Abstract
Dimeric restriction endonucleases and monomeric modification methyltransferases were long accepted as the structural paradigm for Type II restriction systems. Recent studies, however, have revealed an increasing number of apparently dimeric DNA methyltransferases. Our initial characterization of RsrI methyltransferase (M.RsrI) was consistent with the enzyme functioning as a monomer, but, subsequently, the enzyme crystallized as a dimer with 1500 Å2 of buried surface area. This result led us to re-examine the biochemical properties of M.RsrI. Gel-shift studies of M.RsrI binding to DNA suggested that binding cooperativity targets hemimethylated DNA preferentially over unmethylated DNA. Size-exclusion chromatography indicated that the M.RsrI–DNA complex had a size and stoichiometry consistent with a dimeric enzyme binding to the DNA. Kinetic measurements revealed a quadratic relationship between enzyme velocity and concentration. Site-directed mutagenesis at the dimer interface affected the kinetics and DNA-binding of the enzyme, providing support for a model proposing an active enzyme dimer. We also identified a conserved motif in the dimer interfaces of the β-class methyltransferases M.RsrI, M.MboIIA and M2.DpnII. Taken together, these data suggest that M.RsrI may be part of a sub-class of MTases that function as dimers.
INTRODUCTION
Inital studies of Type II restriction-modificiation (R-M) systems showed that the endonucleases acted as homodimers, cleaving both strands of DNA sequentially in a single binding event [Reviewed in (1)]. Each endonuclease subunit was thought to recognize a single strand of the DNA duplex in the duplex palindromic target sequence. The partner MTases (MTases) might also have been expected initially to be dimeric enzymes and recognize DNA in a similar fashion. However, early characterizations of the Type II MTases revealed monomeric enzymes, which add a single methyl group per binding event (2). These findings established the paradigm for the composition of Type II systems as dimeric endonucleases and monomeric MTases (3–5). Eventually, a few MTases, such as M1.DpnII, M.MspI, M.HhaI and M.RsrI, that dimerized in the absence of DNA at high protein concentrations were found, but further biochemical studies suggested that these enzymes functioned as monomers (6–9). Since then, several other β-class MTases have also been reported to dimerize in solution, namely M2.DpnII (6), M.KpnI (10) and M.LlaCI (11). Dimeric DNA MTases, namely T4Dam and M.CcrM, which are not part of an R-M system have also been found (12,13).
M.RsrI dimerization was surprising since, although the enzyme had been shown to dimerize in solution at concentrations greater than 1 µM, biochemical data showed a single methylation event per turnover and no other biochemical data suggested dimerization was anything other than an artifact of high protein concentrations (9). However, the finding of an extensive dimerization interface in the crystal structure (14) [1500 Å2 buried surface area, well above the 600 Å2 observed for a typical crystal contact (15)] suggests the dimerization of M.RsrI might be a functional property of the enzyme. In such a model, one of the two monomers would interact with target DNA and the dimerization would not solely be an artifact of the high enzyme concentrations used for crystallization. Indeed, the subsequent structure of a second β-class, dimeric MTase, M.MboIIA (16) and a recent proposal that many of the crystallized Type II MTases are dimeric (8), suggest that the observed large interfacial area of dimerization of M.RsrI was not an isolated event. Here, we present further biochemical evidence that M.RsrI functions as a dimer.
Data from gel-shift assays of DNA-binding by M.RsrI complexed with the inhibitor sinefungin (17) were re-examined for signs of cooperative binding expected for a dimer binding to DNA. Gel-filtration chromatography was used to estimate the size and stoichiometry of the protein–DNA complex. Kinetic measurements revealed a non-linear relationship between enzyme concentration and catalytic activity. We attempted to disrupt the putative dimer by site-directed mutagenesis, and measured the kinetic and dimerization properties of the mutant. The results of these experiments suggest that a dimer is the functional unit of M.RsrI. Furthermore, sequence alignments between M.RsrI and other dimeric β-class MTases indicate that a dimer interface may be conserved among this group of enzymes.
MATERIALS AND METHODS
Unless otherwise specified, all chemicals were obtained from Sigma-Aldrich Co.
Buffer solutions
Protein Dilution Buffer: 20 mM Tris (pH 7.5), 10% glycerol, 20 mM β-mercaptoethanol (β-ME), 1 mM EDTA, 50 mM KCl and 0.2 mg/ml acetylated BSA (Promega). Storage Buffer: 20 mM Tris (pH 8.0), 300 mM KCl, 10 mM β-ME, 10 mM EDTA. Kinetics Buffer: 100 mM HEPES (pH 7.5), 5 mM EDTA, 5 mM β-ME and 0.5 mg/ml BSA.
Wild-type enzyme
His-tagged M.RsrI was purified as described (14) and stored in Storage Buffer at −80°C. All enzyme concentrations were determined by ultraviolet (UV) absorption at 280 nm using a molar extinction coefficient for the monomer of 48.7 mM−1 cm−1 (18). Enzyme concentrations listed refer to the concentration of monmeric enzyme unless otherwise specified.
Mutagenesis to form S124D and protein production
A S124D mutant was created from the histidine-tagged construct described in Scavetta et al. (14) by a whole-plasmid site-directed PCR mutagenesis method derived from the Quik-Change Site-Directed Mutagenesis Kit (Stratagene). Two mutagenic primers (300 nM, Invitrogen) were mixed with 1 pM of template plasmid pET28a::rsrIM (the histidine-tagged construct of M.RsrI), 100 µM of each of the four dNTPs, 1 mM MgSO4, 1× Pfx Amplification Buffer (Invitrogen) and 2.5 U of Platinum Pfx DNA polymerase (Invitrogen). The sequences of the primers were: forward, d(GGTGATCTGATATCAATCATTGATCACATGAGACAAAACAGC) and reverse, d(GCTGTTTTGTCTCATGTGATCAATGATTGATATCAGATCACC).
The thermocycler protocol was: a 5 min denaturation at 95°C; 25 cycles of a 1 min denaturation at 95°C, a 1 min annealing at 55°C and a 10 min extension at 64°C; finally, a 20 min extension at 64°C. The parental DNA was digested by adding 10 U DpnI endonuclease and incubating for 1 h at 37°C. A dilution (1:10) of the digest was introduced into Escherichia coli DH5α by electroporation and the plasmid DNA was isolated from the resulting clones. The DNA from the clones was screened for the presence of an EcoRV site (underlined above) introduced along with the mutation (boldface). The plasmids identified were subsequently sequenced to confirm presence of the mutation and lack of secondary mutations and introduced by electroporation into E.coli BL21(DE3) for expression. The S124D mutant was purified by nickel-ion metal-affinity chromatography as described previously (14), resulting in >95% pure protein as determined by Coomassie staining of SDS–PAGE gels of the purified protein.
