Abstract
Packaging into nucleosomes results in a global transcriptional repression as a consequence of exclusion of sequence-specific factors. This inhibition can be relieved by using inhibitors of histone deacetylases, acetylation being a major characteristic of transcriptionally active chromatin. Paradoxically, the expression of only ∼2% of the total cellular genes is modulated by histone hyperacetylation. To unravel the potential role of this transcriptional control on BLV expression, we tested the effect of two highly specific inhibitors of deacetylases, trichostatin A (TSA) and trapoxin (TPX). Our results demonstrate that treatment with TSA efficiently enhanced long terminal repeat-directed gene expression of integrated reporter constructs in heterologous D17 stable cell lines. To further examine the biological relevance of these observations made in vitro, we analyzed ex vivo-isolated peripheral blood mononuclear cells (PBMCs) from bovine leukemia virus (BLV)-infected sheep. TSA deacetylase inhibitor induced a drastic increase in viral expression at levels comparable to those induced by treatment with phorbol-12-myristate 13-acetate and ionomycin, the most efficient activators of BLV expression known to date. TSA acted directly on BLV-infected B lymphocytes to increase viral expression and does not seem to require T-cell cooperation. Inhibition of deacetylation after treatment with TSA or TPX also significantly increased viral expression in PBMCs from cattle, the natural host for BLV. Together, our results show that BLV gene expression is, like that of a very small fraction of cellular genes, also regulated by deacetylation.
Bovine leukemia virus (BLV) pathogenesis is characterized by the lack of viral expression in vivo in a large proportion of cells containing an integrated provirus (34, 43). The mechanisms preventing viral expression in these B lymphocytes are poorly understood. Previous reports have demonstrated that BLV expression is regulated at the transcriptional level by the viral protein Tax (18, 66). Tax is a powerful transactivator acting through a triplicate motif of a 21-bp enhancer element, the TxRE (for Tax-responsive element), located in the U3 region of the long terminal repeat (LTR) (19, 20). This viral protein does not bind directly to DNA but rather interacts with cellular proteins of the ATF/CREB family, increasing their affinity for the TxRE motif (2, 37, 67). In the absence of Tax, LTR basal transcription appears to be mediated, at least in part, by ATF1, ATF2, and CREB proteins, as revealed by transient-transfection experiments. These cellular proteins interact with imperfectly conserved CRE elements harboring at least one substitution (underlined) compared with the consensus site (AGACGTCA, TGACGGCA, TGACCTCA), these nucleotide variations being essential for efficient viral replication in vivo (48). The TxREs also contain E-box sequences (CANNTG) overlapping the CREs potentially involved in transcriptional silencing since their mutation significantly increases basal expression (48). In addition to the TxRE motifs, the LTR promoter contains a series of elements potentially involved in the regulation of BLV transcription: a nuclear factor κB (NF-κB) binding site (8), a glucocorticoid responsive element (GRE) (50, 71), and an interferon regulatory factor (IRF) binding site in the U5 region (35).
In ex vivo-isolated peripheral blood mononuclear cells (PBMCs), several activators have been reported to up-regulate BLV expression: fetal calf serum, lipopolysaccharides, anti-immunoglobulin M (anti-IgM) antibodies (43), phytohemagglutinin (5), and phorbol esters (31, 33). The molecular mechanisms by which these agents activate BLV transcription remain misunderstood. Since H7 and calphostin C inhibit viral expression, protein kinase C (PKC) is thought to exert a key role in this regulatory process (31, 33). The calmodulin/calcineurin pathway also seems to be involved in regulation of BLV expression in freshly isolated PBMCs (33). On the other hand, doubts remain concerning the involvement of cyclic AMP (cAMP) and PKA. In vitro, cotransfection of PKA and CREB2 vectors activates LTR-directed expression in D17 osteosarcoma cells, but addition of cyclic AMP ex vivo in cell cultures inhibits viral expression in infected lymphocytes of cattle (33, 67).
Acetylation of histones has been known to closely correlate with transcriptional activation (24, 60, 69, 70). The level of histone acetylation results from an intrinsic balance of histone acetyltransferase (HAT) and histone deacetylase (HDAC) activities (15, 38, 68). By neutralizing the positive charge of lysine residues, the HAT activity reduces the affinity of histones for DNA and consequently leads to transcriptional activation. Several cellular proteins—in particular, CBP/p300, PCAF, Gcn5p, ACTR, SRC-1, and TAFII250—have been shown to possess intrinsic HAT activity (6, 9, 10, 23, 39, 40, 49, 58, 64). Conversely, histone deacetylation causes the formation of tightly packaged nucleosomes which are inaccessible to transcription factors (60, 68). HDACs are known as repressors of transcription and interact with sequence-specific DNA-binding transcriptional repressors like the Mad/Max complex (28, 44, 52). The best characterized cellular factors that establish the link between the DNA-bound repressors and HDACs are Sin3 and N-CoR/SMRT, which are referred to as corepressors (52). However, histones are not necessarily the most important natural targets of HAT and HDAC activities. Indeed, several transcription factors can be acetylated, and in some instances, these modifications have been shown to affect physiological activities of these proteins (7, 25, 29, 38).
In order to further characterize the mechanisms implicated in activation of BLV expression, in this report we aim to unravel the role of deacetylation. Based on the use of two highly specific deacetylase inhibitors, trichostatin A (TSA) and trapoxin (TPX), we demonstrate that inhibition of HDACs induces BLV expression in vitro and in vivo.
MATERIALS AND METHODS
Cell lines and luciferase assays.
D17 canine osteosarcoma cells were grown in minimal essential medium (Life Technologies) supplemented with 10% heat-inactivated fetal calf serum (FCS). D17-LTR WT, D17-LTR Ebox3x, D17-LTR IRF, D17-LTR GRE, D17-LTR NF1, D17-LTR NF2, D17-LTR CRE3x, and D17-LTR Δ21-pb are cell lines derived from D17 fibroblasts after transfection with plasmids pLTR-WT, pLTR-Ebox3x, pLTR-IRF, pLTR-NF1, pLTR-NF2, pLTR-CRE3x, and pLTR-Δ21-pb. These cell lines were also cotransfected with plasmid pRc/CMV, which confers resistance to G418 (Geneticin). Twenty-four hours prior to transfection, the D17 cells were cultured at a density of 3 × 105 cells per well (six-well dish; Nunc). Five micrograms of different pLTR-Luc reporters and 500 ng of pRc/CMV were mixed with CaCl2 and phosphate buffer (as described by the manufacturer), incubated at room temperature for 30 min to allow DNA precipitation, and added to the cells. After incubation for 4 h in the presence of the transfection complexes, the cells were washed with serum-free medium and grown in the presence of 10% FCS. After 48 h of culture at 37°C, cells were washed again and maintained in complete medium supplemented with G418 (800 μg/ml).
The stable cell lines were cultured in complete medium supplemented with TSA (500 nM; Sigma Aldrich) at 37°C for 48 h and washed twice with phosphate-buffered saline (PBS), and luciferase enzyme activity was measured by using the dual-luciferase assay system (Promega) according to the manufacturer's recommendations.
