Abstract
The Rhizobium-legume symbiosis culminates in the exchange of nutrients in the root nodule. Bacteria within the nodule reduce molecular nitrogen for plant use and plants provide bacteria with carbon-containing compounds. Following the initial signaling events that lead to plant infection, little is known about the plant requirements for establishment and maintenance of the symbiosis. We screened 44,000 M2 plants from fast neutron-irradiated Medicago truncatula seeds and isolated eight independent mutant lines that are defective in nitrogen fixation. The eight mutants are monogenic and represent seven complementation groups. To monitor bacterial status in mutant nodules, we assayed Sinorhizobium meliloti symbiosis gene promoters (nodF, exoY, bacA, and nifH) in the defective in nitrogen fixation mutants. Additionally, we used an Affymetrix oligonucleotide microarray to monitor gene expression changes in wild-type and three mutant plants during the nodulation process. These analyses suggest the mutants can be separated into three classes: one class that supports little to no nitrogen fixation and minimal bacterial expression of nifH; another class that supports no nitrogen fixation and minimal bacterial expression of nodF, bacA, and nifH; and a final class that supports low levels of both nitrogen fixation and bacterial nifH expression.
Many species of the legume family form mutually beneficial, symbiotic interactions with Rhizobium and related nitrogen-fixing bacteria, collectively called rhizobia. Following early surface interactions that include an exchange of signaling molecules, rhizobia penetrate plant roots through epidermal cells and are deposited inside cells of a newly formed symbiotic organ, the nodule. Inside the nodule, rhizobia reduce, or fix, molecular dinitrogen into ammonia.
To facilitate this process, the plant must provide an energy source and a permissible environment for nitrogen fixation. While the bacterial enzymes responsible for nitrogen fixation are irreversibly inhibited by oxygen, the environment cannot be fully anaerobic as rhizobia are obligate aerobes. Nitrogen fixation is an energy-intensive process. The plant provides approximately 6 g of carbon in the form of dicarboxylic acids per gram of nitrogen it receives from the bacteria (for review, see Schubert, 1986; Vance and Heichel, 1991). Consequently, bacteria that fail to import dicarboxylic acids are unable to fix nitrogen inside nodules (Ronson et al., 1981; Finan et al., 1983; Udvardi et al., 1988). A precisely controlled developmental and biochemical program must be in place allowing the plant to create a new niche in which all the prerequisites for nitrogen fixation are satisfied.
We have undertaken the genetic analysis of plant factors required for the later stages of the symbiotic program that support active nitrogen fixation by the bacterial partner. Dissection of the Rhizobium-legume symbiosis by mutational analysis has been useful for studying various aspects of the symbiosis. Several genetic screens of rhizobia, including Sinorhizobium meliloti, have identified loci required for the production of symbiotic signaling molecules (Meade et al., 1982), bacterial development (Glazebrook et al., 1993), and for nitrogen fixation (Ruvkun et al., 1982). Plant symbiotic mutants of important crop species (Peterson and Barnes, 1981; Carroll et al., 1985; Walker et al., 2000) and of the model legumes, Lotus japonicus and Medicago truncatula, have been isolated. These mutants are typically grouped into two classes: those unable to initiate nodule development (Nod−) and those that are unable to support nitrogen fixation (Fix−). In L. japonicus, at least eight Nod− and nine Fix− complementation groups have been isolated (Schauser et al., 1998; Szczyglowski et al., 1998; Kawaguchi et al., 2002). In M. truncatula, seven Nod− and three Fix− complementation groups have been isolated (Bénaben et al., 1995; Sagan et al., 1995, 1998; Catoira et al., 2000, 2001; Amor et al., 2003; Oldroyd and Long, 2003). While at least 10 genes essential for nodule formation have been identified (Kalo et al., 2005; Smit et al., 2005; for review, see Oldroyd and Downie, 2004), very little molecular level information has become available about how plant genes facilitate the symbiotic program after nodule inception. Three plant proteins have been shown to be essential for nitrogen fixation: Suc synthase, leghemoglobin, and a sulfate transporter (Gordon et al., 1999; Krusell et al., 2005; Ott et al., 2005). Identification and characterization of new Fix− plant mutants described here in the genetically tractable plant M. truncatula provide a critical genetic framework for more detailed molecular level studies that will follow.
Here we describe the isolation of eight M. truncatula monogenic nitrogen fixation mutants. To characterize the nature of the defect in these mutants, we examine the expression of four bacterial symbiosis genes, which are required at different stages of nodule development. Using microarray analysis, we define patterns of symbiotic gene expression in wild-type plants to provide a reference with which data from mutant plants can be compared. During nodule development, from 6 h to 21 d after inoculation (dai), we identify 584 genes that are differentially expressed. We examine gene expression profiles for three of the Fix− mutants and identify 40 misregulated genes. Based on these data, we place these mutants into three classes.
RESULTS
Isolation of M. truncatula Nitrogen Fixation Mutants
To identify mutants defective in the Rhizobium-legume symbiosis, we screened approximately 44,000 M2 plants derived from 2,862 selfed M1 plants for macroscopic defects in the symbiosis. We visually examined root nodules for the absence of leghemoglobin, which results in a white, rather than a pink, nodule and indicates an inability to support nitrogen fixation (Viands et al., 1979). Thirty Fix− mutants were confirmed in the M3 generation (data not shown). Because mutants isolated from the same M1 bulk may represent siblings, we chose a single mutant of each phenotype from each M1 bulk for further characterization. For the purposes of this study, we define the Fix− phenotype as a defect that results in lower than wild-type levels of nitrogen fixation.
The macroscopic phenotype of mutant nodules allows an initial characterization of the symbiotic defect. Unlike wild-type plants that make larger pink nodules (Fig. 1A), Fix− mutants make small, white nodules (Fig. 1B). All the Fix− plants examined have this small, white-nodule phenotype (data not shown). All mutants described herein are morphologically indistinguishable from wild-type plants when supplemented with a nitrogen-containing fertilizer (data not shown).
