Abstract
Replication of retroviruses requires integration of the linear viral DNA genome into the host chromosomes. Integration requires the viral integrase (IN), located in high-molecular-weight nucleoprotein complexes termed preintegration complexes (PIC). The PIC inserts the two viral DNA termini in a concerted manner into chromosomes in vivo as well as exogenous target DNA in vitro. We reconstituted nucleoprotein complexes capable of efficient concerted (full-site) integration using recombinant wild-type human immunodeficiency virus type I (HIV-1) IN with linear retrovirus-like donor DNA (480 bp). In addition, no cellular or viral protein cofactors are necessary for purified bacterial recombinant HIV-1 IN to mediate efficient full-site integration of two donor termini into supercoiled target DNA. At ∼30 nM IN (20 min at 37°C), approximately 15 and 8% of the input donor is incorporated into target DNA, producing half-site (insertion of one viral DNA end per target) and full-site integration products, respectively. Sequencing the donor-target junctions of full-site recombinants confirms that 5-bp host site duplications have occurred with a fidelity of ∼70%, similar to the fidelity when using IN derived from nonionic detergent lysates of HIV-1 virions. A key factor allowing recombinant wild-type HIV-1 IN to mediate full-site integration appears to be the avoidance of high IN concentrations in its purification (∼125 μg/ml) and in the integration assay (<50 nM). The results show that recombinant HIV-1 IN may not be significantly defective for full-site integration. The findings further suggest that a high concentration or possibly aggregation of IN is detrimental to the assembly of correct nucleoprotein complexes for full-site integration.
Integration of the retrovirus DNA genome into cellular chromosomes requires the viral integrase (IN). In newly virus-infected cells, IN is localized in cytoplasmic nucleoprotein complexes termed preintegration complexes (PIC) (2-4, 7). In the PIC, IN removes two nucleotides from the 3′ OH termini of linear blunt-ended viral DNA (∼10 kbp), producing recessed termini. Purified PIC are capable of inserting their recessed DNA termini in a concerted fashion into exogenous target DNA, mimicking integration in vivo (13, 14, 27, 29, 35, 42). The fidelity of the small host duplications occurring upon concerted integration in vivo is also observed with the PIC in vitro.
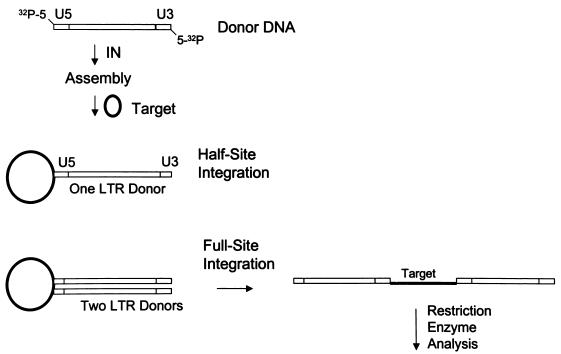
Early reconstitution experiments using purified IN, linear retrovirus-like DNA donors, and target DNA were successful but inefficient for concerted (full-site) integration (10, 15, 25). The efficiency of the full-site integration reactions has been significantly increased by employing purified avian myeloblastosis virus (AMV) IN (32, 39-41), IN in nonionic detergent lysates of human immunodeficiency virus type 1 (HIV-1) virions (6, 17, 18), and recombinant INs of various retroviruses (1, 19, 21, 22, 33). The donor-target products are visualized by agarose gel electrophoresis and characterized by restriction enzyme analysis with radioactively labeled donors (∼300 to 500 bp in length) (Fig. 1). Sequencing the donor-target junctions of individual recombinants verifies the fidelity of the small host duplications observed upon full-site integration in vivo.
FIG. 1.
Schematic for nucleoprotein complex assembly and the half-site and full-site integration reactions. A radioactive 5′-end-labeled linear DNA donor (0.48 kbp) containing 3′ OH recessed U5 and U3 LTR termini is assembled with IN on ice prior to the addition of supercoiled target DNA (2.8 kbp) (40). Half-site integration involves the insertion of one LTR end per target, and full-site integration involves the concerted insertion of two LTR ends per target. Restriction enzyme analysis resolves donor-target products.
The efficiency and fidelity of full-site integration observed with the various reconstitution systems vary significantly. This variance could be due to either the differences between the specific activities of virion and recombinant IN, the requirement of auxiliary viral or cellular proteins to mediate full-site integration, or both. AMV (39-41) and recombinant Rous sarcoma virus (RSV) PragueA (PrA) INs (33, 41) can efficiently mediate the concerted insertion of 3′ OH recessed long terminal repeat (LTR) termini from two linear donors into a circular target (bimolecular reaction) (Fig. 1) with high fidelity (∼95%) for host site duplications. Efficient full-site integration is arbitrarily defined as having ∼10% of the donor incorporated into a target in 20 min at 37°C. The specific activities of AMV and recombinant RSV PrA INs for assembly and full-site integration appear equivalent (33). Recombinant simian immunodeficiency virus (SIV) IN can mediate efficient full-site integration possessing reasonable fidelity (∼84%) for the host site duplications (19). Recombinant murine leukemia virus IN can mediate bimolecular full-site integration with high fidelity but with little efficiency (38). Recombinant RSV Schmitt-RuppinB IN performs full-site integration with high fidelity for the host site duplications by using blunt-ended termini of a single linear donor (∼300 bp; unimolecular reaction) into a DNA target (1, 21, 22). But cellular proteins HMG 2 (high-mobility group) and HMG-I(Y) are required to mediate efficient full-site integration. Recombinant HIV-1 IN also requires the viral nucleocapsid (NC) (6) or HMG-I(Y) (22) to perform efficient full-site integration in vitro. Using similar-size LTR donor substrates described in this report, recombinant HIV-1 IN by itself either fails to perform full-site integration (8, 28) or incorporates <1% of the donor into full-site products in 1- or 2-h reactions at 37°C (6, 22). The fidelity of either recombinant (6, 22) or virion (6, 18) HIV-1 IN for producing 5-bp host site duplications upon full-site integration is ∼70% in vitro. Minor protein-folding defects in recombinant HIV-1 IN or the aggregation of purified IN may prevent the correct assembly of complexes with two viral DNA ends, which is necessary for full-site integration (9).
