Abstract
The ORF74 or vGCR gene encoded by Kaposi's sarcoma-associated herpesvirus (KSHV; also called human herpesvirus 8) has properties of a ligand-independent membrane receptor signaling protein with angiogenic properties that is predicted to play a key role in the biology of the virus. We have examined the expression of vGCR mRNA and protein in primary effusion lymphoma (PEL) cell lines, PEL and multicentric Castleman's disease (MCD) tumors, Kaposi's sarcoma lesions and infected endothelial cell cultures. The vGCR gene proved to be expressed in PEL cell lines as a large spliced bicistronic mRNA of 3.2 kb that also encompasses the upstream vOX2 (K14) gene. This mRNA species was induced strongly by phorbol ester (TPA) and sodium butyrate treatment in the BCBL-1 cell line, but only weakly in the HBL6 cell line, and was classified as a relatively late and low-abundance delayed early class lytic cycle gene product. A complex bipartite upstream lytic cycle promoter for this mRNA was nestled within the intron of the 5′-overlapping but oppositely oriented latent-state transcription unit for LANA1/vCYC-D/vFLIP and responded strongly to both TPA induction and cotransfection with the KSHV RNA transactivator protein (RTA or ORF50) in transient reporter gene assays. A vGCR protein product of 45 kDa that readily dimerized was detected by Western blotting and in vitro translation and was localized in a cytoplasmic and membrane pattern in DNA-transfected Vero and 293T cells or adenovirus vGCR-transduced dermal microvascular endothelial cells (DMVEC) as detected by indirect immunofluorescence assay (IFA) and immunohistochemistry with a specific rabbit anti-vGCR antibody. Similarly, a subfraction of KSHV-positive cultured PEL cells and of KSHV (JSC-1) persistently infected DMVEC cells displayed cytoplasmic vGCR protein expression, but only after TPA or spontaneous lytic cycle induction, respectively. The vGCR protein was also detectable by immunohistochemical staining in a small fraction (0.5 to 3%) of the cells in PEL and MCD tumor and nodular Kaposi's sarcoma lesion specimens that were apparently undergoing lytic cycle expression. These properties are difficult to reconcile with the vGCR protein's playing a direct role in spindle cell proliferation, transformation, or latency, but could be compatible with proposed contributions to angiogenesis via downstream paracrine effects. The ability of vGCR to transactivate expression of both several KSHV promoter-driven luciferase (LUC) reporter genes and an NFκB motif containing the chloramphenicol acetyltransferase (CAT) reporter gene may also suggest an unexpected regulatory role in viral gene expression.
Kaposi's sarcoma-associated herpesvirus (KSHV) is believed to be a major contributing factor to all types of Kaposi's sarcoma (KS), as well as primary effusion lymphoma (PEL) and multicentric Castleman's disease (MCD). Much evidence has been presented that KSHV DNA is consistently present in virtually all KS lesions and that the genomes persist in a latent episomal state in the majority of spindle cells in KS as well as in the in vivo tumor cells of PEL and MCD (13, 19, 22, 42, 62). In addition, viral gene expression has been demonstrated primarily by in situ hybridization for viral mRNA in most KSHV-positive lesions tested.
However, the pattern of viral gene expression in the different types of tumors has not been fully evaluated. In general, at least three latent-state gene products, LANA1 (ORF73), vCYC-D (ORF72), and vFLIP, are believed to be consistently expressed in all cells (24, 27, 65), whereas other gene products, including vIL6 (ORF-K2), ORF26, and the highly abundant T1.1/PAN nuclear RNA, are found to be expressed in only a few cells in vivo and are thought to represent lytic cycle gene products (10).
The establishment of lymphoblastoid cell lines from PEL tumors has revolutionized the study of KSHV gene expression (14, 30, 56, 59, 60). These cell lines carry multicopy KSHV genomes and sometimes also Epstein-Barr virus (EBV) genomes (3, 7, 9, 13, 15, 25). Again these appear to express only the latent-state mRNAs encoding LANA1, vCYC-D, vFLIP, and perhaps T0.7 constitutively in all cells (11, 20, 26, 37, 38, 55, 61, 64). However, other viral genes and even mature extracellular virions can be induced from a variable subfraction of some cell lines in culture after either tetradecanoyl phorbol acetate (TPA) or sodium sodium butyrate treatment (9, 41, 57). A number of the 80 or so virally encoded mRNAs and proteins (46, 58) have been determined to be expressed primarily as lytic cycle class genes under these circumstances, including T1.1/PAN, vIL6, vMIP-B, ORF26, ORF59, ORF65, ORF-K1, and ORF-K8.1 (43, 48, 49, 60, 63, 66, 69).
One of the most interesting genes encoded by KSHV is vGCR (ORF74), which maps in the DL-E region towards the right-hand end of the genome (16, 33, 58). The vGCR protein is predicted to be a 342-amino-acid membrane receptor that has seven transmembrane domains and is a member of the large G-protein-coupled receptor (GPCR) family of related signaling proteins that include the cyclic AMP receptor of Dictyostelium, the mammalian angiotensin, β-adrenergic and rhodopsin/opsin receptors, and the chemokine receptors.
The betaherpesviruses human cytomegalovirus (HCMV), human herpesvirus type 6 (HHV6), and HHV7 each also encode several vGCR proteins, but the KSHV version is believed to have been derived evolutionarily from the mammalian interleukin-8 (IL-8) receptor (IL-8R) family of alpha chemokine receptors (28% identity) and is most closely related to cellular CXCR2 and the positionally equivalent vGCR (ORF74) proteins of other gamma-2 subfamily herpesviruses such as HVS (36% identity) (47), as well as the US28 protein of HCMV (24% identity). Both KSHV vGCR and HCMV US28 have been shown to behave as broad-spectrum low-affinity receptors for both alpha and beta chemokines (1, 2), and US28 can substitute for CCR5 and CXCR4 by acting as a coreceptor for human immunodeficiency virus (HIV) (54). Although there is no vGCR encoded by EBV, a related lymphocyte-specific cellular GCR protein is induced in EBV-transformed lymphoblasts (5).
Cesarman and colleagues have demonstrated that stable expression of the KSHV vGCR gene under the control of the strong HCMV enhancer-promoter region can lead to focus formation in transfected NIH 3T3 cells (2, 4). These cells produce increased levels of biologically active vascular endothelial growth factor (VEGF) and form highly vascular tumors in nude mice. Intriguingly, the vGCR in these cells appears to act as a constitutively active inducer of several mitogen-activated protein kinase (MAPK) signal transduction pathways. The cells express increased levels of inositol phosphate and both p38 MAPK and JNK/SAPK kinases, probably leading to upregulation of the AP1 transcription factor complex.
Despite signaling constitutively in the apparent absence of any specific ligand, vGCR activity can be desensitized by GR kinase and inhibited by the IP-10 chemokine (31, 32). Therefore, vGCR behaves as a ligand-independent potentially transforming angiogenic protein that seems likely to play a major contributing role in pathogenesis of the highly vascular KS lesions. However, exactly how this role is manifested depends greatly on whether the vGCR protein is expressed as a latent or lytic cycle gene product and in what cell types or circumstances this occurs.
The presence of some vGCR mRNA has been detected in the BC1 and BC2 lymphoblastoid PEL cell lines and in KS lesion samples by reverse transcription (RT)-PCR (16, 34), and two previous reports have described a bicistronic spliced vGCR mRNA species in the PEL cell line after sodium butyrate or TPA treatment (40, 68). However, very little is known about the control of vGCR expression, and no one has previously directly detected the vGCR protein product.
In this study, we set out to identify and characterize the KSHV vGCR mRNA and promoter domain and to examine its responsiveness to TPA and viral transactivators. We also generated an antibody to the KSHV-encoded vGCR protein and examined some of its biochemical properties and intracellular distribution in DNA-transfected cells. Finally, we have successfully detected vGCR protein expression in both lymphoid and KS tumor tissue samples as well as in PEL cell lines after lytic cycle induction by TPA and in KSHV-infected cultured endothelial cells.
MATERIALS AND METHODS
KSHV-infected PEL cell lines and tumor specimens.
Six different PEL lymphoblastoid cells were employed in these studies. Three of them, including BCBL-1 (35), BCP1 (7), and BC3 (13), contain KSHV but not EBV genomes, whereas the other three, including HBL6 (29), BC2 (13), and JSC-1 (9), contain both KSHV and EBV genomes. All cultures were grown in RPMI 1640 medium supplemented with either 10 or 20% fetal calf serum (FCS). The MCD specimen was obtained from Johns Hopkins Hospital and was derived from a hyperplastic lymph node with Castleman's-like features from an HIV-positive homosexual man. The PEL tumor specimen derived from an HIV-positive patient was a gift from Ethel Cesarman (Columbia College of Physicians and Surgeons, New York). The KS tumor came from a paraffin block biopsy of a nodular skin lesion from an AIDS patient at Johns Hopkins Dermatology Clinic (referred to elsewhere as AKS1)
KSHV-infected endothelial cell cultures.
A human primary dermal microvascular endothelial cell (DMVEC) culture was obtained from Clonetics Inc. (catalog no. CC2543) and grown and passaged in EGM-2MV medium containing 5% FCS plus endothelial growth factor. The cells were split into two T25 subcultures, one of which (2 × 105 cells) was inoculated with a sample of KSHV virions pelleted from the supernatant medium from 3 × 108 JSC-1 cells (9) that had been treated with TPA for 96 h. Both infected and uninfected cultures were grown continuously for an 8-week period, including five passages (1:3 splits), until the infected culture displayed a completely spindloid phenotype (23). The cells were then grown in LabTek slide chambers for 72 h in the presence or absence of TPA (20 ng/ml) and subjected to single- and double-label immunohistochemistry (immunohistochemical) with the vGCR, vZMP-A (K5), and LANA1 antibodies.
TPA-induced KSHV cDNA Library and vGCR cDNA subclones.
A cDNA library in the λZAPII background, derived from a mixture of BCBL-1 cells treated with TPA for 24, 48, 72, and 96 h, was generated at the University of Kansas Medical Center (17). DNase I-treated mRNA was isolated with an Oligotex mRNA isolation kit (Qiagen, Chatsworth, Calif.), and cDNA was synthesized using an oligo(dT) linker-primer containing an XhoI site (Stratagene, La Jolla, Calif.). Nine vCGR-positive phage lambda cDNA clones (λGCRcl-l to λGCRcl-9) were identified by plaque hybridization with an antisense vGCR (ORF74) riboprobe generated from plasmid pGH398b (vGCR-R) (see below). Positive phages were plaque purified twice, and their DNA was subjected to manual 32P-labeled PCR sequencing with both external and internal primers to determine their structure and DNA sequence.
Northern blot hybridization.
Total cell RNA preparations were extracted from HBL6 or BCBL-1 cell cultures with Trizo1 reagent (Gibco-BRL) at various times either before or after treatment with 5 mM sodium butyrate (sodium butyrate) or 20 ng of TPA) (and, when appropriate, 200 μg of phosphonoacetic acid [PAA]) per ml. The RNA was ethanol precipitated, and the redissolved samples (5 μg per lane) were fractionated by electrophoresis on 1.5% agarose-1.6% formaldehyde gels in MOPS (morpholinepropanesulfonic acid)-EDTA buffer, then transferred to nylon membranes (Nytran; Schleicher & Schuell) by overnight capillary action in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
Confirmation of the effectiveness of transfer and equal loading of RNA samples was carried out by staining with 0.04% methylene blue in acetate buffer (pH 5.2). Strand-specific antisense riboprobes labeled with [32P]UTP were generated with T7 RNA polymerase (Stratagene mCAP kit) from linearized DNA templates of plasmids encoding the simian virus 2 (SV2) vGCR or SV2 vIL6 gene (51) and hybridized to the blots at 42°C for 12 h in the presence of 50% formamide-5× SSPE-5× Denhardt's-0.1% sodium dodecyl sulfate (SDS) plus 25 μg of salmon DNA and 20 μg of poly(dA) per ml as nonspecific carrier nucleic acids (48). Prewashing and blocking were carried out in the same buffer, and multiple posthybridization washes were carried out in 1× SSC-0.1% SDS at 68°C.
Primer extension assays.
Specific identification of mRNA 5′ ends was carried out by primer extension using synthetic oligonucleotide primers that were designed to anneal at positions between 60 and 220 bp downstream of several possible 5′ mRNA start sites. Primers tested included vOX2-1 (LGH3163), 5′-CAGGTGTACATGATTTGTGTTAAGG-3′; vOX2-2 (LGH3008), 5′-GTGTGTATCATTTGGAGGCAGCTGC-3′; vOX2-3 (LGH3164), 5′-CAACCGGGAGCACAGTGGGGGGTAA-3′; vOX2-4 (LGH3165), 5′-CACAGACGACAGGTCGGACCACTG-3′; vGCR (LGH3005), 5′-TTAGAGTTTCATTCCAGGATTCATC-3′; LANA1 (LGH3017), 5′-GTTTATAAGTCAGCCGGACCAAGCTG-3′; vIL6 (LGH2404), 5′-CCAGCACATGGCTGCTAACGCGGCATAC-3′; and vT1.1/PAN (LGH3000), 5′-GAAGGCAAGCAGCGAGCACAAAATC-3′.
