Abstract
We attempted to extend the lifespan of CD34+ stem/progenitor cells in human cord blood (CB) by transduction with entiviral vectors carrying the human telomerase catalytic subunit (hTERT) and/or the human papillomavirus type 16 (HPV16) E6 and E7 oncogenes. We found that hTERT was incapable of prolonging the replicative capacity of CB cells maintained under serum-free conditions in the presence of stem cell factor, Flt3 ligand, thrombopoietin, and inter-leukin-3 beyond 4 months (n = 3). However, transduced CB cells cultured in the same cytokine cocktail constitutively expressing HPV16 E6/E7 alone (n = 2) or in concert with hTERT (n = 9) continued to proliferate, giving rise to permanent (>2 years) cell lines with a CD45+CD34-CD133+/-CD44+CD235a+CD71+CD203+CD33+CD13+ myeloerythroid/mast cell progenitor phenotype. Notably, CB cell cultures expressing only HPV16 E6/E7 went through a crisis period, and the resulting oligoclonal cell lines were highly aneuploid. By comparison, the CB cell lines obtained by coexpression of HPV16 E6/E7 plus hTERT exhibited near-diploid karyotypes with minimal chromosomal aberrations, concomitant with stabilization of telomere length, yet were clonally derived. The immortalized E6/E7 plus hTERT—expressing CB cells were not tumorigenic when injected intravenously or subcutaneously into sublethally irradiated immunodeficient nonobese diabetic/severe combined immunodeficient mice but could be converted to a malignant state by ectopic expression of a v-H-ras or BCR-ABL oncogene. These findings provide new insights into the mechanisms governing the senescence checkpoint of primitive human hematopoietic precursors and establish a paradigm for studies of the multistep process of human leukemogenesis.
Keywords: Cord blood, CD34, Human telomerase catalytic subunit Human papillomavirus E6/E7 oncogenes, v-H-ras, BCR-ABL
Introduction
Human embryonic stem cells (hESCs) circumvent cellular senescence by expressing telomerase reverse transcriptase (hTERT) [1]. hTERT is the catalytic subunit of telomerase, a specialized ribonucleoprotein complex that is responsible for adding telomeric DNA (repetitive TTAGGG sequences) to the ends of chromosomes to prevent shortening during replication [2]. In this context, expression of exogenous hTERT in certain normal human somatic cell types stabilizes telomere length and allows indefinite growth [3-5]. In particular, ectopic expression of hTERT has been reported to extend the lifespan of human mesenchymal stem cells and human neural progenitor cells [4, 5]. Candidate human hematopoietic stem cells express relatively high levels of hTERT [6], and telomere length analysis of human hematopoietic stem/progenitor cell subsets supports the hypothesis that cells with the greatest proliferative potential have the longest telomeres [7]. Conversely, patients with aplastic anemia have short telomeres, and mutations in telomerase have been identified as the cause of hematopoietic failure [8, 9].
Previous efforts to extend the replicative capacity of human CD34+ cord blood (CB) cells by retroviral-mediated expression of the hTERT gene were unsuccessful, perhaps due to transgene silencing [10]. Therefore, to further investigate whether hTERT could be used to immortalize hematopoietic stem/progenitor sub-populations in CB samples, we used a self-inactivating (SIN) lentiviral vector backbone that we developed that directs persistent high-level expression of transgenes in hESCs and primitive human hematopoietic precursors [11, 12]. Besides progressive telomere shortening, it is now apparent that human cells can undergo senescence in response to various types of stress [13]. Regardless of the senescence-initiating stimuli, the signaling pathways triggered converge to varying extents on the p53 and retinoblastoma (Rb) tumor suppressors and the cyclin-dependent kinase inhibitors p21WAF1/CIP1 and p16INK4a. Because other investigators reported that human mesenchymal stem cells could not be immortalized by hTERT alone but required combinatorial expression of the human papillomavirus type 16 (HPV16) E6 and E7 genes [14], which accelerate the degradation of p53 and Rb, respectively [15], we also attempted to prolong the lifespan of CB progenitors by transduction with an HPV16 E6/E7 lentiviral vector, separately and in conjunction with the hTERT lentiviral vector.
Materials and Methods
HPV16 E6/E7 and hTERT Lentiviral Vectors
The HIV-1—based SIN lentiviral vectors used in this study were derived from the SINF-MU3-W-S vector backbone described previously [12], which contains the central polypurine tract of HIV-1 (which creates a central DNAflap) that functions as a cis-acting facilitator of HIV-1 DNA nuclear import, an internal promoter consisting of the U3 region of the murine stem cell virus (MSCV) long-terminal repeat (MU3), the woodchuck hepatitis virus post-transcriptional regulatory element (W) for enhanced expression of transgenes lacking introns, and the human inter-feron-β upstream scaffold attachment region (S) as a potential chromatin domain boundary element. The HPV16 E6/E7 coding region was polymerase chain reaction amplified as a 0.8-kb fragment from the pHPV-16 plasmid (American Type Culture Collection, Manassas, VA, http://www.atcc.org) and inserted upstream of an encephalomyocarditis virus internal ribosome entry site (IRES)—yellow fluorescent protein (YFP) gene cassette into SINF-MU3-W-S to generate SINF-MU3-E6E7-IRES-YFPW-S. The primers used were 5′-CCGGAATTCGCCACCATGCACCAAAAGAGAAC-3′ (sense) and 5′-CCGGAATTCAGCCATGGTAGATTATG-3′ (antisense). The E6/E7 sequence in SINF-MU3-E6E7-IRES-YFP-W-S was verified using the Big-Dye Terminator Cycle Sequencing Kit and an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com). SINF-MU3-hTERT-IRES-GFPW-S was generated by inserting a 3.4-kb EcoRI-SalI fragment containing the hTERT cDNA from the plasmid pCI-Neo-hTERT (kindly provided by Dr. Robert Weinberg, The Whitehead Institute for Biomedical Research, Cambridge, MA) [16] upstream of an IRES—green fluorescent protein (GFP) gene cassette into SINF-MU3-W-S.
Vesicular stomatitis virus (VSV)—G glycoprotein-pseudo-typed lentiviral vector particles were produced by transiently transfecting the lentiviral vector plasmids (15 μg), the packaging plasmid pCMVΔR8.91 (10 μg), and the VSV-G protein envelope plasmid pMD.G (5 μg) into subconfluent human embryonic kidney 293T cells by the calcium phosphate precipitation method; high-titer vector stocks were prepared by ultracentrifugation (×45,000g, 90 minutes), titered on human fibrosarcoma HT1080 cells, and assayed for the presence of replication-competent virus as described previously [17].
