Abstract
Vesicular stomatitis virus glycoprotein G-pseudotyped mouse retroviral vectors have been used as mutagens for a large-scale insertional mutagenesis screen in the zebra fish. To reproducibly generate high-titer virus stocks, we devised a method for rapidly selecting cell lines that can yield high-titer viruses and isolated a producer cell line that yields virus at a high titer on zebra fish embryos. Virus produced from this line, designated GT virus, is nontoxic following injection of zebra fish blastulae and efficiently infects embryonic cells that give rise to the future germ line. Using GT virus preparations we generated roughly 500,000 germ line-transmissible proviral insertions in a population of 25,000 founder fish in about 2 months. The GT virus contains a gene trap, and trap events can be detected in the offspring of almost every founder fish. We discuss potential applications of this highly efficient method for generating germ line-transmissible insertions in a vertebrate
A major task for biologists in the postgenome era is to determine the cellular and organismal functions of an estimated 30,000 human genes (24, 49). A powerful approach for functional determination is forward genetic screens in model organisms. By altering genes at random with mutagens and screening for abnormalities caused by the mutations, genetic screens have yielded large collections of mutants with defects in development, physiology, and/or behavior, and many of these are models of human diseases (6, 11, 15, 16, 20, 34). Identifying the genes mutated in forward genetic screens can be time-consuming, even in invertebrate models such as fly and worm, because most screens use chemical mutagens to induce mutations. Chemical mutagens are efficient, but they usually cause point mutations, the identification of which requires laborious genetic mapping and positional cloning. An alternative and complementary method is insertional mutagenesis (2, 44, 52), in which DNA elements inserted randomly into the genome can disrupt gene function. If an insertion leads to abnormalities, the mutated gene can be readily identified using the insertion as a marker or tag. Efficient insertional mutagenesis using P elements in flies has greatly facilitated the cloning of many mutant genes (44).
Because vertebrates have many more genes than invertebrates and have more complex physiological and behavioral repertoires, genetic screens will undoubtedly play an important role in understanding gene function in vertebrates despite the demanding nature of genetic screens in vertebrate species. Already, two large chemical mutagenesis screens have identified more than 1,000 loci that are essential for zebra fish development (11, 15). Several genetic screens of the mouse are also under way, and several hundred mutants, most of them dominant, have been identified (21, 32, 33, 42). Because most screens in vertebrates employ ethylnitrosourea as the mutagen, cloning of mutated genes usually takes months or even years, even in the postgenome era. In zebra fish for example, the average time to clone a gene responsible for a mutant phenotype induced by ethylnitrosourea is currently about 1.5 years, although it is expected to decrease to about 9 months following completion of the zebra fish genome project. Clearly, efficient methods of insertional mutagenesis in vertebrates could contribute significantly to the task of assigning functions to genes.
Our laboratory developed a method of insertional mutagenesis for zebra fish that involves injecting vesicular stomatitis virus glycoprotein G-pseudotyped retroviruses into blastulae at 3 to 4 h postfertilization (14, 25). Retroviral insertions in the primordial germ cells of injected fish (F0) are transmitted to the offspring, and some cause mutant phenotypes (1, 2, 4, 14, 23). About 1 in 85 proviral insertions inactivates a gene essential for embryonic development. We have reported the identification of several such genes using this technology (1, 2, 4, 14, 23). Given the low frequency of identifiable developmental mutants per insert, we devised a protocol that uses fish with multiple insertions to perform an efficient large-scale screen. This protocol was made possible by the isolation of two cell lines, A7 and F5, which produce vesicular stomatitis virus G-pseudotyped SFG-nLacZ viruses at high titers on zebra fish cells (2, 13). However, in the course of a large experiment that required making many stocks of virus, it proved difficult to reproducibly make high-titer, nontoxic virus preparations from these lines. In an attempt to improve the technology, we devised a method to select a cell line that would consistently yield virus preparations that have high titers on zebra fish cells and that are nontoxic following injection into embryos. The viral vector we used, designated GT, contains a gene trap construct that was introduced in the hope of further facilitating the identification and the expression patterns of mutated genes. Here we report the construction of the GT viral vector and the method we devised for quickly screening a large number of candidate virus producer clones. Using GT virus obtained from the best cell clone we identified, we generated 25,000 injected fish (F0) in 2 months that together carry roughly 500,000 germ line-transmissible insertions. Furthermore, gene trap events can be detected in the progeny of almost all of the F0 fish tested.
MATERIALS AND METHODS
Construction of GT retroviral plasmids.