Oligo(dT)-selected polyadenylated RNA was isolated from HBL6, BCBL-1, or DG75 cells using a Poly(A) pure mRNA kit (Ambion) at various times either before or after treatment with sodium butyrate or TPA. One picomole of 32P-end-labeled primer (10,000 cpm) generated with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]dCTP (3,000 μCi/μmol) was annealed to 20 μg of total RNA by heating at 90°C for 10 min, incubation at 50°C for 20 min, and then slow cooling to room temperature. Extension was carried out using 200 U of Superscript II reverse transcriptase (Gibco-BRL) in the presence of 1× Superscript II buffer, 0.5 mM deoxynucleoside triphosphates, and 0.01 M dithiothreitol (DTT). The products were precipitated at −20°C with 1 μg of carrier DNA in 3 volumes of ethanol and 1/10 volume of 3 M sodium acetate (pH 5.2), resuspended in DNA loading dye buffer, and then fractionated by electrophoresis on 7% acrylamide-7 M urea sequencing gels (1 μg of polyadenylated RNA per lane), followed by autoradiography.
5′RACE analysis.
Polyadenylated RNA was generated from selected PEL cell lines using the Poly(A) Pure mRNA kit (Ambion). 5′ Random amplification of cDNA ends (RACE) was carried out according to the manufacturer's directions for the 5′RACE system, version 2.0 (Gibco-BRL). Polyadenylated RNA (250 ng) was reverse transcribed at 50°C for 1 h using Superscript II, treated with RNase for 30 min at 37°C, and then purified by Glassmax spin column. cDNAs were dC-tailed using terminal deoxytransferase for 20 min at 37°C, followed by heat inactivation.
The first round of PCR consisted of 45 cycles using primer LGH3165 (vGCR 5′RACE experiments) and the abridged anchor primer provided in the kit. The PCR products were diluted 1:100 and subjected to nested PCR consisting of 45 cycles using primer LGH3164 (vGCR) and the AUAP primer provided with the kit. Bands were visualized by 1.5% agarose gel electrophoresis and ethidium bromide staining. Bands were excised and sequenced to verify their structure.
KSHV vGCR and HCMV US28 expression vectors.
The 1,029-bp KSHV vGCR coding region from genomic positions 129371 to 130399 (33, 58) was amplified as an EcoRI-Bg1II fragment by PCR procedures with the primers 5′-CGCATGAATTCCTTGTTATTGTAGGCCATGGCGG-3′ and 5′-CGATGAGATCTGGCTACGTGGTGGCG-3′ (restriction sites are underlined and italicized) directly from DNA of KHSV (BCBL-R) phage λB6-1 (50), then inserted into the simian virus 40 (SV40) enhancer-driven mammalian expression vector plasmid pSG5 in both orientations to generate plasmids pGH398a and pGH398b, containing the wild-type pSV2 vGCR(1-342) gene. A truncated version encoding the SV2 vGCR(1-282) gene in pCJC551 was also generated by BamHI deletion. The vGCR(1-342) open reading frame (ORF) was also moved into a version of pSG5 containing an in-frame 3′-Flag motif to generate the pPSK1 expression plasmid containing the SV2 vGCR/Flag gene.
The HCMV US28 expression vectors were prepared similarly by PCR from HCMV (Towne) template DNA using primers 5′-GCAGAACTGGTGCTATC-3′ (LGH2565) and 5′-GGTTTGTATGAAAAGGC-3′ (LGH2800), followed by insertion into the pCR2.1 TA cloning vector. The inserts were then transferred into pSG5 or the pSG5-based 3′-Flag vector pYW51 to create plasmids pPSK2 and pPSK3, containing the SV2 US28 and SV2 US28/Flag genes, respectively. The amino acid point mutant versions altering the charged residues at the boundary of TM-3 and intracellular loop 2 of pSV2 vGCR/UL74 (V146D) and in pSV2 US28 (D128V) were generated with the Quick Change site-directed mutagenesis kit (Stratagene) and are referred to as pPSK4 and pPSK5, respectively.
The wild-type KSHV vGCR cDNA coding region from pPSK1 was inserted into a pAdlox recombinant plasmid transfer vector under the control of the HCMV major immediate-early (MIE) enhancer-promoter region and used to generate an E1A/B/E3-deleted defective strain constitutively expressing the Ad-vGCR adenovirus vector by cotransfection into 293 CRE8 cells with Adpsi5Lox viral DNA. After four passages on 293/CRE8 cells, the virus was confirmed to lack revertant wild-type adenovirus on A549 cells and grown in mass culture to produce a stock virus vector preparation after purification by CsCl density gradient centrifugation.
Antibodies and IFA.
Rabbit antipeptide polyclonal antibodies (PAb) against KSHV vGCR epitopes between amino acids 4 and 16 (Y)EDFLTIFLDDDES(SC) and 181 and 193 (Y)RHRSRVVKPVSKQ(SC) were generated in rabbits inoculated with the keyhole limpet hemocyanin-conjugated synthetic peptide as described previously (53). These are referred to as PAb vGCR-N and vGCR-L3. Other antibodies used included mouse M2 anti-Flag epitope monoclonal antibody (MAb) (Kodak), mouse MAb against the KSHV ORF59 DNA replication protein (17), mouse MAb LN53 against the LANA1 protein (39), rabbit antipeptide PAbs against KSHV vIL6 (K2) (10) and KSHV ZMP-A (K5) (9), rhodamine-labeled goat anti-mouse immunoglobulin (Ig) secondary MAb, and fluorescein isothiocyanate (FITC)-labeled goat anti-rabbit Ig secondary MAb (Chemicon or Jackson Immunoresearch).
DNA from either the Flag-tagged or untagged pSV2-driven expression vectors was transfected into Vero or HeLa cells by calcium phosphate precipitation as described previously (53). After 48 h, the cells were fixed with 2% paraformaldehyde and permeabilized with 0.2% Triton X-100 in phosphate-buffered saline (PBS). Alternatively, the purified titered Ad-vGCR defective adenovirus vector was used to infect DMVEC cell cultures in four-chamber microwell slide cultures (4 × 104 cells/well) at a multiplicity of infection (MOI) of 10 PFU/cell for 15 days. The protein products were detected by immunofluorescence assays (IFAs) with anti-vGCR-N PAb using FITC-labeled goat anti-rabbit Ig secondary antibody or with anti-Flag MAb using FITC-labeled goat anti-mouse Ig secondary antibody and photographed by double-label FITC plus rhodamine IFA with a Leitz UV-epifluorescence microscope (45). Double-label IFA on cultured PEL cell lines was carried out after drying 2 × 106 cells grown on a polylysine-coated microscope slide and fixing in absolute methanol. FITC staining using vGCR-N PAb was carried out simultaneously with staining for ORF59 using mouse MAb and rhodamine-labeled goat anti-mouse Ig secondary antibody.
In vitro translation and western immunoblot analysis.
In vitro transcription and translation of vGCR and US28 was carried out as described previously (21) using T7 RNA polymerase with SalI-linearized pSG5-based effector plasmid DNAs pGH398a and pPSK3 in rabbit reticulocyte extracts. The [35S]methionine-labeled protein products were fractionated by SDS-10% polyacrylamide gel electrophoresis and detected by autoradiography.
DNA from the 3′ Flag-tagged expression vector pSV2-vGCR/Flag was transfected into 293T cells by the calcium phosphate method. After incubation for 48 h, protein samples were harvested in triple detergent lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholic acid, 0.02% sodium azide, 100 μg of phenylmethylsulfonyl fluoride [PMSF] per ml, 1 μg of aprotinin per ml). Proteins were fractionated by SDS-10% polyacrylamide gel electrophoresis and electroblotted onto a nitrocellulose filter sheet, followed by detection with either rabbit vGCR-N PAb or rabbit anti-Flag PAb (SC-807; Santa Cruz Biotechnology), followed by horseradish peroxidase-coupled donkey anti-rabbit Ig antibody and enhanced chemiluminescence reagent (Amersham).
Immunohistochemistry.
Immunohistochemical staining was used to detect vGCR expression in formalin-fixed, paraffin-embedded sections of PEL, MCD, and KS tissue specimens. Thin sections were deparaffinized in xylene, incubated in citrate buffer (10 mM, pH 6.0) for antigen retrieval by microwaving, and preblocked with 10% normal goat serum-1% bovine serum albumin (BSA)-0.05% Tween-20 in PBS for 30 min. Polyclonal rabbit anti-vGCR-N, anti-vIL6, or anti-zinc finger membrane protein A (ZMP-A or ORF-K5) antibodies were applied at a 1:1,000 dilution in blocking solution and incubated for 18 h.
For single-label experiments, immunodetection was carried out using the streptavidin-phosphatase conjugate followed by Vector Red phosphate chromogens (Vector Laboratories, Burlingame, Calif.). For double-label immunohistochemistry, mouse LN53 anti-LANA1 antibody was used first, with detection by the streptavidin-biotin complex and peroxidase system with diaminobenzidine (DAB) chromogen development, followed by the second primary antibody revealed with streptavidin-phosphatase conjugate and Vector Red chromogen. Hematoxylin counterstain was applied.
JSC-1 cells induced with TPA (20 ng/ml) for 48 h, pSV2-vGCR-transfected 293T cells, or CA46 lymphoblasts were stained in parallel with tissue specimens as positive and negative controls. Immunohistochemical detection in KSHV-infected DMVEC was carried out directly on fixed cells in the Lab-Tek chamber slides using either Vector Red alone or a sequential combination of the peroxidase-DAB (brown) and Vector Red chromogens for double-label experiments.
LUC and CAT reporter genes.
A 565-bp DNA fragment containing the presumed proximal upstream promoter regions for vOX2 and vGCR from genome nucleotide position 127302 to 127867 (58) was amplified from the KSHV (BCBL-R) phage lambda clone λE-C2 (50) by PCR using primers 5′-CTAG AGATCTGGAAATCTGGTCTGACAACC-3′ (LGH2478) and 5′-GATCGGATCCACGAAGCAGTCACGTCC-3′ (LGH2479). This region, containing all three of the potential DL-E divergent P2, P3, and P4 promoter elements, was initially cloned into the pGH56 background (pUC18/BamHI/BglII) to generate plasmid pCJC397. The BamHI- and BglII-bounded insert was then added in both forwards (pCJC396a) and backwards (pCJC396b) orientations into the pGL/Basic luciferase (Promega) gene background to generate the vOX2(F)-LUC and vOX2(R)-LUC reporter genes, respectively.
The control target LUC constructions used included the bidirectional KSHV PAN(T1.1) upstream promoter enhancer region (736 bp from genomic positions 27951 to 28686: boxed in Fig. 2C) inserted in either orientation into plasmid pCJC496a or pCJC496b to generate PAN-LUC and LLP-LUC, respectively. LANA-LUC (containing 381 bp of the DL-E promoter region P1 from positions 127877 to 128667 in pCJC575), vIL6-LUC (pCJC399), and DLBR-LUC (pCJC574) represent our standard latent and weak or strong lytic cycle KSHV promoter controls as described by Ciufo et al. (unpublished data).
FIG. 2.
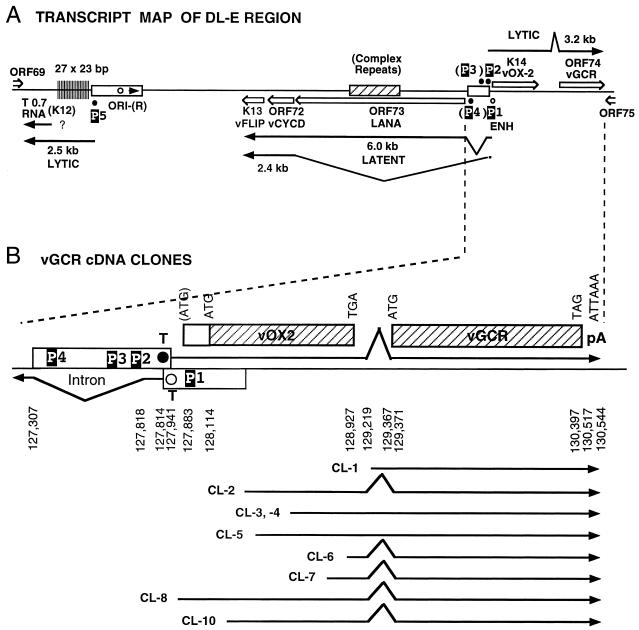
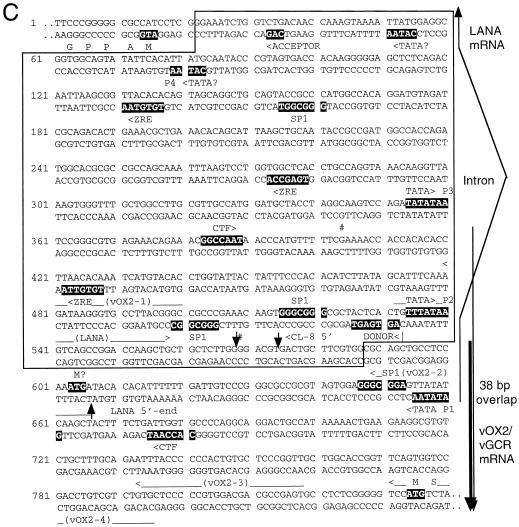
Genomic organization of the DL-E region of KSHV, including the T0.7/K12, vFLIP, vCYCD, LANA1, vOX2, and vGCR genes and their upstream control elements. (A) Structural map of the 13-kb DL-E region, showing sizes and orientations of all known mRNA species and the positions of five known or potential promoters (P1 to P5). (B) Summary map of the nine cloned vGCR cDNAs analyzed, illustrating relative positions of the 5′ termini, their 3′-coterminal features, and the presences or absence of the 147-bp intron between the vOX2 (K14) and vGCR (ORF74) coding regions. (C) Sequence organization of the divergent LANA1 and vOX2/vGCR promoter control regions. The entire 840-bp sequence shown represents genomic nucleotide coordinate positions 127281 to 128120 (58). The divergent 505-bp promoter region contained in the vOX2(F)-LUC and vOX2(R)-LUC reporter genes is boxed. The relative directions and overlap between the 5′ ends of the latent LANA1 mRNA and the lytic cycle vOX2/vGCR mRNA are indicated on the right. Consensus likely transcriptional control motifs such TATA, SP1, and CTF binding sites, potential ZTA binding motifs (ZRE), and LANA1 splice donor and acceptor signals are all denoted in reversed-out type. Solid vertical arrowheads indicate the 5′ ends of LANA1 (position 127887) and vOX2/vGCR (position 127849) mRNA determined here by primer extension analysis, and the open vertical arrowhead indicates the 5′ end (position 127854) of the longest vOX2/vGCR cDNA clone obtained (cl-8). # symbols indicate the two 5′ start sites at positions 127685 and 127849 detected by the 5′RACE technique with uninduced mRNA. Named oligonucleotide primers (LANA1, vOX2-1, vOX2-2, vOX2-3, and vOX2-4) used for RNA extension and 5′RACE analysis are denoted by horizontal arrows underneath the corresponding sequences.
