Abstract
Four kinds of latency-associated transcripts of human herpesvirus 6 were identified which were detected only in latently infected cells. Although they were oriented in the same direction as the immediate-early 1 and 2 (IE1/IE2) genes and shared their protein-coding region with IE1/IE2, their transcription start sites and exon(s) were latency associated.
Human herpesvirus 6 (HHV-6) belongs to the betaherpesvirus subfamily (26), which is represented by human cytomegalovirus (HCMV). HHV-6 infects most individuals at an early age and latently infects throughout life, as do other herpes viruses. It is known that the virus establishes latency in the cells of the monocyte/macrophage lineage (11, 13). To study HHV-6 latency, we identified the latency-associated transcripts of HHV-6 variant B, which is the major subgroup of HHV-6 (1, 4, 6).
To search for HHV-6 latency-associated transcripts (H6LTs), mRNA was collected from this experimental latent infection system by using primary cultures of macrophages (13). (Informed consent was obtained from the blood donors for participation in the study.) Subsequently, we screened the transcripts throughout the immediate-early 1 and 2 (IE1/IE2) regions by reverse transcription (RT)-PCR, because we had previously identified HCMV latency-specific transcripts in the IE1/IE2 region of HCMV (12, 14, 16). PCR primers were designed in accordance with the reported structures of IE1/IE2 (21, 28), the genomic sequence (5, 7, 10), and the cDNA clones of HHV-6B IE1 and IE2 (Fig. 1 and Table 1). As a result, we detected the viral transcripts in the coding region of the IE1/IE2 from the latently infected macrophages (Fig. 2A, lanes 1 and 2).
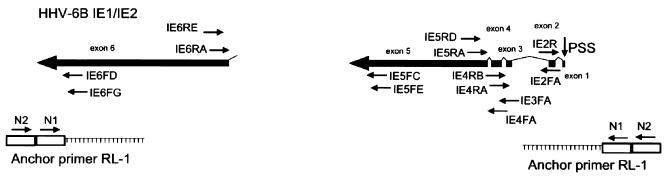
FIG. 1.
HHV-6B and PCR primers. All exons (0 to 6) and introns are drawn to scale. The drawings of the mRNAs are in the same orientation relative to the HHV-6 genome. Thick arrows and boxes represent exons; introns are denoted by the lines connecting the exons. The locations of the PCR primers and a schematic drawing showing the use of the anchor primer RL-1 are also depicted. PSS, productive infection transcription start site.
TABLE 1.
Primer sequencesa
| Primer | Sequence |
|---|---|
| RL-1 | 5′ CTTATGAGTATTTCTTCCAGGGTACTCGAGGCTGGGTAGTCCCCACCTTTCTAGATTTTTTTTTTTTTTTTTT 3′ |
| N1 | 5′ GCTGGGTAGTCCCCACCTTTCTAGA 3′ |
| N2 | 5′ CTTATGAGTATTTCTTCCAGGGTACTCGAG 3′ |
| IE2FA | 5′ GAAACCACCACCTGGAATCAATCTCC 3′ |
| IE2R | 5′ GAGGTTTCTTTGGAGATTGATTCC 3′ |
| IE3FA | 5′ GGAGTCAGCAAAAGATACAACCC 3′ |
| IE4FA | 5′ CGATCCAGTGGTGGAAGAATCC 3′ |
| IE4RA | 5′ GACACATTCTTGGAAGCGATGTCG 3′ |
| IE4RB | 5′ GATGCTCCTTCTTCCACATTACTGG 3′ |
| IE5FA | 5′ GGTGCAT- CAGTGAAGGCTGCCATG 3′ |
| IE5FC | 5′ GACTTGGGAGTCATAACACCAACCAAG 3′ |
| IE5FE | 5′ CACTGCTTCATCAATCACTTTGTCC 3′ |
| IE5RD | 5′ CCATGAGCAAATCTCTGGATTCTGATGCCATC 3′ |
| IE5RA | 5′ TCACTGATGCACCAATACTTTGGATGC 3′ |
| IE5RB | 5′ CAAGACGTGTTTACAACTTTCAAGG 3′ |
| IE5FB | 5′ CCTAACTGTGTGTATGTACAGCCTCAGTG 3′ |
| IE5RC | 5′ CACATCTGTATGCTAATGATTGC 3′ |
| IE6FA | 5′ CCATGATGCGGACGTCACACATAGAC 3′ |
