Abstract
Merle is a pattern of coloring observed in the coat of the domestic dog and is characterized by patches of diluted pigment. This trait is inherited in an autosomal, incompletely dominant fashion. Dogs heterozygous or homozygous for the merle locus exhibit a wide range of auditory and ophthalmologic abnormalities, which are similar to those observed for the human auditory–pigmentation disorder Waardenburg syndrome. Mutations in at least five genes have been identified as causative for Waardenburg syndrome; however, the genetic bases for all cases have not been determined. Linkage disequilibrium was identified for a microsatellite marker with the merle phenotype in the Shetland Sheepdog. The marker is located in a region of CFA10 that exhibits conservation of synteny with HSA12q13. This region of the human genome contains SILV, a gene important in mammalian pigmentation. Therefore, this gene was evaluated as a candidate for merle patterning. A short interspersed element insertion at the boundary of intron 10/exon 11 was found, and this insertion segregates with the merle phenotype in multiple breeds. Another finding was deletions within the oligo(dA)-rich tail of the short interspersed element. Such deletions permit normal pigmentation. These data show that SILV is responsible for merle patterning and is associated with impaired function of the auditory and ophthalmologic systems. Although the mutant phenotype of SILV in the human is unknown, these results make it an intriguing candidate gene for human auditory–pigmentation disorders.
Keywords: short interspersed element, pigmentation, linkage disequilibrium
Merle is a coat pattern in the domestic dog characterized by patches of diluted pigment intermingled with normal melanin. It is a standard coloration for several breeds recognized by the American Kennel Club, including the Shetland Sheepdog, Australian Shepherd, Cardigan Welsh Corgi, and Dachshund. The merle phenotype in the Dachshund is known as dapple. Although merle is not an acceptable color in the Great Dane, the desirable harlequin pattern results from the interaction of the merle locus (M) and a separate harlequin locus (H) (1). In addition, many breeds (e.g., Catahoula Leopard Dog, Bergamasco Sheepdog, and Pyrenean Shepherd) accepted by other kennel clubs present with merle patterning.
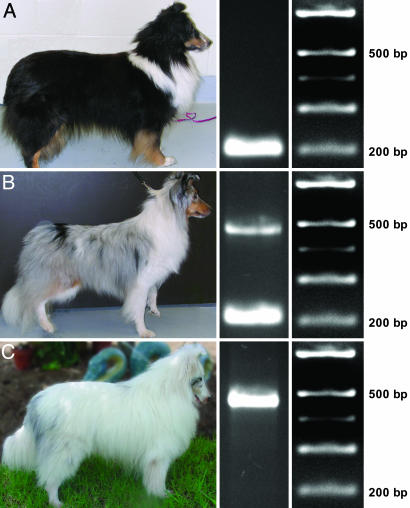
Merle is inherited in an autosomal, incompletely dominant fashion (2). Although rare, a dog that does not present with the overt merle phenotype may possess the merle genotype and subsequently produce merle offspring. Such a dog is termed a cryptic merle. The mechanism for this phenomenon is unknown. Dogs homozygous for merle (MM) are known as double merles and are predominantly white (Fig. 1C).
Fig. 1.
SINE insertion in SILV segregates with merle phenotype. (A) Tricolored (black, sable, and white), nonmerle Shetland Sheepdog (mm). (B) Blue merle Shetland Sheepdog (Mm). (C) Double merle Shetland Sheepdog (MM). (Left) Phenotypes. (Center) Exon 11 PCR products. (Right) Length markers.
Dogs having Mm and MM genotypes typically have blue eyes and often exhibit a wide range of auditory and ophthalmologic abnormalities (3). Reetz et al. (4) studied the auditory capacity of Dachshunds and found that 54.6% of MM and 36.8% of Mm dogs had auditory dysfunction, ranging from mild to severe deafness. All control dogs (mm) in the study had normal hearing. Klinckmann et al. (5, 6) conducted ophthalmologic studies with three groups of Dachshunds (MM, Mm, and mm) and found that merles and double merles had significantly greater frequencies of ocular abnormalities, including increased intraocular pressure and ametropic eyes. Microphthalmia and colobomas are well described in merle and double merle Dachshunds and Australian Shepherds (3, 7, 8). In all breeds, the double merle genotype can be sublethal and is associated with multiple abnormalities of the skeletal, cardiac, and reproductive systems (3, 9, 10). For these reasons, merle-to-merle breedings are strongly discouraged (9).