The plasmid pCLnZ was made by replacing the 3.0-kb BglII/BamHI fragment of pCLMFG-lacZ (31) with the 3.5-kb BglII/BamHI fragment of pSFG-nLacZ (13). GT (gene trap) plasmids were made by inserting a gene trap module(s) into various locations in pCLnZ. The gene trap module consists of a branch point sequence, a splicing acceptor, a splicing enhancer from avian sarcoma and leukosis virus (47), a DNA fragment that encodes the FLAG epitope (DYKDDDDK) in all three reading frames, and a splicing donor. The modules were constructed as follows. A fragment encoding the FLAG epitope in three reading frames followed by a splicing donor (SD) consensus sequence was made by annealing the following two oligonucleotides: pAATTGACTACAAGGACGAGAATACAAGGACGAGACTATAAAGATGAATGAGTGAGGGGTGAGTATCG and pAATTCGATACTCACCCCTCACTCATTCATCTTTATAGTCTCGTCCTTGTATTCTCGTCCTTGTAGTC. The FLAG fragment was inserted into the EcoRI site of pAd3"ENH (8) (a gift from Tom Maniatis of Harvard University) and resulted in pAd3"ENH-FLAG-SD. A synthetic branch point sequence was made by annealing two oligonucleotides, pTCGAGTATACTAACAAGT and pTCGAACTTGTTAGTATAC, and was inserted into the XhoI site of pAd3"ENH-FLAG-SD to make pBPSAd3"ENH-FLAG-SD. The module was excised by digesting with XhoI and BglII and placed into pCLnZ as indicated by blunt-end ligation to generate the GT plasmid.
Production of virus by transient transfection.
To evaluate the suitability of the viral constructs for producing high-titer virus, retroviral preparations were made by transiently cotransfecting pCMV-G (7) and the viral constructs into 293gp/bsr, a packaging cell line from Inder Verma of the Salk Institute. The cells were maintained in Dulbecco's modified Eagle medium-10% normal calf serum-penicillin-streptomycin. For transfection, packaging cells were seeded in a 10-cm-diameter plate at 50% confluence. The next day, cells were transfected using Lipofectamine reagents for 8 h according to the manufacturer's instructions (Life Technologies, Rockville, Md.). For each plate, 4 μg of pCMV-G, 10 μg of viral plasmid, and 100 μl of Lipofectamine reagent were used. Culture medium was changed the next day, and the conditioned medium at day 2 posttransfection was assayed to determine the viral titer on 3T3 cells and zebra fish PAC2 cells (zebra fish embryonic fibroblast cells) as previously described (2, 9).
Selection of candidate virus producer clones.
To select virus producer clones, 293gp/bsr cells were transduced with viruses made by transient transfection. Two days posttransduction, cells were incubated with the fluorescent β-galactosidase substrate fluorescein di-β-d-galactopyranoside (Molecular Probes, Eugene, Oreg.) and cells were sorted by fluorescence-activated cell sorting according to fluorescence intensity. Cells above the 95th percentile were collected individually in 96-well plates. When confluent, close to a third of the cells in each well in the master plates were transferred into a well in three new 96-well plates, and the residual cells were kept in the master plates. The next day, cells in each 96-well plate were transfected by incubating for 8 h with 50 μl of Opti-MEM containing 40 ng of pCMV-G and 3 μl of Lipofectamine reagents. The medium was replaced daily with fresh culture medium.
Determination of viral titers using QPCR.
We used real-time quantitative PCR (QPCR) analysis to compare the number of proviral insertions in transduced cells, similar to that used for selecting multi-insertion F1 fish described previously (2). The PCR assay uses an ABI PRISM 7700 sequence detection system (PE Applied Biosystems, Foster City, Calif.) to quantify PCR product accumulation through a dual-labeled virus-specific fluorogenic probe (i.e., TaqMan Probe) (18, 19). Sequences of PCR primers and the TaqMan probe as well as the detailed method have been reported previously (2). Briefly, PAC2 cells were seeded at 15% confluence in 96-well plates the day after the viral producer candidates were transfected with pCMV-G. The next day, 20 μl of conditioned medium was added into each well of the 96-well plates containing 80 μl of L-15-20% fetal calf serum-penicillin-streptomycin-10-μg/ml Polybrene. After 3 h of incubation, the medium was replaced with normal PAC2 medium. Two days postinfection, PAC2 cells were lysed in 25 μl of GNT buffer (10 mM Tris, 50 mM KCl, 5 mM EDTA, 0.01% gelatin, 0.45% NP-40, 0.45% Tween 20, 200 μg of proteinase K per ml, pH 8.5) (27). Two microliters of lysate from each well was taken for real-time QPCR analysis in which the amount of viral DNA and the zebra fish rag2 gene in each sample were both determined. The relative amount of viral and rag2 DNA, described as ΔCt, was calculated for each sample. The best clones, i.e., the clones that yielded the largest ΔCt, were chosen based on the average of the three data points.
Detection of gene trap events by reverse transcription (RT)-PCR.
To detect gene trap events in the F1 fish, founder fish were out-crossed to uninfected wild-type fish and total RNA was isolated from a pool of 25 embryos from each clutch using Trizol reagents (Life Technologies). cDNAs were synthesized and then used as a template in 5" rapid amplification of cDNA end reactions by using a commercial kit from Life Technologies following the manufacturer's instruction. The nested gene-specific primer was TCGTCGTCCTTGTAATCCCT.
RESULTS
Construction of a novel gene trap virus.