Gel-filtration chromatography
Separations were performed at room temperature using an FPLC system (Pharmacia) coupled to an HR 10/30 (10 mm × 30 cm) column containing Superdex S200 resin (Pharmacia). The flow rate was 0.3 ml/min using a running buffer of 20 mM Tris (pH 8.0), 50 mM KCl and 10 µM S-adenosylhomocysteine (AdoHcy) (Sigma) to help decrease DNA complex dissociation. Buffers were filtered and degassed before use. The absorption of the eluate was monitored at 260 nm or at 260 and 280 nm with a multiwavelength detector. Samples contained 1–4 µM M.RsrI and/or 25 bp duplex DNA oligonucleotide [Invitrogen, purified according to the procedure in reference (17)] with a single, centered RsrI target site (boldface) formed by annealing d(GGCAGACACGGAAT TCCACAGACGG) and its complement. Samples were incubated on ice for 5 min following mixing and dilution in running buffer. Injections were 100 µl. The sizing standards and their respective elution volumes were: Blue Dextran (2000 kDa; 8.3 ml), γ-globulin (300 and 150 kDa; 10.3 and 12.1 ml), BSA (134 and 67 kDa; 12.0 and 13.8 ml), ribonuclease A (13.5 kDa; 20.5 ml), creatine kinase (86 kDa; 13.8 ml) and β-lactoglobulin (35.6 kDa; 16.4 ml), all obtained from Sigma. All standards were diluted to give peaks with absorbances of ∼0.01 AU at 260 nm.
Quantitative kinetic assay
For quantitative measurements of enzyme activity, we adapted the radiokinetic assays used to study M.TaqI (19) and M.MspI (20) to monitor the transfer of tritiated methyl groups from [3H-CH3]S-adenosylmethionine (AdoMet) to the 25 bp unmethylated, duplex DNA oligonucleotide used in the gel-filtration experiments. M.RsrI was diluted to 2 µM in Protein Dilution Buffer before use. [3H-CH3]AdoMet (ICN Biomedicals) was diluted with unlabeled AdoMet (New England Biolabs) to a concentration of 800 µM and a specific activity of 4.7 Ci/mmol. Reactions to determine the Km of AdoMet contained 0.2 µM enzyme, 1 µM DNA and AdoMet concentrations of 0.5, 1, 2, 4 and 8 µM and were incubated for 20 min (WT reactions) or 40 min (S124D reactions). Timepoints were taken to monitor the progress of the reaction. Reactions to determine the Km of DNA contained 20 nM protein (WT) or 60 nM protein (S124D), 8 µM AdoMet and 10, 50, 100 or 1000 nM DNA. All reactions were carried out in Kinetics Buffer at 37°C. Reactions were stopped by diluting 10 µl aliquots of the reaction into 300 µl 1:1 (v/v) DE52 resin:Kinetics Buffer kept on ice. The resin mixture was prepared by washing DE52 resin (Whatman) three times in water and then adding an equal volume of 2× Kinetics Buffer to the sedimented resin. AdoHcy was included as a component of the resin mixture at 33 µM to help quench the reaction. The resin-reaction mixture was washed and sedimented by centrifugation for 2 min at 4 g for six to eight times with 600 µl of 100 mM NH4HCO3 and once with 600 µl of water. The labeled DNA was eluted from the resin with 600 µl of 1 M NaCl and 500 µl was removed and measured in a scintillation counter. DNA recovery was greater than 90% as determined by UV spectrometry. Reactions were performed in triplicate and kinetic constants extracted by non-linear regression fitting to the Michaelis–Menten equation using DynaFit (21).
Sequence alignments
Alignments of dimeric MTases were performed using ClustalW (22) to identify conserved residues in the crystallographic dimer interface. Conserved residues were taken to be identical or structurally similar, namely polar (S, T, C), basic (K, R, H), acids/amides (D, E, N, Q), aromatic (F, W, Y), small aliphatic (A, G, V) or large aliphatic (L, I, M). PSI-BLAST (23) searches starting from the known dimeric MTases identified homologous MTases that were screened manually for the conserved amino acids identified in the dimer interface. PSI-BLAST searches from the final set of putative dimeric MTases were coupled with PHI-BLAST (24) searches of the conserved sequence from the dimer interface to identify additional MTases containing similar sequences.
RESULTS AND DISCUSSION
Analysis of gel-shift data
We published a gel-shift analysis of M.RsrI (without a His-tag) binding to DNA in which complex formation was monitored by titrating a series of differentially methylated DNA oligonucleotides with increasing amounts of protein (17). In these studies, we observed the formation of a single complex irrespective of the DNA methylation state, which could be well described by the dissociation equilibrium of a monomer bound to DNA (F = [E]/([E] + Kd), F = fraction shifted, E = enzyme and Kd = dissociation constant). The data for binding of M.RsrI to unmethylated DNA fit most closely, with a correlation coefficient (R-value) of 0.99 (Figure 1A). Neither the observation of a single complex nor the high correlation between the data and a monomeric model provided any indication of dimer binding to DNA. However, the observation of a crystallographic dimer of M.RsrI prompted a re-examination the gel-shift data to see if the observed binding could be described by a dimeric model.
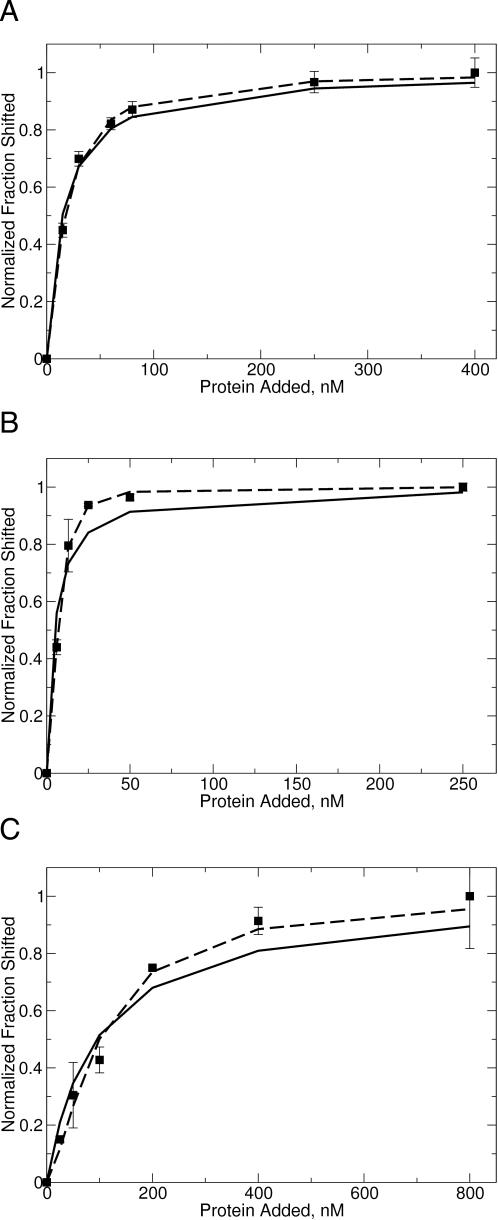
Figure 1.