Figure 1.
Nodule phenotypes of wild-type and mutant plants. Nodules of wild-type (A) and mutant (B) 4A-17 plants 28 dai. Bars = 5 mm.
We determined the number of complementation groups that the eight Fix− mutants represented. Only F1 progeny from a cross of lines 1D-1 and 4A-17 produced Fix− nodules (data not shown), indicating these lines belong to the same complementation group. We crossed mutants from each complementation group to wild-type plants and examined the F2 progeny for segregation of the white-nodule phenotype. Segregation of the Fix− phenotype was in all cases consistent with a mutation at a single locus (data not shown). Segregation ratios and complementation data indicate that the Fix− phenotype is recessive and monogenic in all eight mutants (data not shown). Because the Fix− mutant Mtsym1 (Bénaben et al., 1995) forms very small bump-like nodules under our conditions (data not shown) that are phenotypically different from the nodules of the mutants described here (Fig. 1), we did not examine the allelic relationship between Mtsym1 and these mutants.
Mutants with Small, White Nodules Have Defects in Nitrogen Fixation
To confirm the Fix− defect and quantify its extent, we assayed whether these mutants could support nitrogen fixation using the acetylene reduction assay (Fig. 2). Because all mutants show defects in their ability to support nitrogen fixation, they are named defective in nitrogen fixation (dnf) mutants. The individual loci that these mutants represent, 1D-1, 4A-17, 1B-5, 2C-2, 2E-1, 2F-16, 2H-8, and 4D-5, have been named DNF1 through DNF7, respectively (note that line 1D-1 represents dnf1-1 and line 4A-17 represents dnf1-2). Because dnf3 is most similar to wild-type plants in terms of ability to support nitrogen fixation, we used Student's t test to show that the levels of acetylene reduction were significantly different in dnf3 and wild-type plants (P < 0.001).
Figure 2.
Quantification of the nitrogen fixation defect. Results are pooled from two or more independent experiments. n, Number of individual plants assayed 21 dai. Error bars indicate se.
It is possible that the Fix− phenotype of the dnf mutants is due to an inability by the bacterial symbiont to penetrate the nodule tissue. To test whether the Fix− mutants had defects in early infection, we inoculated plants with S. meliloti strain Rm1021 containing a plasmid-borne hemA∷lacZ fusion and stained inoculated roots for β-galactosidase activity to visualize bacteria within infection threads (Boivin et al., 1990). This assay is useful for examining penetration of root hairs and outer cortical cells early in the establishment of the interaction (1–4 dai), but nodule-cell division later in the symbiosis obscures observation of infection threads, although infected cortical nodule cells are obvious in wild-type plants (data not shown; Boivin et al., 1990). We examined nodules of each dnf mutant 3 and 5 dai, three to five plants per genotype at each time point. Bacterial penetration into epidermal cells and into the outer cortical cells surrounding the nascent nodule of mutant plants was morphologically indistinguishable from those in wild-type plants. Bacterial colonization of nodule parenchyma cells was also indistinguishable between mutants and wild type (data not shown); we conclude that the Fix− phenotype of the dnf mutants is not due to defects in epidermal or cortical cell penetration by S. meliloti.
dnf Mutants Differ in Their Ability to Support and Trigger Nodulation-Related Bacterial Gene Expression
Rhizobium genes are differentially expressed inside the nodule (Oke and Long, 1999b), presumably due to signals and changes in the environment that accompany invasion and nodule development. To define the stage of arrest in nodule development in Fix− mutants, and to characterize the mutants at a molecular level, we examined the expression of several bacterial genes associated with symbiotic development in mutant and wild-type nodules.
We assayed the expression of uidA (encoding β-glucuronidase) fusions to the nodF, exoY, bacA, and nifH promoters, each of which is essential to a successful symbiotic program. The construction of the promoter-uidA fusions that we made created duplications of the promoter in the genome and left an intact copy of the gene, thus allowing bacterial and nodule development to proceed. The nodF operon is required for initiating infection and nodule development (Debellé et al., 1986); the exoY operon is required for infection (Pühler et al., 1991); the bacA gene is required for survival of the bacteria inside the plant cell (Glazebrook et al., 1993); and the nifH operon is required for nitrogen fixation (Ruvkun et al., 1982).
We inoculated dnf mutant plants with S. meliloti strains containing each of the symbiosis gene promoter fusions and assayed uidA expression 21 dai (Fig. 3). The expression of the PnodF∷uidA fusion differed in nodules of dnf2 plants (42.2% of wild type; Fig. 3A), but was statistically the same (P > 0.05) for the other dnf mutants. It is difficult to interpret this result because the role of NodF once bacteria are inside the nodule is unknown. Expression of the PexoY∷uidA fusion differed from wild type in dnf3, dnf5, and dnf6 (66.0%, 69.8%, and 79.4% of wild type, respectively; Fig. 3B). Expression of the PbacA∷uidA fusion in mutant nodules was similar to that of the PnodF∷uidA fusion: dnf2 was the only genotype to differ from wild type (37.7% of wild type; Fig. 3C). Because BacA is required for survival of S. meliloti once bacteria are deposited into the cytoplasm of the plant (Glazebrook et al., 1993), this reduction in bacA expression may indicate that in some nodules the bacteria die upon release from the infection thread. The expression of the PnifH∷uidA fusion showed the largest differences between wild-type and mutant plants, with six of the eight mutants differing significantly from wild type (Fig. 3D). The dnf1, dnf2, and dnf5 mutants were defective in the ability to elicit nifH expression; dnf4 and dnf7 showed reduced numbers of nodules that contained nifH-expressing bacteria (26.0% and 32.0% of wild type, respectively); dnf3 and dnf6 nodules showed an intermediate, although not statistically significant, number of nodules expressing nifH (50.5% and 53.2% of wild type, respectively). This finding correlates well with the finding that these two dnf mutants support the highest level of acetylene reduction activity out of all of the dnf mutants (Fig. 2). To examine the possibility that expression of the PnifH∷uidA fusion is delayed in the dnf mutants, we examined nodules at a later time point (28 dai). Results from this later time point were similar to those at 21 dai (data not shown).
