Abstract
GCN5 is a histone acetyltransferase (HAT) essential for development in mammals and critical to stress responses in yeast. The protozoan parasite Toxoplasma gondii is a serious opportunistic pathogen. The study of epigenetics and gene expression in this ancient eukaryote has pharmacological relevance and may facilitate the understanding of these processes in higher eukaryotes. Here we show that the disruption of T. gondii GCN5 yields viable parasites, which were subsequently employed in a proteomics study to identify gene products affected by its loss. Promoter analysis of these TgGCN5-dependent genes, which were mostly parasite specific, reveals a conserved T-rich element. The loss of TgGCN5 does not attenuate virulence in an in vivo mouse model. We also discovered that T. gondii is the only invertebrate reported to date possessing a second GCN5 (TgGCN5-B). TgGCN5-B harbors a strikingly divergent N-terminal domain required for nuclear localization. Despite high homology between the HAT domains, the two TgGCN5s exhibit differing substrate specificities. In contrast to TgGCN5-A, which exclusively targets lysine 18 of H3, TgGCN5-B acetylates multiple lysines in the H3 tail. We also identify two ADA2 homologues that interact differently with the TgGCN5s. TgGCN5-B has the potential to compensate for TgGCN5-A, which probably arose from a gene duplication unique to T. gondii. Our work reveals an unexpected complexity in the GCN5 machinery of this primitive eukaryote.
Epigenetics plays an important role in the regulation of gene expression in eukaryotic cells; however, very little is understood about this method of transcription modulation in parasitic protozoa. Evidence has been marshaled that histone acetylation in particular is critical to important parasitic processes, including antigenic variation and life cycle stage differentiation (14, 17, 35, 39). In addition, a parasite histone deacetylase has been identified as the target of the broad-spectrum antiprotozoal compound apicidin (9).
Histone acetylation is linked to the activation of gene expression either by attenuating the DNA-histone interaction and/or contributing to a “histone code,” which proposes that patterns of histone modifications constitute a language read by other proteins to bring about a cellular response (43). A variety of histone acetyltransferases (HATs) have been identified in eukaryotes (40). The best characterized is GCN5 (general control nonrepressed), previously identified as a putative transcriptional adaptor in a yeast mutant screen for amino acid biosynthesis deficiencies (6). GCN5 HATs operate in multisubunit complexes (e.g., SAGA and ADA) to acetylate lysine residues in histone tails, preferentially those of histone H3 (40). GCN5 and ADA2, an adaptor protein that binds GCN5 as well as the acidic activation domain of transcriptional activators like GCN4 (2), are nonessential in yeast. However, each is critical for growth under starvation conditions (26, 36) or during temperature stress (31). Consequently, GCN5 has been linked to activating GCN4-dependent genes important to the cellular stress response, controlling ∼10% of yeast genes (3, 21).
The phylum Apicomplexa is home to protozoan parasites of tremendous medical and economical importance, including Plasmodium (malaria), Cryptosporidium (cryptosporidiosis), and Eimeria (coccidiosis). Toxoplasma gondii is an apicomplexan that parasitizes warm-blooded vertebrates, causing congenital birth defects and serious complications in immunocompromised patients (5). GCN5 HATs have been cloned in T. gondii and Plasmodium falciparum (16, 18, 47). Histone H3 is well conserved in these parasites, including each lysine targeted by GCN5 HATs in other species (45). However, in contrast to other GCN5s, the T. gondii homologue (TgGCN5) exhibits an unusual bias to acetylate only lysine 18 of histone H3 (H3 [K18]) (39). Whether another HAT existed in T. gondii that could acetylate the additional lysines in H3 remained unresolved.
The tachyzoite stage of the life cycle causes acute toxoplasmosis as the parasites rapidly invade and replicate inside nucleated cells. To address the role TgGCN5 plays in the context of acute infection, we attempted to disrupt the TgGCN5 locus in the virulent (type I) RH strain of T. gondii, which has largely lost its ability to convert to the bradyzoite cyst form (13). Here we report the successful generation of a TgGCN5 “knockout” clone that proved amenable to the identification of genes that may be regulated by this HAT. Using a mouse model, we also establish that the loss of TgGCN5 does not attenuate virulence. Finally, we identify a second GCN5 in T. gondii (TgGCN5-B) that targets multiple lysines in the H3 tail and clone two distinct ADA2 homologues that interact differently with the two TgGCN5s.
MATERIALS AND METHODS
Parasite culture, methods, and reagents.
Toxoplasma RH strain was maintained in primary human foreskin fibroblasts and filter purified from host cell debris as previously described (38). Parasite mRNA was isolated from filter-purified tachyzoites using the Poly(A)Pure system (Ambion), followed by DNase treatment. Genomic DNA was harvested from purified tachyzoites using sodium dodecyl sulfate (SDS)-proteinase K lysis, phenol-chloroform extraction, and ethanol precipitation. Immunofluorescence assays of T. gondii were carried out as described previously (4). Primary antibodies used for these studies included polyclonal anti-FLAG (Sigma) at 1:1,000 and anti-acetyl H3 [K18] (Abcam) at 1:500. Microneme secretion assays were carried out as described previously (8, 29). Antibody to MIC2 and MIC4 was kindly provided by David Sibley (Washington University, St. Louis, MO); antibody to MIC5 and MIC11 was kindly provided by Vern Carruthers (Johns Hopkins Malaria Research Institute, Baltimore, MD). Anti-SAG1 was purchased from Argene, Inc.
Generation of ΔTgGCN5.
Two λDASH-II clones used to map the TgGCN5 genomic locus (47) provided ample contiguous sequence for the construction of a knockout vector (Fig. 1A). An ∼6.0-kb fragment from pλGCN5Xho was cloned into pminHXGPRT (AIDS reagent no. 2855) at the XhoI site. An ∼6.0-kb SpeI-SmaI fragment from pλGCN5Bgl2 was cloned into the resulting vector at SpeI and a blunted XbaI site (lost). The resulting construct (ΔTgGCN5::HX) contains ∼6.0 kb of continuous genomic sequence on each side of a cDNA-derived HXGPRT minigene, allowing selection in RHΔHXGPRT parasites (AIDS reagent no. 2859). Twenty-five micrograms of the ΔTgGCN5::HX vector was linearized with ScaI for transfection into 2 × 107 parasites by electroporation. Methods of transfection, selection in mycophenolic acid plus xanthine, and cloning by limiting dilution was performed as previously described (11). Clones were prescreened by PCR of genomic DNA using primers designed to amplify the TgGCN5 HAT domain; Southern analysis was used to confirm allelic replacement (Fig. 1B).
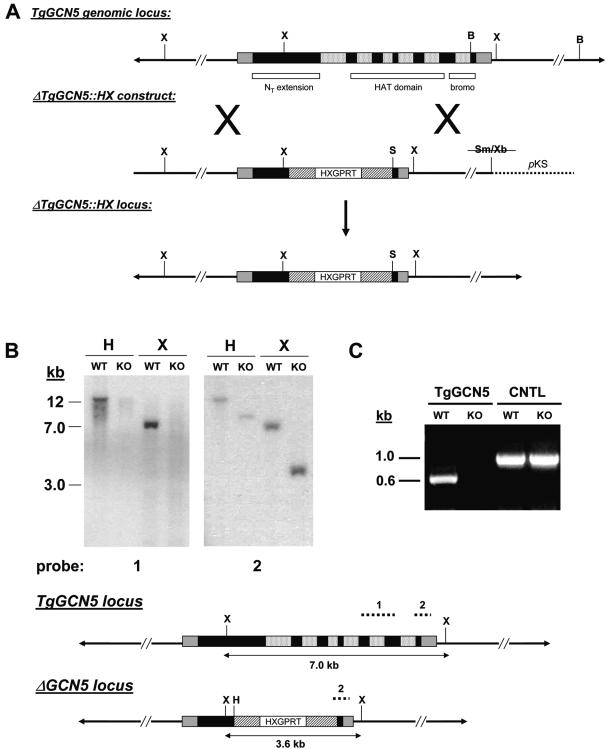
FIG. 1.
Generation of ΔTgGCN5 parasite clone. (A) Schematic diagram comparing the endogenous genomic locus to the replacement allele contained on the ΔTgGCN5::HX “knockout” construct. Black boxes indicate exons, mottled boxes indicate introns, and gray boxes indicate UTRs. In a perfect allelic replacement via a double crossover event, virtually the entire TgGCN5 coding sequence would be replaced by the HXGPRT minigene cassette. Striped boxes denote the dihydrofolate reductase promoter and 3′ UTR regions. B, BglII; S, SpeI; Sm, SmaI; X, XhoI; Xb, XbaI. (B) Southern analysis verifies allelic replacement. Probe 1, a cDNA-derived portion of the HAT domain hybridizes to the expected size fragments in wild-type (WT) genomic DNA digested with indicated restriction enzymes. However, probe 1 does not hybridize to genomic DNA from the ΔTgGCN5 knockout clone (KO). Probe 2, designed to a portion of the final exon plus 3′ UTR, hybridizes to DNA fragments of the expected size in both WT and KO parasites. H, HindIII; X, XhoI. (C) TgGCN5 transcript is absent from ΔTgGCN5. Primers were designed to amplify a portion of the HAT domain of TgGCN5 using total RNA harvested from either wild-type (WT) or “knockout” (KO) parasites. Primers designed to amplify a control transcript (adenosine kinase) validate the RNA purification from each sample. CNTL, control.
Proteomics and analysis.