CD34+ CB Cell Isolation, Transduction, and Culture
Human umbilical CB samples were obtained after informed consent in conformity with an Institutional Review Board—approved protocol. Mononuclear cells were isolated by density-gradient centrifugation on Ficoll-Paque (Amersham Biosciences, Piscataway, NJ, http://www.amersham.com). CD34+ cells were purified from the mononuclear cells by super paramagnetic microbead selection using a varioMACS CD34 progenitor cell isolation kit (Miltenyi Biotec, Auburn, CA, http://www.miltenyibiotec.com), according to the manufacturer's instructions. The CD34+ cells were cultured in 24-well nontissue culture-treated plates coated with 2 μg/cm2 RetroNectin recombinant fibronectin fragment (Takara Mirus Bio, Madison, WI, http://www.takaramirusbio.com) at a density of 1 × 106 cells per well. The cells were prestimulated for 24 hours in X-VIVO-15 serum-free medium (Fisher Scientific, Pittsburgh, PA, http://www.fisherscientific.com) supplemented with 10% BIT 9500 (Stem Cell Technologies, Vancouver, British Columbia, Canada, http://www.stemcell.com), 100 ng/ml stem cell factor (SCF), 100 ng/ml Flt3 ligand (FL), and 20 ng/ml thrombopoietin (TPO), with or without 20 ng/ml interleukin-3 (IL-3) (all from PeproTech, Rocky Hill, NJ, http://www.peprotech.com). The cells were transduced for 24 hours with lentiviral vector particles (2 × 106 TU/ml; multiplicity of infection, 2) in the presence of 6 μg/ml polybrene or 5 μg/ml protamine sul-fate (Sigma-Aldrich, St. Louis, http://www.sigmaaldrich.com) [12]. Fresh medium was added, and the cells were cultured for an additional 48-72 hours before cell sorting.
KG1a human acute myeloid leukemia cells (ATCC CCL-246.1) express the CD34 hematopoietic stem/progenitor cell antigen and approximate a human hematopoietic blast cell arrested at a stage of differentiation at which lineage restrictions to the myeloid and lymphoid pathways occur [18]. KG1a cells constitutively expressing GFP (KG1a-GFP) were described previously [12]; the cells were cultured in Iscove's modified Dulbecco's medium (Invitrogen, Carlsbad, CA, http://www.invitrogen.com) supplemented with 2 mM L-glutamine (Invitrogen) and 10% heat-inactivated fetal bovine serum (Fisher Scientific). All cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Fluorescence-Activated Cell Sorting and Analysis
Lentiviral vector—transduced CD34+ CB cells were pelleted, washed, and resuspended in phosphate-buffered saline plus 2% bovine serum albumin, and GFP+/YFP+ cells were sorted as bulk populations on a FACSVantage SE/FACSDiVa (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com) equipped with an Innova 70C-Spectrum argon-ion laser (Coherent Inc., Santa Clara, CA, http://www.coherentinc.com) tuned to 488 nm. The YFP and GFP signals were separated with a 525-nm shortpass dichroic filter and collected with a 550/30-nm bandpass filter and a 510/20-nm bandpass filter, respectively. Cell sorting, data acquisition, and analysis were performed as previously described [19-21].
GFP+/YFP+ CB cell lines (5 × 105 cells) were immunophenotyped essentially as described [19, 20], with saturating concentrations of allophycocyanin (APC)— or R-phycoerythrin (PE)—conjugated monoclonal antibodies recognizing the following human hematopoietic cell-surface antigens: CD2-PE, CD3-APC, CD11b-APC, CD13-APC, CD14-APC, CD15-PE, CD16-PE, CD19-PE, CD24-PE, CD33-PE, CD34-APC, CD38-APC, CD41a-APC, CD44-PE, CD45-APC, CD56-APC, CD71-PE, CD184 (CXCR4)-PE, CD235a (glycophorin A)-PE and HLADR-APC (all from BD Biosciences Pharmingen, San Diego, http://www.bdbiosciences.com/pharmingen), CD133-APC (Miltenyi Biotec), and CD203c-PE (Beckman Coulter, Inc., Fullerton, CA, http://www.beckmancoulter.com). Corresponding isotype controls were used for each analysis. Flow cytometry data were acquired on a BD LSR instrument and analyzed using Cell-Quest software (BD Biosciences).
Southern Blot Analysis
Southern blotting was carried out by standard procedures as described previously [12]. Structural integrity of the HPV16 E6/E7 and hTERT coding regions was assessed by digestion of genomic DNA (10 μg) with EcoRI plus BglII, the combination of which cleaves sites flanking both of the transgenes. Probes used were a 0.8-kb EcoRI fragment containing the E6/E7 coding region, a 1.2-kb NcoI-BglII fragment containing the 3′ coding region of hTERT, a 0.8-kb fragment of pEGFP-1 as a GFP/YFP-specific probe, and a 1.3-kb EcoRI-AccI fragment of pC△j-989 to detect the endogenous human BCL2 gene [12].
Telomerase Activity and Telomere Length Analysis
Telomerase activity was assessed by the telomere repeat amplification protocol using the TRAPeze XL Telomerase Detection Kit (Chemicon International, Temecula, CA, http://www.chemicon.com) according to the manufacturer's protocol. Telomere lengths were determined by TeloTAGGG Telomere Length Assay (Roche Applied Science, Indianapolis, http://www.roche-applied-science.com) Southern blot analysis of terminal restriction fragments following digestion of genomic DNA with HinfI and RsaI.
Immunoprecipitation and Western Blot Analysis
Immunoprecipitations and Western blotting were performed as described [22]. In brief, cells were lysed on ice in RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.5% Na-deoxycholate, 0.1% SDS, 1 mM PMSF, 1mM benzamidine, 10 μg/ml aprotinin, 10 μg/ml leupeptin) for 30 minutes. Insoluble materials were removed by centrifugation at 14,000 rpm at 4°C for 10 minutes. Whole-cell lysates (500 μg) were precleared with protein A or protein G beads (Invitrogen) for rabbit or mouse antibodies, respectively, for 1 hour, incubated at 4°C with 1-2 μg of antibody for 1-2 hours, and then incubated with protein A or protein G beads for an additional hour. Immunoprecipitates were washed three times with RIPA buffer and once with TBS buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl) and then resolved by 4%-20% gradient SDS-PAGE for immunoblotting. Antibodies used included anti-human Rb (a.a.300-380) and anti-human p16INK4a from BD Biosciences Pharmingen and anti-p53 (FL-393), anti-p21 (C-19), and anti—α-tubulin (B-7) from Santa Cruz Biotechnology (Santa Cruz, CA, http://www.scbt.com). Where noted, the cells were treated for 24 hours with 40 nM actinomycin D (Sigma-Aldrich) before lysis to increase p53 and p21WAF1/CIP1 protein levels [23].
Cell Proliferation and Differentiation Assays
Cell doubling time was determined using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, http://www.promega.com) according to the manufacturer's protocol.
Erythroid potential of the CB cell lines was assessed by transferring the cells into X-VIVO-15 serum-free medium supplemented with 10% BIT 9500, 1 μM hydrocortisone (EMD Biosciences, San Diego, http://www.emdbiosciences.com), 50 ng/ml SCF, 3 U/ml erythropoietin (Stem Cell Technologies), and 50 ng/ml insulin-like growth factor 1 (PreproTech) and culturing for 7 days [24].
To induce monocytic differentiation, the cells were plated into 24-well nontissue culture-treated plates coated with 2 μg/cm2 RetroNectin recombinant fibronectin fragment (Takara Mirus Bio) in X-VIVO-15 serum-free medium supplemented with 10% BIT 9500, 50 ng/ml SCF, 20 ng/ml IL-3, 20 ng/ml macrophage colony-stimulating factor (M-CSF), 20 ng/ml granulocyte-macrophage-CSF (GM-CSF) (all from PeproTech), and 25 ng/ml phorbol 12-myristate 13-acetate (Sigma-Aldrich). After 2 days of culture, 250 μg/ml nitroblue tetrazolium (Sigma-Aldrich) was added to the medium; 4 hours later, the cells were washed with phosphate-buffered saline and cytospin preparations were analyzed by light microscopy as described previously [25].