Figure 3.
Expression of bacterial symbiosis-associated genes inside mutant plant nodules. Expression of nodF (A), exoY (B), bacA (C), and nifH (D) promoter-uidA fusions inside the nodules of wild-type and mutant plants 21 dai. Values are expressed as a percent of wild-type positive-staining nodules ± sd; data are pooled from at least two independent experiments, from five to 10 plants assayed per plant genotype-promoter fusion combination. All nodules from each plant assayed were scored. a, Significant (P < 0.05) difference between mutant and wild type as determined by the Mann-Whitney U test with Bonferroni correction.
584 Genes Are Differentially Expressed in Wild-Type Plants during Nodulation
To characterize Fix− defects in the context of plant symbiotic gene expression, we first needed to determine the gene expression patterns in wild-type plants during nodulation. We used an Affymetrix oligonucleotide microarray consisting of 9,935 tentative consensus (TC) sequences, which are based on cDNA libraries (Mitra et al., 2004). We examined gene expression of wild-type M. truncatula plants after inoculation with wild-type S. meliloti at six time points: 6 h, 1 d, 4 d, 7 d, 14 d, and 21 d. We chose these time points because they span the range of development of the Rhizobium-legume symbiosis, from initiation of the interaction through nitrogen fixation. Plants examined 6 h after inoculation have changes in root hair morphology and calcium signaling (Ehrhardt et al., 1996): At 1 dai, S. meliloti have begun the invasion of root hairs; at 4 dai, cortical cell layers of the root are infected and infection threads have ramified; at 7, 14, and 21 dai, plants are able to support nitrogen fixation (data not shown). Note that gene expression data for 55 genes from the 1-dai time point were previously published (Mitra et al., 2004) and are presented here for comparative purposes only.
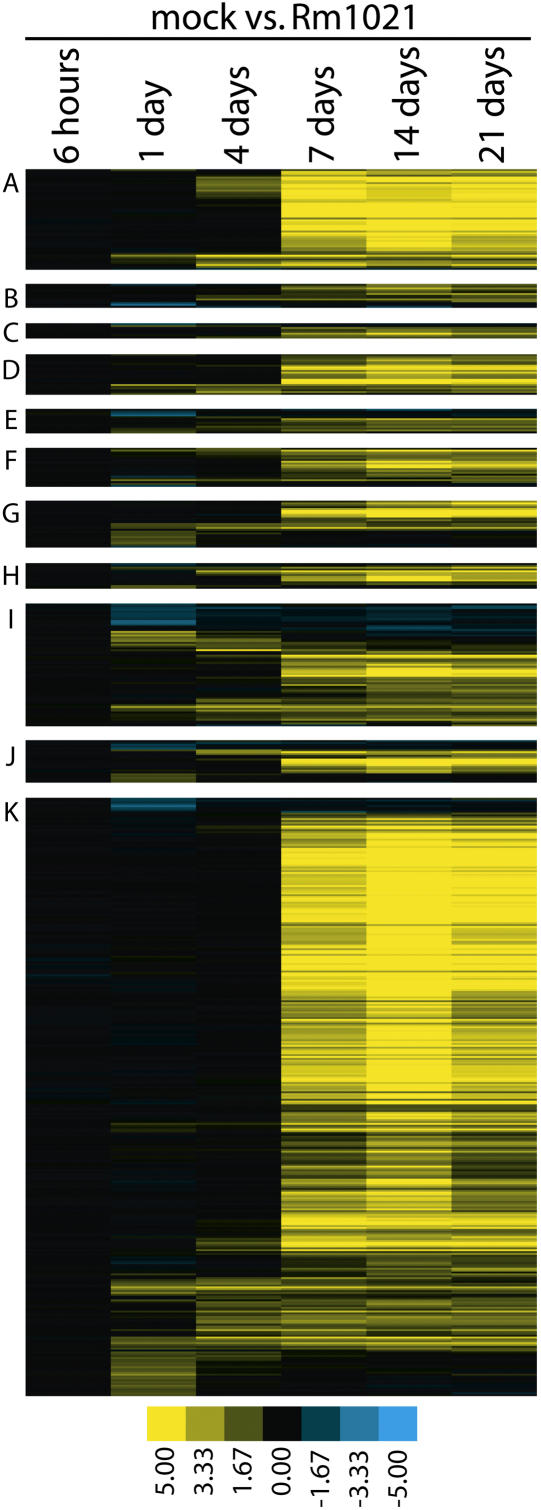
Using analysis criteria of a 2-fold change at 90% confidence level within the software program dChip (Li and Wong, 2001a, 2001b), we found that 584 TCs change in expression during nodule development. At 4 dai, 71 genes are induced and three are repressed upon exposure to Rm1021; at 7 dai, 197 are induced and one is repressed; at 14 dai, 427 are induced and five are repressed; and at 21 dai, 364 are induced and five are repressed (Fig. 4; Supplemental Table I). No TCs fit our criteria as showing significant changes in expression at the 6-h time point. Data from the 1-dai time point are presented elsewhere (Mitra et al., 2004). These data are consistent with data generated by the Significance Analysis of Microarrays (SAM) program (Tusher et al., 2001). SAM identifies over 90% of the TCs identified by dChip, with the exception of data at 7 dai. For this time point, dChip predicts that 164 TCs change expression, while SAM predicts that 111 of these TCs change expression. Fifty of the remaining TCs that were not predicted to change at 7 dai by SAM were predicted to change at 14 dai by both SAM and dChip. We conclude that the list of TCs identified by dChip, using a 2-fold cutoff at 90% confidence, is appropriate.