Total parasite protein was harvested from ∼108 freshly lysed filter-purified tachyzoites from wild-type RH or ΔTgGCN5. After normalizing samples by Bradford assay, first-dimension resolving of proteins was conducted using the Bio-Rad PROTEAN isoelectric focusing cell and ReadyStrips (pH 3 to 10) for 120,000 Vh. Strips were then equilibrated for 20 min in buffer containing 6 M urea, 375 mM Tris-HCl (pH 8.8), 20% glycerol, 2% SDS, and 2% dithiothreitol (DTT). Strips were alkylated with 2.5% iodoacetamide in the same buffer minus DTT for 20 min, washed twice in 1× morpholinepropanesulfonic acid (MOPS), and resolved on Bio-Rad Criterion gels for 3 h at 120 V. Each sample treatment was performed in triplicate. Gels were fixed and stained with Coomassie brilliant blue R250 and scanned using the Fluor-S MAX MultiImager system and analyzed with PDQuest software (version 7.1.1; Bio-Rad). The background values for the gels were subtracted, and the spot peaks were located and quantified. Both the total optical density and the total number of spots correlate to the total concentration of each protein expressed. As a result, each protein's quantity was defined as parts per million of the total integrated optical density. After a reference pattern was prepared, the image analysis software was used to create a match set encompassing all gels of interest and to perform gel matching. Each gel in the match set was matched to the reference pattern, with landmark proteins (those uniformly expressed in all of the gels) used to quickly and accurately match all gels. The abundance of individual proteins was calculated using a quantitative analysis set within the PDQuest software, which allows for the construction of a relative change analysis comparison chart for any or all expressed proteins. Protein spots of interest were excised from the gel robotically using the PROTEAN 2D Spot Cutter (Bio-Rad), destained in 50% acetonitrile-50 mM ammonium bicarbonate, reduced with 10 mM DTT, and alkylated with 55 mM iodoacetamide. Following overnight digestion with trypsin (6 ng/μl) at 37°C, α-cyano-4-hydroxycinnamic acid was used as a matrix and matrix-assisted laser desorption ionization mass spectra were recorded in positive reflectron mode of the matrix-assisted laser desorption ionization-time of flight mass spectrometer (Micromass, Manchester, United Kingdom). Peptide mass profiles were compared to theoretical peptide masses using MS-Fit (http://prospector.ucsf.edu/ucsfhtml4.0/msfit.htm). At least 5 peptides and 15% coverage had to be observed for the identification of a peptide mass profile to be considered unambiguous (30). Two-dimensional gels and mass spectrometry work were performed in conjunction with the Protein Analysis Research Center (Indiana School of Medicine, Indianapolis, IN).
Molecular methods.
Rapid amplification of cDNA ends reactions were performed with the GeneRacer kit (Invitrogen) using T. gondii tachyzoite mRNA as a template for reverse transcription (RT) with random primers. RT-PCRs were performed using the SuperScript one-step kit from Invitrogen. Northern blots of parasite mRNA were produced using the NorthernMax system (Ambion). Probes for Northern blots were cDNA derived and intercalated with psoralen-biotin to allow detection by chemiluminescence following hybridization. All DNA sequencing was carried out by the IUSM Biochemistry Biotechnology Facility (Indianapolis, IN).
Recombinant protein expression and purification.
Full-length FLAGTgGCN5-B was amplified from cDNA using the following primers: sense, 5′-CATATGAAAATGGACTACAAGGACGACGACGACAAGGCGCCTTCAGAGTGTCCCAGCG; antisense, 5′-CCTAGGCTAGAAAAATGTCGGATGCTTCGCGCCC. FLAGΔ528TgGCN5-B was generated using the sense primer 5′-CATATGAAAATGGACTACAAGGACGACGACGACAAGCTCGACAAAGCCCCAACAGGAGCC with the antisense primer above. Both sense primers encode an N-terminal FLAG epitope tag (sequence shown in italics). Inserts were ligated into ptubXFLAG::HX (4) at the NdeI and AvrII sites (underlined above). Plasmid DNA (∼50 μg) was linearized using NotI prior to transfection of RHΔHXGPRT tachyzoites by electroporation as described previously (38). Stably transfected parasites were selected for by using mycophenolic acid and xanthine and cloned by limiting dilution.
To purify FLAG-tagged protein from stable clones, ∼1 × 108 filter-purified parasites were spun at 800 × g at 4°C for 10 min and washed in phosphate-buffered saline. The parasite pellet was resuspended in 1.0 ml of lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100) plus protease inhibitors, and sonicated three times on ice. Lysates were centrifuged at maximum speed for 10 min to remove insoluble material. Preclearing of lysates was performed for 30 min with 60 μl protein A affinity gel (Sigma) and then transferred to a new tube with 60 μl anti-FLAG M2 affinity gel (Sigma) and incubated overnight at 4°C with gentle agitation. Affinity gel was washed three times with wash buffer (50 mM Tris-HCl [pH 7.4r], 300 mM NaCl, and 1 mM EDTA), followed by two washes with 1× HAT assay buffer. After the last wash, ∼10 μl packed resin was transferred into a separate tube for Western blot analysis to insure that immunoprecipitation was successful. The remaining 20 μl packed resin was used in a HAT assay.
Radioactive HAT assay.
Each HAT assay (6) used ∼20 μl of packed resin from a FLAG affinity purification mixed with 2 μl of acetyl [3H]coenzyme A (acetyl-[3H]CoA) (Amersham), 4 μg chicken erythrocyte core histones (Upstate), 6 μl 5× HAT assay buffer (16), and 18 μl of distilled H2O. Polyhistidine-tagged Saccharomyces cerevisiae GCN5 (ScGCN5) was generated in CodonPlus Escherichia coli BL21 (Stratagene) and purified over nickel for use as a positive control (1 μg per HAT assay reaction). HAT reaction mixtures were incubated at 30°C for 60 min and stopped by the addition of 15 μl of 2× NuPAGE loading dye (Invitrogen) containing 2.5 μl of β-mercaptoethanol. After 10 min of incubation at 70°C, the reaction mixture was resolved on a NuPAGE 10% Bis-Tris gel (Invitrogen) using morpholineethanesulfonic acid (MES) buffer. Gel was fixed in an aqueous solution of 5% isopropanol and 5% acetic acid for 2 h followed by gentle washing in distilled water for 1 h. The gel was incubated in Autoflour reagent (National Diagnostic) for 2 h, dried, and processed for autoradiography.
Nonradioactive HAT assay and Western blotting.
The HAT assay was performed as described above, with the following differences: 1 μl of acetyl-CoA (1 mM in 10 mM sodium acetate [pH 4.5]), 1 μg histone H3 (Upstate), and 22 μl of distilled H2O. Gels were transferred to a polyvinylidene difluoride membrane and blocked in 5% nonfat dry milk in Tris-buffered saline-Tween (TBST) overnight at 4°C. Membranes were incubated with anti-AcH3 [K14] or anti-AcH3 [K9] (Upstate) or anti-AcH3 [K18] (Abcam) (all at 1:1,000) in TBST with 5% milk. After three washes with TBST, anti-rabbit antibody conjugated to horseradish peroxidase (Amersham) was incubated with membrane at a 1:2,500 dilution in 5% milk-TBST for 1 h. Results were detected by chemiluminescence using the Amersham ECL detection reagent.
Yeast two-hybrid testing.
Δ950TgGCN5-A was amplified by PCR using the following primers: sense, 5′-CATATGCGTCTCAGCACCCGAATAAATTACC; antisense, 5′-CCCGGGTCAGAAACTCCCGAGAGCCTCGACCTTGG. The insert was ligated in frame to the GAL4 DNA-binding domain of pGBKT7 (Clontech) at the NdeI and XmaI sites (underlined). Δ781TgGCN5-B was ligated into pGBKT7 at the NdeI and BamHI sites following PCR amplification using the following primers: sense, 5′-CATATGCACATCAATCCGCGCATCAATTACC; antisense, 5′-GGATCCCTAGAAAAATGTCGGATGCTTCGCGCCC. Portions of TgADA2-A and TgADA2-B corresponding to the putative GCN5-binding domains were ligated in frame to the GAL4 activation domain of pGADT7 (Clontech) using NdeI and BamHI sites. The Insert was generated using primers 5′-CATATGGAACGGAGAGCGAAACGCGCG (sense) and 5′-GGATCCTTAGCCCATGAGCAGTGCCG (antisense) to amplify TgADA2-A cDNA encoding amino acids 158 to 410 and primers 5′-CATATGCCTGAGGACGGCGGGACTCCTCC (sense) and 5′-GGATCCTTACTCGGGTGGGCTGTGAACTTTTGG (antisense) to amplify TgADA2-B cDNA encoding amino acids 461 to 761. Competent AH109 Saccharomyces cerevisiae cells (Clontech) were cotransformed with 0.25 μg of each vector. Each transformation was plated on double dropout (lacking leucine and tryptophan) and triple dropout (TDO, lacking leucine, tryptophan, and histidine) media. Plates were incubated at 30°C for 3 days. Eight colonies from the TDO plates were spotted onto quadruple dropout plates (QDO, lacking leucine, tryptophan, histidine, and adenine) containing X-α-Gal and incubated as before. Blue colonies larger than 3 mm were considered positive for protein-protein interaction.
Bioinformatics.