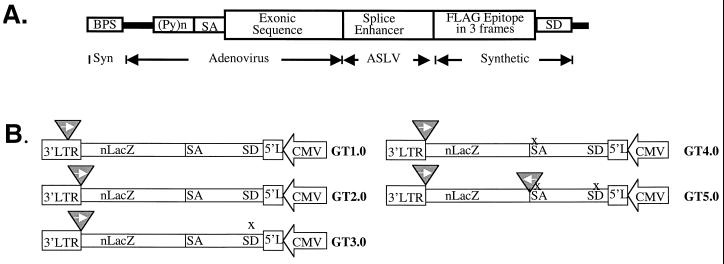
A gene trap virus should facilitate the determination of the identity and the expression patterns of mutated genes. It seemed possible that the gene trap function might also increase the mutagenicity of the virus because it can convert some of the nonconsequential insertions in large introns into mutagenic ones as a consequence of aberrant splicing. Previous unpublished work from our laboratory indicated that conventional gene trap viral constructs containing a terminal exon that encodes a reporter such as β-galactosidase, green fluorescent protein, or neo usually yield virus preparations with titers 10- to 100-fold lower than those of the SFG-nLacZ virus. We therefore sought alternative forms of gene trap constructs by minimally modifying pSFG-nLacZ. Instead of engineering a terminal exon, we inserted an internal exon and other essential RNA splicing elements into the viral vector. To try to prevent exon skipping, we added a splicing enhancer to the exon (47) and kept the exon length under 200 bp (5, 10, 45). A reporter is a nonessential, but convenient, feature for our purpose. Therefore, we incorporated a sequence that encodes the FLAG epitope in all three reading frames. Thus, we generated a 270-bp gene trap fragment that harbors an exon of 172 bp (Fig. 1A). Insertion of such an exon in a transcript downstream of the initiation codon will cause a frameshift mutation.
FIG. 1.
Schematic diagram of the gene trap module and retroviral vectors constructed. (A) Structure of the gene trap module and origins of its components. Exonic sequences are in wider boxes and intronic sequences are in narrower boxes or thick lines. Abbreviations: ASLV, avian sarcoma and leukosis virus; BPS, branch point sequence; (Py)n, polypyrimidine tract; SA, splice acceptor; syn, synthetic. (B) Schematic drawings of GT1.0 to GT5.0. Triangles point to the positions at which the gene trap module is inserted. Arrows in the triangles indicate orientation of the trap exon, and crosses (x) indicate elimination. Abbreviations: SA, splice acceptor; CMV, CMV promoter; LTR, long terminal repeat; L, partial long terminal repeat.
We placed the gene trap fragment in pCLnZ, which was made by replacing the entire 5" long terminal repeat and [psi]+ region of pSFG-nLacZ with that of pCLMFG-lacZ (30). Because the 5" U3 is replaced by a CMV promoter in pCLMFG-lacZ, we can produce reasonably high-titer viral preparations by transient transfection and quickly evaluate the usefulness of different viral constructs. We constructed several gene trap viruses by placing the trap fragment at several locations and determined their titers by transient transfection, testing for ones that can yield high titers. Figure 1 summarizes the schematic structures and the titers of those constructs. Since at best 50% of gene-disrupting insertions can be detected by a unidirectional gene trap, we explored the possibility of a bidirectional gene trap virus. In order to do so, we modified GT2.0 by eliminating the viral splice donor (GT3.0) and the splice acceptor (GT4.0) and placed another gene trap module between [psi]+ and nLacZ (pGT5.0). Comparison of the titers of the viral preparations made by transient transfection suggested that pGT2.0 is as good as pCLnZ, whereas the other four plasmids were not (Table 1); thus, the bidirectional gene trap approach was abandoned. We henceforth refer to GT2.0 as GT. We went on to select producer cell lines for GT in our attempt to routinely obtain viral preparations with higher titers and minimal toxicities.
TABLE 1.
Viral titers of conditioned medium from 293bsr cells 2 days posttransfection
| Viral vector | Titer (cfu/ml) on 3T3 cells |
|---|---|
| pSFG-nLacZ | 1.4 × 107 |
| pGT1.0 | 8 × 105 |
| pGT2.0 | 1.5 × 107 |
| pGT3.0 | 1.8 × 106 |
| pGT4.0 | 2 × 105 |
| pGT5.0 | 2 × 104 |
High-throughput screening of viral producer clones.
Unpublished data from our laboratory have indicated that there is often discordance between virus titers on mouse 3T3 cells and zebra fish PAC2 cells, and thus it is important to select virus producer clones based on titers on PAC2 cells or, ideally, on fish embryos. It seemed possible that the inconsistency of virus production from A7 and F5 might be due, at least in part, to the use of calcium phosphate-mediated transfection, which is known to vary from experiment to experiment. Therefore, we chose to use lipofection as the means of transfection.
