Abstract
Vertically transmitted endogenous retroviruses pose an infectious risk in the course of pig-to-human transplantation of cells, tissues, and organs. Two classes of polytropic type C porcine endogenous retroviruses (PERV) productively infect human cells in vitro. The cloning and characterization of replication-competent PERV-B sequences from infected human cells (F. Czauderna, N. Fischer, K. Boller, R. Kurth, and R. R. Tönjes, J. Virol. 74:4028-4038, 2000) as well as the cloning of functional PERV-A and -B sequences from porcine cell line PK15 (U. Krach, N. Fischer, F. Czauderna, and R. R. Tönjes, J. Virol. 75:5465-5472, 2001) have been previously described. Here we report the isolation of four full-length proviral sequences from a porcine bacterial artificial chromosome (BAC) library that comprises chromosomally assigned PERV. Clones Bac-PERV-A(130A12) and Bac-PERV-A(151B10) map to pig chromosome 1 and demonstrate close homology to PK15-PERV-A(58) in env and to PERV-MSL in long terminal repeat (LTR), gag, and pro/pol sequences. Clone Bac-PERV-A(463H12) is located on pig chromosome 3 and demonstrates close homology to PK15-PERV-A(58) in env and to 293-PERV-B(43) in LTR, gag, and pro/pol (Czauderna et al.; R. R. Tönjes, F. Czauderna, N. Fischer, U. Krach, K. Boller, P. Chardon, C. Rogel-Gailard, M. Niebert, G. Scheef, A. Werner, and R. Kurth, Transplant Proc. 32:1158-1161, 2000). Clone Bac-PERV-B(192B9) is located on pig chromosome 7 in the swine leukocyte antigen region and is highly homologous with but distinct from the previously described functional clone 293-PERV-B(43) and bears the number of repeats initially observed in the LTRs of clone 293-PERV-A(42) (Czauderna et al.; Krach et al.). Clones Bac-PERV-A(130A12), Bac-PERV-A(151B10), and Bac-PERV-A(463H12) were replication competent upon transfection into susceptible 293 and HeLa cells. Bac-PERV-B(192B9), however, bears two stop codons in pro/pol preventing this clone from being replication competent in some individual pigs, but initial screenings indicate that this provirus might be intact in others. The data suggest that the porcine genome harbors a limited number of infectious PERV sequences, allowing for specific screening in different pig breeds.
The therapeutic use of animal cells, tissues, and organs derived from pigs as donors for xenotransplants might help to overcome the growing shortage of human allotransplants. Major concerns regarding infectious risk posed by the possibility of introducing new agents from the animal into the recipient, leading to xenozoonosis, have been raised (7, 8, 9, 15, 25). Breeding and keeping pigs under specific-pathogen-free conditions is considered to reduce the risk of transmitting exogenous agents. However, these methods are not appropriate for avoiding the presence of endogenous retroviruses which are transmitted in the germ line.
Porcine endogenous retroviruses (PERV) released from porcine cells infect human cells in vitro (14, 18, 31, 32). Risk of xenozoonosis is even enhanced if genetically engineered pigs which are produced to reduce the host-versus-graft reaction are used (2, 23, 30).
In a retrospective study, no cross-species transmission of PERV in patients treated with pig tissue was observed (17). On the other hand, in an NOD/SCID mouse model, the diabetic and immunodeficient animals showed infection and expression of PERV in different tissues after xenotransplantation of porcine islet cells, suggesting that PERV are xenozoonotic in vivo (29).
Approximately 30 to 50 integration sites of PERV exist in the genome of different pig breeds (1, 13) and three classes of infectious endogenous gammaretroviruses (PERV-A, PERV-B, and PERV-C) are known (19, 26). These classes display high sequence homology in the genes coding for the group specific antigens (Gag) and the polymerase (Pol) but differ in the genes encoding the envelope proteins (Env) which determine the different host ranges of the classes (13, 19, 26).
We have previously reported the isolation of replication-competent molecular PERV clones derived from human embryonic kidney cells infected with PERV (293 PERV-PK) (5, 27). Subsequently, we described functional proviral PERV isolated from porcine kidney cell line PK15 (12). The level of expression of these proviruses significantly depends on the existence of repeat structures in the PERV long terminal repeats (LTR) (24). In this communication, we describe the cloning and characterization of PERV-A and PERV-B proviral sequences derived from a bacterial artificial chromosome (BAC) library that has been generated using DNA from large white pigs (21). Three proviruses produced infectious and replication-competent virus particles after transfection of different human cell lines. The findings allow us to compare functional PERV from different origins directly on the molecular and cellular level and to map these proviral sequences to chromosomal locations of one specific pig breed. This mapping enables even further analyses, e.g., screening of different pig breeds for replication-competent proviruses or the generation of PERV-free pigs.
MATERIALS AND METHODS
Porcine genomic BAC libraries.
A porcine BAC library that was constructed in a pBeloBAC11 vector using primary fibroblast DNA isolated from a large white pig homozygous for swine leukocyte antigen (SLA) haplotype H01 as described previously (21) was employed for this study. The large white genome harbors at least 20 to 30 copies of PERV as revealed by Southern blot hybridization (21).