Animals.
The three adult cows (animals 01, 51, and 57) used in this study were kept at the National Veterinary Research Institute (Pulawy, Poland). These animals were seropositive for BLV and in persistent lymphocytosis, with lymphocyte counts of 18,400, 28,600, and 16,000 cells/μl, respectively. Uninfected cow 22 and the sheep (BLV-infected sheep 8, 104, 293, 2658, 2659, 2660, 2661, 2662, 2663, 2664, 2665, 2666, 2668, 1084, 2670, and 2671 and uninfected sheep 115) were housed at the Veterinary and Agrochemical Research Center (Machelen, Belgium).
Isolation of PBMCs and cell culture conditions.
Sheep PBMCs were isolated by Percoll gradient centrifugation. Briefly, venous blood was collected by jugular venipuncture and mixed with EDTA. PBMCs were then separated by Percoll density gradient centrifugation (Amersham Pharmacia Biotech) and washed three times with PBS-0.075% EDTA and once with PBS alone. Bovine PBMCs were purified by centrifugation over Histopaque 1077 (Sigma Aldrich) and washed three times with PBS. Cells were resuspended at a concentration of 2 × 106 cells/ml in RPMI 1640 medium supplemented with 10% FCS, 2 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml and cultured in the absence or in the presence of TSA (500 nM; Sigma Aldrich), TPX (100 nM), or phorbol-12-myristate 13-acetate (PMA) (200 nM; Sigma Aldrich) and ionomycin (565 nM; Sigma Aldrich) for 48 h at 37°C. Viable cells were counted by the trypan blue exclusion technique.
B-cell isolation by negative selection.
PBMCs were washed twice in sterile PBS supplemented with 10% FCS and 2 mM EDTA and incubated for 30 min at 4°C with four 1,000-fold-diluted anti-T-cell monoclonal antibodies (anti-CD2, anti-CD4, anti-CD6, and anti-CD8). Cells were washed twice and resuspended at 107 cells/80 μl of sterile buffer (PBS-10% FCS-2 mM EDTA). Twenty microliters of magnetic cell sorting anti-immunoglobulin microbeads were then added, and the mixture was incubated for 30 min at 4°C. After two washes, magnetically labeled cells were resuspended in 500 μl of buffer and separated on a magnetic separation column (Santa Cruz Biotechnology). Selected B cells (levels of purity, 93 to 95%) were then cultured in the absence or in the presence of TSA or PMA and ionomycin for 48 h at 37°C.
Titration of the major capsid protein by ELISA.
Forty eight hours after culture, the cell supernatants were recovered by centrifugation (1,500 × g, 10 min) and analyzed for p24 protein expression using an enzyme-linked immunosorbent assay (ELISA) procedure. Briefly, 96-well microtiter plates (Maxisorb immunoplate; Nunc) were coated with 4′G9 monoclonal antibody (300 ng in PBS per well) for 4 h at room temperature. The plates were washed three times with PBS-Tween 80 (0.2%), and serial threefold dilutions of cell culture supernatants were added to the wells in the presence of bovine serum albumin (0.67%) and Tween 80 (1.33%). After overnight incubation at 4°C and three washes, the presence of the p24 antigen was revealed by using two monoclonal antibodies (2′C1 and 4′F5) conjugated with horseradish peroxidase.
In situ detection of B cell apoptosis and flow cytometry.
For detection of apoptosis in the B-lymphocyte subset, cultured cells were first harvested, washed twice with PBS-10% FCS, and labeled with the anti-IgM 1H4 antibody. Cells were maintained for 30 min at 4°C, washed twice, and incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin (Dako). After two washes in PBS-10% FCS, the cells were fixed with 70% ethanol at −20°C for at least 30 min, washed twice in PBS-10% FCS, incubated with RNase A (50 μg/ml) for 30 min at 37°C, and stained with propidium iodide (PI) (20 μg/ml; Sigma Aldrich), allowing detection of apoptosis in infected cells. A Becton Dickinson FACScan flow cytometer was used to perform the flow cytometry analyses. Doublets were excluded from the analyses by the scatter gating method. Ten thousand events were collected per sample, and data were analyzed with the CELLQUEST software (Becton Dickinson Immunocytometry Systems, San Jose, Calif.).
RESULTS
BLV promoter activity is enhanced in response to TSA in D17 cell lines.
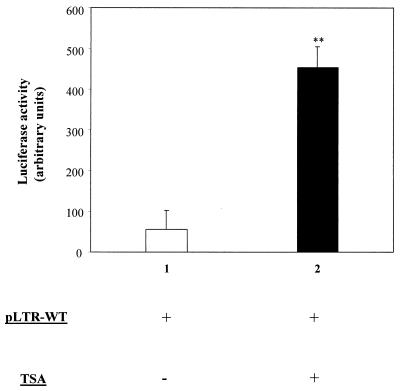
Several reports are available in the literature concerning the response of viral promoters to inhibition of deacetylases (26, 30, 47, 53, 55, 59, 62, 63). However, nothing is known concerning the role of histone acetylation in transcriptional regulation of viruses belonging to the genus Deltaretrovirus, the genus comprising BLV and the related primate T-cell lymphotropic viruses. Therefore, we examined the effect of TSA, a highly specific inhibitor of deacetylases (72), on an integrated BLV promoter. The D17-LTRWT cell line, which contains integrated copies of the wild-type BLV LTR, was established by cotransfection of plasmids pLTR-WT and pRc/CMV into D17 canine osteosarcoma cells. These fibroblasts offer the advantage of yielding low levels of interference with ovine B-lymphocyte factors (1, 67). After transfection and G418 selection, the D17-LTRWT cells were cultured in the presence or in the absence of TSA. Forty-eight hours after addition of the chemical, a marked increase in the basal BLV LTR activity was observed (Fig. 1). Indeed, the luciferase activity was increased by up to 11-fold in the presence of TSA, the induction being highly statistically significant in three independent experiments. This result shows that inhibition of deacetylation modulates the basal transcriptional activity of the BLV promoter.
FIG. 1.
Response of the BLV promoter to TSA in cell culture. D17-LTR WT cells harboring a stably integrated LTR-WT plasmid were cultured in the absence (lane 1) or in the presence (lane 2) of TSA, and the luciferase activities were measured after 48 h. The average values from three independent experiments are presented. Error bars, standard deviations; ∗∗, highly statistically significant (P ≤ 0.01).
In order to identify the sites within the LTR conferring the response to TSA, we next analyzed a series of mutant LTR reporters. Indeed, among these LTRs, some harbor mutations in E-box, NF-κB, and GRE motifs potentially linked to acetylation or deacetylation mechanisms (3, 24, 61). It appeared that in stable cell lines, all LTR mutants responded to TSA as revealed by a strong increase in luciferase activities (Table 1) (between 4.0- and 14.5-fold induction). However, the differences among the TSA inductions observed between the different mutants were not statistically significant. We conclude that none of the individual mutations in the transcription factor binding sites tested was able to eliminate the TSA inducibility of the BLV LTR.