| IE6FB | 5′ CAAGACAGAGTTCTCATATCAAAACATGC 3′ |
| IE6FC | 5′ CGTTTGTTTGAAGTTTTTTCACAAGTTTGAGG3′ |
| IE6FD | 5′ CATATCACGCAACACAACAAAATGC3′ |
| IE6FG | 5′ GGATAACCAAAGTAACATATCACG 3′ |
| IE6RA | 5′ GTATAGTCCGGATAATTTCATGGC 3′ |
| IE6RB | 5′ GATATGAGAACTCTGTCTTGTAGAGAAG 3′ |
| IE6RC | 5′ GGACATTAATTTATTGTTAGGGGTTTTACC 3′ |
| IE6RD | 5′ GTCACCATCAATGATTGAGCAGACC 3′ |
| IE6RE | 5′ GTCTATGTGTGACGTCCGCATCATGG 3′ |
| ULE1F1 | 5′ CGTTACCGAAGATTACTTCGTGCTG 3′ |
| ULE1F2 | 5′ GCATATCCTGGAGTGGCTGCGCTACC 3′ |
| LEF1 | 5′ GAAGTAGTGGATTCAGTGCAGCTGG 3′ |
| LEF2 | 5′ CGTCACAGAATCTAAAAACAAACCATCCGTG 3′ |
| LER1 | 5′ GAAGCATGTAAGCACATCTCTTGCC 3′ |
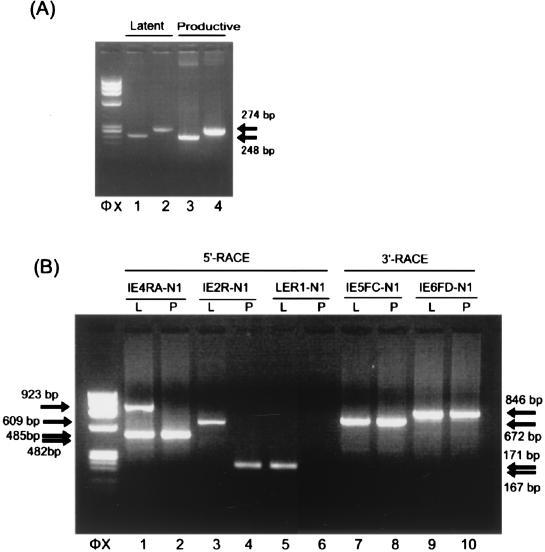
FIG. 2.
Identification of the transcripts arising from the IE1/IE2 locus during latency. (A) Detection of the transcripts arising from the IE1/IE2 locus during latency. HHV-6 gene expression at the IE1/IE2 region in latently infected macrophages and productively infected MT-4 cells was analyzed by RT-PCR. mRNA from 104 latently infected macrophages (lanes 1 and 2) and 102 productively infected MT-4 cells (lanes 3 and 4) was amplified by double-nested RT-PCR with primers IE3FA and IE5RD and IE4FA and IE5RA (lanes 1 and 3) or primers IE3FA and IE6RE and IE4FA and IE6RA (lanes 2 and 4). An ethidium bromide-stained 2% agarose gel is shown. Amplified products of the same size were detected in both the latently infected macrophages and the productively infected MT-4 cells. Arrows indicate the positions of the products. Primer positions are depicted in Fig. 1. φX, HaeIII-digested φX174 DNA. (B) Identification of the 5′ and 3′ ends of the transcripts. mRNA from 105 latently infected macrophages (lanes 1, 3, and 5; labeled L) and from 102 productively infected MT-4 cells (lanes 2, 4, and 6; labeled P) was analyzed by the 5′ RACE method. RACE products from the transcripts are shown. The 5′ end of the cDNA made from each transcript was dA-tailed and annealed with an anchor primer, RL-1. Lanes 1 and 2 show the amplified products obtained with primers N2 and IE4RB followed by primers N1 and IE4RA. The 5′ ends of the transcripts that contained exon 2 of IE1/IE2 (Fig. 3) were detected by using primers N2 and IE4RB and then primers N1 and IE2R (lanes 3 and 4). The 5′ end of the transcript that contained exon LE (Fig. 3) was identified with primers N2 and IE4RB followed by primers N1 and LER1 (lanes 5 and 6). A 3′ RACE study of the latently infected cells (lanes 7 and 9; labeled L) and productively infected cells (lanes 8 and 10; labeled P) was performed. The products amplified with primers N2 and IE5FE and primers N1 and IE5FC (672 bp; lanes 7 and 8) and the products amplified with primers N2 and IE6FG and primers N1 and IE6FD (846 bp; lanes 9 and 10) are shown. Similar results were obtained in the 5′ RACE and 3′ RACE study when umbilical cord blood mononuclear cells or Molt-3 cells were used as productively infected cells. Arrows indicate the positions of the RACE products. HaeIII-digested φX174 DNA fragments were used as size markers (φX). L, latently infected macrophages; P, productively infected MT-4 cells.