Thirty-three BAC clones containing PERV-specific sequences were mapped by fluorescence in situ hybridization to 22 distinct locations on 14 chromosomes (21). All clones were subjected to initial analyses (see below) which revealed that four of these were most likely to harbor intact PERV as deduced from these screening experiments (see below) and were used in this study. These clones are designated Bac-PERV-A(130A12), Bac-PERV-B(192B9), Bac-PERV-A(151B10) and Bac-PERV-A(463H12). Clones derived from cell line 293 PERV-PK are denoted using the prefix 293-, while clones derived from the porcine cell line PK15 are indicated by the prefix PK15-.
Prescreening of BAC library.
All BAC clones were subjected to individual gene-specific PCR amplifications for gag, pro/pol, and env. Resulting PCR fragments were tested in a protein truncation test (PTT; Promega) according to the manufacturer's instructions for the presence of open reading frames (ORFs). A long PCR (see below) was applied to determine if the detected ORFs belong to a single provirus or whether they are scattered across the BAC clone. Only positive clones were subjected to further analyses. The prescreening results are summarized in Table 1.
TABLE 1.
Summary of all chromosomal positions of BAC clones initially screened to harbor PERV sequences
| Clone no. | Chromosome position | PERV class | Gag expressiona | RT test | Full-length PCRb | gagb | polb | envb | Proviral integrationc |
|---|---|---|---|---|---|---|---|---|---|
| 130A12d | 1q2.4 | A | + | + | + | + | + | + | + |
| 242D4 | 13q4.9 | A | + | − | − | + | − | − | − |
| 151B10d | 1q2.3 | A | + | + | + | + | + | + | + |
| 141G12 | 1q2.11 | A | − | − | + | − | − | + | − |
| 463H12d | 3p1.5 | A | + | + | + | + | + | + | + |
| 305F5 | 5q2.3 | A | − | − | + | Trunc | + | + | − |
| 258A11 | 3p1.5 | A | − | − | − | + | + | + | − |
| 135E5 | 8p1.2 | A | − | − | − | + | Trunc | − | − |
| 253B6 | 13q4.2 | A | − | − | − | + | − | Trunc | − |
| 383E10 | 13q4.3 | A | − | − | − | + | + | − | − |
| 1079D8 | 17q1.2 | A | − | − | − | + | + | − | − |
| 192B9d | 7p1.1 | B | + | − | + | + | + | + | − |
| 484G4 | 7p1.2→p1.1 | B | − | − | − | + | − | + | − |
| 161B7 | 4p1.1 | B | − | − | + | Trunc | + | + | − |
| 783D7 | 9q2.6 | B | − | − | + | + | − | Trunc | − |
| 667G4 | 17q2.1 | B | − | − | + | + | Trunc | − | − |
| 534G4 | 17q2.1 | B | − | − | + | + | − | + | − |
| 498D8 | 14q1.1 | B | − | − | + | Trunc | Trunc | + | − |
| 1058D6 | 11q1.2 | B | − | − | − | + | − | − | − |
| 647G4 | 11q1.4 | B | − | − | − | + | − | Trunc | − |
| 80H6 | 14q2.8 | B | − | − | − | + | + | − | − |
Expression of Gag protein was tested by an immunofluorescence assay with specific Gag p10 antibodies after transfection of BAC DNA and was noted as positive when protein expression was detected at 1 to 3 d.p.t. +, positive result; −, negative result; Trunc, a PCR product or an in vitro transcript in PTT experiments was obtained, but the resulting molecular weight was smaller than calculated.
Gene-specific PCR was used to search for PERV genes while long PCR was used to determine if identified genes belong to one ORF. Resulting gene-specific PCR products were subjected to a PTT to test for ORFs.
Genomic DNA of transfected cells was tested for proviral integration.
Functionally investigated clones harboring proviral PERV.
Preparation of BAC DNA.
DNA from individual BAC clones was prepared using conventional alkaline lysis of bacteria followed by CsCl gradient centrifugation of the DNA (50,000 × g overnight) according to standard protocols (22).
Cell lines and replication studies.
The cell line 293 was kindly provided by R. Weiss (London) and HeLa cells were obtained from the European Collection of Cell Cultures (ECACC 93021013). One to 2 μg of BAC DNA was transfected into cells using Lipofectamine (Life Technologies, Karlsruhe, Germany). Due to the size of the BAC constructs, transfections had to be carried out repeatedly to actually result in successful DNA transfer. Viral replication was detected by reverse transcriptase (RT) assay and indirect immunofluorescence microscopy with PERV-specific antibodies. Infectivity was tested by inoculation of semiconfluent cultures of susceptible cell lines with cell-free supernatants of producer cells after filtration through 0.45-μm-pore-size membranes (Sartorius, Göttingen, Germany).
RT assay.
Cell-free membrane-filtered supernatants were tested for RT activity using the C-type RT activity assay (Cavidi Tech Ab, Uppsala, Sweden) according to the instructions of the manufacturer (protocol B).
Immunofluorescence microscopy.
Human 293 and HeLa cells transfected with BAC DNA or infected with PERV present in culture supernatant were fixed at several time points (3, 20, and 31 days posttransfection [d.p.t.] and 22 days postinfection [d.p.i.]) with 2% paraformaldehyde. Indirect immunofluorescence analysis was performed as described previously using PERV-specific antisera directed against Gag p10 (11).
Detection of integrated PERV.
