Abstract
Although considerable progress has been made toward characterizing human DNA sequence variation, there remains a deficiency in information on human phenotypic variation at the single-gene level. We systematically analyzed the function of all protein-altering variants of eleven membrane transporters in heterologous expression systems. Coding-region variants were identified by screening DNA from a large sample (n = 247-276) of ethnically diverse subjects. In total, we functionally analyzed 88 protein-altering variants. Fourteen percent of the polymorphic variants (defined as variants with allele frequencies ≥1% in at least one major ethnic group) had no activity or significantly reduced function. Decreased function variants had significantly lower allele frequencies and were more likely to alter evolutionarily conserved amino acid residues. However, variants at evolutionarily conserved positions with approximately normal activity in cellular assays were also at significantly lower allele frequencies, suggesting that some variants with apparently normal activity in biochemical assays may influence occult functions or quantitative degrees of function that are important in human fitness but not measured in these assays. For example, eight (14%) of the 58 variants for which we had measured the transport of at least two substrates showed substrate-specific defects in transport. These variants and the reduced function variants provide plausible candidates for disease susceptibility or variation in clinical drug response.
Since the completion of the human genome project, considerable progress has been made in characterizing the nature and degree of human DNA sequence variation (Cargill et al. 1999; Halushka et al. 1999; Patil et al. 2001; Stephens et al. 2001; Leabman et al. 2003). Numerous single nucleotide polymorphisms in all regions of the human genome have been identified, including coding and noncoding regions of genes and large intergenic regions. However, there remains a lack of information connecting genetic polymorphisms to human phenotypic variation. As a consequence, current attempts to predict the functional significance of genetic variants from sequence data alone (from interspecies sequence comparisons or degree of chemical change for amino acid substitutions, as examples) (Sunyaev et al. 2000, 2001, 2003; Chasman and Adams 2001; Fay et al. 2001; Muller et al. 2001; Ng and Henikoff 2001, 2003; Wang and Moult 2001; Ramensky et al. 2002) are lacking in support from empirical data. Furthermore, we have little information on the fraction of natural variants with altered or potentially deleterious function that are harbored in healthy populations. In order to understand variation in normal human physiology and responses to the environment (including, for example, drug-response phenotypes), it is important to understand the range of phenotypic variation that is associated with genetic variation in human populations.
Analysis of causal mutations in Mendelian diseases has demonstrated that the majority of these diseases are caused by rare nonsynonymous variants, specifically, amino acid substitutions (Risch 2000). Splice-site mutations and insertions or deletions, although rarer occurrences, account for most other causative mutations in Mendelian diseases. The preponderance of nonsynonymous variants in disease association may also be true for more common disease (Risch 2000). Several recent studies have revealed that among disease-associated polymorphisms with the strongest evidence for true association, almost all are amino acid substitutions (Risch 2000; Hirschhorn et al. 2002). Attempts have been made to estimate the fraction of nonsynonymous variants in the human genome that is functionally deleterious, with estimates ranging from 10% to upward of 50% of all nonsynonymous mutations (Fay et al. 2001; Sunyaev et al. 2001; Ng and Henikoff 2002).
One of the primary goals of current large-scale sequencing projects is to identify SNPs that may be used in candidate gene association studies. However, a major obstacle in designing association studies is choosing appropriate SNPs to genotype. One strategy is to choose SNPs that are expected a priori to affect protein function and are therefore more likely to be associated with an altered phenotype. A variety of algorithms and bioinformatics tools have been developed in recent years to predict the functional consequences of protein-altering variants (Ng et al. 2000; Muller et al. 2001; Ng and Henikoff 2001, 2003). These algorithms attempt to predict the effect of an amino acid substitution on protein function based on the nature of the chemical change, the structural location of the substitution, and/or the evolutionary conservation of the residue, rather than from direct measurement of the function of the variant protein. As amino acid substitutions are particularly amenable to study in biochemical assays, systematic investigation of the functional consequences of these variants in cellular assays represents a first step toward cataloging human phenotypic variation.
