Abstract
The phosphoprotein (P) of human respiratory syncytial virus (RSV) is an essential component of the viral RNA polymerase, along with the large polymerase (L), nucleocapsid (N), and M2-1 proteins. By screening a randomly mutagenized P gene cDNA library, two independent mutations, one with a substitution of glycine at position 172 by serine (G172S) and the other with a substitution of glutamic acid at position 176 by glycine (E176G), were identified to result in the loss of N-P interaction at 37°C in the yeast two-hybrid assay. Both P mutants exhibited greatly reduced activity in supporting the replication and transcription of an RSV minigenome replicon at 37 and 39°C. The G172S and E176G mutations were introduced individually into the RSV A2 (rA2) antigenomic cDNA, and recombinant viruses, rA2-P172 and rA2-P176, were obtained. Both viruses replicate as well as wild-type A2 virus in both Vero and HEp-2 cells at 33°C, but each mutant virus exhibited temperature-sensitive replication in both cell lines. rA2-P176 is more temperature sensitive than rA2-P172. Coimmunoprecipitation of the N protein with each P mutant from virus-infected cells demonstrates that N-P interaction is impaired at 37°C. In addition, the levels of replication of rA2-P172 and rA2-P176 in the lungs of mice and cotton rats were reduced. As is the case with the in vitro assays, rA2-P176 is more restricted in replication in the lower respiratory tract of mice and cotton rats than rA2-P172. During in vitro passage at 37°C, the E176G mutation in rA2-P176 was rapidly changed from glycine to predominantly aspartic acid; mutations to cysteine or serine were also detected. All of the revertants lost the temperature-sensitive phenotype. To analyze the importance of the amino acids in the region from positions 161 to 180 for the P protein function, additional mutations were introduced and their functions were analyzed in vitro. A double mutant containing both G172S and E176G changes in the P gene, substitution of the three charged residues at positions 174 to 176 by alanine, and a deletion of residues from positions 161 to 180 completely abolished the P protein function in the minigenome assay. Thus, the amino acids at positions 172 and 176 and the adjacent charged residues play critical roles in the function of the P protein.
Human respiratory syncytial virus (RSV) is the prototype virus in the Pneumovirus genus of the Paramyxoviridae family. The virus genome consists of a single 15-kb negative-stranded RNA that encodes 11 proteins (for a recent review, see reference 9). The nucleocapsid protein (N), phosphoprotein (P), and large polymerase protein (L) constitute the minimal components for viral RNA replication and transcription in vitro (16, 43). The N protein associates with the genomic RNA to form the nucleocapsid, which serves as the template for RNA synthesis. The L protein is a multifunctional protein that contains RNA-dependent RNA polymerase catalytic motifs and is also probably responsible for capping and polyadenylation of viral mRNAs. However, the L protein alone is not sufficient for the polymerase function; the P protein is also required. Transcription and replication of RSV RNA are also modulated by the M2-1, M2-2, NS1, and NS2 proteins that are unique to the pneumoviruses. M2-1 is a transcription antitermination factor required for processive RNA synthesis and transcription read-through at gene junctions (8, 17-19). M2-2 is involved in the switch between viral RNA transcription and replication (4, 23). NS1 and NS2 have been shown to inhibit minigenome synthesis in vitro (2).
The RSV P protein contains 241 residues, a length which is much shorter than the P proteins of other paramyxoviruses (9, 25, 29). Although the RSV P protein shares no sequence homology with the P proteins of other paramyxoviruses, it shares similar structure and function in viral replication. Recently, it has been determined that the RSV P protein, like that of the Sendai virus (38, 39), forms homotetramers (1). The N, P, L, and M2-1 proteins copurify with nucleocapsids and are colocalized as inclusion bodies in RSV-infected cells (13, 14). Intracellular coexpression of N and P results in the formation of N-P complexes that can be coimmunoprecipitated (13, 14). The N-P interaction has been suggested to promote proper folding of N (5, 22) and specific encapsidation of RNA by N (33). By analogy with the other paramyxovirus P proteins, the P protein of RSV likely acts as a cofactor that serves both to stabilize the L protein and to place the polymerase complex on the N:RNA template (21). Thus, the interactions between the P protein and other RSV proteins play critical roles in virus replication.
The C-terminal six amino acids of the P protein have been identified as the major N protein-binding domain (14, 35). However, several lines of evidence have indicated that other regions in the P protein are also involved in the formation of the N-P complex. The RSV P protein containing a deletion of 10 amino acids from the N-terminal end was coprecipitated with N but failed to induce coaggregation of N (14). The last 54 amino acids in the C terminus are insufficient for N-P complex formation as assayed in a yeast two-hybrid system (35). Studies of the P protein of bovine RSV, a homologue of human RSV, also showed that the C-terminal end and an internal region between residues 161 to 180 are required for N-P complex formation by coimmunoprecipitation analysis (30, 28). Moreover, deletion studies of the bovine RSV P protein shows that only residues 41 to 80 could be removed without significantly impairing bovine RSV minigenome replication (27).
To identify mutations in the P protein that may confer a temperature-sensitive (ts) phenotype on recombinant RSV, we screened a randomly mutagenized P gene cDNA library using a yeast two-hybrid assay for mutations that could confer a temperature-sensitive N-P interaction. Two independent P mutations, one at residue 172 and the other at 176, were identified that resulted in a temperature-sensitive interaction with N. Both mutants were assayed in a minigenome replicon system and were further analyzed by introducing them into recombinant RSV by using reverse genetics (8, 24). The effect of these P mutations on virus replication in cell culture and in the respiratory tracts of mice and cotton rats was investigated.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Monolayer cultures of HEp-2 and Vero cells (obtained from the American Type Culture Collections [ATCC]) were maintained in minimal essential medium containing 5% fetal bovine serum (FBS). Recombinant RSV A2 (rA2) was recovered from an antigenomic cDNA derived from RSV A2 strain, pRSVC4G (24), and grown in Vero cells. The modified vaccinia virus Ankara strain expressing bacteriophage T7 RNA polymerase, MVA-T7 (42), was provided by Bernard Moss and grown in CEK cells. Polyclonal anti-RSVA2 antibodies were obtained from Biogenesis (Sandown, N.H.). Monoclonal anti-RSV P antibodies 1P, 02/021P, and 76P were provided by Jose A. Melero.
Screening N-P protein interaction in the yeast two-hybrid system.
The interaction of the RSV N and P proteins was established by using the yeast two-hybrid system (Clontech). The two hybrid fusion plasmids were constructed as follows. The N open reading frame (ORF) of RSV was fused in frame with the GAL4 DNA-binding domain in the vector pAS2 through NcoI and EcoRI restriction sites. The P ORF was fused in frame with GAL4 activation domain in the pGAD GL vector through the BamHI and XhoI restriction sites. A silent AvrII site was introduced at codon 145 of the P ORF in pGAD GL-P to facilitate the construction of the P cDNA gene library. The mutagenesis was performed with a QuikChange mutagenesis kit (Stratagene) with a pair of primers, 5′-GAAAAATTAAGTGAAATCCTAGGAATGCTTCAC (the AvrII site is underlined) and its complementary sequence.