Fitting of (A) unmethylated, (B) hemimethylated and (C) dimethylated DNA gel-shift data to various binding models. Data were fit as either M.RsrI monomer binding to DNA (solid line) or sequential binding of M.RsrI monomers (dashed line). The DNA used in the gel-shift experiments was a duplex 25 bp oligodexoxyribonucleotide with a single centered GAATTC target site bearing the specified methylation state at a concentration of 5 nM (17).
A dimer of M.RsrI could bind to DNA, such that only a single complex was observed by means of two mechanisms: binding of a preformed dimer, or binding of a monomer which quickly recruits a second subunit so that the level of monomeric complex is below detection. The mathematical descriptions of a monomer binding to DNA {F = [E]/([E] + Kd)}, of two monomers binding in a sequential and cooperative fashion to DNA {F = [E]n/([E]n + Kd)}, and of a preformed dimer binding to DNA {F = [E]2/([E]2 + Kd)} differ only in the power to which the enzyme concentration is raised. This power is a variable, termed the Hill coefficient (n) (25,26), that describes the cooperativity of binding between the subunits. The cooperativity can vary from a lack of cooperativity (n = 1), the equivalent of binding by a monomer, to full cooperativity (n = 2), the equivalent of binding by both subunits of a preformed dimer. Fitting the sequential model to the previously determined gel-shift data would allow discrimination between binding of a monomer or a dimer if a Hill coefficient >1 is obtained because monomeric enzymes cannot exhibit cooperativity (n > 1) in binding measurements, such as gel-shift experiments (27).
Fitting the sequential model to the gel-shift data from reference (17) for all three methylation states (unmethylated, hemimethylated and fully methylated) resulted in a value of n that was between 1 and 2 (Figure 1 and Table 1). The quality of the fit seen in Figure 1 reveals that, while the binding of M.RsrI to unmethylated DNA is fit within the error of the data by both the monomeric and sequential models, a better fit is observed with the sequential model for the binding of M.RsrI to hemimethylated and dimethylated DNA. Since one complex of the same size was formed for all three DNA methylation states (17), the enzyme stoichiometry cannot depend on the methylation state of the DNA, suggesting that the same enzyme complex is binding in all three cases.
Table 1.
Calculated apparent dissociation constants for the M.RsrI–DNA complex and Hill coefficients (nh) for the binding of M.RsrI to DNA of various methylation states [Data for fitting from reference (17)]
| KDapp for monomer binding, nM | KDapp for sequential model, nM | nh for sequential model | |
|---|---|---|---|
| Unmethylated DNA (5 nM) | 15 ± 1.2 | 39 ± 8.8 | 1.3 ± 0.1 |
| Hemimethylated DNA (5 nM) | 4.7 ± 1.0 | 48 ± 7.7 | 2.0 ± 0.1 |
| Hemimethylated DNA (0.5 nM) | 4.8 ± 1.8 | 21 ± 5.0 | 2.2 ± 0.2 |
| Dimethylated DNA (5 nM) | 94 ± 16 | 860 ± 650 | 1.5 ± 0.2 |
The KDapp for the sequential model is the product of the dissociation constant for the binding of the first monomer and the dissociation constant for the binding of the second (25). Model fitting of the sequential and monomer binding models given in the text was performed with EXCEL (Microsoft) and DataFit (Oakdale Engineering) and is shown in Figure1.
The increased cooperativity observed for binding to hemimethylated DNA suggests a functional role for dimerization in preferentially targeting the enzyme, i.e. increasing the affinity of the enzyme for hemimethylated DNA in a cell following replication, or reducing the affinity for unmethylated DNA, which could come from invading bacteriophage. Such a reduction could enhance the resistance of the bacterium to infection by providing the partner endonuclease with increased time to degrade the invading DNA before it was methylated by the MTase.
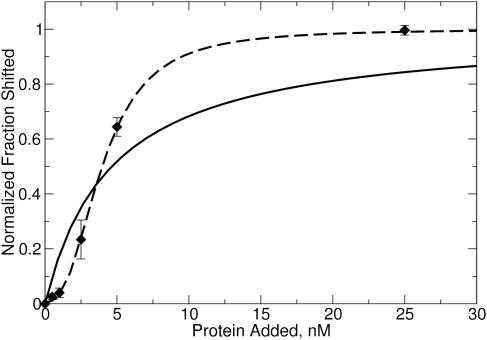
We also examined the binding of enzyme to DNA at low enzyme and DNA (0.5 nM DNA) concentrations to check for the sigmoidal response typical of cooperative systems. These data revealed a sigmoid curve (Figure 2), consistent with the cooperative binding of M.RsrI to hemimethylated DNA. The different Kd observed at this lower DNA concentration (Table 1) is surprising and may reflect a concentration-dependent effect of DNA on binding. On the basis of these data, the binding of M.RsrI to DNA appears to be a complex process of varying cooperativity based on the methylation state of the DNA, from highly cooperative (hemimethylated) to weakly cooperative (unmethylated). This observation raises the issue of whether the mechanism involves binding of a preformed dimer or sequential binding of two monomers to form an active dimer. Because non-integral values of the Hill coefficient were observed for M.RsrI binding to unmethylated and dimethylated DNA (Table 1), the data appear to support two monomers binding in a sequential order. However, non-integral values of the Hill coefficient could be due to an equilibrium between different conformations of a preformed dimer binding to the DNA. In either case, these results are consistent with a model of a dimer of M.RsrI representing the stoichiometry of the enzyme bound to DNA.
Figure 2.
Gel-shift of hemimethylated DNA at low protein concentration. The model of a monomer binding to DNA is the solid curve, the model for sequential binding of monomers is the dashed curve. The same DNA is used as in Figure 1B, but at a concentration of 0.5 nM (17).