Interestingly, many of the abnormalities associated with merle dogs are remarkably similar to those observed in Waardenburg syndrome (WS) (11). WS is an autosomal dominant auditory–pigmentation disorder in humans (1 per 40,000 live births) that accounts for 2% of all cases of congenital deafness (12). Several genes have been implicated in the four clinical varieties of WS: Mutations in PAX3 cause WS type 1 and type 3 (13, 14) and mutations in SOX10, EDNRB,or EDNR3 cause WS type 4 (15–17). Mutations in MITF cause WS type 2; however, the genetic basis for 85% of type 2 cases remains unidentified (18, 19).
To identify a chromosomal region segregating with merle, we carried out a whole-genome scan for the Shetland Sheepdog by using the multiplexed Minimal Screening Set 2 (20, 21). Linkage disequilibrium (LD) for merle was identified with a microsatellite marker in a region of CFA10 that exhibits conservation of synteny with HSA12q13. This region of HSA12 harbors the SILV gene.
SILV (also known as Pmel17; gp100) is a pigment gene best known as the Silver locus responsible for a recessive trait in an inbred strain of black mice in which the hair color dilutes with age (22, 23). Although this gene is known to have a central role in pigmentation, the precise function of SILV remains controversial (24). Multiple studies have provided data to suggest that SILV is involved in the biogenesis of premelanosomes (25–27). Significant expression of the gene is almost exclusive to the skin and eye, providing further evidence to support a role in pigmentation (24). Although mutations in SILV have not been implicated in disease, the aforementioned LD data and the role of SILV in pigmentation made it a candidate gene for merle.
Characterization of SILV in merle and nonmerle Shetland Sheepdogs revealed a short interspersed element (SINE) insertion at the intron 10/exon 11 boundary. The SINE segregates with the merle phenotype in multiple breeds and is absent from dogs representing breeds that do not have merle patterning. All examined harlequin Great Danes harbored the insertion in either a heterozygous or homozygous state. Described herein is LD with merle and characterization of a SINE insertion in SILV that is responsible for merle patterning in the dog.
Results
LD with the Merle Phenotype. Genotype data for 279 Minimal Screening Set 2 markers were generated for 9 merle and 32 nonmerle Shetland Sheepdogs. Only one marker had an allele that appeared to be more common in the merle population. For this marker, FH2537, a statistically significant P value (7.2 × 10-6) was obtained. To validate this result, genotype data were generated for the aforementioned marker by using additional Shetland Sheepdogs: 7 merle, 2 double merle, and 11 nonmerle. These genotypes were combined with the original data set and used to recalculate the P value, which increased in significance (3.0 × 10-8).
Candidate Gene Selection. The above result allowed narrowing of the search to those genes that are (i) important in pigmentation and (ii) also proximal to microsatellite marker FH2537 on CFA10. SILV encodes a melanosomal protein important in pigmentation (28) and maps to HSA12q13–q14, which exhibits conservation of synteny with CFA10 (29). A single base insertion in SILV causes the silver phenotype in the mouse (23), and polymorphisms in this gene are associated with the dominant white, dun, and smoky plumage color variants in chickens (30). The dilute coloration of Charolais cattle has also recently been attributed to a mutation in SILV (24). Furthermore, SILV expression is dependent on MITF (31), which is causative for some cases of WS type 2 (18).
SINE Insertion. PCR was carried out by using genomic DNA from two nonmerle, one blue merle, and one double merle Shetland Sheepdog to obtain amplicons from each exon of SILV. Amplification of exon 11 yielded two products: (i) the expected 206-bp product and (ii) a larger product (slightly smaller than 500 bp). These amplicons segregated with the merle phenotype among the aforementioned dogs: The nonmerle dogs were homozygous for the 206-bp product; the blue merle was heterozygous for the products; and the double merle was homozygous for the larger product (Fig. 1).