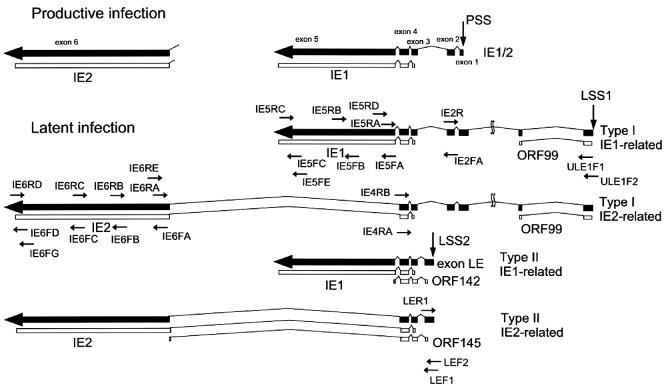
To determine whether the detected transcripts showed latency-specific structures, we identified the structures of the mRNA transcribed during latent infection and productive infection. mRNA isolated from 105 latently infected macrophages and 102 productively infected MT-4 cells was analyzed by the 5′ rapid amplification of cDNA ends (RACE) method described previously (12, 14, 16). Briefly, the 5′ end of the cDNA made from each transcript was dA-tailed and annealed with an anchor primer, RL-1. Subsequently, PCR amplification was performed with primers N2 and IE4RB followed by primers N1 and IE4RA. Two kinds of 5′ ends were detected in latently infected cells upstream of the annealing site of the IE4RA primer (485- and 923-bp products shown in Fig. 2B, lane 1). In contrast, one 5′ end was observed in the productively infected cells (482-bp product shown in Fig. 2B, lane 2). The positions of these three start sites were confirmed with other primer sets (Fig. 2B, lanes 3 to 6). By sequence analysis we located two latent infection transcription start sites (LSSs): one was 9.7 kb upstream of the IE1/IE2 start site (LSS1 in Fig. 3), and the other was between exons 2 and 3 of IE1/IE2 (LSS2 in Fig. 3). The transcription start site identified in productively infected MT-4 cells corresponded to the 5′ end of the IE1/IE2 mRNA (productive infection transcription start site; PSS in Fig. 3). A 3′ RACE study of the latently infected (Fig. 3, lanes 7 and 9; labeled L) and productively infected cells (lanes 8 and 10; labeled P) was also performed. The first-strand cDNA was synthesized with the primer RL-1, and the 3′ ends of IE1-related products were detected by amplification with primers N2 and IE5FE and then with primers N1 and IE5FC (lanes 7 and 8). The 3′ ends of the IE2-related products were amplified with primers N2 and IE6FG and then with primers N1 and IE6FD (lanes 9 and 10). 3′ RACE analysis of the H6LTs indicated the presence of two kinds of 3′ ends (Fig. 2B, lanes 7 and 9). Sequence analysis revealed that each same-sized band had an identical 3′ end. These results identified two types of latent transcripts: type I, originating at LSS1; and type II, originating at LSS2 (Fig. 3). We assumed there were two kinds of transcripts of each type on the basis of the results of the 3′ RACE study. To determine the structures of these four presumed transcripts, multiple RT-PCR analyses were performed with the RNA obtained from 105 HHV-6 latently infected macrophages. cDNA was synthesized with the anchor primer RL-1 and was subjected to PCR amplification. The type I H6LTs were amplified with primers ULE1F1 and N1, and the type II H6LTs were amplified with LEF1 and N1. These amplified products were subjected to nested PCR amplification: the region adjacent to the 5′ ends, the exon 5 region, and the exon 6 region were amplified with the multiple primer sets described in the legends to Fig. 4A, B, and C, respectively. For both the type I and type II H6LTs the amplified regions were electrophoretically homogeneous, and no extra band was detected by these RT-PCR analyses. Furthermore, the sizes of these amplified products from latently infected cells were the same as those from productively infected cells (Fig. 4B, lanes 3 and 6, and 4C, lanes 3, 6, and 9). These data suggested that these transcripts had no alternatively spliced variant species. Thus, the presumed structures of the H6LTs shown in Fig. 3 were verified.
FIG. 3.
Structures of the HHV-6 latent transcripts identified in infected macrophages. Schematic drawings of the H6LT structures are shown. Productive-phase transcripts are also shown. The drawings of the mRNAs are in the same orientation relative to the HHV-6 genome. Thin lines represent introns; thick arrows represent exons. All exons and introns are drawn to scale. Latency-associated exons starting from LSS1 and LSS2 are depicted. The position of the productive start site (PSS) is also shown. The annealing sites of the primers that were used for RT-PCR studies are also depicted. Exon 1 of the type I latent transcript was 138 bp longer than that of IE1/IE2. Two additional exons of the type I latent transcripts were located approximately 7.8 kb and 9.7 kb upstream from the productive transcription start site. ORFs of IE1, IE2, and putative latency-associated proteins ORF99, ORF142, and ORF145 are depicted. ORF99, ORF142, and ORF145 consist of 99, 142, and 145 amino acids, respectively.
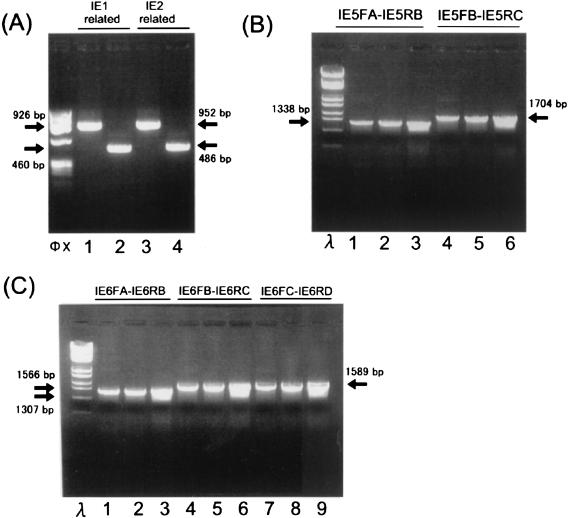
FIG. 4.