Preliminary genomic sequence data was accessed via http://ToxoDB.org. Genomic data were provided by The Institute for Genomic Research (supported by the NIH grant no. AI05093) and by the Sanger Center (Wellcome Trust). Sequence analysis tools included BLAST (http://www.ncbi.nih.gov/BLAST/), SMART (http://smart.embl-heidelberg.de/), and Pfam (http://www.sanger.ac.uk/Software/Pfam/search.shtml). Alignments and phylogenetic tree constructions were performed with Vector NTI 9.0 (Informax).
For the analysis of TgGCN5-dependent promoters, the data set was prepared by extracting ∼1,500 bp upstream of the translational start for most of the gene products in Table 1 (promoter sequences for accession no. 21214754 and 6003679 were disregarded, since the translational start site for these genes is unclear). BLASTX was performed against T. gondii coding sequences to eliminate the inclusion of coding regions within the data set. MEME (1) was used to identify conserved sequence motifs present in the upstream regions for these genes. MEME was run using three different parameters to specify the mode of occurrence of the motif(s), and a motif width of 6 to 20 bp was specified. Sequence logos to represent the motifs were created at http://weblogo.berkeley.edu/.
TABLE 1.
Gene products reported to be down-regulated in ΔGCN5 parasites
| Spot no. | Accession no. | Identityb | % Change |
|---|---|---|---|
| 3804 | 2499221 | NTPase II | −47 |
| 7606 | 22035892 | Glucose-6-phosphate-1-dehydrogenase | −45 |
| 7601 | 22035892 | Glucose-6-phosphate-1-dehydrogenase | −36 |
| 6505 | 13377046 | Glyceraldehyde-3-phosphate dehydrogenase | −59 |
| 5206 | 22035894 | Putative PDI-like protein | −20 |
| 6503 | 12698444 | cAMP-dependent protein kinase catalytic subunit | −34 |
| 8204 | 12698442 | cAMP-dependent protein kinase regulatory subunit | −25 |
| 4401 | 2854042 | Calmodulin domain protein kinase | −33 |
| 7401 | 21214754 | Calcium-dependent protein kinasea | −49 |
| 3302 | 1565285 | Calmodulin | −30 |
| 4802 | 14522886 | Membrane skeletal protein IMC1 | −18 |
| 2203 | 16580144 | Myosin light chain TgMLC1 | −32 |
| 6306 | 28975399 | SAG1 precursor | −64 |
| 6104 | 13957759 | SAG2/p22 | −61 |
| 6501 | 13447088 | SAG3 | −62 |
| 3802 | 4929214 | MIC4 | −34 |
| 1103 | 1806541 | MIC5/H4 | −62 |
| 2403 | 4704627 | MIC6 | −58 |
| 1003 | 23267147 | MIC11 precursor | −29 |
| 8207 | 2506908 | GRA2 precursor | −49 |
| 8202 | 2506908 | GRA2 precursor | −28 |
| 3204 | 2062409 | GRA7 | −55 |
| 7303 | 9909994 | ROP9 | −47 |
| 7605 | 6003679 | Mitochondrial processing peptidase alphaa | −64 |
| 5302 | 28974671 | Ribosomal P protein | −22 |
Excluded from promoter analysis due to low confidence in identified start site.
PDI, protein disulfide isomerase; cAMP, cyclic AMP.
Nucleotide sequence accession number.
New sequence data described in this report is available in the GenBank, EMBL, and DDBJ databases under the accession numbers AY875982 (TgGCN5-B), DQ112184 (TgADA2-A), and DQ112185 (TgADA2-B).
RESULTS
Disruption of the GCN5 gene in Toxoplasma gondii.
To examine the role of TgGCN5 in the context of the acute phase of infection, we generated a “knockout” clone by disrupting the genomic locus in the virulent type I strain. The endogenous TgGCN5 allele in RHΔHX tachyzoites (haploid) was replaced with an HXGPRT selectable marker by homologous recombination (11). As diagrammed in Fig. 1A, the knockout construct (ΔTgGCN5::HX) would remove the bulk of the TgGCN5 locus, including portions encoding the entire catalytic (HAT) domain and bromodomain. Figure 1B shows a Southern analysis of one clone representing a true allelic replacement, showing that the targeted locus is no longer present in the parasite genome. Moreover, mRNA encoding TgGCN5 is no longer detected by RT-PCR in the knockout clone, termed ΔTgGCN5 (Fig. 1C).
Similar to yeast lacking GCN5, no readily observable phenotype was evident in the ΔTgGCN5 clone. No appreciable difference in parasite growth rates between the wild type and ΔTgGCN5 were detected in vitro using parasite doubling or uracil incorporation assays (data not shown). We examined whether the loss of TgGCN5 impaired virulence in vivo in collaboration with David Sibley (Washington University, St. Louis, MO). An equal number of ΔTgGCN5 or wild-type tachyzoites were inoculated (intraperitoneally) into type CD-1 outbred mice. In addition, a T. gondii transgenic clone overexpressing recombinant TgGCN5 (4) was analyzed in the mouse model. Neither the loss nor overexpression of TgGCN5 alters the kinetics of infection in vivo (data not shown), revealing that TgGCN5 is not likely to play a significant role during the acute stage of toxoplasmosis.
Gene products differentially expressed in ΔTgGCN5.
Despite having no discernible phenotype under normal growth conditions, yeast lacking GCN5 were successfully employed to find genes regulated by this factor (20). A viable knockout of TgGCN5 presented a unique opportunity to elucidate the genes that may be controlled by this HAT in protozoa. Thus, differential expression analysis between wild-type and ΔTgGCN5 parasites was performed. As comprehensive microarrays for T. gondii are still in development, a proteomics approach was pursued. Two-dimensional (2D) gels were used to compare the proteomes of wild-type versus ΔTgGCN5 tachyzoites. Protein spots reported to be differentially expressed that could be unambiguously identified are listed in Tables 1 (down-regulated in ΔTgGCN5) and 2 (up-regulated in ΔTgGCN5). In some instances, multiple independent protein spots were assigned the same identity (e.g., GRA2), possibly reflecting differential splice variants or variability in posttranslational processing. As TgGCN5 is associated with transcriptional activation, gene products reported to be up-regulated in ΔTgGCN5 are likely due to an indirect effect caused by the lack of TgGCN5; similar observations were reported for yeast lacking GCN5 (20). Some of the down-regulated products are involved in metabolism and cellular signaling, but most are parasite specific and include microneme (MIC) and rhoptry proteins associated with host cell invasion (Table 1).
TABLE 2.
Gene products reported to be up-regulated in ΔGCN5 parasites
| Spot no. | Accession no. | Identity | % Change |
|---|---|---|---|
| 6806 | 2506932 | Putative NTPase | +42 |
| 4505 | 21912813 | UDP-N-acetyl-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase T1 | +77 |
| 8101 | 5764087 | Enolase I | +61 |
| 7302 | 34555736 | Enoyl-acyl carrier reductase | +48 |
| 3004 | 6984166 | RAB6 golgi GTPase | +82 |
| 3103 | 2507039 | GRA5/p21 | +51 |
| 6002 | 630463 | Peptidylprolyl isomerase | +27 |
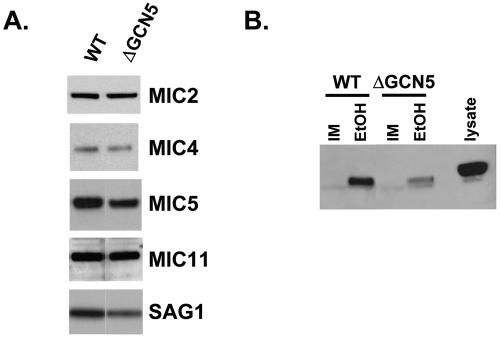
Attempts were made to confirm some of the gene products reported to be differentially expressed. Antibodies to SAG1 and several MIC proteins are available and were used to examine protein levels between wild-type and ΔTgGCN5 parasites (Fig. 2A). Consistent with the proteomics analysis, SAG1, MIC4, MIC5, and MIC11 appear to be down-regulated in the ΔTgGCN5, roughly corresponding to the levels observed on the 2D gels. For example, the 2D gel study suggested that MIC5 and SAG1 are down-regulated ∼60% in ΔTgGCN5, and a difference in protein levels is clear on the immunoblot. In contrast, MIC4 and MIC11 were only reported to be decreased ∼30%, and only modest decreases are observed by immunoblot. MIC2 was included as a control as it was not indicated to be differentially expressed.
FIG. 2.
(A) Western analysis comparing levels of indicated protein between wild-type (WT) and ΔTgGCN5 parasites. Samples were normalized to protein concentration. Antibody dilutions applied were as follows (dilutions are indicated in parentheses): anti-SAG1 (1:500), anti-MIC2 (1:10,000), anti-MIC4 (1:5,000), anti-MIC5 (1:5,000), anti-MIC11 (1:1,000). (B) Immunoblot for MIC2 following a microneme secretion assay. IM, invasion media (control); EtOH, ethanol, used at a final concentration of 1%. Parasite lysate displays the size of unprocessed MIC2 for comparison.
Noting the decreases in certain microneme proteins in ΔTgGCN5, we examined whether there was any functional defect in microneme secretion, which can be stimulated in vitro by ethanol. As shown in Fig. 2B, ΔTgGCN5 exhibits a considerable defect in microneme secretion, as assayed by processed MIC2 levels. Despite the apparent reductions in the expression and secretion of some microneme proteins, we were unable to detect any difference in parasite attachment or invasion in vitro using the red-green invasion assay (22). Either the observed deficiencies are not profound enough to alter the process or other invasion proteins are compensating. It is also possible that ethanol, being an artificial stimulus, is working differently than the native signals that induce microneme secretion.