Genomic DNA was isolated from different cell lines grown to confluence according to standard procedures (22). To avoid amplification of episomal proviral sequences genomic DNA was purified by CsCl gradient centrifugation (22). Integration of PERV was tested by amplification of pro/pol sequences using oligonucleotide primers PK1 (5′-TTG ACT TGG CAG TGG GAC GGG TAA C-3′, nucleotides (nt) 2927 to 2949) and PK6 (5′-GAG GGT CAC CTG AGG GTG TTG GAT-3′, nt 3739 to 3716) for a first PCR and primers PK2 (5′-GGT AAC CCA CTC GTT TCT GGT CA-3′, nt 2954 to 2966) and PK5 (5′-CTG TGT AGG GCT TCG TCA AAG ATG-3′, nt 3696 to 3673) for a nested amplification. Nucleotide positions refer to 293-PERV-B(33) (5, 27).
Cloning and sequence analyses.
Sequences encompassing the proviral genes were amplified from BAC clones 130A12, 151B10, 463H12, and 192B9 using long-distance PCR techniques (ExpandLong; Boehringer Mannheim, Mannheim, Germany) with oligonucleotide primers PK28 (5′-ATC AGC AGA CGT GCT AGG AGG ATC-3′, nt 895 to 918) and PK29 (5′-CCG CAG TCC TCT ACC CCT GCG TGG-3′, nt 8758 to 8781) and a cycle scheme of initial heating at 94°C for 5 min, 35 cycles of 94°C for 1 min, 58°C for 1 min, and 68°C for 10 min, and one final elongation for 20 min at 68°C. Amplification products were subcloned into pGEM-T Easy (Promega, Mannheim, Germany).
The proviral LTR sequences of all BAC clones were amplified separately using oligonucleotide primers 5′LTRfor (5′-TGA AAG GAT GAA AAT GCA ACC TAA-3′, nt 1 to 24) and PBSrev (5′-GTT GGC CGG GAA ATC CTG CG-3′, nt 715 to 735) for the 5′-LTR and PK34 (5′-AAA GGA TGA AAA TGC AAC CTA ACC-3′, nt 8233 to 8210) and 3′LTRrev (5′-ATT TTA ACT CGA CTG GCC TTT CAG-3′, nt 8895 to 8918) for the 3′-LTR. The gap between the 5′-LTR and the proviral gene amplification products (nt 736 to 894) was subsequently bridged by PCR amplification using appropriate primers deduced from both sequences. Primers for sequencing and given nucleotide positions are based on molecular clone 293-PERV-B(33). DNA sequences of both strands were determined as described previously (5) using an ABI 377 DNA sequencing system (Applied Biosystems, Weiterstadt, Germany).
Nucleotide sequence accession numbers.
The proviral sequences of Bac-PERV-A(130A12), Bac-PERV-A(151B10), Bac-PERV-A(463H12), and Bac-PERV-B(192B9) have been deposited in GenBank (accession numbers AJ279056, AF435967, AF435966, and AJ279057, respectively). Sequences used for homology analyses are PK15-PERV-A(58) (AJ293656) (12) and PK15-PERV-B(213) (AJ293657) (12).
RESULTS
Screening for replication-competent PERV.
Due to the size of the clones derived from the BAC library we chose a direct approach to search for replication-competent proviruses. Out of 22 different BAC clones, 4 that were most likely to harbor complete PERV proviral sequences designated Bac-PERV-A(130A12), Bac-PERV-A(151B10), Bac-PERV-A(463H12), and Bac-PERV-B(192B9) were identified by several screening procedures, including Southern blots and PCR analysis (data not shown), and were further investigated. The specific proviral and structural details of each clone are indicated in Fig. 1 and summarized in Table 2.
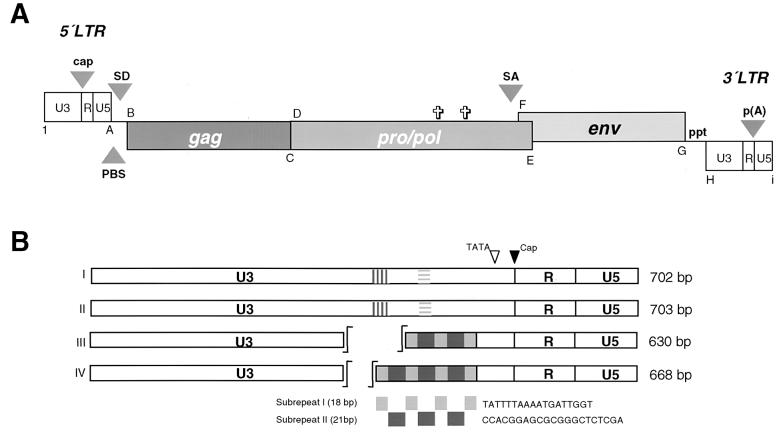
FIG. 1.
Schematic representation of replication-competent PERV proviral structure and LTR organization. (A) Proviral sequences of Bac-PERV-A(130A12), Bac-PERV-A(151B10), Bac-PERV-A(463H12), and Bac-PERV-B(192B9) are 8,918, 8,882, 8,754, and 8,840 bp in length, respectively. Genes are shown as boxes and the first and last nucleotide of LTR and structural genes are shown in bold letters (A to I) which are explained in Table 2. Arrowheads mark the transcriptional start site (cap), the primer binding site (PBS), the splice donor site (SD), the splice acceptor site (SA), the poly(A) addition site [p(A)], and the polypurine tract (ppt). Crosses show the positions of two in-frame stop codons in pol of Bac-PERV-B(192B9) at nt 4687 and 5251. (B) Organization of repetitive sequences in U3 of PERV LTR modulating the transcriptional activity shown for clones Bac-PERV-A(130A12) (I), Bac-PERV-A(151B10) (II), Bac-PERV-A(463H12) (III), and Bac-PERV-B(192B9) (IV) (24). Statistical analysis reveals several binding motifs for transcription factors within the repeat structure. The hatched boxes in sequences I and II indicate homologous but different sequences (24).