Spectral Karyotype Analysis and Chromosome Banding
Metaphase spreads were prepared from exponentially growing cells according to standard techniques using a 0.75-M KCl solution. Spectral karyotyping (SKY) and Giemsa-banding (G-banding) of metaphases were carried out by Chrombios GmbH Molecular Cytogenetics (Raubling, Germany, http://www.chrombios.com). In brief, SKY was performed by fluorescence in situ hybridization (FISH) with human chromosome—specific painting probes labeled with five different fluorochromes in Boolean combinations as described [26]. For each CB cell line, images were obtained for 10 metaphases using an Axioplan II microscope (Carl Zeiss AG, Oberkochen, Germany, http://www.zeiss.de) equipped with narrow-band-pass emission filters (Chroma Technology, Rockingham, VT, http://www.chroma.com) connected to a cooled CCD camera (Roper Scientific, GmbH, Otto-brunn, Germany, http://www.roperscientific.com). Chromosomes were then displayed in false colors and the images merged. Camera control, image capture, and merging were done using SmartCapture VP imaging software (Digital Scientific, Cambridge, U.K., http://www.digitalscientific.co.uk). Inspection of five different image planes resulting from the hybridization signals of the different fluorochromes allowed the detection of trans-locations in each cell line. Karyotyping was done using Smart-Type image processing software (Digital Scientific). G-banded metaphases were similarly recorded by brightfield microscopy using the same instrumentation and software combination. Images were transferred in ICS format into Quips karyotyping software (Vysis, Downers Grove, IL, http://www.vysis.com). For each CB cell line, at least 15 metaphases were recorded and two metaphases were fully karyotyped [27]. For several rearrangements, the origin of the centromere was identified by FISH using centromere-specific repetitive sequence probes [28].
Retroviral Transduction of CB Cell Lines and Assessment of In Vivo Growth Potential
Recombinant MSCV-based retroviral vectors were produced as replication-defective pseudotyped particles carrying the Gibbon ape leukemia virus envelope protein from stably transduced PG13 packaging cells (ATTC CRL-10686) [20, 29]. Producer lines were maintained in Dulbecco's modified Eagle's medium with 4.5 g/L glucose (Invitrogen) supplemented with 2 mM L-glutamine (Invitrogen) and 10% heat-inactivated fetal bovine serum (Fisher Scientific) at 37°C in a humidified atmosphere containing 5% CO2. PG13/MSCVneo-v-H-ras and PG13/MSCVneo producer cells have been described previously [30, 31]. PG13/MSCVneo-BCRABL exports GALV-pseudotyped MSCVneo-BCR-ABL, which was constructed by insertion of a 7.2-kb EcoRI fragment encoding p210BCR-ABL (a gift from Dr. Ruibao Ren, Rosenstiel Basic Medical Sciences Research Center, Brandeis University, Waltham, MA) into MSCVneo [29, 32].
The ET1a and ET2 CB cell lines were transduced with GALV-pseudotyped MSCVneo-v-H-ras, MSCVneo-BCR-ABL, and MSCVneo by spinoculation [17]. Transduced cells were then subjected to selection by culturing in medium containing 800 μg/ml Geneticin (Invitrogen); cells that continued to proliferate after 2 weeks of Geneticin selection were propagated as bulk populations. Geneticin-resistant CB cell lines were evaluated for in vivo growth potential after intravenous (i.v.) injection of 1 × 107 cells into the tail vein (0.5 ml) or subcutaneous (s.c.) injection of 5 × 106 cells into the flank region (0.1 ml) of sublethally irradiated (250-cGy) immunodeficient nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (Jackson Laboratory, Bar Harbor, ME, http://www.jax.org) [33]. Animals were examined daily and were euthanized before they became moribund or the longest diameter of the tumor reached 1 cm. Engraftment of CB cell lines in i.v.-injected mice was characterized by the presence of CD45+GFP+/YFP+ cells in bone marrow, spleen, liver, and peripheral blood [34]. KG1a-GFP cells served as controls. All procedures involving mice followed Institutional Animal Care and Use Committee guidelines.
Results
Extended Lifespan of Human CB-Derived Progenitor Cells Ectopically Expressing HPV16 E6/E7
CD34+ progenitor cells were enriched to >94% purity from mono-nuclear cell preparations of human CB samples by super paramagnetic microbead selection. The CD34+ CB cells were then transduced with VSV-G glycoprotein—pseudotyped lentiviral vectors that express hTERT or HPV16 E6/E7 linked to a downstream GFP or YFP reporter gene on a bicistronic transcript. GFP+ and/or YFP+ CB cells were sorted to >96% purity and maintained under serum-free conditions in the presence of SCF (100 ng/ml), FL (100 ng/ml), and TPO (20 ng/ml) with or without IL-3 (20 ng/ml), conditions demonstrated to transiently support hematopoietic stem cell self-renewal divisions in vitro [35]. Nontransduced CD34+ CB cells served as controls. In all cases (n = 3), control CD34+ CB cells differentiated into macrophage-like cells and underwent senescence-associated proliferation arrest after approximately 4 months in culture (Fig. 1A). Constitutive expression of hTERT failed to extend the proliferative capacity of the CD34+ CB cell—derived cultures beyond this time point in repeated attempts (n = 3), and macrophage-like cells were also the predominant cell type that accumulated in these cultures (Fig. 1B), as previously reported for hTERT retroviral vector—transduced cells [10]. On the other hand, CD34+ CB-derived cells ectopically expressing HPV16 E6/E7 alone or in combination with hTERT continued to proliferate, although the cultures expressing only HPV16 E6/E7 went through a crisis period. In total, 11 CB cell lines were established, some of which have been continuously propagated in culture for more than 2 years. Cell lines obtained by the introduction of the HPV16 E6 and E7 genes were designated by the prefix “E” (two lines), and those originating from the HPV16 E6/E7-hTERT combination by “ET” (nine lines). We restricted most of our analysis to five lines: E1, E2, ET1a, ET1b, and ET2. Examination of the growth factor requirements of these five CB cell lines indicated that they all required SCF for survival and proliferation but grew optimally in the presence of SCF, FL, TPO, and IL-3. The cells were therefore routinely maintained in the four-cytokine combination.
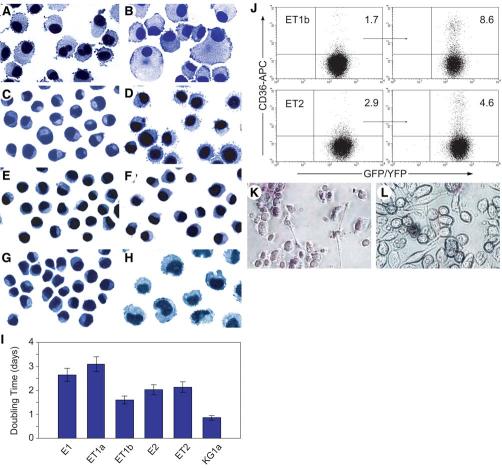
Figure 1.