To generate a P gene cDNA library, random mutagenesis of the C-terminal 96 codons of the P gene was performed by PCR amplification with exonuclease-deficient PFU DNA polymerase (Stratagene) and primers 5′AvrII (5′-GATAATCCCTTTTCTAAACTATAC) and 3′Act2 (5′-CATTTAAAAAATTCTATAGATCAGAGG) with pGAD GL-P as the template. The 5′AvrII primer was designed to anneal to sequences ca. 150 bp upstream of the silent AvrII site in the P ORF, and the 3′Act2 primer anneals to sequences ca. 150 bp downstream of the XhoI site in the pGDL GL vector. The randomly introduced mutations in the PCR cDNA fragments were then transformed into the yeast Saccharomyces cerevisiae Y190 strain, together with pAS2-N and the gapped pGAD GL-P that had the C terminus of the P gene removed by digestion with AvrII and XhoI restriction enzymes. Intracellular recombination of the gapped vector with the random PCR fragments generated a P gene cDNA library. To identify the ts P mutants, the transformants were replica plated on SD-Leu-Trp synthetic media (Bio 101) as follows: two SD-Leu-Trp plates without any additives, two SD-Leu-Trp-His plates containing 50 mM 3 aminotriazole (3-AT), one SD-Leu-Trp-His plate containing 100 mM 3-AT, and one SD-Leu-Trp-His plate containing 150 mM 3-AT. The duplicate plates were incubated at 30 and 37°C, respectively, and the single plate was incubated at 30°C for 3 days. Colonies that showed no growth or highly reduced growth on the SD-Leu-Trp-His plates containing 50 mM 3-AT at 37°C but still showed good growth at least on the SD-Leu-Trp-His plates containing 100 mM 3-AT at 30°C were picked. A total of 64 possible ts mutants were identified in this fashion. The pGAD GL-P mutant plasmids were isolated from the yeast cells, amplified in Escherichia coli and retransformed into the Y190 strain along with pAS2-N to confirm the temperature-dependent N-P interaction on the replica plates as described above. The P gene mutants that exhibited ts interaction were sequenced to identify the mutations.
Functional analysis of P mutants by RSV minigenome replication assay.
The plasmids expressing RSV N, P, and L under the control of the T7 promoter were described previously (24). Mutations were introduced into the P gene by using the QuikChange site-directed mutagenesis kit or ExSite PCR-based site-directed mutagenesis kit (Stratagene). The following changes were made in the pP plasmid: G172S, E176G, G172S/E176G, 174-176A (R174A/E175A/E176A), ΔC6 (deletion of six amino acids from the C terminus) and Δ161-180 (deletion of residues from 161 to 180). The effect of the P mutations on RSV replication was assayed by using a RSV minigenome replicon, pRSV-CAT (37). For minigenome assays, HEp-2 cells in 12-well plates were infected with MVA-T7 at a multiplicity of infection (MOI) of 5 PFU/cell and then transfected with 0.2 μg of pRSV-CAT, together with 0.2 μg of pN, 0.1 μg of pL, and 0.2 μg of wild-type (wt) pP or mutant pP in triplicates. The transfected cells were incubated for 48 h at 33, 37, or 39°C. The amount of chloramphenicol acetyltransferase (CAT) protein expressed in the transfected cells was determined by an enzyme-linked immunosorbent assay assay (Roche Molecular Biochemicals). The protein expression levels of N and P in the transfected cells were determined by Western blotting. Total cellular polypeptides were electrophoresed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 15% acrylamide gels and transferred onto nylon membranes (Amersham Pharmacia Biotech). The blots were incubated with goat anti-RSV antibody (Biogenesis) and subsequently with a horseradish peroxidase-conjugated rabbit anti-goat immunoglobulin G (Dako). The membrane was incubated with the enhanced chemiluminescence substrate (Amersham Pharmacia Biotech). Protein bands were visualized after exposure to BioMAX ML film (Kodak).
Recovery of recombinant RSV.
The G172S and E176G mutations were introduced individually into the RSV antigenomic cDNA clone (24). E176G mutations contained two nucleotide changes from GAA to GAT. Mutations were first introduced into a RSV cDNA subclone, pRSV-(A/S), which contains the RSV A2 sequences from nucleotide 2128 (AvrII) to nucleotide 4485 (SacI), by using the QuikChange site-directed mutagenesis kit (Stratagene). The AvrII-SacI fragment carrying the introduced mutations was then shuttled into the full-length RSV A2 antigenomic cDNA clone, pRSVC4G (24). pRSVC4G contains the C-to-G change at the fourth position of the leader region in the antigenomic sense. Recombinant viruses were recovered from the transfected HEp-2 cells as described previously (24) and designated rA2-P172 and rA2-P176. The recovered viruses were plaque purified and amplified in Vero cells. The virus titer was determined by plaque assay on Vero cells, and the plaques were enumerated after immunostaining them with a polyclonal anti-RSV A2 serum (Biogenesis). The presence of each mutation in the rescued viruses was confirmed by sequence analysis of the P gene cDNA amplified by reverse transcription-PCR by using the viral genomic RNA as a template.
Replication of rA2-P172 and rA2-P176 in HEp-2 and Vero cells.
Plaque formation of each mutant was examined in HEp-2 and Vero cells at 33, 37, 38, and 39°C. Cell monolayers in six-well plates were infected with 10-fold serially diluted virus and incubated under an overlay that consisted of L15 medium containing 2% FBS and 1% methylcellulose in a submerged water bath for 6 days. The plaques were visualized and enumerated after immunostaining with a polyclonal antiserum against RSV A2 (Biogenesis). The plaques were photographed under an inverted microscope for plaque sizes comparisons.
The growth kinetics of rA2-P172, rA2-P176 in comparison with wt rA2 was studied in both HEp-2 and Vero cells. Cells in six-well plates were infected with wt rA2, rA2-P172, or rA2-P176 at an MOI of 1.0 or 0.01 PFU/cell. After 1 h of adsorption at room temperature, the infected cells were washed three times with phosphate-buffered saline, overlaid with 3 ml of Opti-MEM I (Life Technologies), and incubated at either 33 or 38°C. At 24-h intervals, 200 μl of culture supernatant was collected and stored at −80°C in the presence of SPG prior to virus titration (37). Each aliquot taken was replaced with an equal amount of fresh medium. The virus titer was determined by plaque assay on Vero cells at 33°C.
