Abstract
Previous studies have indicated that the UL6, UL15, UL17, UL28, UL32, and UL33 genes are required for the cleavage and packaging of herpes simplex viral DNA. To identify proteins that interact with the UL28-encoded DNA binding protein of herpes simplex virus type 1 (HSV-1), a previously undescribed rabbit polyclonal antibody directed against the UL28 protein fused to glutathione S-transferase was used to immunopurify UL28 and the proteins with which it associated. It was found that the antibody specifically coimmunoprecipitated proteins encoded by the genes UL28, UL15, and UL33 from lysates of both HEp-2 cells infected with HSV-1(F) and insect cells infected with recombinant baculoviruses expressing these three proteins. In reciprocal reactions, antibodies directed against the UL15- or UL33-encoded proteins also coimmunoprecipitated the UL28 protein. The coimmunoprecipitation of the three proteins from HSV-infected cells confirms earlier reports of an association between the UL28 and UL15 proteins and represents the first evidence of the involvement of the UL33 protein in this complex.
The cleavage and packaging of newly synthesized herpesvirus DNA is a highly conserved process occurring late in viral replication. During herpesvirus infection, double-stranded viral DNA genomes accumulate in cell nuclei as head-to-tail concatemers which are subsequently cleaved into single genome lengths and packaged into preformed viral capsids (reviewed in reference 19). Six genes of herpes simplex virus type 1 (HSV-1) are known to be essential for this process, namely, UL6, UL15, UL17, UL28, UL32, and UL33. In cells infected with viruses individually lacking these genes, capsids appear normal and are readily detected, but viral DNA is neither cleaved nor packaged (2, 3, 5, 9, 12, 25, 26, 30, 31, 35, 37, 40–42, 44).
The cleavage and packaging process of herpesviruses is believed to be similar to that employed by double-stranded DNA bacteriophages; consequently, it is useful to consider the mechanisms used by these bacteriophages as a model for the cleavage and packaging events employed by herpesviruses. Such models propose a central role for the viral “terminase,” a complex of at least two proteins that (i) binds the viral DNA and links it with the empty viral capsid (HSV-1) or prohead (bacteriophages); (ii) cleaves the viral DNA at precise internal sites, resulting in the separation of unit-length genomes from concatameric DNA; and (iii) hydrolyzes ATP, providing the energy required to drive the DNA into the capsid (reviewed in reference 10). With this paradigm in mind, efforts have been expended to identify herpesvirus gene products that perform functions expected of the viral terminase, especially DNA binding, ATP hydrolysis, and at least transient association with capsids.
There is a growing body of both direct and indirect evidence suggesting that the UL15 and UL28 gene products comprise two subunits of the HSV-1 terminase. The UL28 gene product has been shown to bind specifically to the HSV-1 DNA sequence pac1, which is found in the a sequence of the genome and is known to be essential for the generation of correct genomic termini (4, 38). The UL15 protein has been hypothesized to hydrolyze ATP, based on limited homology with a putative nucleotide binding motif comprised of Walker boxes A and B within gp17, the larger subunit of the T4 bacteriophage terminase (15). Although the ATPase activity of the UL15 protein has yet to be directly demonstrated, a mutation within the Walker box precludes viral DNA cleavage and packaging (46). The UL28 and UL15 proteins have also been shown to interact in transient-expression assays (1, 22, 23, 45) and in coimmunoprecipitation experiments using nuclear extracts of infected cells (23).
This paper describes the isolation of a protein complex consisting of the UL28, UL15, and UL33 proteins by immunoprecipitation from lysates of infected cells. This suggests for the first time that the UL33 gene product may function as a third subunit of the putative viral terminase.
MATERIALS AND METHODS
Cells and viruses.
Vero and HEp-2 cells and transformed cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum, penicillin, and streptomycin. Sf21 (Spodoptera frugiperda) cells were maintained in Grace's insect cell culture medium (GibcoBRL) supplemented with 10% fetal bovine serum and gentamicin (10 μg/ml). The wild-type virus HSV-1(F) and the mutant HSV-7202 have been previously described (6, 16), and their titers were determined on Vero cell monolayers. The titer of the UL28 deletion virus HSV-gCB was determined by propagation on the transformed cell line C1 (41). The mutant viruses lacking UL15 and UL33 have been described previously (5, 9).
Recombinant baculoviruses encoding the HSV-1 UL28, UL15, and UL33 genes were generated by cotransfecting Sf21 cells with baculovirus (Autographa californica multicapsid nucleopolyhedrosis virus [Ac MNPV]) DNA (Invitrogen) and plasmid DNA encoding the entire open reading frame of UL15, UL28, or UL33 under the control of the baculovirus polyhedron promoter in the pBlueBacIII vector (Invitrogen), followed by plaque purification of recombinant viruses.