To investigate these two possible defects, we utilized different pathways to determine whether recombinant wild-type (wt) HIV-1 IN possesses the ability to mediate efficient full-site integration without the addition of viral or cellular cofactors. To address the aggregation problem, we purified recombinant HIV-1 IN at low protein concentrations (<125 μg/ml) and kept the concentration of IN in the low nanomolar range (<50 nM) in the reaction mixtures. The purification procedure involved lysis of bacteria without nonionic detergent, low-salt washes of “insoluble” IN-DNA pellet fractions followed by high-salt extraction of IN, and further purification by column chromatography (33). In the second approach, we examined the effect of coexpressing protein chaperones (GroEL and GroES) (16, 23, 30) with HIV-1 IN in bacteria to investigate if possible protein-folding defects in IN are corrected. With both approaches, highly purified recombinant HIV-1 IN was capable of efficiently performing both the half-site and full-site integration reactions nearly equivalently without the addition of purified cellular or viral proteins. The apparent key factor for allowing recombinant HIV-1 IN to mediate full-site integration is the avoidance of high IN concentrations in its purification and in the integration assay.
MATERIALS AND METHODS
DNAs.
The linear 480-bp LTR donor fragments were released by NdeI digestion of plasmids and purified on agarose gels. The donor fragments contained 20 bp of wt U5 and wt U3 HIV-1 LTR sequences at the termini (H-2) or only wt U5 sequences at the termini (H-5) (17-19). An additional donor containing 30 bp of wt U5 and wt U3 sequences (P-2) was also used. The DNA donors contain the SupF amber gene for genetic selection of donor-target recombinants using MC1061/P3 cells (Invitrogen) (1, 22, 40). The LTR donors also contain a unique BglII site approximately 50 bp from the U3 terminus. The donors were 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. The plasmid (pGroESL) expressing protein chaperones GroEL and GroES (16) was kindly provided by A. A. Gatenby. The supercoiled target DNA was pGEM3.
Recombinant HIV-1 IN.
Expression vector pET3a (Stratagene) containing the HIV-1 IN coding region (pNY clone) was provided by F. Bushman. The IN coding region was removed from pET3a by NdeI and BamHI digestions and cloned into pET11a. The entire wt HIV-1 IN clone was sequenced to verify the construct. The pET11a-IN vector was transformed into BL21(DE3) cells. Competent BL21(DE3) cells containing pET11a-IN were also cotransformed with pGroESL, and the cells were plated onto agarose plates containing carbenicillin (50 μg/ml) and chloramphenicol (34 μg/ml). Both plasmids replicate in the same cell because they possess different origins of replication. The presence of the two plasmids in individual colonies was verified by restriction enzyme analysis and expression of both sets of proteins. Individual colonies were grown in Luria-Bertani media with both antibiotics to an optical density at 600 nm (OD600) of 0.8 and stored in glycerol at −70°C.
Purification of HIV-1 IN.
Bacterial cells expressing HIV-1 IN, without and with coexpression of pGroESL, were induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside; 0.4 mM) at an OD600 of 0.6 and harvested after 3 h by centrifugation. Purification of HIV-1 IN was very similar to procedures used to purify recombinant RSV PrA IN (33). Briefly, HIV-1 IN was extracted from cells (2 g wet weight) thawed on ice. After a light sonication in 4 ml of extraction buffer per g, the homogenized materials were subjected to high-speed centrifugation and the pellets were suspended in 4 ml of a 0.1 M NaCl buffer. After a series of low-salt (0.1 M NaCl) washes and centrifugation steps, IN was extracted from the pellet with 2 ml of a 1 M NaCl buffer per g wet weight of bacteria. The high-salt extract contained the majority of IN at a reasonable purity (>60%) (see Fig. 2, lane 8). Four-milliliter preparations of HIV-1 IN were loaded unto Pharmacia SP-Sepharose HiTrap (5-ml) columns for further purification and removal of nucleic acids (33). The major nucleic acid peak preceding the elution of IN at 0.38 M NaCl was avoided; the protein peak (OD280) representing the majority of IN was loaded onto a second SP-Sepharose column. This procedure usually resulted in preparations with >90% purity. Heparin affinity (37) HP HiTrap (5-ml) columns were used to purify IN to near homogeneity. HIV-1 IN eluted from heparin-Sepharose columns at 0.68 M NaCl (see Fig. 3). The standard chromatography buffer used for both columns was identical to that previously described (33) (50 mM HEPES-NaOH [pH 7.5], 1 mM dithiothreitol, 1 mM EDTA, 10 mM MgSO4) unless otherwise indicated. Linear gradients with various concentrations of NaCl in the above buffer were used to elute IN from both columns. Fractions (∼0.5 ml) containing IN were divided into aliquots and frozen at −70°C for storage. Protein concentrations were determined by a filter paper technique using Coomassie blue staining for submicrogram quantities (34) with bovine serum albumin as the reference standard.
FIG. 2.
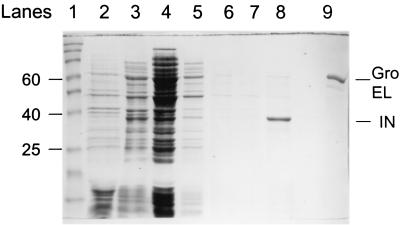
Coomassie blue-stained SDS-PAGE gel showing HIV-1 IN coexpressed with protein chaperones at various steps of induction and purification. Lane 1, molecular weight standards from 15 to 190 kDa; lane 2, total-cell lysate uninduced; lane 3, total-cell lysate induced for 3 h; lane 4, supernatant after sonication and centrifugation of lysate; lane 5, supernatant of the second lysate after sonication of the IN-DNA pellet and centrifugation; lane 6, supernatant of the first wash of the pellet; lane 7, supernatant of the second wash of the pellet; lane 8, 1 M NaCl extraction; lane 9, purified GroEL. For lanes 4 to 7, 4 ml of buffer per g wet weight of bacteria was used; for lane 8, the 1 M NaCl extraction buffer volume was 2 ml per g wet weight of bacteria. A constant volume (10 μl) of extract was applied in lanes 4 to 8. The sizes of several molecular mass markers (in kilodaltons) are indicated on the left. Purified GroEL and IN are indicated on the right.