Coimmunoprecipitation of the N and P proteins.
Coimmunoprecipitation was performed to study the interaction between the N and P proteins. For transient protein expression, MVA-T7-infected HEp-2 cells in 12-well plates were cotransfected with 2 μg each of pN and pP plasmid by using LipofectACE (Life Technologies). To examine the N-P interaction in virus-infected cells, Vero cells were infected with rA2, rA2-P172, or rA2-P176 at MOIs of 1.0 PFU/cell. The transfected or recombinant RSV-infected cells were incubated at 33, 37, or 39°C for 12 h and then exposed to [35S]Cys and [35S]Met (100 μCi/ml) in Dulbecco modified Eagle medium (DMEM) deficient in cysteine and methionine for 4 h. The radiolabeled cell monolayers were lysed in the radioimmunoprecipitation assay buffer (10 mM Tris-HCl, pH 7.5; 150 mM NaCl; 5 mM EDTA; 1% Triton X-100; 1% sodium deoxycholate; 0.1% SDS). The polypeptides were immunoprecipitated with polyclonal goat anti-RSV A2 antibodies or with a mixture of monoclonal antibodies (1P, 021P, and 76P) against the P protein at 4°C for 12 h. The antibody-protein complex was precipitated by the addition of 30 μl of protein G-agarose beads (Life Technologies) at 4°C for 30 min and washed three times with radioimmunoprecipitation assay buffer containing 300 mM NaCl. The immunoprecipitated polypeptides were electrophoresed by SDS-15% PAGE and detected by autoradiography. The N and P proteins detected on the autoradiographs were quantified by densitometry with a Molecular Dynamics densitometer by using ImageQuant 5.0 for Windows NT (Molecular Dynamics).
Virus replication in mice and cotton rats.
Virus replication in vivo was determined in respiratory pathogen-free BALB/c mice and cotton rats (Sigmodon hispidus [Harlan]). Mice or cotton rats in groups of eight were inoculated intranasally under light methoxyflurane anesthesia with 0.1 ml of inoculum containing 106 PFU of virus per animal. At 4 days postinoculation, the animals were sacrificed by CO2 asphyxiation, and their lungs were harvested. The tissues were homogenized in Opti-MEM I (Life Technologies), and the virus titer was determined by plaque assay on Vero cells.
RESULTS
Identification of P mutations that weaken the N-P interaction in the yeast two-hybrid assay.
To identify mutations in the P protein that destabilize its interaction with the N protein, a yeast two-hybrid assay was used to screen a randomly mutagenized P cDNA library with mutations introduced in the C-terminal 96 codons of P for mutants that permitted interaction of P with N at the permissive temperature of 30°C but prevented interaction with N at the nonpermissive temperature of 37°C. The wt N and P proteins interacted with each other in yeast as indicated by the growth of the cotransformed yeast strain on the selective medium at 30°C as well as at 37°C. The transformants were screened for mutants that were capable of activating the yeast two-hybrid reporter gene at the permissive temperature of 30°C but not at 37°C. From approximately 1,300 original transformants, 64 possible ts mutants were identified. These putative ts clones were subjected to a second round of screening in yeast Y190. Two transformants were confirmed for their ts phenotypes. The pGAD GL-P plasmids from these yeast clones were sequenced. The mutations were identified as either Gly at residue 172 replaced by Ser (G172S) or Glu at residue 176 replaced by Gly (E176G) as shown in Fig. 1.
FIG. 1.
Sequences comparison of the P proteins from residues 161 to 180 among several pneumoviruses and the mutants analyzed. RSV-A2, human RSV subgroup A2; RSV-B1, human RSV subgroup B1; ORSV, ovine RSV; BRSV, bovine RSV; APV, avian pneumovirus; PVM, pneumonia virus of mice. Mutants analyzed in this study: G172S, Gly replaced by Ser at position 172; E176G, Glu replaced by Gly at position 176; G172S/E176G, double mutant containing both G172S and E176G; 174-176A, three consecutive charged residues from positions 174 to 176 replaced by Ala; Δ161-180, an internal deletion from residues 161 to 180; ΔC6, a C-terminal deletion from residues 236 to 241.
Immunoprecipitation analysis of N-P interaction in cells transiently expressing N and P.
Sequence alignment of the P proteins of several pneumoviruses revealed that residues 172 and 176 and the adjacent regions are highly conserved and contain several charged residues (Fig. 1). To examine the functional role of the charged residues, a mutant P protein expression plasmid was constructed in which each of the three charged residues, REE at positions 174 to 176, was replaced with alanine. In addition, a plasmid containing both G172S and E176G mutations in the P gene was constructed. Two deletion mutants either lacking the C-terminal six residues (ΔC6) or lacking residues from 161 to 180 (Δ161-180), which both have been shown to interfere with N-P interactions in RSV (14, 28), were made. To examine the effects of the P mutations on the N-P interaction, MVA-T7-infected HEp-2 cells were cotransfected with pN and pP mutant plasmids and incubated at 37 or 39°C. The 35S-labeled polypeptides were immunoprecipitated by anti-P monoclonal antibodies. As shown in Fig. 2, the N protein was only precipitated by anti-P antibodies in the presence of the P protein (lane 1 versus lane 3), demonstrating that the immunoprecipitation of the N protein occurred through its interaction with the P protein. Deletion of the six residues from the C terminus of the P protein (ΔC6) drastically reduced its interaction with the N protein; only a trace amount of N was detected (Fig. 2, lane 8). The N protein was coprecipitated by all of the other P mutants, G172S, E176G, G172S/E176G, 174-176A, and Δ161-180. The amount of Δ161-180 P protein detected on the gel was less than that of wt P, possibly because of the removal of the two potential 35S-labeled methionines in this region. Thus, coimmunoprecipitation of N and P in transiently expressed cells did not reveal any defect in N-P interaction for G172S and E176G mutations.
FIG. 2.
Immunoprecipitation analysis of N-P interaction in cells transiently expressing N and P. MVA-T7-infected HEp-2 cells were transfected with pN and different pP protein expression plasmids under the control of T7 promoters and incubated for 16 h at 37°C (upper panel) or 39°C (lower panel). The proteins were radiolabeled with [35S]Cys and [35S]Met (100 μCi/ml) in DMEM deficient in cysteine and methionine for 4 h, immunoprecipitated by anti-P monoclonal antibodies, separated on a 15% polyacrylamide gel, and exposed to Kodak BioMAX film. The positions of N and P are indicated on the right.
Effects of P mutations on the replication and transcription of the RSV-CAT minigenome.