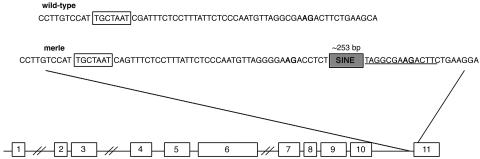
Sequence analysis of exon 11 products revealed an insertion of a tRNA-derived SINE, highly similar to the unique canine SINEs described by Minnick et al. (32). The insertion occurs at the boundary of intron 10 and exon 11 and is flanked by a 15-bp target site duplication (Fig. 2). The SINE insertion is in reverse orientation, with the 5′ end closer to exon 11. Sequence analysis of all coding regions of the gene did not reveal any other mutations that may disrupt the function of SILV.
Fig. 2.
Structure of wild-type canine SILV and sequence of the SINE insertion site in merle dogs. The putative lariat branch point sequence is boxed. Splicing acceptors are indicated by bold type. In merle dogs, the splicing acceptor is located in the 15-bp duplicated sequence (underlined) that flanks the SINE insertion. The average insertion size (not including the duplicated sequence) for the merle dogs analyzed herein is 253 bp.
DNA was available from 50 of the 61 Shetland Sheepdogs used in the linkage analysis. These 50 dogs were analyzed by gel electrophoresis for the insertion. The insert was present in the heterozygous state in 12 merles and in the homozygous state in 2 double merles. Thirty-one nonmerle dogs did not harbor the insertion and four nonmerle dogs were heterozygous for a smaller insertion. Sequence analysis of this smaller insertion from two Shetland Sheepdogs revealed a deletion within the oligo(dA)-rich tail of the SINE. This smaller insertion was also present in a nonmerle that is suspected to be cryptic because it was sired by a double merle; however, no test breedings have been conducted to date to conclusively classify the dog as cryptic.
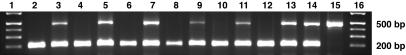
To determine whether the SILV mutation causing merle patterning in the Shetland Sheepdog population was breed specific, merle and nonmerle dogs representing six other breeds (Collie, Border Collie, Australian Shepherd, Cardigan Welsh Corgi, Dachshund, and Great Dane) were analyzed for the insertion. Merle dogs from all six breeds were heterozygous and one double merle Great Dane was homozygous for the insertion (Fig. 3). Sequence analyses of one representative merle dog from each of the aforementioned breeds showed that they have the same SINE insertion in SILV (Fig. 4).
Fig. 3.
Mutation analysis of SILV and its segregation in six breeds. PCR on genomic DNA from a sable/white Collie (lane 2), blue merle Collie (lane 3), black/white Border Collie (lane 4), blue merle Border Collie (lane 5), red Australian Shepherd (lane 6), blue merle Australian Shepherd (lane 7), brindle Cardigan Welsh Corgi (lane 8), blue merle Cardigan Welsh Corgi (lane 9), black/tan Dachshund (lane 10), red dapple Dachshund (lane 11), fawn Great Dane (lane 12), blue merle Great Dane (lane 13), and harlequin Great Danes (lanes 14 and 15).
Fig. 4.
Sequence alignment of the SINE insertion in eight merle dogs from seven breeds (Shetland Sheepdog “Sheltie,” Collie, Border Collie, Australian Shepherd “Aussie,” Cardigan Welsh Corgi, Dachshund, and Great Dane) and three nonmerle dogs from two breeds (Shetland Sheepdog and Great Dane) with the smaller insertion.
Genotypes of 12 harlequin Great Danes were also analyzed by gel electrophoresis: 9 were heterozygous and 3 were homozygous for the insertion. Additionally, seven Great Danes (six harlequin and one black) were heterozygous for a smaller insertion, and sequence analysis from one of these dogs showed a deletion within the oligo(dA)-rich tail, as was observed in the Shetland Sheepdogs (Fig. 4). Gel electrophoresis analysis showed that the SINE insertion also segregated with the merle phenotype in dogs from five additional breeds (American Pit Bull Terrier, Catahoula Leopard Dog, Chihuahua, Miniature Poodle, and Pyrenean Shepherd). Analysis of the intron10/exon 11 segment from 29 dogs representing 26 breeds that do not have merle patterning revealed that they do not have the insertion.
Discussion
Melanocytes are pigment-producing cells present in many tissues, including the epidermis, hair follicle, inner ear, and choroid of the eye (33). Melanocyte cell populations differentiate from unpigmented melanoblasts released from the neural crest during embryogenesis (33). The complex process in which melanoblasts migrate and differentiate into melanocytes is not fully understood; however, the study of pigmentary anomalies may accelerate identification of genes important for normal development (17).
