Abstract
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV; also called human herpesvirus 8) is believed to be the etiologic agent of Kaposi's sarcoma, multicentric Castleman's disease, and AIDS-associated primary effusion lymphoma. KSHV infection of human dermal microvascular endothelial cells (DMVEC) in culture results in the conversion of cobblestone-shaped cells to spindle-shaped cells, a characteristic morphological feature of cells in KS lesions. All spindle-shaped cells in KSHV-infected DMVEC cultures express the latency-associated nuclear protein LANA1, and a subfraction of these cells undergo spontaneous lytic cycle induction that can be enhanced by tetradecanoyl phorbol acetate (TPA) treatment. To study the cellular response to infection by KSHV, we used two different gene array screening systems to examine the expression profile of either 2,350 or 9,180 human genes in infected compared to uninfected DMVEC cultures in both the presence and absence of TPA. In both cases, between 1.4 and 2.5% of the genes tested were found to be significantly upregulated or downregulated. Further analysis by both standard and real-time reverse transcription-PCR procedures directly confirmed these results for 14 of the most highly upregulated and 13 of the most highly downregulated genes out of a total of 37 that were selected for testing. These included strong upregulation of interferon-responsive genes such as interferon response factor 7 (IRF7) and myxovirus resistance protein R1, plus upregulation of exodus 2 β-chemokine, RDC1 α-chemokine receptor, and transforming growth factor β3, together with strong downregulation of cell adhesion factors α4-integrin and fibronectin plus downregulation of bone morphogenesis protein 4, matrix metalloproteinase 2, endothelial plasminogen activator inhibitor 1, connective tissue growth factor, and interleukin-8. Significant dysregulation of several other cytokine-related genes or receptors, as well as endothelial cell and macrophage markers, and various other genes associated with angiogenesis or transformation was also detected. Western immunoblot and immunohistochemical analyses confirmed that the cellular IRF7 protein levels were strongly upregulated during the early lytic cycle both in KSHV-infected DMVEC and in the body cavity-based lymphoma BCBL1 PEL cell line.
Kaposi's sarcoma-associated herpesvirus (KSHV) is a member of the gamma-2 family of herpesviruses and is distantly related to both Epstein-Barr virus (EBV) and herpesvirus saimiri. Subsequent to its initial isolation from Kaposi's sarcoma (KS) tissue by representational difference analysis by Chang et al. (11), KSHV has been found to be associated with all forms of KS, including classical, endemic, AIDS-associated, and transplant-associated forms, as well as rare AIDS-associated body cavity-based lymphomas (BCBL), primary effusion lymphoma (PEL), and multicentric Castleman's disease (MCD).
All stages of KS, from the initial patch lesions to the more advanced plaque and nodular stage lesions, are characterized by common immunological and histopathological findings, including angiogenesis and an increased concentration of proinflammatory cytokines. In late nodular stage lesions, there is evidence of outgrowth of oligoclonal spindle cells, which are thought to be the tumor cells of KS, and true sarcoma formation with invasion of the underlying dermal structure (8, 15, 19, 22, 33, 35).
Initial attempts to elucidate KSHV pathology have focused on the characterization of individual viral genes, including many captured cellular gene homologs, including vIL6, vMIPs, vCYC-D, vGCR, vBcl2, vFLIP, and vIRFs present in the KSHV genome. Of relevance to known KS pathology, LANA1, vCYC, and vFLIP are thought to be expressed constitutively in latently infected spindle cells, and vGCR and vMIP1 were shown to be highly angiogenic, with vGCR also inducing viral epidermal growth factor (VEGF) expression and forming tumors in nude mice (2, 4, 10). Additionally, vIRF1 was shown to be oncogenic, and vBcl-2 and vIL6 were shown to be antiapoptotic (6, 16).
While these studies described the effects of each gene in isolated transfection studies, the need to study numerous genes together under the same conditions and in the same samples has become apparent. The lack of a system for de novo infection of cells with KSHV in culture has previously limited studies of the broader cellular response to infection and establishment of latency. Recently, KSHV viral gene expression in the BC3 PEL cell line at different times before and after tetradecanoyl phorbol acetate (TPA) induction has been evaluated by gene array methodology (20, 29a).
Although the precise origin of KS spindle cells remains unclear, data from a number of studies suggest that they may be derived from an endothelial cell lineage (41). In addition, several groups have recently reported the development of successful infection and propagation methodologies for KSHV in culture using endothelial cells (14, 15, 28). We have shown that early passages of human dermal microvascular endothelial cells (DMVEC) infected with KSHV are converted from a contact-inhibited cobblestone-shaped morphology to a highly elongated, spindle-shaped morphology (14). Most of the spindle-shaped tumor cells of KS lesions are thought to be latently infected with KSHV (15a, 37), and all cultured KSHV-infected spindle-shaped DMVEC express the latent-state LANA1 protein (9, 14). We were interested in using this novel infection system to elucidate changes that occur in cellular gene expression upon the establishment of latent KSHV infection using DNA microarrays for gene expression profiling. Successful use of this type of technology has previously been reported by Zhu et al. for HCMV infection (45).
To compare several of the array technologies available, we used two Clontech Human Atlas arrays and the Incyte Human UniGEM V2.0 array. Clontech Atlas arrays each contain approximately 1,200 DNA fragments corresponding to well-characterized genes. The Clontech arrays are single-channel radioactivity-based nylon arrays which rely on differential hybridization by labeled cDNA samples derived from the RNA of matched infected and uninfected cells to parallel filters. Incyte UniGEM arrays are two-channel fluorescence-based microchip arrays containing approximately 9,000 genes which rely on a pair of differentially dye-labeled cDNA samples prepared from matched infected and uninfected cells being hybridized to a single microarray chip.
We examined the expression profile of genes using the Clontech nylon arrays first and, in an independent experiment, expanded the survey to include the Incyte arrays. The Clontech data were also used to independently examine and compare the effects of the presence or absence of TPA in both uninfected and infected DMVEC cultures. Additionally, we used three different reverse transcription (RT)-PCR procedures, including quantitative real-time PCR, to further evaluate a selected group of 37 highly up- or downregulated genes, which led to confirmation of the gene array data for approximately 75% of the genes tested. Finally, immunoblot and immunohistochemical (IHC) analyses were performed to confirm that upregulated RNA expression for IRF7 was associated with a parallel large increase in IRF7 protein levels, but only in KSHV-infected DMVEC undergoing lytic cycle progression.
MATERIALS AND METHODS
KSHV-infected PEL cell lines and endothelial cells.
Adult human dermal microvascular endothelial cells (CC-2543; Clonetics, Walkersville, Md.) were cultured to passage 5 in EBM-2 medium (Clonetics) supplemented with EGM-2 MV Singlequots (Clonetics) and penicillin-streptomycin (Gibco-BRL, Rockville, Md.) at 37°C and 5% CO2. JSC1 and BCBL1 PEL cells (9) were cultured at 37°C and 5% CO2 in RPMI (Gibco-BRL) supplemented with 10% fetal calf serum (HyClone, Logan, Utah).
Harvest of KSHV virions for infection of endothelial cells.
JSC1 PEL cells were induced into lytic cycle replication with 20 ng of 12-O-tetradecanoyl phorbol 13-acetate (TPA) (Sigma, St. Louis, Mo.) for 120 h. The cells were removed by centrifugation at 1,200 rpm for 10 min. The supernatant containing KSHV virions from six 75-cm2 flasks (108 cells) was centrifuged at 11,000 rpm for 5.5 h at 4°C in a Sorvall RC-5B centrifuge or at 25,000 rpm with a sucrose cushion (40 mM Na2HPO4, 40 mM NaH2PO4, 150 mM NaCl, and 15% sucrose at pH 7.4 and 0.2 μm filtered) in an SW27 rotor and Beckman L7-55 ultracentrifuge for 1 h at 4°C. Pelleted virions were resuspended in 1 ml of phosphate-buffered saline (140 mM NaCl, 8 mM Na2HPO4, 2.7 mM KCl, and 1 mM KH2PO4) and added directly to DMVEC cells. The infected and control (uninfected) cells were cultured for a total of 3 weeks (Clontech, batch 1) or 5 weeks (Incyte, batch 2) and were split concurrently once during the culture period for the Clontech experiment or twice for the Incyte experiment. The growth medium was changed completely every 48 h for both the infected and uninfected cultures.
TPA treatment of infected DMVEC cells.
To examine any changes in the DMVEC cellular gene expression profile after increased KSHV lytic cycle infection compared to the cellular gene expression profile in the primarily latently infected DMVEC culture, half of the matched control and infected cultures were treated with 20 ng of TPA (Sigma) per ml for 60 h following the 3- to 5-week growth period, while the other half of the matched uninfected and infected cultures was harvested in parallel without TPA treatment. For both batches of infected cell cultures, these four separate RNA samples (uninfected −TPA/infected −TPA and uninfected +TPA/infected +TPA) were analyzed as matched pairs on the arrays with the expectation that many genes would be similarly regulated in both comparisons.
Clontech microarray methodology.
Following a 3-week growth period, cells were harvested from all four sets of flasks by scraping, and total RNA was harvested from the cells using the PolyA Pure labeling system (Clontech, Palo Alto, Calif.) for isolation of polyadenylated RNA and the generation of gene-specific [α-32P]ATP-labeled cDNA probes in one procedure (ICN, Costa Mesa, Calif.). Each of the four probes was hybridized to both Clontech Human Array 1.2-I and Clontech Human Array 1.2-II nylon filters overnight as per the manufacturer's directions. The filters, which each represented 1,176 different human genes, were washed and exposed to a Fuji BAS IIIS Phosphorimager screen for 5 days. Each of the resulting eight images was analyzed on a Fuji Phosphorimager (Fujix BAS-1000), and the data were processed using Adobe Photoshop (v. 5.0.2), Clontech AtlasImage software, and Microsoft Excel. Selected results were confirmed by RT-PCR using 19 pairs of primers synthesized from Clontech database sequences as recommended by Clontech (Palo Alto, Calif.).
Incyte microarray methodology.
The ability to continuously passage both the primary uninfected DMVEC and the latently infected spindle cell cultures is severely limited (14). Therefore, to further passage the same infected and uninfected cultures used for the Clontech array experiments without beginning infection de novo as described above, fresh uninfected DMVEC cells at passage 5 were reseeded by cocultivation together with a one-tenth proportion of cells (i.e., 10:1 ratio) from either the already infected DMVEC spindle cell culture or the parallel uninfected DMVEC culture. The supplemented infected and uninfected cultures were then grown for a further 2 weeks in EBM-2 medium (Clonetics) containing EGM-2 MV Singlequots (Clonetics) and penicillin-streptomycin (Gibco-BRL). Again, one half of both the infected and uninfected cultures was treated with TPA for the final 60 h of growth to produce the same four sets of experimental flasks as for the Clontech experiments.
In each case, RNA was harvested from five 75-cm2 flasks (108 cells) using Trizol (Gibco-BRL), and polyadenylated RNA was isolated using two passes through OligoTex mRNA isolation columns (Qiagen, Valencia, Calif.), followed by ethanol precipitation and quantitation using the RiboGreen RNA quantitation kit (Molecular Probes, Eugene, Oreg.). Samples of the polyadenylated RNA (600 ng at 50 ng/μl) were shipped to Incyte Genomics for conversion to cDNA, incorporating indocarbocyanine (Cy3) and indodicarbocyanine (Cy5) fluorescent dye labeling. Samples were then competitively hybridized with two pairs of Human UniGEM V2.0 microarrays containing 9,182 sequence-verified human cDNA clones each (average size, 1,000 bp).
The results were analyzed from a direct comparison of first the infected DMVEC RNA compared to uninfected DMVEC RNA pair in the absence of TPA and second the parallel pair of uninfected with infected cell RNA from the TPA-treated cultures. Data analysis was performed using GemTools 2.4, Microsoft Excel, the Dragon database (7), and Nomad (http://pevsnerlab.kennedykrieger.org/nomad.htm).
Calculation of expression ratios for the Clontech and Incyte data.
Original data obtained from analysis of Adobe Photoshop images of the Clontech arrays with Atlas Image software consisted of a continuum of hybridization intensity values (pixels) for each of the spots corresponding to a gene represented on the arrays. In addition, a global background value for each array was measured. For each of the Clontech experiments, two arrays, one corresponding to the infected or TPA-treated RNA sample, and one corresponding to the uninfected or untreated control RNA sample, were compared to obtain an expression ratio. To calculate expression ratios from the original intensity data, each set of arrays was global mean normalized. Spots in which the intensity value was below background on both the infected and uninfected arrays were removed from the analysis.
For each array, the intensity values for all of the remaining spots were then averaged, resulting in an average intensity for the spots on that particular membrane. In order to calculate the expression ratio, the original intensity value of each spot on the array was divided by the average intensity of all of the spots on the array to yield an adjusted intensity. Finally, the adjusted intensities of the control uninfected arrays were compared with the adjusted intensities of their corresponding infected arrays to calculate ratios and standard deviations (C-SD) (see Tables 2 and 3).
TABLE 2.
Most Up regulated and downregulated genes on the Clontech −TPA arrays after KSHV infectiona
| Regulation | Rank | C-SD | I (%) | I-R | Protein | Unigene | Intensity
|
|
|---|---|---|---|---|---|---|---|---|
| Uninfected | Infected | |||||||
| Up | 1, 3 | +6.5, +5.4 | 96.2 | +1.5 | Neurogranin | Hs.26944 | 2,620 | 16,700 |
| 2 | +5.6 | 98.3 | +1.9 | Calgranulin B (migration inhibitory protein 14) | Hs.112405 | 190 | 12,640 | |
| 4 | +4.9 | 88.4 | +1.3 | CD14 monocyte differentiation protein | Hs.75627 | 2,280 | 12,890 | |
| 5 | +4.6 | 85.2 | +1.2 | Fibrinogen B beta | Hs.7645 | 3,290 | 13,620 | |
| 6 | +4.5 | — | — | Lipopolysaccharide binding protein | Hs.154078 | 140 | 8,280 | |
| 7, 31 | +4.2, +2.2 | 93.2 | +1.4 | IRF7 (interferon regulatory factor 7) | Hs.166120 | 2,580 | 7,110 | |
| 8 | +4.1 | 99.9 | +8.5, +4.5 | RDC1 G-protein-coupled receptor | Hs.23016 | 2,830 | 11,670 | |
| 9 | +3.9 | — | — | Homeobox protein HB24 | M.60721 | 20 | 6,730 | |
| 10 | +3.7 | 0.9 | −2.4 | Ubiquitin-conjugating enzyme E2H10 | Hs.93002 | 2,010 | 10,300 | |
| 11 | +3.6 | — | — | Osteopontin | Hs.313 | 10 | 6,030 | |
| 12 | +3.4 | NR | Estrogen-related receptor alpha | Hs.110849 | 360 | 6,210 | ||
| 13 | +3.2 | NR | TIMP1 (inhibitor of metalloproteinases 1) | Hs.5831 | 21,030 | 36,640 | ||
| 14 | +3.1 | — | — | Steroid 5-alpha reductase 1 | Hs.552 | 2,130 | 8,220 | |
| 15 | +3.0 | — | — | Calgizzarin | Hs.256290 | 3,350 | 9,560 | |
| 16 | +3.0 | 97.1 | +1.6 | RIG-E (stem cell antigen 2) | Hs.77667 | 3,470 | 9,910 | |
| 17 | +2.8 | — | — | CBL-B | Hs.3144 | 1,000 | 6,360 | |
| 18 | +2.8 | — | — | Dual-specificity protein phosphatase-7 | Hs.296938 | 6,420 | 3,380 | |
| 19 | +2.7 | — | — | Leukocyte elastase inhibitor | Hs.183583 | 3,680 | 9,860 | |
| 20 | +2.6 | NR | MIP3 beta (macrophage inflammatory protein) | Hs.50002 | 3,290 | 8,590 | ||
| 21 | +2.5 | — | — | HI05E3 protein | Hs.57698 | 3,700 | 9,030 | |
| 30 | +2.2 | NR | MCM5 (minichromosome maintenance 5) | Hs.77171 | 7,810 | 13,930 | ||
| 33 | +2.1 | NR | Bcl3 (B-cell lymphoma 3 protein) | Hs.31210 | 820 | 4,460 | ||
| 36 | +2.1 | NR | IL8 | Hs.624 | 5,890 | 11,220 | ||
| 43 | +2.0 | NR | TGFβ3 | Hs.2025 | 800 | 4,480 | ||
| Down | 1 | −7.1 | 99.8 | −4.7 | BMP4 (bone morphogenetic protein 4) | Hs.68879 | 23,270 | 2,470 |
| 2 | −6.4 | — | — | MCP1-RA beta chemokine receptor (CCR2) | Hs.395 | 17,150 | 1,090 | |
| 3 | −4.9 | 99.9 | −5.6 | Connexin 37 (gap junction alpha 4) | Hs.296310 | 10,270 | 1,140 | |
| 4 | −4.8 | 99.5 | −3.1 | BMSA1 (bone marrow stromal antigen 1) | Hs.169998 | 13,570 | 1,890 | |
| 5, 23 | −4.6, −2.7 | 99.2 | −2.5 | PRP (major prion protein) | Hs.74621 | 10,110 | 350 | |
| 6 | −4.4 | 98.6 | −1.9 | Skeletal muscle LIM-protein 1 | Hs.239069 | 11,770 | 4,840 | |
| 7 | −4.3 | 96.9 | −1.6 | p57-KIP2 cell cycle inhibitor | Hs.106070 | 12,440 | 2,170 | |
| 8 | −3.8 | 97.1 | −1.6 | β1-Catenin | Hs.171271 | 9,520 | 1,300 | |
| 9 | −3.8 | — | — | Estradiol 17 β-dehydrogenase 2 | Hs.155109 | 9,610 | 2,420 | |
| 10, 26 | −3.6, −2.5 | — | — | GADD45 growth arrest DNA-damage protein 45 | Hs.110571 | 7,380 | 650 | |
| 11 | −3.5 | 99.9 | −11.7 | PAI-1 endothelial plasminogen activator inhibitor-1 | Hs.82085 | 18,850 | 7,720 | |
| 12 | −3.5 | 99.6 | −3.4 | RhoB GDP dissociation inhibitor | Hs.204354 | 11,210 | 2,930 | |
| 13 | −3.3 | NR | Atrophin-1 | Hs.169488 | 11,500 | 3,430 | ||
| 14 | −3.2 | NR | G1S-specific cyclin D1 | Hs.82932 | 11,360 | 3,620 | ||
| 15 | −3.1 | NR | Alzheimer's disease amyloid A4 protein | Hs.177486 | 22,400 | 11,150 | ||
| 16 | −3.1 | 99.4 | −2.9 | BMP6 (bone morphogenetic protein 6) | Hs.285671 | 11,030 | 2,810 | |
| 17 | −3.1 | 99.5 | −3.1 | IGFBP10 cysteine-rich angiogenic inducer, cyr61 | Hs.8867 | 7,410 | 2,000 | |
| 18 | −3.1 | — | — | R-ras2 (TC21) | Hs.206097 | 6,690 | 1,390 | |
| 19 | −3.1 | 94.3 | −1.4 | Endothelial cell multimerin | Hs.268107 | 10,270 | 4,080 | |
| 20 | −3.0 | 21.8 | +1.2 | Elk3 Ets domain protein | Hs.121529 | 10,870 | 3,620 | |
| 22 | −2.7 | — | — | HAT-B2 (histone acetyl transferase B subunit 2) | Hs.31314 | 5,430 | 370 | |
| 25 | −2.5 | NR | MCP1 (macrophage chemotactic protein 1) | Hs.340 | 26,470 | 17,900 | ||
| 28 | −2.5 | 96.5 | −1.5 | Reelin | Hs.12246 | 10,880 | 5,610 | |
| 44 | −2.0 | 99.3 | −2.5 | α3-Integrin (CD49C) | Hs.265820 | 12,320 | 5,770 | |
Genes are listed in rank order 1 through 20, together with several others selected from among the top 50 genes. Hybridization intensity scores are background corrected but not normalized or mean adjusted. R, Clontech numerical rank order for KSHV-regulated genes, comparing each infected RNA to its matching uninfected RNA sample (batch 1). C-SD, Clontech values for standard deviation from the mean (+ for upregulated, − for downregulated). I-R, Incyte ratio values, with + for upregulated and − for downregulated; see Tables 4 and 5; I (%), Incyte rank order as a percentile. Only values above 80% or below 20% are included. —, gene not represented on array. NR, not regulated [i.e., I(%) between 20 and 80% in the Incyte array]. ND, no data.