Immortalization of CB progenitors by HPV16 E6/E7 with or without hTERT. (A—H): Photomicrographs of cytospin preparations after Wright-Giemsa staining (magnification ×60). (A): Nontransduced CB cells at 4 months. (B): hTERT-transduced CB cells at 3 months. (C): E1 cells. (D): ET1a cells. (E): ET1b cells. (F): E2 cells. (G): ET2 cells. (H): KG1a-GFP myeloid leukemia cells. (I): Cell growth rates. The mean and SD of three experiments are shown. (J): Flow cytometric analysis of CD36 expression on ET1b and ET2 cells maintained in the absence (left panels) or presence (right panels) of erythropoietin-supplemented growth medium. The percentages of CD36+ cells are indicated in the upper right quadrants. (K, L): Monocytic differentiation of CB cells. Photomicrographs of (K) ET1a and (L) E1 cells adhering to fibronectin-coated plates after nitroblue tetrazolium staining. Note that most of the ET1a cells contain formazan, the product formed by the reduction of nitroblue tetrazolium by intracellular superoxide (magnification ×20). Abbreviations: APC, allophycocyanin; CB, cord blood; GFP, green fluorescent protein; HPV16, human papillomavirus type 16; hTERT, human telomerase catalytic subunit; KG1a, KG1a-GFP cells; YFP, yellow fluorescent protein.
Morphology and Cell-Surface Phenotype
The CB cell—derived cultures consisted of relatively homogeneous populations of nonadherent cells with round or oval nuclei located eccentrically, generally having scant cytoplasm and, in some cases (e.g., ET1a cells), microvilli-like structures on the cell surface (Figs. 1C-1G). Doubling times of the cultures supplemented with SCF, FL, TPO, and IL-3 ranged from 1.5-3 days (Fig. 1I). The surface phenotype of the CB cell lines was determined by immunofluorescence flow cytometric analysis using a panel of monoclonal antibodies directed against human hematopoietic cell-surface antigens. A summary of the analysis of the E1, E2, ET1a, ET1b, and ET2 cell lines is presented in Table 1. All of the lines expressed the leukocyte common antigen CD45, indicative of a hematopoietic origin (albeit at low levels in the case of ET1a cells). The cell lines also all expressed CD203c (basophilic granulocytes, mast cells, and their progenitors), CD71 (transferrin receptor on early erythroid cells, activated lymphocytes, monocytes, macrophages, and most dividing cells), CD44 (hyaluronan receptor on progenitors of all lineages), CD33 (myeloid progenitors, CFU-Meg, a portion of BFU-E, monocytes and mast cells, as well as activated T cells and dimly on granulocytes), and CD13 (granulocytes, monocytes, mast cells, and their progenitors). Although the CB cell lines were negative for CD34, it is notable that two of the lines (ET1a cells and ET2 cells) expressed the hematopoietic stem/progenitor cell marker CD133 [36]. In addition, all of the cultures contained subpopulations of cells that expressed CD235a/glycophorin A (erythroid progenitor cells). In contrast, the CB cell lines expressed low or background levels of CD184 (CXCR4 “homing receptor” on CD34+ progenitor cells), CD56 (natural killer cells), CD41a (gpIIb/IIIa complex on megakaryocytes), CD38 (early stages of CD34+ hematopoietic stem cell lineage commitment), CD24 (B cells and granulocytes), CD19 (B cells), CD16 (natural killer cells and neutrophils), CD14 (expressed at high levels on monocytes), CD11b (αM chain of the αMβ2 integrin expressed at varying levels on granulocytes, macrophages, myeloid-derived dendritic cells, and natural killer cells), CD3 (T-cell antigen receptor complex), CD2 (T cells and a subset of natural killer cells), and HLA-DR (antigen-presenting B cells, monocytes, and macrophages, as well as activated T cells). With the exception of the E2 cell line, which contained a subpopulation of cells that were positive, the CB cell lines were also negative for CD15 (expressed on granulocytes and to a varying degree on monocytes). Taken together, the cell-surface phenotypic and morphological properties of the CB cell lines suggested that the target cells for immortalization were multipotential progenitors of the granulocyte, monocyte-macrophage, mast cell, and erythroid lineages.
Table 1.
Surface phenotype of cord blood cell lines
| Antigen | E1 | ET1a | ET1b | E2 | ET2 |
|---|---|---|---|---|---|
| CD2 | <1 | <1 | <1 | <1 | <1 |
| CD3 | <1 | <1 | <1 | 1 ± 1 | <1 |
| CD11b | <1 | <1 | <1 | 1 ± 1 | <1 |
| CD13 | 99 ± 1 | 80 ± 1 | 96 ± 2 | 98 ± 2 | 98 ± 1 |
| CD14 | 1 ± 1 | <1 | 1 ± 1 | 2 ± 1 | 3 ± 1 |
| CD15 | <1 | <1 | <1 | 28 ± 2 | <1 |
| CD16 | <1 | <1 | <1 | <1 | <1 |
| CD19 | <1 | <1 | <1 | <1 | <1 |
| CD24 | 1 ± 1 | <1 | <1 | <1 | <1 |
| CD33 | 99 ± 1 | 25 ± 2 | 99 ± 1 | 99 ± 1 | 98 ± 1 |
| CD34 | <1 | <1 | <1 | 1 ± 1 | <1 |
| CD38 | <1 | <1 | <1 | 1 ± 1 | <1 |
| CD41a | 1 ± 1 | <1 | <1 | 1 ± 1 | <1 |
| CD44 | 99 ± 1 | 98 ± 1 | 99 ± 1 | 99 ± 1 | 99 ± 1 |
| CD45 | 95 ± 1 | 4 ± 3 | 85 ± 3 | 33 ± 4 | 47 ± 2 |
| CD56 | <1 | <1 | <1 | <1 | <1 |
| CD71 | 99 ± 1 | 97 ± 3 | 98 ± 1 | 99 ± 1 | 99 ± 1 |
| CD133 | <1 | 10 ± 2 | 1 ± 1 | <1 | 9 ± 2 |
| CD184 | <1 | 1 ± 1 | 1 ± 1 | 2 ± 1 | 1 ± 1 |
| CD203c | 97 ± 1 | 65 ± 1 | 96 ± 1 | 94 ± 1 | 91 ± 2 |
| CD235a | 9 ± 2 | 5 ± 2 | 18 ± 2 | 16 ± 2 | 5 ± 2 |
| HLA-DR | <1 | <1 | 1 ± 1 | 2 ± 1 | 1 ± 1 |
Data represent the average percentages ± SD of two to three independent experiments.
Interference of the p16INK4a/Rb and p53/p21WAF1/CIP1 Pathways
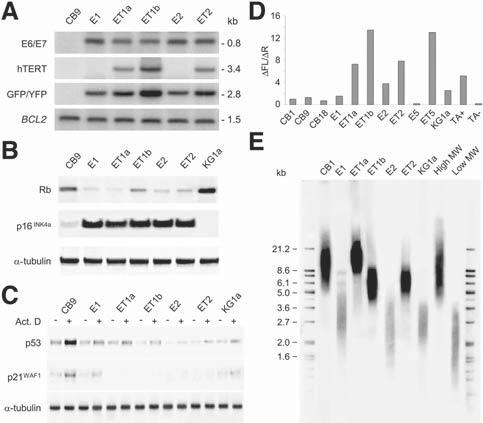
The presence and structural integrity of the HPV16 E6/E7 and hTERT transgenes were documented by Southern blotting after digestion of genomic DNA with EcoRI plus BglII, restriction enzymes which cleave sites flanking the transgenes and the GFP/YFP reporter genes (Fig. 2A). Additional Southern blot analyses with restriction enzymes that cleave once within the vector sequences indicated two to five copies of each lentiviral vector per CB cell line (data not shown).
Figure 2.