Figure 4.
Gene expression changes during nodulation. Color representation of the log2-fold change in expression level of 584 TCs that change during nodulation. TCs are clustered according to classification and expression pattern. TCs are classified into groups based on the The Institute for Genomic Research M. truncatula gene index as nodulins (A), defense related (B), hormone related (C), signaling (D), cell structure (E), transport (F), transcription and translation (G), primary metabolism (H), secondary metabolism (I), function unknown (J), and unknown hypothetical (K). Fold changes represent pooled data from three biological replicates for each condition. Data from the 1-dai time point has previously been published (Mitra et al., 2004).
dnf Mutants Show Altered Gene Expression Patterns
Three dnf mutants, dnf1-1, dnf2, and dnf7, were assayed for large-scale gene expression changes after exposure to S. meliloti. These three mutants represent three different classes of Fix− mutants based on their ability to induce and/or support symbiotic bacterial gene expression (Fig. 3), and on previously identified differences in gene expression (Mitra and Long, 2004). Microarray data show that differences between wild-type and dnf mutant plants fall into three classes (Fig. 5). Seventeen TCs are induced to high levels (from 5.5- to 5.7-fold) in wild-type plants, but are not induced (or are induced only to low levels) in at least one of the dnf mutants examined; 15 TCs are highly induced in wild-type plants and are also induced in the three dnf mutants, but to lower levels than in wild-type plants; and eight TCs are not induced in wild-type plants upon exposure to S. meliloti and are likewise not induced in the three dnf mutants, but their level of expression is significantly different in these mutants compared to wild-type plants (data not shown).
Figure 5.
Gene expression in three Fix− mutants is altered. Color representation of the log2-fold change in expression level. TCs are clustered according to expression pattern. All TCs shown change at least 2-fold when compared to wild-type plants with the same treatment. M, Change in the mutant versus wild-type mock-inoculated treatment; S, change in the mutant versus wild-type S. meliloti comparison. Identity of closest protein and e-value as determined by tBLASTX are shown.
Expression of assayed genes in dnf1-1, dnf2, dnf7, and wild-type plants is similar at 7 dai. Figure 5 represents expression changes of 40 genes from mock-inoculated versus Rm1021-inoculated wild-type plants (at 4, 7, and 14 dai) and dnf1-1, dnf2, and dnf7 plants (at 7 dai). Each of these 40 genes shows a significant difference in expression between wild type and at least one mutant treated similarly (mock inoculated or inoculated with Rm1021) at 7 dai. Of the 164 genes that changed expression in wild-type plants 7 dai, 151 also changed in dnf1-1, 133 changed in dnf2, and 137 changed in dnf7.
Among the TCs not induced in the three dnf mutants are, notably, two genes that have been described previously as being induced during nodulation: leghemoglobin (TC35565) and a calmodulin-like protein (TC35912 [Györgyey et al., 2000]; TC35912 [Fedorova et al., 2002]). Among the TCs that have significant homology to known genes and differ in at least one dnf mutant from wild-type plants are Nodulin27 (TC38783 [Gamas et al., 1996]), a signal peptidase subunit-like protein (TC36736), an invertase/pectin methylesterase inhibitor (TC38721), a formidase-like protein (TC34752), a bimodular Pro-rich-like protein (TC36077), and a wound-induced protein (TC30455; Fig. 5). Twenty-nine TCs that differ between wild-type and dnf plants show limited homology to published sequences (e-values > 2e–6; Fig. 5), indicating that they may encode novel proteins.
DISCUSSION
We report the isolation of eight M. truncatula mutants that are unable to support wild-type levels of nitrogen fixation. We show that the eight mutants represent seven complementation groups and we assign these mutants to three classes using bacterial symbiotic gene expression. Further, we analyze the expression of 9,935 genes at different stages of the symbiotic interaction and examine the expression of these genes in a representation of each of the three Fix− classes.
In our screen for symbiotic mutants of M. truncatula, we isolated mutants from seven complementation groups and, with the exception of the DNF1, only a single mutant represents each locus. We screened F2 progeny from approximately 2,800 F1 plants. Given the large number of Fix− mutants in other legumes, such as Pisum sativum (at least 13; Engvild, 1987; Phillips and Teuber, 1992; Morzhina et al., 2000), and our findings here, we conclude that our screen was not saturating and that many more genes essential to the support of nitrogen fixation remain to be discovered. Further, investigations into the requirements for nitrogen fixation may benefit from more sophisticated genetic screens, such as screens for bacterial suppressor or plant suppressor mutations of mutant phenotypes described here.
The support of nitrogen fixation by the plant is a complex process involving the development of a new organ (the root nodule), a permissive infection process involving the uptake of bacteria into nodule cells and the transport of metabolites between plant and rhizobia symbionts. The DNF genes characterized here may be involved in any of these processes. Our data indicate that all dnf mutants permit the initial nodulation events involved in the establishment of the nodule primordium and nodule and cellular infection by the bacteria (data not shown). Additionally, all dnf mutants are able to support a normal endomycorrhizal symbiosis (M. Harrison, personal communication) unlike some P. sativum Fix− mutants (Gianinazzi-Pearson et al., 1991). Because the dnf mutants show no obvious nonsymbiotic defects in growth, fecundity, or size and shape when supplemented with nitrogen-containing fertilizers (data not shown), we speculate that the DNF genes are required specifically for symbiotic nitrogen fixation.