TABLE 3.
Most upregulated and downregulated genes on the Clontech +TPA arrays after KSHV infectiona
| Regulation | Rank | C-SD | I(%) | I-R | Gene name | Unigene | Intensity
|
|
|---|---|---|---|---|---|---|---|---|
| Uninfected | Infected | |||||||
| Up | 1 | +7.6 | — | — | Metallothionein (MTH1) | Hs.83326 | 4,470 | 25,030 |
| 2 | +6.3 | — | — | Exodus 2 beta chemokine | Hs.57907 | 3,110 | 17,360 | |
| 3 | +5.4 | 93.1 | +1.7 | RAD23A UV excision repair protein (HHR23A) | Hs.180455 | 1,440 | 7,780 | |
| 4 | +4.9 | — | — | Osteopontin | Hs.313 | 740 | 8,200 | |
| 5 | +4.7 | — | — | DCR3 decoy receptor 3 | Hs.278556 | 1,270 | 9,280 | |
| 6 | +4.6 | 99.7 | +2.8, +2.6 | RDC1 G-protein-coupled receptor homolog | Hs.23016 | 1,470 | 11,770 | |
| 7 | +4.2 | — | — | Tripeptidyl-peptidase 1 (TPP) | Hs.20478 | 4,130 | 10,620 | |
| 8 | +4.1 | NR | Proteasome component C9 | Hs.251531 | 990 | 5,110 | ||
| 9 | +3.9 | ND | Calgranulin B (migration inhibitory factor MRP14) | Hs.112405 | 570 | 4,340 | ||
| 10 | +3.6 | 17.4 | −1.2 | TIMP1 inhibitor of metalloproteinases | Hs.5831 | 25,420 | 33,390 | |
| 11 | +3.6 | NR | Cytochrome P450 II A6 | Hs.183584 | 1,020 | 5,890 | ||
| 12 | +3.6 | NR | Bcl3 (B-cell lymphoma 3) | Hs.31210 | 4,570 | 11,400 | ||
| 13 | +3.5 | — | — | TIMP4 inhibitor of metalloproteinases | Hs.190787 | 1,620 | 5,060 | |
| 14 | +3.3 | 86.9 | +1.5 | Interferon-induced leucine zipper protein | Hs.50842 | 4,960 | 8,540 | |
| 15, 27 | +3.2, +2.5 | 89.5 | +1.6 | IRF7 (interferon regulatory factor 7) | Hs.166120 | 940 | 2,960 | |
| 16 | +3.2 | 10.0 | −1.3 | RAMP2 (receptor activation modifier 2) | Hs.155106 | 8,050 | 16,220 | |
| 17 | +3.1 | 96.1 | +1.8 | CDC25B—cell division cycle protein | Hs.153752 | 3,560 | 7,050 | |
| 18 | +3.1 | 00.2 | −4.5 | PAI-1 endothelial plasminogen activator inhibitor-1 | Hs.82085 | 4,960 | 8,550 | |
| 19 | +3.0 | — | — | Smooth muscle cell LIM protein | Hs.10526 | 480 | 3,880 | |
| 20 | +2.8, +1.3 | 94.3 | +1.7 | Neurogranin | Hs.26944 | 3,560 | 6,450 | |
| 22 | +2.7 | NR | ICAM2 (intracellular adhesion molecule 2) | Hs.83733 | 2,730 | 6,300 | ||
| 26 | +2.5 | — | — | H105E3 | Hs.57698 | 11,845 | 18,370 | |
| 28 | +2.5 | NR | Fibrinogen Bβ | Hs.166120 | 15,610 | 23,120 | ||
| 33 | +2.4 | — | — | VWF | Hs.110802 | 11,650 | 17,760 | |
| 36 | +2.4 | — | — | RIG-E (stem cell antigen 2) | Hs.77667 | 6,400 | 11,120 | |
| Down | 1 | −5.0 | NR | Nucleobindin | Hs.172609 | 15,960 | 4,830 | |
| 2 | −4.9 | 81.5 | −1.2 | CXCR4 stromal cell alpha chemokine receptor | Hs.89414 | 13,840 | 4,910 | |
| 3 | −4.6 | NR | IL-8 | Hs.624 | 14,520 | 5,450 | ||
| 4 | −4.4 | — | — | Notch3 homologue | 14,090 | 3,470 | ||
| 5 | −4.0 | 82.7 | −1.2 | GRK4 G-protein-coupled receptor kinase (GPRK2L) | Hs.32959 | 17,090 | 4,980 | |
| 6 | −3.8 | 99.8 | −4.3 | Thrombin receptor (coagulation factor II) | Hs.128087 | 9,670 | 2,890 | |
| 7 | −3.8 | NR | Inhibin β-B subunit | Hs.1735 | 12,070 | 2,250 | ||
| 8 | −3.7 | 97.2 | −1.7 | Cytosolic dynein heavy chain | I.23958 | 17,790 | 6,150 | |
| 9 | −2.9 | NR | Thy1 membrane glycoprotein | Hs.125359 | 26,260 | 13,350 | ||
| 10 | −2.9 | 19.2 | +1.4 | Phosphatidylethanolamine-binding protein | Hs.80423 | 9,810 | 2,380 | |
| 11 | −2.9 | NR | Machado-Joseph disease protein 1 (MJD1) | Hs.66521 | 3,970 | 190 | ||
| 12 | −2.8 | 93.0 | −1.4 | Junction plakoglobin, desmoplakin III | Hs.2340 | 10,110 | 4,760 | |
| 13 | −2.8 | — | — | Glycine amidinotransferase | Hs.75335 | 5,870 | 180 | |
| 14 | −2.8 | NR | Guanine nucleotide exchange factor MSS4 | Hs.90875 | 15,320 | 6,330 | ||
| 15 | −2.7 | 95.8 | −1.5 | c-Maf transcription factor | Hs.30250 | 4,870 | 530 | |
| 16 | −2.6 | — | — | Gastric triacylglycerol lipase | Hs.159177 | 3,910 | 11,360 | |
| 17 | −2.6 | 91.9 | −1.4 | Reelin | Hs.12246 | 7,900 | 1,540 | |
| 18 | −2.6 | 93.2 | −1.4 | LIM and SH3 domain protein LASP-1 | Hs.75080 | 5,380 | 12,260 | |
| 19 | −2.5 | 93.4 | −1.4 | Phospholipase C-gamma-2 | Hs.75648 | 2,710 | 560 | |
| 20 | −2.5 | — | — | Phosphomevalonate kinase | Hs.30954 | 10,110 | 3,250 | |
| 21 | −2.5 | NR | CD40 receptor associated factor | Hs.89676 | 4,870 | 1,620 | ||
| 23 | −2.4 | NR | Cyclin A1 (CNN1) | Hs.86137 | 1,750 | 230 | ||
| 29 | −2.2, −1.8 | — | — | WNT 13 | Hs.258575 | 9,420 | 5,890 | |
| 31 | −2.1 | NR | bFGF-R (basic fibroblast growth factor receptor) | Hs.748 | 9,030 | 5,560 | ||
| 32 | −2.1 | — | — | GRK5 (G-protein receptor kinase 5) | Hs.311569 | 5,140 | 2,700 | |
See Table 2, footnote a.
Incyte expression ratios were analyzed based on the normalized intensity ratios (I-R; see Tables 4 and 5). Background intensity values were subtracted from each intensity signal, and data from Cy3 and Cy5 channels were globally normalized so that total fluorescence intensity in each channel was equal.
TABLE 4.
Most upregulated and downregulated genes on the Incyte −TPA array after KSHV infectiona
| Regulation | Rank | Gene | C-SD | Accession no. | I-R | Unigene |
|---|---|---|---|---|---|---|
| Up | 1 | Incyte EST | + | +13.8 | ||
| 2, 20 | RDC1 orphan G-protein-coupled receptor | +& +4.1 | U67784 | +8.5, +4.5 | Hs.23016 | |
| 3 | Antigen identified by monoclonal antibody MRC OX-2 | AL134591 | +7.9 | Hs.79015 | ||
| 4 | Myx RI (myxovirus resistance I, IFN-inducible p78) | + | AF135187 | +7.7 | Hs.76391 | |
| 5, 42 | SISI-3 (STAT-induced STAT inhibitor 3) | AB006967 | +7.4, +2.9 | Hs.296176 | ||
| 6 | Interferon-induced protein 56 | NM001548 | +7.2 | Hs.20315 | ||
| 7 | Interferon-induced transmembrane protein 3 (1-8U) | + | BF033678 | +6.4 | Hs.182241 | |
| 8 | PLAT (tissue plasminogen activator) | M18182 | +5.8 | Hs.274404 | ||
| 9 | Mal. T-cell differentiation protein | NM002371 | +5.3 | Hs.80395 | ||
| 10 | Interferon alpha-inducible protein (clone IFI-6-16) | + | BE407364 | +5.2 | Hs.265827 | |
| 11 | IGFBP5 (insulin-like growth factor binding protein 5) | + | AU132011 | +5.2 | Hs.103391 | |
| 12 | Thromboinodulin | $ | M16552 | +4.8 | Hs.2030 | |
| 13 | Paired basic amino acid cleaving system 4 | + | D28513 | +4.8 | Hs.170414 | |
| 14 | Complement component 1, r subcomponent | M14058 | +4.6 | Hs.1279 | ||
| 15 | Nucleoside phosphorylase | BE741350 | +4.6 | Hs.75514 | ||
| 16 | α2-Macroglobulin | AU119825 | +4.6 | Hs.74561 | ||
| 17 | TGFβ3 | +& +2.0 | X14885 | +4.5 | Hs.2025 | |
| 18 | γ-Tubulin complex protein 2 | + | AF042379 | +4.5 | Hs.13386 | |
| 19 | Interferon-induced transmembrane protein I (9-27) | BF184283 | +4.5 | Hs.146360 | ||
| 21 | IL-13 | NM002188 | +4.4 | |||
| 26 | Angiopoietin 2 | (+ −2.7) | NM001147 | +4.1 | ||
| 28 | Myx R2 (myxovirus resistance 2) | M30818 | +3.9 | Hs.926 | ||
| 31 | HIF-1α hypoxia-induced factor | AF207601 | +3.8 | Hs.197540 | ||
| 37 | vEts oncogene transcription factor (E26-2) | AF017257 | +3.2 | Hs.85146 | ||
| 40 | STAT-induced STAT inhibitor 2 | NM003877 | +3.0 | |||
| 44 | IL-1 R (interleukin-1 receptor) | M29492 | +2.8 | Hs82112 | ||
| 55 | TGFβ receptor βIII | AJ25610 | +2.6 | Hs.79059 | ||
| Down | 1 | Connective tissue growth factor | +$ | U14750 | −14.1 | Hs.75511 |
| 2 | Fibronectin 1 | + | AW385690 | −12.6 | Hs.287820 | |
| 3 | BARD1 (BRCA1-associated protein 1) | + | NM_006768 | −12.2 | Hs.122764 | |
| 4 | Cardiac ankyrin repeat protein | + | X83703 | −11.8 | Hs.74019 | |
| 5 | PAI-1 plasminogen activator inhibitor, type I | +& −3.5 | J03764 | −11.7 | Hs.82085 | |
| 6, 33 | DROC downregulated in ovarian cancer I | NM_014890 | −7.1, −3.4 | Hs.15432 | ||
| 7 | Keratin 7 | M13955 | −6.7 | Hs.23881 | ||
| 8 | TC-1, function unknown | + | AK026240 | −6.4 | Hs.283683 | |
| 9 | Granzyme K (serine protease, granzyme 3; tryptase II) | $ | NM_002104 | −6.1 | Hs.3066 | |
| 10 | Biglycan | + | U82695 | −6.0 | ||
| 11 | Solute carrier family 21 (member 3) | NM_005075 | −5.8 | Hs.46440 | ||
| 12 | Connexin 37 (gap junction protein α4) | & −4.9 | AL121988 | −5.6 | Hs.296310 | |
| 13 | TSP1 thrombospondin 1 | + | NM_003246 | −5.3 | Hs.87409 | |
| 14 | Endothelin 1 | + | NM_001955 | −5.2 | Hs.2271 | |
| 15 | TGFα (transforming growth factor) | NM_003236 | −5.2 | Hs.170009 | ||
| 16 | Syndecan 2 (heparan sulfate proteoglycan 1) | $ | AK025488 | −5.2 | Hs.1501 | |
| 17 | MMP2 (matrix metalloproteinase 2, type IV collagenase) | +$ | J03210 | −5.1 | Hs.111301 | |
| 18 | Transgelin | BE378718 | −5.0 | Hs.75777 | ||
| 19 | α4-Integrin (α4 subunit of VLA-4 receptor) | + | NM_000885 | −4.8 | Hs.40034 | |
| 20 | BMP4 bone morphogenetic protein 4 | & −7.1 | M22490 | −4.7 | Hs.68879 | |
| 23 | Keratin 7 | AA307373 | −4.1 | Hs.23881 | ||
| 25 | Urokinase plasminigen activator | M15476 | −3.9 | Hs.77274 | ||
| 26 | Stromal cell-derived factor 1 | + | AL137026 | −3.9 | Hs.237356 | |
| 27 | GRK4 G-coupled receptor kinase | (& −4.0) | U27768 | −3.9 | Hs.227571 | |
| 40 | IGFBP 10 cysteine-rich angiogenic inducer, cyr61 | +& −3.1 | Z98053 | −3.1 | Hs.8867 | |
| 41 | BMSA1 (bone marrow stromal antigen 1) | & −4.8 | AI219377 | −3.1 | Hs.169998 | |
| 46 | BMP6 bone morphogenic protein | & −3.1 | AA426586 | −2.9 | Hs.285671 |
Genes are listed in rank order 1 through 20, together with several others selected from among the top 50 genes. R, Incyte numerical rank order for KSHV-regulated genes, comparing each infected RNA to its matching uninfected RNA sample (batch 2). I-R, Incyte ratio values (+ for upregulated, − for downregulated). C-SD, Clontech standard deviation values. +, also found within the top 40 genes regulated in the same direction (up or down) on the +TPA Incyte array. $, also represented on the Clontech −TPA or +TPA array (Tables 2 and 3). &, regulated to a statistically significant level in the same direction on the appropriate Clontech −TPA or +TPA array (Tables 2 and 3). +TPA version standard deviations are in parentheses.
TABLE 5.
Most upregulated and downregulated genes on the Incyte +TPA array after KSHV infectiona
| Regulation | Rank | Gene | C-SD | Accession no. | I-R | Unigene |
|---|---|---|---|---|---|---|
| Up | 1 | Myx R1 (myxovirus resistance 1, IFN-inducible p78) | + | AF135187 | +26.4 | Hs.76391 |
| 2 | Interferon-induced protein 56 (tetratricopeptide repeats 1) | NM_001548 | +24.8 | Hs.20315 | ||
| 3 | Incyte EST | + | +12.1 | |||
| 4 | Interferon alpha-inducible protein (clone IFI-6-16) | + | BE407364 | +9.3 | Hs.265827 | |
| 5 | Interferon-induced protein 54 (tetratricopeptide repeats 2) | A1609624 | +8.2 | Hs.169274 | ||
| 6 | Myx R2 (myxovirus/influenza virus resistance 2) | M30818 | +8.2 | Hs.926 | ||
| 7 | Statherin | M18371 | +4.3 | Hs.37048 | ||
| 8 | γ-Tubulin complex protein 2 | + | AF042379 | +4.2 | Hs.13386 | |
| 9 | ESTs | AW021108 | +3.6 | Hs.16450 | ||
| 10 | Histatin 3 | AW963859 | +3.5 | Hs.177888 | ||
| 11 | Interferon-induced protein 75, 52 kDa | AF280095 | +3.5 | Hs.38125 | ||
| 12 | Lectin, galactoside binding, soluble, 9 (galectin 9) | AB005894 | +3.5 | Hs.81337 | ||
| 13 | Guanylate binding protein 1, interferon inducible, 67 kDa | M55542 | +3.5 | Hs.62661 | ||
| 14 | 2′-5′oligoadenylate synthetase 2 | NM_016817 | +3.4 | Hs.264981 | ||
| 15 | IGFBP5 (insulin-like growth factor binding protein 5) | + | AU132011 | +3.3 | Hs.103391 | |
| 16 | Paired basic amino acid cleaving system 4 | + | D28513 | +3.3 | Hs.170414 | |
| 17 | IFN-induced hepatitis C virus associated microtubular aggregate | NM_006417 | +3.2 | Hs.82316 | ||
| 18 | Interferon-induced transmembrane protein 3 (1-8U) | + | BF033678 | +3.1 | Hs.182241 | |
| 19 | STAT1 (signal transducer and activator of transcription) | $ | AA478534 | +3.0 | Hs.21486 | |
| 20 | RIG-E (lymphocyte antigen 6 complex) | & +2.4 | U42376 | +3.0 | Hs.77667 | |
| 24, 28 | RDC1 orphan chemokine receptor | +& +4.6 | U67784 | +2.8, +2.6 | Hs.23016 | |
| 26 | IFN-induced dsRNA-dependent protein kinase | AW270525 | +2.6 | Hs.274382 | ||
| 38 | SISI3 (STAT-induced STAT inhibitor 3) | + | AB006967 | +2.4 | Hs.296176 | |
| 40 | TGFβ3, transforming growth factor | + | X14885 | +2.3 | Hs.2025 | |
| Down | 1 | Fibronectin 1 | + | AW385690 | −13.6 | Hs.287820 |
| 2 | Connective tissue growth factor | +$ | U14750 | −13.4 | Hs.75511 | |
| 3 | BARDI (BRCA1-associated protein 1) | + | NM_006768 | −11.9 | Hs.122764 | |
| 4 | Matrix Gla protein | $ | AV733652 | −8.4 | Hs.279009 | |
| 5 | Stromal cell-derived factor 1 | + | AL137026 | −6.2 | Hs.237356 | |
| 6 | CD34 antigen | AL035091 | −5.3 | Hs.85289 | ||
| 7 | Cardiac ankyrin repeat protein | + | X83703 | −5.2 | Hs.74019 | |
| 8 | MMP2 (matrix metalloproteinase 2, type IV collagenase) | +$ | J03210 | −5.2 | Hs.111301 | |
| 9 | Endothelin 1 | + | NM_001955 | −5.2 | Hs.2271 | |
| 10 | TC-1, function unknown | + | AK026240 | −5.1 | Hs.283683 | |
| 11 | RhoB GDP dissociation inhibitor | $ | AV704811 | −4.9 | Hs.204354 | |
| 12 | Apolipoprotein D | J02611 | −4.9 | Hs.75736 | ||
| 13 | α4-Integrin (α4 subunit of VLA-4 receptor) | + | NM_000885 | −4.6 | Hs.40034 | |
| 14 | PAI-1 (plasminogen activator inhibitor, type 1) | +$ +3.1 | J03764 | −4.5 | Hs.82085 | |
| 15 | Placental growth factor; VEGF-related protein | $ | A1004656 | −4.4 | Hs.2894 | |
| 16 | Thrombin receptor (coagulation factor II) | & −3.8 | NM_001992 | −4.3 | Hs.128087 | |
| 17 | TSP1 (thrombospondin 1) | + | NM_003246 | −4.2 | Hs.87409 | |
| 18 | β1-Catenin (cadherin-associated protein) | $(& −4.0) | Z19054 | −4.1 | Hs.171271 | |
| 19 | Biglycan | + | U82695 | −4.1 | ||
| 20 | IGFBP10 cysteine-rich angiogenic inducer 61 (cyr61) | +$ | Z98053 | −4.0 | Hs.8867 | |
| 25 | DROC downregulated ovarian cancer | + | NM014890 | −3.6 | ||
| 28 | Rhoβ GDP dissociation inhibitor | & | L07916 | −3.4 | Hs.83656 | |
| 40 | Angiopoietin 2 | NM001147 | −2.7 | Hs.288204 |
See Table 4, footnote a, except + indicates also found within the top 40 genes regulated in the same direction on the −TPA Incyte array. −TPA version standard deviations are in parentheses.