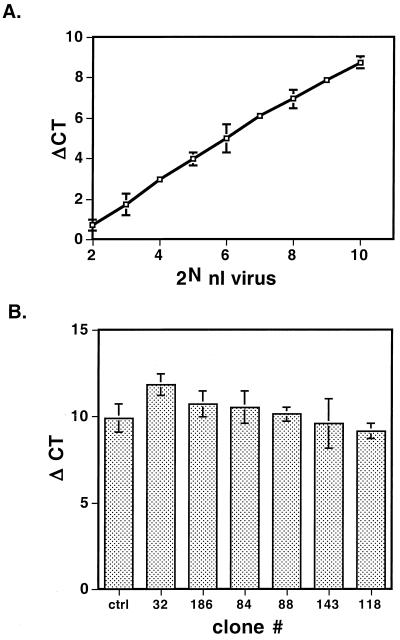
Next, we developed a high-throughput method for screening viral producer lines. Previous producer clones were selected from a very limited number of candidate clones because a labor-intensive colony-counting method was used to select them. It is conceivable that better producer cell lines can be obtained by screening more candidates. Since the relative infectivity, rather than absolute infectivity, of virus from the candidates is important at the screening stage, we decided to select candidate virus producer lines by comparing the relative viral DNA content in infected PAC2 cells using QPCR. To test the feasibility of titrating virus by QPCR, we compared the relative amount of proviral DNA in PAC2 cells after incubating them with 100 μl of 2-fold serial dilutions of a clone F5 produced SFG-nLacZ virus (SFG-nLacZ/F5) preparation in a 96-well plate. We found that the method has a wide linear range (Fig. 2A). Since up to 96 samples can be analyzed at a time and each assay takes only about 2 h, a large number of candidates can be analyzed in a day. We screened 230 candidates, among which 6 were chosen for further characterization because they all yielded titers better than a very good SFG-nLacZ/F5 preparation (10) (Fig. 2B). Clone 186 was selected because of its consistency in producing virus at high titers and its ease of maintenance.
FIG. 2.
High-throughput selection of viral producer clones using QPCR. (A) Correlation between ΔCt and viral load. PAC2 cells were infected with serial dilutions of a concentrated SFG-nLacZ/F5 preparation for 3 h. Transduced cells were lysed 2 days later, and the amounts of proviral DNA and rag2 DNA in the lysates were assayed by QPCR. (B) More than 230 candidate producer lines were screened, and 6 were found to be comparable to a very good SFG-nLacZ/F5 preparation. Error bars are standard deviations.
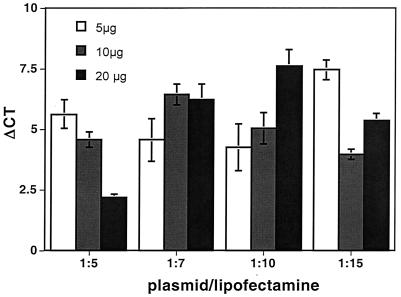
We then optimized the transfection conditions for clone 186 to further improve viral titer and, more importantly, decrease the toxicity of viral preparations exhibited on injected embryos. Of 12 conditions tested, we found that for each 15-cm-diameter plate of cells, the best condition used 7.5 μg of pCMV-G and 50 μl of Lipofectamine (Fig. 3). Using the optimized conditions, we made numerous high-titer stocks of GT virus over 3 months and had no failures.
FIG. 3.
Optimization of transfection conditions. Cells were transfected in a 15-cm-diameter plate with 5, 10, or 20 μg of pCMV-G and 5, 7, 10, or 15 times this amount (vol/wt) of Lipofectamine. Day 2 media were concentrated, and titers were determined in PAC2 cells.
Rapid generation of high-quality F0 founder fish.
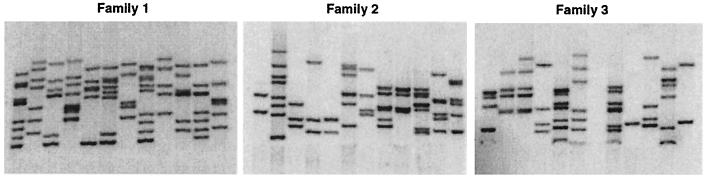
Using GT virus made from clone 186 (GT/186) under the optimized conditions, we generated more than 25,000 F0 fish in less than 3 months. Although the titer of typical GT/186 virus stocks determined on NIH 3T3 cells is only about two- to threefold as high as that of SFG-nLacZ/F5 preparations, its advantages are severalfold. First, GT/186 virus preparations are less toxic. Consequently, a higher fraction of injected embryos survive. Second, GT/186 preparations contain higher concentrations of infectious viral particles. As a result, we only had to inject embryos for one round rather than two rounds, as we had done for SFG-nLacZ/F5 virus preparations, to deliver sufficient virus to generate good founders (see below). This probably also partially explains the increased survival rate of injected embryos. It also doubled the number of embryos one person can inject in a day. On average, a trained injector can inject 1,000 blastulae in about 3 h using GT/186, compared to 450 using SPG-nLacZ/F5. Overall, the survival rate of injected embryos 1 day after injection was 57% using GT/186 versus 37% using SFG-nLacZ/F5. One injector can make about 200 adult founders per day using GT/186 versus about 50 using SFG-nLacZ/F5, a fourfold increase in the rate of F0 production (Table 2). Third, GT/186-injected fish contain higher densities of proviral insertions. We used quantitative Southern blotting and/or QPCR to determine the ratios of viral DNA and endogenous rag2 DNA. The number of proviral insertions per cell was computed by comparing the ratios between fish to be tested and fish with known copy numbers of proviral insertions (2). The average copy number of provirus in injected fish was determined at 2 days postinjection and is called the embryo assay value (EAV). The EAV of GT/186-injected fish is 14.3, compared to 3.6 for SFG-nLacZ/F5-injected fish. Previously, we have reported that injected fish with more than five proviral copies per cell would reliably produce gametes with multiple insertions (10). Such fish are called “good founders.” More than 96% of GT/186-derived fish are good, compared to only 25% using SFG-nLacZ/F5. Overall, compared to SFG-nLacZ/F5, using GT/186 viral preparations increased production of good F0 fish 16-fold (Table 2). Finally, GT/186-injected fish transmit more unique insertions at higher densities. GT/186-derived fish often transmitted gametes with eight or more insertions (Fig. 4) , which is rare in SFG-nLacz/F5-derived founders and has never been seen in SFG-nlacZ/A7-derived founders. The average number of unique inserts an F0 fish transmits has not been systematically analyzed in SFG-nLacZ/F5-injected or GT/186-injected founder fish. After analyzing a large number of SFG-nLacZ/A7-derrived founders, however, our laboratory has found that these fish on average transmit 12 unique germ line-transmissible insertions (21). Based on the F1 insertion profiles of representative F0 fish injected with SFG-nLacZ/A7, SFG-nLacZ/F5, or GT/186, we estimate a 50% increase in insertion density from SFG-nLacZ/A7 to SFG-nLacZ/F5 and an additional 50% increase from SFG-nLacZ/F5 to GT/186. Thus, on average there are 25 or more unique germ line insertions per GT/186-injected F0 fish (Table 2).