Antiserum production.
Plasmid pJB179, predicted to encode the full-length UL28 protein fused to glutathione S-transferase (GST), was generated by ligating a 2.5-kb DNA fragment containing bases 55,682 to 58,158 (27) of HSV-1(F) to the EcoRI site of pGEX-4T-1 (Pharmacia). The plasmid was sequenced to confirm that the UL28-GST junction maintained the open reading frames of the UL28 and GST proteins (data not shown). Escherichia coli BL-21 cells transformed with pJB179 were cultured at 37°C for 1 h followed by induction of the fusion protein with 0.1 mM isopropyl-β-d-thiogalactopyranoside at 25°C for 3 h. The induced UL28-GST fusion protein was isolated from the bacteria as described previously (18) with the following modifications: Triton X-100 (2% [vol/vol]) was added to the clarified bacterial lysate, and the lysate was incubated with glutathione-Sepharose beads overnight at 4°C. Bound protein was eluted from the beads by boiling in buffer containing 2% sodium dodecyl sulfate (SDS) and 5% β-mercaptoethanol, and the UL28-GST fusion protein was separated from contaminating bacterial proteins by SDS-polyacrylamide gel electrophoresis. The separated proteins were visualized by lightly staining with Coomassie blue, and the band containing UL28-GST was removed and minced. To emulsify the acrylamide (containing the fusion protein) for injection, sterile phosphate-buffered saline (PBS) was added to the minced gel and it was passed through successively smaller-gauge needles.
Determination of antibody specificity.
Flasks (25 cm2) of Vero cells were infected at a multiplicity of infection (MOI) of 5.0 with either HSV-1(F) or the mutant gCB virus lacking UL28. Twenty hours postinfection the cells were harvested, pelleted, and resuspended in a buffer consisting of PBS with 0.5% Triton and 0.5% sodium deoxycholate. They were then sonicated for 5 s at low power and clarified at high speed in a microcentrifuge. Fifty microliters of the supernatant was then added to 20 μl of loading buffer, and the sample was boiled for 3 min and electrophoretically separated on an SDS-polyacrylamide gel. The proteins from the gel were transferred to nitrocellulose membranes and probed with either polyclonal rabbit antiserum (diluted in PBS plus 1% bovine serum albumin and 1% Tween 20) directed against ICP8 (36) or the antiserum directed against UL28-GST.
Production of radiolabeled cell lysates.
HEp-2 cell monolayers were infected at an MOI of 5.0 PFU per cell with either HSV-1(F) or the R7202 mutant virus, which is derived from HSV-1(F) but lacks the capacity to produce glycoprotein E (6). After 4 h of incubation, the medium was replaced with labeling medium containing DMEM with 10% of the normal amounts of methionine and cysteine and 50 μCi of [35S]-labeled cysteine and methionine (Translabel; ICN) per ml. Twenty hours after infection, cells were washed four times with ice-cold PBS, scraped off the flask, collected, pelleted and, if necessary, stored at −80°C until use.
Immunoprecipitations.
Immunoprecipitations were performed using radiolabeled lysates of infected HEp-2 cells immediately after thawing on ice (see above). Alternatively, HEp-2 cells were infected with HSV-1(F) or the gE-null mutant R7202 at an MOI of 5.0 PFU per cell and incubated at 37°C for 20 to 24 h. The cells were washed four times with ice-cold PBS, scraped off the flask, collected, pelleted and, if required, stored at −80°C until use.
Cells from the equivalent of a 12.5-cm2 flask were sonicated for 5 s at low power in a 1-ml volume of buffer consisting of PBS containing 1% Tween 20 (PBSTw). Cells were then clarified in an Eppendorf Microfuge for 20 min at 14,000 × g, and the supernatants were transferred to new microcentrifuge tubes. Three microliters of antiserum was added, and the tubes were left on wet ice for 2 h. Fifty microliters of a 50% slurry of Gammabind G-Sepharose beads (Amersham Pharmacia Biotech) in PBS were washed three times in PSBTw and added to each cell lysate, and the tube was rotated at 4°C for 1 h. The beads were then washed four times in excess PBSTw and boiled in 2% SDS, and eluted proteins were separated by electrophoresis on a denaturing polyacrylamide gel. The gel was then used either for immunoblotting or fluorography. For the latter, the gel was incubated for 30 min in a 1.25 M sodium salicylate solution, vacuum dried, and placed next to scientific imaging film (X-Omat; Kodak) at −80°C for a minimum of 24 h, after which the film was developed. The method for immunoblotting has been previously described (6); the antibodies used are listed in Table 1 and detailed below in the text.
TABLE 1.