Analysis of candidate TgGCN5-dependent promoters.
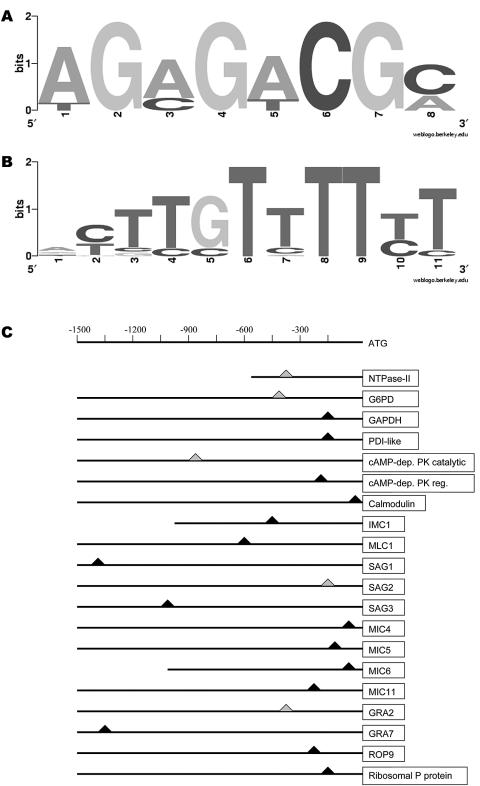
Gene expression facilitated by GCN5 is largely promoter specific (25). GCN5 does not bind DNA directly; rather, it is recruited by a transcriptional activator that is translationally up-regulated during stress. For example, yeast GCN4 recruits the GCN5 complex after binding the general control response element found upstream of target genes. We hypothesized that a similar scenario occurs in Apicomplexa, but bioinformatics analysis of ToxoDB does not reveal any hits for a GCN4 homologue or the human orthologue, ATF4. To assess whether any commonalities exist in the promoters of genes purportedly regulated by TgGCN5, we retrieved ∼1,500 bp of sequence upstream of the translational start from most of the gene products in Table 1. We were unable to detect a palindromic motif akin to the yeast general control response element consensus sequence in the set of T. gondii sequences examined. We could identify the (A/T)GAGACG motif in 16 of these sequences (Fig. 3A), previously reported to be a cis-acting enhancer of T. gondii genes (33). Another significant motif identified with an E-value of 8.1 × 10−2 was an 11-bp T-rich element, (A/G)(C/T)TTGTTTT(T/C)T (Fig. 3B). This motif represents a fairly well conserved sequence present on either strand in all 21 sequences analyzed. To ensure that the T-rich element is not commonplace in T. gondii promoter regions, 16 genes that did not show a change in protein levels in ΔTgGCN5 were analyzed in a similar manner. Importantly, the T-rich motif was not identified in this control set of genes (not shown).
FIG. 3.
Promoter analysis of candidate GCN5-dependent genes. (A) A known cis-acting enhancer of T. gondii genes (33) was identified by MEME in some of the promoters of genes reportedly down-regulated in ΔTgGCN5. The letter size depicts the relative abundance of each nucleotide conserved in the sequence. (B) T-rich element present in GCN5-dependent promoters analyzed in Table 1. (C) Schematic diagram of promoter sequences showing approximate location of the T-rich element. All positions are indicated with respect to the translational start site. A triangle indicates the position of the motif; gray triangles signify that the motif was found on the opposite strand. PDI, protein disulfide isomerase; cAMP, cyclic AMP; PK, protein kinase; dep, dependent; reg, regulatory.
We hypothesize that this T-rich motif may be indicative of a GCN5 requirement for transcriptional activation, possibly through interaction with an unidentified transcriptional activator that associates with this element. We note that this element is located at various positions from the translational start site and is found within the −300 position in 9 of 21 genes examined (Fig. 3C). The lengths of the 5′ untranslated regions (UTRs) for most of these genes are not known, and for at least two of the upstream sequences examined here, the T-rich motif may be within the 5′ UTR. However, yeast GCN5 function is independent of transcriptional initiation (25); thus, it is possible that this motif operates independently of orientation and position to recruit TgGCN5.
T. gondii possesses a second GCN5 family HAT.
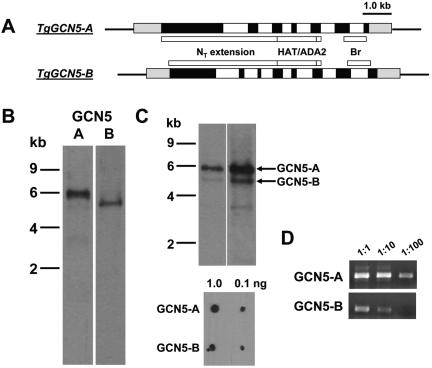
Mammals possess a second GCN5 family member called p300/CBP-associating factor (PCAF), but there are no reports of multiple GCN5 HATs in other animals or plants. While TgGCN5 may simply be nonessential, as in yeast, we monitored the T. gondii genome project (http://ToxoDB.org) to determine whether additional GCN5 homologues are present that could be playing a compensatory role. Database entry TGG_994635 contains significant homology to the nucleic acid sequence of the TgGCN5 previously reported, henceforth referred to as TgGCN5-A. Primers were designed based on genomic sequence data, and the products from multiple overlapping RT-PCRs allowed the verification and construction of the 3,099-bp coding sequence for this second GCN5 homologue in T. gondii (termed TgGCN5-B, accession no. AY875982). 5′ and 3′ rapid amplification of cDNA ends were employed to delineate the corresponding UTRs, which consist of 905 and 818 bp, respectively. The proposed start codon fits Kozak rules and is preceded by an in-frame stop codon 42 bp upstream (24). A comparative diagram of the loci for TgGCN5-A (8.4 kb) and TgGCN5-B (8.7 kb) is shown in Fig. 4A. The structural organization of each genomic locus is most similar in the region spanning the HAT catalytic domain; exons 3 to 5 of TgGCN5-A are highly homologous and identical in length to exons 4 to 6 of TgGCN5-B. According to the ToxoDB, TgGCN5-A is located on chromosome III and TgGCN5-B on chromosome LG14.
FIG. 4.
A pair of GCN5 HATs in T. gondii. (A) Schematic diagram of the genomic loci for TgGCN5-A and -B. TgGCN5-A is comprised of 7 exons (black) and 6 introns (white), while TgGCN5-B contains 8 exons. The approximate location of key domains in the encoded protein are indicated. HAT, histone acetyltransferase domain; ADA2, ADA2-binding domain; Br, bromodomain. Gray boxes represent untranslated regions. (B) Northern blots probed for TgGCN5-A and -B transcripts, respectively. (C) Dual-probing of a single Northern blot with both TgGCN5s. Two different exposure times are shown. Lower panel shows that equivalent amounts of nonisotopically labeled probes emit approximately equal chemiluminescent signals, so the differences observed on the blot are not due to a bias in probe labeling. (D) Equal amounts of cDNA generated from wild-type RNA were used in PCRs to amplify either TgGCN5-A or -B.
Northern blot analysis confirms the predicted transcript length of ∼4.8 kb for TgGCN5-B (Fig. 4B). Similar to TgGCN5-A, there do not appear to be any alternatively spliced messages for TgGCN5-B (4). Hybridizing equivalent levels of probe specific for each GCN5 to the same lane on a Northern blot shows that TgGCN5-A message levels are considerably higher than those for TgGCN5-B in tachyzoites (Fig. 4C). RT-PCR data are consistent with this result (Fig. 4D).
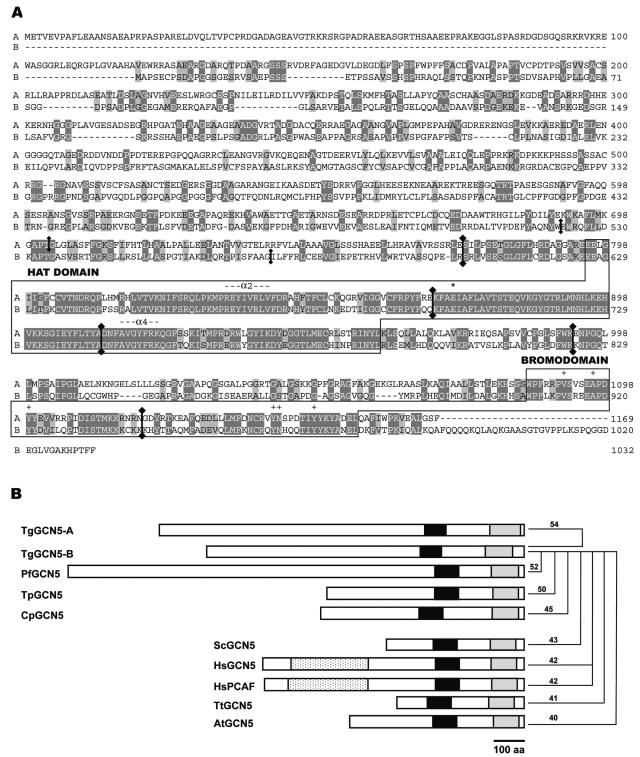
Alignments of the deduced amino acid sequences for TgGCN5-A and -B are shown in Fig. 5A. The TgGCN5-B open reading frame encodes a predicted protein of 1,032 amino acids, 137 residues shorter than TgGCN5-A. The HAT domains are nearly identical, and the putative ADA2-binding domain and bromodomains are less conserved. Most striking is the lack of homology in the lengthy regions upstream of the HAT domain (794 residues in TgGCN5-A; 625 residues in TgGCN5-B). Neither one of these N-terminal extensions has homology to protein sequences in other species, nor are any known protein motifs evident.