The NFκB and chloramphenicol acetyltransferase (CAT) target reporter genes used contained two tandem copies of a 26-bp oligonucleotide representing either the wild-type or a point mutant form of the consensus Ig NFκB motif inserted upstream at position −56 into a c-Fos minimal promoter (−56 to +109)-driven CAT reporter gene (52). The pSG5 vector-based plasmid expressing the intact KSHV-encoded SV2-RTA (ORF50) effector gene in pCJC566 will be described in detail elsewhere (Chiou et al., unpublished data).
Transient cotransfection assays.
Cultured U937 cells (5 × 106) were transfected with one or more reporter and effector plasmid DNAs (1.0 μg each) by the DEAE-dextran procedure described previously (18). Some samples were treated with TPA at 50 ng/ml for 20 h before harvesting. BC2 cells (5 × 106/well) were transfected with 2 μg of sample DNA in 100 μl of RPMI 1640 medium plus 10 μl of Superfect transfection (lipofection) reagent (Qiagen, catalog no. 301305). Lytic cycle induction in BC2 cells for LUC assays involved treatment with 20 ng of TPA per ml for 20 h.
HeLa and Vero cells were transfected by either the calcium phosphate-BBS procedure described previously (53) or with Lipofectamine reagent (Life Technologies, Inc.) for the NFκB-CAT experiment in Fig. 10C. Cells were pelleted for 30 s at 12,000 rpm and washed with 1 ml of PBS buffer containing 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4 (pH 7.3). Cell lysates were prepared by freeze-thawing three times in 150 μl of 250 mM Tris-HCl (pH 7.5) and 1 mM DTT, then clarified by centrifugation at 12,000 rpm for 10 min at 4°C.
For LUC assays, samples of the supernatant (50 μl) were mixed with 350 μl of reaction buffer A (25 mM glycylglycine [pH 7.8], 5 mM ATP, 15 mM Mg2SO4, 4 mM EGTA). Luciferase activity was measured for 10 s in a Lumat LB9501 luminometer (Berthold Systems, Inc.) after injection of 100 μl of 1 mM luciferin (Sigma) in buffer A.
For CAT assays, samples of the supernatants (50 μl) were mixed with 100 μl of 0.7 M Tris-HCl (pH 7.9) containing acetyl-coenzyme A and [14C]chloramphenicol (0.1 μCi) and incubated at 37°C for 45 min. The samples were diluted into 10 volumes of ethyl acetate, vacuum dried, and dissolved in 20 μl of ethyl acetate, then spotted onto thin-layer chromatography plates for separation of the products by 5% methanol-95% chloroform chromatography and measured by scanning with an Instant Imager (Packard).
RESULTS
Unexpectedly large KSHV vGCR mRNA is induced in BCBL-1 cells but not in HBL6 cells.
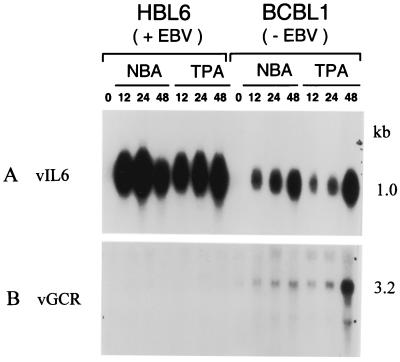
Using an antisense riboprobe specific for the 1,027-bp vGCR coding region, we detected an mRNA of 3.2 kb by Northern blotting in total RNA extracted from the BCBL-1 cell line after induction with either TPA or sodium butyrate (Fig. 1B, lanes 9 to 14). However, this species was not detectable in similarly treated HBL6 cells in parallel lanes in the same experiment (lanes 2 to 7). The 3.2-kb mRNA species was barely detectable in an uninduced BCBL-1 sample (lane 8), but increased somewhat at 12 and 24 h after either sodium butyrate or TPA treatment and reached high abundance at 48 h, especially after TPA treatment (lanes 9 to 14). In comparison, the previously described vIL6 mRNA of 1.0 kb (49) was equally well induced within 12 h by sodium butyrate as well as TPA treatment in both HBL6 cells and BCBL-1 cells, including a characteristic decline by 48 h in the HBL6 sodium butyrate sample (Fig. 1A).
FIG. 1.
Comparison of vGCR mRNA induction in HBL6 cells and BCBL-1 cells by Northern blotting. Total cell RNA samples from PEL cell lines induced for various times by sodium butyrate (NBA) or TPA treatment were fractionated on formaldehyde-agarose gels and transferred to nylon membranes for hybridization with either 32P-labeled vGCR or vIL6 riboprobes. (A) vIL6 probe; (B) vGCR probe. Lanes 1 and 8, untreated; 2 and 9, sodium butyrate, 12 h; 3 and 10, sodium butyrate, 24 h; 4 and 11, sodium butyrate , 48 h; 5 and 12, TPA, 12 h; 6 and 13, TPA, 24 h; 7 and 14, TPA, 48 h. Positions of labeled single-stranded DNA size markers and ribosomal RNAs on an adjacent lane were used to calculate approximate sizes of 1.0 kb and 3.2 kb for vIL6 and vGCR mRNAs, respectively.
There was no significant change in the abundance of either the vIL6 or vGCR mRNA bands in RNA prepared from BCBL-1 cells after treatment with TPA in the presence of PAA for 48 h, whereas the 2.3-kb mRNA band for the late class ORF26 gene was totally abolished under the same condition (data not shown). Although a possible very large minor species (>10 kb) may also be present in the BCBL-1 samples, no obvious smaller mRNA species that might correspond to the vGCR ORF alone was detectable by Northern blotting. Therefore, a relatively late and low-abundance early lytic class mRNA of unexpectedly large size appears to represent the KSHV vGCR transcript in induced BCBL-1 cells. The extremely weak induction of vGCR mRNA in the HBL6 cell line is consistent with our results for other KSHV early mRNAs also (with the striking exception of vIL6) (Chiou et al., unpublished).
Identification of an intact spliced vGCR cDNA clone.
To evaluate the detailed structure of the induced 3.2-kb vGCR mRNA species, we isolated and sequenced nine different molecular clones from a phage lambda cDNA library generated from TPA-treated BCBL-1 RNA (17). These clones were detected and isolated by plaque hybridization with a vGCR riboprobe generated from the cloned ORF74 coding region from within the DL-E segment of the KSHV genome (Fig. 2A). The structures of all of these cDNAs are illustrated in Fig. 2B.
Eight encompassed portions of the upstream vOX2 (K14) ORF as well as the complete vGCR (ORF74) coding region, but all except one (cl-8) appeared to be incomplete at the 5′ end and five were spliced. All nine contained poly(A) runs beginning at genomic position 130544, indicating that they utilized the associated nonconsensus poly(A) addition signal ATTAAA at position 130517. The largest and apparently almost completely intact species of 2.7 kb encompassed both the complete vOX2 and vGCR ORFs and included a small 148-bp intron within the noncoding region between the two ORFs (Fig. 2A and 2B). Four of the other seven cDNAs that included upstream vOX2 sequences utilized the same internal intergenic splice, but three did not. The 5′ end of the intact cl-8 cDNA begins at position 127855, just 41 bp downstream from a probable adjacent consensus TATAA-like promoter motif (TTTATAA) at position 127814 and 28 bp upstream from the predicted vOX2 initiator codon at position 127883 (58). The internal splice spans a reasonable consensus donor motif G′GTGGGT at position 129218 and an acceptor TTGTAG′, which is just 3 bp upstream from the predicted vGCR initiator codon at position 129371.
Confirmation of the 5′ mRNA start site by primer extension analysis.
Curiously, the 5′ end of the largest vGCR cDNA and the predicted upstream promoter region for the vOX2/ORF-K14 region (transcribed rightwards) both overlap in the opposite orientation with the mapped upstream exon and GATATAA promoter element (P1) for the leftwards 6.0- and 2.4-kb latent-state transcripts that encode LANA1 (ORF73), vCYC-D (ORF72), and vFLIP (ORF71) (26, 38, 55, 61). Furthermore, there are two potential TATA-like motifs within the LANA1 intron region that could be associated with the 3.2-kb bicistronic vOX2/cGCR mRNAs (Fig. 2A and 2B). The more distant motif (P3) represents a match to the most efficient TATA motif known (TATATAA), whereas the more proximal one (P2) is a plausible but nonconsensus motif (TTTATAA).
Because of the possibility of there actually being two types of intact mRNAs with different 5′ ends that could not be resolved on the Northern blots, we used primer extension analysis to identify the actual mRNA start sites. Four different primers were designed to attempt to cover all possible variations of leftward start sites and potential 5′ splices associated with these two TATA-like motifs (Fig. 2C).
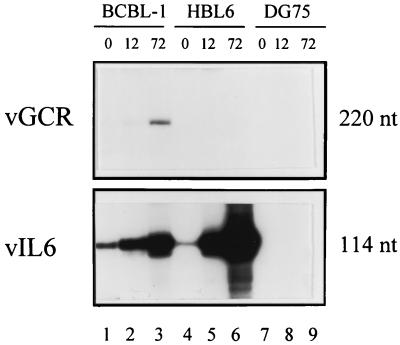
The results with primer probe vOX2-4, which gave rise to a 220-bp extension product, are shown in Fig. 3. This species was most abundant in BCBL-1 RNA at 72 h after TPA treatment (lane 3) but was undetectable at the earlier 12-h time point (lane 2) or with sodium butyrate-induced HBL6 RNA at either 12 or 72 h (lanes 5 and 6). In comparison, the principal 114-bp vIL6 primer extension products were detected in far greater abundance in both the BCBL-1 and HBL6 RNAs as well as at earlier times, as expected. The small amounts of authentic vIL6 mRNA detected in uninduced BCBL-1 and HBL6 cells corresponds to the low levels of spontaneous vIL6 lytic cycle protein expression detected by IFA in 3 and 0.5% of those cells, respectively (compared to 20% after TPA induction of BCBL-1 cells and 30% after sodium butyrate induction of HBL6 cells). No equivalent primer extension products were detectable in either uninduced or TPA-treated DG75 lymphoblastoid cell lines used as a negative control (Fig. 3, lanes 7 to 9).
FIG. 3.
Identification of the vOX-2/vGCR transcription start site by primer extension analysis. (A) Size analysis of vGCR compared to vIL6 primer extension products. Total RNA samples extracted from KSHV-positive PEL cell lines and a control EBV-negative lymphoblast cell line (DG75) were denatured and annealed with 32P-end-labeled antisense strand primers for either vOX2-2 (upper panel) or vIL6 (lower panel). RNA extension products were fractioned by electrophoresis and subjected to autoradiography. Lanes 1, 4, and 7, uninduced BCBL-1, HBL6, and DG75 cultures, respectively; lanes 2 and 3, parallel cultures of BCBL-1 cells treated with TPA for 12 and 72 h, respectively; lanes 5 and 6, HBL6 cells treated with sodium butyrate for 12 and 72 h, respectively; and lanes 8 and 9, Raji cells treated with TPA for 12 and 72 h, respectively. The positions of labeled single-stranded DNA size markers run in parallel lanes were used to measure the sizes (in nucleotides [nt]) of the primer extension products.
Probe OX2-2 gave a single 45-bp product at 72 h in BCBL-1 RNA, and probe OX2-3 gave a single 190-bp product, whereas the further upstream probe OX2-1 failed to produce any detectable discrete primer extension product (data not shown). Therefore, all of these results are compatible with a BCBL-1 vOX2/vGCR transcript utilizing the proximal nonconsensus TTTATAA motif (P2) at position 127814, giving a 2.7-kb mRNA species [not including the poly(A) tail] initiating at position 127848, which is consistent with the cl-8 cDNA 5′ end at position 127855 (Fig. 2C). There was no evidence obtained by this procedure for utilization of the more distal TATATAA motif (P3) in the BCBL-1 cell line after TPA induction.