Provenance of antibodies used in this study
In some experiments, Sf21 cells were infected at an MOI of 5.0 PFU per cell with recombinant baculoviruses expressing the UL15, UL28, or UL33 proteins. The cells were incubated at 30°C for 40 h, after which time the cells were washed four times with ice-cold PBS and stored at −80°C until use. The remainder of the immunoprecipitation was carried out as described above.
RESULTS
UL28 and UL15 proteins form a complex in HSV-1(F) infected cells.
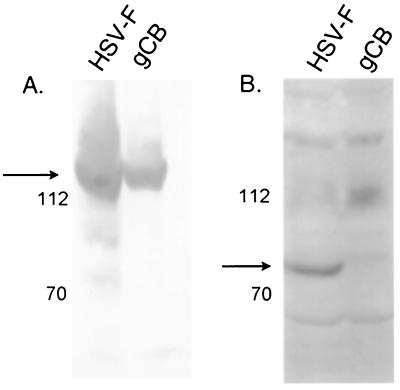
A polyclonal antiserum recognizing the HSV-1 protein UL28 was produced by immunizing rabbits with full-length UL28 fused to the gene encoding GST (see Materials and Methods). To test the specificity of the antiserum, HEp-2 cells were infected with HSV-1(F) (wild-type virus) or the UL28-null mutant HSV-1 virus gCB (41) and harvested after 20 h. Proteins were denatured by boiling in SDS, separated on a denaturing polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with antisera directed against either ICP8, a virally encoded single-stranded DNA binding protein, or the UL28 protein. Figure 1A shows the presence of ICP8 in lysates of cells infected with either virus, indicating that the cells were infected. The antiserum directed against the UL28 protein (Fig. 1B) recognized an 85,000 apparent Mr protein in the HSV-1(F)-infected cells but not in cells infected with gCB, thus indicating recognition of the UL28 protein.
FIG. 1.
Digitally scanned images of immunoblots probed with anti-ICP8 antibody (A) and anti-UL28 antibody (B). Cells were infected with either HSV-1(F) or the UL28 mutant virus gCB, and the proteins were separated on an 8% polyacrylamide gel before being transferred to a nitrocellulose membrane and probed with antiserum. Protein sizes are indicated on the left in thousands.
Immunoprecipitations were performed to identify proteins that directly or indirectly associated with the UL28 protein. Briefly, cells were infected with HSV-1(F) and infected cell proteins were radiolabeled with [35S]methionine and [35S]cysteine as detailed in Materials and Methods. Since the HSV-1 glycoproteins E (gE) and I form an Fc receptor and are normally present in immune complexes derived from HSV-infected cells (20), radiolabeled proteins were also immunoprecipitated from cells infected with the gE-null mutant virus R7202 (6). The infected cells were lysed in PBS supplemented with 0.5% Tween 20, briefly sonicated, and clarified extensively by centrifugation. The lysates were then reacted with anti-UL28 serum as detailed in Materials and Methods. Immune complexes were purified, denatured in SDS, and electrophoretically separated on denaturing polyacrylamide gels, followed by fluorography.
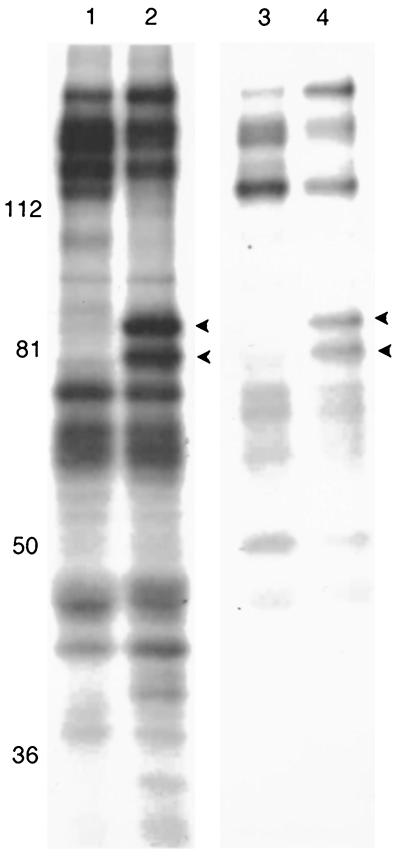
In both the wild-type- and gE− virus-infected cells, the anti-UL28-GST antibody consistently immunoprecipitated two proteins of approximately 80,000 and 85,000 apparent Mr (Fig. 2). On the basis of their respective sizes, these proteins were hypothesized to be derived from UL15 and UL28, respectively. To address this possibility, nonradiolabeled cell lysates were harvested 24 h after infection with HSV-1(F) and immunoprecipitated with the UL28-GST antiserum. The immunoprecipitated proteins were transferred to nitrocellulose and probed with antibodies directed against UL28-GST or the N terminus of UL15 fused to GST (34). As shown in Fig. 3A and B, both the UL28 and UL15 proteins were detected in immune complexes derived from reaction of the anti-UL28-GST antibody with HSV-1(F)-infected cell lysates.
