Abstract
In almost all eukaryotes, mitochondrial (mt) genes are transmitted to progeny mainly from the maternal parent. The most popular explanation for this phenomenon is simple dilution of paternal mtDNA, because the paternal gametes (sperm) are much smaller than maternal gametes (egg) and contribute a limited amount of mitochondria to the progeny. Recently, this simple explanation has been challenged in several reports that describe the active digestion of sperm mtDNA, down-regulation of mtDNA replication in sperm, and proteolysis of mitochondria triggered by ubiquitination. In this investigation, we visualized mt nucleoids in living sperm by using highly sensitive SYBR green I vital staining. The ability to visualize mt nucleoids allowed us to clarify that the elimination of sperm mtDNA upon fertilization is achieved through two steps: (i) gradual decrease of mt nucleoid numbers during spermatogenesis and (ii) rapid digestion of sperm mtDNA just after fertilization. One notable point is that the digestion of mtDNA is achieved before the complete destruction of mitochondrial structures, which may be necessary to avoid the diffusion and transmission of potentially deleterious sperm mtDNA to the progeny.
Keywords: maternal inheritance, medaka, mtDNA
Mitochondria and chloroplasts contain their own genomes, which are believed to be vestiges of their bacterial ancestors (1). Mitochondrial (mt) and chloroplast (cp) genes are inherited in non-Mendelian fashion. A variety of patterns and mechanisms have been observed for the transmission of these genes, but in almost all eukaryotes, they are inherited mainly from the maternal parent (2–4).
Generally, the maternal inheritance of mt (cp) DNA is thought to be a result of the dilution of the paternal contribution, because the paternal gametes (sperm) are much smaller than maternal gametes (egg) and contribute only a limited amount of cytoplasm to the progeny. In animals, a mature sperm carries ≈100 copies of mtDNA, which might be necessary to maintain the integrity of mitochondrial activity (5). In contrast, the animal oocyte contains ≈105 to 108 copies of mtDNA, exceeding that of sperm by a factor of at least 103 (3, 6). Therefore, the paternal contribution of mtDNA or cpDNA would be diluted beyond the limits of detection by using conventional restriction enzyme analysis (7).
This simple explanation, however, has been challenged by many findings (8). For example, there have been several fairly unsuccessful attempts to detect leaky transmission of paternal mtDNA by repetitive backcross experiments using lepidopteran insects (9). On the contrary, in mussels (Mtylus), paternal mtDNA can be transmitted to male progeny at a remarkably high frequency, even though paternal mtDNAs are transmitted by sperm (10–12). A more striking example was found in the unicellular alga Chlamydomonas reinhardtii. In C. reinhardtii, despite the fact that mt+ (female) and mt- (male) gametes contribute equal amounts of cytoplasm to the progeny, cpDNA is transmitted only from the mt+ parent. One biochemical study showed that the amount of mt- cpDNA decreased relative to mt+ cpDNA 6–24 h after mating, suggesting that mt- cpDNA is actively digested at this time (13).
Direct evidence for this active digestion of mt- cpDNA in C. reinhardtii was obtained from cytological studies. mt and cp genomes do not exist in vivo as naked DNA molecules. These genomes are bundled together and organized into discrete DNA–protein bodies called nucleoids, which can be clearly visualized by fluorescent microscopy. mt and cp nucleoids are now thought to be the unit of segregation and inheritance of mt- and cpDNA (2, 14). In C. reinhardtii, 80–90 copies of the cp genome are compacted into 5–8 cp nucleoids (15). In 1982, Kuroiwa et al. (16) found that mt- cp nucleoids disappear preferentially in young zygotes 50 min after mating. In 1998, the preferential disappearance of mt- cp nucleoids was observed in a living zygote (17). This phenomenon is a rapid process that completes within 10 min of initiation. Single zygotes with or without mt- cp nucleoids were then collected by optical tweezers and analyzed by highly sensitive nested PCR, confirming that mt- cpDNA is actively digested during this rapid disappearance of mt- cp nucleoids (18). It is now believed that maternal inheritance in C. reinhardtii is achieved through active processes, including the rapid digestion of mt- cpDNA, protection of mt+ cpDNA, and amplification of mt+ cpDNA (19, 20).
The cytological analysis of mt (cp) nucleoids has been a powerful strategy to determine the sexual transmission of mt (cp) DNA not only in C. reinhardtii but also in higher plants (21), ferns, mosses, fungi, and algae (2). In animals, however, studies on mt nucleoids are rare (22, 23). Although the behaviors of sperm during fertilization have been extensively studied, mainly by transmission or scanning microscopy (24), these methods were not suitable for monitoring mtDNA or nucleoids. Therefore, our aim was to visualize and monitor the behaviors of mt nucleoids during fertilization.