TABLE 2.
Comparison of the three virus producer cell lines used for insertional mutagenesis
| Virus/producer cell line | % Good founders | % Survival of injected embryos at 24 h postinjection | No. of founders/day/person | EAV avg | % with EAV ≥ 5 | No. of insertions/founder | Person-days needed to make 106 GLa insertions |
|---|---|---|---|---|---|---|---|
| SFG/A7 | 15 | 45 | 60 | NAc | NA | 12 | >1,390 |
| SFG/F5 | 75 | 37 | 50 | 3.6 | 25 | 18b | 1,110 |
| GT/186 | 100 | 57 | 200 | 14.3 | 96 | 27b | 185 |
GL, germ line.
Estimate.
NA, not assayed.
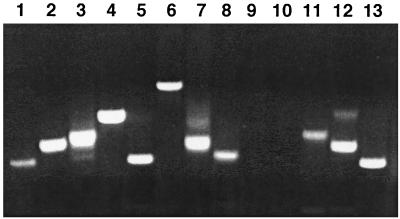
FIG. 4.
Profiles of germ line insertions of three representative founders. Genomic DNA from 5-day-old embryos from matings between a male founder and wild-type females were digested with BglII and analyzed by Southern analysis using a probe derived from the [psi]+ region. Each lane contains DNA from an individual larva, and each band indicates a germ line insertion.
Gene trapping in GT/186-derived F1 fish.
A detectable gene trap is essential for RT-PCR-based identification of mutated genes. Although gene trap events were detected in injected fish (data not shown), it was important to determine if such events could be detected in F1 fish after passage through the germ line since inactivation of gene expression could occur (22, 36). We thus performed 5" rapid amplification of cDNA ends (RACE) to see if gene trap events could be detected in the F1 fish. Our data indicate that, in most cases, at least one gene trap event can be detected in a pool of F1 offspring from each GT/186-injected F0 fish (Fig. 5). We purified 20 randomly selected 5" RACE fragments and determined their sequences. Of the 20, 12 were fusions between the trap exon and genes with known function, four were fusions between the trap exon and zebra fish expressed sequence tags with unknown function, and four were fusions between the trap exon and sequences with no homology in the GenBank. All fusions occurred at the splicing acceptor site of the trap exon as expected (W. Chen and S. Burgess, unpublished results). Due to high background problems, however, FLAG antibodies could not be used to determine the expression patterns of trapped genes.
FIG. 5.
5" RACE analysis of gene trap events in F1 embryos. Total RNA was isolated from 25 2-day-old embryos from a mating between a male founder and two wild-type females. 5" RACE was used to detect the presence of the GT exon in the RNA samples. Each lane contains 5" RACE product from one RNA sample.
DISCUSSION
We have described a high-throughput method for selecting high-titer retrovirus producer cell lines. We used this method to obtain a cell line that consistently yields viral preparations with high titers on zebra fish embryos. These preparations allowed rapid production of founder fish harboring more than 500,000 germ line-transmissible insertions in zebra fish. This number of insertional events has not been reported previously in a vertebrate animal.
Our method for selecting producer lines is independent of viral gene expression, allowing one to select viral producer cell clones for virus without a selectable reporter gene. Many retroviral vectors, including those for gene therapy, contain a selectable marker gene just for selecting a producer cell line. Inclusion of the extra sequence may not only lower the titer and decrease the expression of the therapeutic gene by promoter suppression (12) but also increase the immunogenicity of transduced cells (39). Thus, viral vectors without selectable markers are desirable for gene therapy (35). The method described here should be useful for selecting producer cell lines of such vectors. There are a few other marker gene-independent methods that measure viral RNA content in conditioned medium by various means (30, 35, 38). Our method is more reliable than those, because viral RNA content can be a poor predictor of infection (46). Our method is very similar to one developed independently by Towers et al. (48).