Determination of the significance of expression ratio values.
Genes represented on the Clontech arrays were considered to be significantly regulated if their expression ratio was greater than 2 standard deviations from the mean of the data set. Incyte data were determined to be significant if the expression ratio was equal to or above 1.74 (upregulated) or equal to or below −1.74 (downregulated), as recommended by the company. The expression ratios presented do not correspond to a “fold” up- or downregulation because of the incorporation of reverse transcription and PCR (in the case of Incyte data) in the steps in the preparation for hybridization of the initial polyadenylated samples.
RT-PCR analyses.
Direct confirmation of up- or downregulated RNA expression for selected genes based on the gene array data was carried out using four separate sets of RNA preparations. This included those used for the Clontech arrays (batch 1) and Incyte arrays (batch 2) plus two new preparations (batches 3 and 4) derived from DMVEC cultures infected by cocultivation passaging procedures similar to that described for the Incyte array RNA from frozen stocks of KSHV (JSC1)-infected DMVEC spindle cells. Three separate RT-PCR protocols were employed. Total RNA was harvested from cells using either Trizol (Gibco-BRL) for batches 1 and 2 or an RN-Easy mini kit (Qiagen) and subjected to RNase-free DNase I (Gibco-BRL) treatment and then reverse transcribed to prepare cDNA at 50°C for 50 min using Superscript II (Gibco-BRL).
In the first semiquantitative procedure (see Fig. 6 and Table 7), 10 sets of four single PCR time course reactions with different primer pairs were carried out in parallel with both infected and uninfected RNA samples using Taq DNA polymerase (Promega, Madison, Wis.). In each set of reactions, three test primer pairs were included together with a control primer pair for cytoplasmic β-actin. A total of 19 different gene primer pairs based on the Clontech results were evaluated with either or both of the −TPA or +TPA cDNA sample pairs. For manual quantitation, samples were removed every three cycles, beginning with cycle 18 and continuing through cycle 45. Progressive PCR samples were visualized after agarose gel electrophoresis and ethidium bromide staining, and then the results were quantitated on an AlphaImager 2000 (Alpha Innotech, San Leandro, Calif.) and histograms of fold regulation after normalizing to β-actin were prepared for those samples displaying exponential responses.
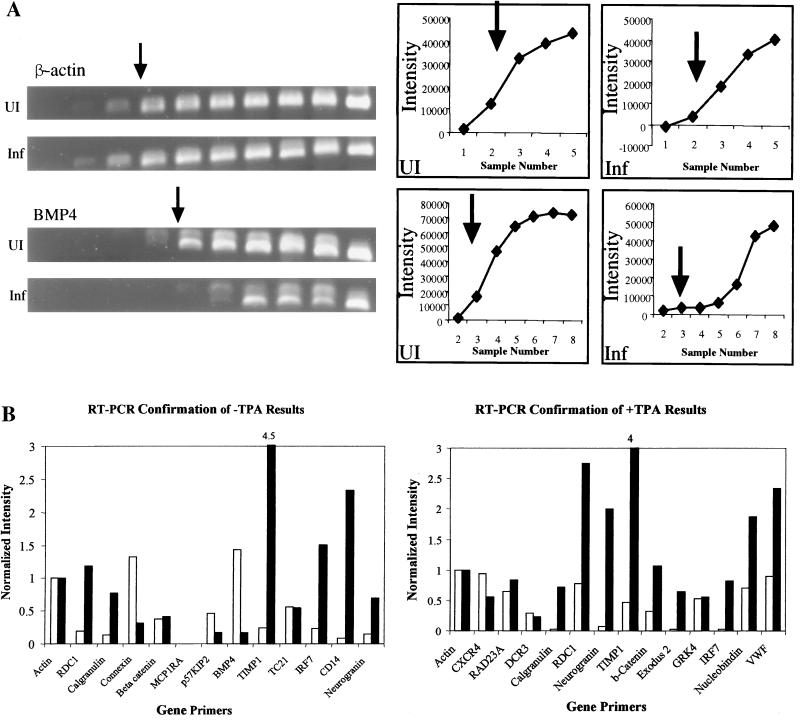
FIG. 6.
Semiquantitative direct RT-PCR for confirmation of selected regulated Clontech genes. (A) Representative ethidium bromide-stained DNA products after agarose gel electrophoresis. Upper panel: control sample amplification of β-actin in uninfected and infected DMVEC RNAs, with IDV quantitation graphed to the right. Lower panel: test sample amplification of BMP4 in the same uninfected and infected DMVEC RNA samples, with IDV quantitation graphed to the right. (B) Histogram presentation of the relative IDV measurements for all 18 gene primer pairs examined, as shown in the example above. Uninfected RNAs are represented by open bars, and infected RNAs are represented by solid bars. Upper panel: KSHV-infected DMVEC in the absence of TPA. Lower panel: KSHV-infected DMVEC in the presence of TPA.
TABLE 7.
Summary of semiquantitative RT-PCR results compared to gene array data for highly regulated Clontech array genesa
| Regulation | Gene | RT-PCR fold regulation (RNA batch 1)
|
Gene array values
|
||||
|---|---|---|---|---|---|---|---|
| Clontech (RNA batch 1)
|
Incyte (RNA batch 2)
|
||||||
| I/U | I+T/U+T | I/U | I+T/U+T | I/U | I+T/U+T | ||
| Up | Calgranulin B | +6.0 | +23.7 | +5.6 | +3.9 | 1.9 | NR |
| CD14 | +26.9 | +4.9 | NR | (+1.3) | NR | ||
| DCR3 | −1.2 | NR | +4.7 | — | — | ||
| Exodus 2 | +20.1 | NR | +6.3 | — | — | ||
| IRF7 | +6.4 | +25.6 | +4.2 | +3.2 | (+1.4) | (+1.6) | |
| Neurogranin | +4.6 | +26 | +6.5 | +2.8 | (+1.5) | (+1.7) | |
| RAD23A | +1.3 | NR | +5.5 | NR | (+1.9) | ||
| RDC1 | +6.1 | +3.6 | +4.2 | +4.6 | +8.5 | +2.8 | |
| TIMP1 | +18.1 | +8.7 | +3.2 | +3.6 | NR | NR | |
| VWF | +2.6 | NR | +2.4 | — | — | ||
| Down | BMP4 | −5.0 | −4.3 | NR | −7.1 | −2.2 | |
| β1-Catenin | −1.1 | +3.3 | −3.8 | NR | (−1.6) | −4.1 | |
| Connexin 37 | −5.0 | −4.9 | NR | −5.6 | NR | ||
| CXCR4 | −1.7 | NR | −4.9 | NR | (−1.2) | ||
| GRK4 | +1.0 | NR | −4.0 | NR | (−1.2) | ||
| Nucleobindin | +2.7 | NR | −5.0 | NR | NR | ||
| p57-KIP2 | −2.5 | −4.3 | NR | NR | NR | ||
| TC21 | +1.0 | −3.1 | NR | — | — | ||
The four cDNAs used, uninfected (U), infected (I), uninfected TPA treated (U+T), and infected TPA treated (I+T), were the same preparations (batch 1) used for the Clontech gene array experiments. The fold regulated values given are normalized to parallel β-actin controls and derived from the histogram results in Fig. 6B. Gene array values are C-SD for Clontech and I-R for Incyte. Values in parentheses were not significant. NR, not regulated, —, not represented.
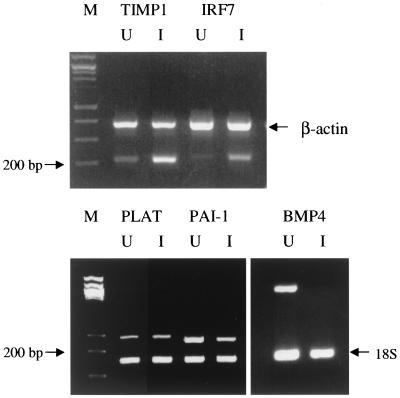
In the second Multiplex RT-PCR procedure (see Fig. 7), primer pairs (50 pmol) for the genes of interest as well as for 18S rRNA (internal reference) were amplified together in the same tube. Each PCR template (50 μl) contained equal amounts of cDNA from either the uninfected DMVEC or KSHV-infected DMVEC RNA sample from batch 3 cells. PCR conditions were 95°C for 60 s, followed by 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s for 30 to 35 cycles.
FIG. 7.
Multiplex RT-PCR results with selected up- and downregulated genes in KSHV-infected DMVEC. The photographs show examples of ethidium bromide-stained DNA products after PCR of cDNA prepared from uninfected (U) and infected (I) DMVEC RNA samples (RNA batch 3). Test primers used included BMP4 (378 bp), TIMP1 (215 bp), IRF7 (223 bp), PAI-1 (239 bp), and PLAT (281 bp). Either 18S rRNA or cytoplasmic β-actin primers were used as internal controls with each PCR template sample. Lane M, size markers.
In the third quantitative real-time procedure (see Tables 8 and 9), DNA analysis and reverse transcription were carried out using Taqman RT reagents (PE Biosystems, Inc.). cDNA was prepared from all four samples of batch 2 RNA, the two samples from batch 3 RNA, and the three samples for batch 4 RNA using RT with 2.5 μM random hexamers at 25°C for 10 min, 37°C for 60 min, and 95°C for 5 min. In each case, minus-RT controls were negative.
TABLE 8.
Summary of quantitative real-time RT-PCR results for confirmation of selected highly regulated Incyte array genes in multiple RNA preparationsa
| Regulation | Gene | RT-PCR fold regulation
|
Gene array values
|
|||||
|---|---|---|---|---|---|---|---|---|
| RNA batch 3
|
RNA batch 4
|
Clontech (RNA batch 1)
|
Incyte (RNA batch 2)
|
|||||
| I/U | I/U | I+T/U | I/U | I+T/U+T | I/U | I+T/U+T | ||
| Up | Angiopoietin 2 | +6.2 | +3.2 | +9.9 | — | — | +4.1 | +2.7 |
| CD14 | +6.3 | +4.9 | NR | (+1.3) | NR | |||
| HIF1α | +1.1 | +1.7 | −1.3 | — | — | +3.5 | NR | |
| IRF7 | +5.4 | +10.4 | +4.2 | +3.2 | (+1.4) | (+1.6) | ||
| Metallothionine H1 | +2.7 | +9.2 | +4.0 | NR | +7.6 | — | — | |
| PLAT | +1.2 | +2.7 | +1.4 | — | — | +5.8 | +2.2 | |
| Myx R1 | +85 | +107 | +229 | — | — | +7.7 | +26.4 | |
| SISI3 | +8.0 | — | — | +7.4 | +2.4 | |||
| TGFβ3 | +5.8 | +2.0 | NR | +4.5 | +2.3 | |||
| Thrombomodulin | +9.2 | +2.0 | +5.3 | NR | NR | +4.8 | NR | |
| Down | BMP4 | −4.2 | −50 | −92 | −7.1 | NR | −4.7 | NR |
| CD34 | −2.6 | −1.2 | −4.6 | — | — | −2.3 | −5.3 | |
| Endothelin 1 | −6.2 | −28 | −89 | — | — | −5.1 | −5.2 | |
| Fibronectin | −40 | −57 | −312 | — | — | −12.6 | −13.6 | |
| α4-Integrin | −450 | −790 | −220 | — | — | −4.8 | −4.6 | |
| IL-8 | −100 | −8.9 | −20 | +2.1 | −4.6 | NR | NR | |
| MMP2 | −87 | NR | NR | −5.1 | −5.2 | |||
| PAI-1 | −56 | −29 | −28 | −3.5 | +3.1 | −11.7 | −4.5 | |
| Thrombospondin 1 | −6.5 | −7.9 | −37 | — | — | −5.3 | −4.2 | |
The fold regulation values given represent corrected average relative abundance calculations using 18S rRNA as an internal control for appropriate pairs of infected (I) and uninfected (U) DMVEC RNA samples as listed. Comparison of real-time RT-PCR results for batch 3 and batch 4 RNA with gene array values for primarily Incyte regulated genes. See Table 7, footnote a, for definitions.
TABLE 9.
Summary of quantitative real-time RT-PCR analysis of Incyte RNA samplesa
| Regulation | Gene name | RT-PCR fold regulation (RNA batch 2)
|
Gene array value
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clontech (RNA batch 1)
|
Incyte (RNA batch 2)
|
|||||||||||
| I/U | I+T/U+T | U+T/U | I+T/I | I+T/U | I/U | I+T/U+T | U+T/U | I+T/I | I/U | I+T/U+T | ||
| Up | Angiopoietin 2 | +3.1 | −5.0 | +3.7 | +2.4 | +7.5 | — | — | — | — | +4.1 | −2.7 |
| CD14 | +1.7 | +1.2 | +1.4 | −2.1 | −1.2 | +4.9 | NR | NR | NR | (+1.3) | NR | |
| Exodus 2 | +19 | +18.4 | −3.8 | −3.2 | +4.8 | (+1.6) | +6.3 | −6.9 | −4.1 | — | — | |
| IRF7 | +2.7 | +2.6 | +1.4 | +1.3 | +4.0 | +4.2 | +3.2 | NR | NR | (+1.4) | (+1.6) | |
| Metallothionine H1 | +1.1 | +1.2 | +1.1 | +1.2 | +1.2 | NR | +7.6 | −6.3 | NR | — | — | |
| Myx R1 | +17 | +80 | −2.5 | +32 | +32 | — | — | — | — | +7.7 | +26.4 | |
| SISI3 | +18 | +25 | −3.7 | −2.0 | +9.2 | — | — | — | — | +7.4 | +2.4 | |
| TGFβ3 | +6.7 | +3.5 | +1.1 | −1.8 | +3.8 | +2.0 | NR | NR | NR | +4.5 | +2.3 | |
| TIMP1 | +9.0 | +2.8 | +3.2 | +3.6 | +15.3 | +3.2 | +3.6 | +2.8 | NR | — | — | |
| RDC1 | +13.5 | +1.9 | +10.6 | +1.5 | +20.5 | +4.1 | +4.6 | NR | NR | +8.5 | +2.6 | |
| Down | BMP4 | −145 | −5.0 | −345 | +2.1 | −69 | −7.1 | NR | −5.6 | NR | −4.7 | NR |
| Connective tissue GF | −33 | −2.0 | −53 | −3.2 | −105 | NR | NR | +2.7 | +3.2 | −14.1 | −13.4 | |
| Fibronectin | −21.6 | −10.6 | −10.0 | −4.9 | −106 | — | — | — | — | −12.6 | −13.6 | |
| IL-8 | −50 | −3.9 | −35 | +5.5 | −9.1 | +2.1 | −4.6 | +5.4 | NR | NR | NR | |
| α4-Integrin | −325 | −139 | −3.4 | −1.5 | −480 | — | — | — | — | −4.8 | −4.6 | |
| MMP2 | −15.8 | −15.2 | +1.2 | +1.1 | −18 | NR | NR | NR | +3.2 | −5.1 | −5.2 | |
| PAI-1 | −17.3 | −1.6 | −4.1 | +2.6 | −6.6 | −3.5 | +3.1 | −3.3 | +3.2 | −11.7 | −4.5 | |
See Table 8, footnote a, except comparison was of real-time RT-PCR results with gene array values for batch 2 (Incyte) RNA in the absence and presence (+T) of TPA.
Real-time PCRs were carried out in triplicate with both 18S rRNA internal controls and no-template controls using cycle conditions of 2 min at 50°C, 95°C for 10 min, and then 40 to 45 cycles of 15 s at 95°C and 1 min at 60°C in 96-well plates on an ABI Prism 7700 sequence detector. Each 50-μl PCR contained 1 μl of relevant cDNA, 50 pmol of gene-specific primers, and 25 μl of SYBR Green I PCR Master Mix (PE Biosystems, Inc.). PCR product intensity data were normalized relative to 18S RNA and averaged and then analyzed with Sequence Detector System software version 1.7a (PE Applied Biosystems) to calculate fold values for the appropriate parallel sample combinations based on exponential-phase measurements (800-fold dynamic range).
IFA.
Indirect immunofluorescence assays (IFA) used uninfected and infected DMVEC cells grown in two-well slides as described above. Slides were fixed in 100% methanol at −20°C for 10 min and allowed to dry. Slides were stored at −20°C. Protein products were detected by single-label IFA with rabbit polyclonal antibodies (PAb) to KSHV-encoded proteins, anti-ZMP-A (zinc finger membrane protein A; ORF-K5), anti-K8 (replication-associated protein [RAP]), anti-vGCR (ORF74), or anti-RTA (RNA transcript activator; ORF50), which were described previously (14, 42), or mouse monoclonal antibodies (MAb) to LANA1 (ORF73) or ORF-K8.1 (ABI Technologies, Gaithersburg, Md.) and appropriate fluorescein isothiocyanate (FITC)-labeled goat or donkey anti-rabbit or anti-mouse immunoglobulin (Ig) secondary antibodies, then viewed and photographed using a Nikon UV-epifluorescence microscope with ImagePro 4 software. In some experiments, DAPI (4′,6′-diamidino-2-phenylindole) stain was included in the mounting solution.