The model system we used in this research was Japanese medaka (Oryzias latipes). O. latipes is a small fish particularly suitable for spermatogenesis and fertilization studies because in vitro spermatogenesis and fertilization techniques have been established (25). A short generation time, small genome size, and the availability of EST information also render O. latipes suitable for molecular genetic analysis (26). Moreover, O. latipes has been bred by Japanese hobbyists for >200 years, during which time varieties of inbred lines have been established. These inbred lines allowed us to obtain detailed linkage maps (27, 28). Among these strains, we found two inbred lines bearing a polymorphism in mtDNA that enabled us to monitor the behaviors of paternal and maternal mtDNA separately during fertilization.
In this research, mt nucleoids were successfully visualized in living sperm during the spermatogenesis and fertilization of O. latipes by vital double staining with DNA-specific fluorochrome SYBR green I and mitochondrial membrane-specific fluorochrome MitoTracker CMTMRos. With this technique, we observed the dramatic reduction of mt nucleoids during spermatogenesis. Upon fertilization, rapid disappearance of sperm mt nucleoids was observed in apparently intact mitochondria. Furthermore, a single sperm with or without mt nucleoids was selectively extracted from the fertilized eggs under direct observation by using optical tweezers and analyzed by highly sensitive nested PCR. The rapid disappearance of observed fluorescent mt nucleoids confirmed that mtDNA is actively digested. These results indicate that the elimination of paternal mtDNA is achieved through two steps: (i) gradual reduction of mtDNA during spermatogenesis and (ii) rapid digestion of sperm mtDNA well before the destruction of mitochondrial structures, which would be necessary to avoid the diffusion and transmission of potentially deleterious sperm mtDNA to the progeny.
Results
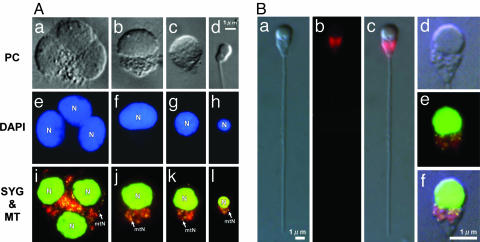
Visualization of mt Nucleoids in Living Sperm. We first stained sperm with DAPI, which has been conventionally used to observe mtDNA. (Fig. 1). Spermatids and sperm were extracted from testes of matured male O. latipes and stained with DAPI after a chemical fixation. We were successful only in observing nuclei with DAPI and were not able to detect cytoplasmic DNA, consistent with a previous observation (29). Another dsDNA-specific fluorochrome, SYBR green I, was used. SYBR green I is a highly penetrative dye appropriate for the noninvasive staining of living cells. This dye has been used to visualize mtDNA in living cells of C. reinhardtii (17). With SYBR green I staining, populations of minute fluorescent dots (≈1 μm) were observed around cell nuclei (Fig. 1). To confirm their localization, the cells were double stained with MitoTracker CMTMRos, a mitochondrial membrane potential-specific fluorochrome that has been used to visualize mitochondria for purposes such as the noninvasive selection of round spermatids (29). As shown in Fig. 1B, the yellow–green fluorescence of SYBR green I precisely overlapped the red fluorescence of MitoTracker CMTMRos at every developmental stage from round spermatids to matured sperm. This result indicates that the yellow spots visualized by SYBR green I are mt nucleoids.
Fig. 1.
mt nucleoids during spermatogenesis. (A) Phase-contrast (PC) (a–d), DAPI (e–h), and SYBR green I/MitoTracker CMTMRos double-stained (i–l) spermatid cells (a–c, e–g, and i–k) at various developmental stages and matured sperm (d, h, and l). DAPI staining allows visualization of only the cell nucleus (N), whereas SYBR green I staining allows visualization of minute yellow spots (≈1 μm) outside of the cell nucleus. The yellow spots precisely overlap with the red fluorescence of MitoTracker CMTMRos at every developmental stage, indicating that they are mt nucleoids (mtN). (B) Phase-contrast (a) and MitoTracker CMTMRos-stained (b) images of matured sperm. (c) An overlay of a and b.(d and e) Magnified phase-contrast (d) SYBR green I/MitoTracker CMTMRos double-stained (e) images of the head part of matured sperm. (f) An overlay of d and e.
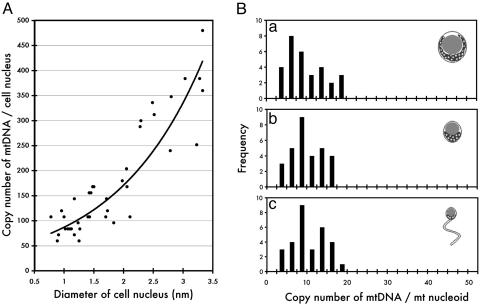
Rapid Reduction of mtDNA During Spermatogenesis. The number and fluorescence intensity of each mt nucleoid were quantified during spermatogenesis (Fig. 2). At the stage of round spermatid, ≈50 mt nucleoids were present per nucleus. During polarization of mitochondria and the condensation of nuclei, the number of mt nucleoids decreased dramatically and finally reached ≈9.8 ± 0.5 mt nucleoids per matured sperm. However, the intensity of the individual mt nucleoids remained unchanged, corresponding to ≈10 copies of mtDNA 16.0 kb in size, despite the dramatic condensation of the cell nucleus and the decreased number of mt nucleoids per cell (Fig. 2). These cytological observations indicate that the copy number of mtDNA decreases ≈5-fold between meiosis and the end of spermatogenesis without changing mt nucleoid organization. Hecht et al. (5) suggested that the amount of mtDNA decreases because mitochondria are discarded during spermatogenesis. Fluorescent and scanning electron microscopic observations support the hypothesis that a large portion of the mitochondria-containing cytoplasm is released from spermatids (25).