TABLE 2.
Nucleotide positions of genes and genetic elements of three replication-competent and one replication-incompetent full-length PERV investigated in this study
| Clone | Nucleotide position of indicated gene or gene elementa
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | Cap | PBS | SD | SA | Poly(A) | |
| Bac-PERV-A (130A12) | 702 | 1153 | 2727 | 2875 | 6309 | 6185 | 8149 | 8216 | 8918 | 540 | 706 | 761 | 5949 | 8810 |
| Bac-PERV-A (151B10) | 703 | 1148 | 2711 | 2859 | 6272 | 6148 | 8112 | 8180 | 8882 | 541 | 707 | 762 | 5912 | 8775 |
| Bac-PERV-A (463H12) | 630 | 1077 | 2660 | 2832 | 6242 | 6118 | 8100 | 8125 | 8754 | 467 | 633 | 689 | 5873 | 8646 |
| Bac-PERV-B (192B9) | 668 | 1115 | 2689 | 2837 | 6277 | 6150 | 8123 | 8173 | 8840 | 506 | 671 | 727 | 5911 | 8744 |
Cap, transcriptional start site; PBS, primer binding site; SD, splice donor site; SA, splice acceptor site; Poly(A), poly(A) addition site; A, end of 5′-LTR; B and C, gag; D and E, pro/pol; F and G, env; H and I, 3′-LTR.
Immunofluorescence analysis of transfected or infected cell lines.
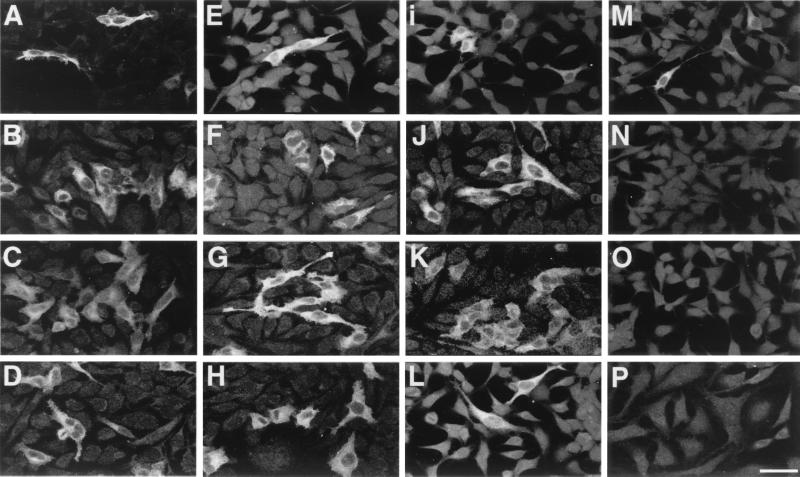
The capacity of proviral PERV sequences in BAC clones to productively infect cells was investigated initially by indirect immunofluorescence analyses using a PERV-specific antiserum against Gag p10 (11). As shown for HeLa cells (Fig. 2A to I), Gag expression in an increasing number of cells was observed for clones Bac-PERV-A(130A12), Bac-PERV-A(151B10), and Bac-PERV-A(463H12) after incubation with p10 antiserum for 3, 20, and 31 d.p.t., respectively, indicating the replication competence of these proviruses (see below). For Bac-PERV-B(192B9), immunoreactivity was detected for up to 3 d.p.t. but diminished when the cells were cultured for longer periods of time (Fig. 2M to O).
FIG. 2.
Detection of PERV Gag expression. Indirect immunofluorescence assay at different time points after transfection and infection of BAC DNA into HeLa cells using an antibody against Gag p10 (11). Expression of clones Bac-PERV-A(130A12) (A to D), Bac-PERV-A(151B10) (E to H), Bac-PERV-A(463H12) (I to L), and Bac-PERV-B(192B9) (M to P). The top three rows show protein expression investigated at 3, 20, and 31 d.p.t. (from top to bottom). The bottom row shows protein expression of HeLa cells at 22 d.p.i. with cell-free supernatant harvested from cells at 32 d.p.t. Scale bar, 50 μm (same magnification for all panels).
The initial immunoreactivity of cells transfected with Bac-PERV-B(192B9) can be explained by transient LTR-mediated expression of Gag shortly after transfection due to the deficiency of this clone to establish productive infection (see below).
Subsequently, transfer of cell-free culture supernatants from transfected HeLa cells (32 d.p.t.) to fresh cells revealed the presence of replication-competent virions by indirect immunofluorescence. Exemplary results for the infectivity of virions produced by clones Bac-PERV-A(130A12), Bac-PERV-A(151B10), and Bac-PERV-A(463H12) but not of Bac-PERV-B(192B9) at 22 d.p.i. are given in Fig. 2D, H, L, and P, respectively. Similar results were obtained for these clones upon transfection and infection into 293 cells (data not shown).
