Abstract
The eukaryotic genome is duplicated exactly once per cell division cycle. A strategy that limits every replication origin to a single initiation event is tightly regulated by a multiprotein complex, which involves at least 20 protein factors. A key player in this regulation is the evolutionary conserved hexameric MCM2-7 complex. From maize (Zea mays) zygotes, we have cloned MCM6 and characterized this essential gene in more detail. Shortly after fertilization, expression of ZmMCM6 is strongly induced. During progression of zygote and proembryo development, ZmMCM6 transcript amounts decrease and are low in vegetative tissues, where expression is restricted to tissues containing proliferating cells. The highest protein amounts are detectable about 6 to 20 d after fertilization in developing kernels. Subcellular localization studies revealed that MCM6 protein shuttles between cytoplasm and nucleoplasm in a cell cycle-dependent manner. ZmMCM6 is taken up by the nucleus during G1 phase and the highest protein levels were observed during late G1/S phase. ZmMCM6 is excluded from the nucleus during late S, G2, and mitosis. Transgenic maize was generated to overexpress and down-regulate ZmMCM6. Plants displaying minor antisense transcript amounts were reduced in size and did not develop cobs to maturity. Down-regulation of ZmMCM6 gene activity seems also to affect pollen development because antisense transgenes could not be propagated via pollen to wild-type plants. In summary, the transgenic data indicate that MCM6 is essential for both vegetative as well as reproductive growth and development in plants.
DNA replication of the eukaryotic genome in S phase is accomplished only once during each cell division cycle. This process is precisely regulated and controlled by the prereplicative complex (pre-RC) consisting of origin recognition complex (ORC), Cdt1, Cdc6, as well as minichromosome maintenance (MCM) proteins (Tye, 1999; Bogan et al., 2000; Hyrien et al., 2003; Arias and Walter, 2005; Blow and Dutta, 2005). The initiation of DNA synthesis is thus regulated as a multistep process involving the binding of ORC to replication origins followed by a stepwise recruitment of Cdc6, Cdt1, and MCM proteins to form the pre-RC. Finally, the pre-RC becomes activated by Cdc7/Dbf4 (DDK) and Cdc28 (Cdk2)/cyclin E protein kinases, leading to Cdc45 binding to the pre-RC to initiate DNA unwinding and DNA synthesis (Labib and Diffley, 2001; Lei and Tye, 2001; Hyrien et al., 2003). After the initiation of DNA replication in yeast (Saccharomyces cerevisiae) and animal cells, MCM proteins are displaced from chromatin and its reassociation is inhibited until cells pass through mitosis (Blow and Dutta, 2005).
Thus, the dynamic changes in the assembly and disassembly of the MCM subcomplex is critical for the regulation of DNA replication. Originally identified as proteins required for MCM in yeast, the evolutionary conserved MCM proteins are now regarded as being essential for both initiation and elongation of DNA replication in eukaryotes and archaebacteria (for review, see Tye, 1999; Forsburg, 2004; Blow and Dutta, 2005). The best known among the MCMs are a family of six structurally related proteins, MCM2 to 7, which assemble at the replication origins during early G1 phase of the cell cycle to form a hexamer. During S phase, MCM proteins bind preferentially to unreplicated DNA rather than to replicating or replicated DNA (Laskey and Madine, 2003) and appear to travel along the chromatin with the replication fork (Claycomb et al., 2002). Based on recent data, the MCM hexamer is currently regarded as the prime candidate for the DNA helicase that unwinds DNA at replication forks (Ishimi, 1997; Labib et al., 2000; Bailis and Forsburg, 2004; Shechter and Gautier, 2004; Blow and Dutta, 2005). Recent experimental data suggest that MCM proteins might also be involved in additional chromosomal processes including transcription, chromatin remodeling, and genome stability (for review, see Forsburg, 2004).
Compared to yeast, animals, and humans, surprisingly little is known about MCM proteins in plants. PROLIFERA (PRL) was the first MCM protein identified in plants and was shown to be required during reproduction for megagametophyte and embryo development, but is also expressed in dividing sporophytic tissues (Springer et al., 1995). PRL is required maternally and not paternally during the early stages of embryogenesis, suggesting that the female gametophyte provides substantial PRL function during the early stages of embryogenesis in Arabidopsis (Arabidopsis thaliana; Springer et al., 2000). A more general role of PRL, which is now called AtMCM7, in cell proliferation and cytokinesis throughout plant development was described by Springer et al. (2000) and Holding and Springer (2002). The maize (Zea mays) homolog of PRL (ZmPRL) was recently identified by differential display as ZmPRL mRNA accumulates in the apical region of the maize immature embryo (Bastida and Puigdomènech, 2002). Up to now, only one additional member of the plant MCM gene family, ZmMCM3 (also called Zea mays Replication Origin Activator [ZmROA]) was described in more detail and was shown to be expressed in proliferative tissues to specific subpopulations of cycling cells (Sabelli et al., 1996). The promoters of ZmROA and its Arabidopsis homolog, AtMCM3, have been cloned (Sabelli et al., 1999; Stevens et al., 2002). The promoter of AtMCM3 was analyzed in more detail because it contains two consensus binding sites for the cell cycle regulator E2F and was shown to be transcriptionally regulated during late G1/S phase of the cell cycle (Stevens et al., 2002).
Here we report the molecular cloning and functional characterization of MCM6 from maize (ZmMCM6). During our investigations about the onset of zygotic gene activation (ZGA)/embryonic gene activation (EGA), we identified a number of genes that are up-regulated or expressed de novo shortly after fertilization in the maize and wheat (Triticum aestivum) zygotes (Dresselhaus et al., 1999; Sprunck et al., 2005). We became especially interested in studying the fertilization-induced gene ZmMCM6 in more detail because the presence of two MCM6 homologs has been reported in frog (Xenopus leavis). The maternal gene XlmMCM6 is expressed until the midblastula transition (MBT) stage of embryo development when embryonic transcription begins. After EGA, the cell cycle is remodeled and a zygotic/embryonic MCM6 (XlzMCM6) that differs from maternal MCM6 assembles into MCM complexes (Sible et al., 1998). ZmMCM6 shows a higher homology to XlzMCM6 and has a similar carboxy-terminal extension that is absent in maternal XlmMCM6. In contrast to frog, we found only one MCM6 gene in databases of other animals and within the genome of Arabidopsis. The other MCM2 to 7 members are encoded by a single-copy gene as well. We have analyzed the gene and protein expression of maize MCM6 in different reproductive and vegetative tissues. Protein localization was investigated using a ZmMCM6-green fluorescent protein (GFP) fusion protein in cell cycle-arrested onion (Allium cepa) epidermal cells and during the cell cycle in maize Black Mexican Sweet (BMS) suspension cells. In addition, overexpressing and antisense transgenic maize plants were generated to investigate ZmMCM6 function and to modify vegetative and reproductive growth by manipulating the cell cycle.
RESULTS
Structural Properties of ZmMCM6 and Other Plant MCM Proteins
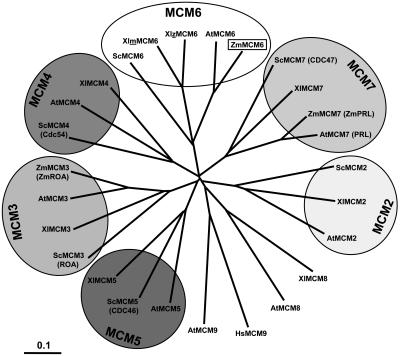
We have compared all MCM proteins of Arabidopsis and the known maize MCM proteins with MCM proteins of budding yeast and African clawed frog. Yeast and frog have been selected because all MCM2 to 7 genes from these two species are known and functions of MCM proteins have been most intensely studied in these species, using genetic approaches in the case of budding yeast and biochemistry in the case of frog. As shown in Figure 1, all MCM2 to 7 proteins are encoded by single-copy genes with the exception of a duplicated MCM6 gene of frog. The expanding tree indicates that MCM proteins of fungi, animals, and plants all evolved from one ancestor molecule. Plant MCMs display a higher homology among each other compared with corresponding MCMs of other organisms. Interestingly, the Arabidopsis genome encodes two non-MCM2 to 7 proteins (MCM8 and MCM9), which show homology to MCM8 of frog and human MCM9, respectively. MCM9 of frog is not known to date.
Figure 1.
Phylogenetic tree of MCM2 to 7 protein sequences. Branch lengths are proportional to phylogenetic distances and scale bar represents 10% substitutions per site. The tree was drawn by TreeView from a ClustalW alignment using all available MCM protein sequences from Arabidopsis (AtMCM2-9), maize (ZmMCM3, 6, and 7), frog (XlMCM2-8), budding yeast (ScMCM2-7), as well as human MCM9. Due to historical reasons, some proteins were given two names (old names in parentheses). Note that a maternal (m) and a zygotic (z) MCM6 protein have been described in frog. MCM6 of maize is described in this study. For GenBank accession numbers of sequences, see Table I.
MCM classes consist of large proteins of 716 to 1,017 amino acid residues (molecular mass between 80 and 113 kD) with the exception of Arabidopsis and human MCM9s, which are 610 and 391 amino acids in length, respectively. Table I shows a summary of characteristic features of MCM proteins. Nuclear localization sequences (NLS) have been predicted by PSORT in MCM2 and MCM3 proteins, but not in maize MCM6 and most other MCMs. We have identified potential zinc-finger motifs that might be involved in protein-protein interactions in the N-terminal regions of all MCM proteins with the exception of the MCM3 class and MCM5 from yeast. The zinc-finger motif CX2CX18-19CX4C was found in all MCM6, MCM7, and MCM8 sequences. MCM4 proteins contain either this or the CX2CX18-19CX2C motif that was also found in the MCM2 protein sequences. Deviations from these classical motifs were found as CX2CX20-24CX5-10C in the MCM5 and MCM9 classes. In addition, cyclin/cyclin-dependent kinase (CDK) phosphorylation sites (S/T)Px(K/R), which might function as cell cycle regulation motifs, were identified in some MCM proteins. Interestingly, two of the hexameric MCM2 to 7 proteins of each organism investigated in our studies contain this CDK box. For yeast, these are MCM3 and MCM4, in African clawed frog, these are MCM2 and MCM4, whereas in Arabidopsis and maize, these are MCM3 and MCM6.
Table I.