Fig. 2.
Fluorescence intensity of mt nucleoids measured by a video-intensified microscope photon-counting system (VIMPCS). (A) Copy number of mtDNA measured by VIMPCS (61) was plotted against the diameter of the cell nucleus. A least-squares fit line calculated by Microsoft excel shows a rapid (≈8- to 10-fold) reduction of the mtDNA copy number along with the condensation of the cell nucleus during the development of sperm cells. (B) Histograms show the distribution of fluorescence intensity of mt nucleoids. The spermatid cells were arbitrarily classified into three groups: before polarization (a), after polarization (b), and matured sperm (c), and the distribution of fluorescence intensity of mt nucleoids was analyzed. Despite the dramatic reorganization of the cell nucleus and the rapid reduction in the number of mt nucleoids per cell along with the development, the fluorescence intensity of mt nucleoids remained almost unchanged.
Active Digestion of Male mtDNA in Natural Fertilization. Even after the rapid reduction of mtDNA during spermatogenesis, mature O. latipes sperm still contained ≈100 copies of mtDNA (Fig. 2), which is consistent with the previous biochemical study in mice (5). In most vertebrates, intact mitochondrial sheath and tail enter oocyte cytoplasm during fertilization; this is also the case in O. latipes (30). The only exception is Chinese hamster (Cricetulus griseus), in which the mitochondria remain outside of the egg (31). Therefore, we next turned our attention to the fate of mtDNA in matured sperm after fertilization.
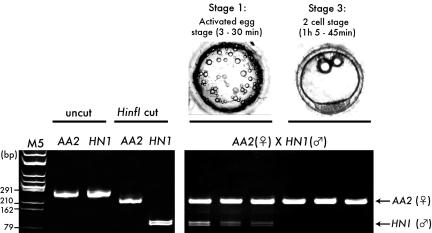
Because fertilized eggs contain two different populations of mtDNA (paternal and maternal), it was necessary to distinguish between these two species of mtDNA. Kaneda et al. (32) used polymorphism of mtDNA for this purpose. In O. latipes, we were able to identify two wild-type strains possessing a polymorphism in mtDNA (AA2 and HNI). Because of this polymorphism, PCR products for NADH dehydrogenase gave different restriction patterns after HinfI digestion, enabling independent observation of paternal and maternal mtDNA (Fig. 3).
Fig. 3.
Polymorphism in NADH dehydrogenase sequence between AA2 and HNI. (Lower Left) Two hundred and forty-nine bp of NADH dehydrogenase sequence was amplified from AA2 and HNI by nested PCR (uncut) and digested by HinfI (Hinf I cut). Because of the polymorphism, AA2 gave 222- + 27-bp fragments (the latter are too small to be detected), and HNI gave 129- + 120-bp fragments. M5, marker 5 (HincII digest of phage φX174 DNA). (Lower Right) Active degradation of sperm mtDNA in natural fertilization. DNA was extracted from single eggs at stage 1, the activated egg stage (3–30 min after fertilization), and stage 3, the two-cell stage (1 h 5 min to 1 h 45 min after fertilization) (AA2, female × HNI, male). (Upper Right) Typical images of the eggs are shown above the gel. Whereas the AA2 (female mtDNA) signal was detected from all of the eggs examined, the HNI (sperm mtDNA) signal was not detected from fertilized eggs at stage 3.
O. latipes spawn eggs at the beginning of light periods and mate immediately after the spawning. The spawned and fertilized eggs adhere to the outside of female bodies. To avoid any contamination of sperm that had not penetrated eggs, we kept only 1 male fish with 10 female fish in a 30-liter water tank. Fertilized eggs were removed by forceps and observed under the microscope (Fig. 3).
For O. latipes, a detailed timeline of developmental events in fertilized eggs is available (33). In the eggs at the early stage after fertilization, the oil droplets become slightly larger (≈50 μm) than unfertilized eggs (10–30 μm), but they are still dispersed evenly throughout the surface of eggs (stage 1, activated egg stage: 3–30 min after fertilization). At stage 2 (blastodisc stage: 30–60 min after fertilization), the oil drops migrate toward the vegetal hemisphere, and a cup-shaped blastodisc is formed at the animal pole. At stage 3 (two-cell stage: 1 h 5 min to 1 h 45 min), two blastomeres are observed after a cleavage. After confirmation of their developmental stages, the eggs at stages 1 and 3 were selected, rinsed in distilled water, and transferred into PCR tubes. We did not use eggs at stage 2 because fertilized eggs stay at stage 2 only for a brief period, and it was difficult to collect eggs at this stage.