TABLE 1.
Induction of luciferase in the presence of TSAa
| Mutant | Fold induction (mean ± SD) |
|---|---|
| pLTR-WT | 11.3 ± 6.1 |
| pLTR-Ebox3x | 4.5 ± 2.9 |
| pLTR-IRF | 7.2 ± 2.3 |
| pLTR-GRE | 4.9 ± 2.8 |
| pLTR-NF1 | 4.0 ± 0.7 |
| pLTR-NF2 | 9.4 ± 3.7 |
| pLTR-CRE3x | 14.5 ± 8.8 |
| pLTR-Δ21-bp | 4.2 ± 2.7 |
Stable cell lines harboring the mutant reporters were established. Cells were cultured for 48 h in the presence or absence of TSA, and luciferase activities were determined at 20 h of induction. The levels of induction (i.e., ratio of luciferase units obtained in the presence of TSA versus that obtained in untreated controls) are indicated. The average values from three independent experiments are presented.
TSA induces BLV expression in sheep PBMCs.
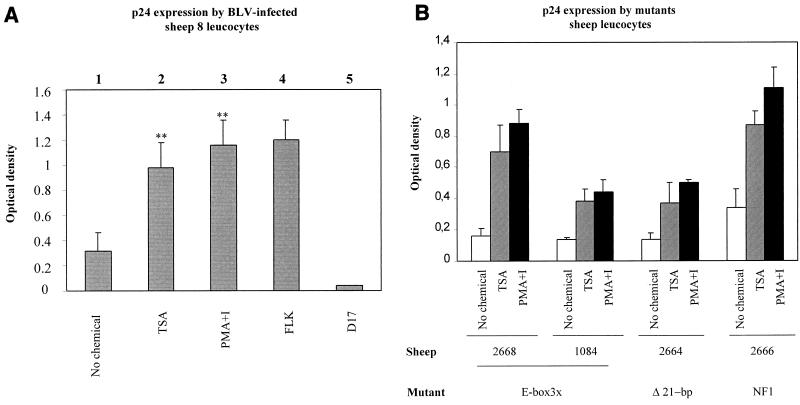
To assess whether inhibition of deacetylases also modulates viral expression ex vivo, PBMCs were isolated from a BLV-infected sheep (sheep 8) and cultured for 48 h in the presence or in the absence of TSA. The major capsid protein p24 was titrated in the cell culture supernatants by using an ELISA. It appeared that viral expression was enhanced in response to TSA treatment (Fig. 2, compare lanes 1 and 2). Remarkably, this increase was almost similar to that observed with PMA-ionomycin (Fig. 2, lane 3), a combination which is among the best inducers of BLV expression (33). As positive and negative controls for the ELISA specificity, supernatants from FLK cells (fetal lamb kidney cells expressing BLV) and D17 fibroblasts were used, respectively.
FIG. 2.
Modulation of p24 expression by TSA in cells isolated from infected sheep. (A) PBMCs were isolated from the circulating blood of sheep 8, purified by Percoll gradient, and cultivated in the absence (lane 1) or in the presence (lane 2) of TSA or PMA-ionomycin (lane 3) for 48 h. The major capsid protein p24 was titrated in the cell culture supernatants by ELISA. The supernatants from BLV-infected FLK cells (lane 4) and D17 fibroblasts (lane 5) were used as positive and negative controls, respectively. (B) p24 expression in PBMCs from sheep. The data, presented as optical densities (means ± standard deviations [error bars]), are from three independent experiments. The statistical evaluation was performed according to Student's t test. ∗∗, highly statistically significant (P ≤ 0.01).
To extend this observation, we next used PBMCs from sheep infected with mutant proviruses (48) (Table 2). As a control, PBMCs from an uninfected sheep (sheep 115) defined the background levels (optical densities of 0.07, 0.09, and 0.06 [Table 2]). Cells from two BLV-infected sheep (sheep 104 and 293) expressed more p24 protein in response to TSA (compare optical densities of 0.81 and 0.31 with optical densities of 0.38 and 0.12), further extending our observations of PBMCs from sheep 8 (optical densities of 0.32 and 0.98). Of note, a significant proportion (50%) of sheep infected with wild-type viruses did not express detectable levels of viral proteins during ex vivo culture (data not shown). Among the other sheep infected with mutant LTR viruses, only four of them (sheep 2668, 1084, 2664, and 2666) expressed enough p24 to be detected by ELISA. In ex vivo cell cultures derived from these animals, p24 levels also increased in response to TSA (graphically represented in Fig. 2B). This enhancement was similar to that induced by PMA-ionomycin. We conclude that three types of mutant (Ebox3x, Δ2-pb, and NF1) are still responsive to TSA.
TABLE 2.
Relative p24 expression in PBMCs from sheep infected with various virusesa
| Virus | Sheep no. | Relative p24 expression (mean ODb ± SD) after treatment with:
|
||
|---|---|---|---|---|
| No chemical | TSA | PMA-ionomycin | ||
| None | 115 | 0.07 ± 0.005 | 0.09 ± 0.04 | 0.06 ± 0.002 |
| WT | 8 | 0.32 ± 0.14 | 0.98 ± 0.21 | 1.16 ± 0.20 |
| 104 | 0.38 ± 0.11 | 0.81 ± 0.11 | 1.18 ± 0.06 | |
| 293 | 0.12 ± 0.03 | 0.31 ± 0.09 | 0.60 ± 0.38 | |
| Ebox3x | 2668 | 0.16 ± 0.05 | 0.70 ± 0.17 | 0.88 ± 0.09 |
| 1084 | 0.14 ± 0.01 | 0.38 ± 0.08 | 0.44 ± 0.08 | |
| NF2 | 2658 | 0.06 ± 0.01 | 0.06 ± 0.01 | 0.07 ± 0.02 |
| 2659 | 0.14 ± 0.01 | 0.11 ± 0.02 | 0.21 ± 0.09 | |
| GRE | 2660 | 0.09 ± 0.01 | 0.07 ± 0.01 | 0.13 ± 0.07 |
| 2661 | 0.09 ± 0.03 | 0.07 ± 0.003 | 0.12 ± 0.06 | |
| CRE3x | 2662 | 0.05 ± 0.01 | 0.05 ± 0.01 | 0.07 ± 0.02 |
| 2663 | 0.07 ± 0.02 | 0.07 ± 0.01 | 0.08 ± 0.02 | |
| Δ21-pb | 2664 | 0.18 ± 0.03 | 0.37 ± 0.13 | 0.50 ± 0.02 |
| 2665 | 0.06 ± 0.02 | 0.07 ± 0.02 | 0.09 ± 0.02 | |
| NF1 | 2666 | 0.34 ± 0.12 | 0.87 ± 0.09 | 1.11 ± 0.13 |
| IRF | 2670 | 0.37 ± 0.08 | 0.53 ± 0.31 | 1.14 ± 0.25 |
| 2671 | 0.50 ± 0.15 | 0.57 ± 0.35 | 1.14 ± 0.10 | |
See reference 48 for a detailed description of the mutants used. Recombinants Ebox3x and CRE3x contain a triplicate mutation of the E-box or the CRE motifs, respectively. In mutants NF1 and NF2, NF-κB sites have been truncated, whereas in Δ21-pb, a TxRE has been deleted. Finally, GRE and IRF are mutated in the glucocorticoid- and interferon-responsive sites, respectively. An uninfected sheep, sheep 115, was used as a control.