FIG. 3.
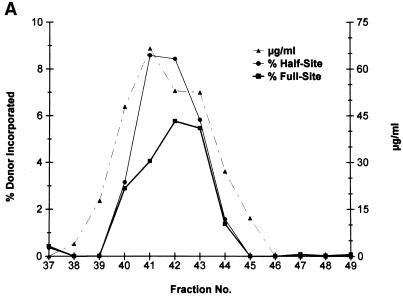
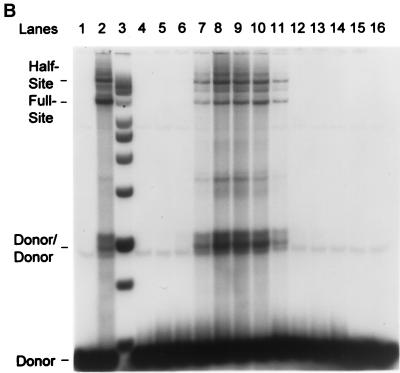
Purification of recombinant HIV-1 IN. (A) HIV-1 IN was purified by SP-Sepharose followed by heparin-Sepharose. Fractions 37 to 49 (heparin column) were analyzed for protein and strand transfer activities. The protein concentrations in fractions 40 to 44 are 48, 66, 53, 52, and 27 μg per ml, respectively (right). For fraction 41, the calculated concentration of IN is 1,030 nM. The percentages of donor incorporated into half-site and full-site strand transfer products are indicated (left; see panel B). (B) Aliquots (1 μl) of each indicated fraction shown in panel A (fractions 37 to 49 are represented in lanes 4 to 16, respectively) were preincubated with wt donor DNA (P-2) at 0°C for 12 min in 50-μl reaction volumes prior to strand transfer for 20 min at 37°C. The NaCl concentration varied from 0.63 to 0.82 M in fractions 40 to 44. Left, half-site, full-site, and donor-to-donor strand transfer products, along with the input donor. DNA products migrating slower than the half-site products contain two or more LTR donors inserted at different locations on the target DNA (40). Lanes 1 to 3, control reaction containing no IN, SIV IN at 100 nM, and molecular weight markers, respectively. The markers range in size from 0.5 to ∼10 kbp (Promega; 1-kbp DNA ladder set) (black dot, 4 kbp). (C) Aliquots (10 μl) of the indicated heparin-Sepharose fractions (lanes 1 to 9, fractions 38 to 46, respectively) were subjected to SDS-PAGE and stained with Coomassie blue.
Integration assay and analysis of full-site donor-target integration products.
Standard integration assay conditions were previously described (18) except for several minor modifications. The MgCl2, NaCl, and polyethylene glycol concentrations were changed to 15 mM, 100 mM, and 10%, respectively. Zinc chloride was added to 25 μM. The donor DNA concentration was decreased from 60 to 15 ng per 20 μl. The reaction volumes were varied between 20 and 100 μl to control the concentration of IN in the reaction mixture. The concentrations of all reactants and buffers were held constant. In addition, the concentration of IN was varied while maintaining constant reaction volumes. Preincubation of IN with the donor was on ice for 12 min prior to the addition of the supercoiled target (100 ng of pGEM3 per 20 μl). The ratio of donor to target molecules was one. The strand transfer reactions were carried out for 20 min at 37°C. Generally, 50% of the reaction volumes was subjected to agarose gel electrophoresis. Scale-up reactions were performed to isolate sufficient quantities of donor-target recombinants for BglII digestion, for isolation of the 3.4-kbp full-site products (see Fig. 4; U5-target-U3), for genetic isolation of recombinants after transformation of the ligated 3.4-kbp product into MC1061/P3 cells, and for the sequencing of the donor-target junctions of individually isolated recombinants (see Table 1) (18, 40).
FIG. 4.
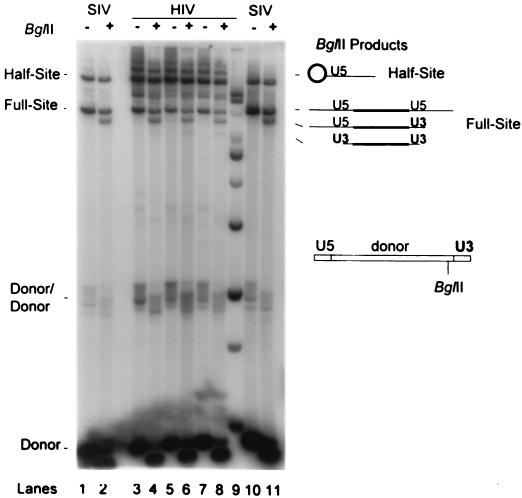
BglII restriction analysis of donor-target products produced by different preparations of HIV-1 IN. The strand transfer products produced by three preparations of HIV-1 IN were subjected to BglII digestion. Lanes 3 and 4, IN coexpressed with protein chaperones and purified through two SP-Sepharose columns; lanes 5 to 8, IN purified identically through heparin-Sepharose that was coexpressed with (lanes 5 and 6) or without (lanes 7 and 8) chaperones. All of the HIV-1 IN preparations were tested at 25 nM in the reaction mixtures. SIV IN concentrations were 25 (lanes 1 and 2) and 50 nM (lanes 10 and 11). The wt donor DNA (P-2; diagram on right) was used. The integration products were divided into two equal samples and were not (−) or were (+) digested by BglII. Left, half-site, full-site, and donor-to-donor products; right, full-site U5-target-U5, U5-target-U3, and U3-target-U3 products produced by BglII digestion. Equivalent quantities of each set were used for comparison purposes. Only the major U5-target half-site product (circle) is apparent on this gel because of the low activity of the U3 LTR end. Lane 9, DNA molecular mass markers as described for Fig. 3.