The function of the P mutants was analyzed by a CAT minigenome replicon assay. The mutant P expression plasmids were transfected, together with pN, pL, and pRSV-CAT, into HEp-2 cells, and CAT expression was measured at 33, 37, or 39°C. As shown in Fig. 3, at 33°C, CAT protein expression was detected in cells expressing the mutant P proteins containing either G172S or E176G, although their activities were reduced by ca. 24 and 45%, respectively. At 37°C, the level of the CAT protein detected was reduced by ca. 80% for G172S and 90% for E176G. The reduction was even greater (>95%) at 39°C. These data indicated that mutations displayed a conditional ts phenotype consistent with the ts interaction phenotype observed in the yeast two-hybrid assays. No CAT expression was detected in cells expressing the P protein containing the combined G172S and E176G mutations, substitution of the three charged residues at positions 174 to 176 by alanine, a deletion of six amino acids from the C-terminal end (ΔC6), or an internal 20 amino acid deletion (Δ161-180).
FIG. 3.
Minigenome replication in cells expressing P mutants. MVA-T7-infected HEp-2 cells were transfected with pRSV-CAT, together with pN, pL, and pP mutant plasmids, and incubated at 33, 37, or 39°C. The levels of N and P protein expression were determined by Western blotting with polyclonal anti-RSV antibodies (insets). CAT reporter gene activities produced by different P mutants were determined by CAT-enzyme-linked immunosorbent assay and are expressed as the percentage of that of wt P at each temperature. The error bars show the standard deviations of three replicate experiments.
To eliminate the possibility that the reduction in reporter gene expression was caused by altered protein expression of these P mutants at higher temperatures, the levels of the P and N proteins produced in the transfected cells were examined by Western blotting. Except for Δ161-180 mutant, all of the other P mutants expressed a comparable level of protein (Fig. 3, insert). Therefore, the reduced ability of the other mutants to support RSV minigenome replication was a direct result of the introduced mutations rather than changes in their protein levels in the transfected cells.
Replication of rA2-P172 and rA2-P176 in cell cultures.
The G172S and E176G mutations were individually introduced into the full-length RSV antigenomic cDNA clone, and recombinant viruses were generated. Both rA2-P172 and rA2-P176 reached peak titers of ca. 2 × 107 PFU/ml in Vero cells at 33°C, a level comparable to that of wt rA2. The plaque formation efficiency of rA2-P172 and rA2-P176 at different temperatures was examined in Vero and HEp-2 cells and is summarized in Table 1 and Fig. 4. Both rA2-P172 and rA2-P176 formed smaller plaques than wt rA2 at 37°C and higher temperatures (Fig. 4). No plaques were visualized for rA2-P172 in Vero cells and HEp-2 cells at 39°C, although RSV-infected single or multiple cells stained by anti-RSV antibody were observed under the microscope (Fig. 4). Likewise, no visible plaques were observed for rA2-P176 in Vero cells or HEp-2 cells at 39°C and in HEp-2 cells at 38°C. rA2-P176 was more temperature sensitive than rA2-P172: the shutoff temperature for rA2-P172 was 39°C in HEp-2 and Vero cells whereas the shutoff temperatures for rA2-P176 were 38°C in HEp-2 cells and 39°C in Vero cells (Table 1).
TABLE 1.
Efficiency of plaque formation of RSV P mutants at various temperatures
| Virus | Mean virus titer (log10 PFU/ml) in Vero or HEp-2 cells ata:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 33°C
|
37°C
|
38°C
|
39°C
|
||||||
| Vero | HEp-2 | Vero | HEp-2 | Vero | HEp-2 | Vero | HEp-2 | ||
| rA2-wt | 6.70 | 6.61 | 6.73 | 6.52 | 6.69 | 6.48 | 6.63 | 6.46 | |
| rA2-P172 | 6.59 | 6.41 | 6.54∗b | 6.33∗ | 6.51∗ | 5.93∗ | -c | - | |
| rA2-P176 | 6.65 | 6.53 | 6.64∗ | 6.24∗ | 5.54∗ | - | - | - | |
Virus titers are the average of two independent experiments from two different virus stocks.
Small plaque size is indicated by an asterisk.
-, No visible plaques.
FIG. 4.
Plaque formation of rA2-P172 and rA2-P176 at different temperatures. Monolayers of Vero cells (upper panel) and HEp-2 cells (lower panel) were infected with wt rA2, rA2-P172, and rA2-P176; overlaid with L15 medium containing 1% methylcellulose and 2% FBS; and incubated at 33, 37, 38, and 39°C for 6 days. The plaques were visualized by immunostaining with polyclonal anti-RSV antibodies. Plaques were photographed on a Nikon inverted microscope. Arrows in the lower panels indicate RSV-infected HEp-2 cells at 38 and 39°C.
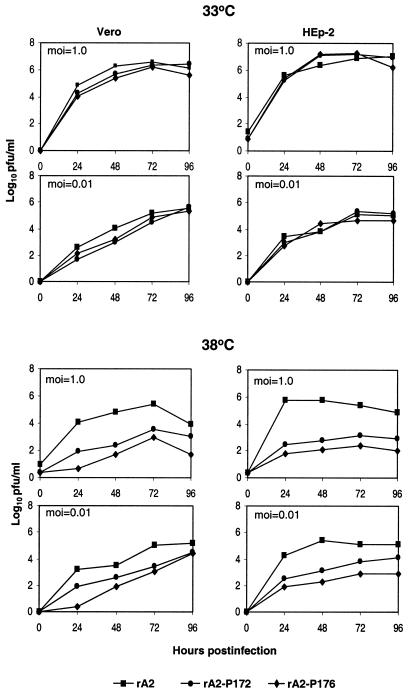
The single-cycle (MOI = 1.0) and multicycle (MOI = 0.01) growth kinetics of rA2-P172 and rA2-P176 were compared to those of rA2 in both HEp-2 and Vero cells (Fig. 5). At 33°C, both rA2-P172 and rA2-P176 had similar replication kinetics and reached peak titers comparable to that of rA2 at both MOIs in both cell lines. At 38°C, rA2-P172 and rA2-P176 reached peak titers much lower than that of wt rA2. At an MOI of 1.0 PFU/cell, rA2-P172 had peak titers ca. 2.0 and 2.3 log10 lower than those of rA2 in Vero cells and HEp-2 cells at 38°C, respectively. The reductions of rA2-P176 in its peak titer relative to wt rA2 at 38°C were even greater: 2.5 and 3.0 log10 in Vero cells and HEp-2 cells, respectively (Fig. 5). The reduction was less pronounced when an MOI of 0.01 was used in infection: 0.6 log10 in Vero cells and 1.0 log10 in HEp-2 cells for rA2-P172 and 0.8 log10 in Vero cells and 2.2 log10 in HEp-2 cells for rA2-P176 (Fig. 5). At 39°C, both rA2-P172 and rA2-P176 replicated to a level below the assay limit (data not shown). These data are consistent with what had been observed in the minigenome assay, in which E176G was more impaired in its functions (Fig. 3).