Predicted protein domains and accessions of MCM proteins
Symbols in parentheses indicate nuclear localization signals (NLS) predicted by PSORT, but deviating from experimental data. Data in the CDK-Box column indicate the number of predicted CDK consensus phosphorylation sites.
| Protein Name | Length (Amino Acids) | NLS | CDK-Box | Zinc Finger | Accession No. |
|---|---|---|---|---|---|
| ScMCM2 | 868 | + | − | + | P29469 |
| XlMCM2 | 886 | + (−) | 2 | + | U44047 |
| AtMCM2 | 928 | + (−) | − | + | NM_103572 |
| ScMCM3 (ROA) | 971 | + | 5 | − | P24279 |
| XlMCM3 | 807 | + | − | − | U26057 |
| AtMCM3 | 776 | + | 1 | − | NM_123997 |
| ZmMCM3 (ZmROA) | 768 | + | 1 | − | AAD48086 |
| ScMCM4 (CDC54) | 933 | − (+) | 2 | + | S56050 |
| XlMCM4 | 858 | − (+) | 2 | + | T47223 |
| AtMCM4 | 743 | − | − | + | NP_179236 |
| ScMCM5 (CDC46) | 775 | − (+) | − | − | P29496 |
| XlMCM5 | 735 | − (+) | − | + | PC4225 |
| AtMCM5 | 727 | − | − | + | NP_178812 |
| ScMCM6 | 1,017 | − (+) | − | + | S64219 |
| XlmMCM6 | 796 | − | − | + | T47222 |
| XlzMCM6 | 824 | − | − | + | AF031139 |
| AtMCM6 | 831 | − | 1 | + | NP_680393 |
| ZmMCM6 | 831 | − | 1 | + | AAW55593 |
| ScMCM7 (CDC47) | 845 | − (+) | − | + | S34027 |
| XlMCM7 | 720 | − (+) | − | + | T47221 |
| AtMCM7 (PRL) | 716 | − | − | + | L39954 |
| ZmMCM7 (ZmPRL) | 720 | − | − | + | CAC44902 |
| AtMCM8 | 777 | − | 2 | + | NM_111800 |
| XlMCM8 | 834 | − | − | + | CAI29793 |
| AtMCM9 | 610 | − | 1 | + | NM_126977 |
| HsMCM9 | 391 | − | 1 | + | NP_694987 |
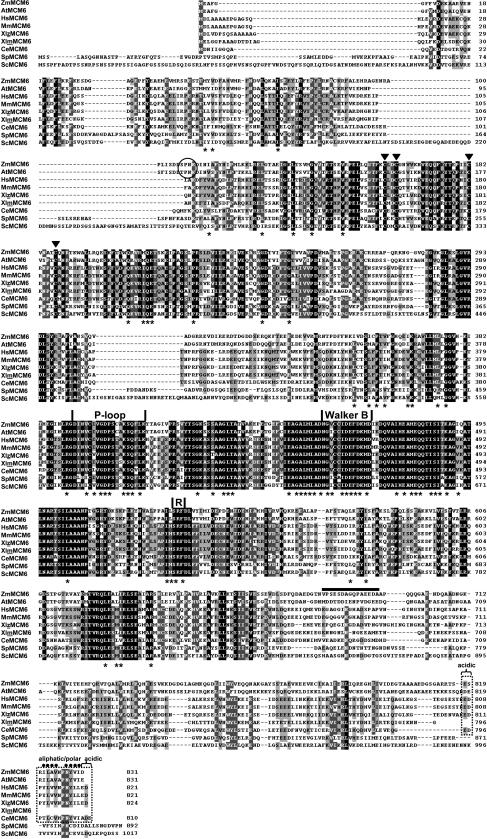
We have analyzed the structural properties of MCM6 proteins in more detail. With the exception of MCM6 from rat, Figure 2 shows the alignment of all MCM6 protein sequences available in public databases. The largest and most conserved stretch of about 153 amino acids in the central region includes elements of the Walker-type nucleoside triphosphate-binding domain (Walker et al., 1982), which is conserved in a number of ATPases. The putative ATP-binding site of MCM6 proteins is shown by a P-loop that includes the Walker A motif (GDPS[C/T][A/S]KS) and the Walker B motif. The B motif is part of a highly conserved domain that begins with the acidic amino acid Glu preceding a stretch of hydrophobic residues predicted to form a β-strand and an acidic stretch. Finally, the conserved central domain also contains the R- or SRF (Ser-Arg-Phe)-finger. The Arg residue within the R-finger probably represents the catalytic activity (Davey et al., 2003).
Figure 2.
ZmMCM6 represents a member of the highly conserved MCM protein family. Homology searches were performed with BLASTP and protein sequences were aligned with ClustalW and drawn using GeneDoc. Identical residues are shaded in black and similar residues are shaded in gray. The alignment shows deduced amino acid sequences of MCM6 from maize (ZmMCM6) and all other eukaryotes available in public databases with the exception of MCM6 from rat, which is almost identical with MCM6 from humans and mouse. GenBank accession numbers for most MCM6 proteins are given in Table I. The human (HsMCM6), mouse (MmMCM6), C. elegans (CeMCM6), and fission yeast (SpMCM6) protein sequence accessions are Q14566, P97311, P34647, and P49731, respectively. Amino acid residues identical/similar in MCM proteins listed in Table I are labeled with asterisks. Highest conserved regions consist of the P-loop, the Walker B motif, and the R- or SRF-finger. The first two motifs are characteristic for ATPases and are essential for NTP binding. The Arg residue within the R-finger probably represents the catalytic activity. The four triangles mark the Cys residues of the zinc-finger-type motif. A predicted cyclin/CDK phosphorylation site that occurs only in plant MCM6 proteins is encircled. A conserved C-terminal region characterized by an aliphatic/polar region of 10 amino acids (dots) flanked on both sides by one to two acidic amino acid residues, which is characteristic for MCM6 of multicellular organisms, but absent in maternal MCM6 of frog and MCM6 proteins of yeast, is boxed with broken lines. The C-terminal region of ZmMCM6 that was used to obtain a peptide antibody is indicated by a horizontal bar.
In addition to an N-terminal zinc-finger motif and central catalytic domain, a cyclin/CDK phosphorylation site (S/T)Px(K/R) was found only in the N-terminal region of plant MCM6 proteins at position 107 to 110 (SPnK) in ZmMCM6 and 102 to 105 (TPnK) in AtMCM6, respectively. Finally, a conserved motif of unknown function was found at the very C terminus of most MCM6 proteins (Fig. 2, boxed region). This region of 14 to 16 amino acids consists of an aliphatic/polar core that is flanked on both sides by acidic amino acids. Although the function of this motif is unknown, it is characteristic for all MCM6 proteins expressed postfertilization in higher eukaryotes, is absent in maternal MCM6 of frog, and is different in fungi.
ZmMCM6 Is Strongly Up-Regulated in the Female Gametophyte after Fertilization and Expression Is Low in Vegetative Proliferating Tissues
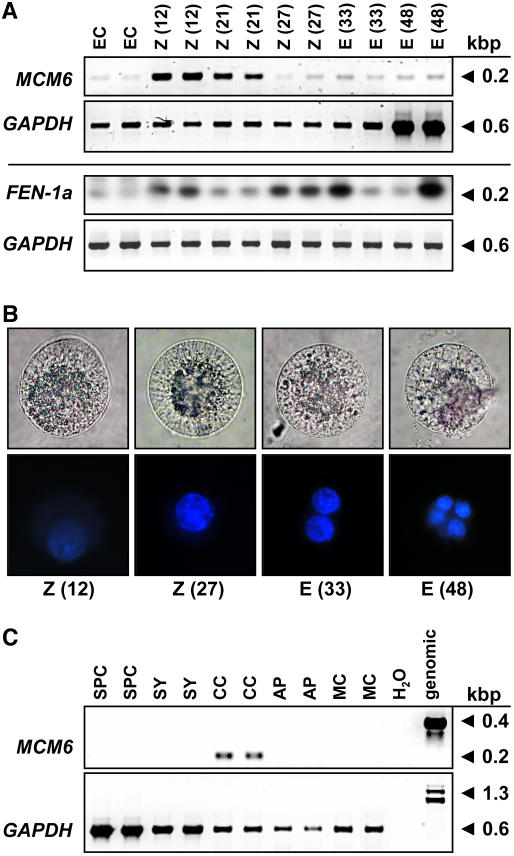
We have analyzed the expression, subcellular localization, and function of the fertilization-induced MCM6 gene of maize in more detail. Low gene expression levels were detected by single-cell (SC) reverse transcription (RT)-PCR in the unfertilized egg cell (Fig. 3A). Twelve hours after in vitro pollination (IVP; this stage corresponds to about 6 h after fertilization; E. Kranz, personal communication), high transcript amounts have been detected in the zygote. Later, during zygote development (21 h after IVP), ZmMCM6 transcript levels decrease and remain low 27 to 48 h after IVP. A significant oscillation of gene expression in a cell cycle-dependent manner was not observed. For comparison, another gene, ZmFEN-1a, which was also identified in our screen for fertilization-induced genes and which encodes a flap endonuclease required for DNA repair, was used to study cell cycle-dependent gene expression during zygote and proembryo development. ZmFEN-1a is a homolog of RAD27 from yeast showing up-regulation of gene expression during late G1 phase (Vallen and Cross, 1995). Similar to ZmMCM6, expression of ZmFEN-1a is induced 12 h after IVP, but additional gene expression peaks have been observed 27 to 33 and 48 h after IVP, indicating that G1 phase occurs in the zygote at about 12 h after IVP, in the two-cell proembryo at about 27 to 33 h, and in the four-cell proembryo at about 48 h. To correlate these findings with zygotic cell division, different developmental stages were stained with 4′,6-diamidino-2-phenylindole (DAPI) and fluorescence quantified by ImageJ software. As shown in Figure 3B, relative DAPI staining of the zygote nucleus 12 h after IVP was about 50% compared with a zygote nucleus 27 h after IVP, indicating that chromatin of the latter cell was in G2 phase, whereas the first cell was in G1 phase. DAPI staining of the egg nucleus was much weaker compared with the zygote nucleus 12 h after IVP (data not shown). Relative DAPI fluorescence measured 33 h after IVP of a two-celled proembryo corresponds to the sum of the zygote nucleus 27 h after IVP, indicating that this proembryo was in G1 phase. A four-celled proembryo was present 48 h after IVP. Please note that the cells used for the expression analysis of ZmFEN-1a were collected in early summer when zygote and embryo development are 10% to 20% accelerated, which explains the accelerated gene expression pattern compared with the DAPI measurements. In summary, ZmMCM6 transcript levels are high about 6 h after fertilization (12 h after IVP) in G1 and continuously decrease throughout zygote and early embryo development (18, 21, 24, 27, 30, 33, and 48 h after IVP; data for stages 18, 24, and 30 h not shown), but significant oscillation of gene expression during early development was not observed. Expression studies of ZmMCM6 in cells of the female gametophyte revealed relatively high transcript amounts in the central cell (Fig. 3C), but not in the other cells of the unfertilized embryo sac. Only one out of six synergids analyzed showed a weak signal (data not shown). Five sperm cells and antipodal clusters of at least 10 cells were collected and used to study ZmMCM6 expression. A very weak signal could be detected in one out of three antipodal clusters tested, but never in sperm cells or in leaf mesophyll protoplasts derived from mature leaves.
Figure 3.
Expression of ZmMCM6 in gametic cells before and after fertilization. A, SC RT-PCR of ZmMCM6 and ZmFEN-1a in egg cells (EC), zygotes (Z), and proembryos (E) at different time points after IVP. The numbers in parentheses indicate hours after pollination (fertilization occurs about 6 h after IVP; top row). Amplification of GAPDH from the same cells is shown for comparison in the corresponding bottom rows. Note that the cells for ZmMCM6 and for DAPI staining (B) were collected in winter and for ZmFEN-1a in summer. ZmFEN1a signals were intensified after blotting and hybridization with a gene-specific radioactive probe. B, Light (top row) and fluorescent images (bottom row) of DAPI-stained zygotes and proembryos. Time points after IVP are indicated. Cell size is about 60 μm. C, SC RT-PCR of ZmMCM6 in sperm cells (SPC), cells of the female gametophyte, including synergid (SY), central cell (CC), and antipodals (AP), as well as leaf mesophyll cell (MC) and controls (water and genomic). Amplification of GAPDH from the same samples is shown in the bottom row.