TABLE 1.
Sequence analysis of donor-target junction sites produced by HIV-1 INa
| Characteristics | No. of recombinants in expt:
|
|
|---|---|---|
| 1 | 2 | |
| Duplications with 5 bp | 23 | 11 |
| Other duplications (0-4 bp) | 2 | 1 |
| Small deletions of 17-20 or 27-30 bp | 4 | 1 |
| Large deletions (>35 bp) | 4 | 2 |
| Total | 33 | 15 |
IN was coexpressed in bacteria with the protein chaperones (GroEL and GroES).
RESULTS
Expression of HIV-1 IN in bacteria.
Recombinant HIV-1 IN was expressed in BL21(DE3) cells without and with coexpression of protein chaperones. Briefly, various quantities of bacterial pellets (0.5 to 7 g wet weight) were processed. The ratios of wet weight bacteria to lysis buffer (4 ml/g) and extraction buffer (2 ml/g) were held constant, which resulted in no apparent differences in either the relative yield or purity of IN with the different-size bacterial preparations (Fig. 2). The expression levels and purity of IN coexpressed with the chaperones and the subsequent lysis, wash, and extraction steps (Fig. 2) were very similar to those for the IN expressed without the chaperones (data not shown). The 1 M NaCl extraction of IN from the IN-DNA pellet generally resulted with IN in the 60 to 70% purity range (Fig. 2, lane 8). IN appears to be stable in the 1 M NaCl extracts at −70°C for at least 6 months.
Purification of HIV-1 IN and strand transfer analysis.
A similarity between the purification procedures used in laboratories for recombinant wt HIV-1 IN is the use of multiple grams wet weight of bacteria to purify milligram quantities of IN. We wanted to avoid nonspecific interactions among recombinant HIV-1 INs which result in aggregation of IN in solution at high protein concentrations (4, 24). We also observed that, upon the use of large amounts of initial 1 M NaCl bacterial extracts containing IN (>3 g [wet weight] of bacteria) (Fig. 2, lane 8) for SP-Sepharose column (5 ml) chromatography, precipitation occurs in the fractions containing the majority of IN due to either nonspecific protein-protein or protein-DNA interactions or both.
To avoid the precipitation of IN with our column purification procedures, we generally used the entire 1 M NaCl extract of IN derived from ∼2 gm or less wet weight of bacteria for the initial SP-Sepharose column step. The column fractions containing the majority of IN (observed by measuring OD280 and performing sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) were pooled and purified further by either an additional passage over another SP-Sepharose column (data not shown) (see Fig. 4, lanes 3 and 4 for IN usage) or by heparin-Sepharose chromatography (see Fig. 3) prior to strand transfer analyses. The last chromatography steps were necessary for further purification of IN, removal of contaminating DNA, and elimination of bacterial DNA endonucleases.
HIV-1 IN expressed in the absence of protein chaperones was purified on a heparin-Sepharose column with the standard chromatography buffer (Fig. 3). Strand transfer analysis demonstrated that HIV-1 IN was capable of producing nearly equivalent amounts of half-site and full-site integration products using the P-2 LTR donor and supercoiled DNA as the target (Fig. 3A and B). PhosphorImager analysis shows that ∼4 to 9% of the donor was inserted into these products in the peak fractions (fractions 41 to 43) (Fig. 3A). SDS-PAGE shows that IN is nearly homogeneous (Fig. 3C). With fractions 41 to 43, the concentrations of IN in the reaction mixtures were calculated to be ∼20, 16, and 16 nM, respectively. A significant amount of donor was also inserted by IN into other donor molecules (∼20% for donor-to-donor insertion) with these same fractions (Fig. 3B). In comparison, purified SIV IN (Fig. 3B, lane 2) at 100 nM incorporates approximately 11 and 23% of the donor into half-site and full-site products, respectively, with supercoiled DNA as the target (19).
In summary, recombinant HIV-1 IN can be purified at low protein concentrations while remaining highly active and possessing the ability to perform full-site integration without the addition of viral or cellular cofactors.
Independence of adding either Zn2+ or Mg2+ to buffers for purifying highly active HIV-1 IN capable of full-site integration.
It was previously shown that Zn2+, Mg2+, and nonionic detergents in the purification buffers differentially affected the interactions between recombinant HIV-1 IN subunits at low concentrations (45 to 200 nM), subsequently affecting their half-site integration properties (11, 28). We tested whether different solution conditions during purification affected the stability and the ability of recombinant HIV-1 IN to perform strand transfer. Our standard chromatography buffer contains 50 mM HEPES, 1 mM dithiothreitol, 1 mM EDTA, and 10 mM MgSO4 with no nonionic detergents present (33). Recombinant HIV-1 IN was passed over two SP-Sepharose columns without MgSO4 but with 10% glycerol in the final column buffer. SDS-PAGE analysis also shows that IN was purified to near homogeneity (data not shown). The SP-Sepharose-purified enzyme (118 μg/ml) (Fig. 4, lanes 3 and 4) was stable for over a year and, in reaction mixtures with two different preparations of HIV-1 IN purified through heparin-Sepharose, had strand transfer activities at 25 nM that were comparable (Fig. 4, lanes 5 to 8). These two preparations of IN were purified with the standard chromatography buffer. IN was coexpressed with chaperones (Fig. 4, lanes 5 and 6) or without them (lanes 7 and 8). The concentrations of the stored enzymes (∼3 months each) were 66 and 101 μg/ml, respectively. The strand transfer activities of the purified IN preparations were also stable over several freeze-thaw cycles.
In summary, the stability of recombinant HIV-1 IN and the ability of IN to efficiently perform half-site and full-site integration at low concentrations appear independent of having added Zn2+, Mg2+, or glycerol in the purification buffers.
Fidelity of the full-site integration products for 5-bp host site duplications.