Analysis of CB cell lines expressing HPV16 E6/E7 with or without hTERT. (A): Southern blot analysis of genomic DNA (10 μg) with the indicated probes after digestion with EcoRI plus BglII. The sizes (kb) of unrearranged transgene sequences determined by comparison with HindIII-digested λ phage DNA are indicated on the right. Also shown is the 1.5-kb fragment corresponding to the endogenous BCL2 gene as restriction enzyme digestion and loading control. (B): Western blot analysis for Rb and p16INK4a. Proteins were immunoprecipitated from whole-cell lysates followed by immunoblotting. The blot was stripped and reprobed with anti—α-tubulin to demonstrate that equal amounts of the respective proteins were loaded. (C): Western blot analysis for p53 and p21WAF1/CIP1 in cells treated with (+) or without (-) actinomycin D for 24 hours. The blot was stripped and reprobed with anti—α-tubulin to demonstrate that equal amounts of the respective proteins were loaded. (D): Analysis of telomerase activity by polymerase chain reaction assay. Values represent the relative ratio of the net increase of fluorescein (△FL) and sulforhodamine (△R) emission determined using a fluorescence plate reader. (E): Expression of hTERT stabilizes telomere length. Mean telo-mere length was assessed by Southern blot analysis of HinfI/RsaI-digested genomic DNA with a telomere-specific probe. The positions of size standards (kb) are indicated on the left. Abbreviations: Act. D, actinomycin D; CB, cord blood; CB1, CB9, and CB18, 1-, 9-, and 18-day cultures of primary CD34+ CB cells, respectively; GFP, green fluorescent protein; high MW, high-molecular-weight control DNA; HPV16, human papillomavirus type 16; hTERT, human telomerase catalytic subunit; KG1a, KG1a-GFP cells; low MW, low-molecular-weight control DNA; Rb, retinoblastoma; TA+, telomerase extract positive control; TA-, telomerase extract negative (heat-inactivated) control; YFP, yellow fluorescent protein.
We next investigated the status of the p16INK4a/Rb pathway and the p53/p21WAF1/CIP1 axis in the CB cell lines. As expected, HPV16 E7 expression in the CB cell lines resulted in reduced levels of hypophosphorylated Rb, rendering them insensitive to the increased levels of p16INK4a that accumulated (Fig. 2B). By comparison with 9-day cultures of primary CD34+ CB cells, all CB cell lines showed decreased expression of the p53 target p21WAF1/CIP1, even after stimulation by actinomycin D treatment (Fig. 2C) [23], indicating that p53 function was compromised by the HPV16 E6 protein.
Increased Telomerase Activity in hTERT-Expressing CB Cell Lines
CB cells stably expressing the exogenous hTERT gene exhibited high levels of telomerase activity as assayed by the telomeric repeat amplification protocol (Fig. 2D). Interestingly, the E2 cell line transduced with HPV16 E6/E7 vector alone had significant telomerase activity, and the E1 cell line exhibited telomerase activity at levels similar to early-passage CD34+ CB cells. It has been reported that HPV16 E6 activates endogenous telomerase activity in precrisis human keratinocytes and mammary epithelial cells [37]. However, analysis of a precrisis culture of HPV16 E6/E7-transduced CB cells (E5 cells at 14 weeks) revealed minimal levels of telomerase activity (Fig. 2D), suggesting that the endogenous enzyme had been reactivated in E1 and E2 cells during the immortalization process. Consistent with the notion that telomerase reactivation was associated with bypass of crisis [13], both E1 and E2 cell lines had substantial erosion of telomeres with mean telomere lengths of 2.9 and 2.6 kb, respectively (Fig. 2E), and widespread karyotypic abnormalities (see below). In this regard, it is noteworthy that the CB cell lines generated by cotransduction with the hTERT vector had stabilized mean telomere lengths >5.9 kb, which exceeded that of 1-day cultured primary CD34+ CB cells (12.3 kb) in the case of ET1a cells (15.8 kb) transduced on the first day of culture (Fig. 2E).
Clonal Outgrowth of HPV16 E6/E7 Plus hTERT Immortalized CB Cells Without Widespread Genomic Instability
After 28-76 weeks of culture, the CB cell lines were analyzed by SKY, chromosome G-banding, and FISH with centromerespecific probes. Although the CB cell populations were not deliberately cloned, these analyses indicated that most of them were clonal (Fig. 3, Table 2). The exceptions were the E1 and E2 cell lines obtained by transduction with HPV16 E6/E7 alone and the ET1b cell line, in which the hTERT gene was introduced at week 44 of culture. All three of these cell lines were highly abnormal and carried additional distinctive karyotypic changes indicating that they were oligoclonal. By comparison, the CB cell lines immortalized by successive coexpression of HPV16 E6/E7 and hTERT within 4 weeks of culture were near diploid and exhibited only one or two structural or numerical chromosomal changes.
Figure 3.
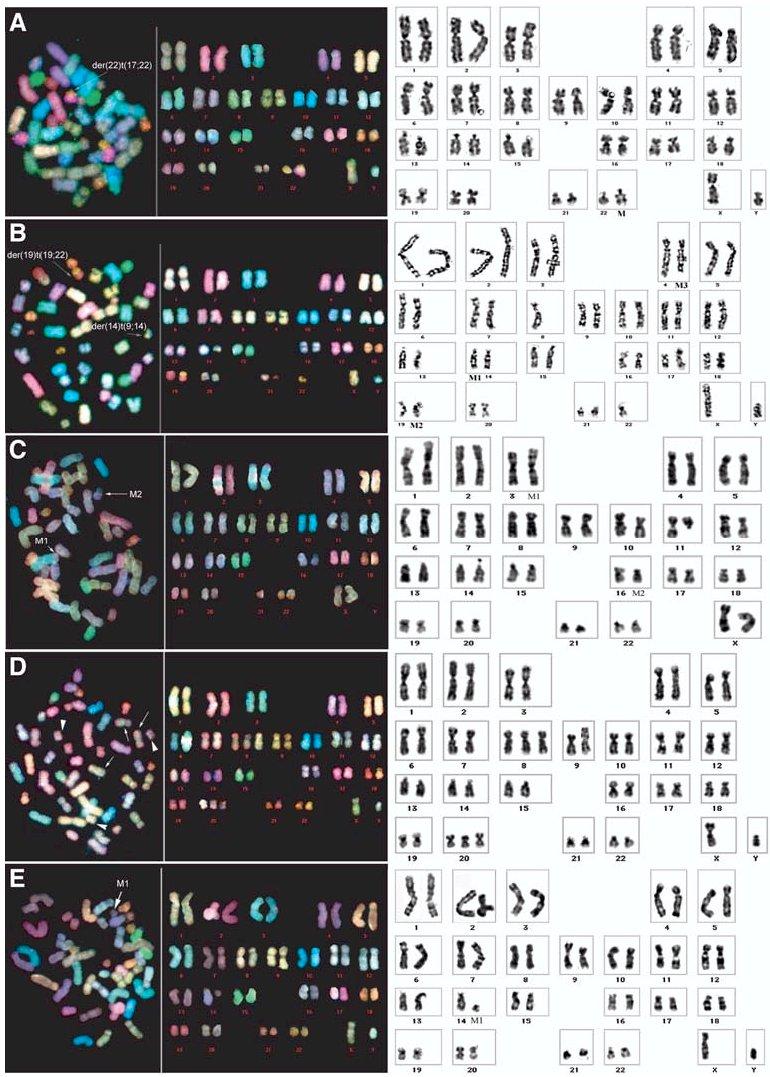
Spectral karyotyping (left panels) and G-banded (right panels) chromosome analysis of cord blood cell lines immortalized by HPV16E6/E7 and hTERT showing limited aneuploidy and rearrangements. (A): ET1a. (B): ET2. (C): ET3. (D): ET4. (E): ET5. Structural and numerical chromosomal abnormalities are indicated by arrows. See Table 2 for a detailed description of the karyotypes. Abbreviations: HPV16, human papillomavirus type 16; hTERT, human telomerase catalytic subunit.