Western immunoblots for detection of viral and cellular proteins.
Protein extracts were harvested from DMVEC cell cultures by scraping the cells into triple detergent lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% sodium dodecyl sulfate [SDS], 1% NP-40, and 0.5% sodium deoxycholic acid). Samples were dissolved in 6× SDS loading buffer, boiled for 10 min, and fractionated by SDS-10% polyacrylamide gel electrophoresis (PAGE). Proteins were transferred by electroblotting onto Protran nitrocellulose filters (Schleicher and Schuell, Inc., Keene, N.H.) and blocked for 1 h in Tris-buffered saline (TBS; 20 mM Tris base and 137 mM sodium chloride, pH 7.6) plus 0.3% Tween 20 and 5% dry nonfat milk.
Primary antibody incubations were performed at dilutions of 1:250 in blocking solution overnight at 4°C. The immunoblots were then washed in TBS plus 0.3% Tween 20 for 25 min, and secondary horseradish peroxidase (HRP)-conjugated antibody incubations were carried out in blocking solution for 1 h followed by washing in TBS plus 0.3% Tween 20 for 35 min. Bound secondary HRP-conjugated donkey anti-goat IgG antibodies were detected using the LumiLight kit (Roche, Indianapolis, Ind.). Primary antibodies used included anti-ZMP-A (ORF-K5), rabbit PAb (14) and mouse MAb to either LANA1 (LN53; ABI Technologies, Gaithersburg, Md.) or cellular cytoplasmic β-actin, and rabbit PAb to cellular IRF7 (H246; Santa Cruz Antibodies).
Immunohistochemical detection of viral and cellular proteins.
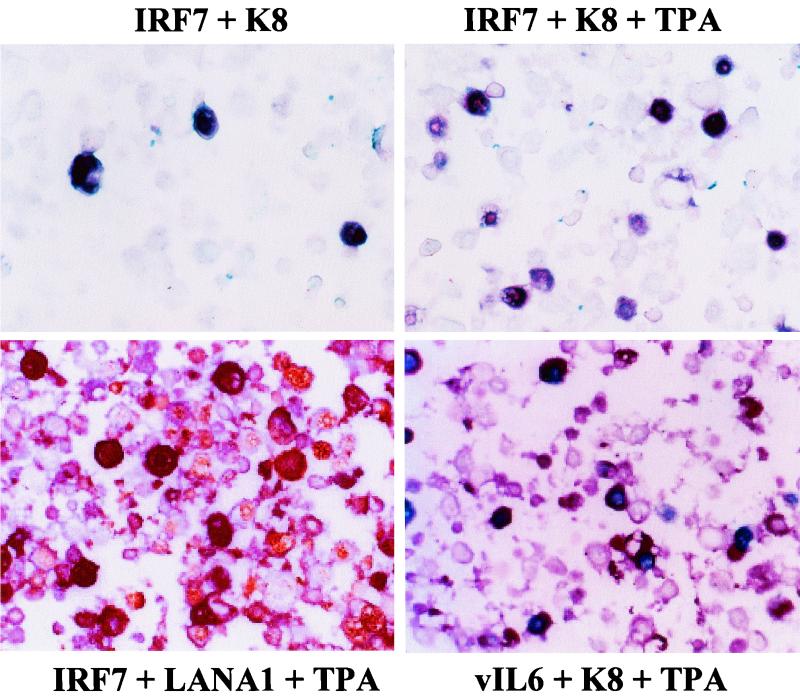
Double-label immunohistochemical staining was used to detect the cellular IRF7 protein in infected DMVEC and PEL cells grown in Labtek slide chamber culture dishes. Cells were washed and fixed with 1:1 methanol-acetone. Half of the cultures were treated with TPA for 60 h before fixing. For double labels, the primary rabbit PAb to cellular IRF7 (H246; Santa Cruz Antibodies) was applied first at an 800-fold dilution for 18 h and detected with biotinylated goat anti-rabbit Ig antibody plus streptavidin-alkaline phosphatase-ABC complex and Vector Red chromagen (Vector Labs, Burlingame, Calif.). After washing, the secondary rabbit PAb against KSHV-encoded K8 (RAP) described previously (42) was applied at an 800-fold dilution in blocking solution, incubated for 18 h, and then detected with biotinylated goat anti-rabbit Ig antibody plus streptavidin-peroxidase-ABC conjugate followed by True Blue peroxidase substrate (Kirkegaard & Perry Laboratories, Gaithersburg, Md.).
For the control viral anti-K8 and anti-vIL6 PAb combination, the blue and red chromogens were reversed. Alternatively, for detection of mouse MAb against LANA1 as secondary antibody, biotinylated goat anti-mouse Ig was used together with streptavidin-peroxidase-ABC conjugate and diaminobenzidine (DAB) chromogen. Finally, hematoxylin counterstain was applied.
RESULTS
Confirmation of DMVEC infection with KSHV and expression of viral proteins.
Two separate batches of RNA were prepared from successive passages of the same culture of KSHV (JSC1)-infected DMVEC cells for examination by both the Clontech and Incyte gene arrays. After the Clontech analysis was completed, the Incyte experiment was intended to provide both a measure of comparison and confirmation and a survey of a much larger sample of human genes. In both gene array experiments, we measured and compared changes in gene expression in the KSHV-infected DMVEC under two experimental conditions: uninfected DMVEC versus KSHV-infected spindle-shaped DMVEC, and a similar comparison in which both the uninfected DMVEC and the KSHV-infected spindle-shaped DMVEC were treated with TPA to enhance lytic viral gene expression.
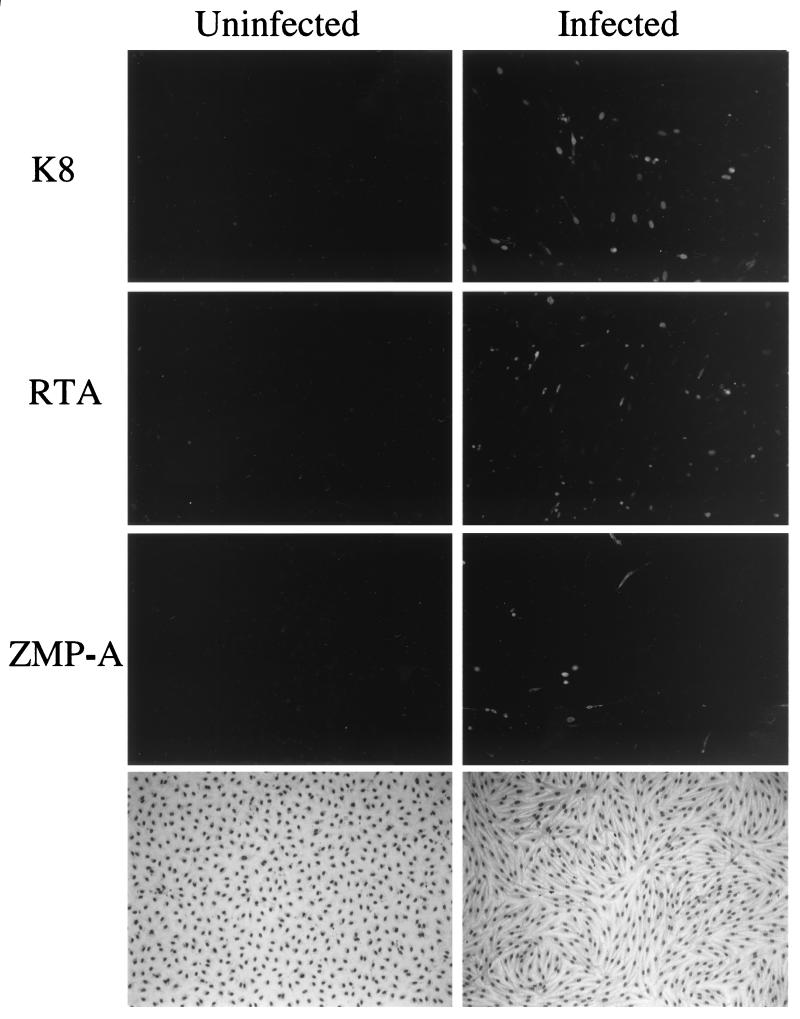
The DMVEC cultures infected with KSHV virions derived from TPA-induced JSC1 PEL cells (9, 14) exhibited a dramatic change in morphology, with 100% of the cells developing a characteristic spindle shape and realigning into clustered swirls after 2 to 3 weeks compared to the cobblestone shape of the parallel control uninfected DMVEC (Fig. 1, lower right-hand panel). As shown previously (9, 14), nearly all of the spindle cells were latently infected with KSHV and expressed the LANA1 protein. A subfraction of the DAPI-stained spindle cells also displayed mitotic figures that were not present in the uninfected DMVEC monolayers (Fig. 2). In addition, at the time of harvesting in both passages of the cultures used, nearly 8% of infected untreated DMVEC spindle cells constitutively expressed the early KSHV-encoded lytic cycle nuclear proteins ORF-K8 (RAP) and ORF50 (RTA) and 4% expressed the ORF-K5 (ZMP-A) lytic cycle cytoplasmic membrane protein, as detected by IFA (Fig. 1, upper right-hand panels), but fewer than 1% of the cells showed cytopathic effects or expressed the late ORF-K8.1 (gpK8.1) protein (not shown).
FIG. 1.
Indirect immunofluorescence demonstrating low level spontaneous lytic cycle KSHV protein expression in the infected DMVEC cultures used for gene array experiments. (Left and right top panels) FITC-labeled ORF-K8 (RAP) antibody; (left and right upper middle panels) FITC-labeled ORF50 (RTA) antibody; (left and right lower middle panels) FITC-labeled ORF K5 (ZMP-A) antibody; (left and right bottom panels) photomicrographs of the cell morphology for Giemsa-stained DMVEC monolayers comparing the uninfected cobblestone pattern with the KSHV-infected spindloid pattern. Virtually all cells in the KSHV infected spindloid cultures expressed the nuclear LANA1 latency protein as detected by IFA (not shown) (14).
FIG. 2.
Effect of KSHV infection and TPA treatment on DMVEC cultures. The photomicrographs show both DAPI-stained cell nuclei (left-hand panels) and IFA with FITC-labeled antibody to the ZMP-A (K5) cytoplasmic membrane protein (right-hand panels) in parallel cultures to those used for the gene array experiments. U, uninfected DMVEC with contact-inhibited cobblestone pattern; I, KSHV-infected DMVEC with rearranged and aligned spindle cell pattern (arrows indicate occasional mitotic cells); U+T, altered uninfected DMVEC pattern after TPA treatment; I+T, KSHV-infected DMVEC spindle cell pattern plus clustered microplaques of cells with rounded CPE.
Treatment of the infected DMVEC cultures with TPA for 60 h only increased the total number of infected DMVEC cells expressing K8 and ZMP-A to approximately 12% (Fig. 2, lower right-hand panel), but this occurred primarily in patches of clustered cells that formed miniplaques in which lytic cycle progression was greatly increased, as judged from cells displaying typical rounding and cytopathic effect (CPE) (Fig. 2, lower panels) together with increased ORF-K8.1 late lytic protein expression detected by IFA and IHC (14). The same cells also contained viral DNA replication compartments detectable both by DAPI staining (Fig. 2, lower left-hand panel) and by IFA for ORF59 (not shown).
Treatment of the control DMVEC culture with TPA also produced some elongation and alignment in the uninfected cobblestone monolayers (Fig. 2, DAPI panels), but without the dramatic spindle shape and clustering seen in the KSHV-infected cultures. Note that the effect of TPA to enhance progression of the lytic cycle primarily in the same subset of cells that have spontaneously entered the lytic cycle is significantly different than in PEL cell cultures, where in addition the total number of cells switching from latent to lytic cycle gene expression (measured by K8 or vIL6 IFA) can be increased 5- to 10-fold depending on the cell line.
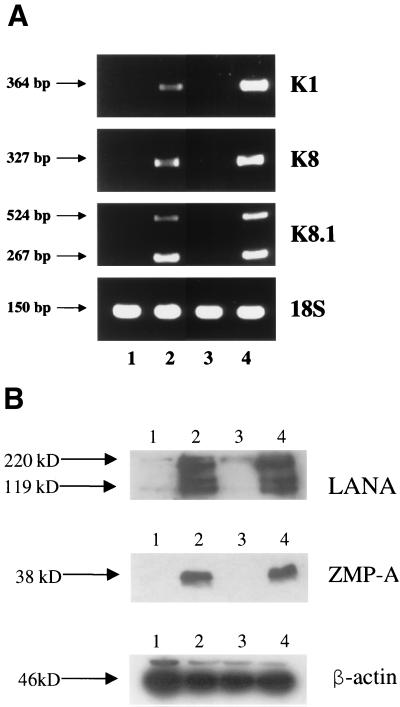
To obtain direct biochemical evidence for lytic cycle gene expression in both the absence and presence of TPA, we carried out standard nonquantitative RT-PCR analysis of the same four RNA samples (batch 1 RNA) used later for the Clontech gene array experiments. Gel electrophoresis analysis of the RT-PCR products for the early lytic cycle KSHV genes ORF-K1 and ORF-K8 and the late lytic cycle gene ORF-K8.1 is shown in Fig. 3A. Each primer pair was chosen to encompass a known intron to assess whether the products represented cDNA derived from spliced mRNA rather than contaminating genomic viral DNA. As expected, infected RNA samples both plus and minus TPA gave spliced RT-PCR products of 364 bp (K1), 327 bp (K8), and 524 bp plus 267 bp (K8.1), whereas there were no products of genomic size (2,007, 407, or 513 bp). After TPA treatment, the relative abundance of the K1 and K8 RT-PCR products was increased (maximum 2.5-fold for K1), and the ratio of the two alternative splice variants of K8.1 changed slightly (47).
FIG. 3.
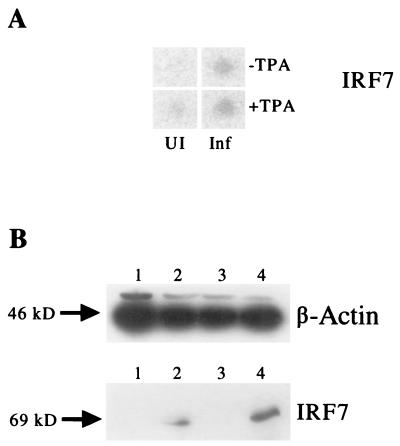
Demonstration of virus-encoded latent and lytic cycle gene expression in KSHV-infected DMVEC cultures. (A) Standard nonquantitative RT-PCR products obtained for selected ORF-K1, ORF-K8, and ORF-K8.1 lytic cycle viral genes in the same set of four DMVEC RNA samples used to generate the probes for the Clontech gene array hybridization experiments (RNA batch 1). Each viral primer pair chosen encompassed known intron loci to confirm that the PCR products were derived from spliced mRNA. Primers for 18S rRNA were used as the loading control. Lane 1, control uninfected DMVEC RNA; lane 2, KSHV-infected DMVEC RNA; lane 3, uninfected DMVEC +TPA RNA; lane 4, KSHV-infected DMVEC +TPA RNA. (B) Western immunoblot comparing the expression of KSHV-encoded LANA1 (latent) and ZMP-A (K5, lytic) proteins in uninfected and infected DMVEC cultures. Lane 1, control uninfected DMVEC extract; lane 2, KSHV-infected DMVEC extract; lane 3, uninfected DMVEC +TPA extract; lane 4, KSHV-infected DMVEC +TPA extract. A β-actin loading control on the same samples is also shown in the lower panel.
Similarly, Western immunoblot analysis (Fig. 3B) of protein extracts from the DMVEC cultures confirmed that high levels of expression of both the KSHV-encoded latent LANA1 (ORF73) and lytic ZMP-A (ORF-K5 or IE-A) proteins occurred in the KSHV-infected spindle cell cultures (lanes 2 and 4). However, no KSHV proteins were detectable either with or without TPA in extracts from the same parallel uninfected DMVEC cultures used to generate control RNA samples for comparison with infected cell RNA samples in both the gene arrays and the later RT-PCR confirmation experiments (lanes 1 and 3). As expected, there was also little quantitative change in the overall levels of these two viral proteins when comparing the infected samples in the presence and absence of TPA (lanes 2 and 4).
DNA microarray experiments.
Both pairs of DMVEC RNA samples from batch 1 were used in hybridization experiments with Clontech Atlas 1.2-I and 1.2-II microarrays (combined total of 2,350 human genes represented) using radioactive cDNA probes prepared in our laboratory, whereas both pairs of batch 2 RNA samples were analyzed on Incyte Human UniGEM V2.0 microarrays (9,180 human genes) using fluorescent dye probe techniques carried out at Incyte laboratories.