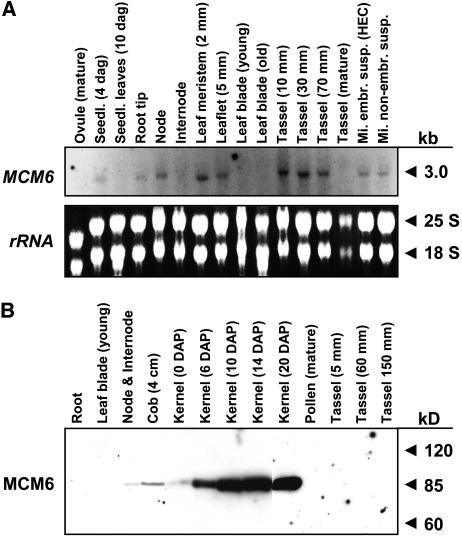
Compared to zygotes shortly after fertilization, expression of ZmMCM6 in vegetative and complex reproductive tissues is very low. To display significant ZmMCM6 transcript signals from a northern blot containing total RNA, hybridization with a radiolabeled probe and exposure of up to 14 d using intensifier screens was necessary (Fig. 4A). A single band slightly smaller than 3.0 kb was detected, indicating that the cloned 2,785 nucleotides of ZmMCM6 represent the full-length transcript. The strongest signals were obtained from tissues containing proliferating cells such as root tips, nodes, leaf meristem, and developing tassels, but also embryonic and nonembryogenic suspension cultures. In developing tassels, signal intensity correlates with developmental stages, while signals were absent in tassels at maturity. Moreover, signals were detected in whole-seedling tissue 4 d after germination, but were absent in leaf tissues of older seedlings (10 d after germination). Leaf meristem displayed relatively strong signals, whereas signals were absent in mature leaves. A ZmMCM6-specific peptide antibody against the less conserved C-terminal part of the protein (see also Fig. 2) was used to detect MCM6 in different tissues and developmental stages (Fig. 4B). A single band of about 90 kD was detected in protein blots, which corresponds to the expected size of 92.5 kD. Minor protein amounts could be detected in nodes, immature cobs, and ovaries, whereas significant ZmMCM6 amounts could not be detected in young and mature leaves and whole roots during tassel maturation as well as in mature pollen. The highest protein amounts are present in developing kernels. While ZmMCM6 protein amounts are relatively low in kernels prior to fertilization (0 days after pollination [DAP]), a strong increase was observed 6 DAP and highest protein levels were detected 10 to 20 DAP.
Figure 4.
Expression of ZmMCM6 gene and protein in vegetative and complex reproductive tissues. A, RNA gel blot showing expression of ZmMCM6 in the vegetative and reproductive tissues indicated. Fifteen micrograms of total RNA from each tissue were hybridized to a radioactively labeled probe containing the 3′ untranslated region as well as 896 bp encoding the C-terminal region of the gene. The film was exposed for 2 weeks using intensifier screens. B, Protein gel blot incubated with a peptide antibody directed against 15 amino acid residues within a ZmMCM6 C-terminal-specific region (see also Fig. 2). Ten micrograms of protein of the tissues indicated were each separated by 8% SDS-PAGE and blotted onto a polyvinylidene difluoride membrane. A single band was obtained showing the specificity of the antibody.
ZmMCM6 Shuttles between Cytoplasm and Nucleoplasm in a Cell Cycle-Dependent Manner
We have used a chimeric ZmMCM6 protein fused to GFP and immunocytochemistry to study the subcellular localization of ZmMCM6. First, onion epidermal cells were bombarded with a construct encoding a ZmMCM6-GFP fusion protein under the control of the strong and constitutively expressing ubiquitin (UBI) promoter of maize. As shown in Figure 5, A and B, relatively strong GFP signals accumulated in cytoplasm around the nucleus and in transvacuolar strands of the cytoplasm surrounding the nucleus. Protein localization in the nucleus was not detectable. Focusing through side views of nuclei displayed more clearly that the fusion protein accumulated in cytoplasm surrounding the nucleus, but was excluded from the nucleoplasm (Fig. 5, C and D). In contrast, the N-terminal 388 amino acids of a maize transcriptional regulator of anthocyanin biosynthesis (Ludwig et al., 1989) that also includes an NLS fused with GFP (Lc-GFP in Fig. 5E) displayed most fluorescence within the nucleus. However, onion epidermal cells are no longer proliferating and might be in G0 phase of the cell cycle. We have therefore used BMS maize suspension cultures to study ZmMCM6-GFP subcellular protein localization during the different stages of the cell cycle. Synchronization of BMS suspension cultures to correlate protein localization with defined cell cycle stages was not successful. We have therefore determined relative DNA amounts of individual BMS cells after DAPI dihydrochloride staining. Strongest DAPI fluorescence was measured from cells in G2 phase. Signals from cells in G1 phase were less than 50%, probably due to higher amounts of heterochromatin and beginning of chromosome condensation in nuclei of cells in G2 in preparation for mitosis. The weakest fluorescence was measured from cells in late G1/S phase, which is likely to result from decondensation of DNA during DNA synthesis. In total, 62 individual cells were analyzed for GFP fluorescence and DAPI staining. As shown in Figure 6, A to C, cells in G1 phase (42% of the cells analyzed) showed localization of the chimeric protein in both cytoplasm and nucleoplasm. A detailed analysis of cells in G1 phase using confocal laser-scanning microscopy (CLSM) displayed a gradient of GFP fluorescence between the cytoplasm and nucleoplasm (Fig. 6, Q and R), indicating precise control of ZmMCM6 import into the nucleus. Highest protein amounts were detected in the nucleus during late G1/S phase (Fig. 6, D–F; 11.5% of the cells studied). At the end of S phase or during early G2 phase, ZmMCM6 became fully excluded from the nucleus, as all cells in G2 phase (45% of the cells analyzed) lacked GFP fluorescence in the nucleus (Fig. 6, G–L). CLSM studies confirmed this observation (Fig. 6M). One cell was observed in early prophase of mitosis (Fig. 6, N–P) showing condensation of chromosomes (Fig. 6O). Similar to G2 cells, GFP signals were not detectable in the nucleus of this cell. Interestingly, we have never detected GFP fluorescence in the nucleolus (Fig. 6, Q and R). In contrast, BMS suspension cells transiently expressing a UBIp:GFP construct always showed equal GFP fluorescence in the cytoplasm, nucleoplasm, and nucleolus independently from the stage of cell cycle (Fig. 6S).
Figure 5.
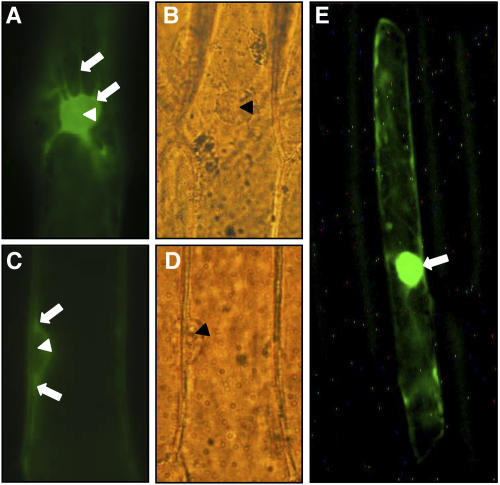
ZmMCM6 protein is excluded from the nucleus in epidermal onion cells. Onion cells were transiently transformed with a UBIp:ZmMCM6-GFP construct and analyzed using epifluorescence (A and C) and light microscopy (B and D). A, Top view of a nucleus showing accumulation of chimeric protein in cytoplasm around the nucleus. The arrows point toward cytoplasm surrounding the nucleus and a transvacuolar cytoplasmic strand. B, Light microscopic image of A. C, Side view of a nucleus showing accumulation of chimeric protein in cytoplasm around the nucleus (arrows), but not inside the nucleus. D, Light microscopic image of C. E, Epifluorescence of an onion epidermal cell bombarded with a 35Sp:Lc-GFP construct encoding the N-terminal 388 amino acids (including the NLS) of a maize transcriptional regulator of anthocyanin biosynthesis in maize (GenBank accession no. A41388) fused with GFP. Most of the fluorescence was detected within the nucleus (arrow). Accumulation of the chimeric protein in cytoplasm around the nucleus, as in A and C, was never observed. Arrowheads in A to D point toward nucleoli.
Figure 6.
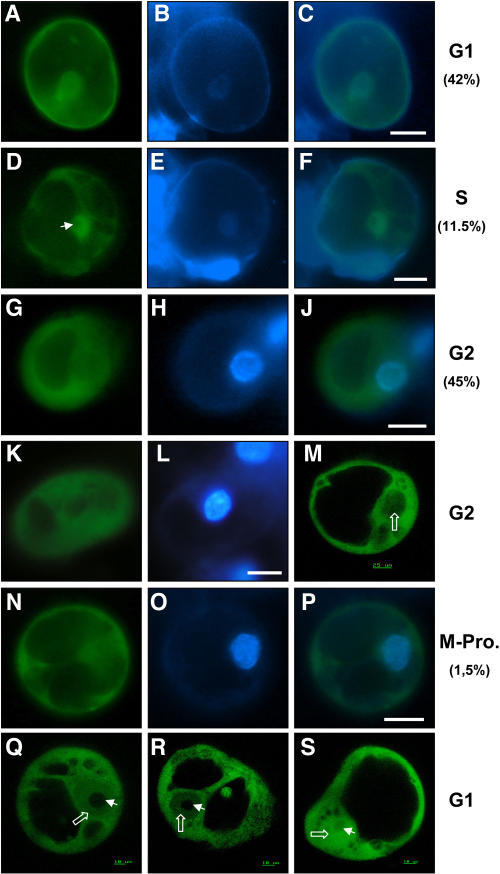
ZmMCM6 shuttles between nucleus and cytoplasm in maize BMS suspension cells in a cell cycle-dependent manner. Suspension cells were transiently transformed with a UBIp:ZmMCM6-GFP construct and analyzed using epifluorescence microscopy (A–L and N–P) as well as CSLM (M, Q–S). Relative DNA content of cells was determined after DAPI staining. A, About 42% cells showed localization of the chimeric protein in both cytoplasm and nucleoplasm. B, DAPI staining of the cell shown in A to display the nucleus. C, Merged image of A and B showing similar DAPI signal in the nucleus compared to GFP fluorescence. These cells were counted for G1 phase. D, About 11.5% of the cells examined accumulated most of the chimeric protein in the nucleoplasm, but not in the nucleolus (arrowhead). E, DAPI staining of the cell shown in D. The image was enhanced to display the weak fluorescence of the nucleus in late G1/S phase. F, Merged image of D and E showing mainly GFP fluorescence in the nucleus. G and K, Majority of cells (about 45%) displayed GFP fluorescence only in the cytoplasm and lacked GFP signals in the nucleus. H and L, Strong DAPI staining of the nuclei of these cells revealed that they were in G2 phase. J, Merged image of G and H showing GFP fluorescence in the cytoplasm and DAPI fluorescence in the nucleus. M, Stack of five CLSM images of a cell in G2 phase confirms lack of GFP fluorescence in the nucleus (open white arrow). N, Cell showing GFP fluorescence exclusively in the cytoplasm displayed condensation of chromosomes (O) during prophase of mitosis. P, Merged image of N and O confirms lack of GFP fluorescence in the nucleus. Q and R, Gradient of the fusion protein between cytoplasm and nucleoplasm was observed in stacks of five images each of two different cells in G1 phase. The weak DAPI staining of these cells is not shown. Note that the chimeric protein is localized in both cytoplasm and nucleoplasm (open white arrows), but never in the nucleolus (arrowhead). S, Control showing a suspension cell bombarded with a UBIp:GFP construct showing GFP fluorescence in the nucleolus, nucleoplasm, and cytoplasm. The arrowhead points toward the nucleolus and the open arrow toward the nucleoplasm. Scale bars are 20 μm, unless otherwise indicated.