RT-PCR analysis of H6LTs. (A) RT-PCR analysis of the regions close to the 5′ ends of the H6LTs. mRNA from the HHV-6 latently infected macrophages was analyzed by RT-PCR amplification. cDNA was synthesized with anchor primer RL-1. The first PCR was performed with primers ULE1F2 and N1 (LSS1; lanes 1 and 3) or primers LEF2 and N1 (LSS2; lanes 2 and 4). The resultant PCR products were then amplified with the following primer sets: ULE1F1 and IE5RA (lane 1, 926-bp predicted product size), LEF1 and IE5RA (lane 2, 460 bp), ULE1F1 and IE6RA (lane 3, 952 bp), and LEF1 and IE6RA (lane 4, 486 bp). φX, HaeIII-digested φX174 DNA. Arrows indicate the positions of PCR products. (B) RT-PCR analysis of the IE1 exon 5 region of the H6LTs. The first PCR products obtained in the experiment depicted in panel A with primers ULE1F2 and N1 (lanes 1 and 4) or LEF2 and N1 (lanes 2 and 5) were amplified with the primer set IE5FA and IE5RB (lanes 1 and 2) or IE5FB and IE5RC (lanes 4 and 5). As a control, the randomly primed cDNA from 105 productively infected MT-4 cells (lanes 3 and 6) was also amplified by a single PCR with the primer set IE5FA and IE5RB (lane 3) or IE5FB and IE5RC (lane 6). The sizes of the amplified products from the type I and II H6LTs were the same as those from productively infected cells. λ, StyI-digested λ DNA. (C) RT-PCR analysis of the IE1/IE2 exon 6 region of the H6LTs. The cDNA amplified with the type I transcript-specific PCR (lanes 1, 4, and 7) and with the type II-specific PCR (lanes 2, 5, and 8) and the cDNA from productively infected cells (lanes 3, 6, and 9) were further amplified by using the primer sets IE6FA and IE6RB (lanes 1, 2, and 3), IE6FB and IE6RC (lanes 4, 5, and 6), and IE6FC and IE6RD (lanes 7, 8, and 9). The sizes of the amplified products from the type I and type II H6LTs were the same as those from productively infected cells. λ, StyI-digested λ DNA.
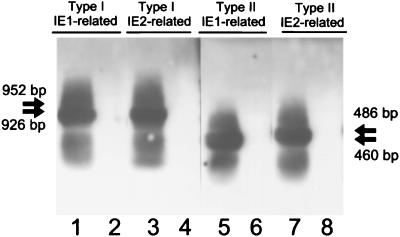
To demonstrate the expression of the H6LTs in vivo, we performed double-nested RT-PCR combined with Southern blot hybridization with cDNA fragments encoding exons 3 and 4 as a probe (Fig. 5). For the type I IE1-related H6LT, nested amplification was carried out with primers ULE1F2 and IE5RD followed by primers ULE1F1 and IE5RA. For the type I IE2-related H6LT, primers ULE1F2 and IE6RE and ULE1F1 and IE6RA were used; for the type II IE1-related H6LT, LEF2 and IE5RD and LEF1 and IE5RA were used; for the type II IE2-related H6LT, LEF2 and IE6RE and LEF1 and IE6RA were used. The expression pattern of each H6LT detected in 107 peripheral blood mononuclear cells (PBMCs) from 20 seropositive healthy adults is shown in Table 2. The type I IE2-related H6LT and the type II IE1-related H6LT were predominantly expressed during HHV-6 latency in vivo.
FIG. 5.
Detection of H6LTs in naturally infected individuals. Representative data of H6LT expression in 107 healthy donor PBMCs examined by double-nested RT-PCR combined with Southern blot hybridization. The predicted sizes of the amplified products are as follows: the type I IE1-related H6LT, 926 bp; the type I IE2-related H6LT, 952 bp; the type II IE1-related H6LT, 460 bp; the type II IE2-related H6LT, 486 bp. Representative positive (lanes 1, 3, 5, and 7) and negative (lanes 2, 4, 6, and 8) cases are shown. Arrows indicate the predicted sizes of the PCR products.
TABLE 2.
H6LT detection in healthy seropositive individualsa
| Transcript | Positive samples (no. positive/total [%])
|
|
|---|---|---|
| Adult PBMCs | CBMCs | |
| Type I IE1-related | 1/20 (5) | 0/20 (0) |
| Type I IE2-related | 7/20 (35) | 0/20 (0) |
| Type II IE1-related | 14/20 (70) | 0/20 (0) |
| Type II IE2-related | 3/20 (15) | 0/20 (0) |
| Productive-phase | 0/20 (0) | 0/20 (0) |
RNA from 107 PBMCs and 107 umbilical cord blood mononuclear cells (CBMCs) was subjected to RT-PCR analysis for each H6LT. The U16/17 and U100 transcripts were examined as productive-phase transcripts.