Additional control primer extension assays for standard lytic T1.1/PAN and latent LANA1 transcripts (not shown) gave the expected sizes in the BCBL-1 RNA sample of 65 nucleotides (predominantly after TPA induction) or 75 nucleotides (only in the presence of cycloheximide), respectively. The latter represents mRNA transcribed rightwards from position 127866, presumably in association with the GATATAA (P1) motif at position 127941 on the opposite strand and overlapping the vOX2/vGCR lytic transcript by 38 bp in a head-to-head (5′ to 5′) fashion (Fig. 2B and 2C). Finally, another primer probe (vGCR), which was designed to detect possible proximal initiation sites within the intron just upstream of the ORF74 coding region itself, failed to give any extension products with either the uninduced or induced BCBL-1 RNA samples.
Identification of an additional 5′ upstream start site by 5′RACE.
Another approach useful for detection of relatively rare transcripts that might otherwise be outcompeted by smaller, more abundant species in primer extension assays is 5′RACE. This technique was carried out with the uninduced BCBL-1 mRNA sample using primer probe vOX2-4 for first-round RT-PCR amplification of 5′ ends, followed by the use of primer probe vOX2-3 for nested PCR. Two approximately equally abundant products were obtained, and the separated bands were isolated and sequenced.
The smaller product proved to have a 5′ end at position 127849, corresponding exactly to the originally detected P2-associated lytic cycle primer extension product. However, the 5′ end of the longer RACE product was determined to be at position 127685, just 50 bp downstream from the predicted distal P3 TATATAA motifs. The relative abundance of this proximal species was evidently far too low to be detectable by the primer extension or cDNA approaches used here, but does suggest the possibility of utilization of the P3 promoter during latency.
Evidence that the proximal upstream region adjacent to the vOX2 (K14) ORF contains functional promoter elements.
Inspection of the DNA sequences upstream from the mapped mRNA start site for the intact 3.2-kb mRNA revealed two excellent but distinct promoter-like domains, one associated with each of the two consensus TATA-like motifs (P2 and P3 above). Both have typical KSHV early lytic cycle promoter elements, including SP1 and CTF sites and ZRE-like motifs (Fig. 2C). No obvious promoter elements are present in or upstream of the 148-bp intergenic region between the vOX2 and vGCR ORFs, suggesting that this bicistronic mRNA species may indeed represent the predominant functional mRNA for vGCR.
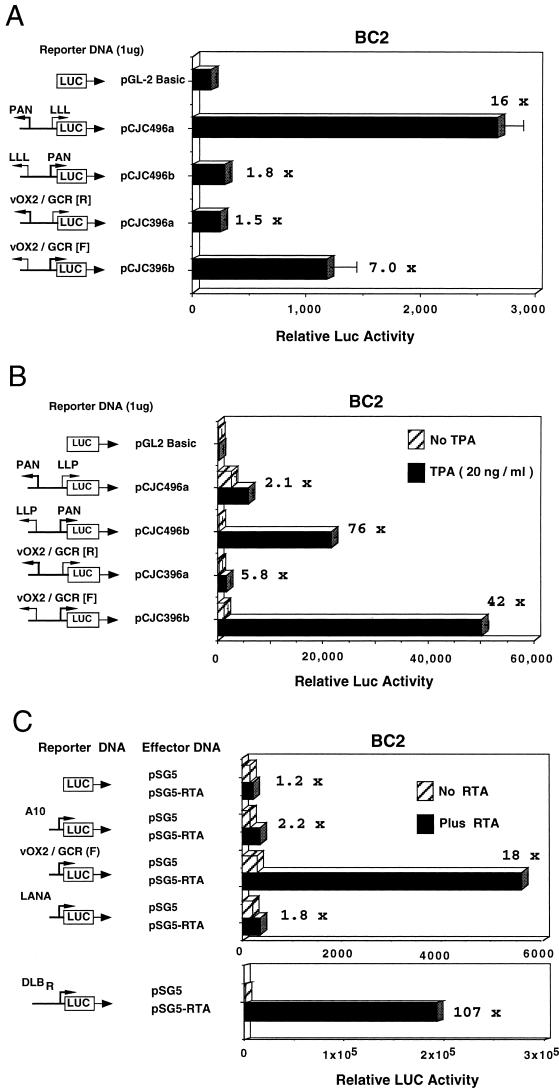
To examine whether this upstream domain functions as a promoter, we inserted a 565-bp DNA segment from genomic positions 127302 to 127867 into a LUC reporter gene cassette and examined its properties in a series of transfection and cotransfection assays in parallel with several previously studied lytic and latent-state KSHV promoter-LUC reporter genes (Chiou et al., unpublished; Ciufo et al., unpublished). Because of the existence of a potential oppositely oriented TATA-like motif (CATAA, P4) in the distal portion of this region, both orientations of the 565-bp fragment were examined.
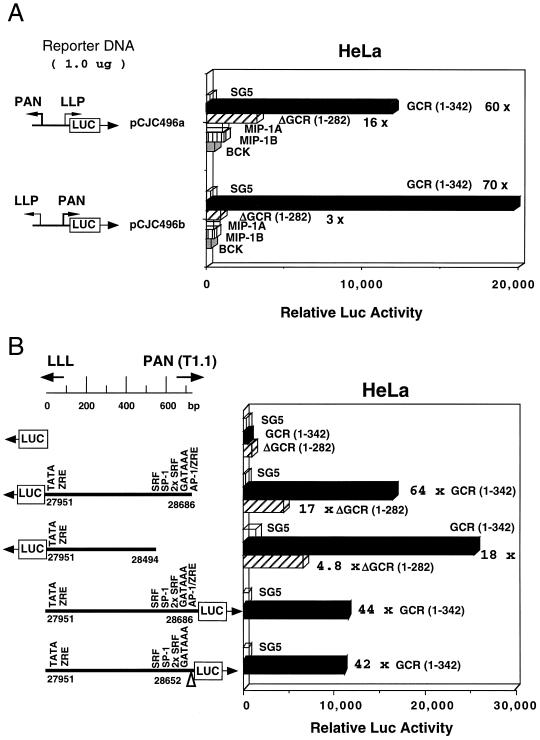
In the first experiment shown (Fig. 4A), basal levels of expression of forwards vOX2(F)-LUC and reverse vOX2(R)-LUC were compared with previously characterized controls PAN-LUC and LLP-LUC (Ciufo et al., unpublished) in transfected BC2 lymphoblastoid cells that are latently infected with KSHV. In our hands, BC2 cells are the most efficiently transfectable of the PEL cell lines (by either lipofection or electroporation) and are considered the most appropriate cells for these experiments. As expected, the LLP-LUC latency promoter (Ciufo et al., unpublished) gave 16-fold higher basal expression than the control promoterless LUC gene, compared to twofold for the lytic cycle PAN-LUC promoter, whereas vOX2(F)-LUC gave an intermediate level (7-fold increased) that was 3- to 4-fold stronger than for the reverse orientation in vOX2(R)-LUC.
FIG. 4.
vOX2/vGCR proximal upstream region behaves as a TPA- and RTA-inducible lytic cycle promoter in cotransfection assays in KSHV latently infected lymphoblast cells. Equal amounts of target and effector DNA were introduced for transient expression assays into BC2 cells by lipofection. Luciferase levels were measured in a luminometer, and average values for basal and induced expression from parallel samples in the same experiment are plotted as histograms. Fold induction values are indicated. (A) Comparison of basal expression properties of the vOX2(F)-LUC reporter gene with those of the vOX2(R)-LUC, LLP-LUC, and PAN-LUC reporter genes. (B) Comparison of induced expression of vOX2(F)-LUC, vOX2(R)-LUC, PAN-LUC, and LLP-LUC target reporter genes in the presence and absence of TPA treatment. (C) Comparison of the responsiveness of vOX2(F)-LUC and the minimal SV40 promoter (A10-LUC) reporter genes to cotransfection with the KSHV RTA (ORF50) effector gene in pSG5-RTA relative to the empty expression vector clone (pSG5). DLBR-LUC and LANA-LUC were used as positive and negative RTA-responsive KSHV promoter controls, respectively.
In a second type of experiment (Fig. 4B), the expression levels were compared in BC2 cells in the presence and absence of TPA treatment. As expected, the positive control lytic cycle PAN-LUC (Ciufo et al., unpublished) was highly inducible (76-fold), whereas the LLP-LUC latency promoter was not (2-fold increase only). In comparison, the forwards direction of vOX2(F)-LUC responded almost as strongly as PAN-LUC to TPA induction (42-fold), but the reverse orientation responded only weakly (6-fold).
To determine whether the vOX2/GCR promoter was a target for the KSHV-encoded RTA (ORF50) transactivator, a cotransfection experiment with an RTA expression vector encoding the wild-type SV2 RTA gene (Chiou et al., unpublished) was also carried out in transfected BC2 cells. The results revealed 18-fold upregulation of vOX2(F)-LUC (Fig. 4C), with no more than 2-fold effects on the activity of the minimal SV40 promoter in pGL2Basic-LUC, the promoterless LUC vector, or the standard KSHV latency promoter LANA-LUC (Fig. 4C). The relative strength of the RTA response compares favorably with 11-fold for vIL6-LUC and 61-fold for PAN-LUC in parallel assays (not shown), although still being 5-fold less than the level obtained with the strongest-responding KSHV promoter in DLBR-LUC (Fig. 4C). Therefore, the immediately adjacent upstream control region for the bicistronic vOX2/vGCR mRNA has the properties of a typical moderately TPA- and RTA-responsive KSHV lytic cycle promoter.
ORF74 encodes a 48-kDa monomeric and dimeric cytoplasmic membrane protein.
To ask whether the ORF74 coding region produces the expected stable vGCR protein product of 342 amino acids, the entire 1,029-bp ORF was inserted into an SV40 early promoter-driven mammalian expression vector (pSG5 based) with or without a Flag epitope marker added in-frame at the C terminus. The Flag-tagged effector plasmid was then transfected into Vero cells and examined by IFA. Up to 10% of the transfected cells produced a typical strongly positive cytoplasmic plus membrane IFA pattern (not shown).
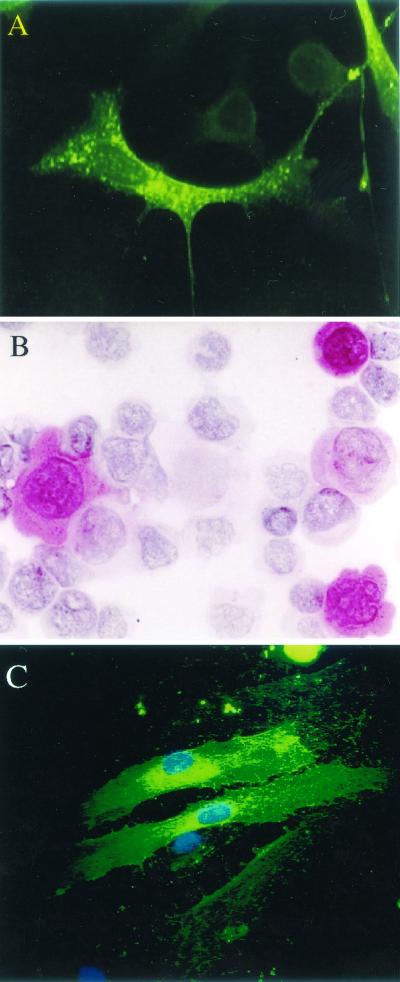
To be able to identify the naturally expressed vGCR protein, we also generated a rabbit antipeptide antiserum directed against epitopes at either the predicted N terminus of vGCR (anti vGCR-N) or the predicted third extracellular loop (anti-vGCR-L3). Both antibodies detected the vGCR protein in a similar cytoplasmic and membrane pattern by IFA in pSV2-vGCR-transfected Vero cells, although the GCR-N antibody produced a superior result (Fig. 5A; green FITC fluorescence). A similar result was obtained by immunohistochemical detection with the vGCR-N PAb in pSV2-vGCR-transfected 293T cells (Fig. 5B; red cytoplasmic stain).
FIG. 5.
Intracellular distribution of vGCR protein in transiently transfected or Ad vector-transduced monolayer cells. Untagged expression vector plasmid DNA in plasmid pCJC398a containing the intact KSHV vGCR coding region under the control of the SV40 enhancer-promoter region was transfected into cells by the calcium phosphate precipitation procedure. Alternatively, DMVEC cultures were transduced with Ad-vGCR vector at an MOI of 10 PFU/cell for 15 days before fixing for IFA. (A) IFA with FITC-labeled anti-vGCR-N PAb in pSV2-vGCR-transfected Vero cells. (B) Immunohistochemistry with Vector Red-stained anti-vGCR-N PAb in pSV2-vGCR-transfected 293T cells. (C) IFA with FITC-labeled anti-vGCR-N PAb in Adv-GCR defective adenovirus vector-transduced DMVEC culture, with DAPI detection of cell nuclei.