Table 2.
Karyotype of cord blood cell lines
| Cells | Transgenesa | Karyotypeb |
|---|---|---|
| E1 | E6/E7 (day 1) | At 76 weeks: 45,XY,der(10)t(9qter→9q12::10cen→10q24::8q13→8qter),der(14)t(9;14),der(22)t(22qter→22cen::17q11.2→17qter),-17 [5]/44-45,XY,der(10)t(9;10),t(10;8)der(14)t(9;14),der(15)t(22qter→22q12::15cen→15qter),-17 [2]/46-48,XY,der(10)t(10pter→10cen::1q21→1qter),+8 [3] |
| ET1a | E6/E7 (day 1) hTERT (day 1) | At 68 weeks: 46,XY,der(22)t(22pter→22q11.2::17q21→17qter) [9] |
| ET1b | E6/E7 (day 1) hTERT (week 44) | At 68 weeks: 43-46,XY,der(2)t(2pter→2q31::7q21→7qter) [8],der(2)t(2q37::12q24.1→12qter) [8], del(7)(q21→qter) [8], -9 [3],der(9)t(9pter→9q34::2?) [6],-11 [4],der(14)t(14pter→14q21::9?) [10], der(19)t(19qter→19cen::2?::12q24→12qter)t(2;12) [9],der(22)t(22qter→22cen::8q13→8qter) [9][cp10] |
| E2 | E6/E7 (day 1) | At 64 weeks: 43-46,XY,+1 [11],i(1q12→qter) [10],der(4)t(17qter→17q21::4pter→4q12::13q12→13qter) [4], der(4)ins(17;4)(17pter→17p11.2::4p12→4q21::17q21→17qter) [4],+5 [2]der(9)t(9q→9p21::21q11→21qter) [9], -13 [5],der(14)t(14pter→14q21::9?) [9],der(19)t(22qter→22q?::19p13.2→19qter) [9]-20 [5],-22 [5][cp10] |
| ET2 | E6/E7 (day 1) hTERT (week 40) | At 64 weeks: 45,XY,der(14)t(14pter→14q21::9?),der(19)t(22qter→22q?::19p13.2→19qter),-22 [8] |
| ET3 | E6/E7 (day 1) hTERT (week 4) | At 40 weeks :46,XX,t(3;16)(p25;p12) |
| ET4 | E6/E7 (day 1) hTERT (week 12) | At 32 weeks: 48, XY, +8,+20 |
| ET5 | E6/E7 (day 1) hTERT (week 2) | At 28 weeks: 46,XY,del(14)(q11.2→qter) [6]/46,XY,der(14)t(14;17)(14pter→14p11.2::17p11.2→17qter) [4] |
Numbers in parenthesis indicate time of transduction.
Karyotypes were determined by analyzing 10 metaphases for each cell line by spectral karyotyping; marker chromosomes were furthercharacterized by chromosome G-banding and by fluorescence in situ hybridization with centromere-specific probes; numbers in square brackets indicate number of cells with marker chromosome.
Differentiation Potential of Immortalized CB Cells
Based on their phenotype and growth factor requirements, the immortalized CB cells most closely resembled myeloerythroid/mast cell progenitors [38]. Although mast cell progenitors have generally been considered to be a separate lineage, CD203c is also expressed on basophilic granulocytes and their progenitors, raising the possibility that the CB cell lines originated from more primitive common committed CD34+ progenitors [39]. Indeed, it was previously shown that TPO played an important role in concert with SCF in the development of mast cells from CD34+ multilineage colony-forming cells that also had the potential to differentiate into neutrophil/macrophage/mast cell/erythroid lineages, neutrophil/macrophage/mast cell lineages, or neutrophil/mast cell lineages [40]. TPO is also known to enhance erythroid progenitor production from CD34+ bone marrow and CB cells [35, 41].
Whether the CB cell lines can be efficiently induced to terminally differentiate into functional mast cells, erythrocytes, granulocytes, and/or monocyte-macrophages by treatment with various physiologic or chemical/pharmacologic agents will require further investigation. However, pilot experiments suggest that the CB cell lines have a certain degree of erythroid and myeloid differentiative potential. Some shifting to an erythroblastic pheno-type was observed when the CB cell lines were cultured under erythropoiesis-supportive conditions [24], as evidenced by slightly increased expression of the CD36 antigen (Fig. 1J) [42]. Moreover, when the immortalized CB cells were subjected to a myeloid differentiation regimen [25], in the best example (ET1a cells) up to 90% of the cells acquired the ability to adhere to fibronectin, approximately 30% of which were capable of superoxide-dependent nitroblue tetrazolium reduction reflective of terminal monocytic differentiation (Figs. 1K, 1L).
In Vivo Growth Potential of E6/E7 Plus hTERT-Expressing CB Cells
The ET1a and ET2 CB cell lines were transduced with retroviral vectors coexpressing the v-H-ras [30, 31] or BCR-ABL [32] oncogenes and the bacterial neomycin phosphotransferase (neo) gene or with a control vector expressing the neo gene alone. Geneticin-resistant bulk populations of ET1a/Neo and ET2/Neo cells did not engraft or grow in sublethally irradiated (250-cGy) immunodeficient NOD/SCID mice after i.v. or s.c. injection, respectively, during observation periods of 28 weeks (Table 3). Disseminated (bone marrow, spleen, liver, and peripheral blood) or solid tumor growth was observed, however, within 14 weeks after i.v. or s.c. injection of v-H-ras—transduced or BCR-ABL—transduced E6/E7 plus hTERT-expressing cell populations. By comparison, malignant growth of i.v.- or s.c.-injected GFP-expressing KG1a myeloid leukemia cells occurred within 8 weeks.
Table 3.
Evaluation of tumor formation and engraftment of CB cell lines in immunodeficient NOD/SCID mice
| Cells | Tumors/injected (s.c.) | Time (wks) | Engrafted/injected (i.v.) | Time (wks) |
|---|---|---|---|---|
| ET1a/Neo | 0/2 | 28 | ND | NA |
| ET1a/v-H-Ras + Neo | 2/2 | 20, 21 | ND | NA |
| ET1a/BCR-ABL + Neo | ND | NA | ND | NA |
| ET2/Neo | 0/4 | 28 | 0/5 | 28 |
| ET2/v-H-Ras + Neo | 2/2 | 14,18 | 2/2 | 14, 20 |
| ET2/BCR-ABL + Neo | 4/4 | 15-20 | 2/2 | 14, 17 |
| KG1a-GFP | 3/3 | 7-8 | 1/1 | 4 |
Results are shown for bulk populations of ET1a and ET2 cells transduced with MSCVneo-v-H-ras, MSCVneo-BCR-ABL, and control (empty) MSCVneo retroviral vectors. Cells were tested for in vivo growth potential by s.c. injection of 5 × 106 cells or i.v. injection of 1 × 107 cells into sublethally irradiated (250-cGy) immunodeficient NOD/SCID mice. Mice were euthanized before the s.c. tumors reached a diameter >1 cm or they became moribund. Engraftment of CB cell lines in i.v. injected mice was characterized by the presence of CD45+GFP+/YFP+ cells in bone marrow and peripheral blood (as well as spleen and liver) as determined by flow cytometric analysis.