The merle phenotype of the dog is a pattern of pigmentation associated with a wide range of developmental defects. A whole genome scan for merle by using 41 Shetland Sheepdogs showed LD with FH2537 on CFA10. No genes previously implicated in WS (PAX3, MITF, SOX10, EDNRB, and EDN3) map to this region. However, another gene important in pigmentation, SILV, is located ≈0.2 Mb from this marker. The SILV protein appears to be necessary for the formation of the fibril matrix upon which melanin intermediates are deposited late in melanosome maturation (24). Other studies have shown that SILV may also participate in melanin biosynthesis by accelerating the conversion of 5,6-dihydroxyindole-2-carboxylic acid to melanin (34, 35). The mutant phenotype of SILV in the human is unknown (28).
A SINE, structurally similar to a class of canine SINEs described by Minnick et al. (32), was identified in SILV for all merle dogs analyzed. This SINE shows high sequence similarity (95–97%) with canine SINEs previously identified in the canine D2 dopamine receptor gene (36), the dystrophin gene (37), and the PTPLA gene, implicated in centronuclear myopathy (38). These SINEs are tRNA-derived and highly abundant in the dog, representing 7% of the genome (39).
It was previously hypothesized that the merle locus contains a transposable element (40). This theory is based, in part, on the finding that matings of homozygous merle dogs have produced nonmerle offspring (40, 41). Subsequent breedings with these offspring produced only nonmerle puppies, providing evidence for a stable germinal reversion (41). It is estimated that the reversion rate in the germ line is 3–4% (41).
In the human, Alu insertions causing disease are usually found in coding exons or near the boundaries of exons and introns (42). These insertions result in disease, presumably by affecting splicing (38, 42). Similarly, SINE insertions at these locations have been reported in the dog. A SINE insertion within exon 2 of the PTPLA gene causes aberrant splicing patterns associated with centronuclear myopathy in the Labrador Retriever (38). Lin et al. (43) identified a SINE insertion in the 5′-flanking intronic region of exon 4 of HCRTR2 that causes narcolepsy. The insertion displaces a putative lariat branch point sequence necessary for proper splicing (43). The SINE insertion reported herein also occurs at an intron/exon boundary and may displace the putative lariat branch point sequence (Fig. 2). This change in sequence of the gene is further complicated by the fact that in humans and presumably in other mammals as well, SILV has alternative splicing patterns (44, 45). Therefore, cDNA transcripts should be analyzed to determine splicing patterns in nonmerle and merle dogs and the effect of the insertion on the encoded protein.
Fifty Shetland Sheepdogs were analyzed for the insertion, which segregated perfectly with the merle phenotype. The mutation also segregated with merle among dogs representing the Collie, Border Collie, Australian Shepherd, Cardigan Welsh Corgi, Dachshund, and Great Dane breeds, and sequence analysis confirmed that they have the same insertion. Aside from a single point mutation in two breeds each, the sequence of the SINE is identical. This finding suggests that the SINE insertion is identical by descent, and that the breeds analyzed in this study share a common ancestor. The occurrence of merle in many breeds and the fact that the first breeds to diverge from the working sheepdog population in the 1800s have merle patterning (Collie, Old English Sheepdog, and Shetland Sheepdog), suggest that the founding mutation may predate the divergence of breeds (46).
Harlequin is a popular coat pattern in the Great Dane and is characterized by black patches on a white background. Studies of the inheritance of harlequin support the hypothesis that it is the result of two genes: M and the dominant gene H (47). A deficiency of white dogs (MM) from harlequin to harlequin matings provides evidence to suggest that the H+ MM genotype has reduced viability (1). The identification of harlequin Great Danes homozygous for the SINE insertion in the present study demonstrates that harlequin dogs may be either H+ Mm or H+ MM. These data suggest that the H gene is dominant to M and that white Great Danes have the ++ MM genotype. This misclassification of phenotype could account for the deficiency of white dogs observed in the aforementioned harlequin studies (1, 47). Further studies are necessary to confirm this hypothesis.