Nodule-specific genes (Enods, nodulins, and late nodulins) were initially defined through examination of protein synthesis and through assays of specific genes. Macro- and microarray studies have identified many more genes that change expression upon inoculation with rhizobia (Barnett et al., 2004; El Yahyaoui et al., 2004; Mitra et al., 2004). Here, we found that 584 genes are differentially expressed in wild-type plants throughout the nodulation process compared to mock-treated controls and, although many genes identified may play roles in nodulation, we cannot rule out the possibility that at least some of the differences we observe are due to nitrogen starvation of the mock-treated plants. The largest number of genes changing expression and the greatest magnitude of change occurs at the onset of the nitrogen-fixation process (7 and 14 dai; Fig. 4). A large number of these genes cannot be assigned any function based on sequence analysis, highlighting the need for additional functional approaches addressing the plant requirements for nodulation.
Our data are somewhat inconsistent with previously published macro- and microarray data. El Yahyaoui et al. (2004) found that approximately one-half of the differentially expressed genes in nodules were down-regulated. In our study, approximately 1% of differentially expressed genes are down-regulated during nodulation. This discrepancy may be due to differences in statistical analysis or to experimental design. For example, in the study by El Yahyaoui et al. (2004), plants were grown aeroponically for 15 d (the last four under nitrogen-starvation conditions) before inoculation with S. meliloti versus this study, where plants were grown for 6 d under nitrogen-starvation conditions on agar plates before inoculation. Additionally, they harvested only nodules (at 4 and 10 dai), whereas in our study, we harvested 5-cm nodule-bearing root segments centered on the site of inoculation, and our mock-inoculated tissues were the same age as the comparable inoculated ones. In this study, primary root tips and lateral roots were removed from all tissues at the time of harvest to reduce the transcript contribution of actively dividing cells not associated with nodules, whereas El Yahyaoui et al. (2004) used whole-root systems of 15-d-old plants as their control. Whereas both sets of conditions have advantages, there are several reasons why the data may not be comparable. It is likely that our method does not identify some genes that are differentially expressed. We chose to minimize the identification of false positives at the expense of increasing sensitivity for those genes that have small differences in expression.
We chose to subject three dnf mutants, dnf1-1, dnf2, and dnf7, to large-scale transcriptional analysis. These mutants were selected based on previously defined transcriptional differences assessed by northern blot with a small number of genes (Mitra and Long, 2004) and on the expression of bacterial symbiotic genes in the dnf mutants (this study). It is remarkable that, out of 164 genes whose expression changes in wild-type plants in the first 7 d of the symbiotic program, only 40 genes, in total, are misregulated in the three dnf mutants examined. Of these, 33 genes of various functions are strongly induced in wild-type roots, but poorly induced in dnf roots. What is the basis for the misregulation of this rather narrow gene set? Perhaps the identification of the DNF gene products themselves will shed light on this question. Due to the similarity of expression profiles of the three mutants we examined, it is difficult to differentiate between these mutants on these data alone. However, using bacterial gene expression at 21 dai (Fig. 3) and northern-blot analysis at 28 dai (Mitra and Long, 2004), we were able to discern differences between these mutants. This suggests that the three dnf mutants have similar transcriptional blocks at earlier time points, but that they are able to proceed with subsequent gene expression and development to different extents. While more data are needed, such a result would be consistent with the idea that the development of nitrogen-fixing plant nodules is more complex than a single linear progression of differentiation events.
The mutants isolated here and in other studies (Bénaben et al., 1995; Kuppusamy et al., 2004; Veereshlingam et al., 2004) can be classified into five groups based on four criteria: nodule size, acetylene reduction activity, support of bacterial symbiotic gene expression, and plant gene expression (Table I). The first group consists of lin, nip, and Mtsym1 (Bénaben et al., 1995; Kuppusamy et al., 2004; Veereshlingam et al., 2004), which have a small bump-like nodule phenotype. The second group contains dnf1-1, dnf1-2, and dnf5. This group displays infection of the inner cortex, little to no acetylene reduction activity, supports no nifH expression, and does not express Nodulin31 and other genes (Mitra and Long, 2004). A third group, which includes dnf4 and dnf7, displays no acetylene reduction, supports no nifH expression, and does express Nodulin31 and other symbiotic genes. The fourth group includes mutants dnf3 and dnf6. Members of this group display a small amount of acetylene reduction activity, support the most nifH expression, and express all nodulation-related genes tested. The reduced ability to support exoY expression in dnf3, dnf5, and dnf6 would seem to link these mutants and disrupts this grouping of mutants; however, we currently have more evidence separating dnf5 from dnf3 and dnf6 than linking them. These four groups can be logically ordered, one to four. The fifth group consists of dnf2. This mutant displays no acetylene reduction activity and does not express Nodulin31. The support of bacterial symbiotic gene expression is most noticeably affected in dnf2 mutants: Fewer nodules possess bacteria expressing nodF and bacA. The dnf2 mutant does not easily fit into a linear ordering of mutants. Microscopic examination of the mutants presented here should further refine these groups and allow us to place them in an order relative to the stage at which nodule development is affected.
Table I.
Classification of Fix− mutants into groups
+, Wild type; −, completely defective; ±, intermediate phenotype; and nd, not determined.
| Group (Mutants)
|
Nodule Size
|
Acetylene Reduction
|
Plant Gene Expression, 28 daidLB1, CAM1, Nodulin31 (Symbiotic)
|
Bacterial Expression
|
|
|---|---|---|---|---|---|
| nodF, bacA | nifH | ||||
| 1 (lina, nipb, Sym1c) | Small/bumpabc | −ac | −e | nd | nd |
| 2 (dnf1, dnf5) | Small | − | − | + | − |
| 3 (dnf4, dnf7) | Small | − | + | + | ± |
| 4 (dnf3, dnf6) | Small | ± | + | + | ± |
| 5 (dnf2) | Small | − | + | ± | − |
| Wild type | Large | + | + | + | + |
Only Sym1 was assayed.