DNA restriction enzyme analysis of the donor-target products produced by HIV-1 IN shows that the U5 terminus is used in preference to the U3 terminus for both the half-site and full-site integration products (Fig. 4, lanes 3 to 8) (4, 6, 18, 31). With the P-2 LTR donor as the substrate, the donor-target integration products produced by the three different purified HIV-1 IN preparations gave the predicted BglII enzyme products (Fig. 4) (18). Digestion of integration products by BglII produced by all three preparations of HIV-1 IN using the LTR donor (H-5) containing two U5 ends also gave the predicted pattern (data not shown) (17, 18). SIV IN at 25 (Fig. 4, lanes 1 and 2) and 50 nM (Fig. 4, lanes 10 and 11) is shown for comparison.
The fidelity of virion (6, 17, 18) and recombinant HIV-1 IN with cellular or viral protein cofactors (6, 22) for producing the 5-bp host site duplications is ∼70%. We wanted to determine if the fidelity of the 5-bp host site duplications was increased if HIV-1 IN was expressed in the presence of protein chaperones (Fig. 4, lane 3 and 4). Table 1 demonstrates the results of two independent experiments involving the sequencing of recombinants produced by HIV-1 IN coexpressed with chaperones. The linear LTR donor (H-5) containing two U5 ends was used for these sequencing experiments (17, 18). The fidelity for producing the 5-bp host site duplications was not increased for HIV-1 IN in comparison to results obtained with HIV-1 virions (6, 17) or recombinant HIV-1 IN with either cellular or viral cofactors (6, 22). Sequence analysis of the host site duplications produced by HIV-1 IN not expressed with chaperones was not performed.
In summary, the results show that HIV-1 IN under appropriate purification and assay conditions has the capacity to efficiently produce the correct full-site integration products without the addition of cellular or viral cofactors. The results also show that coexpression with chaperones does not improve the fidelity of HIV-1 IN for producing 5-bp host site duplications.
Specific activities of the different HIV-1 IN preparations for full-site integration.
We wanted to quantitatively determine if HIV-1 INs expressed with and without protein chaperones have similar specific activities for half-site and full-site integration. Various concentrations of both IN preparations (5 to 50 nM) were assembled with the P-2 LTR donor at 0°C and assayed for strand transfer activities (Fig. 5). The quantities of the half-site and full-site products produced by both IN preparations appear to be similar with respect to protein concentrations. At 30 nM IN, the calculated IN dimer-to-donor end molar ratio is 7 to 1. With both preparations of HIV-1 IN at ∼30 nM, levels of incorporation of donor into both products were ∼12 and 7% for half-site and full-site products, respectively (Fig. 5, lanes 7 and 12). With SIV IN at 25 nM, levels of incorporation for the half-site and full-site products are 3 and 9%, respectively (Fig. 5, lane 2). Generally, under optimum strand transfer conditions for HIV-1 IN, the total amount of input donor used for all strand transfer products is ∼25 to 30% in 20 min, suggesting that a large percentage of IN subunits are enzymatically active (Fig. 5; data not shown). The lack of significant strand transfer activities (Fig. 5, lanes 3 and 9) below ∼8 nM IN in the reaction mixtures is a reproducible result suggesting that a critical concentration of IN subunits is needed for initiation of strand transfer events. Further quantitative analyses of three independent titration experiments using both IN preparations suggest that IN expressed with chaperones is only slightly better than IN not expressed with chaperones for full-site rather than half-site integration (data not shown).
FIG. 5.
Quantitative comparison of strand transfer specific activities of HIV-1 IN expressed without and with protein chaperones. HIV-1 INs purified as described for Fig. 3 (heparin-Sepharose, −chaperones; SP-Sepharose, +chaperones) were simultaneously analyzed for strand transfer efficiencies. The wt donor DNA (P-2) was used. The reaction volume was 100 μl, and one-half of each sample was subjected to 1.5% agarose gel electrophoresis. Equivalent radioactive counts were loaded per lane. Lanes 1 and 2, no IN and SIV IN (25 nM), respectively; lanes 3 to 7, HIV-1 IN expressed without chaperones at 5.2, 10.4, 15.6, 20.8, and 31.2 nM IN, respectively; lane 8, molecular mass markers as described for Fig. 3; lanes 9 to 13, IN coexpressed with chaperones at 8.7, 17.4, 26.1, 34.8, and 52.2 nM, respectively. The wet gel was dried, exposed to X-ray film (6 h without an intensifying screen), and analyzed by a PhosphorImager. The half-site, full-site, and donor-to-donor products and the input donor are indicated on the left.
We investigated the effect of Zn2+ on recombinant HIV-1 IN full-site integration, which had previously been shown to affect the structural and catalytic functions of IN (5, 26, 28, 43). Zn2+ promotes the multimerization of IN subunits and slightly stimulates the Mg2+-dependent 3′ OH processing and half-site strand transfer reactions. Figure 6 shows that Zn2+ also stimulates the half-site and full-site integration reactions of HIV-1 IN (lanes 3 to 6) in comparison to reactions performed without Zn2+ (lanes 10 to 13). At 34 nM IN with Zn2+ (Fig. 6, lane 6), the percentages of donor incorporated into half-site and full-site products were 15 and 8%, respectively. Further quantitative analysis shows that Zn2+ stimulates both reactions from ∼20 to 40%, independent of whether IN was expressed with (compare lanes 5 and 6 to lanes 12 and 13) or without (compare lanes 3 and 4 to lanes 10 and 11) protein chaperones. SIV IN strand transfer activities were only slightly stimulated (Fig. 6, compare lanes 2 to 9). The results show that Zn2+ also enhances the ability of HIV-1 IN to perform full-site integration as well as other strand transfer activities.
FIG. 6.
Effect of Zn2+ on recombinant HIV-1 IN full-site integration. HIV-1 INs coexpressed without protein chaperones (−ESL; purified by heparin-Sepharose) and with chaperones (+ESL; purified by SP-Sepharose) were tested for Zn2+ activation. The wt donor DNA (P-2) was used. Lanes 1 to 6, 25 μM Zn2+; lanes 8 to 13, no Zn2+. Control lanes 1 and 8, no IN; lanes 2 and 9, SIV IN at 25 nM; lanes 3 and 10 and 4 and 11, −ESL IN at 11 (lanes 3 and 10) and 22 nM (lanes 4 and 11); lanes 5 and 12 and 6 and 13, +ESL IN at 17 (lanes 5 and 12) and 34 nM (lanes 6 and 13). The half-site, full-site, and donor-to-donor products as well as the input donor are indicated on the left. The molecular mass markers (lane 7) are described in the legend for Fig. 3.