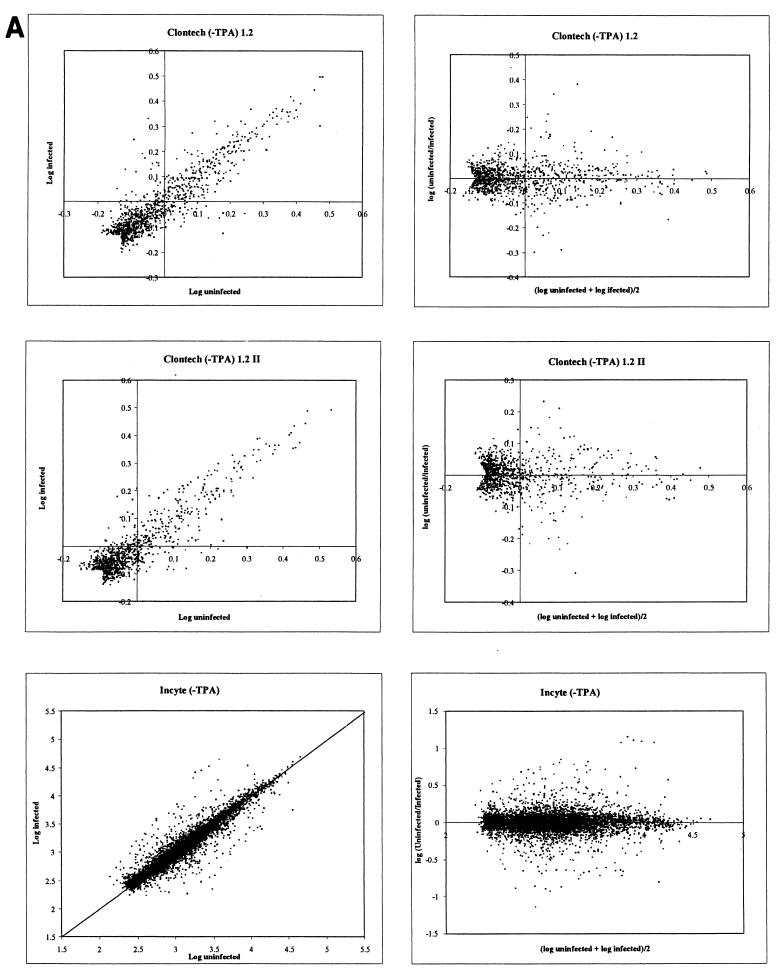
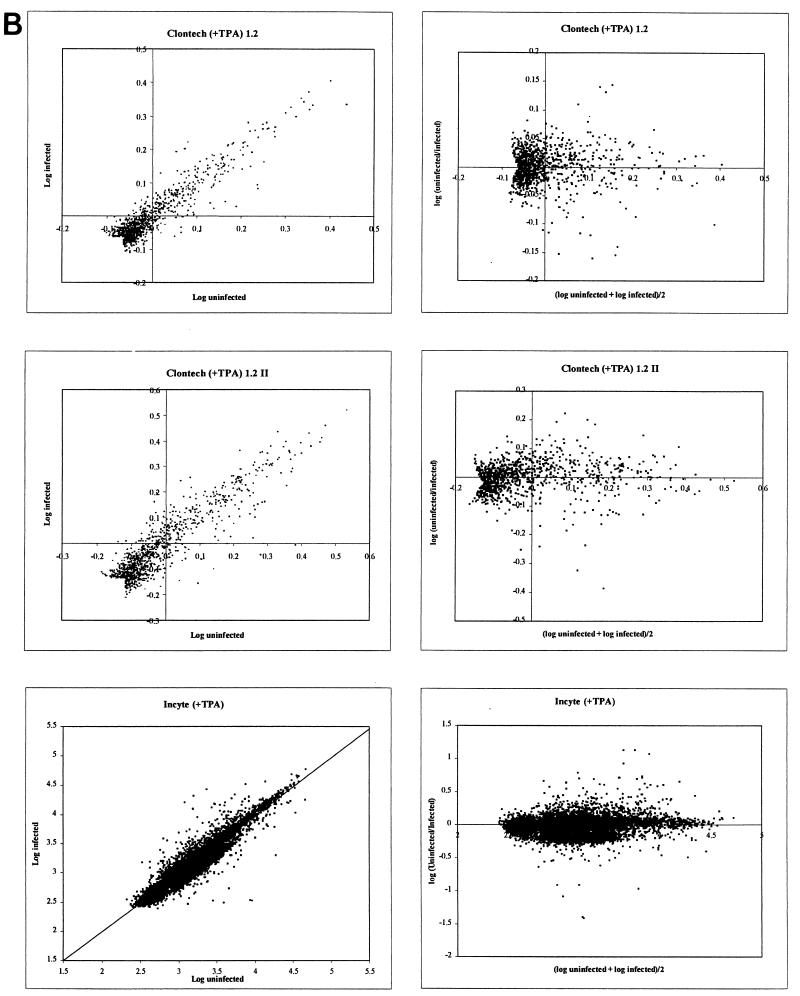
Both the Clontech and Incyte array density values displayed a typical distribution of data points clustered along a linear midpoint of the scatter plot with no obvious distortions (Fig. 4). The results are displayed as both a simple log plot and a horizontal plot for each array set. Nomad program analysis revealed that no curvature or asymmetric adjustments were required. For each Clontech graph, data points that were below the background threshold for both the uninfected and infected blots were removed from the analysis, yielding an arrowhead tail for these data sets. Additionally, phosphorimages obtained for each of the Clontech blots were examined visually, and a small number of adjacent data points that were artificially elevated because of spillover from particularly strong neighboring radioactive signals (e.g., positions adjacent to TIMP1 in the infected plus TPA filter) were removed from graphical analysis. Finally, all control elements on the Incyte arrays were removed. Graphical representation of the data in the horizontal format (Fig. 4A and B, right panels) facilitated calculation and visualization of standard deviation units.
FIG. 4.
Graphical scatter plot representation of gene array data from both the Clontech and Incyte experiments. (A) Results from comparison of IDV for infected and uninfected DMVEC RNA in the absence of TPA. (B) Results from comparison of infected and uninfected DMVEC RNA in the presence of TPA. In each case, the right-hand panels show a horizontal representation of the same data given in the left-hand panels to facilitate the identification of significantly regulated genes. The x axis represents the mean log intensity (i.e., more abundantly expressed genes are to the right), and the y axis represents the log ratio of the compared samples (i.e., upregulated genes have positive y-axis values, and downregulated genes have negative y-axis values).
As one measure of the consistency of our Clontech array results, we have listed 37 of the most abundantly expressed genes in the uninfected DMVEC cultures as determined from the direct absorbance intensity values (Table 1). For each of these genes, both the background corrected absorbance intensity values and their rank order within each experiment are included, but the data are not global mean normalized, in contrast to the later calculated standard adjusted deviation values (Tables 2 and 3). Among these genes are eight that are represented on both the Atlas 1.2-I and 1.2-II arrays, including housekeeping standards glyceraldehyde-3-phosphate dehydrogenase (GAPDH), α1-tubulin, 60S basic ribosomal protein 13A, ubiquitin, and β-actin. These eight were all measured eight times in our data.
TABLE 1.
Most abundantly expressed Clontech genes in KSHV-infected DMVEC culturesa
| Gene | Atlas 1.2-I array intensity values
|
Atlas 1.2-II array intensity values
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −TPA
|
R | +TPA
|
R | −TPA
|
R | +TPA
|
R | |||||
| U | I | U | I | U | I | U | I | |||||
| HSP27 | 40,060 | 36,580 | 1 | 32,600 | 29,770 | 2 | ||||||
| 60SRP13A | 44,040 | 37,480 | 2 | 28,740 | 30,720 | 5 | 33,320 | 31,700 | 2 | 33,410 | 32,570 | 4 |
| Thymosin β10 | 34,120 | 34,220 | 3 | 36,215 | 34,800 | 1 | ||||||
| Thymosin β4 | 31,060 | 27,600 | 4 | 30,670 | 28,100 | 3 | ||||||
| Ubiquitin | 29,780 | 28,560 | 5 | 29,720 | 28,700 | 4 | 25,670 | 27,600 | 5 | 37,270 | 29,420 | 8 |
| RhoGD1α | 28,460 | 30,500 | 6 | 27,040 | 26,050 | 7 | ||||||
| mGST12 | 26,400 | 23,870 | 7 | 21,900 | 18,840 | 20 | ||||||
| GST3 | 26,480 | 23,870 | 8 | 22,360 | 20,840 | 18 | ||||||
| MCP1 | 26,460 | 17,900 | 9 | 11,610 | 12,020 | — | ||||||
| DAD1 | 26,350 | 29,280 | 10 | 22,830 | 23,170 | 15 | ||||||
| NKEnhF | 25,870 | 25,240 | 11 | 26,020 | 23,630 | 19 | ||||||
| Gsα1 | 25,530 | 30,580 | 12 | 20,420 | 20,070 | 22 | ||||||
| Mut-L (MLH1) | 25,600 | 26,620 | 13 | 19,460 | 17,840 | 24 | ||||||
| Hyal-L2 | 25,430 | 25,450 | 14 | 28,160 | 30,720 | 8 | ||||||
| Cathepsin-D | 25,360 | 26,400 | 15 | 20,630 | 23,050 | 21 | 17,970 | 24,500 | 19 | 24,370 | 25,120 | 13 |
| Collagen 1α2 | 25,360 | 23,060 | 16 | 22,370 | 21,640 | 17 | ||||||
| Parathymosin | 24,560 | 22,080 | 17 | 23,970 | 18,440 | 12 | ||||||
| Calpain | 24,100 | 27,120 | 18 | 23,870 | 21,560 | 11 | 22,020 | 24,420 | 9 | 19,280 | 24,820 | 16 |
| HLA-C4 | 23,820 | 29,500 | 19 | 25,630 | 27,140 | 10 | 21,060 | 27,920 | 12 | 28,980 | 31,080 | 5 |
| 40SRDS9 | 23,590 | 22,840 | 20 | 21,560 | 21,330 | 33 | ||||||
| PUF | 23,400 | 28,370 | 21 | 26,730 | 25,100 | 9 | ||||||
| BMP4 | 23,260 | 2,460 | 22 | 6,370 | 5,330 | — | ||||||
| Amyloid α4 | 22,400 | 11,140 | 25 | 23,460 | 20,900 | 13 | 19,300 | 15,160 | 18 | 24,920 | 18,920 | 12 |
| GADPH | 22,200 | 26,940 | 26 | 22,920 | 23,040 | 14 | 33,720 | 38,840 | 1 | 42,470 | 40,720 | 1 |
| β-Actin | 21,400 | 24,000 | 29 | 22,630 | 21,820 | 16 | 28,720 | 21,200 | 3 | 42,470 | 43,580 | 2 |
| TIMP1 | 21,020 | 36,630 | 30 | 28,940 | 38,870 | 6 | ||||||
| Ferritin H | 27,960 | 28,320 | 4 | 22,860 | 23,900 | 14 | ||||||
| Protein di-S isom | 25,120 | 27,000 | 6 | 32,440 | 28,930 | 3 | ||||||
| VWF | 23,940 | 20,400 | 7 | 11,630 | 17,720 | — | ||||||
| Clathrin AP50 | 21,920 | 23,500 | 8 | 29,070 | 22,920 | 6 | ||||||
| Metallothionine | 22,290 | 21,700 | 10 | 4,220 | 15,280 | — | ||||||
| CD81 | 21,860 | 22,400 | 11 | 28,550 | 26,700 | 7 | ||||||
| Exodus 2 | 21,060 | 29,770 | 13 | 2,850 | 17,600 | — | ||||||
| α1-Tubulin | 21,020 | 27,970 | 14 | 25,020 | 27,820 | 11 | ||||||
| Annexin II | 20,820 | 20,100 | 15 | 21,070 | 16,120 | 15 | ||||||
| Cofilin | 20,720 | 23,690 | 16 | 25,340 | 23,950 | 10 | ||||||
| Gsα2 | 19,020 | 19,300 | 17 | 27,170 | 22,320 | 9 | ||||||
Hybridization intensity scores are given separately for each of the top 20 genes in the four Atlas 1.2-I array filter results obtained using cDNA probes prepared from uninfected and infected cell RNA (batch 1) in the absence and presence of TPA and similarly for each of the four Atlas 1.2-II array filters obtained using the same four-cDNA probe set. Averaged background values ranging from 11,780 to 16,540 on different filters have been subtracted, but the intensity values have not been normalized or mean adjusted. Numbers in bold type indicate significantly regulated values. Additional data for eight genes that are common to both filters are included. R, direct numerical rank order for each of the four uninfected DMVEC RNA filters. U = uninfected; I = infected.
Interesting features of the data in Table 1 include first the observations that thymosin β4 and thymosin β10 appear within the top four most abundantly expressed genes in the Atlas 1.2-I array and that monocyte chemotactic protein 1 (MCP1), bone morphogenetic protein 4 (BMP4), parathymosin, tissue inhibitor of metalloproteinases 1 (TIMP1), calpain small subunit, exodus 2 β-chemokine, and Von Willebrandt factor (VWF) all appear on the list. Second, the intensity values for the majority of these genes, whether measured four times or eight times, were highly reproducible. The only significantly regulated genes among these 37 highly abundant species were MCP1, BMP4, amyloid α4, TIMP1, VWF, metallothionine H1, and exodus 2. Six of these genes were downregulated by either KSHV infection, TPA treatment, or both, and only TIMP1 was upregulated. Except for GAPDH and β-actin in the 1.2-II array, none of the other 30 most abundantly expressed genes varied more than 30% in intensity on any sample or filter after KSHV infection and spindle cell conversion, after TPA treatment, or both (Table 1).
The most abundantly expressed individual genes in uninfected DMVEC by the criterion of Clontech array hybridization signal intensity among broad categories of likely interesting genes included those for MCP1, interleukin-10 (IL-10), colony-stimulating factor 1 (CSF-1), placental growth factor, IL-6, and IL-1β in that order among all interleukins and growth factors tested and MCP1-R1 among all G-protein-coupled receptors (GPCR) and chemokine receptors tested. All of these, as well as c-Jun, ICAM1, fibroblast growth factor (FGF)-R1, VEGF-R1, and BMSA1 were among the top 8% in relative abundance on the Clontech arrays. VEGF-R1 was expressed much more strongly than VEGF-R2 or -R3, and VEGF-C was more abundant than VEGF-A, -B, or -D. MCM5, MCM7, CycD1, and MMP12 were the most abundant of the 28 minichromosome maintenance, cyclin, and metalloproteinase genes tested.
Changes in gene expression patterns in KSHV-infected DMVEC.
Importantly, the total number of genes that were up- or downregulated significantly by KSHV infection in both sets of arrays were similar and quite small (mostly between 1.4 and 2.5%), lending credence to the validity of these results. In the Clontech −TPA experiment, a total of 2.1% of the genes surveyed (49 of 2,350) were upregulated by greater than a standard deviation value of 2.0, and 1.8% (45 of 2,400) were downregulated by greater than 2.0 standard deviation units. In the Clontech +TPA experiments, a total of 2.2% (55 of 2,350) and 1.8% (45 of 2,350) of the genes were up- or downregulated by more than 2.0 standard deviations.
For the Incyte −TPA results, overall 1.8% (163 of 9,000) and 1.3% (117 of 9,000) of the genes gave upregulated ratios of >1.74 or ≥2.0 and 1.7% (150 of 9,000) and 1.2% (110 of 9,000) gave downregulated ratios of <1.74 or ≤2.0, respectively. For the Incyte +TPA data, 4.0% (364 of 9,000), 1.7% (156 of 9,000), and 1.0% (86 of 9,000) of the genes gave upregulated values of >1.74, >1.8, and ≥2.0, respectively, with the inconsistent bulge in the TPA upregulated numbers being primarily attributed to more than 200 genes with ratios of 1.8. Downregulated genes in the Incyte +TPA data set were 2.1% (195 of 9,000) at <1.74 and 1.4% (122 of 9,000) at ≤2.0.
The 40 most significantly differentially regulated genes for both sets of experimental conditions are listed in Tables 2 through 5. Regulated genes observed on the combined Atlas 1.2-I and 1.2-II Clontech arrays comparing untreated KSHV-infected DMVEC (−TPA) with uninfected DMVEC (−TPA) are listed in Table 2. Regulated genes observed on the Incyte arrays in the same comparison (in the absence of TPA) are listed in Table 4. In each case, the 20 most upregulated genes (given positive SD values) and the 20 most downregulated genes (given negative SD values) are listed. Similarly, for the TPA-treated samples, the 40 most differentially regulated genes observed on the Clontech arrays in a comparison of KSHV-infected DMVEC (+TPA) with uninfected DMVEC (+TPA) are listed in Table 3, and the most differentially regulated genes observed on the Incyte arrays in the same comparison (in the presence of TPA) are listed in Table 5.
The entire set of gene array results data from both our Clontech and Incyte experiments may be accessed at www.pevsnerlab.kennedykrieger.org/microarraydata/hayward.
Most-regulated genes after infection in the absence of TPA treatment.
The most notable result revealed by these studies was a significant upregulation of a large number of interferon-induced genes, especially in the Incyte array data. However, there were also significant changes in the regulation of some genes involved in cell cycle arrest and cell morphology changes, as well as dysregulation of genes thought to be involved in the processes of tumor formation, angiogenesis, and immune regulation. Among the most heavily upregulated genes in infected cells in the absence of TPA were those for neurogranin, calgranulin B (migration inhibitory factor 14), monocyte differentiation protein CD14, fibrinogen-β, interferon response factor 7 (IRF7), the G protein-coupled receptor RDC1 homolog, the ubiquitin-conjugating enzyme E2 H10, osteopontin, estrogen receptor α, and TIMP1 in the Clontech −TPA array (Table 2), plus RDC1 again, STAT-induced STAT inhibitor 3 (SISI3), tissue plasminogen activator (PLAT), insulin growth factor binding protein IGFBP5, thrombomodulin, and transforming growth factor beta 3 (TGFβ3) in the Incyte −TPA array (Table 4). Five interferon-induced genes, including myxovirus resistance RI (p78), also appeared within the top 20 upregulated genes in the Incyte −TPA array.
The most heavily downregulated genes in infected cells in the absence of TPA included BMP4, CCR2 β-chemokine receptor (MCP1-RA), gap junction protein connexin 37, BMSA1, skeletal muscle LIM-1, p57-KIP2, GADD45β, endothelin 1, plasminogen activator inhibitor 1 (PAI-1), cyclin D1, bone morphogenetic protein 6 (BMP6), and endothelial multimerin in the Clontech arrays (Table 2), plus connective tissue growth factor, fibronectin 1, BRCA1-associated RING protein (BARD1), cardiac ankyrin repeat protein, PAI-1, downregulated in ovarian cancer (DROC1), keratin 7, granzyme K, connexin 37, thrombospondin 1, endothelin 1, transforming growth factor alpha (TGFα), matrix metalloproteinase 2 (MMP2), α4-integrin (CD49D), and both BMP4 and BMSA1 again in the Incyte array data (Table 4).
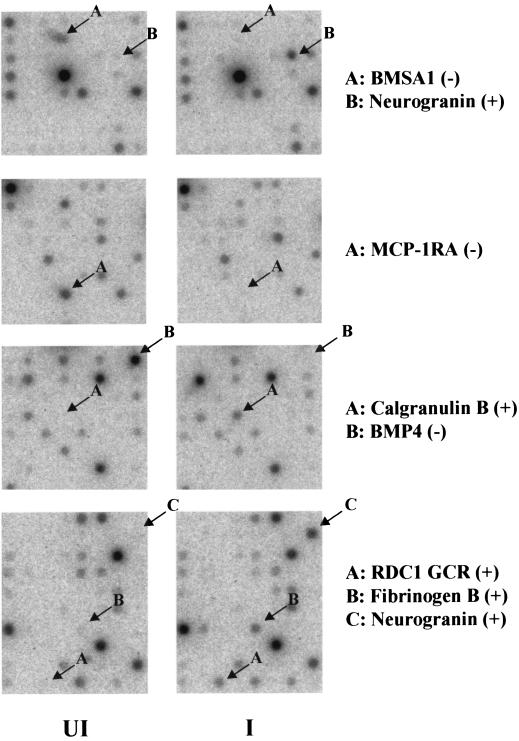
Examples of the direct hybridization radiographic images for several dramatically upregulated genes (neurogranin, RDC1, calgranulin B, and fibrinogen-β) or heavily downregulated genes (BMP4, MCP1-R1, and BMSA1) in the Clontech array in the absence of TPA are shown in Fig. 5.
FIG. 5.
Representative photographs of radioactive spots from sections of the original Atlas 1.2 filters from the Clontech array hybridization experiment to illustrate examples of seven of the most heavily upregulated (+) and downregulated (−) genes, as indicated. Only data for KSHV-infected compared to uninfected filter pairs in the absence of TPA are presented. UI, uninfected; I, infected. Neurogranin appears on both Atlas 1.2I and Atlas 1.2II.
Most-regulated genes after infection in the presence of TPA.
Any additional changes observed after infection in the presence of TPA were anticipated to be related to the increased levels and more advanced stages of lytic virus infection, not to the TPA treatment itself, because the control uninfected cells were also TPA treated. Among the 20 most heavily upregulated genes in infected cells in the presence of TPA were the RDC1 orphan chemokine receptor, metallothionein H1, exodus 2, RAD23A, decoy receptor protein DCR3, calgranulin B, TIMP1, Bcl3, IRF7, RAMP2, CDC25B, and endothelial PAI-1 in the Clontech +TPA arrays (Table 3), and statherin, IGFBP5, and STAT1 plus 10 interferon response genes, including myxovirus resistance I p78 (Myx R1) and 2′-5′-oligoadenylate synthetase 2 in the Incyte +TPA array (Table 5). Overall, a total of 16 interferon-inducible genes placed within the top 42 ranked upregulated positions in the Incyte +TPA array data (compared to 8 of 56 in the absence of TPA).