In the PCR tubes, the single eggs were compressed and heated to 94°C to extract DNA. Nested PCR was performed to amplify the NADH dehydrogenase sequence in mtDNA (Fig. 3). At stage 1, both the female (AA2) and male (HNI) mtDNA were detected. At stage 3, however, we could detect no signal for HNI, whereas the signals of AA2 were still clearly amplified, indicating that the ≈100 copies of mtDNA from sperm had been actively degraded in fertilized eggs before stage 3 (1 h 5 min to 1 h 45 min after fertilization) but no earlier than 30 min, which is consistent with the previous observation in mice (32).
Direct Observation of the Destruction of Paternal mtDNA. With high-resolution fluorescent microscopy, Sutovsky et al. (34) observed that the intact mitochondrial sheath remains attached to the sperm tail, which is maintained in close vicinity to the developing male pronucleus. As pronuclear development advances, the sperm mitochondria become swollen and eventually detach from the disintegrating axoneme. Sutovsky et al. (35) later reported that mitochondrial proteins are ubiquitinated for an efficient degradation mediated by proteasomes.
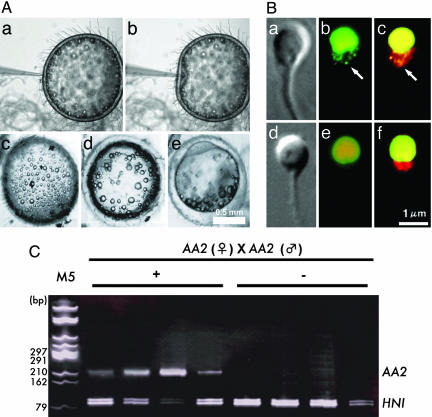
To understand mtDNA behavior during these events, we monitored the activity of sperm mt nucleoids after fertilization. Mature sperm from AA2 were collected and double stained with SYBR green I and MitoTracker CMTMRos and injected into AA2 eggs. After the injection, normal changes in the oil droplets' distribution, fusion, and polarization were observed (Fig. 4). The sperm were extracted from these eggs 60 min after the injection and observed under a fluorescence microscope (Fig. 4). The structure of mitochondria appeared intact under a phase-contrast microscope, and the MitoTracker CMTMRos signal remained unchanged, even if sperm were stained just before observation, showing the intactness of the mitochondrial membrane at least to maintain the active potential. However, the fluorescence of mtDNA could not be detected any longer.
Fig. 4.
Active digestion of sperm mtDNA after injection into eggs. (A) Process of microinjection and the development of injected eggs (a–e). After injection (a and b), the eggs remained intact (c) and progressed through normal developmental stages (d and e). (B) Phase-contrast images (a and d), SYBR green I-stained images (green) (b and e), and SYBR green I/MitoTracker CMTMRos double-stained images (red) (c and f) of sperm before (a–c) and 60 min after (d–f) fertilization. a and b are the identical sperm. d and e are also identical. Sperm mt nucleoids disappeared completely 60 min after fertilization (e and f). The mitochondrial structure visualized by phase-contrast microscopy (d) or by MitoTracker CMTMRos staining (f) remained intact even after the disappearance of fluorescent mt nucleoids. (C) Single sperm with (+) or without (-) mt nucleoids were selectively extracted from fertilized eggs by using optical tweezers (18) and analyzed by nested PCR. In this experiment, sperm and eggs were derived from AA2, and 10-8 nmol of HNI PCR product was added to each reaction as an internal control.
There are several possible explanations for the disappearance of fluorescent mt nucleoids. SYBR green I bleaching could be one possibility, but this explanation is unlikely because nuclear fluorescence was still very clear. Another explanation is that the fluorescent mt nucleoids could not be detected because of mtDNA diffusion throughout the mitochondria. Finally, extensive degradation of mtDNA could have resulted in the disappearance of fluorescent mt nucleoids. To determine which of these two final explanations was correct, we performed optical tweezer-mediated single-cell analysis.
The use of optical tweezers is a technique for manipulating living cells or organelles in suspension without physical contact or damage (36). By using this technique, sperm with or without fluorescent mt nucleoids were extracted from eggs, collected selectively under direct observation, and harvested into PCR tubes by using the method reported in ref. 18. Before performing the nested PCR analysis, 10-8 nmol of PCR products from HNI was added to the reaction mixture as an internal control. The internal control sequence (HNI) was detected in every PCR, but the AA2 NADH dehydrogenase sequence was detected only in sperm with fluorescent mt nucleoids (Fig. 4). These results demonstrate that mtDNA is actively digested well before the complete destruction of mitochondrial structures.
Discussion
Mitochondrial Nucleoids in Animal Cells. The mt (cp) genomes are present at multiple copies per cell and are subject to dynamic regulation with regard to tissue type. Multiple copies of mtDNA or cpDNA are not simply exposed in vivo but are compacted and highly organized into a DNA–protein complex called mt (cp) nucleoids (37). mt nucleoids have been difficult to visualize in animal cells, possibly because mtDNA molecules are highly mobile and flexible, allowing mitochondria to function as a single dynamic unit (38).