FIG. 2.
Scanned digital image of fluorograph of radiolabeled proteins electrophoretically separated on an 8% polyacrylamide gel. HEp-2 cells were infected with either HSV-1(F) (lanes 1 and 2) or the gE-mutant virus strain 7202 (lanes 3 and 4) and immunoprecipitated as described in Materials and Methods by using either preimmune sera (lanes 1 and 3) or anti-UL28 antibody (lanes 2 and 4). Protein sizes are indicated on the left in thousands. Arrowheads indicate proteins of 85,000 and 80,000 apparent Mr, which correspond to the UL28 and UL15 proteins, respectively.
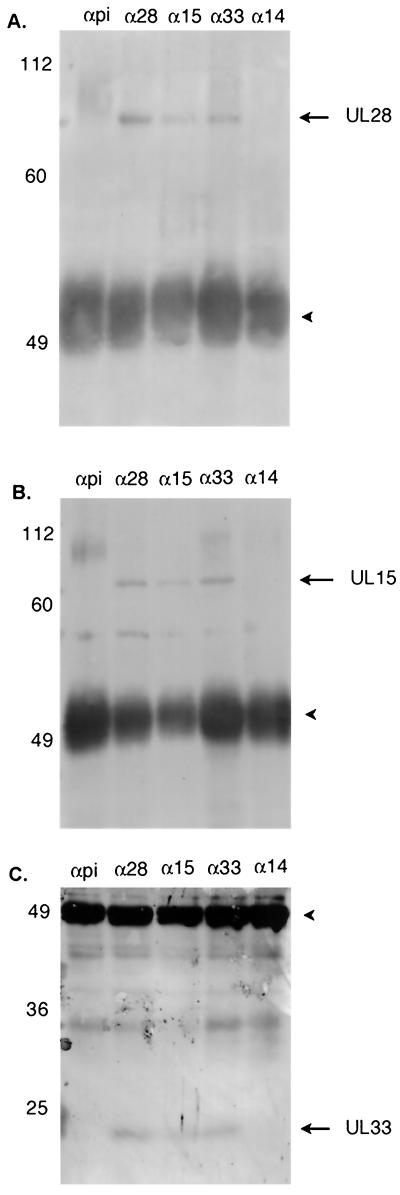
FIG. 3.
Scanned digital images of immunoblots probed with antisera directed against the UL28 (A), UL15 (B), or UL33 (C) proteins. Cell lysates were immunoprecipitated with preimmune serum (αpi) or antibody directed against UL28 (α28), UL15 (α15), UL33 (α33), or UL14 (α14), as described in the text. The position and size of molecular weight standards are indicated on the left in thousands. The heavychain of the immunoprecipitating antibody can be seen as a dark band around 49,000 (arrowhead). (A and B) 8% polyacrylamide gels; (C) 15% polyacrylamide gel.
In order to confirm the putative interaction between UL15 and UL28 proteins, a reciprocal experiment was done. Cell lysates were subjected to immunoprecipitation with the antibody directed against the N terminus of the UL15 protein, followed by immunoblotting with the anti-UL28-GST serum. As shown in Fig. 3, the antiserum directed against the UL15 protein immunoprecipitated both the UL15 and UL28 proteins (Fig. 3A and B). Control reactions containing lysates of HSV-1(F)-infected cells mixed with preimmune sera (Fig. 3) failed to immunoprecipitate either the UL15 or UL28 proteins. These data are therefore consistent with previously published work showing that the UL15 and UL28 proteins interact in HSV-1(F)-infected cells (23).
The UL28 protein forms a complex with the UL15 and UL33 proteins.
The UL28 and UL15 proteins were the most readily detected radiolabeled proteins in the above immunoprecipitations; however, it remained possible that other proteins were coimmunoprecipitated but escaped detection either because they were masked by comigrating nonspecifically immunoprecipitated radiolabeled proteins or because they did not radiolabel efficiently under the experimental conditions. Therefore, the UL28/UL15 complex was immunopurified, and the immunoprecipitated proteins were transferred to nitrocellulose and probed directly with antisera directed against the HSV proteins encoded by UL6, UL14, UL15, UL16, UL17, UL18, UL19, UL21, UL25, UL26.5, UL28, UL29, UL31, UL32, UL33, or UL49 or the host protein actin (see Table 1 for antibody sources). Of these proteins, only the proteins encoded by UL15 and UL33 coimmunoprecipitated with the UL28 protein, as shown in Fig. 3B and C. To confirm that the UL33 protein associated with the UL28/UL15 protein complex, antibody directed against the UL15 protein was used to immunoprecipitate the UL33 protein from infected cell lysates (Fig. 3C), and anti-UL33 antibody was used successfully to immunoprecipitate both the UL28 and UL15 proteins (Fig. 3A and B). All experiments included two negative controls: immunoprecipitations with preimmune sera and antibody directed against the HSV-1 protein UL14 (Fig. 3). In neither case were the UL28, UL15, or UL33 proteins detected in immunoprecipitated material.