The 20 most heavily downregulated genes in infected cells in the presence of TPA included nucleobindin, CXCR4, IL-8, GRK4, thrombin receptor, Thy1, and c-Maf in the Clontech arrays (Table 3), and fibronectin 1, connective tissue growth factor, BARD1, matrix Glα protein, stromal cell-derived factor 1, CD34, cardiac ankyrin repeat protein, MMP2, endothelin 1, α4-integrin, PAI-1, VEGF-related placental growth factor, thrombin receptor, thrombospondin 1, and β1-catenin in the Incyte array (Table 5).
Several other interesting genes appeared among the next group (rank order 21 to 40 in each) of the most up- and downregulated genes in the Incyte arrays (with the I-R values and rank order positions given in parentheses). In the absence of TPA, these included IL-13 (+4.4, up 21), angiopoietin 2 (+4.1, up 27), hypoxia-inducing factor alpha (+3.5 and +3.1, up 31 and up 38), SISI2 (+3.0, up 40), plasminogen activator urokinase (−3.9, down 25), stromal cell-derived factor 1 (−3.9, down 26), and DROC1 (−3.3, down 33). In the presence of TPA, these included RDC1 again (+2.8 and +2.6, up 24 and up 28), SISI3 (+2.4, up 38), TGFβ3 (+2.3, up 40), DROC1 (−3.6, down 25), and angiopoietin 2 (−2.7, down 40).
Again as a measure of reproducibility, the values obtained for several regulated genes that were duplicated, included GADD45 (−3.6, −2.5), IRF7 (+4.2, +2.2), and neurogranin (+6.5, +5.4) for the Clontech −TPA array; IRF7 (+3.2, +2.5) and Bcl3 (+2.1, +1.7) for the Clontech +TPA array; RDC1 (−8.5, −4.5), SISI3 (−7.4, −2.9), IGFBP5 (−5.2, −4.3), HIF1α (−3.5, −3.1), DROC1 (+7.1, +3.4), and keratin 7 (+6.7, +4.1) for the Incyte −TPA array; and interferon-induced tetratricopeptide (−24.8, −8.2), Rhoβ (−4.9, −3.4), interferon-induced p75 (−3.5, −2.4), RDC1 (−2.8, −2.6), apoliprotein D (+4.9, +2.5), keratin 7 (+2.4, +1.7), and MMP2 (+2.1, +2.0) for the Incyte +TPA array.
Additional effects of TPA treatment on KSHV-infected DMVEC.
As hypothesized, a large number of cellular genes were regulated similarly by infection in both the −TPA and +TPA experiments. In the Clontech arrays (Tables 2 and 3), 7 of the top 20 genes (35%) that were upregulated after infection in the absence of TPA were also within the top 30 upregulated genes after infection in the presence of TPA, although only one of the downregulated genes appeared within both the −TPA and +TPA top 25. In the Incyte array (Tables 4 and 5), 9 of the top 20 genes (45%) that were upregulated by infection in the −TPA experiment were also upregulated in the top 40 of the infected +TPA experiment, and 12 of the top 20 genes (60%) and 14 of the top 40 that were downregulated in the infected −TPA experiment were also downregulated in the top 25 of the infected +TPA experiment.
Examination of the effects of TPA on DMVEC in the absence of infection.
To evaluate whether it was valid to consider the effects of TPA to be cancelled out in the infected +TPA versus uninfected +TPA comparisons above, we also compared the Clontech single-channel gene array data for the pair of filters representing uninfected DMVEC in the absence and presence of TPA and for the pair representing KSHV-infected DMVEC in the absence and presence of TPA. Again, in the uninfected cells, 2.0% of the combined Atlas 2.1-I and 2.1-II genes (45 of 2,350) were upregulated, and 2.1% (49 of 2,350) were downregulated by ≥2.0 SD after TPA treatment alone. Similarly, in the infected cultures, overall 2.0% (47 of 2,350) and 1.9% (44 of 2,350) of the Clontech genes were up- or downregulated ≥2.0 SD after TPA treatment.
Some illustrative aspects of the analysis are presented in Table 6, which shows the results (as intensity values, SD ratios, and rank) for both the 20 most upregulated genes and the 20 most downregulated genes in uninfected DMVEC after addition of TPA, together with some selected results from the comparison of TPA-treated versus untreated infected cells.
TABLE 6.
DMVEC genes up- and downregulated by TPA in Clontech arrays without and with KSHV infectiona
| Regulation | Gene | Intensity value
|
C-SD ratio (rank)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Uninfected
|
Infected
|
U+T/U | I+T/I | I/U | I+T/U+T | ||||
| −TPA | +TPA | −TPA | +TPA | ||||||
| Up | CXCR4 | 450 | 17,840 | 2,600 | 8,510 | +7.7 (1) | +3.2 (9) | −4.9 (2) | |
| Thy1 | 750 | 25,980 | 750 | 12,840 | +5.6 (2) | +5.9 (1) | −2.9 (9) | ||
| IL-8 | 4,890 | 18,520 | 10,230 | 9,450 | +5.4 (3) | +2.1 (36) | −4.6 (3) | ||
| MMP14 | 4,270 | 14,560 | 5,260 | 10,710 | +4.4 (4) | +3.4 (6) | |||
| Homeobox HB24 | <0 | 17,890 | 6,750 | 12,610 | +4.4 (5) | +3.9 (9) | |||
| Proteasome inhibitor HP131 | 6,960 | 15,880 | 10,120 | 11,260 | +3.9 (6) | ||||
| β3-Integrin/CD61 | 9,220 | 17,690 | 15,330 | 9,570 | +3.7 (7) | ||||
| GRK4 | 1,240 | 16,800 | 1,080 | 4,960 | +3.5 (8) | +2.3 (33) | −4.0 (5) | ||
| Notch3 | 170 | 13,560 | 1,160 | 2,720 | +3.4 (9) | −4.4 (4) | |||
| OX40L | 9,140 | 16,600 | 11,820 | 14,420 | +3.3 (10) | −2.1 (31) | |||
| Tight junction TJP1 | 3,390 | 10,540 | 6,260 | 8,000 | +2.9 (11) | ||||
| Apoliprotein-D | 12,630 | 33,120 | 10,800 | 20,780 | +2.9 (12) | +2.5 (21) | −2.1 (32) | ||
| TIMP1 | 21,020 | 28,940 | 36,630 | 38,870 | +2.8 (13) | +3.2 (13) | +3.6 (10) | ||
| Inhibin β-B | 370 | 4,580 | 1,880 | 3,480 | +2.9 (14) | −3.8 (7) | |||
| ConnTiss GF | 10,620 | 16,440 | 9,400 | 14,120 | +2.7 (15) | +3.2 (12) | |||
| Nucleotide diP kinase A | 9,390 | 14,900 | 12,960 | 14,320 | +2.7 (16) | ||||
| Oncostatin M | 11,760 | 17,060 | 9,940 | 13,040 | +2.5 (17) | +2.4 (25) | |||
| WNT 13 | 8,390 | 13,900 | 6,940 | 9,420 | +2.5 (18) | −2.2 (29) | |||
| Neuronal pentraxin II | <0 | 2,870 | 1,100 | 5,230 | +2.5 (19) | +4.2 (3) | +2.5 (30) | ||
| Amphiregulin | 5,630 | 11,710 | 5,210 | 8,310 | +2.5 (20) | ||||
| E-cadherin | 3,900 | 8,690 | 3,570 | 10,660 | +4.2 (4) | +2.1 (47) | |||
| MMP2 | 6,600 | 8,600 | 4,080 | 9,950 | +3.2 (10) | ||||
| β1-Catenin | 9,520 | 9,310 | 1,300 | 9,160 | +2.6 (17) | −3.8 (8) | |||
| Amyloid A4 | 22,360 | 23,690 | 11,400 | 20,460 | +4.7 (2) | −3.1 (15) | |||
| DCR3 | 30 | 970 | 2,380 | 9,210 | +3.0 (13) | +4.7 (5) | |||
| Matrix G1a | 11,760 | 11,410 | 8,020 | 12,500 | +3.2 (12) | ||||
| Bcl3 | 570 | 4,470 | 4,030 | 10,840 | +2.3 (28) | +2.1 (31) | +3.6 (12) | ||
| Down | Exodus 2 | 21,060 | 2,850 | 29,970 | 17,600 | −6.9 (1) | −4.1 (4) | +6.3 (2) | |
| Metallothionine H1 | 22,290 | 4,220 | 21,700 | 15,280 | −6.3 (2) | +7.6 (1) | |||
| BMP4 | 23,260 | 6,370 | 2,460 | 5,330 | −5.6 (3) | −7.1 (1) | |||
| MCP-R1 | 17,150 | 6,340 | 4,870 | 2,860 | −5.0 (4) | −6.4 (2) | |||
| RAD23A | 15,090 | 5,550 | 12,530 | 11,780 | −4.6 (5) | +5.4 (3) | |||
| Estradiol 17βdih-2 | 9,530 | 1,370 | 2,420 | 3,330 | −4.6 (6) | −3.8 (9) | |||
| Connexin 37 | 10,190 | 2,720 | 1,140 | 3,780 | −4.3 (7) | −4.9 (3) | |||
| RAMP2 | 17,360 | 8,950 | 19,020 | 16,220 | −4.0 (8) | +3.2 (16) | |||
| EC multimerin | 10,240 | 4,290 | 4,080 | 2,360 | −4.0 (9) | −3.1 (19) | |||
| MCP1 | 26,460 | 11,550 | 17,900 | 12,090 | −3.9 (10) | −2.0 (45) | |||
| VWF | 23,940 | 11,630 | 20,040 | 17,720 | −3.9 (11) | +2.4 (33) | |||
| BMSA1 | 13,580 | 5,690 | 1,890 | 5,490 | −3.8 (12) | −4.8 (4) | |||
| Tropoelastin | 8,100 | 2,160 | 7,780 | 1,080 | −3.8 (13) | −4.5 (3) | |||
| Purinergic receptor P2X4 | 9,950 | 4,020 | 5,070 | 5,530 | −3.7 (14) | −2.4 (31) | |||
| Corticotrophin CRF-R | 14,430 | 6,300 | 6,740 | 5,190 | −3.6 (15) | −2.7 (21) | |||
| Skeletal muscle LIM1 | 11,750 | 5,240 | 2,840 | 1,580 | −3.6 (16) | −4.4 (6) | +3.0 (19) | ||
| Receptor protein PM1 | 10,540 | 5,100 | 11,200 | 10,000 | −3.4 (17) | +2.8 (20) | |||
| Endo PAI-1 | 18,850 | 8,960 | 7,720 | 12,620 | −3.3 (18) | +3.2 (11) | −3.5 (11) | +3.1 (18) | |
| R-ras2 (TC21) | 6,670 | 1,990 | 1,390 | 580 | −3.3 (19) | −3.1 (18) | |||
| α-Actinin 1 | 11,720 | 6,610 | 12,990 | 9,120 | −3.2 (20) | +1.8 (63) | |||
| IGFBP10 | 7,430 | 4,820 | 2,300 | 3,840 | −2.5 (33) | −3.1 (17) | |||
| MIP3β | 3,270 | <0 | 8,590 | <0 | −2.2 (43) | −5.4 (1) | +2.6 (19) | ||
| Neurobindin | 18,060 | 19,960 | 22,100 | 9,830 | −4.0 (5) | −5.0 (1) | |||
| Ubiquitin E2H10 | 2,020 | 3,900 | 10,300 | 4,300 | −3.2 (7) | +3.7 (10) | |||
| TGFβ4 | 2,430 | <0 | 5,190 | 240 | −3.2 (11) | ||||
| β4-Integrin | 7,240 | 4,970 | 8,120 | 4,150 | −2.3 (37) | −2.9 (15) | |||
| MCM5 | 8,190 | 7,120 | 13,880 | 6,840 | −3.2 (8) | +2.2 (28) | |||
| PCNA | 5,860 | 7,150 | 9,530 | 5,330 | −2.4 (22) | ||||
| BMP6 | 10,630 | 6,620 | 2,810 | 5,740 | −2.3 (36) | −3.1 (16) | |||
| IRF7 | 2,580 | 940 | 7,100 | 2,960 | NR | NR | +4.2 (7) | +3.2 (15) | |
Comparison of hybridization intensity scores (combined Atlas 1.2-I and 1.2-II) after background subtraction for RNA batch 1 DMVEC genes regulated by TPA both before and after KSHV infection. Bold numbers indicate significantly regulated values in comparison to the uninfected untreated control. Genes regulated by TPA in the absence of infection are given in rank order 1 through 20, together with selected others in the presence of infection. C-SD ratios and rank orders for regulated genes generated after pairwise comparisons are shown on the right. U, uninfected; +T, with TPA; I, infected. NR, not regulated.
Strikingly, a subset of the Clontech genes that were regulated by TPA alone were the same as those that were regulated in the KSHV-infected DMVEC in the absence of TPA, although there were also many genes regulated by KSHV infection that were not significantly affected by TPA. These dual-regulated genes displayed at least three distinctive patterns. (Note that because it involves direct two-channel comparison of differentially labeled pairs of RNA samples, this type of analysis cannot be carried out with the Incyte data.)
In the first group, both TPA and KSHV infection independently produced similar downregulation results for BMP4 (−5.6 versus −7.1, respectively), MCP1-RA (CCR2) (−5.0 versus −6.4), connexin 37 (−4.6 versus −5.6), BMSA1 (−3.8 versus −4.8), skeletal muscle LIM-1 (−3.6 versus −4.4), endothelial multimerin (−4.0 versus −3.1), MCP1 (−3.9 versus −2.0), endothelial PAI-1 (−3.2 versus −3.5), and BMP6 (−2.3 versus −2.9). In contrast, neurogranin (+2.0 versus +6.5), TIMP1 (+2.8 versus +3.0), and IL-8 (+5.4 versus +2.1) were all upregulated independently by both TPA treatment and KSHV infection.
In the second pattern, metallothionine H1 (−6.3 versus +7.6), exodus 2 (−6.9 versus +6.3), RAD23A (−4.6 versus +5.4), RAMP2 (−4.0 versus +3.2), and VWF (−3.3 versus +3.1) were all downregulated by TPA alone to about the same level that they were measured to be upregulated by infection in the presence of TPA. For this category the most predominant feature was the low intensity obtained in uninfected cells in the presence of TPA, implying either that latent infection blocked the TPA-induced downregulation or that lytically induced infection compensated for it. Endothelial PAI1 seemed to produce a combination of both the first and second patterns.
In the third distinctive pattern, upregulation by TPA appeared to be compensated for by downregulation after infection in the presence of TPA, e.g., CXCR4 (+7.7 versus −4.9), thymosin 1 (+5.6 versus −2.9), IL-8 (+5.4 versus −4.6), and GRK4 (+3.5 versus −4.0). In the case of neurobindin, only the combination of infection and TPA gave downregulation.
Clontech genes that were heavily regulated by KSHV infection but were apparently not subject to TPA regulation included RCD1, calgranulin B, IRF7, neurogranin, fibrinogen-β, CD14, osteopontin, and BMSA1.
Confirmation of regulated transcripts by RT-PCR.
Although the gene array survey data indicated statistically signifcant regulation of many genes likely to be associated with antiviral responses, angiogenesis, transformation, cell cycle regulation, immune regulation, apoptosis, and extracellular matrix or cytoskeletal integrity, these effects needed to be confirmed by additional independent assays. Initially, 19 of the most highly regulated genes were selected from the Clontech results for further analysis by direct semiquantitative RT-PCR from the same RNA samples (batch 1) used for the Clontech array experiment.
For each experiment, three test primer pairs were run together, and cytoplasmic β-actin, which was not significantly regulated in either the Clontech or Incyte array experiments, was used as the control for comparison and normalization purposes. Successive samples (1, 2, 3, etc.) were removed from the RT-PCR mixture every three cycles and electrophoresed on an agarose gel, and then stained with ethidium bromide. For all gene primer sets, integrated density values (IDV), corresponding to the sum of pixel intensities after background corrections, were recorded for both the uninfected and infected samples at linear points on the amplification curve. For each of the 10 β-actin control sets, the sample numbers for the IDV point representing the beginning of the linear portions of the curves for the infected and uninfected RNAs always corresponded closely, indicating that both exhibited linear amplification of β-actin in the same portion of their curves (see Fig. 6A).
To compare amplification of test transcripts against the β-actin controls, the sample number of the RNA PCR product (uninfected or infected) that first entered the linear portion of its amplification curve was used for comparison with the other uninfected or infected RNA PCR product. For example, in Fig. 6A, the BMP4 RT-PCR product from the uninfected RNA was readily visible by sample 4 compared to the RT-PCR product from the infected RNA sample, which was only visible by sample 6. Therefore, the uninfected RNA underwent linear amplification of BMP4 six PCR cycles earlier than did the parallel infected RNA. Finally, the IDV scores for sample 4 from both the infected and uninfected RNAs were compared. This type of comparison was carried out for each of the 19 genes tested, and the results are presented as histograms in Fig. 6B. Only one set of primers (MCP1-RA) failed to give PCR products.
For most of the genes tested (9 of 11 in the absence of TPA and 10 of 13 in the presence of TPA), this initial confirmation by RT-PCR did indeed exhibit matching up- or downregulation in parallel with the results obtained from the Clontech gene arrays (Table 7). However, four genes were either not regulated or discordantly regulated according to the RT-PCR analysis. These included β1-catenin and Ras-related TC21 in the absence of TPA and RAD23A and GRK4 in the presence of TPA. Furthermore, β1-catenin was upregulated (+3.3) by infection but only in the presence of TPA in the RT-PCR analysis, whereas it was recorded as being downregulated (−3.8) only in the absence of TPA in the Clontech array and downregulated (−4.1) only in the presence of TPA in the Incyte array data. A downregulation value of only −1.7 for CXCR4 (in the presence of TPA) was also far less than expected from the Clontech array data (−4.1), although the Incyte result also indicated an insignificant effect (−1.2).
Consistency of RNA preparations.
In a second RT-PCR approach to evaluation and confirmation of the array data, we carried out single time point multiplex PCR on cDNA generated from a new preparation (batch 3) of infected and uninfected DMVEC RNA samples from cells grown nearly a year after the previous two batches. In this case, six selected primer pairs, including four from the Incyte gene set, were generated for direct detection of PCR products by ethidium bromide staining in gels. Within each sample, we also included internal control primers for either 18S rRNA or β-actin.