OD, optical density.
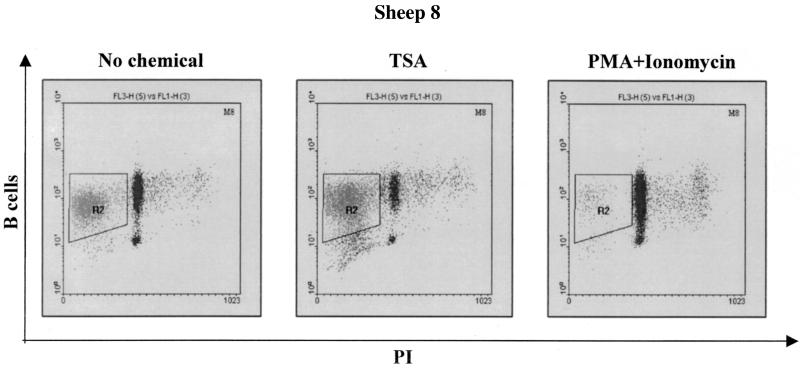
Since a series of studies have previously reported that deacetylase inhibitors induce apoptosis in numerous cell lines and cell types (14, 45, 46, 54), we examined the level of apoptosis in the ovine PBMCs after treatment with TSA. In other words, we aimed to correlate the induction of viral expression in response to TSA with the levels of apoptosis occurring in cells. Therefore, the PBMCs were first labeled with an anti-IgM antibody (1H4) and with an FITC-conjugated secondary antibody. The cells were then fixed in 70% ethanol, stained with PI, and analyzed by flow cytometry. The fluorescence-activated cell sorter analyses revealed that TSA increased the number of cells undergoing apoptosis, as illustrated in Fig. 3 for sheep 8. In contrast, PMA and ionomycin efficiently inhibited cell death under the same culture conditions (Fig. 3, rightmost panel).
FIG. 3.
Detection of apoptosis in B lymphocytes from infected sheep. PBMCs were cultured for 48 h in the absence or in the presence of TSA or PMA-ionomycin and labeled with monoclonal antibody 1H4 directed against surface IgMs and a FITC-coupled conjugate. The cells were then fixed, stained with PI, and analyzed by dual-immunofluorescence flow cytometry. Doublets were excluded using the scatter gating method (R1 [not shown]). Results from a representative experiment corresponding to cultures established from sheep 8 are presented as dot plots. The quadrant (R2) indicates the apoptotic B cells within the PBMC cultures.
The proportions of apoptotic B lymphocytes within the PBMC population of three infected sheep are summarized in Table 3. It appeared that PBMCs cultured in the presence of TSA displayed an increase in apoptosis (53.8, 38.9, and 66.8% versus 41.1, 33.0, and 53.3%). On the other hand, treatment with PMA and ionomycin resulted in a significant decrease in the percentage of apoptotic cells (4.26, 4.95, and 21.7%). Despite the slight increase in cell death consequent to TSA treatment, viral expression was enhanced, indicating that the observed effect was not the consequence of a protection of the B lymphocytes against apoptosis.
TABLE 3.
Apoptotic B cells within total lymphocyte population from various sheepa
| Sheep no. | % Apoptotic B cells (mean ± SD) after treatment with:
|
||
|---|---|---|---|
| No chemical | TSA | PMA-ionomycin | |
| 8 | 41.1 ± 11.0 | 53.8 ± 6.9 | 4.26 ± 0.6 |
| 104 | 33.0 ± 3.6 | 38.9 ± 3.3 | 4.95 ± 0.9 |
| 293 | 53.3 ± 5.8 | 66.8 ± 7.6 | 21.7 ± 5.6 |
The mean values and corresponding standard deviations are derived from three independent experiments.
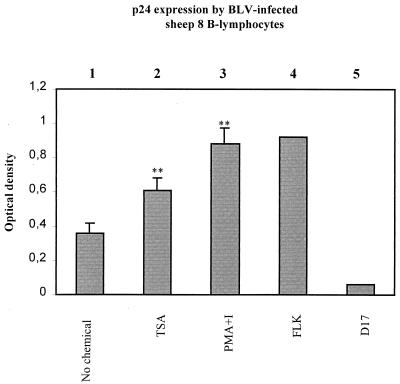
TSA enhances BLV expression in sheep B lymphocytes.
In order to test whether TSA acts directly on B lymphocytes or via T cells to increase BLV expression, T lymphocytes were depleted from PBMCs from sheep 8, and selected B cells were cultured for 48 h in the presence or in the absence of TSA or PMA-ionomycin. As shown in Fig. 4, viral expression was increased in response to TSA even after depletion of T lymphocytes (compare lanes 1 and 2). As reported previously for cattle lymphocytes (33), PMA-ionomycin also directly activated BLV expression in ovine B cells (lane 3). As positive and negative controls for p24 expression, FLK (lane 4) and D17 (lane 5) cell supernatants were used, respectively.
FIG. 4.
Activation of BLV expression in B lymphocytes from infected sheep by TSA. T cells were depleted from PBMCs isolated from the sheep 8, and the selected B lymphocytes were cultured for 48 h in the absence (lane 1) or in the presence (lane 2) of TSA or PMA-ionomycin (lane 3). The BLV p24 antigen was titrated in the cell culture supernatants by ELISA, and the supernatants from BLV-infected FLK cells (lane 4) and D17 fibroblasts (lane 5) were used as positive and negative controls, respectively. The data presented correspond to the mean values from three independent experiments (error bars, standard deviations). The statistical evaluation was performed according to Student's t test. ∗∗, highly statistically significant (P ≤ 0.01).
It thus appears that TSA activation of viral expression efficiently occurs in purified B-lymphocyte populations and does not seem to require cooperation with T cells in the cultures.
BLV expression is increased in response to TSA and TPX in cattle PBMCs.