The UL28/UL15/UL33 protein complex forms in lysates of recombinant baculovirus-infected insect cells.
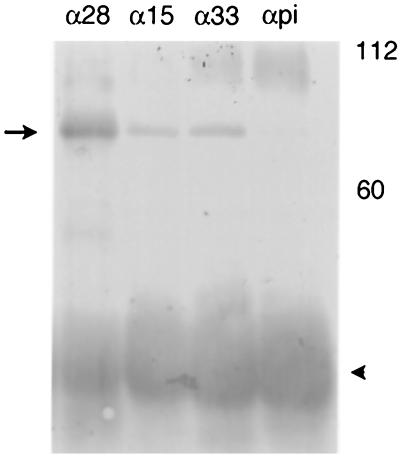
To determine if the three proteins were sufficient to interact with one another in the absence of other HSV-1-encoded proteins, Sf21 cells were infected or coinfected with recombinant baculoviruses expressing the UL28, UL15, and UL33 proteins. In the first experiment, insect cells were coinfected with recombinant baculoviruses expressing all three proteins, and lysates of the infected cells were reacted with antibody directed against either the UL28, UL15, or UL33 protein. Immunoprecipitated material was then denatured and analyzed on immunoblots probed with antibodies directed against UL28-encoded protein. It was found that all three antibodies immunoprecipitated the UL28 protein, thus showing that the three proteins can form a complex in the absence of other HSV proteins when coexpressed in insect cells (Fig. 4).
FIG. 4.
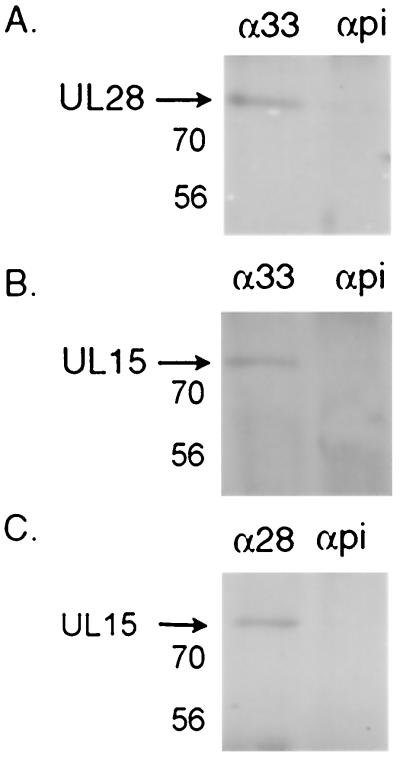
Digitally scanned image of an immunoblot showing the detection of the UL28/UL15/UL33 protein complex from baculovirus-infected Sf21 cells. Cell lysates were immunoprecipitated as described in Materials and Methods with antibody directed against the UL28 (α28), UL15 (α15), or UL33 (α33) proteins or preimmune serum (αpi). The position of the UL28 protein is indicated with an arrow. The heavy chain of the antibody can be seen as a dark band at approximately 50,000 (arrowhead). Molecular weight standards are marked at the right in thousands.
To identify which proteins within the UL28/UL15/UL33 protein complex directly interact, insect cells were coinfected with recombinant baculoviruses expressing either (i) UL28 and UL33, (ii) UL15 and UL33, or (iii) UL28 and UL15. Lysates of the coinfected insect cells were then subjected to immunoprecipitation with antisera as described in Materials and Methods. As had been reported previously (1), UL15 and UL28 were coimmunoprecipitated by the anti-UL28 antibody in the absence of UL33 (Fig. 5C). The anti-UL33 antibody was also found to coimmunoprecipitate the UL28 protein in the absence of UL15 and the UL15 protein in the absence of UL28 protein (Fig. 5A and B). The addition of anti-UL33 antibody to Sf21 cells infected with only the UL28- and UL15-expressing baculoviruses did not result in the coimmunoprecipitation of either protein in the absence of the UL33 protein (data not shown). Further controls included in these experiments were (i) immunoprecipitations using preimmune sera followed by immunoblotting with antisera directed against the UL28, UL15, and UL33 proteins, and (ii) immunoprecipitations from lysates of Sf21 cells infected with wild-type baculovirus followed by immunoblotting with the respective antisera. These samples were uniformly negative, confirming that UL28, UL15, and UL33 form a specific complex when coexpressed by infection of Sf21 cells with recombinant baculoviruses.