Polymorphisms in the 3′ end of the SINE insertion were identified in four nonmerle Shetland Sheepdogs from two families and in seven Great Danes (six harlequin and one black) from one kindred. These Shetland Sheepdogs were heterozygous for the allele in LD with the merle phenotype, suggesting that the deletion may have occurred as a secondary mutation. The 3′ end of retroelements is typically polymorphic because of the presence of an oligo(dA)-rich tail, which is subject to strand slippage during replication and unequal crossing-over (48). Roy-Engel et al. (48) report an association between longer oligo(dA)-rich tails and diseased loci. In general, the oligo(dA)-rich tails decay gradually, with older insertions having shorter tails, although they can also shorten significantly in a single generation (48). Abdelhak et al. (49) report that the oligo(dA)-rich tail of an Alu insertion into the eya1 locus shortened from A97 to A31 in one generation.
The present work identifies variable oligo(dA)-rich tail lengths associated with the SINE insertion: An A91–101 segment present in merle dogs from seven breeds and an A54–65 segment present in the aforementioned nonmerle Shetland Sheepdogs and Great Danes. No oligo(dA)-rich tail lengths intermediate to these were found. These data suggest that the truncation may have occurred in a single generation and could represent a reversion of the merle mutation (41). Additionally, expansion of the oligo(dA)-rich tail in the germ line of a nonmerle dog having the smaller insertion may result in merle offspring and may be the mechanism behind the cryptic merle phenotype. Although the data presented herein suggest that the oligo(dA)-rich tail length determines phenotype (i.e., merle, nonmerle), they are not sufficient to determine the precise threshold for this phenomenon.
Here we have described a mutation in SILV that results in a disease phenotype and presented evidence to suggest a critical role for SILV in normal mammalian development. Phenotypic similarities between merling in the dog and WS in the human suggest that SILV may be involved in human auditory–pigmentation disorders. This work also enables genetic testing to identify dogs that carry the SINE insertion and may produce merle offspring. A genetic test for the merle locus can help responsible breeders of merle dogs prevent undesirable double merle progeny by allowing them to (i) distinguish merle from nonmerle in light-colored dogs that show little contrast between areas of dilution and full pigmentation, (ii) classify harlequin Great Danes as single or double merle, and (iii) identify cryptic merles. We have submitted a provisional patent application (50) for the identification of the mutation causing merle.
Materials and Methods
Sample Collection. DNA samples were obtained during previous studies conducted in the Canine Genetics Laboratory at Texas A&M University and through contributions from participating owners and breeders. Whole blood or buccal cells were collected from all dogs and genomic DNA was isolated by using the Puregene DNA Isolation kit (Gentra Systems).
Genotyping. Fluorescently labeled primers were synthesized and multiplex PCR was performed for Minimal Screening Set 2 markers as described by Clark et al. (21). PCR products were resolved with an internal size standard (GeneScan 500 LIZ, Applied Biosystems) by using an ABI 3730xl DNA Analyzer (Applied Biosystems). Genotypes were determined by using genemapper 3.5 software (Applied Biosystems).
Linkage Analysis. Analyses for LD were carried out for all genotyped Minimal Screening Set 2 markers by using 41 Shetland Sheepdogs. For each marker, the allele more often associated with the merle dogs was identified, and all other alleles were combined into a second independent class. Fisher's exact probability test for 2 × 2 tables was used to evaluate allelic frequencies between the merle and nonmerle dogs. By convention, a P value of <0.0001 provides evidence for LD.
For one marker with evidence of LD, an additional 20 Shetland Sheepdogs were genotyped and the P value was recalculated.
Sequencing. Primers were designed to amplify the complete exon and partial flanking intronic sequences for the 11 exons of SILV (see Table 1) by using the Boxer and the human intron/exon boundaries reported in Bailin et al. (44). Concentrations for an 8.45-μl PCR volume were 0.09 units/μl Taq DNA polymerase with 1.2× buffer B (Fisher Scientific), 3.55 mM MgCl2, 1.2× MasterAmp PCR Enhancer (Epicentre Technologies, Madison, WI), 0.59 mM total dNTPs, 5.9 ng/μl DNA, 0.47 μM each forward and reverse primer, and 2.8 μl of water. All exons were amplified with a single stepdown thermal cycling program: 5 min at 95°C followed by 5 cycles of 30 sec at 95°C, 15 sec at 58°C, and 10 sec at 72°C, and an additional 30 cycles of 20 sec at 95°C, 15 sec at 56°C, and 10 sec at 72°C, with a final extension of 5 min at 72°C.