In our research, the organization of mt nucleoids was clearly visualized in living animal cells by using a highly permeant dsDNA-specific fluorochrome, SYBR green I (Fig. 1). According to the fluorometric analysis of mt nucleoids, each of the nucleoids contained ≈10 copies of the mt genome (Fig. 2). Similar values have been reported for mt nucleoids in mosses, algae (2, 17), and human cells (22, 23), implying a structural similarity of mt nucleoids between these organisms.
It is currently believed that mt (cp) nucleoids serve as sites for replication, recombination, and transcription and that they also function as units of segregation and inheritance (2). A number of mt (cp) nucleoid proteins have been identified in yeast (39–42), algae (43), and plants (44, 45). An intriguing example of a mt nucleoid protein is Glom, which is a structural nucleoid protein that also maintains the replication and transcriptional activity of mtDNA in slime mold (46). mt nucleoids in animals might also function in replication, transcription, and inheritance. The SYBR green I/MitoTracker CMTMRos double-staining technique for visualization of mt nucleoids in living animal cells will be a simple and invaluable tool for the identification and characterization of mt nucleoids.
Active Destruction of Sperm DNA upon Fertilization. SYBR green I vital staining techniques, in combination with the utilization of optical tweezers (18), allowed us to visualize mt nucleoids and further clarify that the elimination of sperm mtDNA upon fertilization is achieved through two steps: (i) gradual decrease in the number of mt nucleoids during spermatogenesis (Figs. 1 and 2) and (ii) rapid digestion of sperm mtDNA just after fertilization (Figs. 3 and 4).
One notable point is that the disappearance of mt nucleoids was observed before the destruction of sperm mt structure (Fig. 4). The mt structure appeared to be intact with either phase-contrast or fluorescence microscopy with MitoTracker CMTMRos staining, although we cannot exclude the possibility that a slight degradation of the mitochondrial membrane might initiate the digestion of mtDNA. There is also a possibility that degradation of mt nucleoid proteins might be required to expose mtDNA to nucleolytic attack. Because of the absence of proteasome and components required for ubiquitination in mitochondria, this degradation might be based on a ubiquitin-independent pathway. Mitochondrial intrinsic proteases, such as ATPases associated with diverse cellular activities (AAA) proteases (47), might be involved in this initial protein degradation.
According to Sutovsky and Schatten (24), paternal mtDNA can be deleterious because it might be heavily damaged and accumulate mutations. The most likely explanation for this damage would be oxidative stress during spermatogenesis. It is known that oxidative mechanisms play an important role in the pathophysiology of mammalian sperm function, such as by creating redox activity to induce sperm capacitation (48). However, oxidative stress can also induce peroxidation of the plasma membrane and DNA damages that would cause male infertility (49).
It is likely that the sperm nuclear genome is protected from oxidative stress by dramatic reorganization, because histones and other nonhistone chromosomal proteins are replaced by protamines (50), but no possible defense mechanisms have been proposed for sperm mtDNA so far. We observed no changes in mt nucleoid organization, whereas the nucleus experiences dramatic reorganization during spermatogenesis (Figs. 1 and 2). Indeed, it has been reported that sperm mtDNA is more susceptible to oxidative damage than nuclear DNA (51). One suggestion is that mtDNA may serve a “sentinel” role as a sensitive biomarker of oxidative stress in the male germ line (52). It therefore appears reasonable to digest sperm mtDNA before the destruction of mitochondrial structures because it will avoid the diffusion, recombination, and transmission of the potentially deleterious sperm mtDNA to the progeny.
Evidence for recombination in mtDNA has been reported for plants and fungi (53), and it has been regarded as a driving force to create subgenomic molecules that confer an extreme fluidity to the organization of plant mt genomes (54). Homologous recombination is well studied in chloroplasts, and cpDNA transformation is performed taking advantage of this activity (55, 56). Furthermore, direct evidence for homologous recombination in animal mtDNA was recently reported (57). These observations validate the necessity to eliminate paternal mt (cp) DNA before the destruction of mt (cp) structures. Active digestion of uniparental mt (cp) DNA has been observed in various species, including higher plants (2, 21), algae (13, 16), and slime mold (58, 59). Also, in these cases, mt (cp) DNA molecules are digested, whereas the structures of mitochondria or chloroplasts remain apparently intact, which might be necessary to avoid the potential hazard caused by sexual recombination of mt (cp) genomes in these species.
Mechanism underlying the active digestion of mt (cp) genomes is still an open question. It is likely that eggs play an important role in this process, because the active digestion of sperm mtDNA was not observed without the exposure to ooplasm. Ooplasm might execute all of the processes of mtDNA digestion, ubiquitination, and protein degradation or it might just initiate the self-destruction program encoded by sperm. Careful experiments are required to answer these questions. Furthermore, by performing a more comprehensive analysis, similar phenomena and mechanisms may be found in diverse taxa of animals and plants. It would also be necessary to compare and contrast various eukaryotes to understand the mechanisms and significance of maternal inheritance.