FIG. 5.
Digitally scanned images of Western blots showing the proteins immunoprecipitated from dual-infected Sf21 cells. Cells were infected with baculoviruses expressing UL28 and UL33 (A), UL15 and UL33 (B), or UL28 and UL15 (C) and processed as described in Materials and Methods. Proteins were immunoprecipitated with antibody directed against the UL33 protein (α33) or the UL28 protein (α28), or with preimmune serum (αpi). An equivalent amount of lysate was used in each lane. Membranes were blotted with antibody directed against the UL28 protein (A) or UL15 protein (B and C). Protein sizes are indicated on the left in thousands.
DISCUSSION
This paper describes the immunopurification of a complex of proteins containing the UL28, UL15, and UL33 gene products. These data support the conclusions of others that the UL28 and UL15 proteins interact (1, 22, 23, 45), and they represent the first report to show a specific interaction between the UL33 protein and other cleavage and packaging proteins. These results are supported by previously reported immunofluorescence assays that detailed a similar distribution of all three proteins in both the cytoplasm and replication compartments of the nucleus in infected cells (8, 23, 32, 45).
The UL33 protein is the smallest of the seven cleavage and packaging proteins, with an apparent Mr of 19,000. Since it contains only two methionine and two cysteine codons in its open reading frame, the UL33 protein does not radiolabel to high specific activities under the conditions used in these studies and is therefore difficult to detect by fluorography. To overcome this problem, immunoprecipitated material was subjected to immunoblotting with the UL33-GST antisera. Using this method, the UL33 protein was readily detected in lysates of infected cells immunoprecipitated with antiserum directed against the UL28, UL15, or UL33 proteins (Fig. 3C).
The UL33 protein is expressed as a late gene product and, using indirect immunofluorescence, it can be identified in the cytoplasm as well as nuclear replication compartments late in infection (32). Viruses lacking a functional UL33 gene are unable to carry out cleavage and packaging of the HSV genome (5), but until recently no evidence revealing a specific role for the protein in DNA cleavage and packaging reactions had been reported. However, the interaction with the UL28 and UL15 proteins described in this paper suggests that the UL33 protein may serve in some way that is relevant to terminase function. Although the precise function of UL33 protein remains unknown, some possibilities include (i) ensuring that the terminase is correctly folded or assembled, (ii) ensuring that the terminase complex is correctly transported from the cytoplasm into the nucleus, and (iii) a role as part of the terminase holoenzyme. In light of the small size of UL33, it is intriguing that HSV proteins of 21,000 and 22,000 apparent Mr have been shown to bind the a sequence of HSV-1 DNA (14). Although it is not known if these proteins are encoded by UL33, the discovery that the UL33 protein binds to the UL28 and UL15 proteins indicates that further work into its potential role as a member of the HSV terminase, possibly as a DNA-binding protein, is warranted.
The presence of a third protein in the UL28/UL15 protein complex reveals that, if this tripartite complex does comprise the terminase, it is structurally more complex than its counterpart in bacteriophage λ, where the C-terminal end of gpNu1 simply interacts with the N terminus of gpA (17). The full extent of the HSV terminase may in fact be larger still. Sixteen HSV proteins and one host protein were screened in the present work for inclusion in the putative terminase complex (Table 1). This work represents a thorough search of likely interacting candidates, including all of the cleavage-packaging proteins; however, it is not an exhaustive search of all HSV proteins. It is also possible that the antibodies directed against the UL28 protein in the immunoprecipitation reactions could disrupt interactions with additional proteins, or that the experimental technique is not sensitive enough to detect the presence of additional proteins. Finally, there remains the possibility that a host protein may be included in the complex, as suggested by previous workers (21).
A recent report has suggested that the UL33 protein of HSV-2 is transported into the nuclei of cells transiently coexpressing the UL14 and UL33 proteins, implying that the proteins interact (43). Such a conclusion was not corroborated in this study. The results of immunoprecipitation with antibody directed against the UL14 protein followed by immunoblotting with monospecific antisera failed to detect either the UL28, UL15, or UL33 proteins in immunoprecipitated material (Fig. 3). In addition, the UL14 protein was not detected when material immunoprecipitated by the UL28 antibody was probed with UL14-specific antiserum, whereas the UL33 protein was readily detected in this assay.
The anti-UL28 antibody immunoprecipitated all three proteins from lysates of Sf21 cells coinfected with baculoviruses expressing UL28, UL15, and UL33, thus mirroring the results obtained in lysates of HEp-2 cells infected with HSV-1(F). This indicated that other HSV proteins were not necessary for the interactions of the tripartite complex. When Sf21 cells were infected with subsets of the recombinant baculoviruses, it was revealed that none of the possible interactions were dependent on the presence of the third protein in the complex, implying direct interactions between all three proteins.