In this work, we describe seven complementation groups that are essential for nitrogen fixation, required for bacterial symbiotic gene expression, and deficient in plant symbiosis-associated gene expression. These dnf mutants will provide a framework for future functional studies and will greatly enhance our understanding of the nodulation process.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions
Sinorhizobium meliloti cultures were grown on Luria-Bertani or TY medium at 30°C with appropriate antibiotics.
The PexoY∷uidA fusion was created using PCR amplification of the exoY genomic region from S. meliloti strain Rm1021 DNA with the primers 5′-cgccgtTcTagaactAgacgagggccatgatgagc-3′ and 5′-gttgttgccGGAtcctcctgcctggccac-3′ (capitalized bases are altered from Rm1021 sequence to introduce restriction sites and an in-frame stop codon). The PCR product was first cloned into pCR2.1 (Invitrogen) and then subcloned into pVO155 (Oke and Long, 1999a) after digestion with BamHI and XbaI. The resulting plasmid was transferred into Rm1021 via triparental mating. Integration of this plasmid into the genome created a duplication of the exoY promoter: one exoY promoter was fused to uidA, while the other remained intact with the exoY gene (strain CSB365). PnifH∷uidA was created similarly, using primers 5′-ccaggtcTaGaagcgcggcgagtgtattttAggagg-3′ and 5′-aagcgttaagcagGaTccggaatggtcc-3′ to create strain CSB357. Each fusion was transduced using N3 phage (Martin and Long, 1984) into an unmarked Rm1021 background and the chromosomal location of each fusion was verified by PCR (data not shown). Strains CSB365, CSB357, and VO2196, which contain a PbacA∷uidA fusion (V. Oke, unpublished data), and DW386, which contains a PnodF∷uidA fusion (Wais et al., 2002) form Fix+ nodules on wild-type plants (data not shown).
Plant Growth Conditions
In vitro plant growth conditions were described previously (Mitra and Long, 2004). Additionally, plants were grown in commercial potting mix in either a glass house (natural light supplemented with 400 W high-pressure sodium bulbs to achieve a 16-h day) or in a growth chamber (Mitra and Long, 2004). Plants grown in potting soil were inoculated at the time of planting by adding approximately 1 mL of an overnight culture of S. meliloti to approximately 10 cm3 of potting mix.
For microarray experiments, plants were grown on buffered nodulation medium (BNM; Ehrhardt et al., 1992), pH 6.5, with 1 mm α-aminoisobutyric acid and either 1.15% (w/v) agar (tissue harvested 6 h, 1 dai, and 4 dai) or 2.5% (w/v) agar (tissue harvested 7, 14, and 21 dai). Plants were inoculated 6 d after planting with either 1 μL of a suspension of Rm1021 (optical density at 600 nm [OD600] = 0.05) or 1 μL of 0.5× BNM as a mock control. Harvested root segments (5 cm) centered on the spot of bacterial or mock treatment. Root tips (if within the 5-cm segment) and lateral roots were removed.
Plant Mutagenesis and Primary Screen
Medicago truncatula Gaertn. cv Jemalong seeds (Purkiss Seeds) were mutagenized by bombardment with fast neutrons at the International Atomic Energy Agency, Plant Breeding Section (Vienna). Irradiation levels of 15, 20, 25, 35, and 50 Gray (Gy) were used on separate seed sets. M1 plants from the higher irradiation levels (35 and 50 Gy) did not survive to set seed. Mutagenized seeds (M1) were grown, 50 per bulk, in a glass house. Eighty-six seed bulks representing 2,862 M1 plants were harvested. Germination frequencies for the progeny of M1 plants irradiated at 15, 20, and 25 Gy were 68%, 63%, and 67%, respectively. All mutants described herein originated from a radiation dose of 15 Gy. The non-nodulating mutants nsp2-1, nsp2-2 (Oldroyd and Long, 2003), and dmi1-5 (Ane et al., 2004) also were isolated via this screen.
M2 plants were inoculated with Rhizobium field isolate strain B1 and screened 3.5 to 4 weeks postinoculation. In addition to normal watering, plants were also watered once with 0.5× BNM. Strain B1 was used in the initial screen of M2 plants and to confirm mutant phenotypes of M3 plants grown in potting soil.
Segregation Analysis
To assess segregation of Fix− mutants, F2 plants from a cross between wild-type plants and mutant plants were grown in potting soil and inoculated with Rm1021 (OD600 = 0.05). Typically, root systems were examined 2 months after inoculation and plants with only white nodules were repotted and reexamined 3 weeks later. If, upon reexamination, all nodules were white, the plant was scored as Fix−. Plants with any pink nodules were scored as Fix+, regardless of the presence of white nodules. F2 plants from the line 2C-2 and wild-type cross were scored differently from other F2 plants, as the 2C-2 mutant line often has a small percentage of Fix+ nodules in a background of Fix− nodules. All plants from this segregating F2 population were examined and the percentage of Fix− nodules per plant was estimated based on the observation of whole-root systems. Plants for which >80% of nodules were Fix− were scored as carrying the mutation.
Acetylene Reduction Assays
The acetylene reduction assay (Turner and Gibson, 1980) was performed as described (Oke and Long, 1999a) and used to determine whether plants could support nitrogen fixation. To assay complementation, several nodule-bearing root segments were excised from Rm1021-inoculated plants grown in potting soil and placed in culture tubes on strips of cellulose chromatography paper (Whatman) wetted with 1.5 mL 0.5× BNM.
Bacterial Gene Expression
Plants grown in glass tubes were flood inoculated with the appropriate bacterial strain at OD600 = 0.05. Twenty-one days after inoculation nodules were bisected transversely with respect to the root axis and left attached to the root to facilitate data quantification. Staining for β-glucuronidase activity was performed as described (Swanson et al., 1993).