FIG. 5.
Growth kinetics of rA2-P172 and rA2-P176 in Vero and HEp-2 cells. Vero or HEp-2 cells were infected with virus at an MOI of 1.0 or 0.01 PFU/cell and incubated at 33 or 38°C. Aliquots of culture supernatants (200 μl) were harvested at 24-h intervals for 5 days, and the virus titers were determined by plaque assay on Vero cells. Each virus titer is an average of two experiments.
Replication of rA2-P172 and rA2-P176 in mice and cotton rats.
The replication of rA2-P172 and rA2-P176 in the lower respiratory tracts of mice and cotton rats was examined (Table 2). The replication of rA2-P172 and rA2-P176 in the lungs of mice was reduced by 2.7 and 3.7 log10, respectively. The replication of rA2-P172 and rA2-P176 in the lungs of cotton rats was reduced by 1.5 and 2.5 log10, respectively. Consistent with the result from the minigenome assay at 37°C and the growth kinetics in cell culture at 38°C, rA2-P176 was more attenuated than rA2-P172 as measured by replication in the lower respiratory tracts of mice and cotton rats.
TABLE 2.
Replication of recombinant RSV in mice and cotton rats
| Virus | Virus titer in lungs (mean log10 PFU/g ± SE)a in:
|
|
|---|---|---|
| Mice | Cotton rats | |
| rA2 | 4.64 ± 0.08 | 4.72 ± 0.08 |
| rA2-P172 | 1.97 ± 0.99 | 3.29 ± 0.39 |
| rA2-P176 | 0.90 ± 1.20 | 2.21 ± 0.11 |
Groups of eight BALB/c mice or cotton rats were inoculated with 106 PFU of virus intranasally under light anesthesia on day 0 and sacrificed on day 4. Virus titers from the lung tissues were determined by plaque assay.
Analysis of N-P interaction in virus-infected cells by immunoprecipitation.
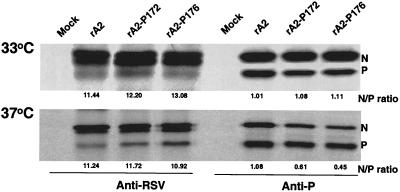
To examine whether the G172S and E176G mutations in P affected their interaction with the N protein in virus-infected cells, viral proteins from cells infected with rA2, rA2-P172, and rA2-P176 were immunoprecipitated with either polyclonal anti-RSV antibodies or a mixture of monoclonal anti-P antibodies (Fig. 6). Anti-P monoclonal antibodies precipitated both N and P, demonstrating the formation of N-P complex in the infected cells. The N protein precipitated by anti-RSV antibody appeared as a double band but as a single band when precipitated together with the P protein by anti-P antibodies. The faster-migrating species of N may represent an unmodified form of N, which was not coimmunoprecipitated with P. There was an overall reduction in the total amount of viral proteins produced in rA2-P172- and rA2-P176-infected cells at 39°C, as expected from the observed growth kinetics in Vero cells. Therefore, coimmunoprecipitation was performed for the infected cells that were incubated at 33 and 37°C. Anti-RSV antibody did not react well with the P protein, but rA2-P172 and rA2-P176 had an N/P ratio similar to that of wt rA2 when precipitated by anti-RSV antibody at 33 and 37°C. The amounts of the N and P proteins immunoprecipitated by anti-P antibodies on the autographs were quantified by densitometry, and their relative ratios are indicated in Fig. 6. At 33°C, the N/P ratios of rA2-P172, and rA2-P176 were similar to that of rA2, indicating that the N-P interaction was not affected at the lower temperature. However, at 37°C, the amount of N coprecipitated by P was reduced in cells infected with rA2-P172 and rA2-P176. The average ratio of the N and P proteins for wt rA2 was 1.08 at 37°C. The N/P ratio of rA2-P172 was 0.61 or at a level of 56% of wt rA2; rA2-176 had an even lower N/P ratio of 0.45 or at a level of 42% of wt rA2. The reduced N/P ratio for rA2-172 and rA2-176 at 37°C was reproducible, demonstrating that the G172S and E176G mutations decreased the interaction between N and P at high temperatures with the E176G mutation being more impaired than G172S.
FIG. 6.
Immunoprecipitation of viral proteins from RSV-infected cells. Vero cells were infected with wt rA2, rA2-P172, or rA2-P176 at an MOI of 1.0 and incubated at 33 and 37°C for 18 h. Proteins were then radiolabeled with [35S]Cys and [35S]Met (100 μCi/ml) in DMEM deficient in cysteine and methionine for 4 h, immunoprecipitated by either anti-RSV or anti-P monoclonal antibodies, separated by SDS-15% PAGE, and autoradiographed. The positions of the N and P proteins are indicated on the right. The N and P ratio for each mutant was determined from four independent experiments.
Stability of the P ts mutations in rA2-P172 and rA2-P176.
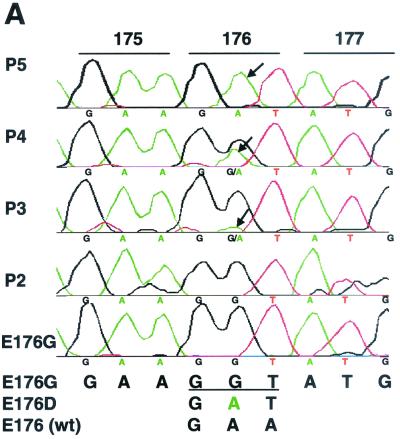
To examine the stability of the G172S and E176G mutations in the P protein, rA2-P172 and rA2-P176 were passaged in Vero cells in duplicate at 33 and 37°C five consecutive times. Viral RNA was extracted from the infected cell culture supernatant, and the P gene cDNA was amplified by reverse transcription-PCR and sequenced. The G172S mutation was maintained at both 33 and 37°C. The E176G mutation, however, rapidly changed from Gly to Asp starting from passage 3 at 37°C in one set of the passage samples. More than 95% of the virus population contained the E176D change at passage 5 (Fig. 7A). No changes were detected at position 176 when the infected cells were incubated at 33°C. The E176D virus was then examined for replication at various temperatures. As shown in Fig. 7B, only a slight reduction in virus titer was observed at 39°C compared to that seen at 33°C. Thus, virus bearing the E176D change was no longer temperature sensitive at 39°C. Sequence analysis of the second set of rA2-P176 passaged five times at 37°C indicated mixed residues at the 176 position. Virus was then plaque purified, and the P gene cDNA was sequenced. Of eight plaque isolates, four contained Asp changes, two contained Cys, and the remaining two had Ser changes. Substitutions of Gly by Cys or Ser also resulted in the loss of the virus ts phenotype (data not shown). From these results, it appeared that the negatively charged residue at position 176 was preferred by virus, with Cys or Ser as the second choice. Cys and Ser each contain side chains that can form a disulfide bond or a hydrogen bond, respectively, implying that the residue at 176 of P is involved in protein interaction.