Although insertional mutagenesis has been extensively used in large-scale forward genetic screens in bacteria, fungi, and plants, it has rarely been used for forward screens in vertebrates. Many insertional mouse mutants are found in transgenic lines by serendipity (50). These mutants have sometimes helped in the identification of some long-sought mutant genes (see reference 28, for example). However, large-scale forward genetic screens using insertional mutagenesis have not been done in mice because they require such large resources. In zebra fish, however, because of the smaller body size and superior fecundity, our laboratory was able to conduct an insertional mutagenesis screen for genes essential for early development; this screen is expected to yield about 500 to 600 mutants, most of them induced by the GT provirus (2). This number of mutants should correspond to roughly 15 to 20% of the genes that can be mutated to cause embryonic lethality in zebra fish. The current screen employs a conventional three-generation F3 scheme, which is space- and time-consuming. As a result, only a small fraction of the insertions transmitted by the F0 fish are homozygosed and screened. However, gynogenesis-based screens in zebra fish have been successfully carried out and require less space and fewer resources (3, 37). If combined with insertional mutagenesis, this approach could allow a moderate-sized laboratory to screen hundreds of thousands of insertions per year and to identify both embryonic and later-appearing mutant phenotypes.
The mutant frequency per F3 family in our insertional mutagenesis screen is about seven- to eightfold lower than that in chemical mutagenesis screens. We had hoped that the gene trap feature of GT virus could increase the mutagenicity because it could convert some of the otherwise inconsequential insertions in large introns into mutagenic ones. Unfortunately, our data suggest that GT virus did not increase the mutagenicity significantly if at all relative to SFG-nLacZ virus (unpublished results). It is possible that the gene trap module is not as effective as hoped. In fact, we have observed skipping of the GT exon in many trap events (Chen and Burgess, unpublished). Alternatively, very large introns may be rare in the zebra fish genome. Nevertheless, even in the absence of the gene trap, the ease of cloning mutated genes makes insertional mutagenesis a highly attractive approach in zebra fish.
Besides their utility as mutagens, retroviral insertions have the potential to provide other useful forms of genetic manipulation. One obvious potential application is transgenesis. Currently, transgenic fish are made by injecting DNA into single-cell-stage embryos. In most cases, only 5 to 10% of injected fish transmit the injected DNA through the germ line, a frequency far below that which was seen by injecting GT/186 virus into early blastulae. Another potential application of this methodology is to express transgenes in a subset of cells in a specific tissue or organ if the virus is applied to a specific region of the animal at later stages of development, as has been routinely done with chicken embryos (29). The insertional mutagenesis method may also be used for gain-of-function screens (17, 40, 41), using retroviral vectors that contain transcriptional regulatory sequences such as the tetracycline-responsive promoter (TetOR) similar to the enhanced retroviral mutagen (ERM) vectors (26), and the Saccharomyces cerevisiae UASgal4. The inclusion of the TetOR and/or the UASgal4 promoters in the virus, in conjunction with transgenic lines that express their cognate transcriptional activators in tissue-specific patterns, may allow one to screen for gain-of-function phenotypes in specific organs or tissues. Such vectors can also be used in loss-of-function screens in the absence of the expression of exogenous transcriptional activators, allowing one to determine the function a virus-tagged gene in multiple ways.
Insertional mutagenesis can be used for reverse genetics as well as forward genetics. For example, hundreds of thousands of Tc1 insertion lines in Caenorhabditis elegans have been created, and their genomic DNA has been arrayed for PCR screens to find mutations in specific genes (52). In Drosophila melanogaster, the flanking genomic sequences of more than 1,000 P-element insertions have been determined (43). In the mouse, more than one-third of all the genes in the genome have been disrupted in ES cells by proviral insertion, and the sequences of those genes have been determined (51). These insertion libraries are valuable resources for determining gene function and will become increasingly so as fully annotated genome sequences become available. Since zebra fish sperm can be frozen and in vitro fertilization is a routine laboratory practice, the rapid production of 0.5 × 106 to 1 × 106 germ line-transmissible insertions carried by about 2.5 × 104 adult founder fish makes it possible to create a PCR-screenable sperm library similar to that in C. elegans, mouse ES cell clones, or P-element-tagged fly lines.
REFERENCES
- 1.Allende, M. L., A. Amsterdam, T. Becker, K. Kawakami, N. Gaiano, and N. Hopkins. 1996. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev. 10:3141-3155. [DOI] [PubMed] [Google Scholar]
- 2.Amsterdam, A., S. Burgess, G. Golling, W. Chen, Z. Sun, K. Townsend, S. Farrington, M. Haldi, and N. Hopkins. 1999. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 13:2713-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beattie, C. E., D. W. Raible, P. D. Henion, and J. S. Eisen. 1999. Early pressure screens. Methods Cell Biol. 60:71-86. [PubMed] [Google Scholar]
- 4.Becker, T. S., S. M. Burgess, A. H. Amsterdam, M. L. Allende, and N. Hopkins. 1998. not really finished is crucial for development of the zebrafish outer retina and encodes a transcription factor highly homologous to human nuclear respiratory factor-1 and avian initiation binding repressor. Development 125:4369-4378. [DOI] [PubMed] [Google Scholar]
- 5.Berget, S. M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270:2411-2414. [DOI] [PubMed] [Google Scholar]
- 6.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns, J. C., T. Friedmann, W. Driever, M. Burrascano, and J. K. Yee. 1993. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. USA 90:8033-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiara, M. D., and R. Reed. 1995. A two-step mechanism for 5" and 3" splice-site pairing. Nature 375:510-513. [DOI] [PubMed] [Google Scholar]
- 9.Culp, P. 1994. Ph.D. thesis. Massachusetts Institute of Technology, Cambridge, Mass.