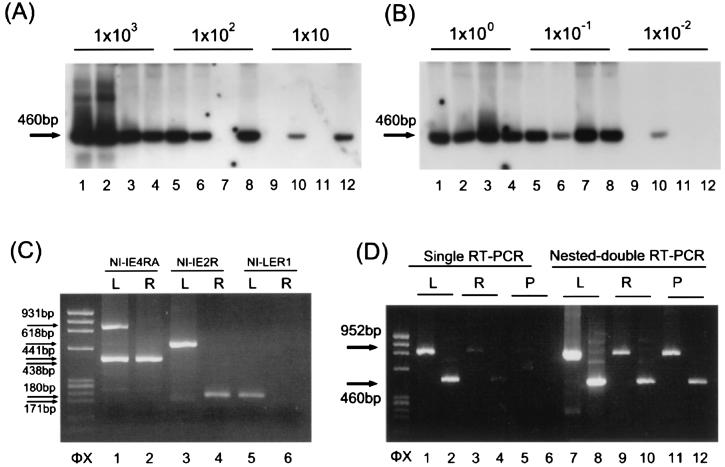
To examine the absolute levels of the H6LTs, we first studied the percentage of H6LT-expressiong cells in HHV-6 DNA-positive cells. Latently infected macrophages were detached from the plates as described previously (13) and were serially diluted (104 to 10 cells) into sample tubes, with 4 tubes for each dilution. Representative data are shown in Fig. 6A. As a control MT-4 cells were infected with HHV-6, and these cells were also diluted. RNA isolated from each sample tube was evaluated by double-nested RT-PCR analysis with each H6LT-specific primer set, as for the experiments shown in Fig. 5. Productive-phase IE1 in MT-4 cells was examined by double-nested RT-PCR with primers IE5RD and IE2FA and IE5RA and IE3FA. Each dilution of the infected macrophages was also amplified by DNA-PCR (15), and the percentage of HHV-6 DNA-positive cells was examined. The percentages of the transcript-expressing cells were calculated by the Reed-Muench method (Table 3). The prevalence of cells that were positive for the H6LTs was several percent, which is similar to the prevalence of HCMV latent transcript-positive cells (12, 29). This situation is analogous to that of Epstein-Barr virus, where not all latent gene products are expressed all the time (22).
FIG. 6.
H6LT expression during latency, reactivation, and productive infection. (A) Percentage of H6LT-positive cells. We investigated the expression of the type II IE1-related H6LT in HHV-6-infected macrophages. RNA from serially diluted HHV-6 latently infected macrophages was amplified by double-nested RT-PCR with primers LEF2 and IE5RD and LEF1 and IE5RA. PCR products were hybridized with the cDNA fragments encoding exons 3 and 4 of IE1. The numbers above the lanes indicate the number of cells in each sample tube. Each dilution was also amplified by DNA-PCR, and the percentage of HHV-6 DNA-positive cells was calculated. Four tests at each dilution were performed to estimate the percentage of H6LT-positive cells by using the Reed-Muench method. In the experiment shown, the estimated percentage of H6LT-positive cells was 5.0%. The arrow indicates the predicted size of the PCR products. (B) Copy number of H6LT. mRNA from latent cells was reverse transcribed and serially diluted, and the type II IE1-related H6LT was amplified by double-nested RT-PCR with primers LEF2 and IE5RD and LEF1 and IE5RA. The template cDNA was equivalent to the amount of cDNA from a single H6LT-positive cell (lanes 1, 2, 3, and 4), 10−1 cell (lanes 5, 6, 7, and 8), or 10−2 cell (lanes 9, 10, 11, 12). Four tests at each dilution were performed to estimate the copy number of H6LT by using the Reed-Muench method. In the experiment shown, the estimated copy number of the type II IE1 related H6LT was 46.8 copies/cell. (C) 5′ RACE amplification of the latent and reactivation-induced cells. mRNA from 105 latently infected macrophages (lanes 1, 3, and 5) and 105 reactivation-induced macrophages (lanes 2, 4, and 6) was analyzed by the 5′ RACE method. The RACE method used was the same as that used for the experiment depicted in Fig. 2B. Lanes 1 and 2 show the products amplified first with primers N2 and IE4RB and then with primers N1 and IE4RA. The 5′ ends of the productive IE1/IE2 or type I H6LTs were detected with primers N2 and IE4RB and N1 and IE2R (lanes 3 and 4). The 5′ end of the type II H6LTs was identified with primers N2 and IE4RB and N1 and LER1 (lanes 5 and 6). L, latently infected macrophages; R, reactivation-induced macrophages. (D) RT-PCR amplification of the latent, reactivation-induced, and productive cells. mRNA from 105 latently infected macrophages (lanes 1, 2, 7, and 8; labeled L), from 105 reactivation-induced macrophages (lanes 3, 4, 9, and 10; labeled R), and from 105 productively infected MT-4 cells (lanes 5, 6, 11, and 12; labeled P) was analyzed by single RT-PCR (lanes 1 to 6) and double-nested RT-PCR (lanes 7 to 12). For the single PCR amplification, the primers were those used as inner primers for the experiments depicted in Fig. 5. The expression of the type I IE2-related H6LT (primers ULE1F1 and IE6RA; lanes 1, 3, and 5) and the type II IE1-related H6LT (primers LEF1 and IE5RA; lanes 2, 4, and 6) was examined. For the double-nested PCR amplification, the outer and inner primers were those used for the experiment depicted in Fig. 5, and the expression of the type I IE2-related H6LT (primers ULE1F2 and IE6RE and ULE1F1 and IE6RA; lanes 7, 9, and 11) and the type II IE1-related H6LT (primers LEF2 and IE5RD and LEF1 and IE5RA; lanes 8, 10, and 12) was examined. Arrows indicate the positions of the PCR products. HaeIII-digested φX174 DNA fragments were used as size markers (φX).