Gel-filtration of protein and protein–DNA complex
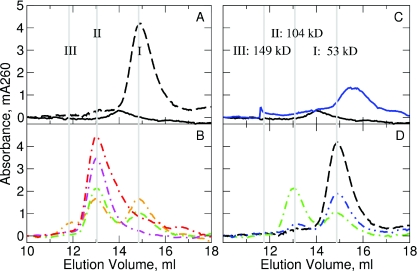
Since the analysis of the gel-shift data suggested that the protein could bind to the DNA as a dimer, we attempted to assess directly the size of the complex to confirm the stoichiometry of the complex. Gel-filtration chromatography, which separates molecules on the basis of their hydrodynamic or Stokes radius, allowed us to examine this property. A series of protein standards ranging from 14 to 300 kDa was used to create a calibration curve that related elution volume and molecular weight (data not shown). Deriving a molecular weight determination using these standards assumes that the unknown macromolecules have globular shapes similar to the macromolecules used to construct the calibration curve. DNA, because it is not globular, will elute at volumes corresponding to molecular weights significantly greater than its actual molecular weight. Consequently, gel-filtration of a 25 bp duplex oligonucleotide in the absence of M.RsrI showed a peak eluting at a much larger apparent molecular weight (53 kDa) (I, dashed black line, Figure 3A) with respect to the actual size (16 kDa) of the DNA. In the absence of DNA, gel-filtration of samples of His-tagged RsrI MTase at a concentration of 4 µM resulted a peak which had an elution volume indicating a MW of 74.4 kDa (solid black line, Figure 3A), consistent with the size of a dimer (calculated MW 71.2 kDa). Unfortunately, M.RsrI failed to show a reproducible absorption peak during elution at concentrations below 4 µM, presumably because the protein denatured at these low salt and protein concentrations (9). The M.RsrI peak could be observed at concentrations as low as 1 µM with increased salt concentrations (100 mM KCl); however, the higher salt concentrations prevented the formation of a protein::DNA complex (data not shown).
Figure 3.
Gel-filtration of M.RsrI–DNA complexes. Elution profiles at 260 nm are: (A) native M.RsrI (solid black) and DNA (dashed black) alone; (B) native M.RsrI–DNA complexes (dash-dot lines) with protein:DNA ratios of 0.5:1 (orange), 1:1 (green), 2:1(magenta) and 4:1 (red); (C) S124D M.RsrI (solid blue) and native M.RsrI (solid black) alone; (D) 1:1 complexes of S124D M.RsrI–DNA (blue dash-dot-dot), native M.RsrI–DNA (green dash-dot) and DNA (dashed black) alone. Elution peaks were marked with gray lines, labeled I, II and III with their corresponding molecular weight in (C).
When M.RsrI was mixed with DNA at a 1:1::protein:DNA molar ratio, the peak of DNA alone decreased in amplitude, and a second peak appeared with an estimated molecular weight of 104 kDa (II, Figure 3B, green curve). From this molecular weight alone, we could not predict the exact stoichiometry of the complex because of the non-globular nature of DNA. We expect the estimated molecular weight to overestimate the actual size due to the non-globular shape of the complex. Consequently, a dimer bound to one DNA (87 kDa) and a monomer bound to one DNA (52 kDa) are the most likely complexes. The binding of M.RsrI to DNA should result in a complex that is more globular than DNA alone. Consequently, the difference between the actual and observed molecular weights of the complex should be smaller than the 37 kDa difference measured for the DNA alone (53 − 16 kDa = 37 kDa). A monomer of M.RsrI bound to DNA would have a larger (52 kDa) rather than smaller size difference, suggesting that the stoichiometry of the complex is not 1:1.
A free DNA peak observed along with the complex peak (Figure 3B, green curve) from a separation of a 1:1::protein:DNA mixture offered additional insight into the stoichiometry of the M.RsrI–DNA complex. The area under the free DNA peak was consistent with the expected fraction of free DNA given complete formation of a 2:1 complex. Other explanations for this peak include incomplete formation of the complex or its dissociation. The first of these possibilities is highly unlikely given the nanomolar dissociation constant previously determined for the M.RsrI–DNA complex under similar conditions (17). Complex dissociation, unlikely due to its low dissociation constant, cannot be easily observed because the expected free protein peak would overlap with the tail of the complex peak due to their similar elution volumes (Figure 3).
Separations of protein:DNA ratios from 0.5:1–4:1::protein:DNA revealed a smooth increase in the size of the complex peak and a smooth decrease in that of the free DNA peak. At a 2:1 molar ratio, only the peak respresenting the protein::DNA complex was observed (II, magenta curve, Figure 3B). The presence of a free DNA peak at a 1:1::protein:DNA ratio and its disappearance at a 2:1 ratio further implies that the observed complex is two monomers of M.RsrI and one DNA molecule. While it is possible that the complex was 1:1 and the excess protein became denatured on the column and, hence was not observed, separation of a 4:1::protein:DNA mixture resulted in an increased tailing of the complex, as would be expected for an excess of free protein due to the overlap of the protein and complex peaks (red curve, Figure 3).
A second, smaller, peak eluting with a calculated molecular weight of 149 kDa (III, orange curve in Figure 3B) may indicate a dimer bound to two DNA molecules (expected MW 103 kDa). This peak was only observed as a significant fraction of the observed complexes at a 0.5:1::protein:DNA ratio, although it was detectable at a 1:1 ratio as well. Only one complex was observed in the gel-shift experiments, but the conditions and protein:DNA ratios used in those experiments may have interfered with formation of the larger, second complex.
The absorbance spectra of the peaks eluting during separation of a 1:1::protein:DNA mixture were monitored using a multiwavelength detector to estimate the composition of the complex from the A260/A280 ratio (data not shown). The absorbance profile of the complex showed an increase in relative absorbance at 280 nm over that of the DNA alone, as would be expected for the contribution of a protein, which absorbs more at 280 nm than 260 nm, to the overall absorbance of the complex (Table 2). The A260/A280 ratio of the complex could be estimated at 1.5, while that of the free DNA peak is approximately 2.0. The expected A260/A280 ratios for different complex compositions can be calculated from the molar extinction coefficients of the protein, DNA and AdoHcy ligand, which in turn are either known or can be calculated based on their chromophore composition (See Table 2 legend for details). Comparing the observed A260/A280 ratio of the complex (1.5) and the expected ratios (Table 2) for 1:1 and 2:1::protein:DNA complexes, and assuming that minimal chromophore quenching occurs upon complex formation, suggested that the complex contains 2:1::protein:DNA.
Table 2.
Calculated absorbance ratios of M.RsrI, DNA and complex
| ɛ260, mM−1 cm−1 | ɛ280, mM−1 cm−1 | A260/A280 | |
|---|---|---|---|
| M.RsrI | 22.7 | 48.7 | 0.47 |
| DNA | 336 | 177 | 1.90 |
| AdoHcy | 15.1 | 2.11 | 7.00 |
| 1:1::Protein: DNA complex | 374 | 228 | 1.64 |
| 2:1::Protein: DNA complex | 412 | 279 | 1.48 |
| Observed complex | 1.5 |
Complexes are theoretical 1:1 or 2:1::protein:DNA complexes with 1 bound AdoHcy/protein monomer. A260/A280 is calculated as the ratio of the molar absorptivities. The molar absorptivities of M.RsrI was calculated according to the method of Gill and von Hippel (18). The molar absorptivities of the oligodeoxyribonucleotide used were calculated from the molecular weight of the DNA, an absorbance of 20 OD/(mg/ml) at 260 nm for double-strand DNA (35), and the observed A260/A280 ratio. The molar absorptivities of AdoHcy were taken to be equal to that of adenosine (36). The molar absorptivities of complexes were calculated as the sum of the component parts.