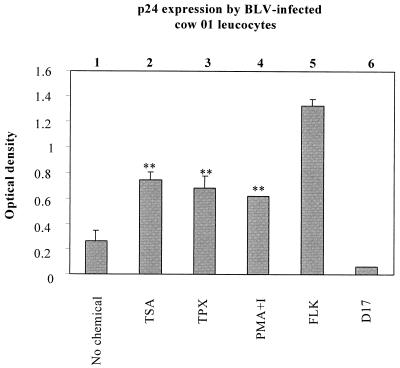
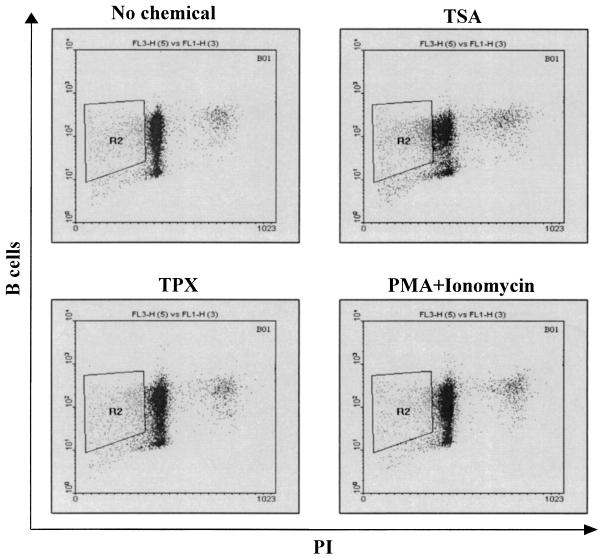
The sheep is considered to be a convenient experimental model for BLV since this species develops leukemia at higher frequencies and after shorter latency periods than cattle. However, sheep is not the natural host for BLV, and some aspects of virus-induced disease differ in these two ruminants (17, 65). To determine whether TSA has a stimulating effect on BLV expression in cattle lymphocytes, PBMCs from three BLV-infected animals (cows 01, 51, and 57) and from an uninfected cow (cow 22) were isolated by Histopaque gradient centrifugation. The cells were then cultured for 48 h in the presence or in the absence of TSA, TPX (another specific inhibitor of deacetylases), and PMA-ionomycin. The expression of BLV, as estimated by the p24 levels, was enhanced when TSA or TPX was added to culture medium (illustrated for cow 01 in Fig. 5 and summarized in Table 4). Indeed, both chemicals increased viral expression by at least twofold in PBMCs from the three BLV-infected cows. However, neither TSA nor TPX had any effect on PBMCs from the uninfected cow (cow 22) used as a negative control. The rate of induction by TSA was even greater than that by PMA-ionomycin. In parallel, the apoptotic rates within the B-lymphocyte populations were calculated. As shown in Fig. 6 and Table 5, TSA and TPX treatment resulted in an increase in the levels of apoptosis, whereas an incubation with PMA-ionomycin induced a slight decrease in the percentage of apoptotic B lymphocytes.
FIG. 5.
Modulation of p24 expression in infected cows by TSA and TPX. PBMCs were isolated from the circulating blood of BLV-infected cattle (cow 01), purified by Histopaque gradient, and cultivated for 48 h in the absence (lane 1) or in the presence of TSA (lane 2), TPX (lane 3), or PMA-ionomycin (lane 4). The major capsid protein p24 in the cell culture supernatants was titrated by ELISA. The supernatants from BLV-infected FLK cells (lane 5) and D17 fibroblasts (lane 6) were used as positive and negative controls, respectively. The data, presented as mean optical densities (means ± standard deviations [error bars]), are from three independent experiments. The statistical evaluation was performed according to Student's t test. ∗∗, highly statistically significant (P ≤ 0.01).
TABLE 4.
p24 expression in three BLV-infected cows and one control animala
| Virus | Cow no. | Relative p24 expression (mean OD ± SD) after treatment with:
|
|||
|---|---|---|---|---|---|
| No chemical | TSA | TPX | PMA-ionomycin | ||
| WT | 01 | 0.26 ± 0.08 | 0.74 ± 0.07 | 0.68 ± 0.08 | 0.61 ± 0.005 |
| 51 | 0.33 ± 0.19 | 0.75 ± 0.08 | 0.67 ± 0.09 | 0.61 ± 0.07 | |
| 57 | 0.20 ± 0.08 | 0.74 ± 0.12 | 0.53 ± 0.14 | 0.64 ± 0.06 | |
| None | 22 | 0.07 ± 0.01 | 0.06 ± 0.02 | ND | 0.06 ± 0.01 |
Results are from three independent experiments. Abbreviations: OD, optical density; WT, wild type; ND, not done.
FIG. 6.
Detection of apoptosis in B lymphocytes from BLV-infected cows. PBMCs isolated from three cows (cows 01, 51, and 57) were cultured for 48 h in the presence or in the absence of TSA, TPX, or PMA-ionomycin and labeled with the monoclonal antibody 1H4 IgM and a FITC-conjugated secondary antibody. The cells were then fixed, stained with PI, and analyzed by dual-immunofluorescence flow cytometry. Doublets were excluded using the scatter gating method (R1 [not shown]). Shown are representative experiments corresponding to PBMC cultures isolated from cow 01, presented as dot plots. The quadrants (R2) indicate the apoptotic B cells within the PBMC cultures.
TABLE 5.
Apoptotic B cells in cultures from three cowsa
| Cow no. | % Apoptotic B cells (mean ± SD) after treatment with:
|
|||
|---|---|---|---|---|
| No chemical | TSA | TPX | PMA-ionomycin | |
| 01 | 11.7 ± 1.60 | 19.8 ± 4.41 | 12.9 ± 1.43 | 8.96 ± 1.10 |
| 51 | 23.0 ± 12.6 | 35.7 ± 10.3 | 28.2 ± 8.25 | 22.6 ± 11.4 |
| 57 | 11.3 ± 1.50 | 15.6 ± 4.02 | 13.5 ± 3.90 | 14.2 ± 3.32 |
The mean values and corresponding standard deviations are derived from three independent experiments.
We conclude that treatment with TSA or TPX induces BLV expression in cattle PBMCs and that this increase in viral expression is not the consequence of a reduction in apoptosis.
DISCUSSION
In human lymphoid cells lines, it has been predicted that the expression of only 2% of cellular genes is modulated in response to histone acetylation (62). The expression of some genes, like c-myc, is down-regulated by TSA or TPX, whereas GAPDH (glyceraldehyde-3-phosphate dehydrogenase) is not affected by any of these chemicals. In contrast, human immunodeficiency virus (HIV) expression is induced following histone hyperacetylation (55, 59, 62, 63). Induction of HIV type 1 expression is associated in part with the disruption of a specific nucleosome positioned at the transcriptional start in the viral promoter in response to TSA or TPX (62, 63). Transcriptional silencing of other viruses like Epstein-Barr virus, human cytomegalovirus, and Kaposi's sarcoma-associated herpesvirus also appears to involve deacetylation mechanisms (11, 26, 30, 47).
In the present report, we examined the role of deacetylase inhibitors (TSA and TPX) in BLV expression in vitro and in ex vivo-isolated sheep and cattle PBMCs. We show that inhibition of HDACs induced BLV expression as observed for a minority (2%) of cellular genes. In pools of D17 cell lines, treatment with TSA resulted in an increase in BLV promoter activity when the LTR reporter was stably integrated in the chromosomal DNA. Since these in vitro observations might be devoid of biological relevance, we next extended our assays in vivo using two different species susceptible to BLV infection and pathogenesis. We have demonstrated that, in cattle and sheep, inhibition of deacetylation caused a marked activation of BLV expression. This induction of expression was not due to a decrease in the levels of apoptosis of the B lymphocytes. Enhancement of viral expression induced by TSA was correlated with a slight increase in the percentage of apoptotic B lymphocytes, as is frequently reported for other cell types (14, 45, 46, 54). Incubation with PMA-ionomycin, however, almost completely impeded cell death in sheep PBMCs cultures. In this case, increase in viral expression could simply be the result of enhanced cell viability. It should be mentioned that TSA also activates viral expression directly in BLV-infected lymphocytes without apparent cooperation of T cells in the cultures (Fig. 4).