Materials and Methods
Strains and Preparation of Eggs and Spermatogenic Cells. The genetically divergent AA2 and HNI inbred strains were established from native medaka (O. latipes) populations in the southern and the northern part of Japan, respectively. The medaka were raised in separate tanks at 27°C under light (cool white light ≈35.5 μE·s-1·m-2; 14 h) (E, einstein, 1 mol of photons) and dark (10 h) conditions. O. latipes spawned daily within 1 h of the onset of light periods. Eggs and spermatogenic cells were kept in medaka saline (111.2 mM NaCl/5.4 mM KCl/1.1 mM CaCl2/0.6 mM MgSO4,pH 7.3, adjusted with NaHCO3) (60).
Vital Staining of DNA and Mitochondrial Membranes and Microscopic Quantification of mtDNA. The fluorescent probe (SYBR green I and MitoTracker CMTMRos) was purchased from Molecular Probes. A primary stock of 1 mmol/liter MitoTracker CMTMRos was prepared in methanol. To stain the DNA in active spermatogenic cells, SYBR green I was added to give a final dilution of 1:1,000 (17). To stain mitochondrial membranes, MitoTracker CMTMRos was used to give a final concentration of 200 nmol/liter (29). The fluorescence intensity of mt nucleoids was quantified by using a video-intensified microscope photon-counting system (VIMPCS) (61). Monochrome images of SYBR green I-stained sperm were obtained by using a photomultiplier tube. The number of photons emitted from mt nucleoids was counted and converted into a DNA amount by using SYBR green I-stained T4 phage (170 kb) as a control.
Microinjection. Nonfertilized eggs were collected from strain AA2. Attached filaments were removed with fine forceps. Approximately 100 vital spermatogenic cells stained with SYBR green I and MitoTracker CMTMRos were microinjected into eggs according to Kinoshita et al. (62). The injected eggs were incubated at 25°C and allowed to develop further. After the first cell division, the eggs were punctured and sperm were released from the eggs. In some experiments, cells were stained just before observation.
Optical Isolation and Nested PCR Analysis. The optical tweezers used were similar to those described by Ashkin et al. (36). The main components of the optical tweezer device are a laser and a microscope system. The laser was a diode-pumped neodymium–yttrium/aluminum garnet (YAG) laser (ADLAS DPY 421, Adlas, Lubeck, Germany) that emits continuous infrared (IR) light at 1,064 nm with a maximum power of 2,000 mW.
Single sperm in the egg extracts were obtained selectively under direct microscopic observation by using the method described in ref. 18. In this experiment, medaka saline containing 10% Percoll was used as isolation buffer to avoid the random diffusion and contamination of mtDNA molecules. The single sperm obtained were subjected to nested PCR for mtDNA sequences (NADH dehydrogenase).
Primers were designed as follows: NADH_F0 (5′-TTTAATTCTTATAATTGCACATGGC-3′), NADH_R0 (5′-GCTCATCCTCAGATTAAGGAAGGTT-3′), NADH_F1 (5′-AATTGCACATGGCCTGACTTCTTCC-3′), and NADH_R1 (5′-CCAGCCCCAGTCAAGAGTAAGGTTC-3′). Twenty-five cycles of PCR were repeated twice, first with F0 and R0 primers and twice with F1 and R1 primers by using 1 μl of the product of the first PCR as template. The second PCR product was cut by a restriction enzyme (HinfI) to detect the polymorphism between AA2 and HNI. For the optical isolation experiment, five eggs were used and four sperm with or without mt nucleoids were extracted from each of the eggs and analyzed to confirm reproducibility.
Acknowledgments
We thank Sara L. Zimmer and Gregory T. Smaldone for helpful comments and suggestions. This work was supported by Japan Society for the Promotion of Science by Young Scientists Research Fellowship 13-5096 and a Francis Goelet Fellowship (to Y.N.); Ministry of Education, Culture, Sports, Science, and Technology of Japan Grants 17051029, 12440222, and 13206011 (to T.K.); and a Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) grant (to T.K.).
Author contributions: Y.N. and T.K. designed research; Y.N., T. Yoshinari, T. Yamada, and K.S. performed research; Y.N., K.N., H.M., T.H., and T.K. contributed new reagents/analytic tools; Y.N., T. Yoshinari, and K.N. analyzed data; and Y.N. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: cp, chloroplast; mt, mitochondrial.