RT activity studies.
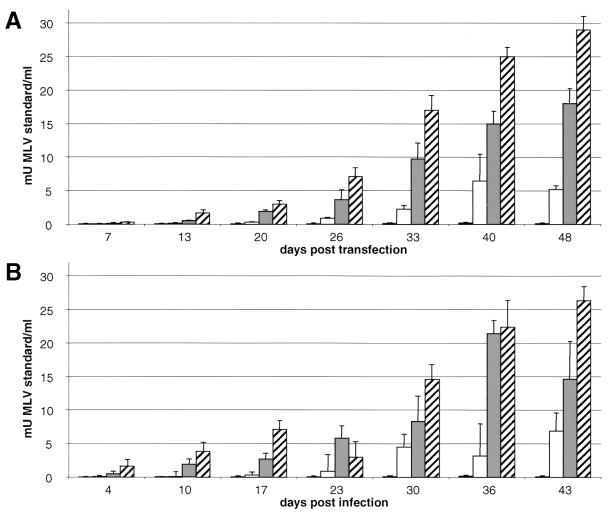
To confirm the results obtained by immunofluorescence analyses the activity of the viral RT was measured. Cell-free culture supernatants from human HeLa and 293 cells transfected or infected with clones Bac-PERV-A(130A12), Bac-PERV-A(151B10), Bac-PERV-A(463H12), and Bac-PERV-B(192B9) were collected for up to 48 d.p.t. and 43 d.p.i. (Fig. 3). The supernatant used to infect fresh cells was harvested at 32 d.p.t. For clones Bac-PERV-A(130A12), Bac-PERV-A(151B10), and Bac-PERV-A(463H12), RT activity was detected in 293 cells up to 28 mU of murine leukemia virus (MLV) standard per ml (Fig. 3A) and could be transferred to new cells (Fig. 3B), while no RT activity was observed for clone Bac-PERV-B(192B9) posttransfection or postinfection. Experiments with HeLa cells revealed similar results (data not shown).
FIG. 3.
Replicative properties of PERV. (A) Detection of RT activity in cell-free culture supernatants of 293 cells upon transfection of BAC DNA. Results are average values from three independent experiments. (B) Detection of RT activity in cell-free culture supernatants of 293 cells upon infection with cell-free supernatant from respective clones harvested at 32 d.p.t. Results are average values from two independent experiments. Gray bars, Bac-PERV-A(130A12); white bars, Bac-PERV-A(151B10); hatched bars, Bac-PERV-A(463H12); black bars, Bac-PERV-B(192B9).
Detection of proviral integration.
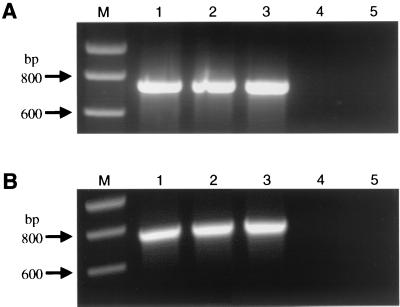
Genomic DNA extracted from cell lines transfected (Fig. 4A) or infected (Fig. 4B) with PERV was investigated at 40 d.p.t. and d.p.i. for integration of proviral sequences by PCR, revealing a pro/pol amplification product of 729 bp. HeLa cells (Fig. 4) and 293 cells (data not shown) used for infection studies were integration positive for Bac-PERV-A(130A12), Bac-PERV-A(151B10), and Bac-PERV-A(463H12) (Fig. 4B, lanes 1 to 3). No proviral Bac-PERV-B(192B9) sequences were detectable in genomic DNA either of cells that showed PERV Gag expression (Fig. 2M) after transfection with BAC DNA (Fig. 4A, lane 4) or of cells treated with supernatant (Fig. 4B, lane 4).
FIG. 4.
Proviral integration of functional PERV. (A) Genomic DNA of HeLa cells transfected with DNA of BAC clones was isolated at 40 d.p.t. by CsCl gradient centrifugation to avoid amplification of episomal proviral sequences. This DNA was tested for integration of PERV by PCR (for details, see text). The PCR amplification product was calculated to be 729 bp in length. (B) Same experiment as for panel A conducted with genomic DNA isolated from HeLa cells at 40 d.p.i. that had been treated with cell-free supernatant of HeLa cells harvested at 32 d.p.t. Genomic DNA of uninfected HeLa cells was prepared from the same stock used to perform the transfection experiment. Lane 1, Bac-PERV-A(130A12); lane 2, Bac-PERV-A(151B10); lane 3, Bac-PERV-A(463H12); lane 4, Bac-PERV-B(192B9); lane 5, uninfected HeLa cells; M, molecular size standard (Smart ladder; Eurogentec, Brussels, Belgium).
Analyses of full-length PERV sequences.
For further characterization of the viral genome, the sequences of all four proviral clones were determined. As the sequences of the LTRs and viral genes were determined separately, they were assembled for this analysis.
Sequence analyses revealed two different structures of LTRs. One LTR shows a distinct repeat pattern in U3 [Bac-PERV-A(463H12) and Bac-PERV-B(192B9)] and one LTR is devoid of these repeats [Bac-PERV-A(130A12) and Bac-PERV-A(151B10)]. The organization of these repeat patterns is shown in Fig. 1B.