The relatively mild conditions used to produce the HSV-1(F)-infected cell lysates in these studies suggest that these lysates contain predominantly cytoplasmic rather than nuclear proteins. It is therefore hypothesized that the UL28/UL15/UL33 complex forms in the cytoplasm of infected cells and is subsequently imported into the nucleus, where all three proteins are present late in infection (8, 23, 32). Further studies are necessary to address this possibility. The data also suggest that the putative terminase complex forms independently from capsids, since none of the major or minor capsid proteins, including the putative portal vertex protein encoded by UL6 (29), were detected within the complex. Assembly of the terminase as a distinct process from assembly of the capsid might serve to (i) promote proper assembly of the terminase prior to import into the nucleus where DNA cleavage and packaging takes place, (ii) promote diffusion of the terminase within the nucleoplasm to increase the likelihood that genomic ends are engaged, or (iii) regulate the initiation of DNA cleavage and packaging.
Although the identification of the genes required for HSV-1 DNA cleavage and packaging was completed a number of years ago, elucidating the exact functions of these proteins has proved challenging. While the models of bacteriophage cleavage and packaging have been useful for heuristic purposes, this and other studies suggest that the analogous processes of herpesviruses are likely to be more complex.
Acknowledgments
We gratefully acknowledge the technical assistance from Jarek Okulicz-Kozaryn. We also thank J. Blaho for UL49 antiserum, W. Ruyechan for ICP8 antiserum, F. Homa for the gCB deletion virus, and G. Cohen and R. Eisenberg for antibodies against HSV-1 capsid proteins.
These studies were supported by grant R01 GM50740 from the National Institutes of Health.
REFERENCES
- 1.Abbotts, A. P., V. G. Preston, M. Hughes, A. H. Patel, and N. D. Stow. 2000. Interaction of the herpes simplex virus type 1 packaging protein UL15 with full-length and deleted forms of the UL28 protein. J. Gen. Virol. 81:2999-3009. [DOI] [PubMed] [Google Scholar]
- 2.Addison, C., F. J. Rixon, J. W. Palfreyman, M. O'Hara, and V. G. Preston. 1984. Characterisation of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology 138:246-259. [DOI] [PubMed] [Google Scholar]
- 3.Addison, C., F. J. Rixon, and V. G. Preston. 1990. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J. Gen. Virol. 71:2377-2384. [DOI] [PubMed] [Google Scholar]
- 4.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 98:3086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.al-Kobaisi, M. F., F. J. Rixon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology 180:380-388. [DOI] [PubMed] [Google Scholar]
- 6.Baines, J. D., and B. Roizman. 1993. The UL10 gene of herpes simplex virus 1 encodes a novel glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J. Virol. 67:1441-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baines, J. D., A. H. Koyama, T. Huang, and B. Roizman. 1994. The UL21 gene of herpes simplex virus 1 is dispensable for replication in cell culture. J. Virol. 68:2929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baines, J. D., A. P. Poon, J. Rovnak, and B. Roizman. 1994. The UL15 gene of herpes simplex virus encodes two proteins and is required for cleavage of viral DNA. J. Virol. 68:8118-8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baines, J. D., C. Cunningham, D. Nalwanga, and A. J. Davison. 1997. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J. Virol. 71:2666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catalano, C. E. 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell. Mol. Life Sci. 57:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen, G. H., M. Ponce de leon, H. Diggelmann, W. C. Lawrence, S. K. Vernon, and R. Eisenberg. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham, C., and A. J. Davison. 1993. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology 197:116-124. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham, C., A. J. Davison, A. R. MacLean, N. S. Taus, and J. D. Baines. 2000. Herpes simplex virus type 1 gene UL14: phenotype of a null mutant and identification of the encoded protein. J. Virol. 74:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalziel, R. G., and H. S. Marsden. 1984. Identification of two herpes simplex virus type 1-induced proteins (21K and 22K) which interact specifically with the a sequence of herpes simplex virus DNA. J. Gen. Virol. 65:1467-1475. [DOI] [PubMed] [Google Scholar]
- 15.Davison, A. J. 1992. Channel catfish virus: a new type of herpesvirus. Virology 186:9-14. [DOI] [PubMed] [Google Scholar]
- 16.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 17.Frackman, S., D. A. Siegele, and M. Feiss. 1985. The terminase of bacteriophage lambda: functional domains for cosB binding and multimer assembly. J. Mol. Biol. 183:225-238. [DOI] [PubMed] [Google Scholar]