Table 1. Primer sequences for the 11 exons of canine SILV.
| Exon | Forward | Reverse |
|---|---|---|
| 1 | GTAGCGGGATGTCCAGGG | GAGAAAAATCAGAGCAGGTGTG |
| 2 and 3 | ATGGTGCTGTCCCCTGA | ATCTGAGCCCTTGGAATAA |
| 4 | GGTTTGAGGGTGACTCTGTGT | GGGCAGTGAAGATTTAGGGAA |
| 5 | TTCCCTATGCTCAGTTCTTCC | GCTTTGCCCCTTCCCA |
| 6a | GGTGTGCCTGTGAAAGAAG | CAAGCGTAGTGCCTGTGAC |
| 6b | GCAGATGACGACCACGG | GTCCCACCTCAATGAACCT |
| 7 | GCCTCTTCAATCCTCTCC | CAAGGTATGCTTTCACTGG |
| 8 | GAAGCAGCCTTACGGTTTT | CGGAGTTCTCAGGACAATCA |
| 9 | CCATTGCCCTGACCTAAGC | AGCCTGTCCAACGCCTG |
| 10 | TGGCGGGGAGCAGACA | AAGAATGAGCAGTGGCAAGAG |
| 11 | CAGTTTCTCCTTTATTCTCCCA | CCTCGGCAAATCACAGCA |
PCR products were analyzed by electrophoresis on a 3% agarose gel. The Gel Extract kit was used to purify amplicons (Qiagen, Valencia, CA). Products were ligated into pCR4.0-TOPO (Invitrogen) and transformed into chemically competent Escherichia coli TOP-10 cells (Invitrogen). Two clones for each dog were selected for sequencing. Nucleotide sequencing was performed by using the Big Dye Terminator version 1.1 Cycle Sequencing kit (Applied Biosystems) and an ABI 3130 Genetic Analyzer (Applied Biosystems). Sequences were aligned by using clustal w (www.ebi.ac.uk/clustalw).
Acknowledgments
We thank the breeders and owners that submitted samples for this study. We thank Sherry Lindsey, who kindly provided a large majority of the samples and phenotype information used in this study.
Author contributions: L.A.C., J.M.W., and K.E.M. designed research; L.A.C. and J.M.W. performed research; C.A.R. and K.E.M. contributed new reagents/analytic tools; L.A.C. and J.M.W. analyzed data; and L.A.C., J.M.W., and K.E.M. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LD, linkage disequilibrium; SINE, short interspersed element; WS, Waardenburg syndrome.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ306880).
See Commentary on page 1157.
References
- 1.O' Sullivan, N. & Robinson, R. (1989) Genetica (The Hague) 78, 215-218. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell, A. L. (1935) J. Hered. 26, 425-430. [Google Scholar]
- 3.Sorsby, A. & Davey, J. B. (1954) J. Genet. 54, 425-440. [Google Scholar]
- 4.Reetz, I., Stecker, M. & Wegner, W. (1977) Dtsch. Tierarztl. Wochenschr. 84, 273-277. [PubMed] [Google Scholar]
- 5.Klinckmann, V. G., Koniszewski, G. & Wegner, W. (1987) Dtsch. Tierarztl. Wochenschr. 94, 338-341. [PubMed] [Google Scholar]
- 6.Klinckmann, V. G. & Wegner, W. (1987) Dtsch. Tierarztl. Wochenschr. 94, 337-338. [PubMed] [Google Scholar]
- 7.Gelatt, K. N. & McGill, L. D. (1973) J. Am. Vet. Med. Assoc. 162, 393-396. [PubMed] [Google Scholar]
- 8.Dausch, D., Wegner, W., Michaelis, M. & Reetz, I. (1977) Dtsch. Tierarztl. Wochenschr. 84, 468-475. [PubMed] [Google Scholar]
- 9.Little, C. C. (1957) The Inheritance of Coat Color in Dogs (Howell Book House, New York).