TABLE 3.
Percentage of H6LT-expressing cells and copy number of each H6LTa
| Transcript | % Positive | Copy number/cell (copy number/H6LT-positive cell) |
|---|---|---|
| Type I IE1-related | 1.4 ± 0.4 | 0.49 ± 0.36 (35 ± 26) |
| Type I IE2-related | 4.7 ± 3.8 | 2.8 ± 0.75 (60 ± 16) |
| Type II IE1-related | 7.4 ± 3.1 | 5.3 ± 3.2 (72 ± 43) |
| Type II IE2-related | 2.3 ± 1.1 | 0.87 ± 0.62 (38 ± 27) |
| Productive-phase IE1 | 4.3 ± 0.75b | 35.8 ± 14.4 (833 ± 335)c |
Performed on four independent macrophage cultures. RT-PCR was performed at least four times for each cell dilution, and the percentages of H6LT-positive cells in the HHV-6 DNA-positive cells were estimated by the Reed-Muench method shown in Fig. 6A. The copy number of the H6LT was also estimated by the cDNA dilution study (Fig. 6B). The copy number of each H6LT per H6LT-positive cell is shown. The average of four independent cultures ± standard deviation is given for each latent transcript.
Percentage of IE1-positive cells in total cells.
Copy number of IE1 in a single IE1-positive cell.
Subsequently, we examined the copy numbers of the H6LTs. mRNAs from 104 HHV-6-infected macrophages and MT-4 cells were reverse transcribed and serially diluted into sample tubes with tRNA as a carrier (4 tubes for each dilution), and the cDNA in each tube was amplified by double-nested PCR as described above. The adapted PCR was able to detect a single copy of the cDNA (data not shown). Representative data are shown in Fig. 6B. Copy numbers of each mRNA per transcript-positive cell were calculated by the Reed-Muench method (Table 3). The copy number of the H6LTs per transcript-positive cell was in the double digits, an order of magnitude lower than the copy number of productive-phase IE1 that was known to be an abundant transcript. Although we cannot yet conclude that these amounts of the H6LTs are sufficient for their hypothesized function, there is support for this idea and for their biological importance. Recent reports suggest that a small number of viral mRNAs, such as the number of mRNAs that are incorporated into the virion, may be sufficient for function (2, 8).
To show that the H6LTs are specific for latency rather than for cell type, we examined the expression of H6LTs and productive IE1/IE2 in macrophages during the latent phase and reactivation phase. We performed the 5′ RACE amplification used for the experiment depicted in Fig. 2 on the mRNAs collected from latently infected macrophages and from macrophages that were treated with 20 ng of tetradecanoyl phorbol acetate/ml for 7 days to induce viral reactivation (13).
In the case of latently infected cells, two kinds of 5′ ends were detected upstream of the annealing site of the IE4RA primer (Fig. 6C). In contrast, one 5′ end was observed in the reactivation-induced cells, and amplification with primers N1 and IE2R and N1 and LER1 showed that the product corresponded to the productive-phase IE1/IE2 (Fig. 6C).
To examine the expression of the H6LTs during viral reactivation and productive infection, the predominantly expressed H6LTs (that is, the type I IE2-related H6LT and the type II IE1-related H6LT) were tested for their expression by single RT-PCR and double-nested RT-PCR (Fig. 6D). In the latently infected cells, the H6LTs were detected with the single RT-PCR. However, in the reactivated or productively infected cells, the H6LTs were not detected with the single amplification alone but they were detected with the double-nested amplification (Fig. 6D). Thus, the observed predominance of the H6LT expression in the latent cells may indicate that the transcripts mainly play their role(s) during latency rather than during productive infection. Although low-level expression of the latent transcripts was detectable in the productively infected cells, this phenomenon is also observed in HCMV and herpes simplex virus (19, 30).