Affymetrix GeneChip Experiments
Construction of a M. truncatula-S. meliloti oligonucleotide chip has been described (Barnett et al., 2004; Mitra et al., 2004). RNA was purified using TRIzol (Invitrogen) as described (Mitra et al., 2004). For each preparation, total RNA was isolated from approximately 90 root segments. Three independent biological replicates were used for each time point and each treatment. Either 30 μg (root segments harvested 1 dai) or 40 μg (root segments harvested 4, 7, 14, and 21 dai) of total RNA were used for double-stranded cDNA synthesis (SuperScript II; Invitrogen). Biotin-labeled cRNA was synthesized using the BioArray high yield RNA transcription labeling kit (Enzo Diagnostics). Because samples inoculated with Rm1021 and harvested 7, 14, and 21 dai contain large amounts of bacterial RNA, 20% more cDNA (by volume; versus mock treated) was used from these samples for biotin-labeled cRNA synthesis. Twenty micrograms of biotin-labeled cRNA were hybridized to Affymetrix DNA chips for all samples. Data analysis was performed as described (Mitra et al., 2004).
Supplementary Material
Acknowledgments
We would like to thank Ann Morrison (Stanford University) for greenhouse help, Derek Wells (Stanford University) and Valerie Oke (University of Pittsburgh) for bacterial strains, Maria Harrison (Noble Foundation) for assaying mycorrhizal phenotypes, Fumiaki Katagiri (University of Minnesota) for statistical assistance, and current and former members of the Long laboratory, particularly Robert Fisher, Giles Oldroyd, Joel Griffiths, David Almassian, and Sidney Shaw, for useful discussions.
This work was supported by the Howard Hughes Medical Institute and the U.S. Department of Energy (grant no. DE–FG03–90ER20010 to S.R.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sharon R. Long (srl@stanford.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.072132.
References
- Amor BB, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long JD Sr, Gough C (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34: 495–506 [DOI] [PubMed] [Google Scholar]
- Ane JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Levy J, Debelle F, Baek JM, Kalo P, et al (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Barnett MJ, Toman CJ, Fisher RF, Long SR (2004) A dual-genome Symbiosis Chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc Natl Acad Sci USA 101: 16636–16641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénaben V, Duc G, Lefebvre V, Huguet T (1995) TE7, an inefficient symbiotic mutant of Medicago truncatula Gaertn. cv Jemalong. Plant Physiol 107: 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin C, Camut S, Malpica CA, Truchet G, Rosenberg C (1990) Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell 2: 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll BJ, McNeil DL, Gresshoff PM (1985) A supernodulation and nitrate-tolerant symbiotic (Nts) soybean mutant. Plant Physiol 78: 34–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J (2000) Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Timmers AC, Maillet F, Galera C, Penmetsa RV, Cook D, Dénarié J, Gough C (2001) The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development 128: 1507–1518 [DOI] [PubMed] [Google Scholar]
- Debellé F, Rosenberg C, Vasse J, Maillet F, Martinez E, Dénarié J, Truchet G (1986) Assignment of symbiotic developmental phenotypes to common and specific nodulation (nod) genetic loci of Rhizobium meliloti. J Bacteriol 168: 1075–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt DW, Atkinson EM, Long SR (1992) Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256: 998–1000 [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR (1996) Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- El Yahyaoui F, Kuster H, Ben Amor B, Hohnjec N, Puhler A, Becker A, Gouzy J, Vernie T, Gough C, Niebel A, et al (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136: 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engvild KC (1987) Nodulation and nitrogen fixation mutants of pea, Pisum sativum. Theor Appl Genet 74: 711–713 [DOI] [PubMed] [Google Scholar]
- Fedorova M, Van De Mortel J, Matsumoto PA, Cho J, Town CD, VandenBosch KA, Gantt JS, Vance CP (2002) Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol 130: 519–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan TM, Wood JM, Jordan DC (1983) Symbiotic properties of C4-dicarboxylic acid transport mutants of Rhizobium leguminosarum. J Bacteriol 154: 1403–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P, De Carvalho-Niebel F, Lescure N, Cullimore J (1996) Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol Plant Microbe Interact 9: 233–242 [DOI] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V, Gianinazzi S, Guillemin JP, Trouvelot A, Duc G (1991) Genetic and cellular analysis of resistance to vesicular arbuscular (VA) mycorrhizal fungi in pea plants. In H Hennecke, DP Verma, eds, Advances in Molecular Genetics of Plant-Microbe Interactions, Vol 1. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 336–342
- Glazebrook J, Ichige A, Walker GC (1993) A Rhizobium meliloti homolog of the Escherichia coli peptide-antibiotic transport protein SbmA is essential for bacteroid development. Genes Dev 7: 1485–1497 [DOI] [PubMed] [Google Scholar]
- Gordon AJ, Minchin FR, James CL, Komina O (1999) Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiol 120: 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györgyey J, Vaubert D, Jiménez-Zurdo JI, Charon C, Troussard L, Kondorosi A, Kondorosi E (2000) Analysis of Medicago truncatula nodule expressed sequence tags. Mol Plant Microbe Interact 13: 62–71 [DOI] [PubMed] [Google Scholar]
- Kalo P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Imaizumi-Anraku H, Koiwa H, Niwa S, Ikuta A, Syono K, Akao S (2002) Root, root hair, and symbiotic mutants of the model legume Lotus japonicus. Mol Plant Microbe Interact 15: 17–26 [DOI] [PubMed] [Google Scholar]