Immunocytochemistry with isolated BMS nuclei was performed to measure the cell cycle dependency of nuclear ZmMCM6 localization more precisely and to prove that the difference of DAPI signal intensity is not originating from problems of dye uptake. DNA and ZmMCM6 content of isolated BMS nuclei were measured after DAPI staining and by using a fluorescein isthiocyanate (FITC)-coupled secondary antibody against the ZmMCM6-specific peptide antibody described above. FITC signals of nuclei showing endoreduplication have not been measured. As shown in Figure 7, A and B, all nuclei displaying strong FITC signals were in G1 or early S phase of the cell cycle. Nuclei in late S or G2 (Fig. 7, C and D) never showed significant signals. Those signals were in the range of background signals that were also obtained after using preimmune serum instead of the serum containing the specific antibody (Fig. 7, E and F). Figure 7G shows a summary of measurements obtained from 44 nuclei. A relative DNA content of 2C (±15%) was considered as G1, a DNA content of 2C (+16% to 25%) as G1/S, and a DNA content of 4C (±15%) as G2. To determine FITC background fluorescence, DNA and FITC signal intensities of 16 randomly chosen nuclei were measured after incubation with preimmune serum (Fig. 7H). In contrast to the control, more than 50% of nuclei in G1 showed significant ZmMCM6 amounts. All nuclei in late G1/early S phase contain high ZmMCM6 levels, which decrease during S phase progression and are no longer measurable at later stages of S phase or in G2. In summary, immunocytochemistry data confirm the above finding that ZmMCM6 is taken up by the nucleus during G1 phase and protein levels are highest during late G1/S phase, while ZmMCM6 is excluded from the nucleus during late S, G2, and mitosis. Attempts to measure ZmMCM6 levels of nuclei from isolated cells of the female gametophyte were not successful, probably because only nuclei of zygotes at defined stages contain sufficient detectable protein amounts and the number of zygote nuclei was not sufficient. A very high number of nuclei, similar to the approach with the BMS suspension cells, will be necessary to determine relative ZmMCM6 protein amounts in female gametophyte nuclei.
Figure 7.
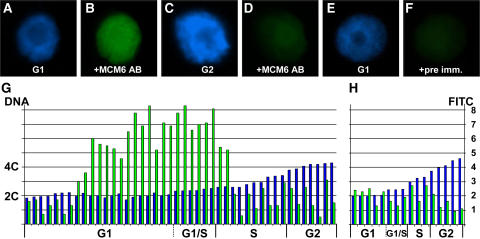
ZmMCM6 is taken up by the nucleus during G1 phase and highest protein levels are detectable during late G1/early S phase of the cell cycle. Relative DNA content of isolated BMS nuclei were determined after DAPI staining. ZmMCM6 protein amounts were measured after incubation with an FITC-coupled antibody against a ZmMCM6 peptide antibody. DAPI staining (A) and FITC signals (B) of a nucleus in G1; DAPI staining (C) and FITC signals (D) of a nucleus in G2; DAPI staining of a G1 phase nucleus (E) and the same nucleus after incubation with preimmune serum showing minor background signals (F). G, Summary of measurements of 44 nuclei. After DAPI measurements, nuclei were classified as G1 (2C ± 15%), G1/S (2C + 16%–25%), S (2C + 26%–84%), and as G2 (4C ± 15%). Histograms show relative DNA (blue bar) and FITC (green bar) levels of individual nuclei. Relative DNA content is indicated at the left and relative FITC signal intensity at the right of image H. H, To show antibody specificity and to determine background signals, nuclei have been incubated with preimmune serum. Classification of nuclei was as described in G.
Phenotypes of Transgenic Maize after ZmMCM6 Up- and Down-Regulation
A transgenic approach was chosen to increase and to down-regulate ZmMCM6 gene expression in maize. One hundred-fifty immature hybrid embryos have been bombarded with a sense construct (UBIp:ZmMCM6) to increase ZmMCM6 transcript amounts by expressing the full-length ZmMCM6 cDNA under the control of the strong and constitutively expressing maize UBI promoter. Four plants have been regenerated (transformation efficiency of 2.7%), all representing one clonal line as they displayed the identical transgene integration pattern (plants SE1a–SE1d). The Southern blot in Figure 8A shows that this line contains multiple transgene integrations, including one or more full-length integrations. However, quantification of ZmMCM6 transcript amounts showed that there was no significant difference between the four transgenic lines compared to wild-type plants (Table II). An obvious phenotype was not observed and plants were both fully male and female fertile. We have therefore bombarded another 400 immature hybrid embryos with a sense construct carrying a GFP conjugate (UBIp:ZmMCM6-GFP) both to increase ZmMCM6 transcript amounts and to simultaneously study ZmMCM6 protein localization. Five independent transgenic lines (G1–G5) have been generated (transformation efficiency of 1.2%) containing one to two transgene integrations. Unfortunately, none of these lines contained a full-length and thus functional integration of the construct (Fig. 8A). GFP expression was therefore not detectable in any of the tissues investigated. The observation that these plants were small (Table II) was probably an effect of longer regeneration periods and growth in winter. Reproductive organs were fully developed and seed set was obtained after selfing.
Figure 8.
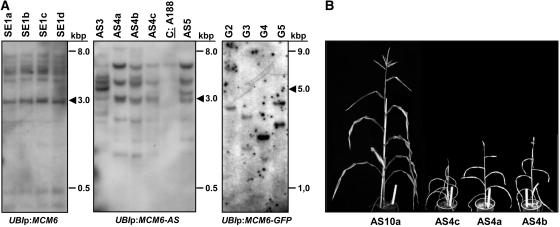
Molecular and phenotypical analysis of transgenic maize containing the ZmMCM6 gene under control of the strong and constitutive maize UBI promoter. A, Examples of Southern blots containing 10 μg genomic DNA of transgenic lines with ZmMCM6 constructs in sense (SE, left) and antisense (AS, middle) orientation, or fused with GFP (G, right). Genomic DNA was digested with enzyme combinations to display full-copy integrations (arrowheads). Left and middle blots were hybridized with a ZmMCM6-specific probe and the right blot with a GFP-specific probe. The control (A188) on the blot in the middle shows the endogenous ZmMCM6 signal. B, Transgenic plants containing a functional ZmMCM6 AS construct were small in size and did not develop cobs to maturity. Tassels contained little pollen. Here, the clonal line AS4a to AS4c is shown. The line AS10a lacking a functional transgene is shown at the left. Plant height of AS10a was comparable to wild-type plants (A188; 160–190 cm) and both a cob and tassel were fully developed. The white ruler in flower pots is 25 cm.
Table II.
Summary of molecular and phenotypical analysis of transgenic maize (T0 and T1 generations, respectively) containing the ZmMCM6 gene under control of the strong and constitutive maize UBI promoter in sense and AS orientation or fused to GFP
n.d., Not determined.
| Construct | Plant No. | Functional Transgenesa | Transgene Quantityb | Plant Heightc | Fertilityd |
|---|---|---|---|---|---|
| − | Wild type (A188) | − | 100/25/0.68 | 160–190 | + |
| Overexpression | |||||
| UBIp:MCM6 (T0) | SE1a | ≥5 (+) | 0.66 | 158 | + |
| UBIp:MCM6 (T0) | SE1b | ≥5 (+) | 0.33 | 185 | + |
| UBIp:MCM6 (T0) | SE1c | ≥5 (+) | 0.27 | 183 | + |
| UBIp:MCM6 (T0) | SE1d | ≥5 (+) | 0.26 | 172 | + |
| UBIp:MCM6-GFP (T0) | G1 | 1 (−) | n.d. | (150) | + |
| UBIp:MCM6-GFP (T0) | G2 | 2 (−) | n.d. | (100) | + |
| UBIp:MCM6-GFP (T0) | G3 | 2 (−) | n.d. | (60) | + |
| UBIp:MCM6-GFP (T0) | G4 | 1 (−) | n.d. | (60) | + |
| UBIp:MCM6-GFP (T0) | G5 | 2 (−) | n.d. | (96) | + |
| Down-regulation | |||||
| UBIp:MCM6-AS (T0) | AS1 | ≥3 (−) | 0.63 | 175 | + |
| UBIp:MCM6-AS (T0) | AS2 | ≥4 (−) | 0.96 | 182 | + |
| UBIp:MCM6-AS (T0) | AS3 | ≥7 (+) | 0.39 | 130 | FS |
| UBIp:MCM6-AS (T0) | AS4a | ≥5 (+) | 0.73 | 83 | FS/(MS) |
| UBIp:MCM6-AS (T0) | AS4b | ≥5 (+) | 0.48 | 86 | FS/(MS) |
| UBIp:MCM6-AS (T0) | AS4c | ≥5 (+) | 0.36 | 58 | FS/MS |
| UBIp:MCM6-AS (T0) | AS5 | ≥6 (+) | 1.10 | 66 | FS/MS |
| UBIp:MCM6-AS (T0) | AS6 | ≥5 (+) | 0.42 | 68 | FS/(MS) |
| UBIp:MCM6-AS (T0) | AS7 | ≥4 (−) | 0.90 | 182 | + |
| UBIp:MCM6-AS (T0) | AS8a | ≥4 (−) | 0.55 | 161 | + |
| UBIp:MCM6-AS (T0) | AS8b | ≥4 (−) | 0.78 | 159 | + |
| UBIp:MCM6-AS (T0) | AS8c | ≥4 (−) | 0.96 | 175 | + |
| UBIp:MCM6-AS (T0) | AS9a | ≥4 (−) | 0.08 | 174 | + |
| UBIp:MCM6-AS (T0) | AS9b | ≥4 (−) | 1.10 | 158 | + |
| UBIp:MCM6-AS (T0) | AS10a | ≥6 (−) | 0.42 | 187 | + |
| UBIp:MCM6-AS (T0) | AS10b | ≥6 (−) | 0.49 | 169 | + |
| [UBIp:MCM6-AS] (T1)e | AS4a/1–20 | − | n.d. | 155–185 | + |
| [UBIp:MCM6-AS] (T1)e | AS4b/1–22 | − | n.d. | 155–185 | + |
| [UBIp:MCM6-AS] (T1)e | AS6/1 | − | n.d. | 150 | + |
Number of transgene integration events, functional transgene integrations (+), and lack of functional transgene (−), respectively.