Histone acetylation results from an equilibrium between opposing activities of HAT and HDAC. Previous reports have shown that increase in HIV expression in response to TSA correlates with enhancement in the level of histone acetylation (55, 59, 63). Hence, the induction of BLV expression by TSA reported here could be due to an increase in histone acetylation. In mammals, several coactivators harbor an intrinsic HAT activity: CBP and p300 and associated cofactors (e.g., PCAF, p/CIP-ACTR, and SRC-1). Coactivators bearing HAT activity are recruited to cis-regulatory regions by interaction with transcription factors, and they can enhance expression of the corresponding genes by acetylating nearby nucleosomes (24, 38). These coactivators thus act as molecular links between transcription factors and the basal transcriptional machinery. A number of studies have shown that CBP interacts with CREB and NF-κB (12, 42, 61, 73). CREB and NF-κB could thus target HATs to the BLV promoter since these transcriptional factors specifically bind to the LTR. Furthermore, the implication of CBP and PCAF in the process of transcriptional activation by human T-cell leukemia virus (HTLV) Tax has been previously described (21, 27, 32, 41, 56). We therefore speculate that a similar mechanism could also mediate BLV Tax transactivation. In contrast to acetyltransferases, the role of deacetylases in BLV or HTLV transcription has, however, not been described.
Deacetylases interact with sequence-specific DNA-binding transcriptional repressors, like Mad/Max, and inhibit transcription by deacetylating chromatin (38, 52). Factors Sin3 and N-CoR/SMRT appear to establish a link between deacetylases and the Mad/Max complex, which recognize specifically the regulatory E-box sequence (CANNTG) (4, 22, 24, 28, 44, 57). Interestingly, the BLV promoter possesses three E-box motifs located within the three TxREs enhancers. We therefore hypothesized that these E-box motifs could be involved in TSA response by recruiting HDACs to the LTR since our results demonstrate that deacetylase inhibitors activate BLV expression. Supporting this model, we have previously demonstrated that, in transient transfection experiments, mutation of all three E-boxes located in the U3 region provokes a slight but reproducible increase in LTR basal activity, suggesting the repressor role of the E-box element (48). However, our data show that a mutation (in boldface type) within the E-box sequences (CANNTG→CANNGA) did not, at least under our experimental conditions, alleviate the activation of the BLV promoter by TSA either in D17 stable cell lines (Table 1) or in sheep PBMCs (Table 2). Nevertheless, in other cell lines, mutations in the E-box motifs have been shown to decrease the TSA induction of the BLV promoter (C. Calomme and C. Van Lint, unpublished data), suggesting that TSA might have different effects depending on the cell types. Furthermore, mutation in the other transcription factor binding sites (IRF, GRE, NF1, NF2, CRE3x, and Δ21-pb) did not seem to impede the induction of BLV expression by TSA. Most importantly, this could be clearly demonstrated for the Ebox3x, Δ21-pb, and NF1 mutants in the context of primary lymphocytes isolated from sheep (Fig. 2B).
Histones are not the sole natural substrates for acetylation, since HATs are able to efficiently acetylate a growing list of cellular proteins. Indeed, CBP and PCAF can acetylate p53 (25) as well as general transcription factors such as TFIIF and TFIIEβ (29). Additionally, CBP, PCAF, and human GCN5 are also able to directly acetylate the HIV type 1 transactivator protein, Tat, leading to an increase of its transcriptional activity (13, 16, 36, 51). It is thus possible that the increase in BLV expression occurring ex vivo in response to TSA was in part a consequence of Tax acetylation. However, it should be stressed that our in vitro data demonstrate an activating effect of TSA in cell lines containing an integrated LTR in the absence of BLV proteins (Fig. 1). TSA could thus induce BLV expression through acetylation of histones and/or nonhistone proteins such as Tax and other cellular factors. In this regard, we have demonstrated in a detailed separate study that TSA synergistically enhanced Tax-mediated transcriptional activation of the BLV promoter (in several cell lines), suggesting that Tax could be functionally regulated by posttranslational acetylation (T. L.-A. Nguyen and C. Van Lint, unpublished results).
In summary, the results presented here demonstrate that inhibition of deacetylation plays an important role in the regulation of BLV transcription. Deacetylation mechanisms could be implicated in the silencing strategy adopted by BLV to escape from the host immune response. In this context, induction of viral expression by TSA or others deacetylase inhibitors (such as valproic acid) could open novel prospects for therapeutical approaches not only for BLV but, more importantly, for the related HTLV type 1 virus.
Acknowledgments
C.M. is a fellow of the “Pôle d'Attraction interuniversitaire” (SSTC P4/30). R.K., C.V.L., and L.W. are members of the “Fonds national de la Recherche scientifique” (FNRS). We thank the “Fédération belge contre le Cancer,” the “Action de Recherche concertée du Ministère de la Communauté française de Belgique,” the “Fortis Banque Assurance,” the FNRS, Maria Sklodowska-Curie Joint Fund II (PL-AES-284), the Commissariat général aux relations internationales de la Communauté Wallonie-Bruxelles, and the “Service de Programmation pour la Politique scientifique” (SSTC P4/30) for financial support.
We are grateful to M. Nuttinck, M. Zaborna, T. Peremans, J. M. Londes, and G. Vandendaele for skillful technical help. We thank M. Yoshida (University of Tokyo, Tokyo, Japan) for his generous gift of TPX.