- Krusell L, Krause K, Ott T, Desbrosses G, Kramer U, Sato S, Nakamura Y, Tabata S, James EK, Sandal N, et al (2005) The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17: 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy KT, Endre G, Prabhu R, Penmetsa RV, Veereshlingam H, Cook DR, Dickstein R, Vandenbosch KA (2004) LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections. Plant Physiol 136: 3682–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH (2001. a) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH (2001. b) Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol 2: RESEARCH0032.1–RESEARCH0032.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MO, Long SR (1984) Generalized transduction in Rhizobium meliloti. J Bacteriol 159: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade HM, Long SR, Ruvkun GB, Brown SE, Ausubel FM (1982) Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol 149: 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Long SR (2004) Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the Medicago truncatula/Sinorhizobium meliloti symbiosis. Plant Physiol 134: 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Shaw SL, Long SR (2004) Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc Natl Acad Sci USA 101: 10217–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morzhina EV, Tsyganov VE, Borisov AY, Lebsky VK, Tikhonovich IA (2000) Four developmental stages identified by genetic dissection of pea (Pisum sativum L.) root nodule morphogenesis. Plant Sci 155: 75–83 [DOI] [PubMed] [Google Scholar]
- Oke V, Long SR (1999. a) Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol Microbiol 32: 837–849 [DOI] [PubMed] [Google Scholar]
- Oke V, Long SR (1999. b) Bacteroid formation in the Rhizobium-legume symbiosis. Curr Opin Microbiol 2: 641–646 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA (2004) Calcium, kinases, and nodulation signaling in legumes. Nat Rev Mol Cell Biol 5: 566–576 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Long SR (2003) Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in nod factor signaling. Plant Physiol 131: 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott T, van Dongen JT, Gunther C, Krusell L, Desbrosses G, Vigeolas H, Bock V, Czechowski T, Geigenberger P, Udvardi MK (2005) Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr Biol 15: 531–535 [DOI] [PubMed] [Google Scholar]
- Peterson MA, Barnes DK (1981) Inheritance of ineffective nodulation and non-nodulation traits in alfalfa. Crop Sci 21: 611–616 [Google Scholar]
- Phillips DA, Teuber LR (1992) Plant genetics of symbiotic nitrogen fixation. In G Stacey, RH Burris, HJ Evans, eds, Biological Nitrogen Fixation. Chapman and Hall, New York, pp 625–647
- Pühler A, Arnold W, Buendia-Claveria A, Kapp D, Keller M, Niehaus K, Quandt J, Roxlau A, Weng WM (1991) The role of Rhizobium meliloti exopolysaccharide EPSI and EPSII in the infection process of alfalfa nodules. In H Hennecke, DPS Verma, eds, Advances in Molecular Genetics of Plant-Microbe Interactions, Vol 1. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 189–194
- Ronson CW, Lyttleton P, Robertson JG (1981) C4-dicarboxylate transport mutants of Rhizobium trifolii form ineffective nodules on Trifolium repens. Proc Natl Acad Sci USA 78: 4284–4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun GB, Sundaresan V, Ausubel FM (1982) Directed transposon Tn5 mutagenesis and complementation analysis of Rhizobium meliloti symbiotic nitrogen fixation genes. Cell 29: 551–559 [DOI] [PubMed] [Google Scholar]
- Sagan M, deLarambergue H, Morandi D (1998) Genetic analysis of symbiosis mutants in Medicago truncatula. In C Elmerich, A Kondorosi, WE Newton, eds, Biological Nitrogen Fixation for the 21st Century. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 317–318
- Sagan M, Morandi D, Tarenghi E, Duc G (1995) Selection of nodulation and mycorrhizal mutants in the model plant Medicago truncatula (Gaertn.) after gamma-ray mutagenesis. Plant Sci 111: 63–71 [Google Scholar]
- Schauser L, Handberg K, Sandal N, Stiller J, Thykjaer T, Pajuelo E, Nielsen A, Stougaard J (1998) Symbiotic mutants deficient in nodule establishment identified after T-DNA transformation of Lotus japonicus. Mol Gen Genet 259: 414–423 [DOI] [PubMed] [Google Scholar]
- Schubert KR (1986) Products of biological nitrogen fixation in higher plants: synthesis, transport, and metabolism. Annu Rev Plant Physiol 37: 539–574 [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debelle F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Swanson JA, Mulligan JT, Long SR (1993) Regulation of syrM and nodD3 in Rhizobium meliloti. Genetics 134: 435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczyglowski K, Shaw RS, Wopereis J, Copeland S, Hamburger D, Kasiborski B, Dazzo FB, de Bruijn F (1998) Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol Plant Microbe Interact 11: 684–697 [Google Scholar]
- Turner GL, Gibson AH (1980) Measurement of nitrogen fixation by indirect means. In FJ Bergersen, ed, Methods for Evaluating Biological Nitrogen Fixation. John Wiley & Sons, Chichester, UK, pp 111–138
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M, Price GD, Gresshoff PM, Day DA (1988) A dicarboxylate transporter on the peribacteroid membrane of soybean nodules. FEBS Lett 231: 36–40 [Google Scholar]
- Vance CP, Heichel GH (1991) Carbon in N2 fixation: limitation of exquisite adaptation. Annu Rev Plant Physiol Plant Mol Biol 42: 373–392 [Google Scholar]
- Veereshlingam H, Haynes JG, Penmetsa RV, Cook DR, Sherrier DJ, Dickstein R (2004) nip, a symbiotic Medicago truncatula mutant that forms root nodules with aberrant infection threads and plant defense-like response. Plant Physiol 136: 3692–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viands DR, Vance CP, Heichel GH, Barnes DK (1979) An ineffective nitrogen fixation trait in alfalfa. Crop Sci 19: 905–908 [Google Scholar]
- Wais RJ, Keating DH, Long SR (2002) Structure-function analysis of Nod factor-induced root hair calcium spiking in Rhizobium-legume symbiosis. Plant Physiol 129: 211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SA, Viprey V, Downie JA (2000) Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by Nod factors and chitin oligomers. Proc Natl Acad Sci USA 97: 13413–13418 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.