- 18.Frangioni, J. V., and B. G. Neel. 1993. Solubilization and purification of enzymatically active glutathione-S-transferase (pGEX) fusion proteins. Anal. Biochemistry 210:179-187. [DOI] [PubMed] [Google Scholar]
- 19.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, D. C., M. C. Frame, M. W. Ligas, A. M. Cross, and N. D. Stow. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J. Virol. 62:1347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemble, G. W., and E. S. Mocarski. 1989. A host cell protein binds to a highly conserved sequence element (pac-2) within the cytomegalovirus a sequence. J. Virol. 63:4715-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koslowski, K. M., P. R. Shaver, X.-Y. Wang, D. J. Tenny, and N. Pedersen. 1997. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J. Virol. 71:9118-9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koslowski, K. M., P. R. Shaver, J. T. I. Casey, T. Wilson, G. Yamanaka, A. K. Sheaffer, D. J. Tenny, and N. E. Pedersen. 1999. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J. Virol. 73:1704-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotsakis, A., L. E. Pomeranz, A. Blouin, and J. A. Blaho. 2001. Microtubule reorganization during herpes simplex virus type 1 infection facilitates the nuclear localization of VP22, a major virion tegument protein. J. Virol. 75:8697-8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamberti, C., and S. K. Weller. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226:403-407. [DOI] [PubMed] [Google Scholar]
- 26.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 28.Nalwanga, D., S. Rempel, B. Roizman, and J. D. Baines. 1996. The UL16 gene product of herpes simplex virus is a virion protein that colocalizes with intranuclear capsid proteins. Virology 226:236-242. [DOI] [PubMed] [Google Scholar]
- 29.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel, A. H., F. J. Rixon, C. Cunningham, and A. J. Davison. 1996. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology 217:111-123. [DOI] [PubMed] [Google Scholar]
- 31.Poon, A. P. W., and B. Roizman. 1993. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J. Virol. 67:4497-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynolds, A. E., Y. Fan, and J. D. Baines. 2000. Characterization of the UL33 gene product of herpes simplex virus 1. Virology 266:310-318. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salmon, B., and J. D. Baines. 1998. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of UL15-encoded proteins with B capsids requires at least the UL6, UL17 and UL28 genes. J. Virol. 72:3045-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shelton, L. S. G., A. G. Allbright, W. T. Ruyechan, and F. J. Jenkins. 1994. Retention of the herpes simplex virus type 1 (HSV-1) UL37 protein on single stranded DNA columns requires the HSV-1 ICP8 protein. J. Virol. 68:521-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman, G., and S. L. Bachenheimer. 1987. DNA processing in temperature-sensitive morphogenetic mutants of HSV-1. Virology 158:427-430. [DOI] [PubMed] [Google Scholar]
- 38.Stow, N. D., and P. D. Hodge. 2001. Effects of mutations within the herpes simplex virus type 1 DNA encapsidation signal on packaging efficiency. J. Virol. 75:8977-8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taus, N. S., and J. D. Baines. 1998. Herpes simplex virus DNA cleavage and packaging: the UL28 gene product is a minor component of B capsids. Virology 252:443-449. [DOI] [PubMed] [Google Scholar]
- 40.Taus, N. S., B. Salmon, and J. D. Baines. 1998. The herpes simplex virus 1 UL17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology 252:115-125. [DOI] [PubMed] [Google Scholar]
- 41.Tengelsen, L. A., N. E. Pedersen, P. R. Shaver, M. W. Wathen, and F. L. Homa. 1993. Herpes simplex virus type 1 DNA cleavage and capsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J. Virol. 67:3470-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weller, S. K., E. P. Carmichael, D. P. Aschman, D. J. Goldstein, and P. A. Schaffer. 1987. Genetic and phenotypic characterization of mutants in four essential genes that map to the left half of HSV-1 UL DNA. Virology 161:198-210. [DOI] [PubMed] [Google Scholar]
- 43.Yamauchi, Y., K. Wada, F. Goshima, H. Takakuwa, T. Daikoku, M. Yamada, and Y. Nishiyama. 2001. The UL14 protein of herpes simplex virus type 2 translocates the minor capsid protein VP26 and the DNA cleavage and packaging UL33 protein into the nucleus of coexpressing cells. J. Gen. Virol. 82:321-330. [DOI] [PubMed] [Google Scholar]
- 44.Yu, D., A. K. Shaeffer, D. J. Tenny, and S. K. Weller. 1997. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J. Virol. 71:2656-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu, D., and S. K. Weller. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, D., and S. K. Weller. 1998. Genetic analysis of the UL15 gene locus for the putative terminase of herpes simplex virus type 1. Virology 243:32-44. [DOI] [PubMed] [Google Scholar]