- 10.Dietz, H. C., D. Valle, C. A. Francomano, R. J. Kendzior, Jr., R. E. Pyeritz, and G. R. Cutting. 1993. The skipping of constitutive exons in vivo induced by nonsense mutations. Science 259:680-683. [DOI] [PubMed] [Google Scholar]
- 11.Driever, W., L. Solnica-Krezel, A. F. Schier, S. C. Neuhauss, J. Malicki, D. L. Stemple, D. Y. Stainier, F. Zwartkruis, S. Abdelilah, Z. Rangini, J. Belak, and C. Boggs. 1996. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123:37-46. [DOI] [PubMed] [Google Scholar]
- 12.Emerman, M., and H. M. Temin. 1984. Genes with promoters in retrovirus vectors can be independently suppressed by an epigenetic mechanism. Cell 39:449-467. [PubMed] [Google Scholar]
- 13.Gaiano, N., M. Allende, A. Amsterdam, K. Kawakami, and N. Hopkins. 1996. Highly efficient germ-line transmission of proviral insertions in zebrafish. Proc. Natl. Acad. Sci. USA 93:7777-7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaiano, N., A. Amsterdam, K. Kawakami, M. Allende, T. Becker, and N. Hopkins. 1996. Insertional mutagenesis and rapid cloning of essential genes in zebrafish. Nature 383:829-832. [DOI] [PubMed] [Google Scholar]
- 15.Haffter, P., M. Granato, M. Brand, M. C. Mullins, M. Hammerschmidt, D. A. Kane, J. Odenthal, F. J. van Eeden, Y. J. Jiang, C. P. Heisenberg, et al. 1996. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123:1-36. [DOI] [PubMed] [Google Scholar]
- 16.Hartwell, L. H., J. Culotti, and B. Reid. 1970. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. USA 66:352-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hay, B. A., R. Maile, and G. M. Rubin. 1997. P element insertion-dependent gene activation in the Drosophila eye. Proc. Natl. Acad. Sci. USA 94:5195-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 19.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5"-3" exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horvitz, H. R., and J. E. Sulston. 1980. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96:435-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hrabe de Angelis, M. H., H. Flaswinkel, H. Fuchs, B. Rathkolb, D. Soewarto, S. Marschall, S. Heffner, W. Pargent, K. Wuensch, M. Jung, et al. 2000. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat. Genet. 25:444-447. [DOI] [PubMed] [Google Scholar]
- 22.Jahner, D., and R. Jaenisch. 1985. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature 315:594-597. [DOI] [PubMed] [Google Scholar]
- 23.Kawakami, K., A. Amsterdam, N. Shimoda, T. Becker, J. Mugg, A. Shima, and N. Hopkins. 2000. Proviral insertions in the zebrafish hagoromo gene, encoding an F-box/WD40-repeat protein, cause stripe pattern anomalies. Curr. Biol. 10:463-466. [DOI] [PubMed] [Google Scholar]
- 24.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 25.Lin, S., N. Gaiano, P. Culp, J. C. Burns, T. Friedmann, J. K. Yee, and N. Hopkins. 1994. Integration and germ-line transmission of a pseudotyped retroviral vector in zebrafish. Science 265:666-669. [DOI] [PubMed] [Google Scholar]
- 26.Liu, D., X. Yang, D. Yang, and Z. Songyang. 2000. Genetic screens in mammalian cells by enhanced retroviral mutagens. Oncogene 19:5964-5972. [DOI] [PubMed] [Google Scholar]
- 27.Malumbres, M., R. Mangues, N. Ferrer, S. Lu, and A. Pellicer. 1997. Isolation of high molecular weight DNA for reliable genotyping of transgenic mice. BioTechniques 22:1114-1119. [DOI] [PubMed] [Google Scholar]
- 28.Miao, G. G., R. J. Smeyne, G. D'Arcangelo, N. G. Copeland, N. A. Jenkins, J. I. Morgan, and T. Curran. 1994. Isolation of an allele of reeler by insertional mutagenesis. Proc. Natl. Acad. Sci. USA 91:11050-11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan, B. A., J. C. Izpisua-Belmonte, D. Duboule, and C. J. Tabin. 1992. Targeted misexpression of Hox-4.6 in the avian limb bud causes apparent homeotic transformations. Nature 358:236-239. [DOI] [PubMed] [Google Scholar]
- 30.Murdoch, B., D. S. Pereira, X. Wu, J. E. Dick, and J. Ellis. 1997. A rapid screening procedure for the identification of high-titer retrovirus packaging clones. Gene Ther. 4:744-749. [DOI] [PubMed] [Google Scholar]
- 31.Naviaux, R. K., E. Costanzi, M. Haas, and I. M. Verma. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70:5701-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nolan, P. M., D. Kapfhamer, and M. Bucan. 1997. Random mutagenesis screen for dominant behavioral mutations in mice. Methods 13:379-395. [DOI] [PubMed] [Google Scholar]