While all proviruses display structural homology, Bac-PERV-B(192B9) is described in close detail as this clone showed mutations in its coding sequence. The details of proviral clones Bac-PERV-A(130A12), Bac-PERV-A(151B10), and Bac-PERV-A(463H12) as well as of clone Bac-PERV-B(192B9) are given in Fig. 1A, while the nucleotide positions of genes and genetic elements are summarized in Table 2.
The LTR of clone Bac-PERV-B(192B9) is 668 bp long, while the gag gene starts at nt 1115 and is colinear with the pro/pol ORF (nt 2837 to 6277). The stop codon at nt 2689 separating both genes is suppressed by tRNAGln; however, two stop codons at nt 4687 and 5251 within the pro/pol sequence disrupt the ORF, and as a consequence, prevent this clone from replication (Fig. 2M to P, 3, and 4). The env gene partially overlaps with pro/pol and forms a new ORF (nt 6150 to 8123). Clone Bac-PERV-B(192B9) has been chromosomally assigned and maps to 7p1.1 within the SLA cluster (21).
All clones showed a close relationship to proviral PERV sequences described previously (5, 12, 27). In particular, Bac-PERV-A(130A12), Bac-PERV-A(151B10), Bac-PERV-A(463H12), and Bac-PERV-B(192B9) were compared to PK15-PERV-A(58) and PK15-PERV-B(213) (5, 12). Results of the homology analysis are summarized in Table 3.
TABLE 3.
Comparison of nucleotide and amino acid sequences of Bac-PERV-A(130A12), Bac-PERV-A(151B10), Bac-PERV-A(463H12), and Bac-PERV-B(192B9) with native PERV proviral sequences isolated from the PK15 cell line (10)a
| Native PERV sequence | Gene | % Nucleotide Homology (% amino acid homology) with clone:
|
|||
|---|---|---|---|---|---|
| Bac-PERV-A(130A12) | Bac-PERV-A(151B10) | Bac-PERV-A(463H12) | Bac-PERV-B(192B9) | ||
| PK15-PERV-A(58) | LTR | 99.9 (NA)b | 98.4 (NA) | 70.1 (NA) | 63.4 (NA) |
| gag | 99.8 (98.9) | 92.6 (84.0) | 96.7 (72.7) | 95.2 (95.0) | |
| pro/pol | 99.7 (98.4) | 81.5 (65.9) | 85.1 (66.9) | 96.7 (NA) | |
| env | 99.8 (98.0) | 99.4 (98.0) | 98.6 (98.7) | 71.6 (65.6) | |
| PK15-PERV-B(213) | LTR | 67.5 (NA) | 66.8 (NA) | 94.8 (NA) | 82.9 (NA) |
| gag | 95.2 (95.6) | 88.7 (70.3) | 96.1 (84.0) | 99.2 (98.3) | |
| pro/pol | 96.5 (95.9) | 80.4 (63.4) | 83.3 (65.4) | 99.3 (NA) | |
| env | 71.5 (64.6) | 71.5 (64.6) | 71.9 (65.5) | 99.4 (99.1) | |
Low homology scores are due to the fact that different PERV classes are compared. Homology scores were revealed using sequence analysis DNASIS software (Hitachi).
NA, not applicable, as the nucleotide sequence is either not translated into protein (LTRs) or stop codons prevent the sequence from being translated [Bac-PERV- B(192B9) pol gene].
DISCUSSION
In this report we describe the characterization of proviral PERV-A and PERV-B sequences isolated from the genome of large white pigs. We thus are the first to compare native proviral PERV sequences derived from the pig genome with PERV derived from infected human cells on the molecular and cellular level.
Recently, full-length replication-competent PERV from pig cell line PK15 were isolated and characterized in our laboratory (12). These data combined with the four full-length PERV sequences described in this report demonstrate that the pig genome harbors a limited number of intact proviruses that form virions which can be transmitted to human cells in vitro as previously reported (14, 18, 31, 32).
As we have identified six unique PERV proviruses from two sources of porcine genomes, we are confident to have identified almost all PERV in pig breeds analyzed so far, since all other chromosomal locations harboring PERV sequences have been shown to contain defective proviruses represented in at least one viral gene (Table 1). The differences in length of the PERV genes described to date have no impact on the highly conserved structural motifs for mammalian type C retroviruses. Particularly, the env genes bear motifs such as the variable regions A and B (VRA and VRB) and the proline-rich region (PRO) of the gp70 protein that determine the host range via receptor interaction (3, 4, 13, 20). While the role of the Env protein in this interaction is well known, other more subtle viral features, some of which are located within the PERV LTR, may be responsible for the replicative performance of different PERV on a series of cell lines (24).
We have discovered two different LTR structures, one harboring a U3 repeat structure and one without these repeats (Fig. 1B). We assume that these differences account in part for the differences in replication performance observed in the various clones (Fig. 3) (24). A main feature of these repeats represents several binding motifs for transcription factors, including NF-Y, according to digital analysis. Generally, the more repeats are present in one given LTR, the higher is the transcriptional activity (24). Furthermore, the number of U3 repeats probably represents an adaptation of the virus to its human host cells, as they are dynamically regulated during serial passaging (24). Nevertheless, different numbers of repeats also occur in the natural and proviral state of PERV and account for many of the observed differences in length.
Although Bac-PERV-B(192B9) is defective, it demonstrated transcriptional activity, as revealed by transient expression of Gag after transfection of BAC DNA (Fig. 2).