FIG. 7.
Reversion of rA2-P176 during in vitro passage at 37°C. (A) Sequence of the P gene in the region of residue 176 from rA2-P176 passaged in Vero cells. The introduced E176G mutation was progressively reverted to E176D starting from passage 3 (P3). Arrows indicate the G-to-A change in the 176 codon. (B) Replication of the E176D revertant at different temperatures. Monolayers of Vero and HEp-2 cells were infected with rA2 P-E176D; overlaid with L15 medium containing 1% methylcellulose and 2% FBS; and incubated at 33, 37, and 39°C.
DISCUSSION
Using a yeast two-hybrid system to screen a RSV P gene cDNA library, two mutations (G172S and E176G) were identified that affected the N-P interaction in a temperature-dependent manner. The replication of recombinant viruses bearing either the G172S or the E176G mutation was shown to exhibit a ts phenotype in tissue culture. Interestingly, the G172S mutation coincides with the ts mutation identified in the RSV subgroup B RSN-2 strain (6, 12). It was later demonstrated that the introduction of G172S mutation into the P gene of the RSV subgroup A RSS-2 strain also resulted in much-reduced replication of a RSV minigenome at 37 and 39°C (32).
The E176G and G172S mutations identified in this study are only three residues apart. The E176G mutation had a more severe effect on the P protein function than the G172S mutation in the minigenome assay. rA2-P176 virus was more temperature sensitive in tissue culture and more restricted in replication in the respiratory tracts of mice and cotton rats than rA2-P172 virus. Sequence alignment of several different strains of pneumoviruses revealed that the region flanking 172 to 176 is highly conserved. One of the notable features in this region is the abundance of charged residues (Fig. 1). A triple mutant, 174-176A was constructed to remove the three consecutive charged residues from the P protein to evaluate their impacts on the protein function. This triple mutant was found to be nonfunctional in a minigenome system, indicating a critical role of these charged residues. This may explain the more severe defect manifested by E176G mutation compared to G172S mutation. Introduction of both the G172S and E176G mutations in the P gene resulted in a synergistic effect that completely abolished the P protein function in the minigenome assay, and virus was not recovered from the cDNA bearing a combination of these two mutations. Taken together, the 172 to 176 residues of P are critical for P protein function.
Recombinant virus, rA2-P176, rapidly reverted when the virus-infected cells were incubated at 37°C, which led to the loss of its ts phenotype. Since Gly (GGT) contained two nucleotide changes compared to Glu (GAA), it was difficult for virus to revert back to the original residue. Instead, the introduced Gly was predominantly changed to Asp (GAT), also a negatively charged residue, suggesting that virus favors the charged residue at position 176. The Asp mutation was introduced into the P protein expression plasmid so that we could examine its function in the RSV CAT minigenome assay. At 37°C, the CAT activity detected in cells expressing P-E176D was ca. 50% of that of the wt (data not shown), which was much higher than the 5% activity of E176G. Interestingly, the second set of passage samples of rA2-P176 at 37°C also contained substitution of Cys and Ser, although the majority had the Asp change. Since Cys and Ser each may also interact with other protein residues through disulfide or hydrogen bond, the data support the notion that the residue at the 176 position is very important that is likely involved in protein-protein interaction (possibly the N-P interaction). It is possible that substitution of E176 by other amino acids could also be tolerated by virus. For example, replacement of E176 with Ala did not significantly reduce the P protein function in a minigenome assay (data not shown), indicating that the 176 residue has an important structural role in the P protein function.
G172S and E176G mutations identified by the yeast two-hybrid assay were shown to be sensitive to temperature with respect to their interaction with N in yeast. The minigenome replication assay showed that the function of each mutant was only slightly reduced at 33°C, was greatly reduced at 37°C, and was further reduced at 39°C. The expression level of G172S and E176G protein in transfected cells at 37 and 39°C was similar to that of wt P, indicating that the temperature sensitivity was not due to the thermolability of the protein. However, coimmunoprecipitation of cells transfected with pN and the mutant pP at 37 and 39°C did not reveal any alteration in N-P complex formation. This could be due to the overexpression of the two proteins in the vaccinia virus-T7 expression system so that the subtle difference in N-P interaction was not detected. Since the C terminus of the P protein is the major N protein binding site and only a trace amount of the N protein was coimmunoprecipitated with the P protein that had the C-terminal 6 amino acids deleted, the contribution of 161 to 180 amino acids in interaction with the N protein must be very trivial. It was thus possible that the effect of the mutation in the region of 161 to 180 of P was difficult to detect by the cotransfection assay. It was also found that N and P interaction took place during the immunoprecipitation reaction when the cell lysates from individually transfected N and P proteins were mixed prior to the immunoprecipitation process (data not shown). As had been reported earlier, G172S mutation introduced in the P protein of RSS-2 strain also did not show reduction in N-P interaction in a mammalian two-hybrid system at 39°C (32). We also used the mammalian two-hybrid system to analyze the interaction of P-E176G and N and did not find reduced N-P interaction in that system at 39°C (data not shown). Nevertheless, the reduced N-P interaction was observed in cells infected with rA2-P172 or rA2-P176 at 37°C. The amount of the N protein coimmunoprecipitated with the P protein was reduced by at least twofold as shown in Fig. 6. The reduction in the N/P ratios of rA2-P172 and rA2-P176 at 37°C is reproducible, and the difference in the N/P ratio between the mutants and the wt rA2 is probably not due to the experimental errors. On the other hand, the reduction in the N/P ratio of rA2-172 and rA2-176 at the nonpermissive temperature could be >2-fold if the immunoprecipitation process could be performed at 37°C to reduce any N-P interaction that may have taken place in the lysis buffer during the immunoprecipitation process. However, it was difficult to perform immunoprecipitation at 37°C due to protein degradation (data not shown). Since functional differences between soluble N-P and the nucleocapsid-bound N-P complex exist (11, 36), it is possible that the N-P interaction detected in virus-infected cells is different from that in the N/P cotransfected cells. Thus, it is most likely that residues at positions 172 and 176 are involved in N-P interaction as originally identified in the yeast two-hybrid system and that rA2-P176 has a more severe defect in N-P interaction than rA2-P172 at nonpermissive temperatures. The rapid reversion of Gly at residue 176 to Asp or Ser and Cys during in vitro passages of rA2-P176 at a nonpermissive temperature also indicates a more critical role of E176. The reduced ability of G172S and E176G mutations to interact with N is likely the reason why the recombinant viruses containing these mutations have their ts phenotypes.