REFERENCES
- 1.Adam, E., P. Kerkhofs, M. Mammerickx, A. Burny, R. Kettmann, and L. Willems. 1996. The CREB, ATF-1, and ATF-2 transcription factors from bovine leukemia virus-infected B lymphocytes activate viral expression. J. Virol. 70:1990-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam, E., P. Kerkhofs, M. Mammerickx, R. Kettmann, A. Burny, L. Droogmans, and L. Willems. 1994. Involvement of the cyclic AMP-responsive element binding protein in bovine leukemia virus expression in vivo. J. Virol. 68:5845-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adcock, I. M. 2001. Glucocorticoid-regulated transcription factors. Pulm. Pharmacol. Ther. 14:211-219. [DOI] [PubMed] [Google Scholar]
- 4.Ayer, D. E., Q. A. Lawrence, and R. N. Eisenman. 1995. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80:767-776. [DOI] [PubMed] [Google Scholar]
- 5.Baliga, V., and J. F. Ferrer. 1977. Expression of the bovine leukemia virus and its internal antigen in blood lymphocytes. Proc. Soc. Exp. Biol. Med. 156:388-391. [DOI] [PubMed] [Google Scholar]
- 6.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 7.Bannister, A. J., and E. A. Miska. 2000. Regulation of gene expression by transcription factor acetylation. Cell. Mol. Life Sci. 57:1184-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks, P. A., J. K. Nyborg, and G. L. Cockerell. 1995. Identification of an NF-κB binding site in the bovine leukemia virus promoter. J. Virol. 69:6005-6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 11.Chen, W. Y., and T. M. Townes. 2000. Molecular mechanism for silencing virally transduced genes involves histone deacetylation and chromatin condensation. Proc. Natl. Acad. Sci. USA 97:377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chrivia, J. C., R. P. Kwok, N. Lamb, M. Hagiwara, M. R. Montminy, and R. H. Goodman. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365:855-859. [DOI] [PubMed] [Google Scholar]
- 13.Col, E., C. Caron, D. Seigneurin-Berny, J. Gracia, A. Favier, and S. Khochbin. 2001. The histone acetyltransferase, hGCN5, interacts with and acetylates the HIV transactivator, Tat. J. Biol. Chem. 276:28179-28184. [DOI] [PubMed] [Google Scholar]
- 14.Dangond, F., and S. R. Gullans. 1998. Differential expression of human histone deacetylase mRNAs in response to immune cell apoptosis induction by trichostatin A and butyrate. Biochem. Biophys. Res. Commun. 247:833-837. [DOI] [PubMed] [Google Scholar]
- 15.Davie, J. R., and M. J. Hendzel. 1994. Multiple functions of dynamic histone acetylation. J. Cell. Biochem. 55:98-105. [DOI] [PubMed] [Google Scholar]
- 16.Deng, L., C. de la Fuente, P. Fu, L. Wang, R. Donnelly, J. D. Wade, P. Lambert, H. Li, C. G. Lee, and F. Kashanchi. 2000. Acetylation of HIV-1. Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones. Virology 277:278-295. [DOI] [PubMed] [Google Scholar]
- 17.Dequiedt, F., G. H. Cantor, V. T. Hamilton, S. M. Pritchard, W. C. Davis, P. Kerkhofs, A. Burny, R. Kettmann, and L. Willems. 1999. Bovine leukemia virus-induced persistent lymphocytosis in cattle does not correlate with increased ex vivo survival of B lymphocytes. J. Virol. 73:1127-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derse, D. 1987. Bovine leukemia virus transcription is controlled by a virus-encoded trans-acting factor and by cis-acting response elements. J. Virol. 61:2462-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derse, D., S. J. Caradonna, and J. W. Casey. 1985. Bovine leukemia virus long terminal repeat: a cell type-specific promoter. Science 227:317-320. [DOI] [PubMed] [Google Scholar]
- 20.Derse, D., and J. W. Casey. 1986. Two elements in the bovine leukemia virus long terminal repeat that regulate gene expression. Science 231:1437-1440. [DOI] [PubMed] [Google Scholar]
- 21.Gachon, F., S. Thebault, A. Peleraux, C. Devaux, and J. M. Mesnard. 2000. Molecular interactions involved in the transactivation of the human T-cell leukemia virus type 1 promoter mediated by Tax and CREB-2 (ATF-4). Mol. Cell. Biol. 20:3470-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grandori, C., J. Mac, F. Siebelt, D. E. Ayer, and R. N. Eisenman. 1996. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 15:4344-4357. [PMC free article] [PubMed] [Google Scholar]
- 23.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 24.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 25.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 26.Gwack, Y., H. Byun, S. Hwang, C. Lim, and J. Choe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrod, R., Y. L. Kuo, Y. Tang, Y. Yao, A. Vassilev, Y. Nakatani, and C. Z. Giam. 2000. p300 and p300/cAMP-responsive element-binding protein associated factor interact with human T-cell lymphotropic virus type-1 Tax in a multi-histone acetyltransferase/activator-enhancer complex. J. Biol. Chem. 275:11852-11857. [DOI] [PubMed] [Google Scholar]
- 28.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89:341-347. [DOI] [PubMed] [Google Scholar]
- 29.Imhof, A., X. J. Yang, V. V. Ogryzko, Y. Nakatani, A. P. Wolffe, and H. Ge. 1997. Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol. 7:689-692. [DOI] [PubMed] [Google Scholar]
- 30.Jenkins, P. J., U. K. Binne, and P. J. Farrell. 2000. Histone acetylation and reactivation of Epstein-Barr virus from latency. J. Virol. 74:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen, W. A., B. J. Wicks-Beard, and G. L. Cockerell. 1992. Inhibition of protein kinase C results in decreased expression of bovine leukemia virus. J. Virol. 66:4427-4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kashanchi, F., J. F. Duvall, R. P. Kwok, J. R. Lundblad, R. H. Goodman, and J. N. Brady. 1998. The coactivator CBP stimulates human T-cell lymphotrophic virus type I Tax transactivation in vitro. J. Biol. Chem. 273:34646-34652. [DOI] [PubMed] [Google Scholar]
- 33.Kerkhofs, P., E. Adam, L. Droogmans, D. Portetelle, M. Mammerickx, A. Burny, R. Kettmann, and L. Willems. 1996. Cellular pathways involved in the ex vivo expression of bovine leukemia virus. J. Virol. 70:2170-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kettmann, R., G. Marbaix, Y. Cleuter, D. Portetelle, M. Mammerickx, and A. Burny. 1980. Genomic integration of bovine leukemia provirus and lack of viral RNA expression in the target cells of cattle with different responses to BLV infection. Leuk. Res. 4:509-519. [DOI] [PubMed] [Google Scholar]
- 35.Kiermer, V., C. Van Lint, D. Briclet, C. Vanhulle, R. Kettmann, E. Verdin, A. Burny, and L. Droogmans. 1998. An interferon regulatory factor binding site in the U5 region of the bovine leukemia virus long terminal repeat stimulates Tax-independent gene expression. J. Virol. 72:5526-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiernan, R. E., C. Vanhulle, L. Schiltz, E. Adam, H. Xiao, F. Maudoux, C. Calomme, A. Burny, Y. Nakatani, K. T. Jeang, M. Benkirane, and C. Van Lint. 1999. HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J. 18:6106-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiss-Toth, E., S. Paca-uccaralertkun, I. Unk, and I. Boros. 1993. Member of the CREB/ATF protein family, but not CREB alpha plays an active role in BLV tax trans activation in vivo. Nucleic Acids Res. 21:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo, M. H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 39.Kuo, M. H., J. E. Brownell, R. E. Sobel, T. A. Ranalli, R. G. Cook, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383:269-272. [DOI] [PubMed] [Google Scholar]