We also determined that the optimum NaCl concentration for assembly and full-site integration by recombinant HIV-1 IN was ∼150 mM (data not shown). The titration experiments showed that recombinant HIV-1 IN at 32 nM displayed nearly equal responses to NaCl between 110 and 170 mM for both half-site and full-site integration activities. Below ∼80 mM NaCl, only a minor population of full-site integration products are produced. In comparison, ∼40 mM NaCl was observed to be optimum for half-site and full-site integration with nonionic detergent HIV-1 lysates (6, 18) or recombinant HIV-1 IN (6, 22). In addition, results from expression and purification of recombinant HIV-1 IN (strain HXB2) in bacteria showed that HIV-1 IN derived from another wt HIV-1 IN gene possesses very similar specific activities for half-site and full-site integration (data not shown).
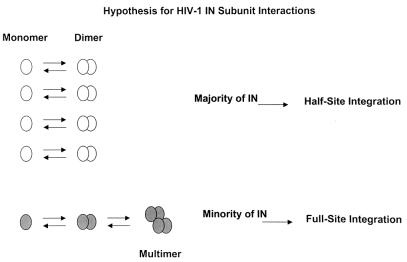
In summary, the results suggest that HIV-1 IN possesses similar specific activities for performing full-site integration when purified at low protein concentrations with different buffer systems. The specific activities of HIV-1 IN preparations appear equivalent to those for SIV IN with respect to the quantity of full-site integration product produced. However, significant quantities of half-site integration products are produced by the various HIV-1 IN preparations in comparison to what is found for SIV IN, suggesting that a majority of the HIV-1 IN subunits are still not assembling correctly on the viral DNA ends to perform full-site integration (see the model in Fig. 7).
FIG. 7.
Model depicting the interactions of recombinant HIV-1 IN subunits in solution. White circles, IN monomers that interact to form dimers. This population of IN subunits possibly forms the majority of IN in recombinant HIV-1 IN preparations. The majority of these subunits can perform half-site integration but not full-site integration due to their inability to assemble two viral DNA ends correctly. Gray circles, IN monomers possessing the ability to form dimers and multimers which can correctly assemble on two viral DNA ends allowing full-site integration.
DISCUSSION
Recombinant wt HIV-1 IN purified from bacteria has the capacity to promote the half-site and full-site integration reactions nearly equivalently. The efficient concerted insertion of two LTR DNA ends into a circular target by recombinant HIV-1 IN without the addition of viral or cellular protein cofactors suggests that IN is not significantly defective for promoting full-site integration. Further improvement in the assembly of nucleoprotein complexes capable of full-site integration may be related to the state of aggregation of HIV-1 IN, as suggested by the avoidance of high IN concentrations during both its purification and its use in the integration assay. Recombinant HIV-1 IN appears not to possess major protein-folding defects, as shown by our protein chaperone coexpression studies. Our reconstitution studies do not exclude the possibility that other proteins assist IN with integration in vivo.
With LTR donor substrates similar in size to those described in this report, recombinant HIV-1 IN either failed to perform full-site integration (8, 28) or incorporated <1% of the donor DNA into full-site integration products after 1- (6) or 2-h (22) incubations at 37°C. We demonstrated that recombinant HIV-1 IN at similar protein concentrations (∼30 nM) is capable of incorporating 8 to 10% of the donor substrate into target DNA, producing full-site integration products in 20 min. The fidelity of the 5-bp host site duplication for full-site integration catalyzed by recombinant HIV-1 IN is comparable to that observed with nonionic detergent lysates of HIV-1 virions (6, 17) or recombinant HIV-1 IN enhanced by cellular or viral cofactors (6, 22). The reported amounts of half-site integration products produced by recombinant HIV-1 IN also vary significantly from ∼2 to 20% (6, 8, 22, 28). The total amounts of circular half-site and donor-to-donor integration products produced by recombinant HIV-1 IN under optimum conditions are ∼25% (Fig. 5 and 6). As discussed later, the majority of IN subunits are apparently highly capable of performing half-site integration but only a minority are capable of full-site integration (Fig. 7, model). Our report suggests that the concentrations of IN during purification and in the reaction mixture play an important role in the assembly of IN-DNA complexes favorable to full-site integration. It is possible that nonspecific aggregation of IN results in incorrect nucleoprotein complexes for full-site integration (9).
What are the factors that influence the ability of IN to assemble on two viral DNA ends allowing the nucleoprotein complexes to perform full-site integration? Detailed biochemical analyses of recombinant HIV-1 IN (45 to 200 nM) derived from different purified preparations and in different solutions required for strand transfer activities have given critical insights into factors affecting the multimeric state of recombinant IN (11, 28). Nonionic detergents appear to promote dissociation of IN subunits to monomers, while Zn2+ promotes multimerization to dimers and tetramers (11, 28), protein states required for strand transfer activities (4, 12, 24, 26, 36, 43). The minimum IN structure for 3′ OH processing and half-site integration is a dimer (4), with the predicted minimum structure of one to two tetramers for full-site integration (5, 9, 20, 39).