- 10.Sponenberg, D. P. & Bowling, A. T. (1985) J. Hered. 76, 393-394. [PubMed] [Google Scholar]
- 11.Schaible, R. H. & Brumbaugh, J. A. (1976) Pigm. Cell 3, 191-200. [Google Scholar]
- 12.Nayak, C. S. & Isaacson, G. (2003) Ann. Otol. Rhinol. Laryngol. 112, 817-820. [DOI] [PubMed] [Google Scholar]
- 13.Baldwin, C. T., Hoth, C. F., Amos, J. A., da-Silva, E. O. & Milunsky, A. (1992) Nature 355, 637-638. [DOI] [PubMed] [Google Scholar]
- 14.Tassabehji, M., Read, A. P., Newton, V. E., Harris, R., Balling, R., Gruss, P. & Strachan, T. (1992) Nature 355, 635-636. [DOI] [PubMed] [Google Scholar]
- 15.Puffenberger, E. G., Hosoda, K., Washington, S. S., Nakao, K., deWit, D., Yanagisawa, M. & Chakravarti, A. (1994) Cell 79, 1257-1266. [DOI] [PubMed] [Google Scholar]
- 16.Pingault, V., Bondurand, N., Kuhlbrodt, K., Goerich, D. E., Prehu, M. O., Puliti, A., Herbarth, B., Hermans-Borgmeyer, I., Legius, E., Matthijs, G., et al. (1998) Nat. Genet. 18, 171-173. [DOI] [PubMed] [Google Scholar]
- 17.McCallion, A. S. & Chakravarti, A. (2001) Pigm. Cell Res. 14, 161-169. [DOI] [PubMed] [Google Scholar]
- 18.Tassabehji, M., Newton, V. E. & Read, A. P. (1994) Nat. Genet. 8, 251-255. [DOI] [PubMed] [Google Scholar]
- 19.Choi, J. H., Moon, S., Lee, K. H., Lew, H. M. & Chang, Y. (2004) Korean J. Ophthalmol. 18, 185-189. [DOI] [PubMed] [Google Scholar]
- 20.Guyon, R., Lorentzen, T. D., Hitte, C., Kim, L., Cadieu, E., Parker, H. G., Quignon, P., Lowe, J. K., Renier, C., Gelfenbeyn, B., et al. (2003) Proc. Natl. Acad. Sci. USA 9, 5296-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark, L. A., Tsai, K. L., Steiner, J. M., Williams, D. A., Guerra, T., Ostrander, E. A., Galibert, F. & Murphy, K. E. (2004) Genomics 84, 550-554. [DOI] [PubMed] [Google Scholar]
- 22.Dunn, L. C. & Thigpen, L. W. (1930) J. Hered. 21, 495-498. [Google Scholar]
- 23.Kwon, B. S., Halaban, R., Ponnazhagan, S., Kim, K., Chintamaneni, C., Bennett, D. & Pickard, R. T. (1995) Nucleic Acids Res. 23, 154-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Theos, A. C., Truschel, S. T., Raposo, G. & Marks, M. S. (2005) Pigm. Cell Res. 18, 322-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi, T., Urabe, K., Orlow, S. J., Higashi, K., Imokawa, G., Kwon, B. S., Potterf, B. & Hearing, V. J. (1994) J. Biol. Chem. 269, 29198-29205. [PubMed] [Google Scholar]
- 26.Zhou, B. K., Kobayashi, T., Donatien, P. D., Bennett, D. C., Hearing, V. J. & Orlow, S. J. (1994) Proc. Natl. Acad. Sci. USA 91, 7076-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berson, J. F., Harper, D., Tenza, D., Raposo, G. & Marks, M. S. (2001) Mol. Biol. Cell. 12, 3451-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sturm, R. A., Teasdale, R. D. & Box, N. F. (2001) Gene 277, 49-62. [DOI] [PubMed] [Google Scholar]
- 29.Kwon, B. S., Chintamaneni, C., Kozak, C. A., Copeland, N. G., Gilbert, D. J., Jenkins, N., Barton, D., Francke, U., Kobayashi, Y. & Kim, K. K. (1991) Proc. Natl. Acad. Sci. USA 88, 9228-9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerje, S., Sharma, P., Gunnarsson, U., Kim, H., Bagchi, S., Fredricksson, R., Schutc, K., Jensen, P., von Heijne, G., Okimoto, R. & Andersson, L. (2004) Genetics 168, 1507-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baxter, L. L. & Pavan, W. J. (2003) Gene Expr. Patterns 3, 703-707. [DOI] [PubMed] [Google Scholar]