Quantification of relative sense and AS transcript amounts. Expression in immature wild-type tassels (10 mm; see also Fig. 4A) was set as 100%. Relative expression in wild-type roots was 25% and 0.68% in mature leaves. No distinction between sense and AS transcript levels.
Plant height in centimeters. G1 to 5 plants were generated in winter after a long tissue culture stage.
Fully developed and fertile cobs as well as tassels (+). FS, Female sterility; MS, male sterility; parentheses, tassels mostly sterile, development of little pollen.
Pollen of transgenic lines were back crossed to wild-type (A188) plants to generate T1 progeny. One selfed kernel was obtained from a tassel flower of plant AS6, which developed into a female spikelet.
In addition, we have generated transgenic maize with the aim of decreasing ZmMCM6 transcript levels. Three hundred-eighty immature embryos of the inbred line A188 have been bombarded with an antisense (AS) construct (UBIp:ZmMCM6-AS). Sixteen plants containing ZmMCM6 AS integrations were regenerated (transformation efficiency of 4.2%) representing 10 independent lines (Table II, plants AS1–AS10b). The transgene integration pattern of five plants is shown in Figure 8A. The genomic Southern blot shows multiple transgene integrations for each plant. AS4a to AS4c displayed the same pattern, indicating that they represent a clonal line. The other two plants (AS3 and AS5) show a different integration pattern. Full-length integrations could be observed in these two lines as well as in line AS6, while none of the lines AS1, AS2, and AS7 to AS10b, respectively, contained full-copy transgene integrations (Table II). Surprisingly, expression of the AS transcript could not be detected in a single line in northern blots (data not shown). The more sensitive RT-PCR method was therefore applied and showed weak AS expression after 38 PCR cycles in lines AS5 and AS6, respectively (Table II). Quantification of both sense and AS transcript amounts in transgenic AS plants by quantitative real-time RT-PCR were in the range of wild-type background sense signals, indicating that the AS transcript amounts were extremely low and not increased above wild-type sense transcript amounts. Nevertheless, those plants that showed a very weak expression of the AS transgene (AS5 and AS6) and/or contained functional copies of the transgene (AS3 and AS4a–c) were strongly reduced in size (Fig. 8B) and did either develop only immature cobs (lines AS3, AS4b, AS4c, AS5, and AS6) or no cob at all (plant AS4a). As shown in Table II, these plants were additionally male sterile due to the lack of anthers or whole male florets (AS4c and AS5). Three plants produced little pollen (AS4a, AS4b, and AS6). Pollen of the plants AS4a and AS4b was used to pollinate wild-type plants. Forty-two progeny plants (plants AS4a/1–20 and AS4b/1–22) were used to study transgene transmission and expression. Surprisingly, none of these plants contained a transgene (Table II). These findings suggest that even mild ZmMCM6 down-regulation affects both male and female gametophyte development and thus transgene transmission.
DISCUSSION
Structure and Domains of Plant MCM Proteins
Similar to fungi and animals, plants seem to possess a single gene for each subunit of the MCM hexamer complex. In addition to the classical MCM2 to 7 genes, Arabidopsis contains two additional MCM genes (AtMCM8 and AtMCM9). Homologs of these genes are not present in the yeast genome (Forsburg, 2004), but MCM8 homologs have recently been reported in humans (Gozuacik et al. 2003; Johnson et al., 2003) as well as in frog (Maiorano et al., 2005). While HsMCM8 is involved in the assembly of the pre-RC (Volkening and Hoffmann, 2005), XlMCM8 was shown to function as a DNA helicase during replication elongation, but not during initiation of DNA replication. A function of MCM9 has not been elucidated to date (Yoshida, 2005).
All MCM proteins are likely to have evolved from a single gene, as the archeon Methanobacterium thermoautotrophicum contains a single MCM gene that is able to form a homohexamer complex and that possesses both a DNA-dependent ATPase and a 3′ to 5′ helicase activity to unwind 500 bp of DNA (Kelman et al., 1999). It is thus not surprising that the central domains of MCM proteins for this activity are most highly conserved. Interestingly, only the trimeric complex of MCM4, MCM6, and MCM7 has been shown to possess in vitro helicase activity (Ishimi, 1997; Lee and Hurwitz, 2001). In mouse, it was recently reported that it is the ATP-binding activity of MCM6 that is critical for DNA helicase activity (You et al., 1999). As in plant MCM6 proteins, the putative catalytic Arg residue is embedded in a conserved SRF motif (Davey et al., 2003), suggesting a similar protein activity.
The other domains of MCM proteins are less conserved. Although the known MCM functions are within the nucleus, only a few MCM proteins were predicted to contain NLS. Experimentally, nuclear import of MCM monomers has been functionally demonstrated only for MCM2 of fission yeast (Schizosaccharomyces pombe), budding yeast, and mice, as well as for MCM3 of yeast and humans (for review, see Forsburg, 2004). However, probably all MCM proteins contain domains to interact with other proteins. The zinc-finger motif that was found in almost all MCMs (with the exception of the MCM3 class) is likely to be involved in protein-protein interactions, which are necessary to form the hexamer, but may also be required to bind either to MCM2 and/or MCM3 for cotranslocation into the nucleus. Reconstitution experiments have demonstrated recently that yeast MCM6 physically interacts with MCM2 (Davey et al., 2003). Thus a major function of MCM2 might be to shuttle the enzymatically active MCM6 and other MCM proteins into the nucleus. Finally, a motif for cell cycle regulation (Cdk-box) was identified in some MCM proteins, indicating that the periodic association of the MCM complex with chromatin, nuclear import-export, and/or protein-protein interactions might be regulated via phosphorylation by CDKs. In yeast, nuclear export of MCM4 was shown to be regulated by CDK activity (Labib et al., 1999; Nguyen et al., 2000). Whether a similar mechanism exists for MCM6 as well as MCM8 and MCM9 of plants that contain such motifs remains to be shown. MCM3 from Arabidopsis, which also contains a putative Cdk-box, was shown to localize nuclear throughout interphase and prophase (Sabelli et al., 1999), indicating a role of this motif not involved in nuclear import and export.
Expression of Plant MCM Genes and Cell Cycle-Dependent Nuclear Localization
Little is known about the expression and subcellular protein localization of plant MCM genes. Abundance in the expression of ZmMCM3, ZmMCM7, and AtMCM7 genes has been correlated with cell proliferation throughout vegetative plant development (Sabelli et al., 1996; Springer et al., 2000; Bastida and Puigdomènech, 2002; Holding and Springer, 2002). MCM7 from Arabidopsis and maize was shown to be strongly expressed in developing embryo and endosperm, respectively (Springer et al., 2000; Bastida and Puigdomènech, 2002). Expression of ZmMCM6 was found to be similar and only detectable in tissues containing proliferating cells. The highest protein amounts were detected in the developing embryo and endosperm. Expression of AtMCM7 in the female gametophyte was indirectly demonstrated by Springer et al. (1995), as gene function is required for female gametophyte development. We have shown that the MCM6 from maize is only expressed in those cells of the mature female gametophyte, which are the precursor cells of both embryo and endosperm (egg and central cell), but not in mature synergids and antipodals or mature male gametes (sperm cells). The high supply of maternal ZmMCM6 transcript amounts in the central cell correlates well with the observation that mitosis after fertilization is initiated much faster during endosperm development compared with embryogenesis (Kranz et al., 1998; Faure et al., 2002).
Compared with frog, where distinct maternal and zygotic genes encode MCM6 (Sible et al., 1998), a single gene seems to exist in maize and Arabidopsis, which is expressed at a low level in maize egg cells, but strongly induced shortly after fertilization. A cell cycle-dependent expression of ZmMCM6 after fertilization was not observed. We have therefore used ZmFEN-1a, the maize homolog of rice (Oryza sativa) OsFEN-1a (Kimura et al., 2001), as a cell cycle marker. The yeast homolog RAD27 (a flap endonuclease required for DNA repair) was previously shown to be up-regulated during late G1 phase (Vallen and Cross, 1995). In contrast to ZmFEN-1a, a significant cell cycle-dependent gene expression pattern after fertilization was not observed for ZmMCM6, suggesting that either the strong induction of gene expression after fertilization generated enough protein for the next cell cycles or the low level of gene expression is sufficient to generate enough protein for the first cell division cycles and regulation of ZmMCM6 activity is mainly posttranscriptional. The finding that MCM protein amounts in the nucleus during G1/S phase have been estimated to be approximately 10- to 100-fold higher than the number of replication origins and replication forks (MCM paradox; Walter and Newport, 1997; Blow and Dutta, 2005) favors the latter hypothesis and suggests that regulation at the level of gene expression is less important. However, the strong induction after fertilization might also be correlated with the highly heterochromatic sperm chromatin, which becomes decondensed 2 to 3 h after fertilization in maize (Scholten et al., 2002). Thus, without a larger maternal supply, a bulk of MCM proteins will be required to saturate the numerous replication origins of the sperm genome for initiation of DNA synthesis during zygote development. Additionally, ZmMCM6 might also be required for chromatin remodeling and transcription of the sperm chromatin, which might explain this strong increase of gene activity. The lack of ZmMCM6 expression and probably of MCM6 protein in sperm cells supports this hypothesis.
In yeast, it was shown that MCM proteins shuttle in and out of the nucleus during a single cell cycle. In contrast, MCM proteins in metazoans remain in the nucleus throughout the cell cycle (Blow and Dutta, 2005). In yeast, MCM proteins are taken up by the nucleus during G1 phase of the cell cycle and the bulk of protein is present in the nucleus shortly before S phase begins. During S phase, MCM proteins are exported to the cytoplasm rather than degraded and excluded from the nucleus by G2 and M phase (Yan et al., 1993; Dalton and Whitbread, 1995; Nguyen et al., 2000). We have used a gene fusion of MCM6 with GFP and immunocytochemistry to study subcellular protein localization. Our data show a cell cycle-dependent nuclear localization of ZmMCM6 similar to observations in yeast, but completely different for metazoans. A similar result was described for MCM7 from Arabidopsis, as it localizes transiently to the nucleus (Springer et al., 2000).
ZmMCM6-GFP was evenly distributed in the nucleoplasm and we never found accumulation at certain spots that might represent pre-RC sites. It is thus likely that ZmMCM6 not only binds pre-RC sites, but is also capable of binding chromatin from all parts of the genome, excluding genomic regions containing rDNA genes, as we never observed ZmMCM6 protein in the nucleolus. Perhaps MCM6 is replaced in this chromosomal region by another MCM protein, for example, MCM8 or MCM9. Binding of human and frog MCM proteins to genomic DNA outside of ORCs, and not specifically to many sites of the chromatin, has been reported recently (Edwards et al., 2002; Schaarschmidt et al., 2002).