The H6LTs in this study contained open reading frames (ORFs) that were common to the productive-phase transcripts IE1/IE2. However, they used novel transcription start sites, and the structures of their 5′ non-coding regions were different from those of the productive-phase transcripts. As a result, novel short ORFs were located at the 5′ proximal region of the H6LTs (ORF99, ORF142, and ORF145 shown in Fig. 4). A similar situation is observed in HCMV latent transcripts; they carry IE1/IE2 ORFs, and short ORFs appeared in the latency-specific exon. Furthermore, in the case of the HCMV latent transcripts the translation of IE1/IE2 protein was prevented probably by the existence of the latency-specific ORFs upstream of the IE1/IE2 ORFs (16). Similarly, the HHV-6 IE1/IE2 protein was not detectable in the latently infected macrophages (data not shown). These findings suggest that viral replication of HCMV and HHV-6 may be suppressed at the point of the translation of major immediate-early proteins during latency. Comparable upstream short ORFs are observed in the mRNAs of cellular proteins, and they play important roles in the regulation of cellular functions (17, 31). We hypothesize that the reactivation signal of HCMV and HHV-6 may upregulate the translation of the downstream IE1/IE2 ORF and may stimulate viral replication and that the H6LTs may function as the source of the IE1/IE2 proteins that contribute to the efficient reactivation of these viruses. This hypothesized function is comparable to the possible function of herpes simplex virus latency-associated transcripts, which may enhance viral reactivation (9, 18).
Our findings revealed that the structures, the encoded proteins, and the expression of the HHV-6 latency-associated transcripts were similar to those of HCMV latency-specific transcripts (14, 16). The H6LTs and the HCMV latency-specific transcripts may have some common function during latency. It has previously been reported that the HHV-6 late gene U94 (23, 25) is also expressed during latency (27). Because other betaherpesviruses, such as HCMV and human herpesvirus 7, do not carry homologues of U94 in their genome (3, 20, 24), the U94 gene may play some specific role in HHV-6. The conserved features of the HHV-6 and HCMV latent transcripts that we observed in this study may assist us in understanding betaherpesvirus latency further.
Nucleotide sequence accession numbers.
EMBL/GenBank/DDBJ accession numbers of HHV-6B IE1 and IE2 are AB075773 and AB075774, respectively. EMBL/GenBank/DDBJ accession numbers of H6LTs are as follows: type I IE1-related H6LT, AB075775; type I IE2-related H6LT, AB075776; type II IE1-related H6LT, AB075777; type II IE2-related H6LT, AB075778.
Acknowledgments
We thank Takako Yamada and her colleagues for their kind help in obtaining blood samples.
This study was partially supported by a Special Coordination Fund and a grant-in-aid for general scientific research of the Ministry of Education, Culture, Sports, Science, and Technology, of the Japanese Government.
REFERENCES
- 1.Ablashi, D. V., N. Balachandran, S. F. Josephs, C. L. Hung, G. R. Krueger, B. Kramarsky, S. Z. Salahuddin, and R. C. Gallo. 1991. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology 184:545-552. [DOI] [PubMed] [Google Scholar]
- 2.Bresnahan, W. A., and T. Shenk. 2000. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 288:2373-2376. [DOI] [PubMed] [Google Scholar]
- 3.Chee, M. S., A. T. Bankier, S. Beck, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 4.Dewhurst, S., K. McIntyre, K. Schnabel, and C. B. Hall. 1993. Human herpesvirus 6 (HHV-6) variant B accounts for the majority of symptomatic primary HHV-6 infections in a population of U.S. infants. J. Clin. Microbiol. 31:416-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez, G., T. R. Dambaugh, F. R. Stamey, S. Dewhurst, N. Inoue, and P. E. Pellett. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73:8040-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frenkel, N., G. C. Katsafanas, L. S. Wyatt, et al. 1994. Bone marrow transplant recipients harbor the B variant of human herpesvirus 6. Bone Marrow Transplant 14:839-843. [PubMed] [Google Scholar]
- 7.Gompels, U. A., J. Nicholas, G. Lawrence, M. Jones, B. J. Thomson, M. E. Martin, S. Efstathiou, M. Craxton, and H. A. Macaulay. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29-51. [DOI] [PubMed] [Google Scholar]