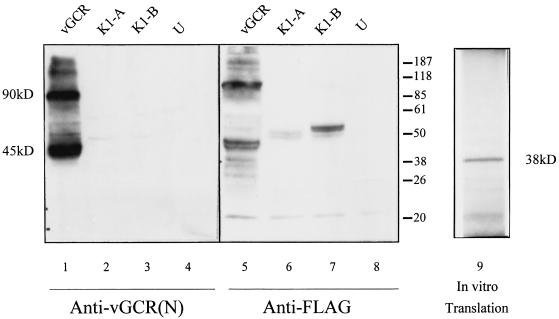
To assess whether the expression vectors were likely producing an intact protein product, they were also transfected into 293T cells, and protein extracts were evaluated by Western immunoblotting. The results of a comparison of the extracts from cells transfected with plasmids expressing either SV2-vGCR/Flag, SV2-ORF-K1A/Flag, SV2-ORF-K1B/Flag, or the empty vector only are shown in Fig. 6. Two major protein products of approximately 45 and 90 kDa that were present only in the SV2-vGCR extracts were detected with both the anti-Flag and anti-vGCR antibodies (lanes 1 and 5). The specificity of the antibodies was confirmed by detection of the 55-kDa ORF-K1 control proteins only with the anti-Flag antibody (lanes 6 and 7), but not with the anti-vGCR antibody (lanes 2 and 3).
FIG. 6.
Detection of the KSHV vGCR protein by Western immunoblotting in transfected cells and after in vitro translation. Expression vector plasmid DNA was transfected into 293T cells, and protein extracts were fractionated by gel electrophoresis, transferred to a nitrocellulose membrane, and incubated with either rabbit anti-vGCR-N antibody (lanes 1 to 4) or anti-Flag epitope antibody (lanes 5 to 8). Lanes 1 and 5, extract from cells receiving SV2-vCGR/Flag DNA; 2 and 6, SV2-ORF-K1A/Flag; 3 and 7, SV2-ORF-K1B/Flag; 4 and 8, untransfected 293T cell controls. Lane 9, autoradiograph of gel electrophoretically fractionated 35S-labeled protein products following in vitro transcription-translation from the SV2-vGCR expression plasmid (pGH398a). Positions of stained protein size markers are indicated (in kilodaltons).
Comparison with in vitro-translated product from the same SV2-vGCR template plasmid (lane 9), which gave a 35S-labeled protein band of 37 kDa (close to the expected size for a 342-amino-acid protein) suggested that the vGCR proteins in transfected cells may be both modified (by glycosylation or phosphorylation) and aggregated into dimers. Loss of all of these protein bands after boiling of the extracts (not shown) further supported the presumption that they are all aggregatable membrane proteins.
Detection of vGCR protein expressed after induction in PEL cell lines.
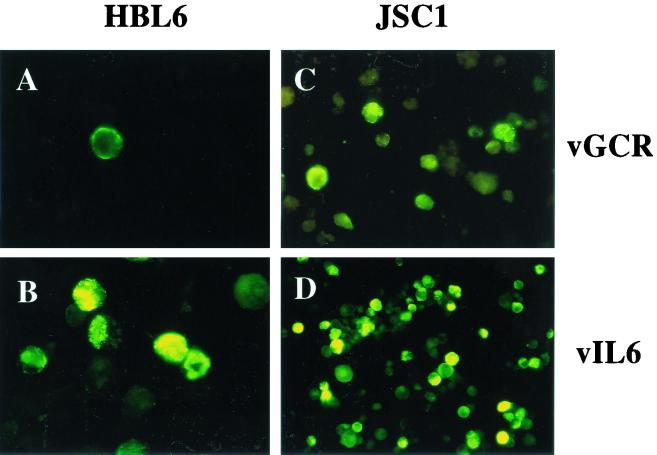
The possibility that the native vGCR protein might be expressed and detectable in several different uninduced and TPA-induced PEL cell lines was then examined by IFA. Expression of vGCR was inducible by TPA in some cells in most cultures, but was generally only detected much later than vIL6 and in significantly fewer cells. Only one of the six PEL cell lines tested (JSC-1) gave any significant constitutive expression of vGCR as detected by IFA, although all six cultures could be induced to express it in at least a few cells.
A comparison of vGCR and vIL6 expression levels in the HBL6 and JSC-1 cell lines at 48 h after TPA induction is shown in Fig. 7. In HBL6 cells, only 0.2% were positive for vGCR, compared to 15% for vIL6 even after TPA treatment, whereas for JSC-1 cells 8% were positive for vGCR and 42% for vIL6 after TPA treatment. To examine whether the vGCR-N PAb antibody could also be used for diagnostic purposes (see later), we carried out an immunohistochemical assay with a cell pellet of fixed sodium butyrate-induced JSC-1 cells embedded in paraffin. Again, up to 10% of the cells proved to be positive for vGCR protein expression as a cytoplasmic and membrane pattern (not shown).
FIG. 7.
Induction of vGCR protein by TPA in PEL cell lines as detected by IFA. Cell cultures were incubated in the presence of TPA for 48 h and fixed on lysine-coated glass slides by the formaldehyde-Triton X-100 procedure. Induced vGCR or vIL6 cytoplasmic and membrane proteins were detected with FITC-labeled anti-vGCR-N or anti-vIL6 rabbit PAb. (A) TPA-induced HBL6 cells (×100, vGCR PAb); (B) TPA-induced HBL6 cells (×100, vIL6 PAb); (C) TPA-induced JSC-1 cells (×60, vGCR PAb); (D) TPA-induced JSC-1 cells (×40, vIL6 PAb).
KSHV vGCR protein is also expressed in a small subfraction of cells in PEL and MCD tumors and in KS lesions.
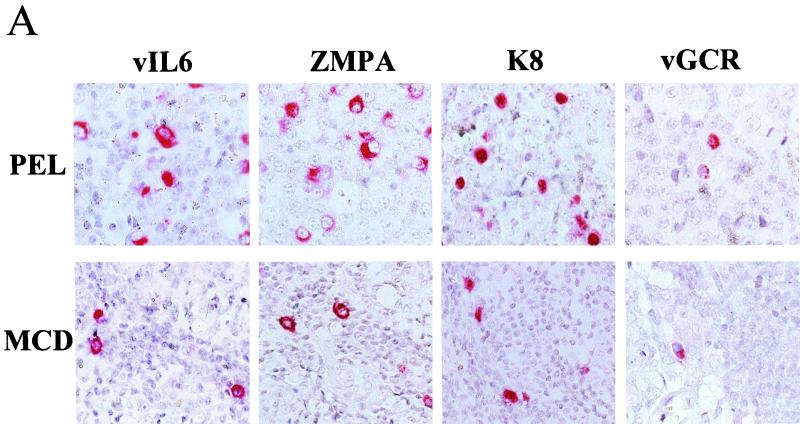
To ask whether the vGCR is also expressed in original uncultured lymphoid tumors or in KS lesions carrying KSHV genomes, we used the immunohistochemical assay with rabbit anti-vGCR-N-specific PAb to attempt to detect the viral protein in tissue sections from diagnostic paraffin blocks. A primary PEL tumor sample (Fig. 8A, top panel), and an MCD tumor sample (Fig. 8A, bottom panel) were both examined. Just as in the BCBL cell lines, occasional tumor cells gave moderate to strong positive staining predominantly localized to the cytoplasm. The much more abundant cytoplasmic immunohistochemical signals obtained for vIL6 and ZMP-A and nuclear immunohistochemical signal for K8 in the same PEL and MCD samples are shown for comparison. Overall, vGCR expression in the PEL tumor specimen was detected in 2 to 5% of the cells, compared to 10 to 20% for vIL6 (10).
FIG. 8.
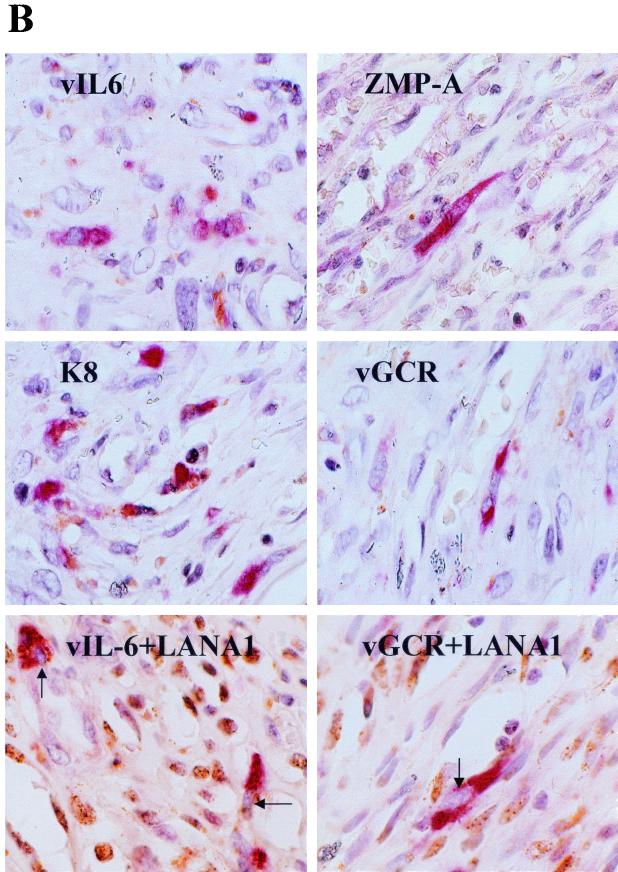
Detection of lytic cycle vGCR expression by single and double label immunohistochemistry in KSHV-associated lymphoma tumors and a Kaposi's sarcoma lesion. Polyclonal rabbit vIL6, ZMP-A, ORF-K8, or vGCR-N antibodies and Vector Red-labeled goat anti-rabbit Ig antibodies or mouse monoclonal LN53 antibody and peroxidase-DAB chromogen were used to detect expression in formalin-fixed paraffin-embedded sections of archival tumor specimens. Hematoxylin counterstain was also applied. (A) Top layer, positive B cells of a primary effusion lymphoma; lower layer, scattered positive cells in the mantle layer of the germinal center of an HIV-positive MCD tumor. (B) Lytic cycle KSHV antigens in spindle cells of a nodular KS lesion. The lower two frames show double-label immunohistochemistry in the same nodular KS lesion showing punctate nuclear LANA1 (brown spots) in many cells with vIL6, as well as vGCR expression (red cytoplasm) in a few scattered lytic cells with nuclei that are devoid of LANA1 (arrows).
Expression of vGCR in the MCD tissue was localized to the mantle zone of germinal centers in approximately 0.5 to 1.0% of the cells, as determined by the average of several fields. This compares with up to 5% LANA1 positivity in the same region of the same sample. In contrast, paraffin sections of KSHV-negative, EBV-positive control CA46 lymphoblasts and a Hodgkin's lymphoma sample were negative for both LANA1 and vGCR detection in parallel experiments (not shown). Again, the proportion of vGCR-positive cells in the MCD sample was approximately fivefold lower than for vIL60-positive cells in adjacent sections from the same paraffin block.
Scattered vGCR-positive spindle cells were also detected among the tumor cells in a nodular KS skin lesion biopsy specimen (Fig. 8B). In this case, we also carried out double-label immunohistochemical staining with DAB-peroxidase detection to show latently infected LANA1-positive nuclei (brown spots) in greater than 50% of the spindle cells. Interestingly, most (but not all) of the spindle cells that were positive for vGCR were negative for LANA1, providing further evidence that vGCR expression is not associated with the latent state. This particular KS sample (AKS1) was also positive for cytoplasmic vIL6 and ZMP-A (K5) as well as the nuclear K8 protein in a similar fraction (approximately 5%) of the spindle cells in some areas (Fig. 8B), but none of 18 patch and plaque type KS lesions examined were positive for even vIL6 (10).
Detection of vGCR protein in Ad vGCR-infected and KSHV lytic cycle-infected endothelial cells.
To evaluate whether KSHV vGCR could also be expressed efficiently in primary human DMVEC, we generated a defective adenovirus (Ad) vector encoding vGCR (ORF74) under the control of the HCMV MIE enhancer-promoter region. This virus was used to transduce the DMVEC culture at an MOI of 2, 10, or 50 PFU/cell (293 T-cell titer), and the cells were fixed and examined by IFA with FITC-labeled anti-vGCR-N PAb at 5, 10, and 15 days. Between 10 and 20% of the cells proved to express vGCR protein as a typical cytoplasmic membrane pattern at both the 10- and 15-day time points at an MOI of 10 (Fig. 5C, green cytoplasm in two of the three cells shown). The number of positive DMVEC cells was highly input virus dependent and reached 35 to 40% at an MOI of 50.
Expression of vGCR was also examined by immunohistochemical staining in DMVEC cultures persistently infected with KSHV (23, 28, 44). Parallel cultures of KSHV (JSC-1)-infected and uninfected DMVEC were passaged five times over an 8-week period after addition of virus. Unlike the uninfected cells, the infected cell culture consisted of latently infected spindle-shaped cells that were all expressing the LANA1 protein (9, 23). Without TPA treatment, a small subset of the infected DMVEC cells were positive for the early lytic nuclear KSHV-encoded K8 protein and for the early lytic cytoplasmic ZMP-A (K5) protein, and an even smaller subset were positive for the cytoplasmic vGCR protein (not shown).