Gel electrophoresis images of the RT-PCR products obtained with uninfected and infected cell RNA (all in the absence of TPA) are shown in Fig. 7. Quantitatively, IRF7 gave 8-fold upregulation compared to +4.2 and +3.2 (Clontech) or +1.4 (Incyte), and TIMP1 gave 3.7-fold upregulation compared to +3.2 (Clontech) in the array results. Similarly, BMP4 gave 23-fold downregulation compared to −7.1 (Clontech) and −4.7 (Incyte), and PAI-1 gave 3.7-fold downregulation compared to −11.7 (Incyte), but PLAT gave only 1.2-fold upregulation compared to +5.8 (Incyte), and β1-catenin (not shown) continued to be variable, giving 1.9-fold downregulation here compared to −3.8 (Clontech), −4.1(Incyte +TPA), and +3.3 by RT-PCR in batch 1 RNA. However, overall, except for PLAT and the somewhat weaker or inconsistent effects for PAI-1 and β1-catenin, these data closely resembled the gene array trends obtained with the previous KSHV-infected DMVEC RNA samples.
Real-time quantitative RT-PCR.
In a third and more quantitative approach to confirming some of the principal results of the gene arrays by RT-PCR procedures, we designed and synthesized a new set of 20 selected gene-specific primer pairs (based primarily on the Incyte data), which all generated PCR products in the size range of 80 to 140 bp, appropriate for quantitative real-time PCR. All of these primers were used initially with the same set of uninfected DMVEC and fully spindle-shaped infected DMVEC cDNAs (batch 3) used above in the multiplex experiments. However, to further examine the biological reproducibility, a fourth batch of RNA samples were prepared from new cultures of uninfected DMVEC, fully spindle-shaped infected DMVEC, and the same spindle cells after 60 h of TPA treatment. Note that the infected +TPA to uninfected untreated comparison (I+T/U) in this case (batch 4) differs from that in the gene arrays (I+T/U+T) because the control sample represents untreated uninfected DMVEC RNA, not TPA-treated uninfected DMVEC RNA.
The results for 15 of the 19 genes tested (presented in Table 8) were highly supportive of the Incyte gene array screening data. Only four of these genes gave inconsistent results compared to the gene array data, with IL-8 being strongly downregulated even in the absence of TPA in batch 3 and 4 RNAs by real-time RT-PCR, and metallothionine HI was apparently upregulated even the absence of TPA, whereas neither HIF1α nor PLAT was significantly regulated in batch 3 or 4 RNAs when assayed by this procedure.
Some quantitative but not qualitative discordance or variation also occurred between the real-time PCR fold values and the Incyte array ratios for BMP4 (−145 compared to −7.1), fibronectin 1 (−40.3 compared to −12.6), MMP2 (−87 compared to −5.1), PAI-1 (−55.7 compared to −11.7), Myx R1 (+85 compared to +7.7), and especially α4-integrin (−455 compared to −4.8). In general, the fold regulated values produced by real-time PCR were much higher than either the Clontech standard deviation or Incyte ratio standard deviation values.
Importantly, the results for Myx R1 (+85 versus +107), angiopoietin 2 (+6.2 versus +3.2), IL-8 (−100 versus −8.9), and α4-integrin (−455 versus −790) using batch 4 RNA in the absence of TPA were fully consistent with the results for batch 3 RNA and with the Incyte gene arrays (Table 8). The RT-PCR results for batch 4 RNA in the presence of TPA were also highly consistent with the other two infected RNA samples (Table 8).
As a final assessment to compare the different RNA preparations and to link the RT-PCR results to the gene array data, we carried out real-time PCR for 17 of the identified most highly regulated genes using the same four RNA samples (batch 2 RNA) that were used for the original Incyte array experiments (Table 9). The results revealed overall very high consistency between the three different RNA preparations and between the different RT-PCR assays, as well as more resemblance between RT-PCR results for infected DMVEC plus and minus TPA than either within the gene array data or between the two sets of gene array data.
Once again in KSHV-infected cells, Myx R1, exodus 2, RDC1, TIMP1, and SISI3 were highly upregulated, and IRF7 and angiopoietin 2 were weakly upregulated, but in contrast to the previous results, in this case CD14 and metallothionine H1 were unaffected by infection in either the absence or presence of TPA. Similar to the previous assays, BMP4 and α4-integrin were heavily downregulated (145- to 325-fold), and connective tissue growth factor, fibronectin, IL-8, PAI-1, and MMP2 were moderately downregulated (15- to 50-fold).
BMP4, connective tissue growth factor, IL-8 and fibronectin were also all downregulated to similar levels as in infected cells by TPA treatment alone, whereas α4-integrin, MMP2, PAI-1, and TGFβ3 were all essentially unaffected by TPA treatment in uninfected cells. Furthermore, Myx R1 was further upregulated by TPA in infected cells, but the effects of TPA on angiopoietin 2, BMP4, connective tissue growth factor, exodus 2, IL-8, PAI-1, RDC1, SISI3, TGFβ3, and TIMP1 in infected cells were heavily reduced, abolished, or even reversed in TPA-treated infected cells.
Surprisingly, while confirming most of the gene array data, the real-time RT-PCR results nevertheless indicated that the +7.6 and +4.9 values for metallothionine H1 and CD14 in infected compared to uninfected RNA in the Clontech gene array with batch 1 RNA (and matching upregulation by RT-PCR in batch 3 and 4 RNA) were not reproducible in the Incyte batch 2 RNA preparation or in the Incyte array data for CD14. The real-time RT-PCR results for IL-8 (−50 and −35) also resolved the previously inconsistent array data, where only the Clontech +TPA value of −4.6 appears to be accurate, compared to the Clontech +TPA value of +2.1 and the not significantly regulated results for both of the Incyte surveys. Similarly, the real-time RT-PCR results for connective tissue growth factor with batch 2 RNA are far more consistent with the Incyte array result than with the Clontech array results.
Overall, a total of 14 cellular genes were confirmed to be consistently upregulated in KSHV-infected DMVEC by either direct semiquantitative RT-PCR or real-time RT-PCR or both, including angiopoietin 2, exodus 2, calgranulin B, CD14, IRF7, Myx R1, neurogranin, RDC1, SISI3, TGF-β3, TIMP1, and thrombomodulin in the absence of TPA, and also nucleobindin and VWF in the presence of TPA. Similarly, 13 genes overall were confirmed to be downregulated by infection, including BMP4, connective tissue growth factor, CD34, connexin 37, endothelin 1, fibronectin 1, α4-integrin, IL-8, MMP2, p57-KIP2, PAI-1, and thrombospondin 1 in the absence of TPA, and also CXCR4 weakly in the presence of TPA. In contrast, seven genes tested, including DCR3, GRK4, HIF1α, PLAT, RAD23, STAT1, and TC21, were not confirmed to be regulated, and the results for β1-catenin, CD14, and metallothionine H1 were inconsistent between different RNA preparations.
Evaluation of cellular IRF7 protein expression in infected DMVEC and PEL cells.
To begin to address whether the observed RNA-level regulation is also manifested at the protein level, we performed a Western immunoblot analysis of the cellular IRF7 protein as a representative of the many interferon response genes that were upregulated. The results showed that the moderate upregulation of the IRF7 transcript seen in both types of arrays and in the RT-PCR experiments correlated with a large increase in IRF7 protein levels in infected cells in both the presence and absence of TPA, compared to almost undetectable levels in uninfected cells (Fig. 8). Importantly, there was also no detectable IRF7 protein in the TPA-treated uninfected DMVEC cell extract, which correlates with the absence of increased IRF7 RNA after TPA treatment alone in both the RT-PCR and gene array data.
FIG. 8.
Induction of cellular IRF7 RNA and protein expression in DMVEC cultures by KSHV infection but not by TPA treatment. (A) Comparison of Clontech gene array hybridization results for the IRF7 gene with 32P-labeled cDNA representing uninfected (UI) DMVEC, KSHV-infected (Inf) DMVEC, TPA-treated uninfected DMVEC, and TPA-treated KSHV-infected DMVEC. (B) Western blot analysis of IRF7 protein expression in the same uninfected and infected DMVEC cultures used for the Clontech gene array experiments. Lane 1, control uninfected DMVEC extract; lane 2, KSHV-infected DMVEC extract; lane 3, uninfected DMVEC +TPA extract; and lane 4, KSHV-infected DMVEC +TPA extract.
To ask whether the IRF7 activation occurred in all latently infected cells or just in the subpopulation of cells that had entered the lytic cycle, we also carried out IHC analysis of both infected DMVEC and PEL cell cultures in the presence and absence of TPA. Somewhat surprisingly, in single-label experiments, IRF7 protein proved to be strongly expressed in the cytoplasm but in just a small fraction of the cells (not shown). Furthermore, subsequent double-label IHC experiments revealed that high levels of IRF7 protein were detectable in the cytoplasm of only the same few cells that also expressed the early nuclear lytic cycle protein K8.
This is illustrated for the BCBL1 PEL cell line in double-label IHC in Fig. 9. In the absence of TPA, both the K8 (red nuclear) and IRF7 (blue cytoplasmic) proteins were detected together in approximately 1% of the cells (top left-hand panel). At 48 h after TPA treatment, the fraction of both K8- and IRF7-positive cells increased to nearly 10% (top right-hand panel), and again, the majority of those cells showed coexpression of both proteins. Parallel control IHC staining revealed that both the K8 (blue nuclear) and virus-encoded vIL6 (red cytoplasmic) proteins were also being expressed together in 10% of the cells after TPA treatment (lower right-hand panel), but that vIRF7 protein expression (red cytoplasmic) occurred in many fewer and usually different cells than those that expressed the latent-state LANA1 protein (brown nuclei, lower left-hand panel).
FIG. 9.
Cellular IRF7 protein expression is induced by KSHV only in lytically infected cells. The photomicrographs show double-label immunohistochemical detection of cellular IRF7 protein only in infected PEL cells expressing lytic cycle viral proteins, but not in the majority of LANA1-positive latently infected PEL cells. (Upper left) Viral ORF-K8 (red nuclear chromogen) and cellular IRF7 (blue cytoplasmic chromogen) in untreated KSHV-positive BCBL1 cells; (upper right) ORF-K8 (red nuclei) and IRF7 (blue cytoplasm) in BCBL1 cells treated with TPA for 60 h; (lower left) IRF7 (red cytoplasm) and LANA1 (brown nuclei) in TPA-treated BCBL1 cells; (lower right) ORF-K8 (blue nuclei) and vIL6 (red cytoplasm) in TPA-treated BCBL1 cells.
DISCUSSION
KSHV has been found to be associated with all forms and stages of KS, as well as BCBL and MCD. Characteristic KS pathology includes angiogenesis and immune dysregulation, and in late stages, invasive potential. We have been interested in characterizing changes that occur in cellular gene expression profiles upon infection with KSHV. Previously, these studies have been limited to the use of KS biopsy specimens and latently infected PEL cell lines derived from AIDS patients. However, these systems have proven to be suboptimal because of the lack of matched, noninfected cells and the frequent additional presence of EBV in PEL cell lines. Therefore, the recent development of an effective method for de novo KSHV infection of human DMVEC promises to greatly facilitate characterization of changes in gene expression upon KSHV infection (9, 14, 28).
There are several important considerations for the study of KSHV infection of DMVEC. First, evidence suggests that KS spindle cells may be derived from vascular or lymphatic endothelial cells; hence, the changes in endothelial cell gene expression may relate directly to the pathology of KS lesions. Second, the changes in morphology observed upon KSHV infection of DMVEC are consistent with the cellular morphology of KS spindle cell biopsy samples. Finally, the demonstration of expression of known latent KSHV gene products in spindle-shaped, infected DMVEC and the expression of known lytic KSHV gene products in rounded-up infected DMVEC found in plaques indicates that both latent and productive lytic infection processes occur in this cell type (14, 29).
DNA microarray experiment results are extraordinarily sensitive to a number of factors that may influence mRNA expression and stability, including the age and passage number of cells. Control cells in all experiments were grown from the same initial stock of DMVEC cells as the cells that were used for infection. Additionally, control and infected cells were passaged in parallel to ensure matching passage number and age of culture at the time of harvest.
To explore the validity of performing array experiments to characterize gene expression in a cell culture system, as well as to screen a much larger set of human genes, the experiments were performed using two different types of arrays, radioactive, user-hybridized, single-channel Clontech arrays (2,350 genes) and nonradioactive, company-hybridized, two-channel Incyte arrays (9,180 genes). Polyadenylated RNA used for these experiments was derived from two independent sets of RNA preparations from cultured infected and uninfected DMVEC cultures, each in the presence and absence of TPA, and the subsequent RT-PCR confirmation experiments used a total of four independent DMVEC RNA batches.
Our initial screen of the Clontech and Incyte arrays yielded a number of regulated genes consistent with both the natural history of KS and the current hypotheses regarding function of KSHV genes. Overall, significant changes occurred in the expression of between 1.4 and 2.5% of the genes screened, including many genes likely to be involved in cytoskeletal arrangement, interferon regulation, apoptosis, angiogenesis, and immune regulation. A number of the most highly regulated of these genes were then examined by both standard and real-time RT-PCR in multiple RNA preparations to confirm their status.
There was a reasonable correlation between the most highly regulated genes on the Clontech arrays and the most highly regulated genes on the Incyte arrays. For example, 7, 12, 6, and 7 of the top 20 genes in the four Clontech regulated lists also appeared within the top 15% of the similarly regulated Incyte lists (I-R approximately ≥1.4), although at the more stringent significance level of I-R ≥ 1.74 the numbers fell to only 2 of 12, 8 of 16, 2 of 12, and 1 of 16 among those that were also represented within the Incyte array data. Genes that behaved similarly in both the Clontech top 20 list and top 50 Incyte ranking lists included RDC1, BMP4, PAI-1, connexin 37, BMS1, IGFBP10, BMP6, and thrombin receptor. However, a robust correlation of this type was confounded by the numerous genes that were represented on only one of the arrays, as well as by distortions attributed to the large number of upregulated interferon response genes in the Incyte array (which were largely absent from the Clontech array).
Nevertheless, the success of our RT-PCR confirmation results suggests that both our model cell culture system for KSHV infection and the arrays themselves produced repeatable results for 27 of the 37 genes examined carefully. The other 10 appear to represent a combination of either errors or misidentification in the gene array data or as yet inconsistent or indeterminate data. This level of nearly 75% reproducibility by RT-PCR analysis appeared to be similar for both the Clontech and Incyte survey data. Obviously, at least one layer of RNA-level confirmation is necessary beyond array screening to confirm DNA microarray results. Extensive RT-PCR analysis of four different RNA batches by a variety of approaches revealed substantial inconsistency between them for only two genes (CD14 and metallothionine H1), implying that biological variation was only likely to explain a small subset of the cases in which the two arrays and/or the RT-PCR results did not correlate.
We had expected that at least some of the genes that responded differently to infection in the presence of TPA compared to in its absence might represent the effects of increased KSHV lytic cycle expression, because the design of the gene array experiments (i.e., comparison of infected cells plus TPA with parallel uninfected cells plus TPA) was intended to eliminate any direct effects of TPA. However, the flexibility provided in the single-channel Clontech data of also being able to compare uninfected cells in the presence and absence of TPA as well as infected cells in the presence and absence of TPA revealed that the situation was more complicated. In particular, a significant subset of the genes that responded to KSHV infection also responded similarly and independently to TPA treatment alone (e.g., BMP4, BMSA1, connexin 37, CXCR4, MCP-R1, multimerin, Myx R1, PAI-1, and TIMP1. Furthermore, some cellular genes responded very differently to TPA in the presence of a KSHV latent state than in the absence of the virus, and in some cases the response to both KSHV infection and TPA treatment was additive, while in others the two effects appeared to be cancelled out when in combination. In addition, because of the low levels (10%) of spontaneous lytic gene expression in the DMVEC cultures even before TPA treatment, it is probable that at least some upregulated genes, but presumably not any of the downregulated genes, in the uninduced latently infected cells may actually represent cellular genes that are activated only in the lytic cycle (e.g., IRF7).
Interestingly, one of the confirmed upregulated genes in both the Clontech array and most of the RT-PCR analysis in the absence of TPA was CD14, which agrees with a previous report that KS cells coexpress both endothelial cell and macrophage antigens, including CD14 (41). In contrast, CD34 was downregulated in the absence of TPA in both the Incyte array and the RT-PCR data. Similarly, VWF mRNA expression was recorded as modestly downregulated by both Clontech gene array and RT-PCR. VWF-positive cytoplasmic Weible-Palade bodies are a prominent endothelial cell-specific feature of the uninfected DMVEC, and we have already reported that the level of VWF protein as detected by IFA is greatly reduced in KSHV-infected DMVEC (14). Some other typical endothelial markers such as E-selectin, E-cadherin, and VE-cadherin did not show significant regulation at the RNA level.
The interferon-regulated genes were a major group of upregulated genes in our experiments. Among the top 20 genes in the Incyte array were four (STAT1, IFI 6-16, Myx R1 [MxA] p78, and IFI 9-27) that were also found to be significantly upregulated by Renne et al. in their array analysis of LANA1-expressing B-cell lines (34). In fact, 28 of 29 interferon-inducible genes represented in the Incyte array were significantly upregulated in KSHV-infected cells. Myx R1 proved to be one of the most highly upregulated genes that we encountered, giving real-time RT-PCR values ranging between +17 and +107.
Both our Clontech and Incyte gene array results indicated a significant increase in the amount of IRF7 mRNA, possibly consistent with cellular upregulation of the gene to compensate for interference by the viral IRF3 (or vice versa). All of the RT-PCR results also confirmed a modest upregulation of IRF7 in infected but not in TPA-treated cells, and our Western blot analysis confirmed a massive increase in cellular IRF7 protein expression upon KSHV infection of DMVEC. Finally, the IHC photographs indicated that highly upregulated levels of cytoplasmic IRF7 protein occurred in all KSHV-infected DMVEC and PEL cells undergoing early lytic cycle induction, as judged from coexpression of cytoplasmic cellular IRF7 and nuclear virus-encoded ORF-K8. However, the IRF7 protein was not detectable by IHC in the majority of latently infected LANA1-positive cells. Additionally, our array results confirmed a lack of significant upregulation of IFN-α, IFN-β, or IFN-γ or their receptors in either untreated or TPA-treated infected compared to uninfected DMVEC mRNA samples.
Differential gene display and gene array evidence from lytic cycle human cytomegalovirus (HCMV)-infected fibroblasts also indicates that numerous interferon responses are upregulated by HCMV, although some are directly related to virion surface contacts and do not require de novo HCMV gene expression (45, 46). Zhang and Pagano have previously reported the induction of cellular IRF7 in response to EBV infection and, specifically, LMP1 expression (43, 44). KSHV, in turn, encodes at least three viral interferon regulatory factors (vIRFs), which have been shown to interfere with the cellular interferon responses induced upon infection.
Li et al. have reported that vIRF1 interferes directly with the cellular transcriptional coactivator p300 and displaces p300/CBP-associated factor, thereby inhibiting the histone acetyltransferase activity of p300 (25). According to the work of Lubyova and Pitha, KSHV encodes another viral interferon regulatory factor, vIRF3, with homology to cellular IRF4 and KSHV vIRF2, which is transcriptionally upregulated in KSHV-infected BCBL-1 cells upon treatment with TPA (26). vIRF3 may function as a dominant negative version of both cellular IRF3 and IRF7, inhibiting transcription from the IFN-α promoter and preventing the synthesis of biologically active IFN-α and IFN-β.