- 32.Minnick, M. F., Stillwell, L. C., Heineman, J. M. & Stiegler, G. L. (1992) Gene 110, 235-238. [DOI] [PubMed] [Google Scholar]
- 33.Steingrimsson, E., Copeland, N. G. & Jenkins, N. A. (2004) Annu. Rev. Genet. 38, 365-411. [DOI] [PubMed] [Google Scholar]
- 34.Lee, Z. H., Hou, L., Moellmann, G., Kuklinska, E., Antol, K., Fraser, M., Halaban, R. & Kwon, B. S. (1996) J. Invest. Dermatol. 106, 605-610. [DOI] [PubMed] [Google Scholar]
- 35.Chakraborty, A. K., Platt, J. T., Kim, K. K., Kwon, B. S. Bettett, D. C. & Pawelek, J. M. (1996) Eur. J. Biochem. 236, 180-188. [DOI] [PubMed] [Google Scholar]
- 36.Jeoung, D., Myeong, H., Lee, H., Ha, J., Galibert, F., Hitte, C. & Park, C. (2000) Anim. Genet. 31, 334-335. [DOI] [PubMed] [Google Scholar]
- 37.Fletcher, S., Carville, K. S., Howell, J. M., Mann, C. J. & Wilton, S. D. (2000) Am. J. Vet. Res. 12, 1964-1968. [DOI] [PubMed] [Google Scholar]
- 38.Pele, M., Tiret, L., Kessler, J., Blot, S. & Panthier, J. (2005) Hum. Mol. Genet. 14, 1417-1427. [DOI] [PubMed] [Google Scholar]
- 39.Kirkness, E. F., Bafna, V., Halpern, A. L., Levy, S., Remington, K., Rusch, D. B., Delcher, A. L., Pop, N., Wang, W., Fraser, C. M. & Venter, J. C. (2003) Science 301, 1898-1903. [DOI] [PubMed] [Google Scholar]
- 40.Whitney, J. B. & Lamoreux, M. L. (1982) J. Hered. 73, 12-18. [DOI] [PubMed] [Google Scholar]
- 41.Sponenberg, D. P. (1984) J. Hered. 75, 78. [DOI] [PubMed] [Google Scholar]
- 42.Druker, R. & Whitelaw, E. (2004) J. Inherited Metab. Dis. 27, 319-330. [DOI] [PubMed] [Google Scholar]
- 43.Lin, L., Faraco, J., Li, R., Kadotani, H., Rogers, W., Lin, X., Qui, X., de Jong, P. J., Nishino, S. & Mignot, E. (1999) Cell 98, 365-376. [DOI] [PubMed] [Google Scholar]
- 44.Bailin, T., Lee, S. & Spritz, R. A. (1996) J. Invest. Dermatol. 106, 24-27. [DOI] [PubMed] [Google Scholar]
- 45.Nichols, S. E., Harper, D. C., Berson, J. F. & Marks, M. S. (2003) J. Invest. Dermatol. 121, 821-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neff, M. W., Robertson, K. R., Wong, A. K., Safra, N., Broman, K. W., Slatkin, M., Mealey, K. L. & Pedersen, N. C. (2004) Proc. Natl. Acad. Sci. USA 101, 11725-11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sponenberg, D. P. (1985) J. Hered. 76, 224-225. [DOI] [PubMed] [Google Scholar]
- 48.Roy-Engel, A. M., Salem, A., Oyeniran, O. O., Deininger, L., Hedges, D. J., Kilroy, G. E., Batzer, M. A. & Deininger, P. L. (2002) Genome Res. 12, 1333-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdelhak, S., Kalatzis, V., Heilig, R., Compain, S., Samson, D., Vincent, C., Levi-Acobas, F., Cruaud, C., Le Merrer, M., Mathieu, M., et al. (1997) Hum. Mol. Genet. 6, 2247-2255. [DOI] [PubMed] [Google Scholar]
- 50.Murphy, K. E., Clark, L. A., Wahl, J. M & Rees, C. A. (2005) U.S. Patent Appl. U.S.S.N. 60/708,589.