At 96 h after TPA treatment, the fraction of vGCR-positive cells (Fig. 9B, red signal) was measured at 15%, and a similar fraction of the cells were positive for ZMP-A (Fig. 9C, red signal). Double-label staining for LANA1 expression (Fig. 9C, brown nuclei) confirmed that all cells in the culture were infected with KSHV. No vGCR (or LANA1)-positive signals were detected in the uninfected culture (Fig. 9A). Some of the vGCR-positive cells showed rounding and cytopathic effects, and indeed all cells with cytopathic effects were vGCR positive. Therefore, the KSHV vGCR protein is expressed as an abundant cytoplasmic and membrane protein in lytically induced DMVEC cells, but it is produced in fewer cells and is more TPA dependent than are the very early lytic proteins ZMP-A and K8. Furthermore, it was virtually undetectable in most LANA1-positive spindle cells in the uninduced latent state.
FIG. 9.
Expression of vGCR in primary human DMVEC cells infected with KSHV. DMVEC cultures were infected with supernatant virus from TPA-treated JSC-1 cells and passaged five times over an 8-week period in the presence of endothelial cell growth factors. Cells were then seeded onto glass slides and grown in the presence of TPA for 96 h, then fixed for immunohistochemistry. Cells positive for vGCR were detected with Vector Red-stained anti-vGCR-N PAb. (A) TPA-treated mock-infected DMVEC cell monolayer. (B) Cytoplasmic vGCR protein expression in TPA-treated KSHV-infected DMVEC spindle cells. (C) Double-label immunohistochemistry for Vector Red-stained lytic cycle cytoplasmic ZMP-A (K5) protein and DAB-stained nuclear latent-state LANA1 protein (brown) in the same culture of TPA-treated KSHV-infected DMVEC spindle cells as shown in B. Magnification, ×160.
Transactivation of viral lytic cycle promoters by vGCR.
While evaluating a number of KSHV gene products for the ability to either reactivate lytic viral gene expression after transfection into latently infected BC2 cells or activate a PAN-LUC reporter gene after cotransfection, we discovered that in addition to the known RTA (ORF50) lytic cycle triggering protein (67; Chiou et al., unpublished), the vGCR (ORF74) effector gene also functioned as a transactivator in the latter studies. Among a number of KSHV-encoded genes tested, only RTA, but not ZMP-A (K5, IE-A), MTA (ORF57), vIRF1 (K9), or vGCR (ORF74) gave 5-fold induced vIL6 expression (2.5% positive) as measured by IFA with specific antibody to vIL6 in electroporated BC2 cells, compared to 0.5% before transfection or with the other genes (Chiou et al., unpublished). In contrast, in preliminary studies SV2 vGCR (but not SV2 K8) induced PAN-LUC target gene expression 15-fold in cotransfected HeLa cells, which compared favorably with the 30-fold positive effect produced by SV2 RTA cotransfection (not shown).
To compare the effects on both orientations of the divergent enhancer-promoter domain upstream from the T1.1/PAN RNA region in DL-B, the lytic PAN-LUC and latent LLP-LUC target reporter genes were each cotransfected with wild-type SV2 vGCR(1-342), truncated SV2 vGCR(1-282), or the pSG5 empty vector DNA (Fig. 10A). The results showed nearly equal responsiveness of these two oppositely oriented promoters to cotransfection with wild-type vGCR (70- and 60-fold upregulation), with truncation at amino acid 282 reducing the transactivation on the LLP and PAN promoter targets by 4-fold and 23-fold, respectively.
FIG. 10.
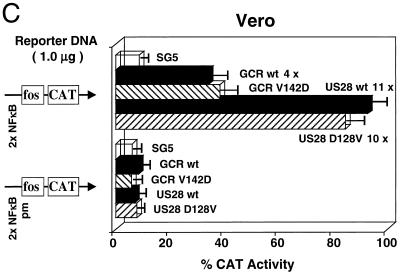
Evidence that vGCR expression leads to upregulation of viral promoters in cotransfection assays. (A) Comparison of the transactivator properties of three KSHV-encoded chemokines and the wild-type and truncated vGCR chemokine receptor in cotransfection studies. Both orientations of the DL-B divergent promoter region containing the LLP-LUC (pGH496a) and T1.1-LUC (pGH496b) target reporter genes were cotransfected with pSG5 vector-based effector plasmids encoding either wild-type SV2 vGCR(1-342), truncated SV2 vGCR(1-282), SV2-vMIP-1A, SV2-vMIP-1B, or SV2-BCK in transiently transfected HeLa cells. (B) Effects of SV2 vGCR on expression of LLP-LUC and ΔLLP-LUC in transiently transfected HeLa cells. Two versions of LLP-LUC reporter genes including or lacking the distal domain associated with the oppositely oriented T1.1 promoter were compared together with the minimal promoter control in pGLBasic. Levels of LUC activity in the presence or absence of pSG5 empty vector (open bars), wild-type SV2 vGCR(1-342) (solid bars), or the truncation mutant vGCR(1-282) (hatched bars) are plotted as a histogram. (C) Transactivation of an NFκB reporter gene by both KSHV vGCR/ORF74 and HCMV US28 in Vero cells. The histogram compares the averaged results from three experiments after cotransfecting either the wild-type NFκB-CAT target reporter gene (upper panel) or a mutant ΔNFκB-CAT reporter gene (lower panel) with effector genes consisting of the empty pSV2 vector (pSG5) or versions containing wild-type SV2 vGCR, the SV2 vGCR V142D point mutant, wild-type SV2 US28, or the SV2 US28 D128V point mutant.
In dramatic contrast in parallel experiments (not shown) (Chiou et al., unpublished) the PAN-LUC promoter responded 200-fold to cotransfected KSHV RTA, whereas the latent LLP-LUC promoter failed to respond to KSHV RTA (less than 2-fold effect). Therefore, the vGCR protein, which is known to upregulate AP1 activity (2), appears to be functionally equivalent to a viral transactivator in these assays. We also asked whether the three KSHV-encoded chemokine proteins referred to as vMIP-1A (vMIP-I), vMIP-1B (vMIP-II), and vBCK (vMIP-III) (48, 49, 58), two of which have also been shown to induce angiogenesis (6), might have similar transactivator effects to vGCR. However, this was not the case (Fig. 10A), with neither SV2 vMIP-1A, SV2 vMIP-1B, nor SV2 vBCK having any significant effects on either orientation of the divergent promoter.
To further examine this activity, we compared the ability of wild-type SV2 vGCR(1-342) and mutant SV2 vGCR(1-282) to activate deleted versions of both the LLP-LUC latent-state promoter and the PAN-LUC lytic cycle promoter in cotransfected HeLa cells (Fig. 10B). In this experiment, LLP-LUC responded 64-fold to cotransfection with wild-type vGCR, and the truncated version of the effector was again nearly 4-fold less effective. Removal of the far upstream T1.1/PAN promoter and its associated AP1 and serum response element-containing domain to create ΔLLP-LUC boosted basal expression 5-fold, but this version still responded 18-fold to vGCR upregulation. As a control, there was no significant effect of cotransfected vGCR on the minimal promoter in pGL2-LUC. Furthermore, removal of the proximal AP1 motif at +10 from PAN-LUC also failed to reduce the transactivation significantly (Fig. 10B, 42-fold) compared to the intact PAN-LUC target (Fig. 10B, 44-fold).
Two other KSHV promoter-driven reporter genes tested showed similar upregulation when cotransfected with intact vGCR(1-342) (not shown). These effects were measured at 25-fold for DSBR-LUC in HeLa cells and 52-fold for K1-LUC in U937 cells, with the latter being reduced to just 4-fold with truncated vGCR(1-282). Therefore, these results suggest that many KSHV promoters may respond to vGCR-induced transactivation in a relatively nonspecific fashion and that the targeting by vGCR does not rely only on upregulation of AP1 activity.
Finally, we asked whether a minimal promoter containing inserted upstream NFκB motifs might respond to vGCR cotransfection in Vero cells. As shown in Fig. 10C, the added wild-type 2xNFκB motif did not significantly increase basal Fos-CAT activity, but it did mediate a 4-fold response to cotransfected wild-type KSHV vGCR and an 11-fold positive response to cotransfected HCMV US28, another herpesvirus-encoded chemokine GPCR receptor family protein. Neither affected the control reporter gene containing two copies of a mutated NFκB motif.
Previous studies with the cellular CXCR2 protein have shown that altering a conserved VRY motif in intracellular loop 3 to DRY (matching the motif found in vGCR) converted CXCR2 from a ligand-dependent form to a constitutively signaling ligand-independent protein like vGCR (8). Therefore, mutant forms of both vGCR and US28 in which the DRY/VRY motifs were altered to attempt to convert vGCR into a ligand-dependent form (V142D mutant) or to convert US28 into a ligand-independent form (D128V) were also employed in this experiment. However, neither mutation had any significant effect on the NFκB-dependent transactivation properties. Therefore, these broad-spectrum upstream promoter element targeting properties may be common features of both herpesvirus-encoded vGCR proteins and may be independent of ligand engagement in both vGCR/ORF74 and US28.
DISCUSSION
The KSHV-encoded vGCR (ORF74) protein has remarkable properties that appear likely to contribute to the unique features of KSHV pathogenicity, including spindle cell proliferation and angiogenesis. Unlike its homologues US28 in HCMV and ORF74 in HVS, the transforming KSHV version of vGCR apparently has acquired the ability both to signal in a ligand-independent manner and to induce VEGF-mediated angiogenesis via MAPK signaling pathways (2, 4).
The initial goals of our studies were, first, to determine whether vGCR mRNA represents a latent or lytic class transcript, as well as to identify its promoter and to study the control of its expression at the transcriptional level; second, to identify the predicted protein product, generate a specific antibody, and examine the pattern of expression of the viral protein product in latently infected or induced PEL and endothelial cell lines; and third, to ask directly whether the vGCR protein is expressed in human tumor samples associated with KSHV.
Our results revealed that (i) vGCR mRNA was primarily synthesized as both spliced and unspliced versions of a delayed early class bicistronic species of approximately 3.0 kb in size in either TPA- or sodium butyrate-induced BCBL-1 cells, but was not induced at all even by 72 h after either sodium butyrate or TPA treatment in the more restrictive HBL6 cell line. However, even in BCBL-1 cells, significant expression did not occur until the 48- and 72-h time points, with kinetics that matched those of vSSB (ORF6), vTS (ORF70), and vDHFR (ORF2), which is much later than for vIL6 and other members of the very early class of genes that also include the ZMP-A(K5), vMIP-1A, T1.1/PAN, and MTA (ORF57) mRNA species (9; Ciufo et al., unpublished).
(ii) The bipartite proximal upstream vOX2/vGCR promoter control region functioned as an RTA- and TPA-inducible lytic cycle promoter that overlaps head to head with the latent-state LANA1 promoter, which in contrast does not respond to RTA or TPA.
(iii) The predicted vGCR coding region expressed under the control of the SV40 enhancer produced a 48-kDa monomeric and apparently aggregated dimeric 90-kDa form in DNA-transfected Vero cells. Comparison with the 38-kDa form produced by in vitro transcription and translation suggested that it may be modified by either glycosylation or phosphorylation or both.
(iv) The expected cytoplasmic and membrane-associated vGCR protein encoded by ORF74 was detected as a TPA-induced product by IFA and immunohistochemistry in many BCBL-1 cells but rarely in HBL6 cells, as well as in a subset of spontaneously lytic cells in the JSC-1 PEL cell line and in the subset of persistently infected DMVEC cells that are undergoing lytic cycle progression, especially after TPA treatment.
(v) Cytoplasmic vGCR expression was also detected by immunohistochemistry in archival paraffin block sections of both primary PEL and MCD lymphoid tumor samples, as well as in a nodular KS lesion, but again only within a small percentage of apparently lytically activated cells.
(vi) An intact vGCR effector expression plasmid strongly upregulated the viral T1.1/PAN, DSBR, and K1 lytic cycle promoter-driven reporter genes as well as both the LLP latent-state promoter reporter genes and an NFκB motif-containing minimal heterologous c-Fos promoter in cotransfection assays, suggesting that the vGCR protein may potentially play a role in upregulation of either cellular or viral gene expression in a relatively nonspecific fashion.
The same predominant 2.8- to 3.0-kb spliced bicistronic vOX2/vGCR mRNA species initiating at position 127848 was also described both by Talbot et al. (68) in BCP1 cells after sodium butyrate treatement and by Kirschner et al. (40) in BCBL-1 cells after TPA treatment. The bicistronic nature of the vGCR cDNA accounts for the unexpectedly large size of the vGCR mRNA species detected in Northern blotting experiments in all three studies, but a satisfactory explanation for translation of the vGCR protein itself as a downstream ORF is not yet available. Whether vOX2 or vGCR represents the predominant protein product encoded by the intact bicistronic mRNA species remains to be determined.
The originally predicted upstream initiator codon for vOX2 (K14) of CAAATGA at position 127883 is likely to be relatively weak, whereas there is another, probably better potential internal initiator codon of TCC ATGT at position 128114. Furthermore, it is curious that the proposed promoter elements for LANA1 are seemingly embedded within the predicted N-terminal coding region for vOX2 (37, 38, 55, 58, 61). At present there is no direct evidence that all of the predicted ORF-K14 coding region is indeed used to make the vOX2 protein. In fact, comparison with the equivalent ORF in the HVS and RRV genomes suggests instead that only this second TCCATG motif (amino acid 78) in ORF-K14 is conserved and therefore may be the actual initiator codon for the KSHV vOX2 protein.