Abbreviations: CB, cord blood; NA, not applicable; ND, not done; NOD/SCID, nonobese diabetic/severe combined immunodeficient.
Discussion
In this study, we explored the possibility that lentiviral-mediated expression of the exogenous hTERT gene or the HPV16E6 and E7 genes would permit the immortalization of human hematopoietic progenitor cells in CB samples. We found that constitutive hTERT expression was incapable of increasing the replicative capacity of CD34+ CB cells and did not prevent the cells from undergoing terminal differentiation under the culture conditions examined. This finding is in accord with a previous report that used a retroviral vector to express hTERT in CD34+ CB cells, where it was not possible to rule out vector silencing as the reason for the lack of effect [10]. In contrast, we demonstrated that ectopic expression of the HPV16 E6 and E7 genes was capable of extending the lifespan of certain myeloerythroid/mast cell progenitors present in CB samples. Notably, however, the HPV16 E6/E7-immortalized cell lines were highly aneuploid and exhibited numerous chromosomal alterations, as noted previously in other cell types immortalized by these oncogenes [43]. Importantly, when hTERT was expressed concomitantly with E6/E7, telomere-length stabilization was observed and the immortalized CB cell lines established exhibited much less genomic instability. The latter observation is in agreement with recent work indicating that hTERT regulates the DNA damage response pathway and participates in chromatin maintenance independent of telomere length maintenance [44]. Although we can neither rule in nor rule out a role, it is tempting to speculate that persistent hTERT expression also may have contributed in some manner to the presence of the CD133 hematopoietic stem/progenitor cell marker on ET1a and ET2 cells.
All of the CB cell lines derived demonstrated clonal outgrowth of one or a few cells, suggesting acquisition of a proliferative advantage and implicating the specific chromosomal changes identified in the immortalization process. Selective outgrowth was observed even in the case of the CB cell lines established by the combination of HPV16 E6/E7 and hTERT, which did not undergo crisis. The mechanisms by which the particular genetic changes observed conferred a growth advantage to the CB cells are unknown. It is intriguing, however, that hESC lines passaged as bulk cultures frequently exhibit increased dosage of the long arm of chromosome 17 and that in a subclone of H1 hESCs, gain of 17q involved translocation of the same region—17q11.2→17qter—observed in the E1 CB cell line [45]. Trisomy of chromo-some 20, which was found in the ET4 CB cell line, has also been reported in a subclone of BG02 hESCs [46]. Cloning of the trans-location breakpoints present in the CB cell lines with only one or two chromosomal translocations may provide insights into the mechanisms operative in these instances.
Abnormal karyotypes notwithstanding, the ET1a and ET2 cell lines were nontumorigenic in immunodeficient NOD/SCID mice. Recent evidence obtained for other normal human cell types implicates abrogation of senescence as a critical step in experimental transformation, with frank malignancy requiring perturbation of six to seven signaling pathways [47]. Ectopic expression of the v-H-ras or BCR-ABL oncogenes resulted in the tumorigenic conversion of ET1a and ET2 cells, but no attempt was made to determine whether the resulting tumor cell populations were polyclonal or the outgrowth of variant sub-populations that had acquired additional genetic alterations [48, 49]. Further systematic studies will therefore be required to define the minimal number of biochemical pathways that need to be disrupted to convert normal human hematopoietic precursors to leukemogenicity.
In conclusion, these findings establish the feasibility of bypassing senescence in human hematopoietic progenitors through genetic engineering, providing proof of principle for approaches that might eventually allow establishment of permanent human hematopoietic stem cell lines. Accordingly, our future efforts will focus on extending these results by using reversible gene delivery systems [50] and a variety of growth regulatory genes (e.g., TLX1/HOX11) [22, 51] toward the conditional immortalization of other human hematopoietic stem/progenitor cell subpopulations.
Acknowledgments
We thank Joseph Molete for technical assistance. This work was supported in part by National Institutes of Health grants R01HL65519, R01HL66305, and R24RR16209.
Footnotes
Disclosures: R.G.H. receives royalties derived from the licensing of gene transfer technology (MSCV Retroviral Expression System) for research purposes to Clontech.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 3.Bodnar AG, Ouellette M, Frolkis M, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 4.Simonsen JL, Rosada C, Serakinci N, et al. Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat Biotechnol. 2002;20:592–596. doi: 10.1038/nbt0602-592. [DOI] [PubMed] [Google Scholar]
- 5.Roy NS, Nakano T, Keyoung HM, et al. Telomerase immortalization of neuronally restricted progenitor cells derived from the human fetal spinal cord. Nat Biotechnol. 2004;22:297–305. doi: 10.1038/nbt944. [DOI] [PubMed] [Google Scholar]
- 6.Yui J, Chiu CP, Lansdorp PM. Telomerase activity in candidate stem cells from fetal liver and adult bone marrow. Blood. 1998;91:3255–3262. [PubMed] [Google Scholar]
- 7.Van Ziffle JA, Baerlocher GM, Lansdorp PM. Telomere length in subpopulations of human hematopoietic cells. STEM CELLS. 2003;21:654–660. doi: 10.1634/stemcells.21-6-654. [DOI] [PubMed] [Google Scholar]
- 8.Vulliamy T, Marrone A, Goldman F, et al. The RNA component of telom-erase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi H, Calado RT, Ly H, et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med. 2005;352:1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 10.Elwood NJ, Jiang XR, Chiu CP, et al. Enhanced long-term survival, but no increase in replicative capacity, following retroviral transduction of human cord blood CD34+ cells with human telomerase reverse transcriptase. Haematologica. 2004;89:377–378. [PubMed] [Google Scholar]
- 11.Ma Y, Ramezani A, Lewis R, et al. High-level sustained transgene expression in human embryonic stem cells using lentiviral vectors. STEM CELLS. 2003;21:111–117. doi: 10.1634/stemcells.21-1-111. [DOI] [PubMed] [Google Scholar]
- 12.Ramezani A, Hawley TS, Hawley RG. Performance- and safety-enhanced lentiviral vectors containing the human interferon-β scaffold attachment region and the chicken β-globin insulator. Blood. 2003;101:4717–4724. doi: 10.1182/blood-2002-09-2991. [DOI] [PubMed] [Google Scholar]
- 13.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Okamoto T, Aoyama T, Nakayama T, et al. Clonal heterogeneity in differentiation potential of immortalized human mesenchymal stem cells. Biochem Biophys Res Commun. 2002;295:354–361. doi: 10.1016/s0006-291x(02)00661-7. [DOI] [PubMed] [Google Scholar]