Taken together, the gel-filtration experiments estimate the size of the complex to be consistent with two molecules of M.RsrI and one of DNA. Unfortunately, the observed molecular weight is only an upper limit of the actual molecular weight. To accurately measure the mass and dimensions of the complex, a method that offers more definitive data, such as ultracentrifugation, would be required. Furthermore, from our data alone, we cannot rule out non-specific binding of two monomers to one DNA. However the following kinetic analysis supports the gel-filtration data that M.RsrI binds and is active as a dimer on DNA.
M.RsrI kinetics
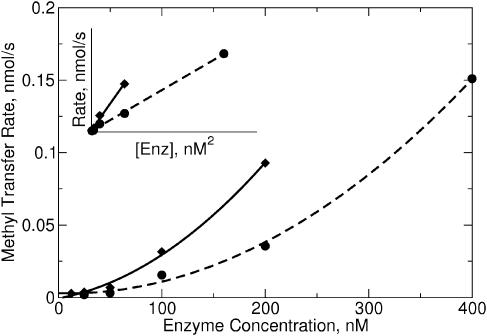
If, as the gel-filtration and gel-shift data suggested, M.RsrI binds to DNA as a dimer, the dimer could be the active form of M.RsrI. The observed rate of an enzyme varies with the concentration of the enzyme; the rate varies linearly with enzyme concentration for enzymes acting as monomers, whereas enzymes that act as dimers show a quadratic rate dependence of activity on the enzyme concentration (10). Increasing the concentration of M.RsrI resulted in a non-linear variation of methylation activity on initially unmethylated DNA with increasing enzyme concentration (Figure 4). Enzyme activity at each data point was constant over time, suggesting the variation in activity was not due to enzyme instability. Plotting the activity versus the square of the enzyme concentration resulted in a linear plot (inset, Figure 4), indicating the activity varies quadratically with the enzyme concentration, as would be expected if a dimer were the active unit of M.RsrI. A similar profile was observed for the dimeric MTase M.KpnI (10).
Figure 4.
Enzyme activity as a function of protein concentration. The reactions contained 0.5 µM 25 bp unmethylated oligonucleotide and 8 µM AdoMet and were stopped after 20 min for native enzyme (diamonds) and after 40 min for S124D (circles). A quadratic function was fit to each dataset. The data are replotted against the square of the protein concentration in the inset.
Dimer disruption by S124D mutant
Examination of the crystallographic dimer revealed the serine residue at position 124 is located closest to the 2-fold axis of rotation of the dimer, and forms a hydrogen bond to the same residue on the opposite monomer (Figure 5). Introducing a negative charge at this position should lead to strong charge-charge repulsions and either disrupt the dimer completely or cause a significant structural change in the monomer–monomer interactions. Because it is located away from the ligand-binding site, active-site and putative target recognition domain (TRD), any effects observed should be due to disruption of the dimer. During purification of the S124D mutant, we noticed reduced solubility of the protein relative to that of the wild-type enzyme, suggesting an increase in hydrophobic surface area. Furthermore, the mutant failed to crystallize under the same conditions used to crystallize wild-type enzyme, indicating the structure of the mutant differed significantly from that of the wild-type enzyme.
Figure 5.
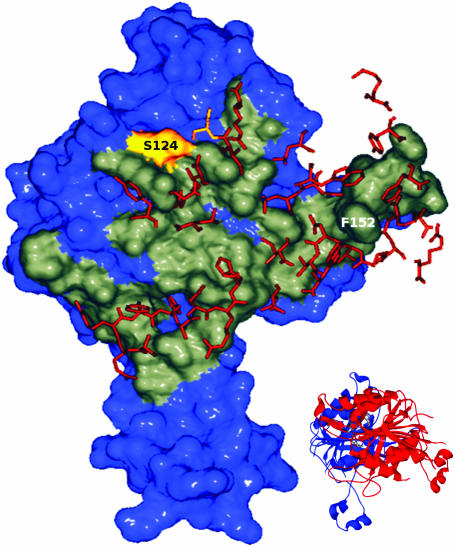
Dimerization interface of RsrI MTase. One monomer of M.RsrI is shown in surface representation with the dimer interface highlighted in gray. Residues within 4 Å of this interface coming from the opposite monomer are shown as orange sticks. The S124 residue is colored yellow. The figure was created using Swiss PDB Viewer (34) and POV-RAY (www.povray.org). A ribbon diagram showing the orientation of the dimer is inset in the figure.
The S124D mutant was active and showed a non-linear dependence of activity on enzyme concentration similar to that seen with the wild-type enzyme (Figure 4), suggesting the mutation does not completely inhibit dimer formation. Quantitative determination of the kinetic parameters for the mutant revealed the kcat for the mutant (0.01 ± 0.001 min−1) was reduced 20-fold from that of His-tagged wild-type (0.21 ± 0.015 min−1), under the conditions used. The kcat previously determined for the untagged wild-type enzyme was 1.14 min−1 (17); the difference could be due to differences in reaction composition or an effect of the His-tag.
The Km for AdoMet was reduced 3-fold by the mutation (wild-type 2.7 ± 0.47 µM, S124D 0.86 ± 0.31 µM). Determination of the Km for DNA was hampered by the large activity reduction of both wild-type and S124D enzymes at low enzyme concentrations. Consequently, the lowest usable enzyme concentrations were 20 nM for wild-type and 60 nM for the mutant. Measurements at these concentrations suggested that the Km for DNA was increased by 2-fold for the S124D mutant (∼20 nM) relative to the wild-type enzyme (∼10 nM). The increased affinity of the S124D mutant for AdoMet was unexpected and may indicate that the structural changes to the enzyme are not localized to the dimer interface. Overall, these results suggest that the dimer plays a role in enzyme activity.