Kirshner et al. (40) point out that because vGCR is toxic in supposedly overexpressed-DNA-transfected cells, it may only be synthesized at very low abundance in vivo, as would be expected for the downstream ORF in a bicistronic mRNA. However, one could also argue that the toxicity is not much of an issue once the lytic cycle is initiated, and we point out that the relative abundance of the vGCR protein in lytically induced PEL and DMVEC cells, as judged from the strength of the IFA and immunohistochemical signals in individual positive cells, is very similar to that obtained in both DNA-transfected and Ad-vGCR vector-transduced cells.
The alternative possibility that vGCR is instead translated only from a very minor monocistronic mRNA initiating within the intron between the ORF-K14 and ORF74 coding regions remains plausible, although there are no obvious TATA-like or other promoter elements nearby. One of the nine cDNA clones that we examined (cl-1) had a 5′ end at position 129301 and could potentially be interpreted as such a species rather than as just a truncated fragment of the larger 3.2-kb mRNA. However, no evidence for a significant amount of an mRNA species with the appropriate size of 1.2 kb was obtained in the Northern blotting experiment with TPA-induced BCBL-1 RNA, and our primer extension analysis also failed to detect an appropriate ORF74-proximal mRNA initiation site. Kirshner et al. (40) and Talbot et al. (68) also failed to detect any mRNA initiating in the region immediately proximal to the ORF74 coding region in BCBL-1 or BCP1 cells.
The apparent bimodal structure of the proposed rightwards vOX2/vGCR promoter region and its position as a overlapping divergent promoter with the previously characterized leftwards upstream LANA1 latent-state promoter (P1) are both extremely unusual features. In fact, all of the predicted elements of the two potential rightwards lytic cycle promoters (P3 and P2) lie within the 5′ intron of the tricistronic leftwards LANA1/vCYC-D/vFLIP latent-state 6.0-kb transcript, which is itself both 5′ and 3′ coterminal with the 2.4-kb bicistronic leftwards latent-state vCYC-D/vFLIP transcript. We have demonstrated here that this TATA box-containing region (565 bp in size) lying just upstream of the vOX2/vGCR coding sequences does indeed function as an inducible promoter in transient reporter gene expression assays in BC2 PEL cells in the forwards but not the backwards orientation and that it responds strongly and directly to both TPA induction and cotransfection with the KSHV-encoded RTA (ORF50) transactivator protein. These properties differ greatly from those of the high basal but noninducible latent-state LLP and LANA1 promoters in parallel studies and are consistent with the proposed classification of both vOX2 and vGCR as delayed-early lytic cycle genes.
Talbot et al. (68) also compared the properties of the LANA1 and vOX2/GCR promoter regions by transfection of reporter genes in both KSHV-positive and KSHV-negative cell lines but, in contrast to the specific latent and lytic properties that we describe, found similar basal activity and sodium butyrate responsiveness in different cell types. However, in our experience, and in contrast to TPA, sodium butyrate is a nonspecific transactivator of many target reporter genes in transient-cotransfection assays. Very recently, a paper by Jeong et al. (36) has described RTA- and TPA-responsive properties of a vOX2/GCR promoter-driven reporter gene similar to those that we have presented here.
Although there are two consensus TATA-like motifs, each with its own associated SP1 and CTF-like motifs in the proximal vOX2 upstream region (P3 and P2), we were able to detect only one predominant mRNA start site by primer extension analysis and cDNA sequencing of TPA-induced BCBL-1 PEL cell mRNA. This implies that the more proximal nonconsensus P2 TTTATAA motif (position 127814) rather than the more distal consensus P3 TATATAA motif (position 127634) is operative during the lytic cycle in B cells. Nevertheless, in our studies, 5′RACE analysis detected a second, presumably very low abundance, rightwards transcript in uninduced BCBL-1 mRNA that appeared to originate from the distal far upstream P3 TATATAA motif region. We were unable to isolate any vGCR-positive cDNA clones from either an uninduced BCBL-1 or PEL tumor cDNA library despite being able to detect vIL6 cDNAs very readily in both. Therefore, confirmation that this novel potential distinct promoter drives the expression of a true latent-state transcript, and that this second rare mRNA does indeed encode either vOX2 or vGCR or both, will require both dissection of the complex P3 and P2 promoter region and additional more sensitive RACE analysis approaches in the future.
The overlapping nature of the divergent vOX2/vGCR and LANA1 promoters produces oppositely oriented 5′ ends in the two classes of mRNAs that overlap by at least 38 bp and might be expected to be mutually exclusive or antagonistic. Interestingly, the levels of both LANA1 protein (by IFA) and LANA1 transcription (by primer extension) decrease after lytic cycle induction in both BCBL-1 PEL cells and in JSC-1-infected DMVEC spindle cell cultures (not shown), which could be partly accounted for by interference between the two overlapping oppositely oriented transcription units. Overlap of the 5′ end of the potential second minor latent P3-driven transcript with that of LANA1 would be by as much as 200 bases and would complicate the scenarios for differential control even further.
A role for vGCR in transformation or immortalization in PEL and in angiogenesis or KS could be relatively easily imagined if it was expressed constitutively as a latent-state gene product in all lymphoid tumor cells or in all KS spindle cells. However, this is evidently not the case. Both Cesarman et al. (16) and Guo et al. (33) originally reported preliminary RT-PCR evidence for expression of vGCR RNA in both PEL and KS tumors, but could not address the fraction of positive cells or the kinetic class of the RNA. Kirshner et al. (40) also demonstrated vGCR RNA expression in a KS lesion and in induced BCBL-1 cells by in situ hybridization, but no one has previously detected the vGCR protein directly either in vitro or in vivo. The number and the pattern of vGCR mRNA-positive cells that they observed was very similar to those that we have detected here directly by immunohistochemical staining for the vGCR protein itself in KS, PEL tumor, and MCD specimens. The fraction of constitutively expressing vGCR-positive cells both in the tumor samples and in uninduced PEL cells is even lower than for other early lytic cycle gene products such as vIL6, ZMP-A, and RAP (K8).
Our preliminary 5′RACE evidence suggested that a very low level of latent-state expression of vGCR mRNA from the P3 promoter may occur in PEL cells that is below the threshold of detection by most of our present methods. This obviously requires further intensive investigation, but clearly lytic cycle upregulation from the P2 promoter leads to far more abundant expression in just a small subset of the cells. Nevertheless, despite the fact that lytic cycle gene expression of the early cytokine and vGCR genes occurs in only a very small fraction of KS or PEL tumor cells, this could still contribute in a downstream paracrine fashion to pathogenic effects on adjacent cells (28). Furthermore, expression of just some of the early-class captured cellular genes may represent a true semipermissive or extended “pseudolatent” class of gene expression that may be compatible with cell survival and somewhat similar to the class II and III EBV gene expression found in cultured immortalized but nontumorigenic B lymphoblasts.
Although we believe that the concept of a pseudolatent state or superearly class of gene expression may be valid, as exemplified here by the expression of vIL6 and ZMP-A (K5), but not vGCR, vSSB (ORF6), or vPPF (ORF59), in HBL6 cells even at 48 h after either sodium butyrate or TPA treatment, we have not observed any PEL cells that express high levels of vGCR without also expressing the ORF59 lytic cycle DNA replication protein. Therefore, the evidence at present favors vGCR protein expression only in those relatively rare cells that enter the full lytic cycle.
Our studies also demonstrated that the intact vGCR protein has the ability to behave as a somewhat nonspecific or promiscuous transactivator of KSHV promoter-driven reporter genes in transient-cotransfection assays. Although others have interpreted that vGCR is a ligand-independent MAPK signaling protein that leads to upregulation of AP1 activity and VEGF expression in particular (2, 4), the property of strongly upregulating both orientations of the divergent T1.1/PAN lytic promoter and the LLP latency leader promoter region was quite surprising. Furthermore, other viral promoter-driven reporter genes such as K1-LUC that do not contain any obvious AP1 sites also responded similarly, and the vGCR-induced responses by both the latent LLP-LUC and lytic PAN-LUC reporter genes still occurred after removal of all of the classic AP1 motifs present.
Interestingly, an engineered CAT reporter gene containing two NFκB motifs inserted into a minimal promoter background also responded to vGCR transactivation in an NFκB motif-dependent manner. The related HCMV US28 vGCR family protein also transactivated the NFκB-CAT reporter gene in parallel assays, in support of a recent report by Casarosa et al. (12). Curiously, despite dramatic effects on ligand-dependent versus ligand-independent activity of CXCR2 (8), mutations converting the charged conserved TM2/loop3 motif DRY to VRY in vGCR and vice versa in US28 failed to influence the NFκB-activating ability of either protein.
Some recent studies suggest that oligomerization of GPCR receptor family proteins contribute to their signaling and regulation (51), and Casarosa et al. (12) have argued that HCMV US28 signaling may also be constitutive or ligand independent, like that of vGCR/ORF74. Therefore, both our NFκB response data and our observations that both vGCR/ORF74 (Fig. 6) and US28 (data not shown) appear to form dimers as well as monomers on Western immunoblots with extracts from DNA-transfected cells would be consistent with the notion that these two herpesvirus vGCRs have very similar properties.
Overall, these results suggest that KSHV vGCR/ORF74 may activate other transcriptional pathways (including NFκB) in addition to AP1, and that these reporter gene assays provide a convenient way to measure its activity. Conceivably, this activity may play some as yet unknown role in viral gene regulation, perhaps in global activation of late viral or cellular genes during the lytic cycle. However, in contrast to the RTA (ORF50) effector gene, we were unable to activate KSHV lytic cycle gene expression (as measured by vIL6 IFA) with the vGCR effector DNA plasmid alone when introduced into BC2 PEL cells. Perhaps significantly, herpes simplex virus, HCMV, EBV, and HVS also all encode proteins that behave as nonspecific transactivators of reporter genes, i.e., the IE110 (ICP0), IE2 (IE86), MTA (BMLF1 or MS), and IE-G (ORF57) proteins, respectively. Although there are at least three different mechanisms of transcriptional or posttranscriptional regulation involved here, they all nevertheless contribute in important ways to the overall efficiency of herpesvirus lytic cycle gene expression.
ADDENDUM IN PROOF
Schwarz and Murphy (M. Schwarz and P. M. Murphy, J. Immunol. 167:505-513, 2001) and Pati et al. (S. Pati, M. Cavrois, H.-G. Guo, J. S. Foulke, Jr., J. Kim., R. A. Feldman, and M. Reitz, J. Virol. 75:8660-8673, 2001) have also recently described NFκB motifs as targets for KSHV vGCR transactivation.
Acknowledgments
These studies were supported by National Institutes of Health research grants R01 CA73585 and R01 CA84122 awarded to G.S.H. and P01 CA70062 awarded to R.F.A. J.S.C. and P.S.K. were partially supported by Johns Hopkins Pharmacology Training Grant 5T32-GM08763, and L.J.P. was partially supported by Johns Hopkins Biochemistry, Cell and Molecular Biology Training Grant 2T32-GM07445.
We thank Marvin S. Reitz (Institute of Human Virology, University of Maryland, Baltimore) and John Nicholas (Oncology Department, Johns Hopkins School of Medicine) for gifts of the original KSHV vGCR coding region clone and the Flag-tagged expression vector version, respectively. Ethel Cesarman (Columbia University, New York, N.Y.) kindly provided the KSHV-positive MCD and PEL tumor paraffin sections used for immunohistochemistry. Steve Hardy (Somatix Therapy Corporation, Alameda, Calif.) provided a seed culture of Adpsi5/Lox virus.