ZmMCM6 Is Essential for Plant Growth and Development
In fungi and animals, loss of MCM function causes severe effects, including DNA damage and genome instability. In Caenorhabditis elegans, for example, reduction of MCM5 and MCM6 function using RNAi resulted in failure of nuclear reassembly following mitosis (Gönczy et al., 2000). An essential role for MCM proteins during meiosis was shown for fission yeast, as mcm4 mutants were unable to carry out premeiotic DNA replication (Lindner et al., 2002). Mutations in the Drosophila MCM2 and MCM4 genes resulted in DNA replication and developmental defects in late embryo and larval stages. These studies also indicated that a maternal supply of MCM proteins is sufficient for early embryo development (Treisman et al., 1995; Su et al., 1997). Large quantities of maternal MCM proteins are also present in frog egg extracts (Kubota et al., 1997) and members of this family have been shown to be involved in restricting DNA replication to a single cell cycle throughout embryo development until the MBT stage (Leno et al., 1992; Sible et al., 1998). However, this mechanism seems to be different in higher plants. In maize and wheat, ZGA occurs shortly after fertilization (Sauter et al., 1998; Dresselhaus et al., 1999; Scholten et al., 2002; Sprunck et al., 2005; this study) and the strong induction of ZmMCM6 expression after fertilization suggests that MCM6 function is required already shortly after fertilization in the zygote, as gene expression in the egg cell is low and the existence of a large maternal MCM6 supply is unlikely.
We have chosen an AS approach to decrease, but not to eliminate, ZmMCM6 transcript levels. Under control of the strong maize UBI promoter, transgenic maize lines usually display a broad level of transgene expression, sometimes exceeding endogenous transcript levels up to 20 times (Dresselhaus et al., 2005). Surprisingly, this was completely different for ZmMCM6 because we have not been able to generate plants displaying significant AS transcript levels. The few transgenic plants containing functional transgene integrations were strongly reduced in growth of vegetative tissues and reproductive organs, respectively. In addition, attempts to increase ZmMCM6 transcript levels have been unsuccessful as well. Taken together, these data indicate that expression of the MCM6 gene in maize is strictly regulated and every deregulation is likely to cause severe effects because ZmMCM6 function is important throughout the plant life cycle. In yeast, MCM2 and MCM6 are the main factors limiting the initiation of replication (Lei et al., 1996). This might explain the strong phenotypes obtained for the ZmMCM6 AS plants.
A correlation of cell cycle control and plant growth as well as morphogenesis has already been reported for a number of plant cell cycle regulators including CDKs and cyclins (for review, see Hemerly et al., 1999). However, these proteins are encoded by multigene families and knockouts of individual cell cycle regulators rarely resulted in lethality. Gene functions probably have been compensated by other members of the corresponding gene family. This is obviously different for MCM genes. Each member might be required to form a functional hexamer. In addition, each member might have evolved specific functions, and thus is not capable of fully complementing another member of the hexamer. For future analyses of MCM genes and proteins in plants, we suggest using their own or inducible promoters to avoid lethality. Yeast two-hybrid screens, for example, might be useful to identify interacting partners of MCM proteins. Developing seeds, especially where MCM proteins amount to high levels, seem to be the right source for such screens. T-DNA and transposon insertion in C-terminal regions of plant MCM genes, as was the case for AtMCM7, might elucidate specific functions of domains in this region. N-terminal or central insertions in the catalytic domain are likely to be lethal.
MATERIALS AND METHODS
Plant Material, Isolation of Cells from the Female Gametophyte, and In Vitro Cultures
Maize (Zea mays) inbred lines A188 (Green and Phillips, 1975) and H99 (D'Halluin et al., 1992) as well as transgenic lines were grown under standard greenhouse conditions at 26°C with 16 h light and a relative air humidity of about 60%. Cells of the maize embryo sac were isolated according to Kranz et al. (1991) and zygotes according to Cordts et al. (2001). Microspore-derived suspension cultures were obtained from B. Krautwig (described in Krautwig and Lörz, 1995). The BMS maize cells were cultivated as a suspension culture in Murashige and Skoog medium in the dark with centrifugation at 60 to 70 rpm at 26°C according to Kirihara (1994). Onion (Allium cepa) bulbs were obtained from the supermarket.
Differential Plaque Screening, 5′-RACE, DNA Sequencing, and Bioinformatic Analysis
A cDNA library of maize in vitro zygotes (Dresselhaus et al., 1996) was screened by differential plaque screening against a cDNA library of unfertilized egg cells (Dresselhaus et al., 1994) to identify fertilization-induced genes. Double plaque lifts were prepared from 15-cm plates of the zygote library at a density of 500 plaque-forming units (pfu). The filters were hybridized with PCR-amplified [32P]cDNA from the egg cell and [32P]cDNA of in vitro zygotes. Candidate cDNA clones were further analyzed by reverse northern hybridization (Dresselhaus et al., 1999). Among the putative fertilization-induced genes, 1,123 bp of a cDNA encoding the 3′ end of the ZmMCM6 transcript and 273 bp of the ZmFEN-1a transcript (GenBank accession no. DQ138311) were identified. The 5′ region of the ZmMCM6 transcript was isolated from zygote cDNA as follows: 2.5 μg Eco-Adaptor was ligated to PCR-amplified cDNA from in vitro zygotes (Dresselhaus et al., 1996), ligated products purified using a low-melting agarose gel, cloned into calf intestine alkaline phosphatase-treated EcoRI restriction sites of the λ-vector UniZAP II, and packed using the Gigapack-Gold II extract (Stratagene) to generate a cDNA library from zygotes containing full-length cDNAs. The 5′ region (2 kb) of ZmMCM6 was amplified from this library using the T3 vector primer and the gene-specific primer 19A (5′-CATGATGTAGACCAGATCAA-3′) in a standard PCR reaction with Taq DNA polymerase. PCR products were blunted, purified, and cloned into the pPCR-Script vector (Stratagene), according to the manufacturer's specifications. The full-length cDNA of ZmMCM6 was amplified from 1 μg poly(A)+ RNA extracted from root tips. Twenty nanograms of the primer 19U2 (5′-GTCAGACTACAGATGCTAATT-3′) derived from the 3′ end of the ZmMCM6 transcript was incubated with poly(A)+ RNA and SuperScript reverse transcriptase according to the protocol of the 5′-RACE system (Life Technologies). PCR was performed with proofreading pfu DNA polymerase (Stratagene) as well as primers 19U2 and GB19A (5′-TGCCAATCTCCAACTCATACCC-3′), the latter derived from the 5′ region of the ZmMCM6 transcript. The PCR product was cloned into the pPCR-script vector (see above) generating the plasmid pK19U2 and fully sequenced. The full-length ZmMCM6 nucleotide sequence data reported in this article is available in the EMBL, GenBank, and DNA Data Bank of Japan (DDJB) nucleotide sequence databases under accession number AY862320 (ZmMCM6 cDNA). The protein sequence is available under accession number AAW55593.
Sequence data were compiled and compared online with EMBL, GenBank, DDBJ, Swiss-Prot, Protein Information Resource, and Protein Research Foundation databases with FASTA and BLAST algorithms (Pearson, 1990). MCM protein sequences were obtained from GenBank and aligned online by ClustalW (Higgins et al., 1994; http://www.ebi.ac.uk/clustalw). The alignment data obtained were used to generate a phylogram (Fig. 1) with Treeview, version 1.6.6 (Page, 1996). Protein alignments were drawn by GeneDoc, version 2.6.02 (Nicholas et al., 1997), using protein alignments generated by ClustalW. Prediction of protein localization sites was performed online using PSORT (http://psort.nibb.ac.jp), iPSORT (http://www.HypothesisCreator.net/iPSORT), and SignalP version 2.0 (http://www.cbs.dtu.dk/services/SignalP-2.0). Protein motifs such as putative CDK posphorylation sites and zinc-finger motifs were searched online using Prosite (http://www.expasy.org/tools/scanprosite).
DNA and RNA Extraction, Northern and Southern Blots, SC RT-PCR, and Quantitative RT-PCR
Extraction of genomic DNA from plant tissues was performed according to Dellaporta et al. (1983). Plant material for northern-blot analyses was either collected in the greenhouse from different tissues and organs of the maize inbred line A188 or from in vitro cultures. Total RNA was extracted from all samples with TRIzol (Life Technologies) and poly(A)+ RNA from total RNA using the PolyATtract mRNA isolation system (Promega), according to the manufacturer's specifications.
Capillary Southern and northern blots, as well as labeling, hybridization, washing, and autoradiographic exposures, were performed as described in Dresselhaus et al. (2005). The 1,123 bp representing the 3′ region of ZmMCM6 was labeled as a probe for northern blots. Probes to detect sense and AS transgene integrations were isolated from plasmids that have been used for maize transformations. DNA fragments were purified using the Gel Band isolation kit (Pharmacia Biotech) and radioactively labeled as described (Dresselhaus et al., 2005), after digestion with enzyme combinations that were also used to restrict genomic DNA. Genomic DNA of transgenic sense and AS lines was restricted using BamHI/SalI and BamHI/SstI, respectively. These enzyme combinations cut out the full-length ZmMCM6 cDNA. A GFP-specific probe was prepared as follows. The GFP sequence was amplified in a standard PCR using the primers GFP1for (5′-GAGGAACTGTTCACTGGCGT-3′) and GFP3rev (5′-GTTCATCCATGCCATGCGTG-3′). After amplification, the PCR product was purified and labeled as described above. Genomic DNA of putative transgenic ZmMCM6-GFP maize plants was restricted with DraI, which cuts out the complete construct except for the first 135 bp of the UBI promoter.
SC RT-PCR analysis was performed as described by Cordts et al. (2001) using the primers MCM6rev (5′-GAACACCACCCAAAAGCATAAGAA-3′) or FENrev (5′-GGACTCCCTTACTTTTGGG-3′) in addition to Gap2 (5′-GTAGCCCCACTCGTTGTCGTA-3′) for cDNA synthesis. After RT, reactions were split into two reaction tubes and 38 PCR cycles were conducted in separate reactions with ZmMCM6-specific primers MCM6for (5′-GCAGGTCGCAGATGGTAGGAGAG-3′) and MCM6rev, ZmFEN-1a-specific primers FENfor (5′-CCAAGATGCTTTCTATGGAC-3′) and FENrev, as well as GAPDH-specific primers Gap1 (5′-AGGGTGGTGCCAAGAAGGTTG-3′) and Gap2. Twenty-five microliters of the ZmMCM6 and ZmFEN-1a, as well as 15 μL GAPDH PCR products, were each separated in agarose gels. The size of the amplified ZmMCM6 transcript was 219 bp (genomic approximately 550 bp), 241 bp for ZmFEN-1a (genomic approximately 450 bp), and 622 bp for GAPDH (genomic approximately 1.2–1.3 kb). Quantitative RT-PCR analyses were performed as described by Dresselhaus et al. (2005) using 1 μg total RNA extracted from leaf material. The ZmMCM6-specific primers MCM6for and MCM6rev (400 nm each primer) or GAPDH-specific primers Gap1 and Gap2 (400 nm each primer; Richert et al., 1996) were used in an iCycler iQ machine with iQ SYBR Green Supermix (Bio-Rad), according to the manufacturer's recommendations. PCR results were controlled by agarose gel electrophoresis. Samples showing both ZmMCM6- and GAPDH-specific amplifications were further processed with the iCycler iQ real-time detection system software, version 3.0 (Bio-Rad). GAPDH-specific PCR products were used to normalize ZmMCM6 transcript amounts.