Solute Carrier (SLC) transporters maintain cellular and total body homeostasis by importing nutrients and exporting cellular waste products and toxic compounds. These transporters also play a critical role in drug response, serving as drug targets and facilitating drug absorption, metabolism, and elimination. The SLC superfamily is comprised of transporters from a wide range of functional classes, including neurotransmitter, nutrient, heavy metal, and xenobiotic transporters. Genetic defects in SLC transporters have been associated with a variety of Mendelian diseases, including metabolic disorders such as glucose-galactose malabsorption (Pascual et al. 2004) and neurologic disorders such as peripheral neuropathy with agenesis of the corpus callosum (ACCPN) (Howard et al. 2002), demonstrating the diverse physiological functions of these proteins.
In this study, we characterized the function of protein-altering variants of 11 SLC transporters belonging to three different families: SLC22, SLC28, and SLC29. These transporters are present in a variety of epithelial tissues and have diverse biological roles. Although several of these transporters have specific functions that are important for normal human physiology, they are all capable of transporting xenobiotic small molecules (i.e., drugs), and were selected for screening as candidate genes to explain variability in drug response. In addition to identifying and functionally characterizing all naturally occurring protein-altering variants of these transporter genes, we attempted to identify characteristics of protein-altering variants that are predictive of alterations in protein function, both in biochemical assays and in vivo.
Results
Fourteen percent of all protein-altering polymorphisms identified in 11 SLC transporters have decreased function in biochemical assays
We systematically analyzed the function of all protein-altering variants of 11 membrane transporters in the Solute Carrier families SLC22, SLC28, and SLC29 in heterologous expression systems. Coding-region variants were identified by screening many DNA samples (n = 247-276) from ethnically diverse human populations. The transporters are dispersed throughout the human genome on five chromosomes, although some pairs (OCT1-OCT2, OAT1-OAT3, OCTN1-OCTN2) are found in tandem at a single locus and presumably arose by gene duplication (Table 1). The amino acid diversity (πNS) of the nine transporters ranges from 0.11 × 10-4 to 8.7 × 10-4. Previous large-scale sequencing studies have found that the average amino acid diversity in the human genome is ∼2.0 × 10-4 (Cargill et al. 1999; Leabman et al. 2003). Therefore, this subset of genes includes a representative sampling of genetic diversity within the human genome. We expressed all protein-altering variants in heterologous systems and determined activity by measuring the uptake of radiolabeled probe substrates (Fig. 1). We include in the analysis new information on functional variation in four membrane transporter genes and pooled analysis of variants of seven membrane transporters for which functional data have been reported previously (Leabman et al. 2002; Osato et al. 2003; Shu et al. 2003; Gray et al. 2004; Badagnani et al. 2005; Fujita et al. 2005; Owen et al. 2005). In total, functional analysis of 88 protein-altering variants is presented, including 80 amino acid substitutions, two insertions, four deletions, and two nonsense mutations.
