. 270:15022-15028. [DOI] [PubMed] [Google Scholar]
- 10.Macrae, D. R., V. Bruss, and D. Ganem. 1991. Myristylation of a duck hepatitis B virus envelope protein is essential for infectivity but not for virus assembly. Virology 181:359-363. [DOI] [PubMed] [Google Scholar]
- 11.Norkin, L. C. 1995. Virus receptors: implications for pathogenesis and the design of antiviral agents. Clin. Microbiol. Rev. 8:293-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigg, R. J., and H. Schaller. 1992. Duck hepatitis B virus infection of hepatocytes is not dependent on low pH. J. Virol. 66:2829-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothman, K., M. Schnoelzer, G. Radziwill, E. Hildt, K. Moelling, and H. Schaller. 1998. Host cell-virus cross talk: phosphorylation of the duck hepatitis B virus large envelope protein mediates intracellular signaling. J. Virol. 72:10138-10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlicht, H. J., C. Kuhn, B. Guhr, R. J. Mattaliano, and H. Schaller. 1987. Biochemical and immunological characterization of the duck hepatitis B virus envelope proteins. J. Virol. 61:2280-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sprengel, R., E. F. Kaleta, and H. Will. 1988. Isolation and characterization of a hepatitis B virus endemic in herons. J. Virol. 62:3832-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong, S., J. Li, and J. R. Wands. 1995. Interaction between duck hepatitis B virus and a 170-kilodalton cellular protein is mediated through a neutralizing epitope of the pre-S region and occurs during viral infection. J. Virol. 69:7106-7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuttleman, J. S., J. C. Pugh, and J. W. Summers. 1986. In vitro experimental infection of primary duck hepatocyte cultures with duck hepatitis B virus. J. Virol. 58:17-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urban, S., K. M. Breiner, F. Fehler, U. Klingmüller, and H. Schaller. 1998. Avian hepatitis B virus infection is initiated by the interaction of a distinct pre-S subdomain with the cellular receptor gp180. J. Virol. 72:8089-8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urban, S., C. Kruse, and G. Multhaup. 1999. A soluble form of the avian hepatitis B virus receptor: biochemical characterization and functional analysis of the receptor ligand complex. J. Biol. Chem. 274:5707-5715. [DOI] [PubMed] [Google Scholar]
- 21.Urban, S., C. Schwarz, U. C. Marx, H. Zentgraf, H. Schaller, and G. Multhaup. 2000. Receptor recognition by a hepatitis B virus reveals a novel mode of high affinity virus-receptor interaction. EMBO J. 19:1217-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wimmer, E. (ed.). 1994. Cellular receptors for animal viruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.