A recent study has shown that the residues between 161 and 180 of the bovine RSV P protein constitute an N protein-binding domain, in addition to the domain located in the C-terminal end between residues 221 and 241 (28). The removal of residues 161 to 180 from bovine RSV P protein abolished the N-P complex formation as determined by coimmunoprecipitation assay (28). The P protein of bovine RSV shares 81% sequence identity with that of the human RSV (31). Furthermore, the region between residues 161 and 180 of these two proteins shares 95% homology. However, the human RSV P protein with a deletion of 161 to 180 residues generated in this study was able to coimmunoprecipitate with N, although it lost its function in supporting the RSV minigenome replication. This result is consistent with that reported by Slack and Easton in which the human RSV P protein with residues 168 to 198 deleted retained its interaction with N in a two-hybrid assay (35). Thus, the contribution of this internal region to N-P interaction for the P protein of human RSV is different from that of bovine RSV, which is less likely due to the assay variations. It should be noted that the assay performed in the in vitro system may not reflect what was observed in virus-infected cells since the reduced N-P interaction was indeed detected in cells infected with rA2-P172 and rA2-P176, whereas the in vitro assay failed to show the reduced interaction. Previously, it has been shown that the N-terminal deletion of 10 or 14 residues resulted in the loss of colocalization of P with N in transfected cells, although N-P complex formation was detected by coimmunoprecipitation (14). The available data suggested that the formation of N-P complexes involves subtle interactions of multiple regions in the N protein.
Although the P proteins are very diverse among different paramyxoviruses, they share similar features in their structure and function. Studies from the N-P interactions of other paramyxoviruses have indicated that multiple N-binding sites are often present in the P protein. For example, both the N terminus and the C terminus of P are involved in N-P interaction for pneumonia virus of mice (3), measles virus (20), Sendai virus (11, 34), vesicular stomatitis virus (15), and rabies virus (7). For human parainfluenza virus type 3, the C-terminal residues and the region adjacent to the C terminus of P are required for its binding to N (44). Although our current data demonstrated the involvement of the region flanking 172 to 176 of P in N-P interaction, it is not clear whether these residues directly participate in N protein binding or are responsible for the correct folding of the protein structure in order to enable its binding to the N protein. It is not known whether this region also affects other protein-protein interactions, such as the binding of P to L and P to P oligomerization.
Conditional lethal mutations are important for the development of live attenuated vaccines. The temperature-sensitive lesions previously identified in chemically mutagenized or cold-passaged RSV have mostly been mapped to the L protein (10, 26, 40, 41), possibly due to its large size. The identification of protein-protein interactions in a heterologous system, such as the yeast two-hybrid system, should be very useful in identifying mutations in other viral components such as P that will produce attenuated virus.
Acknowledgments
We thank Aviron's animal facility for assistance with the mouse and cotton rat experiments; the tissue culture facility for supplying cells; Helen Zhou and Mary Munoz for technical assistance; and Harry Greenberg, George Kemble, and Roderick Tang for useful discussions and critical reviews of the manuscript. We are grateful to Jose Melero for providing anti-P monoclonal antibodies. The suggestions and comments from the reviewers are also greatly appreciated.
This work was supported in part by NIH SBIR grants (2R44A145267-01/02).
REFERENCES
- 1.Asenjo, A., and N. Villanueva. 2000. Regulated but not constitutive human respiratory syncytial virus (HRSV) P protein phosphorylation is essential for oligomerization. FEBS Lett. 467:279-284. [DOI] [PubMed] [Google Scholar]
- 2.Atreya, P. L., M. E. Peeples, and P. L. Collins. 1998. The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication. J. Virol. 72:1452-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr, J., and A. J. Easton. 1995. Characterization of the interaction between the nucleoprotein and phosphoprotein of pneumonia virus of mice. Virus Res. 39:221-235. [DOI] [PubMed] [Google Scholar]
- 4.Bermingham, A., and P. L. Collins. 1999. The M2-2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc. Natl. Acad. Sci. USA 96:11259-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman, M. C., S. Smallwood, and S. A. Moyer. 1999. Dissection of individual functions of the Sendai virus phosphoprotein in transcription. J. Virol. 73:6474-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caravokyri, C., A. J. Zajac, and C. R. Pringle. 1992. Assignment of mutant tsN19 (complementation group E) of respiratory syncytial virus to the P protein gene. J. Gen. Virol. 73:865-873. [DOI] [PubMed] [Google Scholar]
- 7.Chenik, M., K. Chebli, Y. Gaudin, and D. Blondel. 1994. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. J. Gen. Virol. 75:2889-2896. [DOI] [PubMed] [Google Scholar]
- 8.Collins, P., M. Hill, E. Camargo, H. Grosfeld, R. Chanock, and B. Murphy. 1995. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc. Natl. Acad. Sci. USA 92:11563-11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, p. 1443-1485. In D. M. Knipe et al. (ed.), Fields virology, 4th ed. Lippincott, Philadelphia, Pa.