Protein Extraction, Western Blots, and Immunodetection
Plant tissue was ground in liquid nitrogen. One volume of extraction buffer (250 mm KCl, 20 mm Tris-HCl, pH 6.8, 50% v/v glycerol, 2.5% w/v polyvinylpyrrolidone, 5 mm dithiothreitol, and one mini protease inhibitor tablet [Roche] in 10 mL extraction buffer) was added and mixed until material thawed. Samples were centrifuged at 13,000 rpm for 30 min at 4°C. This step was repeated twice with the supernatant. Protein concentrations were measured after adding 100 μL Bradford reagent (Bradford, 1976; Bio-Rad), according to the manufacturer's recommendations.
SDS-PAGE in a discontinuous Tris-Gly buffer system was performed according to Sambrook et al. (1989) using mini gels (5% stacking gel and 8% resolving gel). Protein samples (each 10 μg) were mixed 1:1 in 2× Laemmli sample buffer and denatured at 96°C for 12 min before loading on the gel. Gels were blotted onto Immobilon-P polyvinylidene difluoride membranes from Millipore with transfer buffer (25 mm Tris, 192 mm Gly, 10% v/v methanol) in a semidry method according to Frey (2002) using the Transblot synthetic dextrose system (Bio-Rad) followed by a Ponceau-S staining.
For immunodetection, a rabbit peptide antibody (anti-MCM6-Ab) was generated by BioTrend against a ZmMCM6 C-terminal-specific region (VPSESDAGQPAEEDA) between position 680 and 694 (Fig. 2) and tested for specificity by ELISA. Protein blots were blocked overnight in 5% phosphate-buffered saline (PBS)-Blotto at 4°C or for 1 h at room temperature. Blots were incubated with a 1:500 dilution of anti-MCM6-Ab in 5% PBS-Blotto for 2 h at room temperature, rinsed twice with PBS for 5 min each, followed by a 1-h treatment at room temperature with a 1:5,000 dilution of the secondary antibody, a mouse monoclonal anti-rabbit IgG (γ-chain-specific) horseradish peroxidase conjugate clone RG-96 (Sigma). Blots were rinsed twice with PBS for 5 min, incubated for 5 min in 1:1 luminol:peroxide solution from Pierce in the dark, followed by exposure to autoradiographic films, according to the manufacturer's instructions.
Immunocytochemistry to determine ZmMCM6 levels during the cell cycle were performed as follows. Each 2-mL BMS cell was collected by centrifugation at 1,000 rpm for 4 min. Supernatants were removed and cell pellets immediately fixed in 1 mL 4% paraformaldehyde and 0.25% glutaraldehyde in PBS, and incubated for 1 h at room temperature. Fixed cells were centrifuged at 1,000 rpm, supernatant discarded, and pellets rinsed four times with PBS (containing 1% Triton) and each time centrifuged for 10 min at 1,000 rpm to collect cells. Cell walls were degraded for 30 min at room temperature after adding 500 μL of enzyme mixture, which contained 1.5% pectinase, 0.5% pectolyase, 1.0% hemicellulase, and 1.0% cellulase in mannitol solution (570 milliosmolar; pH 4.9–5.0). Digested cells were resuspended with a pipette, nuclei collected after centrifugation at 1,000 rpm, and supernatants removed. Nuclei were washed four times in 1 mL PBS containing 0.1% Triton for 10 min during centrifugation at 1,000 rpm at 4°C. The pellets were resuspended in 500 μL PBS containing the 1:250 diluted anti-MCM6-Ab and incubated at 4°C overnight. Nuclei were washed three times in 1 mL PBS and centrifuged at 1,000 rpm for 10 min. After a final wash, pellets containing nuclei were resuspended in 500 μL PBS containing an FITC-coupled anti-rabbit antibody (1:500) and incubated for 4 h at 4°C. Nuclei were collected after centrifugation at 1,000 rpm for 10 min and washed five times each in 1 mL PBS as described above. Finally, nuclei were resuspended in 500 μL PBS and centrifuged at 500 rpm for 1 min. Twenty-microliter fractions containing nuclei were collected from the bottom of the tubes and transferred to microscopic slides after adding 0.25 μL DAPI solution.
Generation of Constructs, Biolistic Transformation, and Regeneration of Transgenic Plants
To generate an AS construct (UBIp:MCM6-AS) of ZmMCM6, the full-length cDNA of ZmMCM6 was excised from pK19U2 (see above) using the enzymes SstI and BamHI and cloned into the corresponding restriction sites of the vector pUbi.Cas (Christensen and Quail, 1996). The open reading frame of ZmMCM6 was amplified from pK19U2 using the primers MCM6 (5′-GTCGACCCTGATTCTTCCAC-3′) and MCMB (5′-GGATCCATGTTAAGATGCCGTTGC-3′) containing SalI and BamHI restriction sites, respectively, to generate the sense construct (UBIp:MCM6). After PCR amplification using pfu DNA polymerase, the PCR product was restricted with SalI and BamHI, cloned in the corresponding restriction sites of the vector pUbi.Cas, and fully sequenced. A construct encoding a MCM6-GFP fusion protein (UBIp:MCM6-GFP) was prepared as follows. The full-length ZmMCM6 sequence was amplified from the plasmid pK19U2 using the primers M6F-Spe (5′-CGACACTAGTTGTCGGTGATG-3′) and M6R-Bam (5′-CGTGGATCCAATCAATAACATAGTTCG-3′) containing SpeI and BamHI restriction sites, respectively, which were then used for cloning the fragment between the SpeI and BamHI sites of the vector pLNU-GFP (UBIp:GFP:NOSt; unpublished data). This vector contains a multicloning site between the maize UBI promoter and the GFP gene, the latter containing the ST-LS1 intron (derived from pMon30049; Pang et al., 1996), followed by the nopaline synthase terminator. The vector pLNU-GFP was also used as a positive control for transient biolistic transformation experiments.
Epidermal onion cell layers were bombarded with 2 to 5 μg plasmid DNA, according to the procedure described by Scott et al. (1999), except that inner onion peels (2 × 2.5 cm) were placed with the concave side up on 0.5% agar plates. The condition of bombardment was 1,100-psi rupture discs under a vacuum of 28 mm Hg with 6-cm target distance using the particle gun model PDS100/He (Bio-Rad). Bombarded peel halves were placed after transformation with the concave side down and the cut surface in sterile 0.6% agar (Fluka) petri dishes for about 17 to 22 h in the dark before removing the epidermis for observation using a fluorescence microscope. For bombardment of maize BMS cells, a uniform layer was spread on solid Murashige and Skoog medium and incubated at 26°C for 1 to 2 h before biolistic transformation. After transformation, plates were incubated overnight in the dark at 26°C. Cells were transferred to fresh liquid medium in 35-mm petri dishes and cultivated in darkness using a shaker (60 rpm) for at least 4 h before microscopic observations. Photos were taken immediately after a transfer of 100 μL of medium containing individual cells or cell clusters showing GFP fluorescence onto glass slides followed by an addition of 1 μL DAPI staining solution.
Embryos of the maize inbred line A188 were used 12 to 14 DAP for stable transformation using the construct UBIp:MCM6-AS. Hybrid embryos from both lines A188 and H99 were used for stable transformation experiments with the constructs UBIp:MCM6 and UBIp:MCM6-GFP. Constructs were cotransformed with the plasmid construct 35Sp:PAT carrying the selectable marker PAT for glufosinate ammonium resistance. Particle bombardment, tissue culture, and selection of transgenic maize plants were performed according to Brettschneider et al. (1997).
Microscopy
Axiovert 35 m or Axiovert 200 fluorescence microscopes (Zeiss) with the filter set 01 (FITC filter with excitation at 450–490 nm; emission at >515 nm) or filter set 38 (GFP filter with excitation at 470–495 nm; emission at 525 nm) were used to observe GFP fluorescence in onion epidermal and maize BMS cells, as well as FITC fluorescence of isolated BMS nuclei after immunostaining. A DAPI filter (Zeiss, excitation at 359–371 nm and emission >397 nm) was used to visualize DNA and cell wall material. Samples were excited with UV light produced by a HBO 50/Ac lamp and images taken with a Nikon DS-5Mc camera. Nikon software EclipseNet plug in MCF was used to obtain and merge fluorescence images. ImageJ software was used to measure DAPI, GFP, and FITC fluorescence. CLSM was performed using the Leica TCS 4D CLSM (Leica-Laser-Technologie). Samples were excited by 488 nm with an Argon laser as described in Knebel et al. (1990).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY862320 (ZmMCM6 cDNA), AAW55593 (ZmMCM6 protein), and DQ138311 (ZmFEN-1a cDNA).
Acknowledgments
We are grateful to Stefanie Sprunck for critical comments on the manuscript and to Gislind Bräcker for excellent technical support. We thank Natascha Techen for the 35Sp:Lc-GFP construct and Hartmut Quader for help with the CLSM studies.
This work was supported by the Deutsche Forschungsgemeinschaft (grant Dr334/2 to T.D.), the Südwestdeutsche Saatzucht, Rastatt, Germany (to D.N.S.), a Roman Herzog research fellowship (to D.L.-L.), and a postgraduate Hamburg (HmbNFG) fellowship (to K.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Thomas Dresselhaus (dresselh@botanik.uni-hamburg.de).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.074294.