FIG. 5.
TgGCN5-B protein analysis. (A) Pairwise alignment of the deduced protein sequences for TgGCN5-A and -B. Dark gray highlights show identical residues; light gray denotes conserved differences. Locations of introns in the genomic sequence corresponding to the protein are indicated by vertical lines with diamondheads at junction sites. HAT and bromodomains are boxed and labeled. The conserved glutamic acid residue critical for HAT activity is marked with an asterisk, and conserved bromodomain residues important for interacting with acetylated lysines are designated with plus signs. (B) Cartoon depicting protein structures of GCN5 family proteins from Apicomplexa (top 5) and other eukaryotes (lower 5). Black boxes indicate HAT domains, gray boxes indicate bromodomains, and mottled boxes indicate PCAF homology domains. ADA2-binding domains are located between HAT and bromodomains. Numerical values represent the percent identities between TgGCN5-B and the indicated protein. Tg, Toxoplasma gondii; Pf, Plasmodium falciparum; Tp, Theileria parvum; Cp, Cryptosporidium parvum; Sc, Saccharomyces cerevisiae; Hs, Homo sapiens; Tt, Tetrahymena thermophila; At, Arabidopsis thaliana. It should be noted that structures for C. parvum and T. parvum were drawn based on predicted open reading frames in genome sequencing databases.
Crystal structures of HAT domains of GCN5 orthologues have been resolved for Tetrahymena (28) and yeast (49). Consistent with all other GCN5 family member HATs, both TgGCN5s possess the strictly conserved glutamic acid residue whose carboxyl side chain has been proposed to serve as the proton acceptor for the substrate lysine (28, 49). This corresponds to Glu-872 in TgGCN5-A and Glu-703 in TgGCN5-B (Fig. 5A) and is located in the conserved motif A shown to bind CoA (32). Histone H3 peptide substrate binds to a pronounced cleft formed by the loop-α2 and -α4, both of which are present in the T. gondii GCN5 homologues (37). The structure of the bromodomain has also been delineated and demonstrated to be a module that can interact with acetylated lysine residues (10). Both TgGCN5 bromodomains contain the 6 residues that form the acetyl lysine binding pocket (10), suggesting a conserved function in the parasite (Fig. 5A). Not as well conserved is the region between the HAT and bromodomain, which in other GCN5 family members has been shown to interact with the SAGA component ADA2.
We conducted surveys of completed apicomplexan genomes to check if others possessed a pair of GCN5 homologues. Plasmodium (16), Cryptosporidium (CpIOWA_EAK89017), and Theileria (TP01_0465) harbor only one GCN5 each, making T. gondii unique in being the only apicomplexan reported to possess two GCN5s. Figure 5B compares the structural features of apicomplexan GCN5s to those of other eukaryotic organisms. Curiously, apicomplexans universally possess N-terminal extensions of various lengths and compositions. N-terminal regions of GCN5 in other lower eukaryotes such as yeast and fellow alveolate Tetrahymena thermophila are greatly abbreviated.
TgGCN5-B localizes to the nucleus via a different signal.
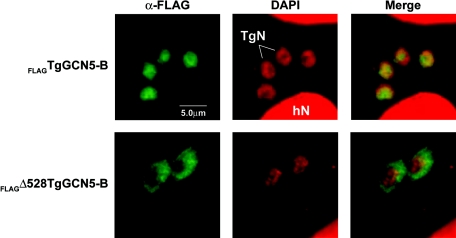
To begin delineating the functions of each TgGCN5 in T. gondii, we examined whether TgGCN5-B was cytosolic (type B HAT) or nuclear (type A HAT). We have previously shown that TgGCN5-A is imported into the parasite nucleus by virtue of a nuclear localization signal (NLS) capable of associating with T. gondii importin-α (4). To test the subcellular localization of TgGCN5-B, we fused a recombinant version of the full-length protein to the FLAG peptide for expression in T. gondii. Immunofluorescent assays reveal that TgGCN5-B completely localizes to the parasite nucleus (Fig. 6). The TgGCN5-A NLS (RKRVKR) is a part of the unusual N-terminal extension, but this NLS is not present in TgGCN5-B. However, removal of the N-terminal extension of TgGCN5-B causes it to accumulate in the cytoplasm (Fig. 6). Precisely how TgGCN5-B gains entry into the nucleus remains unresolved, but like TgGCN5-A, its N-terminal extension plays a critical role.
FIG. 6.
TgGCN5-B localizes to the parasite nucleus via the N-terminal extension. Recombinant protein in parasites expressing either full-length FLAGTgGCN5-B or FLAGΔ528TgGCN5-B was detected by immunofluorescence assay. Antibody recognizing the FLAG epitope is shown in green. Infected cells were also stained with 4′,6′-diamidino-2-phenylindole (DAPI), shown in red. hN, host cell nucleus; TgN, parasite nucleus.
Substrate specificity differences between TgGCN5-A and -B.
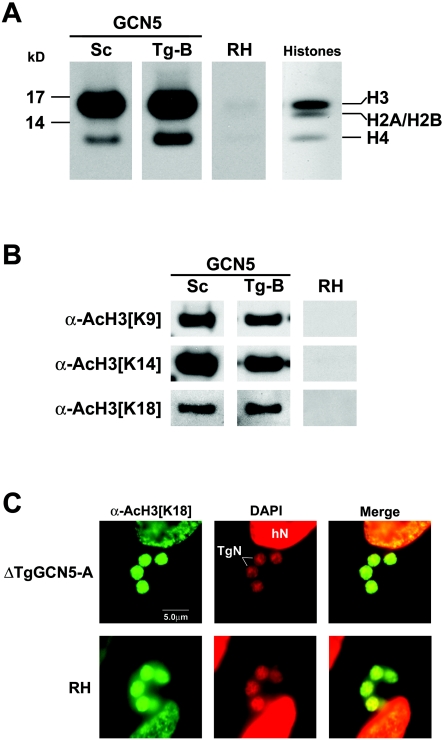
We examined whether there is a difference in substrate specificity between the TgGCN5 family members. GCN5 HATs from other species universally target histone H3, particularly at lysine 14 (H3 [K14]). We have previously shown that TgGCN5-A has a surprising bias to acetylate H3 [K18], with no detectable activity on K9 or K14 (39). We expressed and purified recombinant FLAG-tagged TgGCN5-B in the same fashion and found that it also has a strong preference to acetylate H3 in HAT assays using free histone substrates (Fig. 7A). However, when individual lysine residues are examined for acetylation, we find that TgGCN5-B is more like a prototypical GCN5 HAT because it is capable of targeting H3 [K9], [K14], and [K18] (Fig. 7B). These differences in substrate specificity are not due to the divergent N-terminal extensions, as their removal does not alter the histone acetylation profile (data not shown).
FIG. 7.
TgGCN5-B preferentially acetylates histone H3 at multiple lysine residues. (A) Autoradiogram of HAT assays using core histones and either recombinant yeast GCN5 (Saccharomyces cerevisiae, Sc) or FLAGTgGCN5-B (Tg-B). An equivalent amount of untransfected parasite lysate was used as a negative control. The right panel is a Coomassie-stained SDS-polyacrylamide gel showing the positions of the core histones. (B) Western blot of similar HAT assays performed on recombinant histone H3, probed with antibodies (α) to specific acetylated (Ac) H3 lysine residues. (C) Immunofluorescence assay showing acetylation of H3 [K18] in ΔTgGCN5 and wild-type (RH) parasites (green). 4′,6′-Diamidino-2-phenylindole (DAPI) was applied to stain nuclei (red). hN, host cell nucleus; TgN, parasite nucleus.
Given the highly specific nature of substrate specificity seen with TgGCN5-A, we evaluated whether this modification is altered in the ΔTgGCN5 parasites using an antibody specific to acetylated H3 [K18] in an immunofluorescence assay. As shown in Fig. 7C, H3 [K18] is still being acetylated despite the lack of TgGCN5-A. Since TgGCN5-B is capable of targeting H3 [K18], it therefore has the potential to compensate for the loss of TgGCN5-A.
Two homologues of ADA2 exist in T. gondii.
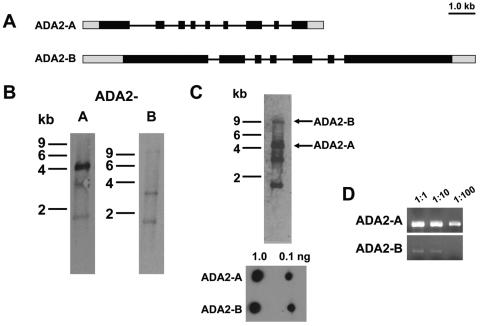
In other species, GCN5 HATs associate with the transcriptional coactivator ADA2. Lower eukaryotes are not known to possess more than one ADA2, but given that two GCN5 HATs exist in T. gondii, we sought to determine whether two ADA2 proteins were present. Drawing on data harnessed from the T. gondii genome sequence, we were able to clone two distinct ADA2 homologues and map the genomic loci (Fig. 8A). TgADA2-A refers to the shorter one (DQ112184), a ∼4.0-kb transcript as determined by Northern analysis, predicted to give rise to a 958-amino-acid protein (Fig. 8B). The TgADA2-A genomic locus is ∼8 kb, comprised of 9 exons interrupted by 8 introns. In contrast, TgADA2-B is much longer (DQ112185), hybridizing to a barely detectable ∼10-kb transcript (Fig. 8B). The open reading frame is 8,094 kb and encodes a protein of 2,697 amino acids. The ∼13-kb TgADA2-B genomic locus consists of 7 exons and 6 introns. According to ToxoDB, TgADA2-A resides on chromosome XI and TgADA2-B on chromosome VI. As observed for the ADA2 homologue in Plasmodium falciparum (15), TgADA2s are considerably larger than those in other species (yeast and metazoan ADA2s are comprised of 400 to 550 amino acids). Regions flanking the ADA2 domain in these apicomplexan ADA2s are not conserved with other species or one another.