Table 1.
Summary of population genetic statistics of eleven membrane transporters in the SLC family
| Gene | HGNC name | Chromosome | PAVa(#) | RFVb(#) | πcoding (× 104)c | πS (× 104) | πNS (× 104) | πNS/πS | Orthologsd |
|---|---|---|---|---|---|---|---|---|---|
| OCT1 | SLC22A1 | 6q26 | 15 | 5 | 6.58 ± 4.78 | 11.20 ± 11.00 | 5.11 ± 4.40 | 0.46 | 7 |
| OCT2 | SLC22A2 | 6q26 | 9 | 1 | 6.99 ± 4.99 | 22.54 ± 17.50 | 2.23 ± 2.64 | 0.10 | 7 |
| OCTN1 | SLC22A4 | 5q31.1 | 6 | 3 | 8.13 ± 5.58 | 13.98 ± 12.65 | 6.25 ± 5.04 | 0.45 | 5 |
| OCTN2 | SLC22A5 | 5q31.1 | 8 | 1 | 6.58 ± 4.77 | 23.93 ± 18.01 | 1.01 ± 1.70 | 0.04 | 6 |
| OAT1 | SLC22A6 | 11q13.1-q13.2 | 6 | 1 | 1.25 ± 1.70 | 3.87 ± 5.88 | 0.37 ± 1.02 | 0.10 | 7 |
| OAT3 | SLC22A8 | 11q11 | 10 | 5 | 4.25 ± 3.57 | 15.20 ± 13.42 | 0.74 ± 1.46 | 0.05 | 6 |
| CNT1 | SLC28A1 | 15q25-26 | 12 | 2 | 11.96 ± 7.24 | 22.55 ± 16.51 | 8.56 ± 5.98 | 0.38 | 5 |
| CNT2 | SLC28A2 | 15q15 | 5 | 0 | 7.64 ± 5.09 | 7.61 ± 8.22 | 7.64 ± 5.47 | 1.00 | 6 |
| CNT3 | SLC28A3 | 9q22.2 | 10 | 1 | 5.54 ± 3.97 | 18.13 ± 14.19 | 1.81 ± 2.13 | 0.10 | 5 |
| ENT1 | SLC29A1 | 6p21.1-p21.2 | 2 | 0 | 0.38 ± 0.97 | 0.60 ± 2.47 | 0.30 ± 1.00 | 0.50 | 6 |
| ENT2 | SLC29A2 | 11q13 | 5 | 3 | 0.54 ± 1.18 | 1.81 ± 4.27 | 0.11 ± 0.61 | 0.06 | 4 |
PAV: protein-altering variants
RFV: reduced-function variants
Values of π are listed as mean ± standard deviation. πcoding is the nucleotide diversity in the entire coding region of each gene, πS is the nucleotide diversity at synonymous sites and πNS is the nucleotide diversity at non-synonymous sites
Number of orthologs used in the alignments for manual scoring of evolutionarily conserved residues
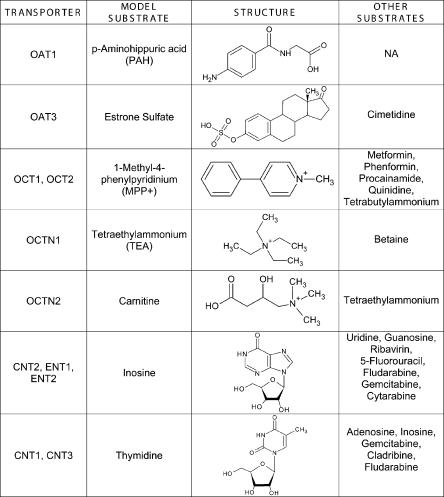
Figure 1.
Model substrates of 11 SLC transporters. The names and chemical structures of the model substrates used to functionally characterize each transporter are shown.
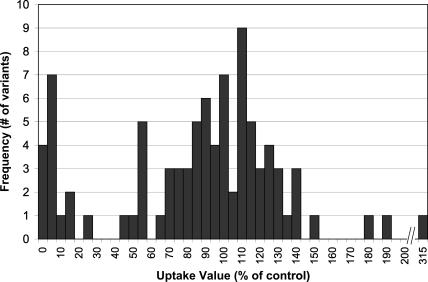
The distribution of uptake values for all variants analyzed is shown in Figure 2. The uptake values show a multimodal distribution, with breaks in the distribution at 25%-40%, 55%-60%, and 150%-175% of the activity of the control. Twenty-two (25%) of the 88 variants tested exhibited decreased transport function (defined as uptake <60% of control) (Table 2). Of the 88 protein-altering variants tested, 50 (57%) of the variants were polymorphic (defined as allele frequency ≥1% in at least one ethnic population), and seven (14%) of those 50 polymorphisms had decreased transport function. Interestingly, three variants appeared to be hyperfunctional, that is, had uptake values >150% of the control. These three variants shared the properties (discussed below) of the other variants with greater than 60% activity. Therefore, we used a bimodal retained-function vs. reduced-function model (as opposed to a trimodal normal-function vs. altered-function model) to analyze the data.
Figure 2.
Distribution of uptake values for protein-altering variants in 11 SLC transporters. Initial rate of uptake of radiolabeled probe substrate was measured and the results expressed as a percent of the activity of the reference sequence clone after subtracting background uptake. Uptake values used to construct the histogram reflect the mean of several experiments.
Table 2.