References
- 1.Gray, M. W. (1992) Int. Rev. Cytol. 141, 233-357. [DOI] [PubMed] [Google Scholar]
- 2.Kuroiwa, T. (1991) Int. Rev. Cytol. 128, 1-60. [Google Scholar]
- 3.Birky, C. W., Jr. (2001) Annu. Rev. Genet. 35, 125-148. [DOI] [PubMed] [Google Scholar]
- 4.Birky, C. W., Jr. (1995) Proc. Natl. Acad. Sci. USA 92, 11331-11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hecht, N. B., Liem, H., Kleene, K. C., Distel, R. J. & Ho, S. M. (1984) Dev. Biol. 102, 452-461. [DOI] [PubMed] [Google Scholar]
- 6.Jansen, R. P. & de Boer, K. (1998) Mol. Cell. Endocrinol. 145, 81-88. [DOI] [PubMed] [Google Scholar]
- 7.Gyllensten, U., Wharton, D., Josefsson, A. & Wilson, A. C. (1991) Nature 352, 255-257. [DOI] [PubMed] [Google Scholar]
- 8.Ankel-Simons, F. & Cummins, J. M. (1996) Proc. Natl. Acad. Sci. USA 93, 13859-13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lansman, R. A., Avise, J. C. & Huettel, M. D. (1983) Proc. Natl. Acad. Sci. USA 80, 1969-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skibinski, D. O., Gallagher, C. & Beynon, C. M. (1994) Genetics 138, 801-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zouros, E., Oberhauser Ball, A., Saavedra, C. & Freeman, K. R. (1994) Proc. Natl. Acad. Sci. USA 91, 7463-7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao, L., Kenchington, E. & Zouros, E. (2004) Genetics 166, 883-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sager, R. & Lane, D. (1972) Proc. Natl. Acad. Sci. USA 69, 2410-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs, H. T., Lehtinen, S. K. & Spelbrink, J. N. (2000) BioEssays 22, 564-572. [DOI] [PubMed] [Google Scholar]
- 15.Kuroiwa, T., Suzuki, T., Ogawa, K. & Kawano, S. (1981) Plant Cell Physiol. 22, 381-396. [Google Scholar]
- 16.Kuroiwa, T., Kawano, S., Nishibayashi, S. & Sato, C. (1982) Nature 298, 481-483. [DOI] [PubMed] [Google Scholar]
- 17.Nishimura, Y., Higashiyama, T., Suzuki, L., Misumi, O. & Kuroiwa, T. (1998) Eur. J. Cell Biol. 77, 124-133. [DOI] [PubMed] [Google Scholar]
- 18.Nishimura, Y., Misumi, O., Matsunaga, S., Higashiyama, T., Yokota, A. & Kuroiwa, T. (1999) Proc. Natl. Acad. Sci. USA 96, 12577-12582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishimura, Y., Misumi, O., Kato, K., Inada, N., Higashiyama, T., Momoyama, Y. & Kuroiwa, T. (2002) Genes Dev. 16, 1116-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umen, J. G. & Goodenough, U. W. (2001) Genes Dev. 15, 2585-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagata, N., Saito, C., Sakai, A., Kuroiwa, H. & Kuroiwa, T. (1999) Planta 209, 53-65. [DOI] [PubMed] [Google Scholar]
- 22.Satoh, M. & Kuroiwa, T. (1991) Exp. Cell Res. 196, 137-140. [DOI] [PubMed] [Google Scholar]
- 23.Legros, F., Malka, F., Frachon, P., Lombes, A. & Rojo, M. (2004) J. Cell Sci. 117, 2653-2662. [DOI] [PubMed] [Google Scholar]
- 24.Sutovsky, P. & Schatten, G. (2000) Int. Rev. Cytol. 195, 1-65. [DOI] [PubMed] [Google Scholar]
- 25.Saiki, A., Tamura, M., Matsumoto, M., Katowgi, J., Watanabe, A. & Onitake, K. (1997) Dev. Growth Differ. 39, 337-344. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa, Y. (2000) Bioessays 22, 487-495. [DOI] [PubMed] [Google Scholar]
- 27.Naruse, K., Fukamachi, S., Mitani, H., Kondo, M., Matsuoka, T., Kondo, S., Hanamura, N., Morita, Y., Hasegawa, K., Nishigaki, R., et al. (2000) Genetics 154, 1773-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura, T., Jindo, T., Narita, T., Naruse, K., Kobayashi, D., Shin, I. T., Kitagawa, T., Sakaguchi, T., Mitani, H., Shima, A., et al. (2004) Mech. Dev. 121, 915-932. [DOI] [PubMed] [Google Scholar]
- 29.Sutovsky, P., Ramalho-Santos, J., Moreno, R. D., Oko, R., Hewitson, L. & Schatten, G. (1999) Hum. Reprod. 14, 2301-2312. [DOI] [PubMed] [Google Scholar]
- 30.Ohta, T. & Iwamatsu, T. (1983) J. Exp. Zool. 227, 109-119. [DOI] [PubMed] [Google Scholar]
- 31.Pickworth, S., Yerganian, G. & Chang, M. C. (1968) Anat. Rec. 162, 197-208. [DOI] [PubMed] [Google Scholar]