Several factors appear to impact the ability of recombinant wt HIV-1 IN in this report to perform efficient full-site integration. The concentration of HIV-1 IN upon purification was kept in the 125 μg/ml or less range to prevent apparent aggregation of IN in solution. Our purification procedure (33) also lacks nonionic detergents, possibly allowing dimers and tetramers to be formed at low nanomolar concentrations (11, 28). Divalent metal ion Mg2+ can be removed during the second SP-Sepharose chromatography step for HIV-1 IN, and it still remains fully active, even when stored in the presence of 1 mM EDTA (Fig. 4, lane 3; Fig. 5, lanes 9 to 13). The lack of added Zn2+ during purification does not appear to be critical for IN stability although it enhances strand transfer activities (Fig. 6) (5, 26, 28, 43). The multimeric state of purified IN in this report is unknown. However, a minimum concentration of IN (∼8 nM) in the reaction mixture (Fig. 5, lanes 3 and 9) is required for detectable strand transfer activities, suggesting that multimerization of IN on DNA ends is required for all strand transfer events. The actual concentrations of IN in our reaction mixtures are unknown because of the molecular crowding effects of polyethylene glycol (10%), which significantly enhances full-site integration activity in vitro (3, 6, 40). In summary, our data suggest that recombinant HIV-1 IN can be purified at low concentrations in different buffers without a major effect on its ability to perform full-site integration.
Figure 7 predicts a general assembly process of IN subunits in solution (11, 28). Conformational microheterogeneity of IN subunits may produce different populations of assembled subunits. The model may define the population of IN structures observed in our preparations as determined by the population of half-site and full-site integration products produced by these preparations (Fig. 4 to 6). The majority of IN subunits (as dimers) (Fig. 7, top) appear to be highly active for performing half-site integration because ∼25% of the input donor is incorporated in 20-min reactions. It is currently not possible to define whether all of the IN molecules are enzymatically active because of the high nonspecific DNA binding capabilities of IN (4). A minority population of IN (Fig. 7, bottom) in our preparations appear to have the capability of forming the higher-order multimers predicted to be required for assembly of IN on two viral DNA ends for full-site integration. The nonspecific interactions of defective dimers (Fig. 7, top) with correctly formed dimers (Fig. 7, bottom), either in solution or in the assembly of nucleoprotein complexes, may significantly influence the specific activities of different IN preparations for full-site integration. This minority population of HIV-1 IN subunits (Fig. 7, bottom) prefer ∼150 mM NaCl for correct assembly of two viral DNA ends, significantly higher than the ∼40 mM NaCl previously reported for HIV-1 virion lysates (6, 18) and recombinant IN (6, 22). It is not possible to accurately compare the specific activities of recombinant HIV-1 IN to those of virion lysates because the lysates contain nonionic detergents, which appear to be detrimental for correct IN subunit interactions (11, 28). However, under optimum conditions, recombinant HIV-1 IN appears to produce larger quantities of all integration products (Fig. 5 and 6) than virion lysates by severalfold (6, 17, 18) (data not shown).
In summary, further studies are necessary to improve the population of IN subunits capable of assembling nucleoprotein complexes possessing the capacity to perform full-site integration. Subsequent site-directed mutagenesis of HIV-1 IN may allow further insights into the multiple structure-functional relationships necessary for IN (2, 4, 7, 20) to mediate correct assembly of nucleoprotein complexes capable of full-site integration.
Acknowledgments
This work was supported by United States Public Health Services grant AI31334 from NIAID and in part by grant 02750-30-RGT from the American Foundation for AIDS Research (amfAR).
We thank T. Mueser and C. C. Hyde for development of the procedure to purify RSV PrA IN (33) that was used to purify recombinant HIV-1 IN. We thank Jim Taylor for his studies expressing the HXB2 IN gene, Daria Hazuda for purified SIV IN, and Carl Frieden for purified GroEL and GroES.
REFERENCES
- 1.Aiyar, N., P. Hindmarsh, A. M. Skalka, and J. Leis. 1996. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J. Virol. 70:3571-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, H. E., H. Chen, and A. Engelman. 1999. Structure-based mutagenesis of the HIV-1 DNA attachment site: effects on integration and cDNA synthesis. J. Virol. 73:9011-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, P., B. Bowerman, H. Varmus, and J. Bishop. 1987. Correct integration of retroviral DNA in vitro. Cell 49:347-356. [DOI] [PubMed] [Google Scholar]
- 4.Brown, P. O. 1998. Integration, p. 161-203. In J. M. Coffin, S. J. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 5.Cai, M., R. Zheng, M. Caffrey, R. Craigie, G. M. Clore, and A. M. Gronenborn. 1997. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat. New Biol. 4:567-577. [DOI] [PubMed] [Google Scholar]
- 6.Carteau, S., R. J. Gorelick, and F. D. Bushman. 1999. Coupled integration of human immunodeficiency virus type 1 cDNAs ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 73:6670-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., S. Q. Wei, and A. Engelman. 1999. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type 1 intrasome. J. Biol. Chem. 274:17358-17364. [DOI] [PubMed] [Google Scholar]
- 8.Cherepanov, P., D. Surratt, J. Toelen, W. Pluymers, J. Griffith, E. De Clercq, and Z. Debyser. 1999. Activity of recombinant HIV-1 integrase on mini-HIV DNA. Nucleic Acids Res. 27:2202-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craigie, R. 2001. HIV integrase, a brief overview from chemistry to therapeutics. J. Biol. Chem. 276:23213-23216. [DOI] [PubMed] [Google Scholar]
- 10.Craigie, R., T. Fujiware, and F. D. Bushman. 1990. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 62:829-837. [DOI] [PubMed] [Google Scholar]
- 11.Deprez, E., P. Tauc, H. Lev, J.-F. Mouscadet, C. Auclair, and J.-C. Brochon. 2000. Oligomeric states of the HIV-1 integrase as measured by time-resolved fluorescence anisotropy. Biochemistry 39:9275-9284. [DOI] [PubMed] [Google Scholar]