FIG. 8.
A pair of ADA2s in T. gondii. (A) Schematic diagram of the genomic loci for TgADA2-A and -B. Black boxes represent exons, and the lines in between are introns. Gray boxes represent untranslated regions. (B) Northern blots probed for TgADA2-A and -B transcripts, ∼4 kb and ∼10 kb, respectively. Bands at ∼1.7 and 3.5 kb overlap with rRNA contamination. (C) Dual probing of a single Northern blot with both TgADA2s. The lower panel shows that equivalent amounts of nonisotopically labeled probes emit approximately equal chemiluminescent signals. (D) Equal amounts of cDNA generated from wild-type RNA were used in PCRs to amplify either TgADA2-A or -B.
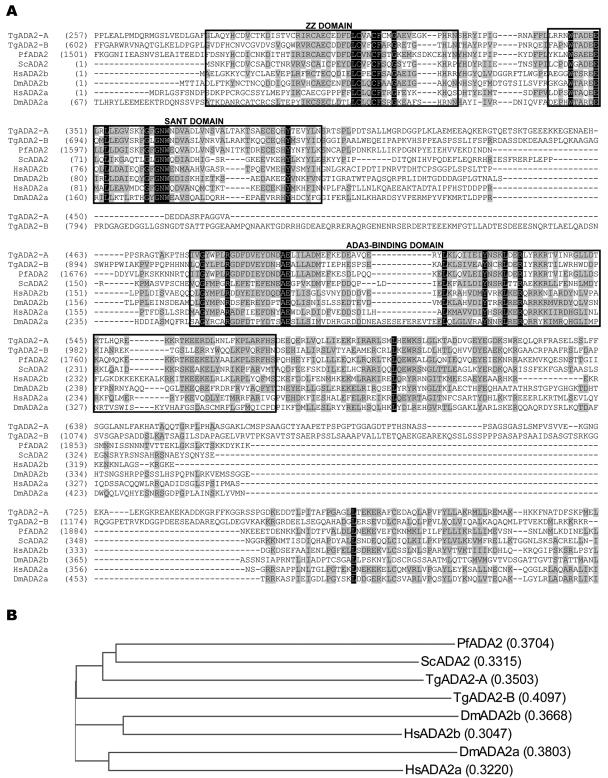
When probes specific to each TgADA2 are hybridized simultaneously to the same lane on a Northern, it is clear that TgADA2-A is much more abundant than TgADA2-B in tachyzoites (Fig. 8C). Data from RT-PCRs using tachyzoite RNA is consistent with the Northern result (Fig. 8D). The ADA2 domain of each T. gondii homologue contains the trademark regions present in other species, including the zinc finger ZZ domain, SANT domain, and ADA3-binding domain (Fig. 9A). While the ADA3-binding domain is fairly well conserved, it may be of note that no clear homologue of ADA3 is evident in any apicomplexan database. The SWIRM protein-protein interaction domain seen at the extreme C terminus of several ADA2 proteins is not present on the apicomplexan homologues. TgADA2-A most resembles the singular ADA2 homologue found in other lower eukaryotes, while TgADA2-B is phylogenetically closer to those in higher eukaryotes (Fig. 9B).
FIG. 9.
Comparative analysis of ADA2 domains. (A) ADA2 domains from TgADA2-A and -B are aligned for comparison to those found in other species. Black boxes denote identity of residues across species; gray boxes denote residues that are chemically similar. The ZZ, SANT, and ADA3-binding domains are boxed. Numerals in parentheses refer to amino acid numbers. Tg, Toxoplasma gondii; Pf, Plasmodium falciparum; Sc, Saccharomyces cerevisiae; Hs, Homo sapiens; Dm, Drosophila melanogaster. (B) Phylogenic analysis of ADA2 orthologues from representative species with the calculated distance values in parentheses.
As seen for GCN5, only one ADA2 homologue is present in the apicomplexan genomes of Plasmodium (15), Cryptosporidium (CpIOWA_EAK87649), and Theileria (TP01_0779).
Interactions between TgGCN5s and TgADA2s.
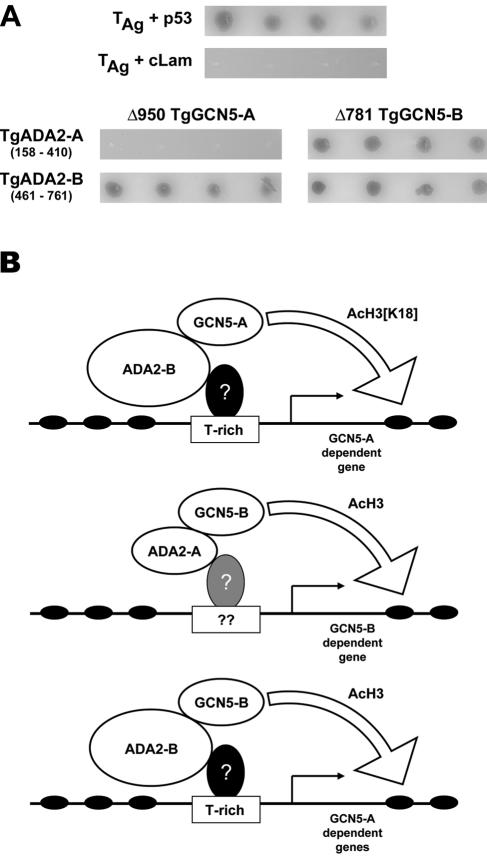
It has been noted above that the putative ADA2-binding domain is not very conserved between the two TgGCN5s. Conventional biochemistry is difficult to perform on this organism, so to test whether either TgADA2 binds to either TgGCN5 we employed a directed yeast two-hybrid test. Based on well-characterized corresponding regions elucidated in other species, the putative GCN5-binding domains of TgADA2-A (amino acids 158 to 410) and -B (amino acids 461 to 761) were amplified and fused to the GAL4 activation domain (7, 15, 41). C-terminal portions of each TgGCN5, starting with the putative ADA2-binding domain (Δ950TgGCN5-A and Δ781TgGCN5-B, respectively), were fused to the GAL4 DNA-binding domain. Combinations of TgGCN5 plus TgADA2 were cotransformed into AH109 yeast and plated onto QDO media with a colorimetric substrate to test for protein-protein interactions. This system utilizes three independent reporters (two nutritional genes plus galactosidase) to minimize false-positive results; no growth argues that the test proteins do not interact. The growth of blue colonies on QDO plates indicates that all three independent reporter genes were activated by virtue of a strong interaction between the test proteins. As shown in Fig. 10A, TgGCN5-B has the ability to interact with either TgADA2-A or -B, while TgGCN5-A only interacts with TgADA2-B. These results suggest that three GCN5-containing complexes may exist in T. gondii: TgGCN5-A plus TgADA2-B, TgGCN5-B plus TgADA2-A, and TgGCN5-B plus TgADA2-B. It is worth noting that TgGCN5-B can associate with the only TgADA2 capable of interacting with TgGCN5-A. Considered with its enzymatic abilities, it is plausible that TgGCN5-B can partially complement the function of TgGCN5-A. This complementation may not be complete, as reflected in the proteomics study comparing ΔTgGCN5-A versus wild-type tachyzoites. In contrast, the more exclusive nature of TgADA2-binding and substrate preference makes it unlikely that TgGCN5-A could compensate for a loss of TgGCN5-B.
FIG. 10.
Protein-protein interactions between TgGCN5s and TgADA2s. (A) Top panels: AH109 yeast were cotransformed with plasmids expressing simian virus 40 large-T antigen (TAg) and murine p53 (positive control) or TAg and human lamin C (cLam) (negative control). Four independent colonies initially plated on TDO plates were spotted onto QDO plates containing X-α-gal and grown for 3 days at 30°C. Growth of blue colonies represents protein-protein interaction. Bottom panels: AH109 yeast were cotransformed with the plasmids indicated and plated for incubation as described above. All colonies shown are blue in color. (B) Working model for TgGCN5 HAT complexes. Based on yeast two-hybrid data, at least three possible complexes containing GCN5 may exist in T. gondii. The lowest panel depicts a complex comprised of TgGCN5-B and TgADA2-B that could compensate for TgGCN5-A. Based on what is observed in other eukaryotes, a question mark refers to a putative DNA-binding transcription factor with the capacity to recruit the GCN5 complexes by virtue of binding ADA2. Note that the position of the T-rich motif is approximate, as its actual location varies as shown in Fig. 3C.