- 32.Kaneda, H., Hayashi, J., Takahama, S., Taya, C., Lindahl, K. F. & Yonekawa, H. (1995) Proc. Natl. Acad. Sci. USA 92, 4542-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwamatsu, T. (2004) Mech. Dev. 121, 605-618. [DOI] [PubMed] [Google Scholar]
- 34.Sutovsky, P., Navara, C. S. & Schatten, G. (1996) Biol. Reprod. 55, 1195-1205. [DOI] [PubMed] [Google Scholar]
- 35.Sutovsky, P., Moreno, R. D., Ramalho-Santos, J., Dominko, T., Simerly, C. & Schatten, G. (1999) Nature 402, 371-372. [DOI] [PubMed] [Google Scholar]
- 36.Ashkin, A., Dziedzic, J. M. & Yamane, T. (1987) Nature 330, 769-771. [DOI] [PubMed] [Google Scholar]
- 37.Kuroiwa, T. (1982) Int. Rev. Cytol. 75, 1-59. [DOI] [PubMed] [Google Scholar]
- 38.Hayashi, J., Takemitsu, M., Goto, Y. & Nonaka, I. (1994) J. Cell Biol. 125, 43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyakawa, I., Sando, N., Kawano, S., Nakamura, S. & Kuroiwa, T. (1987) J. Cell Sci. 88, 431-439. [DOI] [PubMed] [Google Scholar]
- 40.Kaufman, B. A., Newman, S. M., Hallberg, R. L., Slaughter, C. A., Perlman, P. S. & Butow, R. A. (2000) Proc. Natl. Acad. Sci. USA 97, 7772-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, X. J., Wang, X., Kaufman, B. A. & Butow, R. A. (2005) Science 307, 714-717. [DOI] [PubMed] [Google Scholar]
- 42.Meeusen, S., Tieu, Q., Wong, E., Weiss, E., Schieltz, D., Yates, J. R. & Nunnari, J. (1999) J. Cell Biol. 145, 291-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi, T., Takahara, M., Miyagishima, S. Y., Kuroiwa, H., Sasaki, N., Ohta, N., Matsuzaki, M. & Kuroiwa, T. (2002) Plant Cell 14, 1579-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato, N., Albrieux, C., Joyard, J., Douce, R. & Kuroiwa, T. (1993) EMBO J. 12, 555-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakano, T., Murakami, S., Shoji, T., Yoshida, S., Yamada, Y. & Sato, F. (1997) Plant Cell 9, 1673-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sasaki, N., Kuroiwa, H., Nishitani, C., Takano, H., Higashiyama, T., Kobayashi, T., Shirai, Y., Sakai, A., Kawano, S., Murakami-Murofushi, K. & Kuroiwa, T. (2003) Mol. Biol. Cell 14, 4758-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langer, T. (2000) Trends Biochem. Sci. 25, 247-251. [DOI] [PubMed] [Google Scholar]
- 48.Leclerc, P., de Lamirande, E. & Gagnon, C. (1997) Free Radical Biol. Med. 22, 643-656. [DOI] [PubMed] [Google Scholar]
- 49.Aitken, R. J., Baker, M. A. & Sawyer, D. (2003) Reprod. Biomed. Online 7, 65-70. [DOI] [PubMed] [Google Scholar]
- 50.Aoki, V. W. & Carrell, D. T. (2003) Asian J. Androl. 5, 315-324. [PubMed] [Google Scholar]
- 51.Bennetts, L. E. & Aitken, R. J. (2005) Mol. Reprod. Dev. 71, 77-87. [DOI] [PubMed] [Google Scholar]
- 52.O'Connell, M., McClure, N. & Lewis, S. E. (2002) Hum. Reprod. 17, 1565-1570. [DOI] [PubMed] [Google Scholar]
- 53.Gray, M. W., Burger, G. & Lang, B. F. (2001) Genome Biol. 2, reviews1018.1-1018.5. [DOI] [PMC free article] [PubMed]
- 54.Hanson, M. R. (1991) Annu. Rev. Genet. 25, 461-486. [DOI] [PubMed] [Google Scholar]
- 55.Boynton, J. E., Gillham, N. W., Harris, E. H., Hosler, J. P., Johnson, A. M., Jones, A. R., Randolph-Anderson, B. L., Robertson, D., Klein, T. M., Shark, K. B., et al. (1988) Science 240, 1534-1538. [DOI] [PubMed] [Google Scholar]
- 56.Staub, J. M. & Maliga, P. (1992) Plant Cell 4, 39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ladoukakis, E. D. & Zouros, E. (2001) Mol. Biol. Evol. 18, 1168-1175. [DOI] [PubMed] [Google Scholar]
- 58.Moriyama, Y. & Kawano, S. (2003) Genetics 164, 963-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moriyama, Y., Yamazaki, T., Nomura, H., Sasaki, N. & Kawano, S. (2005) Curr. Genet. 48, 334-343. [DOI] [PubMed] [Google Scholar]
- 60.Iwamatsu, T. (1978) J. Exp. Zool. 204, 401-408. [DOI] [PubMed] [Google Scholar]
- 61.Kuroiwa, T., Miyamura, S., Kawano, S., Hizume, M., Tho, E. A., Miyakawa, I. & Sando, N. (1986) Exp. Cell Res. 165, 199-206. [DOI] [PubMed] [Google Scholar]
- 62.Kinoshita, M., Kani, S., Ozato, K. & Wakamatsu, Y. (2000) Dev. Growth Differ. 42, 469-478. [DOI] [PubMed] [Google Scholar]