Structural changes present in the mutant were also examined by gel-filtration chromatography. In contrast to the wild-type enzyme, gel-filtration chromatography revealed a peak with a calculated MW of 37 kDa, corresponding to a monomeric form of the enzyme, at a sample concentration of 4 µM S124D (Figure 3C, blue line). No monomer peak was observed at any of the concentrations of wild-type enzyme tested (down to 1 µM in the presence of increased salt levels necessary to maintain solubility), indicating that, as expected, the S124D mutant appears to interfere with dimerization. Gel-filtration of a 1:1::protein:DNA ratio of the S124D mutant with DNA revealed formation of a complex eluting at the same position as the complex of the wild-type enzyme with DNA (Figure 3D, blue curve). However, the amount of complex formed was reduced relative to that observed for the wild-type enzyme (Figure 3D, green curve), suggesting that the mutation interferes with complex formation, but does not prevent it. Formation of a 2:1::protein:DNA complex by the mutant is consistent with the enzyme dilution kinetics observed earlier that suggested S124D acted as a dimer. Additionally, because S124D appeared to be a monomer at the concentrations used to form the complex, the formation of a 2:1 complex suggests the dimerization of the enzyme is essential for DNA binding.
Sequence studies of the dimer interface of M.RsrI
The buried surface area at the dimer interface includes more than 10% of the protein residues (see Supplementary data). Sequence comparison of MTases has identified nine globally conserved sequence motifs (28), which have been assigned structural and functional roles. The dimer interface observed here involves motifs VII and VIII, as well as a portion of motif X (28). The interface included a number of hydrogen bonds and salt bridges between residues; however, close examination (Figure 5) also revealed the presence of a hydrophobic pocket. This hydrophobic pocket was formed primarily by residues Trp161, Phe170, Pro204 and Trp208 on one monomer, and is filled by Phe152 from the other monomer. The large hydrophobic surface involved in this interaction might be responsible for the tendency of M.RsrI to denature at low protein concentrations as the exposed hydrophobic surface on the monomers could promote non-specific aggregation (9).
A second dimeric MTase, M.MboIIA, was recently crystallized (16). This Type IIS β-class amino MTase recognizes the non-symmetric DNA sequence GAAGA and shares 33% sequence identity with M.RsrI. Overlaying the structures of M.MboIIA and M.RsrI revealed an overall similarity (1.4 Å r.m.s.d. between the backbone atoms of the dimers), as well as conservation of a number of residues in the dimer interfaces. Among the residues of the dimer interface, the Phe inserted into the opposite monomer (M.RsrI numbering: Phe152) was conserved, as were five of the residues comprising the hydrophobic pocket (M.RsrI numbering: Asn136, Phe170, Pro204, Trp208 and Ala238). Other conserved residues formed hydrogen bonds to the opposite monomer (M.RsrI numbering: Ala148, Arg150, Lys166 and Asn203) or other contacts (M.RsrI numbering: Arg176 and Val207). This conservation suggests that the hydrophobic pocket is an important part of the dimer interface in both enzymes.
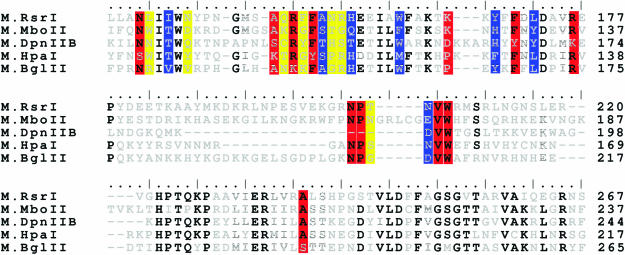
The conservation of the dimer interface between M.RsrI and M.MboIIA prompted us to compare the sequences of other MTases that have been reported to dimerize. Due to the sequence permutations between the various MTase classes, only β-class amino MTases, which comprise the majority of the MTases reported to dimerize, were compared. These MTases are: M.RsrI (9), M2.DpnII (25% identity to M.RsrI) (6), M.MboIIA (33% identity) (16), M.KpnI (18% identity) (10), M.LlaCI (24% identity) (11) and M.PvuII (24% identical) (8). M.KpnI was excluded from the sequence alignments because of its lower sequence identity with M.RsrI. Sequence alignments using ClustalW (22) revealed that the dimer interface amino acids conserved between M.RsrI and M.MboIIA extend to M2.DpnII as well (Figure 7), but not to M.LlaCI or M.PvuII (data not shown). Of the conserved amino acids, six of the twelve (M.RsrI numbering: Asn136, Arg150, Phe152, Val207, Trp208 and Ala238) are identical in M2.DpnII and three of the remaining six amino acids are conservative substitutions (M.RsrI→M2.DpnII: Ala148→Ser, Phe170→Tyr and Arg176→Lys). This high level of similarity in the dimer interface between M.RsrI/M.MboIIA and M2.DpnII suggests that M2.DpnII shares a similar dimer interface.
The sequence alignment of these three MTases known to dimerize should predict the dimerization of other, less well characterized MTases. Aligning other highly-homologous MTases identified by a PSI-BLAST (23) search with these three known dimers and looking for MTases with at most four substitutions of the ten conserved amino acids in the dimer interface (the number found in M2.DpnII) revealed that the MTases M.BglII (1 substitution) and M.HpaI (4 substitutions) may share this interface and be dimeric (Figure 7). This is not surprising, given that they are the most closely related to M.RsrI and M.MboIIA [See Figure 5 in (29)]. M.HpaI has been previously biochemically characterized (30), and appears to be a monomer at concentrations up to 2.5 µM, suggesting that one or more of the amino acid changes present in that protein (such as K108, which is A or S in the other MTases) may prevent dimerization.
We previously defined the conserved residues in the dimer interface as those identical in M.RsrI and M.MboIIA. However, it is likely that both of these MTases contain amino acid substitutions at residues which are conserved in the other, presumably dimeric, MTases. Examining all the amino acids at the dimer interface in M.RsrI (see Supplementary data) identified an additional seven amino acids which are conserved in all five MTases (Figure 6). An amino acid was considered conserved if it was identical or replaced by a structurally conservative substitution in four of the five MTases. Using the same criteria, Lys166, which was originally taken to be conserved because it appears in both M.RsrI and M.MboIIA, appears not to be conserved over this whole group. Considering all of the amino acids identified to be conserved in the dimer interface, the motif NXXT(X9–11)AXRXFSXXH(X4)W(X6–9)YXFXL(X3)R(X9–26)NP(X1–6)NVW(X29–34)A, where X is any amino acid, defined the dimer interface. The sequence alignment of the five MTases revealed other conserved residues as well; however, these do not participate in the dimer interface in M.RsrI or M.MboIIA and are presumed to serve scaffolding or other roles.
Figure 6.
Dimer interface sequence alignments of putative dimeric MTases. The sequences of M.RsrI, M.MboIIA, M2.DpnII, M.HpaI and M.BglII were aligned using ClustalW with its default settings; only the region of the dimer interface is shown. Black letters indicate completely conserved amino acids, striped letters are similar amino acids and light gray letters are unconserved. Conserved amino acids participating in the dimer interface identified by comparison of M.RsrI and M.MboIIA are in red boxes; additional interfacial amino acids identified as conserved in all five MTases are colored blue. Unconserved amino acids involved in the dimer interface of M.RsrI are colored yellow.