- 40.Kuo, M. H., J. Zhou, P. Jambeck, M. E. Churchill, and C. D. Allis. 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12:627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwok, R. P., M. E. Laurance, J. R. Lundblad, P. S. Goldman, H. Shih, L. M. Connor, S. J. Marriott, and R. H. Goodman. 1996. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature 380:642-646. [DOI] [PubMed] [Google Scholar]
- 42.Kwok, R. P., J. R. Lundblad, J. C. Chrivia, J. P. Richards, H. P. Bachinger, R. G. Brennan, S. G. Roberts, M. R. Green, and R. H. Goodman. 1994. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370:223-226. [DOI] [PubMed] [Google Scholar]
- 43.Lagarias, D. M., and K. Radke. 1989. Transcriptional activation of bovine leukemia virus in blood cells from experimentally infected, asymptomatic sheep with latent infections. J. Virol. 63:2099-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 45.McBain, J. A., A. Eastman, C. S. Nobel, and G. C. Mueller. 1997. Apoptotic death in adenocarcinoma cell lines induced by butyrate and other histone deacetylase inhibitors. Biochem. Pharmacol. 53:1357-1368. [DOI] [PubMed] [Google Scholar]
- 46.Medina, V., B. Edmonds, G. P. Young, R. James, S. Appleton, and P. D. Zalewski. 1997. Induction of caspase-3 protease activity and apoptosis by butyrate and trichostatin A (inhibitors of histone deacetylase): dependence on protein synthesis and synergy with a mitochondrial/cytochrome c-dependent pathway. Cancer Res. 57:3697-3707. [PubMed] [Google Scholar]
- 47.Meier, J. L. 2001. Reactivation of the human cytomegalovirus major immediate-early regulatory region and viral replication in embryonal NTera2 cells: role of trichostatin A, retinoic acid, and deletion of the 21-base-pair repeats and modulator. J. Virol. 75:1581-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merezak, C., C. Pierreux, E. Adam, F. Lemaigre, G. G. Rousseau, C. Calomme, C. Van Lint, D. Christophe, P. Kerkhofs, A. Burny, R. Kettmann, and L. Willems. 2001. Suboptimal enhancer sequences are required for efficient bovine leukemia virus propagation in vivo: implications for viral latency. J. Virol. 75:6977-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizzen, C. A., X. J. Yang, T. Kokubo, J. E. Brownell, A. J. Bannister, T. Owen-Hughes, J. Workman, L. Wang, S. L. Berger, T. Kouzarides, Y. Nakatani, and C. D. Allis. 1996. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 87:1261-1270. [DOI] [PubMed] [Google Scholar]
- 50.Niermann, G. L., and G. C. Buehring. 1997. Hormone regulation of bovine leukemia virus via the long terminal repeat. Virology 239:249-258. [DOI] [PubMed] [Google Scholar]
- 51.Ott, M., M. Schnolzer, J. Garnica, W. Fischle, S. Emiliani, H. R. Rackwitz, and E. Verdin. 1999. Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr. Biol. 9:1489-1492. [DOI] [PubMed] [Google Scholar]
- 52.Pazin, M. J., and J. T. Kadonaga. 1997. What's up and down with histone deacetylation and transcription? Cell 89:325-328. [DOI] [PubMed] [Google Scholar]
- 53.Radkov, S. A., R. Touitou, A. Brehm, M. Rowe, M. West, T. Kouzarides, and M. J. Allday. 1999. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 73:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salminen, A., T. Tapiola, P. Korhonen, and T. Suuronen. 1998. Neuronal apoptosis induced by histone deacetylase inhibitors. Brain Res. Mol. Brain Res. 61:203-206. [DOI] [PubMed] [Google Scholar]
- 55.Sheridan, P. L., T. P. Mayall, E. Verdin, and K. A. Jones. 1997. Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev. 11:3327-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shuh, M., and D. Derse. 2000. Ternary complex factors and cofactors are essential for human T-cell leukemia virus type 1 Tax transactivation of the serum response element. J. Virol. 74:11394-11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sommer, A., S. Hilfenhaus, A. Menkel, E. Kremmer, C. Seiser, P. Loidl, and B. Luscher. 1997. Cell growth inhibition by the Mad/Max complex through recruitment of histone deacetylase activity. Curr. Biol. 7:357-365. [DOI] [PubMed] [Google Scholar]
- 58.Spencer, T. E., G. Jenster, M. M. Burcin, C. D. Allis, J. Zhou, C. A. Mizzen, N. J. McKenna, S. A. Onate, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1997. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194-198. [DOI] [PubMed] [Google Scholar]
- 59.Steger, D. J., A. Eberharter, S. John, P. A. Grant, and J. L. Workman. 1998. Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc. Natl. Acad. Sci. USA 95:12924-12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 61.Vanden Berghe, W., K. De Bosscher, E. Boone, S. Plaisance, and G. Haegeman. 1999. The nuclear factor-κB engages CBP/p300 and histone acetyltransferase activity for transcriptional activation of the interleukin-6 gene promoter. J. Biol. Chem. 274:32091-32098. [DOI] [PubMed] [Google Scholar]
- 62.Van Lint, C., S. Emiliani, and E. Verdin. 1996. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 5:245-253. [PMC free article] [PubMed] [Google Scholar]
- 63.Van Lint, C., S. Emiliani, M. Ott, and E. Verdin. 1996. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15:1112-1120. [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, L., C. Mizzen, C. Ying, R. Candau, N. Barlev, J. Brownell, C. D. Allis, and S. L. Berger. 1997. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol. Cell. Biol. 17:519-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Willems, L., A. Burny, D. Collete, O. Dangoisse, F. Dequiedt, J. S. Gatot, P. Kerkhofs, L. Lefebvre, C. Merezak, T. Peremans, D. Portetelle, J. C. Twizere, and R. Kettmann. 2000. Genetic determinants of bovine leukemia virus pathogenesis. AIDS Res. Hum. Retrovir. 16:1787-1795. [DOI] [PubMed] [Google Scholar]
- 66.Willems, L., A. Gegonne, G. Chen, A. Burny, R. Kettmann, and J. Ghysdael. 1987. The bovine leukemia virus p34 is a transactivator protein. EMBO J. 6:3385-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Willems, L., R. Kettmann, G. Chen, D. Portetelle, A. Burny, and D. Derse. 1992. A cyclic AMP-responsive DNA-binding protein (CREB2) is a cellular transactivator of the bovine leukemia virus long terminal repeat. J. Virol. 66:766-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolffe, A. P. 1996. Histone deacetylase: a regulator of transcription. Science 272:371-372. [DOI] [PubMed] [Google Scholar]
- 69.Wolffe, A. P., and D. Pruss. 1996. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell 84:817-819. [DOI] [PubMed] [Google Scholar]
- 70.Wolffe, A. P., J. Wong, and D. Pruss. 1997. Activators and repressors: making use of chromatin to regulate transcription. Genes Cells 2:291-302. [DOI] [PubMed] [Google Scholar]
- 71.Xiao, J., and G. C. Buehring. 1998. In vivo protein binding and functional analysis of cis-acting elements in the U3 region of the bovine leukemia virus long terminal repeat. J. Virol. 72:5994-6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]
- 73.Zhong, H., R. E. Voll, and S. Ghosh. 1998. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell 1:661-671. [DOI] [PubMed] [Google Scholar]