Since the library used to characterize the proviruses presented in this communication is derived from a defined SLA haplotype (H01), it remains to be shown whether the haplotype has an impact on the distribution of replication-competent proviruses. If this correlation exists, pig breeds could be screened for this particular proviral PERV more easily.
A recent publication demonstrated PERV expression and subsequent infection of different tissues after transplantation of porcine pancreatic islets into immunodeficient NOD/SCID mice (29). These in vivo data indicated that a risk for infection by PERV exists in highly immunosuppressed transplant recipients during xenotransplantation. Even if the PERV clones derived from the porcine genome so far appear to be barely active, they have the capacity to productively infect different cell lines and thus bear xenozoonotic potential. Moreover, molecular virus clones turned out to mutate after serial passages on susceptible cells, indicating that PERV has the capacity to adapt to the host cell on the molecular level (24).
For the full characterization of the four proviruses, their genomic flanking sequences were determined by inverse PCR (Niebert et al., submitted). Knowledge of these sequences allowed us to screen individual pigs as well as different pig breeds for the prevalence of those four specific proviruses and for polymorphisms in different copies of the same provirus present within different individuals. Analyses indicated that individual proviruses showed polymorphisms in different regions of their genes as well as the varying presence of the localized proviruses between individual pigs and different breeds (Niebert et al., submitted).
In particular, the point mutations influencing the replicative properties of clone Bac-PERV-B(192B9) were of major interest. Therefore, we have initiated analyses of whether the defective sequence of this provirus located in the SLA is polymorphic in other individual pigs or different pig breeds (Niebert et al., submitted). Since we have detected several copies displaying an intact pol ORF, we assume that the related proviruses can encode replication-competent viruses compared to known infectious PERV sequences (5, 12); however, experimental evidence has not been provided yet. Corresponding single nucleotide polymorphisms have been shown to have great impact on genetic variability in several other cases (6, 10, 16, 28, 33).
Therefore, we cannot rule out the possibility that polymorphic PERV, both as mutational variants in known locations and as complete proviruses in new locations, exist in other pig breeds not analyzed so far.
As available data suggest that the number of replication-competent proviral PERV sequences within the porcine genome is rather limited, it appears feasible to generate PERV-free strains of pigs for xenotransplantation.
Acknowledgments
This study was supported by a grant from the German Ministry of Health (Bundesministerium für Gesundheit), Bonn, Germany.
REFERENCES
- 1.Akiyoshi, D. E., M. Denaro, H. Zhu, J. L. Greenstein, P. T. Banerjee, and J. A. Fishman. 1998. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J. Virol. 72:4503-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bach, F. H., S. C. Robson, H. Winkler, C. Ferran, K. M. Stuhlmeier, C. J. Wrighton, and W. W. Hancock. 1995. Barriers to xenotransplantation. Nat. Med. 1:869-873. [DOI] [PubMed] [Google Scholar]
- 3.Battini, J. L., O. Danos, and J. M. Heard. 1995. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J. Virol. 69:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battini, J. L., J. E. J. Rasko, and D. Miller. 1999. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G-protein-coupled signal transduction. Proc. Natl. Acad. Sci. USA 96:1385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czauderna, F., N. Fischer, K. Boller, R. Kurth, and R. R. Tönjes. 2000. Establishment and characterization of molecular clones of porcine endogenous retroviruses replicating on human cells. J. Virol. 74:4028-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deal, C., J. Ma, F. Wilkin, J. Paquette, F. Rozen, B. Ge, T. Hudson, M. Stampfer, and M. Pollak. 2001. Novel promoter polymorphism in insulin-like growth factor-binding protein-3: correlation with serum levels and interaction with known regulators. J. Clin. Endocrinol. Metab. 86:1274-1280. [DOI] [PubMed] [Google Scholar]
- 7.Fishman, J. A. 1994. Miniature swine as organ donors for man: strategies for prevention of xenotransplant-associated infections. Xenotransplantation 1:47-57. [Google Scholar]
- 8.Fishman, J. A. 1997. Xenosis and xenotransplantation: addressing the infectious risks posed by an emerging technology. Kidney Int. 51:41-45. [PubMed] [Google Scholar]
- 9.Hunkeler, D., A. M. Sun, S. Korbutt, R. V. Rajotte, R. G. Gill, R. Calafiore, and P. Morel. 1999. Bioartificial organs and acceptable risk. Nat. Biotechnol. 17:1045. [DOI] [PubMed]
- 10.Kerb, R., S. Hoffmeyer, and U. Brinkmann. 2001. ABC drug transporters: hereditary polymorphisms and pharmacological impact in MDR1, MRP1 and MRP2. Pharmacogenetics 2:51-64. [DOI] [PubMed] [Google Scholar]