- 10.Crowe, J. E., Jr., C. Y. Firestone, S. S. Whitehead, P. L. Collins, and B. R. Murphy. 1996. Acquisition of the ts phenotype by a chemically mutagenized cold-passaged human respiratory syncytial virus vaccine candidate results from the acquisition of a single mutation in the polymerase (L) gene. Virus Genes 13:269-273. [DOI] [PubMed] [Google Scholar]
- 11.Curran, J., J. B. Marq, and D. Kolakofsky. 1995. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J. Virol. 69:849-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faulkner, G. P., P. V. Shirodaria, E. A. Follett, and C. R. Pringle. 1976. Respiratory syncytial virus ts mutants and nuclear immunofluorescence. J. Virol. 20:487-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia, J., B. Garcia-Barreno, A. Vivo, and J. A. Melero. 1993. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195:243-247. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Barreno, B., T. Delgado, and J. A. Melero. 1996. Identification of protein regions involved in the interaction of human respiratory syncytial virus phosphoprotein and nucleoprotein: significance for nucleocapsid assembly and formation of cytoplasmic inclusions. J. Virol. 70:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, T. J., S. Macpherson, S. Qiu, J. Lebowitz, G. Wertz, W., and M. Luo. 2000. Study of the assembly of vesicular stomatitis virus N protein: role of the P protein. J. Virol. 74:9515-9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosfeld, H., M. G. Hill, and P. L. Collins. 1995. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J. Virol. 69:5677-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardy, R. W., S. B. Harmon, and G. W. Wertz. 1999. Diverse gene junctions of respiratory syncytial virus modulate the efficiency of transcription termination and respond differently to M2-mediated antitermination. J. Virol. 73:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, R. W., and G. W. Wertz. 2000. The Cys3-His1 motif of the respiratory syncytial virus M2-1 protein is essential for protein function. J. Virol. 74:5880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy, R. W., and G. W. Wertz. 1998. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J. Virol. 72:520-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harty, R. N., and P. Palese. 1995. Measles virus phosphoprotein (P) requires the NH2- and COOH-terminal domains for interactions with the nucleoprotein (N) but only the COOH terminus for interactions with itself. J. Gen. Virol. 76:2863-2867. [DOI] [PubMed] [Google Scholar]
- 21.Horikami, S. M., J. Curran, D. Kolakofsky, and S. A. Moyer. 1992. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 66:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber, M., R. Cattaneo, P. Spielhofer, C. Orvell, E. Norrby, M. Messerli, J. C. Perriard, and M. A. Billeter. 1991. Measles virus phosphoprotein retains the nucleocapsid protein in the cytoplasm. Virology 185:299-308. [DOI] [PubMed] [Google Scholar]
- 23.Jin, H., X. Cheng, H. Z. Zhou, S. Li, and A. Seddiqui. 2000. Respiratory syncytial virus that lacks open reading frame 2 of the M2 gene (M2-2) has altered growth characteristics and is attenuated in rodents. J. Virol. 74:74-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin, H., D. Clarke, H. Z. Zhou, X. Cheng, K. Coelingh, M. Bryant, and S. Li. 1998. Recombinant human respiratory syncytial virus (RSV) from cDNA and construction of subgroup A and B chimeric RSV. Virology 251:206-214. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, P. R., and P. L. Collins. 1990. Sequence comparison of the phosphoprotein mRNAs of antigenic subgroups A and B of human respiratory syncytial virus identifies a highly divergent domain in the predicted protein. J. Gen. Virol. 71:481-485. [DOI] [PubMed] [Google Scholar]
- 26.Juhasz, K., S. S. Whitehead, P. T. Bui, J. M. Biggs, J. E. Crowe, C. A. Boulanger, P. L. Collins, and B. R. Murphy. 1997. The temperature-sensitive (ts) phenotype of a cold-passaged (cp) live attenuated respiratory syncytial virus vaccine candidate, designated cpts530, results from a single amino acid substitution in the L protein. J. Virol. 71:5814-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khattar, S. K., A. S. Yunus, P. L. Collins, and S. K. Samal. 2001. Deletion and substitution analysis defines regions and residues within the phosphoprotein of bovine respiratory syncytial virus that affect transcription, RNA replication, and interaction with the nucleoprotein. J. Virol. 285:253-269. [DOI] [PubMed] [Google Scholar]
- 28.Khattar, S. K., A. S. Yunus, and S. K. Samal. 2001. Mapping the domains on the phosphoprotein of bovine respiratory syncytial virus required for N-P and P-L interactions using a minigenome system. J. Gen. Virol. 82:775-779. [DOI] [PubMed] [Google Scholar]
- 29.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe et al. (ed.), Fields virology, 4th ed. Lippincott, Philadelphia, Pa.
- 30.Mallipeddi, S. K., B. Lupiani, and S. K. Samal. 1996. Mapping the domains on the phosphoprotein of bovine respiratory syncytial virus required for N-P interaction using a two-hybrid system. J. Gen. Virol. 77:1019-1023. [DOI] [PubMed] [Google Scholar]
- 31.Mallipeddi, S. K., and S. K. Samal. 1992. Sequence comparison between the phosphoprotein mRNAs of human and bovine respiratory syncytial viruses identifies a divergent domain in the predicted protein. J. Gen. Virol. 73:2441-2444. [DOI] [PubMed] [Google Scholar]
- 32.Marriott, A. C., S. D. Wilson, J. S. Randhawa, and A. J. Easton. 1999. A single amino acid substitution in the phosphoprotein of respiratory syncytial virus confers thermosensitivity in a reconstituted RNA polymerase system. J. Virol. 73:5162-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masters, P. S., and A. K. Banerjee. 1988. Complex formation with vesicular stomatitis virus phosphoprotein NS prevents binding of nucleocapsid protein N to nonspecific RNA. J. Virol. 62:2658-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryan, K. W., E. M. Morgan, and A. Portner. 1991. Two noncontiguous regions of Sendai virus P protein combine to form a single nucleocapsid binding domain. Virology 180:126-134. [DOI] [PubMed] [Google Scholar]
- 35.Slack, M. S., and A. J. Easton. 1998. Characterization of the interaction of the human respiratory syncytial virus phosphoprotein and nucleocapsid protein using the two-hybrid system. Virus Res. 55:167-176. [DOI] [PubMed] [Google Scholar]
- 36.Spehner, D., R. Drillien, and P. M. Howley. 1997. The assembly of the measles virus nucleoprotein into nucleocapsid-like particles is modulated by the phosphoprotein. Virology 232:260-268. [DOI] [PubMed] [Google Scholar]
- 37.Tang, R., N. Nguyen, X. Cheng, and H. Jin. 2001. Requirement of cysteines and length of the human respiratory syncytial virus m2-1 protein for protein function and virus viability. J. Virol. 75:11328-11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarbouriech, N., J. Curran, C. Ebel, R. W. Ruigrok, and W. P. Burmeister. 2000. On the domain structure and the polymerization state of the Sendai virus P protein. Virology 266:99-109. [DOI] [PubMed] [Google Scholar]
- 39.Tarbouriech, N., J. Curran, R. W. Ruigrok, and W. P. Burmeister. 2000. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat. Struct. Biol. 7:777-781. [DOI] [PubMed] [Google Scholar]
- 40.Tolley, K. P., A. C. Marriott, A. Simpson, D. J. Plows, D. A. Matthews, S. J. Longhurst, J. E. Evans, J. L. Johnson, P. A. Cane, V. B. Randolph, A. J. Easton, and C. R. Pringle. 1996. Identification of mutations contributing to the reduced virulence of a modified strain of respiratory syncytial virus. Vaccine 14:1637-1646. [DOI] [PubMed] [Google Scholar]
- 41.Whitehead, S. S., C. Y. Firestone, P. L. Collins, and B. R. Murphy. 1998. A single nucleotide substitution in the transcription start signal of the M2 gene of respiratory syncytial virus vaccine candidate cpts248/404 is the major determinant of the temperature-sensitive and attenuation phenotypes. Virology 247:232-239. [DOI] [PubMed] [Google Scholar]
- 42.Wyatt, L. S., B. Moss, and S. Rozenblatt. 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210:202-205. [DOI] [PubMed] [Google Scholar]
- 43.Yu, Q., R. W. Hardy, and G. W. Wertz. 1995. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J. Virol. 69:2412-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao, H., and A. K. Banerjee. 1995. Interaction between the nucleocapsid protein and the phosphoprotein of human parainfluenza virus 3. Mapping of the interacting domains using a two-hybrid system. J. Biol. Chem. 270:12485-12490. [DOI] [PubMed] [Google Scholar]