A second important category of regulated genes included those involved in cell signaling and angiogenesis. The human homolog of RDC1, an orphan G-protein-coupled receptor, was found to be upregulated on both arrays and by RT-PCR. RDC1 is a CXC alpha chemokine receptor that is present in endothelial cell precursors and has been implicated as a human immunodeficiency virus (HIV) coreceptor molecule (24). KSHV encodes a novel G-protein-coupled receptor (vGCR, ORF74) which has been shown to induce tumor formation and neovascularization in nude mice (2, 4).
The upregulation of the RDC1 homolog gene may play a role in angiogenesis or tumor formation in KS. Additionally, GPCR receptor kinases (GRKs) have been shown to inhibit vGCR signaling (18). GRK4 was found to be significantly downregulated in KSHV-infected cells treated with TPA in the Clontech array, but not in the infected cells that were not treated with TPA. Hypoxia-induced transcription factor 1α (HIF1α) was also detected as an upregulated gene in the infected cells in the absence of TPA in the Incyte array and has recently been shown to be upregulated as part of the mechanism that upregulates VEGF in response to KSHV vGCR (ORF74) expression (36). However, the apparent regulation of both GRK4 and HIF1α detected in the gene arrays was not confirmed by initial RT-PCR analysis in later RNA preparations and requires further evaluation.
Significant regulation of the tissue plasminogen activator (PLAT) and endothelial plasminogen activator inhibitor (PAI-1) system occurred on both the −TPA and +TPA Incyte arrays, but only the PAI-1 result was consistently confirmed by RT-PCR. Plasminogen activators cleave the zymogen plasminogen to form plasmin, an enzyme which is involved in the degradation of extracellular matrix proteins and the activation of growth factors (1). High levels of plasminogen activator and, paradoxically, its inhibitor (PAI) have been associated with tumor invasion and angiogenesis (1, 5, 30, 31). However, there is contradictory evidence that recombinant PAI-1 inhibits angiogenesis and reduces the size of tumors (38). Interestingly, basal levels of both PLAT and PAI-1 RNA were high in uninfected DMVEC, whereas basal levels of the urokinase form of plasminogen activator and PAI-2 were undetectable in the Clontech arrays.
There was also a significant RT-PCR-confirmed downregulation of the angiogenesis inhibitor thrombospondin 1 (TSP1). TSP1 has been shown to be present in lower amounts in the supernatants of cultured KS cells compared with the supernatants of cultured endothelial cells, and also is sparsely expressed in KS lesions by immunohistochemical analysis (39). TSP1 and TSP2 have also both been reported to be decreased by HCMV infection in fibroblasts (13). Dysregulation of TSP1, angiopoietin 2, Cys-rich angiogenesis inducer (IGFBP10), and thrombomodulin by KSHV may all potentially contribute to angiogenesis in KS lesions, a prominent and defining feature of the disease. Matrix metalloproteinase functions are also implicated in angiogenesis (21), and, potentially consistent with this notion, both the −TPA and +TPA Clontech arrays indicated upregulation of the TIMP1 inhibitor of metalloproteinases in infected cells, which was confirmed by the RT-PCR results. However, the Incyte array data and real-time PCR results show that MMP2 itself was downregulated by KSHV infection and that most other matrix metalloproteinases were unaffected.
Noticeably, some of the most potent angiogenic growth factors and their receptors, including VEGF-A, VEGF-C, VEGF-R1, VEGF-R2, VEGF-R3, basic FGF, TGFβ1, and platelet-derived growth factor, were not strongly influenced by KSHV infection in DMVEC at the transcriptional level. Only VEGF-related placental growth factor was among the highly regulated genes, being downregulated by infection in the presence of TPA in the Incyte array. Similarly, endothelin 1, which was previously reported to be highly expressed in a KS cell line lacking KSHV (3), was downregulated in the Incyte array and RT-PCR assay in infected cell samples in both the presence and absence of TPA.
Bone morphogenetic protein 4 (BMP4) was confirmed to be heavily downregulated by RT-PCR in all assays in the absence of TPA (real-time RT-PCR fold values of −42, −50, and −145) and, together with the two other confirmed most strongly downregulated genes, namely, α4-integrin (−450, −325, and −790) and fibronectin I (−22, −40, and −57), appears almost certain to represent a latency-associated effect (because no more than 10% of the cells show evidence of lytic cycle viral gene expression in the absence of TPA). BMP4 is an important developmental control protein in the tumor necrosis factor alpha (TNF-α) receptor family that is abundantly expressed in neural embryonic endoderm tissue and certain other endothelial cells. It is evidently one of the most strongly expressed genes in the cultured uninfected DMVEC cells (Table 1), but its expression was nearly completely obliterated in KSHV-infected spindle cell RNA (Fig. 7).
α4-Integrin (CD49D) plays a role in cell-cell interactions and cell adhesion to the extracellular matrix. In particular, CD49D forms the α4 subunit of the T-cell VLA-4 complex that acts as both a fibronectin receptor and a ligand for VCAM-1 and also binds to thrombospondin. Because α4-integrin, fibronectin, and thrombospondin are all downregulated, this may reflect a coordinated response associated with the change from the cobblestone contact-inhibited morphology of uninfected DMVEC to the aligned elongated spindle cell morphology of KSHV latently infected DMVEC.
A third category of regulated genes included those involved in cell cycle progression and apoptosis, although the effects here appear more subtle. KSHV is known to encode its own D-type cyclin (vCYC/ORF72) as well as functional vFLIP and vBcl2 proteins (12). Among many genes in this category that were apparently specifically but only modestly regulated after infection in the Clontech and Incyte arrays were CDC25B and p57-KIP2. CDC25B is thought to be the “starter” phosphatase which sends the cell into mitosis at the G2/M checkpoint, although there is contradictory information that it may play a role in progression of the cell into S phase (17, 23). Its upregulation may indicate that cells infected with KSHV undergo uncontrolled mitosis, leading to cell proliferation and tumor formation.
We found p57-KIP2 to be modestly downregulated on both the Clontech −TPA array and by standard RT-PCR techniques (−2.5-fold). p57-KIP2 has been described as a potent inhibitor of cell cycle progression in the G1 phase (27) and has been shown to induce senescence in human astrocytomas (40). Again, it is possible that KSHV downregulates expression of p57-KIP2 during latency to prevent cell cycle arrest, although a block in cell cycle progression would be expected to occur only after lytic cycle induction. c-Jun expression was both initially high (basal Clontech rank 60) and further upregulated (+2.0 C-SD) by infection, but PCNA was less affected.
Cellular gene arrays will be of considerable value in the analysis of the complex transcriptional regulation of cellular genes that occurs during herpesvirus infection and have already been used successfully for HCMV infection of fibroblast cell cultures (45). While isolated gene studies are important for the elucidation of individual pathways that are involved in the regulation of cell growth, they cannot match the environment that exists when the complete viral gene expression and gene regulation pathways occur in vivo.
Because of their long evolutionary history (32), herpesviruses are likely to modulate cellular gene expression in multiple, complex, and additive ways to help maintain lifelong latent infections in human cells. The results obtained in this initial gene array survey are closely aligned with our current knowledge of KSHV biology and will undoubtedly lead to more extensive future evaluation of the roles of the genes identified here. Although RT-PCR analysis confirmed the array results for 14 upregulated and 13 downregulated genes, the discordance between some of the data from the two different array techniques used and between the array data and RT-PCR results for at least 7 of the 37 genes examined carefully here emphasizes the need for caution and confirmation by other techniques.
Overall, these results represent a strong starting point for the analysis of links between KSHV infection and cellular gene dysregulation. Extensive additional studies will be required to differentiate between which of these cellular genes respond only to virion surface contacts or only to KSHV latent state gene products, compared to those, like IRF7, that are evidently regulated preferentially during progression into the viral lytic cycle.
Acknowledgments
This work was supported by research grants R01 CA73585, R01 CA81422, and P01 CA81400 to G.S.H. L.J.P. was partially supported by a Training Grant in Biochemistry, Cellular and Molecular Biology (5T32-GM07445), and P.S.K. was partially supported by a Training Grant in Pharmacology (T32-GM08763).
We thank Amy E. Purcell, Christopher M. L. Bouton, and Carlo Colantuoni for their assistance with array analysis procedures.
REFERENCES
- 1.Andreasen, P. A., L. Kjoller, L. Christensen, and M. J. Duffy. 1997. The urokinase-type plasminogen activator system in cancer metastasis: a review. Int. J. Cancer 72:1-22. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis, L., E. Geras-Raaka, A. Varma, M. C. Gershengorn, and E. Cesarman. 1997. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature 385:347-350. [DOI] [PubMed] [Google Scholar]
- 3.Bagnato, A., L. Rosano, V. Di Castro, A. Albini, D. Salani, M. Varmi, M. R. Nicotra, and P. G. Natali. 2001. endothelin receptor blockade inhibits proliferation of Kaposi's sarcoma cells. Am. J. Pathol. 158:841-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bais, C., B. Santomasso, O. Coso, L. Arvanitakis, E. G. Raaka, J. S. Gutkind, A. S. Asch, E. Cesarman, M. C. Gershengorn, E. A. Mesri, and M. C. Gerhengorn. 1998. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature 391:86-89. [DOI] [PubMed] [Google Scholar]
- 5.Bajou, K., A. Noel, R. D. Gerard, V. Masson, N. Brunner, C. Holst-Hansen, M. Skobe, N. E. Fusenig, P. Carmeliet, D. Collen, and J. M. Foidart. 1998. Absence of host plasminogen activator inhibitor 1 prevents cancer invasion and vascularization. Nat. Med. 4:923-928. [DOI] [PubMed] [Google Scholar]
- 6.Bellows, D. S., B. N. Chau, P. Lee, Y. Lazebnik, W. H. Burns, and J. M. Hardwick. 2000. Antiapoptotic herpesvirus Bcl-2 homologs escape caspase-mediated conversion to proapoptotic proteins. J. Virol. 74:5024-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouton, C., and J. Pevsner. 2000. Dragon: database referencing of array genes online. Bioinformatics 16:1038-1039. [DOI] [PubMed] [Google Scholar]
- 8.Buchbinder, A., and A. E. Friedman-Kien. 1991. Clinical aspects of epidemic Kaposi's sarcoma. Cancer Surv. 10:39-52. [PubMed] [Google Scholar]
- 9.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cesarman, E., R. G. Nador, F. Bai, R. A. Bohenzky, J. J. Russo, P. S. Moore, Y. Chang, and D. M. Knowles. 1996. Kaposi's sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi's sarcoma and malignant lymphoma. J. Virol. 70:8218-8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, E. H., J. Nicholas, D. S. Bellows, G. S. Hayward, H. G. Guo, M. S. Reitz, and J. M. Hardwick. 1997. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc. Natl. Acad. Sci. USA 94:690-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cinatl, J., Jr., M. Bittoova, S. Margraf, J. U. Vogel, J. Cinatl, W. Preiser, and H. W. Doerr. 2000. Cytomegalovirus infection decreases expression of thrombospondin-1 and -2 in cultured human retinal glial cells: effects of antiviral agents. J. Infect. Dis. 182:643-651. [DOI] [PubMed] [Google Scholar]
- 14.Ciufo, D., J. S. Cannon, L. J. Poole, F. Wu, P. Murray, R. F. Ambinder, and G. S. Hayward. 2001. Spindle cell conversion by Kaposi's Sarcoma-Associated Herpesvirus: Formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J. Virol. 75:5614-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delabesse, E., E. Oksenhendler, C. Lebbe, O. Verola, B. Varet, and A. G. Turhan. 1997. Molecular analysis of clonality in Kaposi's sarcoma. J. Clin. Pathol. 50:664-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flore, O., S. Rafii, S. Ely, J. J. O'Leary, E. M. Hyjek, and E. Cesarman. 1998. Transformation of primary human endothelial cells by Kaposi's sarcoma-associated herpesvirus. Nature 394:588-592. [DOI] [PubMed] [Google Scholar]
- 16a.Gao, S. J., C. Boshoff, S. Jayachandra, R. A. Weiss, Y. Chang, and P. S. Moore. 1997. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene 15:1979-1985. [DOI] [PubMed] [Google Scholar]
- 17.Garner-Hamrick, P. A., and C. Fisher. 1998. Antisense phosphorothioate oligonucleotides specifically downregulate cdc25B causing S-phase delay and persistent antiproliferative effects. Int. J. Cancer 76:720-728. [DOI] [PubMed] [Google Scholar]
- 18.Geras-Raaka, E., L. Arvanitakis, C. Bais, E. Cesarman, E. A. Mesri, and M. C. Gershengorn. 1998. Inhibition of constitutive signaling of Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor by protein kinases in mammalian cells in culture. J. Exp Med. 187:801-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermans, P. 1998. Epidemiology, etiology and pathogenesis, clinical presentations and therapeutic approaches in Kaposi's sarcoma: 15-year lessons from AIDS. Biomed. Pharmacother. 52:440-446. [DOI] [PubMed] [Google Scholar]
- 20.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene statement as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.John, A., and G. Tuszynski. 2001. The role of matrix metalloproteinases in tumor angiogenesis and tumor metastasis. Pathol. Oncol. Res. 7:14-23. [DOI] [PubMed] [Google Scholar]
- 22.Judde, J. G., V. Lacoste, J. Briere, E. Kassa-Kelembho, E. Clyti, P. Couppie, C. Buchrieser, M. Tulliez, J. Morvan, and A. Gessain. 2000. Monoclonality or oligoclonality of human herpesvirus 8 terminal repeat sequences in Kaposi's sarcoma and other diseases. J. Natl. Cancer Inst. 92:729-736. [DOI] [PubMed] [Google Scholar]
- 23.Lammer, C., S. Wagerer, R. Saffrich, D. Mertens, W. Ansorge, and I. Hoffmann. 1998. The cdc25B phosphatase is essential for the G2/M phase transition in human cells. J. Cell Sci. 111:2445-2453. [DOI] [PubMed] [Google Scholar]
- 24.Law, N. M., and S. A. Rosenzweig. 1994. Characterization of the G-protein linked orphan receptor GPRN1/RDC1. Biochem. Biophys. Res. Commun. 201:458-465. [DOI] [PubMed] [Google Scholar]
- 25.Li, M., H. Lee, J. Guo, F. Neipel, B. Fleckenstein, K. Ozato, and J. U. Jung. 1998. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor. J. Virol. 72:5433-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lubyova, B., and P. M. Pitha. 2000. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. J. Virol. 74:8194-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuoka, S., M. C. Edwards, C. Bai, S. Parker, P. Zhang, A. Baldini, J. W. Harper, and S. J. Elledge. 1995. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 9:650-662. [DOI] [PubMed] [Google Scholar]
- 28.Moses, A. V., K. N. Fish, R. Ruhl, P. P. Smith, J. G. Strussenberg, L. Zhu, B. Chandran, and J. A. Nelson. 1999. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J. Virol. 73:6892-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orenstein, J. M., D. M. Ciufo, J. P. Zoeteweij, A. Blauvelt, and G. S. Hayward. 2000. Morphogenesis of HHV8 in primary human dermal microvascular endothelium and primary effusion lymphomas. Ultrastruct. Pathol. 24:291-300. [DOI] [PubMed] [Google Scholar]
- 29a.Paulose-Murphy, M., H. Nguyen-Khoi, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen, H., J. Grondahl-Hansen, D. Francis, K. Osterlind, H. H. Hansen, K. Dano, and N. Brunner. 1994. Urokinase and plasminogen activator inhibitor type 1 in pulmonary adenocarcinoma. Cancer Res. 54:120-123. [PubMed] [Google Scholar]
- 31.Pedersen, H., N. Brunner, D. Francis, K. Osterlind, E. Ronne, H. H. Hansen, K. Dano, and J. Grondahl-Hansen. 1994. Prognostic impact of urokinase, urokinase receptor, and type 1 plasminogen activator inhibitor in squamous and large cell lung cancer tissue. Cancer Res. 54:4671-4675. [PubMed] [Google Scholar]
- 32.Poole, L. J., J. C. Zong, D. M. Ciufo, D. J. Alcendor, J. S. Cannon, R. Ambinder, J. M. Orenstein, M. S. Reitz, and G. S. Hayward. 1999. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J. Virol. 73:6646-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabkin, C. S., S. Janz, A. Lash, A. E. Coleman, E. Musaba, L. Liotta, R. J. Biggar, and Z. Zhuang. 1997. Monoclonal origin of multicentric Kaposi's sarcoma lesions. N. Engl. J. Med. 336:988-993. [DOI] [PubMed] [Google Scholar]
- 34.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simonart, T., P. Hermans, L. Schandene, and J. P. Van Vooren. 2000. Phenotypic characteristics of Kaposi's sarcoma tumour cells derived from patch-, plaque- and nodular-stage lesions: analysis of cell cultures isolated from AIDS and non-AIDS patients and review of the literature. Br. J. Dermatol. 143:557-563. [DOI] [PubMed] [Google Scholar]
- 36.Sodhi, A., S. Montaner, V. Patel, M. Zohar, C. Bais, E. A. Mesri, and J. S. Gutkind. 2000. The Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor upregulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1α. Cancer Res. 60:4873-4880. [PubMed] [Google Scholar]
- 37.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swiercz, R., R. W. Keck, E. Skrzypczak-Jankun, S. H. Selman, and J. Jankun. 2001. Recombinant PAI-1 inhibits angiogenesis and reduces size of LNCaP prostate cancer xenografts in SCID mice. Oncol. Rep. 8:463-470. [DOI] [PubMed] [Google Scholar]
- 39.Taraboletti, G., R. Benelli, P. Borsotti, M. Rusnati, M. Presta, R. Giavazzi, L. Ruco, and A. Albini. 1999. thrombospondin-1 inhibits Kaposi's sarcoma (KS) cell and HIV-1 Tat-induced angiogenesis and is poorly expressed in KS lesions. J. Pathol. 188:76-81. [DOI] [PubMed] [Google Scholar]
- 40.Tsugu, A., K. Sakai, P. B. Dirks, S. Jung, R. Weksberg, Y. L. Fei, S. Mondal, S. Ivanchuk, C. Ackerley, P. A. Hamel, and J. T. Rutka. 2000. Expression of p57KIP2 potently blocks the growth of human astrocytomas and induces cell senescence. Am. J. Pathol. 157:919-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uccini, S., L. P. Ruco, F. Monardo, A. Stoppacciaro, E. Dejana, I. L. La Parola, D. Cerimele, and C. D. Baroni. 1994. Coexpression of endothelial cell and macrophage antigens in Kaposi's sarcoma cells. J. Pathol. 173:23-31. [DOI] [PubMed] [Google Scholar]
- 42.Wu, F. Y., J. H. Ahn, D. J. Alcendor, W. J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, L., and J. S. Pagano. 1997. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol. Cell. Biol. 17:5748-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, L., and J. S. Pagano. 2000. Interferon regulatory factor 7 is induced by Epstein-Barr virus latent membrane protein 1. J. Virol. 74:1061-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 94:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, L., V. Puri, B. Chandran. 1999. Characterization of human herpesvirus-8 K8.1A/B glycoproteins by monoclonal antibodies. Virology 262:237-249. [DOI] [PubMed] [Google Scholar]