A PHI-BLAST (24) search using the entire dimer-interface motif identified identified M1.BsmI and several putative MTases which contain the motif. Additionally, M2.Ssu8074I, M2.Ssu4109I and M2.LlaII were very similar to M2.DpnII and shared much of the dimerization motif, although all of those enzymes, like M2.DpnII, replaced the conserved NP with a conserved QM. The MTases identified in these sequence searches have been incompletely characterized, with only M.HpaI having been biochemically purified (30). Further characterization of these potentially dimeric MTases will be necessary to establish the essential amino acids present in the dimerization motif.
Biological relevance of RsrI methyltransferase dimer
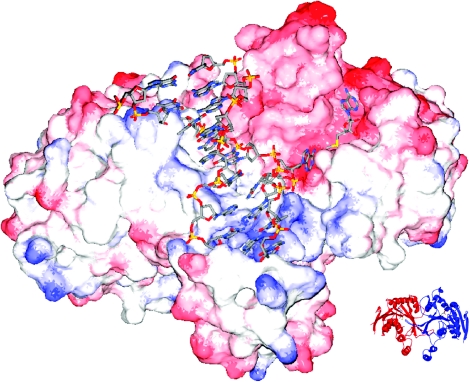
The data presented above provide evidence that M.RsrI can bind and methylate DNA as a dimer. Indeed, much of the data cannot be reconciled with the model of a monomer binding to DNA. What might be the biological function of dimerization? Using the coordinates from the M.TaqI-DNA co-crystal structure (31), the conserved MTase fold in M.TaqI was overlaid on the equivalent portion of the M.RsrI dimer. This superposition resulted in the positioning of the extrahelical target base of the DNA in the active-site pocket of the M.RsrI enzyme across from the bound AdoMet cofactor (Figure 7). Furthermore, superimposing the proteins positioned the DNA along the only significantly positively charged portion of the RsrI MTase dimer, namely a groove at the border of the monomer–monomer interface. Part of this interface (159–200 amino acid) has been suggested to be involved in site-specific binding (29), and is exposed at the edge of the dimer interface so that such binding may be possible. Because cooperativity is often the result of an interfacial binding site (27), the dimer-binding model explains the observed cooperative binding of DNA, and also how DNA-binding may stabilize the dimer as was seen with the S124D mutant. The model also suggests how the dimer might bind two DNA molecules simultaneously (one along each groove formed by the monomer–monomer interface), consistent with the posited 2:2 complex observed at the low protein::DNA ratio in the gel-filtration experiments (Figure 3B). The observed modification of a single DNA strand per catalytic cycle (9) is also explained by the model, since the active sites of the two monomers are on opposite sides of the dimer and would interact with separate DNA recognition sites. Such a possibility is consistent with the recent proposition that M.HhaI may bind as a dimer to two DNAs simultaneously (8).
Figure 7.
RsrI MTase dimer modeled with bound DNA. The model was created by superimposing the β-strand cores of RsrI MTase and the M.TaqI-DNA structures. M.RsrI is shown as a charged surface from red (negative) to blue (positive). The DNA is from the M.TaqI structure and shown as sticks colored by atom type (red = O, blue = N, yellow = P, gray = C). AdoMet bound to M.RsrI is also shown as sticks colored similar to the DNA except that yellow = S. A ribbon diagram of the orientation of the protein dimer alone is shown in the inset. The figure was created using Swiss PDB Viewer (34) and POV-RAY (www.povray.org).
A DNA-binding site at the border of the dimer interface positions the DNA to cross the putative TRD stalk of the partner monomer, implicating this portion of the TRD, not so much in specific binding, but rather for non-specifically increasing the affinity of the MTase for the DNA. This hypothesis is supported by the observation that the D173A mutant, which disrupts the hydrogen bond network responsible for maintaining the stalk structure of the TRD, retains specific DNA MTase activity while losing the ability to form stable complexes in an in vivo measure of DNA complex formation, the challenge-phage assay (29). Furthermore, when the D173A mutation is combined with the L72P mutant, which forms a stable complex with DNA, the affinity of the double mutant for DNA is reduced relative to the L72P mutant alone, suggesting a decrease of DNA-binding affinity with the D173A mutation.
Taken together, our observations suggest that the dimer stabilizes and enhances site-specific DNA-binding by the MTase, because the DNA-binding site may require an interfacial structure formed by both monomers. Combined with the observed processivity differences between M.Dam and Type II MTases (32), such binding may serve to preferentially target the enzyme to the cellular hemimethylated DNA with respect to invading unmethylated phage DNA, supporting the proposed biological role of Type II MTases as defense systems against phage. The dimer may also serve to reorient the enzyme to the productive strand as seen in burst experiments for both M.RsrI (17) and T4Dam (33).
CONCLUSIONS
Only a few dimeric DNA MTases have been identified. The large buried surface area seen in the crystal structure of M.RsrI suggested that the crystallographic dimer might be more than an artifact of crystallization and allowed identification of a conserved motif for the dimer interface among β-group amino MTases. On the basis of these results, we have re-examined our published gel-shift data and determined they are consistent with a dimer binding to DNA. Gel-filtration data suggested the size and stoichiometry of the protein–DNA complex are consistent with a ratio of two protein monomers to one dsDNA. Furthermore, enzyme activity showed a non-linear dependence on enzyme concentration, revealing the possibility that M.RsrI, while depositing a single methyl group per binding reaction (9), acts as a dimer. Additionally, site-directed mutagenesis in an attempt to disrupt the dimer created the S124D mutant that showed reduced activity relative to that of wild-type. The effect of this mutation on dimerization was confirmed by gel-filtration experiments which revealed a monomeric enzyme. Taken together, these data suggest that the dimer observed in the crystal structure may be the functional unit of RsrI MTase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported, in part, by NIH Grant GM25621 to R.I.G., the Department of Biochemistry of the University of Illinois, and the Mrs Louise Cornelie Vermey Leonard Trust. The authors gratefully acknowledge the contributions of the reviewers for helpful comments that improved the clarity of the presentation. Funding to pay the Open Access publication charges for this article was provided by the Leonard Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pingoud A., Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubin R., Modrich P. EcoRI methylase. Physical and catalytic properties of the homogeneous enzyme. J. Biol. Chem. 1977;252:7265–7272. [PubMed] [Google Scholar]