REFERENCES
- 1.Ahuja, S. K., and P. M. Murphy. 1993. Molecular piracy of mammalian interleukin-8 receptor type B by herpesvirus saimiri. J. Biol. Chem. 268:20691-20694. [PubMed] [Google Scholar]
- 2.Arvanitakis, L., E. Geras-Raaka, A. Varma, M. C. Gershengorn, and E. Cesarman. 1997. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature 385:347-350. [DOI] [PubMed] [Google Scholar]
- 3.Arvanitakis, L., E. A. Mesri, R. G. Nador, J. W. Said, A. S. Asch, D. M. Knowles, and E. Cesarman. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648-2654. [PubMed] [Google Scholar]
- 4.Bais, C., B. Santomasso, O. Coso, L. Arvanitakis, E. G. Raaka, J. S. Gutkind, A. S. Asch, E. Cesarman, M. C. Gerhengorn, and E. A. Mesri. 1998. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature 391:86-89. [DOI] [PubMed] [Google Scholar]
- 5.Birkenbach, M. 1993. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J. Virol. 67:2209-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boshoff, C. 1998. Coupling herpesvirus to angiogenesis. Nature 391:24-25. [DOI] [PubMed] [Google Scholar]
- 7.Boshoff, C., S.-J. Gao, L. E. Healy, S. Matthews, A. J. Thomas, L. Coignet, R. A. Warnke, J. A. Strauchen, E. Matutes, O. W. Kamel, P. S. Moore, R. A. Weiss, and Y. Chang. 1998. Establishing a KSHV positive cell line (BCP-1) from peripheral blood and characterizing its growth in Nod/SCID mice. Blood 91:1671-1679. [PubMed] [Google Scholar]
- 8.Burger, M., J. A. Burger, R. C. Hoch, Z. Oades, H. Takamori, and I. U. Schraufstatter. 1999. Point mutation causing constitutive signaling of CXCR2 leads to transforming activity similar to Kaposi's sarcoma herpesvirus-G protein-coupled receptor. J. Immunol. 163:2017-2022. [PubMed] [Google Scholar]
- 9.Cannon, J. S., D. Cuifo, A. L. Hawkins, C. A. Griffin, M. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields highly infectious Kaposi's sarcoma associated herpesvirus supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon, J. S., J. Nicholas, J. M. Orenstein, R. B. Mann, P. J. Browning, J. A. DiGiuseppe, E. Cesarman, G. S. Hayward, and R. F. Ambinder. 1999. Heterogeneity of viral IL6 expression in HHV-8-associated diseases. J. Infect. Dis. 180:824-828. [DOI] [PubMed] [Google Scholar]
- 11.Carbone, A., A. M. Cilia, A. Gloghini, D. Capell, L. Fassone, T. Perin, D. Rossi, V. Canzonieri, P. De Paoli, E. Vaccher, U. Tirelli, R. Volpe, and G. Gaidano. 2000. Characterization of a novel HHV-8-positive cell line reveals implications for the pathogenesis and cell cycle control of primary effusion lymphoma. Leukemia 14:1301-1309. [DOI] [PubMed] [Google Scholar]
- 12.Casarosa, P., R. A. Bakker, D. Verzijl, M. Navis, H. Timmerman, R. Leurs, and M. J. Smit. 2001. Constitutive signaling of the human cytomegalovirus-encoded chemokine receptor US28. J. Biol. Chem. 276:1133-1137. [DOI] [PubMed] [Google Scholar]
- 13.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 14.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two AIDS-related lymphoma cell lines containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 15.Cesarman, E., R. G. Nador, K. Aozasa, G. Delsol, J. W. Said, and D. M. Knowles. 1996. Kaposi's sarcoma-associated herpesvirus in non-AIDS-related lymphomas occurring in body cavities. Am. J. Pathol. 149:53-57. [PMC free article] [PubMed] [Google Scholar]
- 16.Cesarman, E., R. G. Nador, F. Bai, R. A. Bohenzky, J. J. Russo, P. S. Moore, Y. Chang, and D. M. Knowles. 1996. Kaposi's sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi's sarcoma and malignant lymphoma. J. Virol. 70:8218-8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan, S. R., C. Bloomer, and B. Chandran. 1998. Identification and characterization of human herpesvirus-8 lytic cycle-associated ORF 59 protein and the encoding cDNA by monoclonal antibody. Virology 240:118-126. [DOI] [PubMed] [Google Scholar]
- 18.Chan, Y.-J., C.-J. Chiou, Q. Huang, and G. S. Hayward. 1996. Synergistic interactions between overlapping binding sites for the serum response factor and ELK-1 proteins mediate both basal enhancement and phorbol ester responsiveness of primate cytomegalovirus major immediate-early promoters in monocyte and T-lymphocyte cell types. J. Virol. 70:8590-8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 20.Chang, Y., P. S. Moore, S. J. Talbot, C. H. Boshoff, T. Zarkowska, D. Godden-Kent, H. Paterson, R. A. Weiss, and S. Mittnacht. 1996. Cyclin encoded by KS herpesvirus. Nature 382:410.. [DOI] [PubMed] [Google Scholar]
- 21.Chang, Y.-N., D. Dong, G. S. Hayward, and S. D. Hayward. 1990. The Epstein-Barr virus Zta transactivator: a member of the B/Zip family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 64:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chuck, S., R. M. Grant, E. Katongole-Mbidde, M. Conant, and D. Ganem. 1996. Frequent presence of a novel herpesvirus genome in lesions of human immunodeficiency virus-negative Kaposi's sarcoma. J. Infect. Dis. 173:248-251. [DOI] [PubMed] [Google Scholar]
- 23.Ciufo, D. M., J. S. Cannon, L. J. Poole, F. Wang, P. Murray, R. F. Ambinder, and G. S. Hayward. 2001. Spindle cell conversion by Kaposi's sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J. Virol. 75:5614-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis, M. A., M. Sturzl, C. Blasig, A. Schrier, H.-G. Guo, M. Reitz, S. R. Opalenik, and P. J. Browning. 1997. Expression of human herpesvirus 8-encoded cyclin D in Kaposi's sarcoma spindle cells. J. Natl. Cancer Inst. 89:1868-1874. [DOI] [PubMed] [Google Scholar]
- 25.Decker, L. L., P. Shankar, G. Khan, R. B. Freeman, B. J. Dezube, J. Lieberman, and D. A. Thorley-Lawson. 1996. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J. Exp. Med. 184:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. v. Marck, D. Salmon, I. Gorin, J.-P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flore, O., S. Rafil, S. Ely, J. J. O'Leary, E. M. Hyjek, and E. Cesarman. 1998. Transformation of primary human endothelial cells by Kaposi's sarcoma-associated herpesvirus. Nature 394:588-592. [DOI] [PubMed] [Google Scholar]
- 29.Gaidano, G., K. Cechova, Y. Chang, P. S. Moore, D. M. Knowles, and R. Dalla-Favera. 1996. Establishment of AIDS-related lymphoma cell lines from lymphomatous effusions. Leukemia 10:1237-1240. [PubMed] [Google Scholar]
- 30.Gaidano, G., C. Pastore, A. Gloghini, D. Capello, U. Tirelli, G. Saglio, and A. Carbone. 1997. Microsatellite instability in KSHV/HHV-8 positive body-cavity-based lymphoma. Hum. Pathol. 28:748-750. [DOI] [PubMed] [Google Scholar]
- 31.Geras-Raaka, E., L. Arvanitakis, C. Bais, E. Cesarman, E. A. Mesri, and M. C. Gershengorn. 1998. Inhibition of constitutive signaling of Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor by protein kinases in mammalian cells in culture. J. Exp. Med. 187:801-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geras-Raaka, E., A. Varma, H. Ho, I. Clark-Lewis, and M. C. Gershengorn. 1998. Human interferon-γ-inducible protein 10 (IP10) inhibits constitutive signaling of Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor. J. Exp. Med. 188:405-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo, H.-G., P. Browning, J. Nicholas, G. S. Hayward, Y. W. Jiang, M. Sadowska, E. Tschachler, M. Raffeld, S. Columbini, R. C. Gallo, and M. Reitz. 1997. Characterization of a chemokine receptor-related gene in human herpesvirus 8 and its expression in Kaposi's sarcoma. Virology 228:371-378. [DOI] [PubMed] [Google Scholar]
- 34.Guo, W. X., T. Antakly, M. Cadotte, Z. Kachra, R. Kunkel, R. Masood, and P. Gill. 1996. Expression and cytokine regulation of glucocorticoid receptors in Kaposi's sarcoma. Am. J. Pathol. 148:1999-2008. [PMC free article] [PubMed] [Google Scholar]
- 35.Herndier, B. G., A. Werner, P. Arnstein, N. W. Abbey, F. Demartis, R. L. Cohen, M. A. Shuman, and J. A. Levy. 1994. Characterization of a human Kaposi's sarcoma cell line that induces angiogenic tumors in animals. AIDS 8:575-581. [DOI] [PubMed] [Google Scholar]
- 36.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kellam, P., C. Boshoff, D. Whitby, S. Matthews, R. A. Weiss, and S. J. Talbot. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum Virol. 1:19-29. [PubMed] [Google Scholar]
- 39.Kellam, P., D. Bourboulia, N. Dupin, C. Shotton, C. Fisher, S. Talbot, C. Boshoff, and R. A. Weiss. 1999. Characterization of monoclonal antibodies raised against the latent nuclear antigen of human herpesvirus 8. J. Virol. 73:5149-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirshner, J. R., K. Staskus, A. Haase, M. Lagunoff, and D. Ganem. 1999. Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi's sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J. Virol. 73:6006-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, G., L. Heston, E. Grogan, L. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and those without HIV infection. N. Engl. J. Med. 332:1182-1185. [DOI] [PubMed] [Google Scholar]
- 43.Moore, P. S., S.-J. Gao, G. Dominguez, E. Cesarman, O. Lungu, D. Knowles, R. Garber, P. E. Pellett, D. J. McGeoch, and Y. Chang. 1996. Primary characterization of a herpesvirus agent associated with Kaposi's sarcoma. J. Virol. 70:549-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moses, A. V., K. N. Fish, R. Ruhl, P. P. Smith, J. G. Strussenberg, L. Zhu, B. Chandran, and J. A. Nelson. 1999. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J. Virol. 73:6892-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mullen, M.-A., S. Gersberger, D. M. Ciufo, J. D. Mosca, and G. S. Hayward. 1995. Evaluation of colocalization interactions between the IE110, IE175, and IE63 transactivator proteins of herpes simplex virus within subcellular punctate structures. J. Virol. 69:476-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neipel, F., J.-C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicholas, J., K. R. Cameron, and R. W. Honess. 1992. Herpesvirus saimiri encodes homologues of G protein-coupled receptors and cyclins. Nature 355:362-365. [DOI] [PubMed] [Google Scholar]
- 48.Nicholas, J., V. Ruvolo, J.-C. Zong, D. Ciufo, H.-G. Guo, M. S. Reitz, and G. S. Hayward. 1997. A single 13 kilobase divergent locus in Kaposi sarcoma-associated herpesvirus (HHV8) contains at least nine genes homologous to or related to cellular proteins. J. Virol. 71:1963-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicholas, J., V. R. Ruvolo, W. H. Burns, G. Sandford, X. Wan, D. Ciufo, S. B. Hendrickson, H.-G. Guo, G. S. Hayward, and M. S. Reitz. 1997. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat. Med. 3:287-292. [DOI] [PubMed] [Google Scholar]
- 50.Nicholas, J., J.-C. Zong, D. J. Alcendor, D. M. Ciufo, L. J. Poole, R. T. Sarisky, C. J. Chiou, X. Zhang, X. Wan, H.-G. Guo, M. S. Reitz, and G. S. Hayward. 1998. Novel organizational features, captured cellular genes and strain variability within the genome of KSHV/HHV8. J. Natl. Cancer Inst. Monogr. 23:79-88. [DOI] [PubMed] [Google Scholar]
- 51.Overton, M. C., and K. J. Blumer. 2000. G-protein-coupled receptors function as oligomers in vivo. Curr. Biol. 10:341-344. [DOI] [PubMed] [Google Scholar]
- 52.Pierce, J. W., M. Lenardo, and D. Baltimore. 1988. Oligonucleotide that binds nuclear factor NF-κB acts as a lymphoid-specific and inducible enhancer element. Proc. Natl. Acad. Sci. USA 85:1482-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pizzorno, M. C., M.-A. Mullen, Y.-N. Chang, and G. S. Hayward. 1991. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J. Virol. 65:3839-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pleskoff, O., C. Treboute, A. Brelot, N. Heveker, M. Seman, and M. Alizon. 1997. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science 276:1874-1878. [DOI] [PubMed] [Google Scholar]
- 55.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S.-J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Renne, R., M. Lagunoff, W. Zhong, and D. Ganem. 1996. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J. Virol. 70:8151-8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 58.Russo, J. J., R. A. Bohenzky, M.-C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Said, J. W., M. R. Rettig, K. Heppner, R. A. Vescio, G. Schiller, H. J. Ma, D. Belson, A. Savage, I. P. Shintaku, H. P. Koeffler, H. Asou, G. Pinkus, J. Pinkus, M. Schrage, E. Green, and J. R. Berenson. 1997. Localization of Kaposi's sarcoma-associated herpesvirus in bone marrow biopsy samples from patients with multiple myeloma. Blood 90:4278-4282. [PubMed] [Google Scholar]
- 60.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 73:1438-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 63.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sturzl, M., C. Blasig, A. Schreier, F. Neipel, C. Hohenadl, E. Cornali, G. Ascherl, S. Esser, N. H. Brockmeyer, M. Ekman, E. E. Kaaya, E. Tschachler, and P. Biberfeld. 1997. Expression of HHV-8 latency-associated T0.7 RNA in spindle cells and endothelial cells of AIDS-associated, classical and African Kaposi's sarcoma. Int. J. Cancer 72:68-71. [DOI] [PubMed] [Google Scholar]
- 65.Sturzl, M., C. C. Hohenadi, C. Zeitz, E. Castanos-Velez, A. Wunderlich, G. Ascheri, P. Biberfeld, P. Monini, P. J. Browning, and B. Ensoli. 1999. Expression of K13/v-FLIP gene of human herpesviruses 8 and apoptosis in Kaposi's sarcoma spindle cells. J. Natl. Cancer Inst. 91:1725-1733. [DOI] [PubMed] [Google Scholar]
- 66.Sun, R., S.-F. Lin, L. Gradoville, and G. Miller. 1996. Polyadenylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 93:11883-11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun, R., S.-F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Talbot, S. J., R. A. Weiss, P. Kellam, and C. Boshoff. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84-94. [DOI] [PubMed] [Google Scholar]
- 69.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 93:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]