- 8.Greijer, A. E., C. A. J. Dekkers, and J. M. Middeldorp. 2000. Human cytomegalovirus virions differentially incorporate viral and host cell RNA during the assembly process. J. Virol. 74:9078-9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill, J. M., H. H. Garza, Jr., Y. H. Su, R. Meegalla, L. A. Hanna, J. M. Loutsch, H. W. Thompson, E. D. Varnell, D. C. Bloom, and T. M. Block. 1997. A 437-base-pair deletion at the beginning of the latency-associated transcript promoter significantly reduced adrenergically induced herpes simplex virus type 1 ocular reactivation in latently infected rabbits. J. Virol. 71:6555-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isegawa, Y., T. Mukai, K. Nakano, M. Kagawa, J. Chen, Y. Mori, T. Sunagawa, K. Kawanishi, J. Sashihara, A. Hata, P. Zou, H. Kosuge, and K. Yamanishi. 1999. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J. Virol. 73:8053-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kempf, W., V. Adams, N. Wey, et al. 1997. CD68+ cells of monocyte/macrophage lineage in the environment of AIDS-associated and classic-sporadic Kaposi sarcoma are singly or doubly infected with human herpesviruses 7 and 6B. Proc. Natl. Acad. Sci. USA 94:7600-7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo, K., H. Kaneshima, and E. S. Mocarski. 1994. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc. Natl. Acad. Sci. USA 91:11879-11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo, K., T. Kondo, T. Okuno, M. Takahashi, and K. Yamanishi. 1991. Latent human herpesvirus 6 infection of human monocytes/macrophages. J. Gen. Virol. 72:1401-1408. [DOI] [PubMed] [Google Scholar]
- 14.Kondo, K., and E. S. Mocarski. 1995. Cytomegalovirus latency and latency-specific transcription in hematopoietic progenitors. Scand. J. Infect. Dis. Suppl. 99:63-67. [PubMed] [Google Scholar]
- 15.Kondo, K., H. Nagafuji, A. Hata, C. Tomomori, and K. Yamanishi. 1993. Association of human herpesvirus 6 infection of the central nervous system with recurrence of febrile convulsions. J. Infect. Dis. 167:1197-1200. [DOI] [PubMed] [Google Scholar]
- 16.Kondo, K., J. Xu, and E. S. Mocarski. 1996. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc. Natl. Acad. Sci. USA 93:11137-11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Law, G. L., A. Raney, C. Heusner, and D. R. Morris. 2001. Polyamine regulation of ribosome pausing at the upstream open reading frame of s-adenosylmethionine decarboxylase. J. Biol. Chem. 276:38036-38043. [DOI] [PubMed] [Google Scholar]
- 18.Leib, D. A., C. L. Bogard, M. Kosz-Vnenchack, K. A. Hicks, D. M. Coen, D. M. Knipe, and P. A. Schaffer. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J. Virol. 63:2893-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lunetta, J. M. 2000. Latency-associated sense transcripts are expressed during in vitro human cytomegalovirus productive infection. Virology 278:467-476. [DOI] [PubMed] [Google Scholar]
- 20.Megaw, A. G., D. Rapaport, B. Avidor, N. Frenkel, and A. J. Davison. 1998. The DNA sequence of the RK strain of human herpesvirus 7. Virology 244:119-132. [DOI] [PubMed] [Google Scholar]
- 21.Mirandola, P., P. Menegazzi, S. Merighi, T. Ravaioli, E. Cassai, and D. Di Luca. 1998. Temporal mapping of transcripts in herpesvirus 6 variants. J. Virol. 72:3837-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyashita, E. M., B. Yang, G. J. Babcock, and D. A. Thorley-Lawson. 1997. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J. Virol. 71:4882-4891. [Erratum, 72:9419, 1998.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mori, Y., P. Dhepakson, T. Shimamoto, K. Ueda, Y. Gomi, H. Tani, Y. Matsuura, and K. Yamanishi. 2000. Expression of human herpesvirus 6B rep within infected cells and binding of its gene product to the TATA-binding protein in vitro and in vivo. J. Virol. 74:6096-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholas, J. 1996. Determination and analysis of the complete nucleotide sequence of human herpesvirus. J. Virol. 70:5975-5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rapp, J. C., L. T. Krug, N. Inoue, T. R. Dumbaugh, and P. E. Pellett. 2000. U94, the human herpesvirus 6 homolog of the parvovirus nonstructural gene, is highly conserved among isolates and is expressed at low mRNA levels as a spliced transcript. Virology 268:504-516. [DOI] [PubMed] [Google Scholar]
- 26.Roizman, B., R. C. Desrosiers, et al. 1992. The family herpesviridae: an update. Arch. Virol. 123:425-449. [DOI] [PubMed] [Google Scholar]
- 27.Rotola, A., T. Ravaioli, A. Gonelli, et al. 1998. U94 of human herpesvirus 6 is expressed in latently infected peripheral blood mononuclear cells and blocks viral gene expression in transformed lymphocytes in culture. Proc. Natl. Acad. Sci. USA 95:13911-13916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiewe, U., F. Neipel, D. Schreiner, and B. Fleckenstein. 1994. Structure and transcription of an immediate-early region in the human herpesvirus 6 genome. J. Virol. 68:2978-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slobedman, B., and E. S. Mocarski. 1999. Quantitative analysis of latent human cytomegalovirus. J. Virol. 73:4806-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spivack, J. G. 1987. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J. Virol. 61:3841-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, Z., P. Fang, and M. S. Sachs. 1998. The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it. Mol. Cell. Biol. 18:7528-7536. [DOI] [PMC free article] [PubMed] [Google Scholar]