DISCUSSION
Chromatin constitutes a formidable barrier to the expression of genes. One mechanism cells have evolved to circumvent this obstacle to transcription involves the covalent modification of histones. Certain modifications, such as acetylation, may attenuate the histone-DNA interaction by neutralizing the nucleosome's positive charge, thereby exposing regulatory elements to transcription factors. Combinatorial modifications may even represent a “histone code,” an epigenetic marking system that adds another dimension to gene regulation (23). Studies of the protozoan Tetrahymena led to the groundbreaking discovery that linked histone acetyltransferase activity to the activation of gene expression (6). Our study of this phenomenon in another lower eukaryote, the pathogenic apicomplexan T. gondii, may lead to further insights into eukaryotic gene regulation while simultaneously identifying new avenues of chemotherapy. Indeed, apicidin exhibits antiprotozoal activity by interfering with histone modification machinery (9).
Genes under control of GCN5 family HATs.
The nature of the genes controlled by GCN5 HATs is incompletely characterized. Studies with yeast show that GCN5 modulates the expression of tightly regulated genes governing the stress response (21). By virtue of being recruited to the yeast master regulator GCN4, GCN5 activates expression of genes for amino acid biosynthesis and transport and for vitamin and purine biosynthesis as well as peroxisomal genes (19). In plants, similar evidence argues that GCN5 regulates stress remediation genes (42). Serving to ameliorate stress would also explain why GCN5 is nonessential for organisms free of environmental insult. GCN5 plays a vital role in development in some plants (50); but in mammals, which have two GCN5s, only one seems essential for development (51, 52). Our data show that the subset of genes under TgGCN5-A control is remarkably different than other eukaryotes. Our proteomic analysis of ΔTgGCN5-A shows most of the affected products to be parasite specific, including surface antigens and micronemes, proteins that participate in attachment and invasion into host cells. Despite the differential expression of these genes in ΔTgGCN5, the phenotypic impact in vitro and in vivo appears to be inconsequential. Considering the critical nature of parasite invasion, it is conceivable that redundancies exist that adequately compensate for the reduced expression of such gene products. It is also possible that there is a basal level of transcription of the GCN5-dependent genes that provides adequate amounts of the respective gene products.
Our aim in this study was to examine the impact of TgGCN5-A on the tachyzoites responsible for the acute stage of infection, for which type I strains like RH are ideal. The discovery of TgGCN5-B creates the possibility that it is more important in regulating gene expression during acute infection. Moreover, histone acetylation was recently found to correlate with stage-specific changes in gene expression in T. gondii (39). To study the potential roles of TgGCN5s in this process, we are attempting to disrupt each locus in an avirulent type II strain, which is better suited for differentiation studies. It is also possible that TgGCN5-A and/or -B governs gene expression changes that are pertinent to transcriptional changes occurring while in the definitive (feline) host or during sporulation.
Multiple GCN5 HATs in T. gondii.
We have previously shown that T. gondii has a GCN5-type HAT that exhibits an uncharacteristic proclivity to acetylate strictly H3 [K18] (39, 47). Now we show that T. gondii is also in possession of a more prototypical GCN5 family HAT, TgGCN5-B, capable of acetylating multiple lysine residues in the H3 tail. This is significant for several reasons. First, T. gondii is unique in being the only invertebrate described to date harboring two GCN5 HATs. Second, unlike the dual GCN5 HATs in mammalian cells, TgGCN5-A and -B have different histone acetylation profiles. Third, the two TgGCN5s contain lengthy N-terminal extensions that are not similar to one another. In contrast, mammalian GCN5 and PCAF possess N-terminal domains that are highly analogous and associate with another HAT, p300/CBP (CREB-binding protein). p300/CBP is not present in apicomplexan genomes or other lower eukaryotes, but it remains possible that the N-terminal extensions of TgGCN5s are involved in protein-protein interactions. We have discerned that the N-terminal extensions of each TgGCN5 are dispensable for HAT activity in vitro but are required for nuclear localization.
Why two HATs?
T. gondii appears to be unique among its fellow apicomplexans in harboring more than one GCN5 family HAT. We speculate that the two TgGCN5s are paralogues that probably arose from a gene duplication event occurring exclusively in T. gondii prior to their segregation into three different clonal lineages ∼10,000 years ago (44). TgGCN5-B resembles archetypal GCN5s with regard to its sequence homology as well as its behavior in HAT assays, making it more likely to be the original gene. The vastly different N-terminal extensions between TgGCN5-A and -B might suggest that only the HAT domain through the bromodomain was copied. Notably, removal of the N-terminal extensions does not alter in vitro substrate specificity of the TgGCN5s. The more restrictive nature of TgGCN5-A HAT activity may involve one or more of the four residues previously noted to be conserved in all other GCN5s except TgGCN5-A (47). Further study will have to be performed to confirm this hypothesis.
Mammalian GCN5 and PCAF are much more similar to one another in both sequence and substrate preference than the two TgGCN5 HATs. Our inability to discern a clear phenotype for ΔTgGCN5-A mirrors the results seen in mice that showed that GCN5 could compensate for the loss of PCAF (52). Interestingly, PCAF could not compensate for the loss of GCN5, making it of interest to investigate whether a knockout of TgGCN5-B is possible in T. gondii. We imagine that TgGCN5-A, given its more restricted enzymatic ability and exclusivity for one ADA2, will not be able to adequately compensate for the loss of TgGCN5-B.
It is unclear at present why T. gondii has two GCN5 HATs. It is possible that the two GCN5s regulate genes in different stages of the parasite life cycle. The availability of comprehensive microarrays for T. gondii will permit gene expression profiling experiments designed to identify the complement of genes associated with each TgGCN5 in different life cycle stages.
TgGCN5 complexes: simple or complex?
ADA2 proteins associate with GCN5 and act as coactivators bridging the GCN5 complex to the acidic activation domains of DNA-binding transcription factors and TATA-binding protein (2, 12). As is the case for GCN5, it was unexpected to find two novel ADA2 homologues in a lower eukaryotic cell. While most metazoans and land plants have two ADA2s, our search results indicate that most protozoa, like yeast, have only one. T. gondii appears to be an exception to this observation, possibly because it contains two GCN5s, but precisely how and why a second ADA2 locus arose remains an intriguing issue. Moreover, like PfADA2 (15), TgADA2-A and -B are much longer than other species, containing large regions flanking the ADA2 domain that have no similarity to other protein sequences or known motifs.
Our studies using the yeast two-hybrid assay determined that either TgADA2 can interact with TgGCN5-B. However, TgGCN5-A can only interact with TgADA2-B. It remains possible that TgGCN5-A may interact with TgADA2-A in vivo, but the data at present suggest that at least three potential GCN5-containing complexes may exist in T. gondii (Fig. 10B). It should be mentioned that yeast two-hybrid results do not always reflect what truly occurs in vivo in the parasite, and further studies are under way to confirm the result suggested by the yeast two-hybrid test. It is also possible that the TgADA2 homologues operate independently of TgGCN5. Drosophila has two ADA2 homologues, but only one (dADA2b) is incorporated into the SAGA complex (27). Data argue that dADA2a may have novel roles in transcription that are independent of GCN5 (27, 34).
ADA3 is another component found in all GCN5 complexes, but no homologue of it is readily apparent in apicomplexan databases. Either the ADA3 homologue is very divergent in these parasites or they have evolved an unprecedented means to nucleate the GCN5 complex. If the latter is true, the lengthy extensions on TgGCN5 and TgADA2 proteins may mimic the role of ADA3 in other eukaryotes.
One of the most striking features of apicomplexan genomes is their lack of conventional transcription factors (48). The elucidation of a T-rich motif may assist the effort to identify a DNA-binding factor that subsequently recruits chromatin remodeling complexes to the target promoters. Given the paucity of conventional transcription factors but robust repertoire of chromatin remodeling enzymes (39, 46, 47), a plausible case can be made that T. gondii has streamlined gene expression regulation, relying more heavily on epigenetics. On a related note, in addition to the lack of a conserved ADA3, we have also noted a lack of other SAGA components in the Apicomplexa (W. J. Sullivan, Jr., unpublished data), arguing further that these ancient eukaryotic cells have simplified GCN5 complexes. The unique attributes of their gene regulatory machinery, as well as the potential for pharmacological applications (9), makes T. gondii an increasingly attractive organism for the study of these phenomena.
Acknowledgments
The research was funded by an IUSM Showalter grant and NIH GM065051 (to W.J.S.).
We are grateful to David Sibley (Washington University, St. Louis, MO) and Vern Carruthers (Johns Hopkins Malaria Research Institute, Baltimore, MD) for MIC antibodies and Blima Fux (Sibley lab) for assistance with the virulence study. We thank Tony Sinai (University of Kentucky, Lexington, KY) and M. A. Hakimi (Joseph Fourier University, France) for a critical reading of the manuscript and Chunling Jiang (Sullivan lab) for technical assistance. Preliminary genomic sequence data were accessed via http://ToxoDB.org. Genomic data were provided by The Institute for Genomic Research (supported by NIH grant no. AI05093) and by the Sanger Center (Wellcome Trust).