- 15.Munger K, Baldwin A, Edwards KM, et al. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Counter CM, Hahn WC, Wei W, et al. Dissociation among in vitro telom-erase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci U S A. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramezani A, Hawley RG. Generation of HIV-1-based lentiviral vector particles. In: Ausubel F, Brent R, Kingston B, et al., editors. Current Protocols in Molecular Biology. John Wiley & Sons; Hoboken, NJ: 2002. pp. 16.22.1–16.22.15. [DOI] [PubMed] [Google Scholar]
- 18.Claudio JO, Liew C-C, Dempsey AA, et al. Identification of sequence-tagged transcripts differentially expressed within the human hematopoietic hierarchy. Genomics. 1998;50:44–52. doi: 10.1006/geno.1998.5308. [DOI] [PubMed] [Google Scholar]
- 19.Cheng L, Du C, Murray D, et al. A GFP reporter system to assess gene transfer and expression in viable human hematopoietic progenitors. Gene Ther. 1997;4:1013–1022. doi: 10.1038/sj.gt.3300507. [DOI] [PubMed] [Google Scholar]
- 20.Dorrell C, Gan OI, Pereira DS, et al. Expansion of human cord blood CD34+CD38- cells in ex vivo culture during retroviral transduction without a corresponding increase in SCID repopulating cell (SRC) frequency: dissociation of SRC phenotype and function. Blood. 2000;95:102–110. [PubMed] [Google Scholar]
- 21.Hawley TS, Herbert DJ, Eaker SS, et al. Multiparameter flow cytometry of fluorescent protein reporters. Methods Mol Biol. 2004;263:219–238. doi: 10.1385/1-59259-773-4:219. [DOI] [PubMed] [Google Scholar]
- 22.Riz I, Hawley RG. G1/S transcriptional networks modulated by the HOX11/ TLX1 oncogene of T-cell acute lymphoblastic leukemia. Oncogene. 2005;24:5561–5575. doi: 10.1038/sj.onc.1208727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hietanen S, Lain S, Krausz E, et al. Activation of p53 in cervical carcinoma cells by small molecules. Proc Natl Acad Sci U S A. 2000;97:8501–8506. doi: 10.1073/pnas.97.15.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neildez-Nguyen TM, Wajcman H, Marden MC, et al. Human erythroid cells produced ex vivo at large scale differentiate into red blood cells in vivo. Nat Biotechnol. 2002;20:467–472. doi: 10.1038/nbt0502-467. [DOI] [PubMed] [Google Scholar]
- 25.Akimov SS, Belkin AM. Cell surface tissue transglutaminase is involved in adhesion and migration of monocytic cells on fibronectin. Blood. 2001;98:1567–1576. doi: 10.1182/blood.v98.5.1567. [DOI] [PubMed] [Google Scholar]
- 26.Schrock E, du MS, Veldman T, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 27.Veldman T, Vignon C, Schrock E, et al. Hidden chromosome abnormalities in haematological malignancies detected by multicolour spectral karyo-typing. Nat Genet. 1997;15:406–410. doi: 10.1038/ng0497-406. [DOI] [PubMed] [Google Scholar]
- 28.Muller S, Neusser M, Wienberg J. Towards unlimited colors for fluorescence in-situ hybridization (FISH) Chromosome Res. 2002;10:223–232. doi: 10.1023/a:1015296122470. [DOI] [PubMed] [Google Scholar]
- 29.Hawley RG, Lieu FHL, Fong AZC, et al. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 30.Hawley RG, Fong AZC, Ngan B-Y, et al. Hematopoietic transforming potential of activated ras in chimeric mice. Oncogene. 1995;11:1113–1123. [PubMed] [Google Scholar]
- 31.Dorrell C, Takenaka K, Minden MD, et al. Hematopoietic cell fate and the initiation of leukemic properties in primitive primary human cells are influenced by Ras activity and farnesyltransferase inhibition. Mol Cell Biol. 2004;24:6993–7002. doi: 10.1128/MCB.24.16.6993-7002.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Ren R. Bcr-Abl efficiently induces a myeloproliferative disease and production of excess interleukin-3 and granulocyte-macrophage colony-stimulating factor in mice: a novel model for chronic myelogenous leukemia. Blood. 1998;92:3829–3840. [PubMed] [Google Scholar]
- 33.Hawley TS, Lach B, Burns BF, et al. Expression of retrovirally transduced IL-1α in IL-6-dependent B cells: a murine model of aggressive multiple myeloma. Growth Factors. 1991;5:327–338. doi: 10.3109/08977199109000296. [DOI] [PubMed] [Google Scholar]
- 34.Ramezani A, Hawley TS, Hawley RG. Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Mol Ther. 2000;2:458–469. doi: 10.1006/mthe.2000.0190. [DOI] [PubMed] [Google Scholar]
- 35.Petzer AL, Zandstra PW, Piret JM, et al. Differential cytokine effects on primitive (CD34+CD38-) human hematopoietic cells: novel responses to Flt3-ligand and thrombopoietin. J Exp Med. 1996;183:2551–2558. doi: 10.1084/jem.183.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin AH, Miraglia S, Zanjani ED, et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–5012. [PubMed] [Google Scholar]
- 37.Klingelhutz AJ, Foster SA, McDougall JK. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 38.Kirshenbaum AS, Akin C, Goff JP, et al. Thrombopoietin alone or in the presence of stem cell factor supports the growth of KIT(CD117)low/ MPL(CD110)+ human mast cells from hematopoietic progenitor cells. Exp Hematol. 2005;33:413–421. doi: 10.1016/j.exphem.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 39.Buhring HJ, Simmons PJ, Pudney M, et al. The monoclonal antibody 97A6 defines a novel surface antigen expressed on human basophils and their multipotent and unipotent progenitors. Blood. 1999;94:2343–2356. [PubMed] [Google Scholar]
- 40.Sawai N, Koike K, Mwamtemi HH, et al. Thrombopoietin augments stem cell factor-dependent growth of human mast cells from bone marrow multipotential hematopoietic progenitors. Blood. 1999;93:3703–3712. [PubMed] [Google Scholar]
- 41.Kobayashi M, Laver JH, Kato T, et al. Recombinant human thrombopoietin (Mpl ligand) enhances proliferation of erythroid progenitors. Blood. 1995;86:2494–2499. [PubMed] [Google Scholar]
- 42.Scicchitano MS, McFarland DC, Tierney LA, et al. In vitro expansion of human cord blood CD36+ erythroid progenitors: temporal changes in gene and protein expression. Exp Hematol. 2003;31:760–769. doi: 10.1016/s0301-472x(03)00185-1. [DOI] [PubMed] [Google Scholar]
- 43.White AE, Livanos EM, Tlsty TD. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 44.Masutomi K, Possemato R, Wong JM, et al. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc Natl Acad Sci U S A. 2005;102:8222–8227. doi: 10.1073/pnas.0503095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Draper JS, Smith K, Gokhale P, et al. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004;22:53–54. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- 46.Mitalipova MM, Rao RR, Hoyer DM, et al. Preserving the genetic integrity of human embryonic stem cells. Nat Biotechnol. 2005;23:19–20. doi: 10.1038/nbt0105-19. [DOI] [PubMed] [Google Scholar]
- 47.Rangarajan A, Hong SJ, Gifford A, et al. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 48.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 49.Warner JK, Wang JC, Takenaka K, et al. Direct evidence for cooperating genetic events in the leukemic transformation of normal human hematopoietic cells. Leukemia 2005; Aug 11 [Epub ahead of print] [DOI] [PubMed]
- 50.Salmon P, Oberholzer J, Occhiodoro T, et al. Reversible immortalization of human primary cells by lentivector-mediated transfer of specific genes. Mol Ther. 2000;2:404–414. doi: 10.1006/mthe.2000.0141. [DOI] [PubMed] [Google Scholar]
- 51.Keller G, Wall C, Fong AZC, et al. Overexpression of HOX11 leads to the immortalization of embryonic precursors with both primitive and definitive hematopoietic potential. Blood. 1998;92:877–887. [PubMed] [Google Scholar]