References
- Arias EE, Walter JC (2005) Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev 19: 114–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis JM, Forsburg SL (2004) MCM proteins: DNA damage, mutagenesis and repair. Curr Opin Genet Dev 14: 17–21 [DOI] [PubMed] [Google Scholar]
- Bastida M, Puigdomènech P (2002) Specific expression of ZmPRL, the maize homolog of MCM7, during early embryogenesis. Plant Sci 162: 97–106 [Google Scholar]
- Blow JJ, Dutta A (2005) Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan JA, Natale DA, Depamphilis ML (2000) Initiation of eukaryotic DNA replication: conservative or liberal? J Cell Physiol 184: 139–150 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities utilising the principle of protein dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brettschneider R, Becker D, Lörz H (1997) Efficient transformation of scutellar tissue of immature maize embryos. Theor Appl Genet 94: 737–748 [Google Scholar]
- Christensen AH, Quail PH (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5: 213–218 [DOI] [PubMed] [Google Scholar]
- Claycomb JM, MacAlpine DM, Evans JG, Bell SP, Orr-Weaver TL (2002) Visualization of replication initiation and elongation in Drosophila. J Cell Biol 159: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordts S, Bantin J, Wittich PE, Kranz E, Lörz H, Dresselhaus T (2001) ZmES genes encode peptides with structural homology to defensins and are specifically expressed in the female gametophyte of maize. Plant J 25: 103–114 [DOI] [PubMed] [Google Scholar]
- Dalton S, Whitbread L (1995) Cell cycle-regulated nuclear import and export of CDC47, a protein essential for initiation of DNA replication in budding yeast. Proc Natl Acad Sci USA 92: 2514–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MJ, Indiani C, O'Donnell M (2003) Reconstitution of the Mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J Biol Chem 278: 4491–4499 [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Report 4: 19–21 [Google Scholar]
- D'Halluin K, Bonne E, Bossut M, De Beuckeleer M, Leemans J (1992) Transgenic maize plants by tissue electroporation. Plant Cell 4: 1495–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T, Amien S, Márton ML, Strecke A, Brettschneider R, Cordts S (2005) TRANSPARENT LEAF AREA1 encodes a secreted proteolipid required for anther maturation, morphogenesis, and differentiation during leaf development in maize. Plant Cell 17: 730–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T, Cordts S, Heuer S, Sauter M, Lörz H, Kranz E (1999) Novel ribosomal genes from maize are differentially expressed in the zygotic and somatic cell cycles. Mol Gen Genet 261: 416–427 [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Hagel C, Lörz H, Kranz E (1996) Isolation of a full-length cDNA encoding calreticulin from a PCR-library of in vitro zygotes of maize. Plant Mol Biol 31: 23–34 [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Lörz H, Kranz E (1994) Representative cDNA libraries from few plant cells. Plant J 5: 605–610 [DOI] [PubMed] [Google Scholar]
- Edwards MC, Tutter AV, Cvetic C, Gilbet CH, Prokhorova TA, Walter JC (2002) MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J Biol Chem 277: 33049–33057 [DOI] [PubMed] [Google Scholar]
- Faure JE, Rotman N, Fortuné P, Dumas C (2002) Fertilization in Arabidopsis thaliana wild type: developmental stages and time course. Plant J 30: 481–488 [DOI] [PubMed] [Google Scholar]
- Forsburg SL (2004) Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev 68: 109–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A (2002) Protein-Blotting und Nachweis membrangebundener Proteine. In G Schrimpf, ed, Gentechnische Methoden: eine Sammlung von Arbeitsanleitungen für das molekularbiologische Labor, Ed 3. Spektrum Akademischer Verlag, Heidelberg, pp 343–368
- Gönczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, et al (2000) Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331–336 [DOI] [PubMed] [Google Scholar]
- Gozuacik D, Chami M, Lagorce D, Faivre J, Murakami Y, Poch O, Biermann E, Knippers R, Brechot C, Paterlini-Brechot P (2003) Identification and functional characterization of a new member of the human Mcm protein family: hMcm8. Nucleic Acids Res 31: 570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CE, Phillips RL (1975) Plant regeneration from tissue cultures of maize. Crop Sci 15: 417–421 [Google Scholar]
- Hemerly AS, Ferreira PCG, Van Montagu M, Inzé D (1999) Cell cycle control and plant morphogenesis: is there an essential link? Bioessays 21: 29–37 [Google Scholar]
- Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding DR, Springer PS (2002) The Arabidopsis gene PROLIFERA is required for proper cytokinesis during seed development. Planta 214: 373–382 [DOI] [PubMed] [Google Scholar]
- Hyrien O, Marheineke K, Goldar A (2003) Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays 25: 116–125 [DOI] [PubMed] [Google Scholar]
- Ishimi Y (1997) A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem 272: 24508–24513 [DOI] [PubMed] [Google Scholar]
- Johnson EM, Kinoshita Y, Daniel DC (2003) A new member of the MCM protein family encoded by the human MCM8 gene, located contrapodal to GCD10 at chromosome band 20p12.3-13. Nucleic Acids Res 31: 2915–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman Z, Lee J-K, Hurwitz J (1999) The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum H contains DNA helicase activity. Proc Natl Acad Sci USA 96: 14783–14788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Suzuki T, Yanagawa Y, Yamamoto T, Nakagawa H, Tanaka I, Hashimoto J, Sakaguchi K (2001) Characterization of plant proliferating cell nuclear antigen (PCNA) and flap endonuclease-1 (FEN-1), and their distribution in mitotic and meiotic cell cycles. Plant J 28: 643–653 [DOI] [PubMed] [Google Scholar]
- Kirihara JA (1994) Selection of stable transformants from Black Mexican Sweet maize suspension cultures. In M Freeling and V Walbot, eds, The Maize Handbook. Springer, New York, pp 690–694
- Knebel W, Quader H, Schnepf E (1990) Mobile and immobile endoplasmic reticulum in onion bulb epidermis cells: short- and long-term observations with a confocal laser scanning microscope. Eur J Cell Biol 52: 328–340 [PubMed] [Google Scholar]
- Kranz E, Bautor J, Lörz H (1991) In vitro fertilization of single, isolated gametes of maize mediated by electrofusion. Sex Plant Reprod 4: 12–16 [Google Scholar]
- Kranz E, von Wiegen P, Quader H, Lörz H (1998) Endosperm development after fusion of isolated, single maize sperm and central cells in vitro. Plant Cell 10: 511–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautwig B, Lörz H (1995) Single androgenic structures of maize (Zea mays L.) for the initiation of homogeneous cell suspension and protoplast cultures. Plant Cell Rep 14: 477–481 [DOI] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, Takisawa H (1997) Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J 16: 3320–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Diffley JF (2001) Is the MCM2-7 complex the eukaryotic DNA replication fork helicase? Curr Opin Genet Dev 11: 64–70 [DOI] [PubMed] [Google Scholar]
- Labib K, Diffley JFX, Kearsey SE (1999) G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat Cell Biol 1: 415–422 [DOI] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JFX (2000) Uninterrupted MCM2-7 function required for DNA replication fork progression. Science 288: 1643–1647 [DOI] [PubMed] [Google Scholar]
- Laskey RA, Madine MA (2003) A rotary pumping model for helicase function of MCM proteins at a distance from replication forks. EMBO Rep 4: 26–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-K, Hurwitz J (2001) Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, 7 complex requires forked DNA structures. Proc Natl Acad Sci USA 98: 54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Tye BK (1996) Physical interactions among MCM proteins and effects of MCM dosage on DNA replication in Saccharomyces cerevisiae. Mol Cell Biol 16: 5081–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Tye BK (2001) Initiating DNA synthesis: from recruiting to activating the MCM complex. J Cell Sci 114: 1447–1454 [DOI] [PubMed] [Google Scholar]
- Leno GH, Downes CS, Laskey RA (1992) The nuclear membrane prevents replication of human G2 nuclei but not G1 nuclei in Xenopus egg extract. Cell 69: 151–158 [DOI] [PubMed] [Google Scholar]
- Lindner K, Gregan J, Montgomery S, Kearsey S (2002) Essential role of MCM proteins in pre-meiotic DNA replication. Mol Biol Cell 13: 435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR (1989) Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci USA 86: 7092–7096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano D, Cuvier O, Danis E, Mechali M (2005) MCM8 is an MCM2-7-related protein that functions as a DNA helicase during replication elongation and not initiation. Cell 120: 315–328 [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Irie K, Li JJ (2000) Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr Biol 10: 195–205 [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: analysis and visualization of genetic variation. Embnew News 4: 14 [Google Scholar]
- Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Pang S, DeBoer DL, Wan Y, Ye G, Layton JG, Neher MK, Armstrong CL, Fry JE, Hinchee M, Fromm ME (1996) An improved green fluorescent protein gene as a vital marker in plants. Plant Physiol 112: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR (1990) Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol 183: 63–98 [DOI] [PubMed] [Google Scholar]
- Richert J, Kranz E, Lörz H, Dresselhaus T (1996) A reverse transcriptase polymerase chain reaction assay for gene expression studies at the single cell level. Plant Sci 114: 93–99 [Google Scholar]
- Sabelli P, Parker J, Barlow P (1999) cDNA and promoter sequences for MCM3 homologues from maize, and protein localization in cycling cells. J Exp Bot 50: 1315–1322 [Google Scholar]
- Sabelli PA, Burgess SR, Kush AK, Young MR, Shewry PR (1996) cDNA cloning and characterisation of a maize homologue of the MCM proteins required for the initiation of DNA replication. Mol Gen Genet 252: 125–136 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sauter M, von Wiegen P, Lörz H, Kranz E (1998) Cell cycle regulatory genes from maize are differentially controlled during fertilization and first embryonic cell division. Sex Plant Reprod 11: 41–48 [Google Scholar]
- Schaarschmidt D, Ladenburger EM, Keller C, Knippers R (2002) Human Mcm proteins at a replication origin during the G1 to S phase transition. Nucleic Acids Res 30: 4176–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholten S, Lörz H, Kranz E (2002) Paternal mRNA and protein synthesis coincides with male chromatin decondensation in maize zygotes. Plant J 32: 221–231 [DOI] [PubMed] [Google Scholar]
- Scott A, Wyatt S, Tsou P-L, Robertson D, Allen NS (1999) Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26: 1125–1132 [DOI] [PubMed] [Google Scholar]
- Shechter D, Gautier J (2004) MCM proteins and checkpoint kinases get together at the fork. Proc Natl Acad Sci USA 101: 10845–10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sible JC, Erikson E, Hendrickson M, Maller JL, Gautier J (1998) Developmental regulation of MCM replication factors in Xenopus laevis. Curr Biol 8: 347–350 [DOI] [PubMed] [Google Scholar]
- Springer PS, Holding DR, Groover A, Yordan C, Martienssen RA (2000) The essential Mcm7 protein PROLIFERA is localized to the nucleus of dividing cells during the G1 phase and is required maternally for early Arabidopsis development. Development 127: 1815–1822 [DOI] [PubMed] [Google Scholar]
- Springer PS, McCombie WR, Sundaresan V, Martienssen RA (1995) Gene trap tagging of PROLIFERA, and essential MCM2-3-5 like gene in Arabidopsis. Science 268: 877–880 [DOI] [PubMed] [Google Scholar]
- Sprunck S, Baumann U, Edwards K, Langridge P, Dresselhaus T (2005) The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L.). Plant J 41: 660–672 [DOI] [PubMed] [Google Scholar]
- Stevens R, Mariconti L, Rossignol P, Perennes C, Cella R, Bergounioux C (2002) Two E2F sites in the Arabidopsis MCM3 promoter have different roles in cell cycle activation and meristematic expression. J Biol Chem 277: 32978–32984 [DOI] [PubMed] [Google Scholar]
- Su TT, Yakubovich N, O'Farrell PH (1997) Cloning of Drosophila MCM homologs and analysis of their requirement during embryogenesis. Gene 192: 283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman JE, Follette PJ, O'Farrell PH, Rubin GM (1995) Cell proliferation and DNA replication defects in a Drosophila MCM2 mutant. Genes Dev 9: 1709–1715 [DOI] [PubMed] [Google Scholar]
- Tye BK (1999) MCM proteins in DNA replication. Annu Rev Biochem 68: 649–686 [DOI] [PubMed] [Google Scholar]
- Vallen EA, Cross FR (1995) Mutations in RAD27 define a potential link between G1 cyclins and DNA replication. Mol Cell Biol 15: 4291–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkening M, Hoffmann I (2005) Involvement of human MCM8 in prereplication complex assembly by recruiting hcdc6 to chromatin. Mol Cell Biol 25: 1560–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1: 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport JW (1997) Regulation of replicon size in Xenopus egg extracts. Science 275: 993–995 [DOI] [PubMed] [Google Scholar]
- Yan H, Merchant AM, Tye BK (1993) Cell cycle-regulated nuclear localization of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes Dev 7: 2149–2160 [DOI] [PubMed] [Google Scholar]
- Yoshida K (2005) Identification of a novel cell-cycle-induced MCM family protein MCM9. Biochem Biophys Res Commun 331: 669–674 [DOI] [PubMed] [Google Scholar]
- You Z, Komamura Y, Ishimi Y (1999) Biochemical analysis of the intrinsic Mcm4-Mcm6-Mcm7 DNA helicase activity. Mol Cell Biol 19: 8003–8015 [DOI] [PMC free article] [PubMed] [Google Scholar]