REFERENCES
- 1.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. [PubMed] [Google Scholar]
- 2.Barlev, N. A., R. Candau, L. Wang, P. Darpino, N. Silverman, and S. L. Berger. 1995. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J. Biol. Chem. 270:19337-19344. [DOI] [PubMed] [Google Scholar]
- 3.Berger, S. L., B. Pina, N. Silverman, G. A. Marcus, J. Agapite, J. L. Regier, S. J. Triezenberg, and L. Guarente. 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70:251-265. [DOI] [PubMed] [Google Scholar]
- 4.Bhatti, M. M., and W. J. Sullivan, Jr. 2005. Histone acetylase GCN5 enters the nucleus via importin-alpha in protozoan parasite Toxoplasma gondii. J. Biol. Chem. 280:5902-5908. [DOI] [PubMed] [Google Scholar]
- 5.Black, M. W., and J. C. Boothroyd. 2000. Lytic cycle of Toxoplasma gondii. Microbiol. Mol. Biol Rev. 64:607-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 7.Candau, R., J. X. Zhou, C. D. Allis, and S. L. Berger. 1997. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 16:555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carruthers, V. B., O. K. Giddings, and L. D. Sibley. 1999. Secretion of micronemal proteins is associated with toxoplasma invasion of host cells. Cell. Microbiol. 1:225-235. [DOI] [PubMed] [Google Scholar]
- 9.Darkin-Rattray, S. J., A. M. Gurnett, R. W. Myers, P. M. Dulski, T. M. Crumley, J. J. Allocco, C. Cannova, P. T. Meinke, S. L. Colletti, M. A. Bednarek, S. B. Singh, M. A. Goetz, A. W. Dombrowski, J. D. Polishook, and D. M. Schmatz. 1996. Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc. Natl. Acad. Sci. USA 93:13143-13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 11.Donald, R. G., D. Carter, B. Ullman, and D. S. Roos. 1996. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J. Biol. Chem. 271:14010-14019. [DOI] [PubMed] [Google Scholar]
- 12.Drydale, C. M., B. M. Jackson, R. McVeigh, E. R. Klebanow, Y. Bai, T. Kokubo, M. Swanson, Y. Nakatani, P. A. Weil, and A. G. Hinnebusch. 1998. The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme TFIID, and the Adap-Gcn5p coactivator complex. Mol. Cell. Biol. 18:1711-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubey, J. P., S. K. Shen, O. C. Kwok, and J. K. Frenkel. 1999. Infection and immunity with the RH strain of Toxoplasma gondii in rats and mice. J. Parasitol. 85:657-662. [PubMed] [Google Scholar]
- 14.Duraisingh, M. T., T. S. Voss, A. J. Marty, M. F. Duffy, R. T. Good, J. K. Thompson, L. H. Freitas-Junior, A. Scherf, B. S. Crabb, and A. F. Cowman. 2005. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 121:13-24. [DOI] [PubMed] [Google Scholar]
- 15.Fan, Q., L. An, and L. Cui. 2004. PfADA2, a Plasmodium falciparum homologue of the transcriptional coactivator ADA2 and its in vivo association with the histone acetyltransferase PfGCN5. Gene 336:251-261. [DOI] [PubMed] [Google Scholar]
- 16.Fan, Q., L. An, and L. Cui. 2004. Plasmodium falciparum histone acetyltransferase, a yeast GCN5 homologue involved in chromatin remodeling. Eukaryot. Cell 3:264-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freitas-Junior, L. H., R. Hernandez-Rivas, S. A. Ralph, D. Montiel-Condado, O. K. Ruvalcaba-Salazar, A. P. Rojas-Meza, L. Mancio-Silva, R. J. Leal-Silvestre, A. M. Gontijo, S. Shorte, and A. Scherf. 2005. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121:25-36. [DOI] [PubMed] [Google Scholar]
- 18.Hettmann, C., and D. Soldati. 1999. Cloning and analysis of a Toxoplasma gondii histone acetyltransferase: a novel chromatin remodelling factor in apicomplexan parasites. Nucleic Acids Res. 27:4344-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinnebusch, A. G., and K. Natarajan. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 1:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 21.Huisinga, K. L., and B. F. Pugh. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13:573-585. [DOI] [PubMed] [Google Scholar]
- 22.Huynh, M. H., K. E. Rabenau, J. M. Harper, W. L. Beatty, L. D. Sibley, and V. B. Carruthers. 2003. Rapid invasion of host cells by Toxoplasma requires secretion of the MIC2-M2AP adhesive protein complex. EMBO J. 22:2082-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 24.Kozak, M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 263:19867-19870. [PubMed] [Google Scholar]
- 25.Kuo, M. H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 26.Kuo, M. H., J. Zhou, P. Jambeck, M. E. Churchill, and C. D. Allis. 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12:627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kusch, T., S. Guelman, S. M. Abmayr, and J. L. Workman. 2003. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol. Cell. Biol. 23:3305-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, Y., C. M. Fletcher, J. Zhou, C. D. Allis, and G. Wagner. 1999. Solution structure of the catalytic domain of GCN5 histone acetyltransferase bound to coenzyme A. Nature 400:86-89. [DOI] [PubMed] [Google Scholar]
- 29.Luo, S., F. A. Ruiz, and S. N. Moreno. 2005. The acidocalcisome Ca2+-ATPase (TgA1) of Toxoplasma gondii is required for polyphosphate storage, intracellular calcium homeostasis and virulence. Mol. Microbiol. 55:1034-1045. [DOI] [PubMed] [Google Scholar]
- 30.Mann, M., S. E. Ong, M. Gronborg, H. Steen, O. N. Jesnsen, and A. Pandey. 2002. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 20:261-268. [DOI] [PubMed] [Google Scholar]
- 31.Marcus, G. A., N. Silverman, S. L. Berger, J. Horiuchi, and L. Guarente. 1994. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 13:4807-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marmorstein, R. 2001. Structure of histone acetyltransferases. J. Mol. Biol. 311:433-444. [DOI] [PubMed] [Google Scholar]
- 33.Mercier, C., S. Lefebvre-Van Hende, G. E. Garber, L. Lecordier, A. Capron, and M. F. Cesbron-Delauw. 1996. Common cis-acting elements critical for the expression of several genes of Toxoplasma gondii. Mol. Microbiol. 21:421-428. [DOI] [PubMed] [Google Scholar]
- 34.Muratoglu, S., S. Georgieva, G. Papai, E. Scheer, I. Enunlu, O. Komonyi, I. Cserpan, L. Lebedeva, E. Nabirochkina, A. Udvardy, L. Tora, and I. Boros. 2003. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol. Cell. Biol. 23:306-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarro, M., G. A. Cross, and E. Wirtz. 1999. Trypanosoma brucei variant surface glycoprotein regulation involves coupled activation/inactivation and chromatin remodeling of expression sites. EMBO J. 18:2265-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts, S. M., and F. Winston. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147:451-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rojas, J. R., R. C. Trievel, J. Zhou, Y. Mo, X. Li, S. L. Berger, C. D. Allis, and R. Marmorstein. 1999. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 401:93-98. [DOI] [PubMed] [Google Scholar]
- 38.Roos, D. S., R. G. Donald, N. S. Morrissette, and A. L. Moulton. 1994. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 45:27-63. [DOI] [PubMed] [Google Scholar]
- 39.Saksouk, N., M. M. Bhatti, S. Kieffer, A. T. Smith, K. Musset, J. Garin, W. J. Sullivan, Jr., M. F. Cesbron-Delauw, and M.-A. Hakimi. 2005. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol. Cell. Biol. 25:10301-10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sterner, D. E., X. Wang, M. H. Bloom, G. M. Simon, and S. L. Berger. 2002. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem. 277:8178-8186. [DOI] [PubMed] [Google Scholar]
- 42.Stockinger, E. J., Y. Mao, M. K. Regier, S. J. Triezenberg, and M. F. Thomashow. 2001. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29:1524-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 44.Su, C., D. Evans, R. H. Cole, J. C. Kissinger, J. W. Ajioka, and L. D. Sibley. 2003. Recent expansion of Toxoplasma through enhanced oral transmission. Science 299:414-416. [DOI] [PubMed] [Google Scholar]
- 45.Sullivan, W. J., Jr. 2003. Histone H3 and H3.3 variants in the protozoan pathogens Plasmodium falciparum and Toxoplasma gondii. DNA Seq. 14:227-231. [DOI] [PubMed] [Google Scholar]
- 46.Sullivan, W. J., Jr., M. A. Monroy, W. Bohne, K. C. Nallani, J. Chrivia, P. Yaciuk, C. K. Smith II, and S. F. Queener. 2003. Molecular cloning and characterization of an SRCAP chromatin remodeling homologue in Toxoplasma gondii. Parasitol. Res. 90:1-8. [DOI] [PubMed] [Google Scholar]
- 47.Sullivan, W. J., Jr., and C. K. Smith II. 2000. Cloning and characterization of a novel histone acetyltransferase homologue from the protozoan parasite Toxoplasma gondii reveals a distinct GCN5 family member. Gene 242:193-200. [DOI] [PubMed] [Google Scholar]
- 48.Templeton, T. J., L. M. Iyer, V. Anantharaman, S. Enomoto, J. E. Abrahante, G. M. Subramanian, S. L. Hoffman, M. S. Abrahamsen, and L. Aravind. 2004. Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 14:1686-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trievel, R. C., J. R. Rojas, D. E. Sterner, R. N. Venkataramani, L. Wang, J. Zhou, C. D. Allis, S. L. Berger, and R. Marmorstein. 1999. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc. Natl. Acad. Sci. USA 96:8931-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vlachonasios, K. E., M. F. Thomashow, and S. J. Triezenberg. 2003. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15:626-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu, W., D. G. Edmondson, Y. A. Evrard, M. Wakamiya, R. R. Behringer, and S. Y. Roth. 2000. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26:229-232. [DOI] [PubMed] [Google Scholar]
- 52.Yamauchi, T., J. Yamauchi, T. Kuwata, T. Tamura, T. Yamashita, N. Bae, H. Westphal, K. Ozato, and Y. Nakatani. 2000. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 97:11303-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]