- 33.Nolan, P. M., J. Peters, L. Vizor, M. Strivens, R. Washbourne, T. Hough, C. Wells, P. Glenister, C. Thornton, J. Martin, et al. 2000. Implementation of a large-scale ENU mutagenesis program: towards increasing the mouse mutant resource. Mamm. Genome 11:500-506. [DOI] [PubMed] [Google Scholar]
- 34.Nusslein-Volhard, C., and E. Wieschaus. 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287:795-801. [DOI] [PubMed] [Google Scholar]
- 35.Onodera, M., A. Yachie, D. M. Nelson, H. Welchlin, R. A. Morgan, and R. M. Blaese. 1997. A simple and reliable method for screening retroviral producer clones without selectable markers. Hum. Gene Ther. 8:1189-1194. [DOI] [PubMed] [Google Scholar]
- 36.Pannell, D., and J. Ellis. 2001. Silencing of gene expression: implications for design of retrovirus vectors. Rev. Med. Virol. 11:205-217. [DOI] [PubMed] [Google Scholar]
- 37.Pelegri, F., and S. Schulte-Merker. 1999. A gynogenesis-based screen for maternal-effect genes in the zebrafish, Danio rerio. Methods Cell Biol. 60:1-20. [DOI] [PubMed] [Google Scholar]
- 38.Persons, D. A., M. G. Mehaffey, M. Kaleko, A. W. Nienhuis, and E. F. Vanin. 1998. An improved method for generating retroviral producer clones for vectors lacking a selectable marker gene. Blood Cells Mol. Dis. 24:167-182. [DOI] [PubMed] [Google Scholar]
- 39.Riddell, S. R., M. Elliott, D. A. Lewinsohn, M. J. Gilbert, L. Wilson, S. A. Manley, S. D. Lupton, R. W. Overell, T. C. Reynolds, L. Corey, and P. D. Greenberg. 1996. T-cell mediated rejection of gene-modified HIV-specific cytotoxic T lymphocytes in HIV-infected patients. Nat. Med. 2:216-223. [DOI] [PubMed] [Google Scholar]
- 40.Rorth, P. 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93:12418-12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rorth, P., K. Szabo, A. Bailey, T. Laverty, J. Rehm, G. M. Rubin, K. Weigmann, M. Milan, V. Benes, W. Ansorge, and S. M. Cohen. 1998. Systematic gain-of-function genetics in Drosophila. Development 125:1049-1057. [DOI] [PubMed] [Google Scholar]
- 42.Soewarto, D., C. Fella, A. Teubner, B. Rathkolb, W. Pargent, S. Heffner, S. Marschall, E. Wolf, R. Balling, and M. Hrabe de Angelis. 2000. The large-scale Munich ENU-mouse-mutagenesis screen. Mamm. Genome 11:507-510. [DOI] [PubMed] [Google Scholar]
- 43.Spradling, A. C., D. Stern, A. Beaton, E. J. Rhem, T. Laverty, N. Mozden, S. Misra, and G. M. Rubin. 1999. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153:135-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spradling, A. C., D. M. Stern, I. Kiss, J. Roote, T. Laverty, and G. M. Rubin. 1995. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc. Natl. Acad. Sci. USA 92:10824-10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sterner, D. A., T. Carlo, and S. M. Berget. 1996. Architectural limits on split genes. Proc. Natl. Acad. Sci. USA 93:15081-15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tafuro, S., L. Zentilin, A. Falaschi, and M. Giacca. 1996. Rapid retrovirus titration using competitive polymerase chain reaction. Gene Ther. 3:679-684. [PubMed] [Google Scholar]
- 47.Tanaka, K., A. Watakabe, and Y. Shimura. 1994. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol. Cell. Biol. 14:1347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Towers, G. J., D. Stockholm, V. Labrousse-Najburg, F. Carlier, O. Danos, and J. C. Pages. 1999. One step screening of retroviral producer clones by real time quantitative PCR. J. Gene Med. 1:352-359. [DOI] [PubMed] [Google Scholar]
- 49.Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans, R. A. Holt, et al. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 50.Woychik, R. P., and K. Alagramam. 1998. Insertional mutagenesis in transgenic mice generated by the pronuclear microinjection procedure. Int. J. Dev. Biol. 42:1009-1017. [PubMed] [Google Scholar]
- 51.Zambrowicz, B. P., G. A. Friedrich, E. C. Buxton, S. L. Lilleberg, C. Person, and A. T. Sands. 1998. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature 392:608-611. [DOI] [PubMed] [Google Scholar]
- 52.Zwaal, R. R., A. Broeks, J. van Meurs, J. T. Groenen, and R. H. Plasterk. 1993. Target-selected gene inactivation in Caenorhabditis elegans by using a frozen transposon insertion mutant bank. Proc. Natl. Acad. Sci. USA 90:7431-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]