- 11.Krach, U., N. Fischer, F. Czauderna, R. Kurth, and R. R. Tönjes. 2000. Generation and testing of a highly specific antiserum directed against porcine endogenous retrovirus nucleocapsid. Xenotransplantation 7:1-10. [DOI] [PubMed] [Google Scholar]
- 12.Krach, U., N. Fischer, F. Czauderna, and R. R. Tönjes. 2001. Comparison of replication-competent molecular clones of porcine endogenous retrovirus class A and class B derived from pig and human cells. J. Virol. 75:5465-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeTissier, P., J. P. Stoye, Y. Takeuchi, C. Patience, and R. A. Weiss. 1997. Two sets of human-tropic pig retroviruses. Nature 389:681-682. [DOI] [PubMed] [Google Scholar]
- 14.Martin, U., V. Kiessig, J. H. Blusch, A. Haverich, K. von der Helm, T. Herden, and G. Steinhoff. 1998. Expression of pig endogenous retrovirus by primary porcine endothelial cells and infection of human cells. Lancet 352:692-694. [DOI] [PubMed] [Google Scholar]
- 15.Michaels, M. G., and L. R. Simmons. 1994. Xenotransplant-associated zoonoses. Transplantation 57:1-7. [DOI] [PubMed] [Google Scholar]
- 16.Moulds, J. M., P. A. Zimmermann, O. K. Doumbo, L. Kassambara, I. Sagara, D. A. Diallo, J. P. Atkinson, M. Krych-Goldberg, R. E. Hauhart, D. E. Hourcade, D. T. McNamara, D. J. Birmingham, J. A. Rowe, J. J. Moulds, and L. H. Miller. 2001. Molecular identification of knops blood group polymorphisms found in long homologous region D of complement receptor 1. Blood 97:2879-2885. [DOI] [PubMed] [Google Scholar]
- 17.Paradis, K., G. Langford, Z. Long, W. Heneine, P. Sandstrom, W. M. Switzer, L. E. Chapman, C. Lockey, D. Onions, The XEN 111 Study Group, and E. Otto. 1999. Search for cross-species transmission of porcine endogenous retrovirus in patients treated with liver pig tissue. Science 285:1236-1241. [DOI] [PubMed] [Google Scholar]
- 18.Patience, C., Y. Takeuchi, and R. A. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3:282-286. [DOI] [PubMed] [Google Scholar]
- 19.Patience, C., W. M. Switzer, Y. Takeuchi, D. J. Griffiths, M. E. Goward, W. Heneine, J. P. Stoye, and R. A. Weiss. 2001. Multiple groups of novel retroviral genomes in pigs and related species. J. Virol. 75:2771-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasko, J. E. J., J. L. Battini, R. J. Gottschalk, I. Mazo, and A. D. Miller. 1999. The RD114/simian type D retrovirus receptor is a neutral amino acid transporter. Proc. Natl. Acad. Sci. USA 96:2129-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogel-Gaillard, C., N. Bourgeaux, A. Billault, M. Vaiman, and P. Chardon. 1999. Construction of a swine BAC library: application to the characterization and mapping of the porcine type C endoviral elements. Cytogenet. Cell Genet. 85:205-211. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Sandrini, M. S., W. L. Fodor, E. Mouhtouris, N. Osman, S. Cohney, S. A. Rollins, E. R. Guilmette, E. Setter, S. P. Squinto, and I. F. McKenzie. 1995. Enzymatic remodelling of the carbohydrate surface of a xenogeneic cell substantially reduces human antibody binding and complement mediated cytolysis. Nat. Med. 1:1261-1267. [DOI] [PubMed] [Google Scholar]
- 24.Scheef, G., N. Fischer, U. Krach, and R. R. Tönjes. 2001. The number of a U3 repeat box acting as an enhancer in long terminal repeats of polytropic replication-competent porcine endogenous retroviruses dynamically fluctuates during serial virus passages in human cells. J. Virol. 75:6933-6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoye, J. P., and J. M. Coffin. 1995. The danger of xenotransplantation. Nat. Med. 1:1100.. [DOI] [PubMed] [Google Scholar]
- 26.Takeuchi, Y., C. Patience, S. Magre, R. A. Weiss, P. T. Banerjee, P. LeTissier, and J. P. Stoye. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 72:9986-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tönjes, R. R., F. Czauderna, N. Fischer, U. Krach, K. Boller, P. Chardon, C. Rogel-Gaillard, M. Niebert, G. Scheef, A. Werner, and R. Kurth. 2000. Molecularly cloned porcine endogenous retroviruses replicate on human cells. Transplant Proc. 32:1158-1161. [DOI] [PubMed] [Google Scholar]
- 28.Turner, G., M. Barbulescu, M. Su, M. I. Jensen-Seaman, K. K. Kidd, and J. Lenz. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531-1535. [DOI] [PubMed] [Google Scholar]
- 29.Van der Laan, L. J. W., C. Lockey, B. C. Griffeth, F. S. Fraisier, C. A. Wilson, D. E. Onions, B. J. Hering, Z. Long, E. Otto, B. E. Torbett, and D. R. Salomon. 2000. Infection by porcine endogenous retrovirus after islet xenotransplantation in SCID mice. Nature 407:90-94. [DOI] [PubMed] [Google Scholar]
- 30.Weiss, R. A. 1998. Transgenic pigs and virus adaptation. Nature 391:327-328. [DOI] [PubMed] [Google Scholar]
- 31.Wilson, C. A., S. Wong, J. Muller, C. E. Davidson, T. M. Rose, and P. Burd. 1998. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J. Virol. 72:3082-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson, C. A., S. Wong, M. Van Brocklin, and M. Federspiel. 2000. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J. Virol. 74:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, X., J. Erdmann, V. Regitz-Zagrosek, S. Kurzinger, H. W. Hense, and H. Schunkert. 2000. Evaluation of three polymorphisms in the promoter region of the angiotensin II type I receptor gene. J. Hypertens. 18:267-272. [DOI] [PubMed] [Google Scholar]