- 12.Engelman, A., and R. Craigie. 1992. Identification of conserved amino acids residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66:6361-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farnet, C., and F. D. Bushman. 1997. HIV-1 cDNA integration: requirement of HMG 1(Y) protein for function of preintegration complexes in vitro. Cell 88:483-492. [DOI] [PubMed] [Google Scholar]
- 14.Farnet, C. M., and W. A. Haseltine. 1991. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 65:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzgerald, M., A. C. Vora, W. Zeh, and D. P. Grandgenett. 1992. Concerted integration of viral DNA termini by purified avian myeloblastosis virus integrase. J. Virol. 66:6257-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goloubinoff, P., A. A. Gatenby, and G. Lorimer. 1989. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature 337:44-47. [DOI] [PubMed] [Google Scholar]
- 17.Goodarzi, G., R. Chiu, K. Brackmann, K. Kohn, Y. Pommier, and D. P. Grandgenett. 1997. Host site selection for concerted integration by human immunodeficiency virus type-1 virions in vitro. Virology 231:210-217. [DOI] [PubMed] [Google Scholar]
- 18.Goodarzi, G., G. Im, K. Brackmann, and D. P. Grandgenett. 1995. Concerted integration of retrovirus-like DNA by HIV-1 integrase. J. Virol. 69:6090-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodarzi, G., M. Pursley, P. Felock, M. Witmer, D. Hazuda, K. Brackmann, and D. P. Grandgenett. 1999. Efficiency and fidelity of full-site integration reactions using recombinant simian immunodeficiency virus integrase. J. Virol. 73:8104-8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heuer, T., and P. O. Brown. 1998. Photo-crosslinking-studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry 37:6667-6678. [DOI] [PubMed] [Google Scholar]
- 21.Hindmarsh, P., M. Johnson, R. Reeves, and J. Leis. 2001. Base-pair substitutions in avian sarcoma virus U5 and U3 long terminal repeat sequences alter the process of DNA integration in vitro. J. Virol. 75:1132-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hindmarsh, P., T. Ridky, R. Reeves, M. Andrake, A. M. Skalka, and J. Leis. 1999. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J. Virol. 73:2994-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, Y.-S., and D. T. Chuang. 1999. Mechanisms of GroEL/GroES-mediated folding of a large 86-kDa fusion polypeptide in vitro. J. Biol. Chem. 274:10405-10412. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins, T. M., A. Engelman, R. Ghirlando, and R. Craigie. 1996. A soluble active mutant of HIV-1 integrase. J. Biol. Chem. 271:7712-7718. [DOI] [PubMed] [Google Scholar]
- 25.Katz, R. A., J. Merkel, J. Kulkosky, J. Leis, and A. M. Skalka. 1990. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell 63:87-95. [DOI] [PubMed] [Google Scholar]
- 26.Lee, S. P., J. Xiao, J. R. Knutson, M. Lewis, and M. Han. 1997. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry 36:173-180. [DOI] [PubMed] [Google Scholar]
- 27.Lee, Y., and J. Coffin. 1991. Relationship of avian retrovirus DNA synthesis to integration in vitro. Mol. Cell. Biol. 11:1419-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leh, H., P. Brodin, J. Bischerour, E. Deprez, P. Tauc, J.-C. Brochon, E. LeCam, D. Coulaud, C. Auclair, and J.-F. Mouscadet. 2000. Determinants of Mg2+-dependent activities of recombinant human immunodeficiency virus type 1 integrase. Biochemistry 39:9285-9294. [DOI] [PubMed] [Google Scholar]
- 29.Li, L., C. M. Farnet, W. F. Anderson, and F. D. Bushman. 1998. Modulation of activity of Moloney murine leukemia virus preintegration complexes by host factors in vitro. J. Virol. 72:2125-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maier, G., E. Manakova, and H. Heumann. 2000. Effect of Escherichia coli chaperonin GroELS on heterologously expressed human immunodeficiency virus type 1 reverse transcriptase in vivo and in vitro. Appl. Biochem. Biotechnol. 87:103-115. [DOI] [PubMed] [Google Scholar]
- 31.Masuda, T., M. J. Kuroda, and S. Harada. 1998. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. J. Virol. 72:8396-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCord, M., R. Chiu, A. C. Vora, and D. P. Grandgenett. 1999. Retrovirus DNA termini bound by integrase communicate in trans for full-site integration in vitro. Virology 259:392-401. [DOI] [PubMed] [Google Scholar]
- 33.McCord, M., J. Stahl, T. C. Mueser, C. C. Hyde, A. C. Vora, and D. P. Grandgenett. 1998. Purification of recombinant Rous sarcoma virus integrase possessing physical and catalytic properties similar to virion-derived integrase. Protein Expr. Purif. 14:167-177. [DOI] [PubMed] [Google Scholar]
- 34.McKnight, S. 1977. A colorimetric method for the determination of submicrogram quantities of protein. Anal. Biochem. 78:86-92. [DOI] [PubMed] [Google Scholar]
- 35.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petit, C., O. Schwartz, and F. Mammano. 1999. Oligomerization within virions and subcellular localization of human immunodeficiency virus type 1 integrase. J. Virol. 73:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman, P., and J. Fyfe. 1990. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl. Acad. Sci. USA 87:5119-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh, I., R. Crowley, and P. Brown. 1997. High-resolution functional mapping of a cloned gene by genetic footprinting. Proc. Natl. Acad. Sci. USA 94:1304-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vora, A. C., and D. P. Grandgenett. 2001. DNase protection analysis of retrovirus integrase at the viral DNA ends for full-site integration in vitro. J. Virol. 75:3556-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vora, A. C., M. McCord, M. Fitzgerald, R. B. Inman, and D. P. Grandgenett. 1994. Efficient concerted integration of retrovirus-like DNA in vitro by avian myeloblastosis virus integrase. Nucleic Acids Res. 22:4454-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vora, A. C., M. McCord, G. Goodarzi, S. J. Stahl, T. C. Mueser, C. C. Hyde, and D. P. Grandgenett. 1997. Avian retrovirus U3 and U5 DNA inverted repeats: role of nonsymmetrical nucleotides in promoting full-site integration by purified virion and bacterial recombinant integrases. J. Biol. Chem. 272:23938-23945. [DOI] [PubMed] [Google Scholar]
- 42.Wei, S. Q., K. Mizuuchi, and R. Craigie. 1997. A large nucleoprotein assembly at the viral DNA mediates retroviral DNA integration. EMBO J. 16:7511-7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng, R., T. M. Jenkins, and R. Craigie. 1996. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc. Natl. Acad. Sci. USA 93:13659-13664. [DOI] [PMC free article] [PubMed] [Google Scholar]