- 3.Modrich P. Studies on sequence recognition by type II restriction and modification enzymes. CRC Crit. Rev. Biochem. 1982;13:287–323. doi: 10.3109/10409238209114231. [DOI] [PubMed] [Google Scholar]
- 4.Reich N., Mashhoon M. Presteady state kinetics of an S-adenosylmethionine-dependent enzyme. Evidence for a unique binding orientation requirement for EcoRI DNA methyltransferase. J. Biol. Chem. 1993;268:9191–9193. [PubMed] [Google Scholar]
- 5.Modrich P., Zabel D. EcoRI endonuclease. Physical and catalytic properties of the homogenous enzyme. J. Biol. Chem. 1976;251:5866–5874. [PubMed] [Google Scholar]
- 6.de laCampa A., Kale P., Springhorn S., Lacks S. Proteins encoded by the DpnII restriction gene cassette. Two methylases and an endonuclease. J. Mol. Biol. 1987;196:457–469. doi: 10.1016/0022-2836(87)90024-6. [DOI] [PubMed] [Google Scholar]
- 7.Dubey A., Mollet B., Roberts R. Purification and characterization of the MspI DNA methyltransferase cloned and overexpressed in E.coli. Nucleic Acids Res. 1992;20:1579–1585. doi: 10.1093/nar/20.7.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong A., Zhou L., Zhang X., Stickel S., Roberts R., Cheng X. Structure of the Q237W mutant of HhaI DNA methyltransferase: an insight into protein–protein interactions. Biol. Chem. 2004;385:373–379. doi: 10.1515/BC.2004.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaszubska W., Webb H., Gumport R. Purification and characterization of the M.RsrI DNA methyltransferase from Escherichia coli. Gene. 1992;118:5–11. doi: 10.1016/0378-1119(92)90242-h. [DOI] [PubMed] [Google Scholar]
- 10.Bheemanaik S., Chandrashekaran S., Nagaraja V., Rao D. Kinetic and catalytic properties of dimeric KpnI DNA methyltransferase. J. Biol. Chem. 2003;278:7863–7874. doi: 10.1074/jbc.M211458200. [DOI] [PubMed] [Google Scholar]
- 11.Mruk I., Cichowicz M., Kaczorowski T. Characterization of the LlaCI methyltransferase from Lactococcus lactis subsp. cremoris W15 provides new insights into the biology of type II restriction-modification systems. Microbio. 2003;149:3331–3341. doi: 10.1099/mic.0.26562-0. [DOI] [PubMed] [Google Scholar]
- 12.Shier V., Hancey C., Benkovic S. Identification of the active oligomeric state of an essential adenine DNA methyltransferase from Caulobacter crescentus. J. Biol. Chem. 2001;276:14744–14751. doi: 10.1074/jbc.M010688200. [DOI] [PubMed] [Google Scholar]
- 13.Malygin E.G., Sclavi B., Zinoviev V.V., Evdokimov A.A., Hattman S., Buckle M. Bacteriophage T4Dam DNA-(adenine-N(6))-methyltransferase. Comparison of pre-steady state and single turnover methylation of 40mer duplexes containing two (un)modified target sites. J. Biol. Chem. 2004;279:50012–50018. doi: 10.1074/jbc.M409786200. [DOI] [PubMed] [Google Scholar]
- 14.Scavetta R., Thomas C., Walsh M., Szegedi S., Joachimiak A., Gumport R., Churchill M. Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases. Nucleic Acids Res. 2000;28:3950–3961. doi: 10.1093/nar/28.20.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janin J. Specific versus non-specific contacts in protein crystals. Nature Struct. Biol. 1997;4:973–974. doi: 10.1038/nsb1297-973. [DOI] [PubMed] [Google Scholar]
- 16.Osipiuk J., Walsh M., Joachimiak A. Crystal structure of MboIIA methyltransferase. Nucleic Acids Res. 2003;31:5440–5448. doi: 10.1093/nar/gkg713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szegedi S., Reich N., Gumport R. Substrate binding in vitro and kinetics of RsrI (N6-adenine) DNA methyltransferase. Nucleic Acids Res. 2000;28:3962–3971. doi: 10.1093/nar/28.20.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill S., vonHippel P. Calculation of protein extinction coefficients from amino-acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 19.Pues H., Bleimling N., Holz B., Wolcke J., Weinhold E. Functional roles of the conserved aromatic amino acid residues at position 108 (motif IV) and position 196 (motif VIII) in base flipping and catalysis by the N6-adenine DNA methyltransferase from Thermus aquaticus. Biochemistry. 1999;38:1426–1434. doi: 10.1021/bi9818016. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharya S., Dubey A. Kinetic mechanism of cytosine DNA methyltransferase MspI. J. Biol. Chem. 1999;274:14743–14749. doi: 10.1074/jbc.274.21.14743. [DOI] [PubMed] [Google Scholar]
- 21.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 22.Thompson J., Higgins D., Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul S., Madden T., Schaffer A., Zhang J., Zhang Z., Miller W., Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z., Schaffer A.A., Miller W., Madden T.L., Lipman D.J., Koonin E.V., Altschul S.F. Protein sequence similarity searches using patterns as seeds. Nucleic Acids Res. 1998;26:3986–3990. doi: 10.1093/nar/26.17.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss J. The Hill equation revisited: uses and misuses. FASEB J. 1997;11:835–841. [PubMed] [Google Scholar]
- 26.Hill A. The possible effects of aggregation of the molecules of haemoglobin on its dissociation curves. J. Physiol. 1910;40:iv–vii. [Google Scholar]
- 27.Neet K. Cooperativity in enzyme function: equilibrium and kinetic aspects. Methods Enzymol. 1980;64:139–192. doi: 10.1016/s0076-6879(80)64009-9. [DOI] [PubMed] [Google Scholar]
- 28.Malone T., Blumenthal R., Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 29.Szegedi S., Gumport R. DNA binding properties in vivo and target recognition domain sequence alignment analyses of wild-type and mutant RsrI [N6-adenine] DNA methyltransferases. Nucleic Acids Res. 2000;28:3972–3981. doi: 10.1093/nar/28.20.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoo O., Dwyer-Hallquist P., Agarwal K. Purification and properties of the HpaI methylase. Nucleic Acids Res. 1982;10:6511–6519. doi: 10.1093/nar/10.20.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goedecke K., Pignot M., Goody R., Scheidig A., Weinhold E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nature Struct. Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 32.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. ChemBioChem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 33.Malygin E.G., Evdokimov A.A., Zinoviev V.V., Ovechkina L.G., Lindstrom W.M., Reich N.O., Schlagman S.L., Hattman S. A dual role for substrate S-adenosyl-L-methionine in the methylation reaction with bacteriophage T4 Dam DNA-[N6-adenine]-methyltransferase. Nucleic Acids Res. 2001;29:2361–2369. doi: 10.1093/nar/29.11.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guex N., Peitsch M. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J., Fritsch E., Maniatis T. Molecular Cloning. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. p. 1.41. [Google Scholar]
- 36.O'Neil M., editor. 13th edn. Whitehouse Station, NJ, USA: The Merck Index, Merck & Co., Inc.; 2001. p. 29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.