Characteristics of variants that exhibit reduced function in biochemical assays
| Variant | Total freq.a | AA freq.b | EA freq. | AS freq. | ME freq. | Grantham | EC/EUc | Loop/TMDd |
|---|---|---|---|---|---|---|---|---|
| OCT1-R61C | 0.036 | 0.000 | 0.072 | 0.000 | 0.056 | 85 | EC | L |
| OCT1-G220V | 0.002 | 0.005 | 0.000 | 0.000 | 0.000 | 109 | EC | T |
| OCT1-P341L | 0.041 | 0.082 | 0.000 | 0.117 | 0.000 | 98 | EU | L |
| OCT1-G401S | 0.009 | 0.007 | 0.011 | 0.000 | 0.000 | 56 | EC | L |
| OCT1-G465R | 0.020 | 0.000 | 0.040 | 0.000 | 0.000 | 125 | EC | L |
| OCT2-F45Ins | 0.002 | 0.000 | 0.005 | 0.000 | 0.000 | n/a | n/a | T |
| OCTN1-D165G | 0.002 | 0.000 | 0.000 | 0.008 | 0.000 | 94 | EC | L |
| OCTN1-M205I | 0.002 | 0.006 | 0.000 | 0.000 | 0.000 | 10 | EC | T |
| OCTN1-R282X | 0.002 | 0.006 | 0.000 | 0.000 | 0.000 | n/a | n/a | L |
| OCTN2-F17L | 0.004 | 0.000 | 0.000 | 0.017 | 0.000 | 22 | EC | L |
| OAT1-R454Q | 0.002 | 0.006 | 0.000 | 0.000 | 0.000 | 43 | EC | L |
| OAT3-R149S | 0.004 | 0.000 | 0.006 | 0.008 | 0.000 | 177 | EC | L |
| OAT3-Q239X | 0.002 | 0.000 | 0.000 | 0.008 | 0.000 | n/a | n/a | T |
| OAT3-I260R | 0.002 | 0.000 | 0.000 | 0.008 | 0.000 | 97 | EC | L |
| OAT3-R277W | 0.002 | 0.007 | 0.000 | 0.000 | 0.000 | 101 | EU | L |
| OAT3-I305F | 0.009 | 0.000 | 0.000 | 0.035 | 0.011 | 21 | EC | L |
| CNT1-V385Del | 0.015 | 0.030 | 0.000 | 0.000 | 0.000 | n/a | n/a | L |
| CNT1-S546P | 0.003 | 0.005 | 0.000 | 0.000 | 0.000 | 74 | EC | T |
| CNT3-G367R | 0.002 | 0.000 | 0.000 | 0.008 | 0.000 | 125 | EU | T |
| ENT2-D5Y | 0.003 | 0.005 | 0.000 | 0.000 | 0.000 | 160 | EU | L |
| ENT2-S184Del | 0.003 | 0.000 | 0.005 | 0.000 | 0.000 | n/a | n/a | L |
| ENT2-S282Del | 0.003 | 0.005 | 0.000 | 0.000 | 0.000 | n/a | n/a | L |
For OCT1, OCT2, CNT1, and ENT2 variants, total population includes 100 AA and 100 EA samples. For OCTN1, OCTN2, OAT1, OAT3, and CNT3, total population includes 80 AA, 80 EA, 60 AS, and 50 ME samples
(AA) African American; (EA) European American; (AS) Asian American; (ME) Mexican American
(EC) evolutionarily conserved; (EU) evolutionarily unconserved
Variant residue is located in predicted transmembrane domain (T) or loop region (L) of protein
Variants with decreased function are more likely to alter evolutionarily conserved amino acid residues
To learn about the characteristics of variants that decrease function and thus aid in the development of prediction tools, we evaluated the amino acid substitutions in our data set using several measures (based on degree of chemical change, evolutionary conservation, and/or location in the protein) and examined relationships between the nature of the amino acid substitution and protein function. For these analyses, only amino acid substitutions were considered, due to problems inherent in quantifying the chemical change or evolutionary conservativeness of insertions, deletions, and nonsense mutations. Of the five frameshift and nonsense mutations in the dataset, all showed virtually no activity.
The amino acid substitution matrix of Grantham (1974) is commonly used to measure the degree of chemical similarity or difference between alternative residues. The variants that retained function had lower Grantham values than the variants that decreased function (65 ± 48 vs. 87 ± 48, respectively), though this difference narrowly missed significance (P = 0.052 by one-tailed t-test).
The amino acid residues found in the transmembrane regions of proteins are highly conserved throughout evolution, owing to unique physical constraints on membrane-spanning helices (Leabman et al. 2003). Transmembrane regions had a lower fraction of reduced-function variants than loop regions (18% vs. 30%). However, the difference was not significant (χ2 = 1.60, P = 0.66).
We have used two methods to evaluate evolutionary conservation of the variant sites in our dataset. In the first method, based on multiple sequence alignment with known vertebrate orthologs, each amino acid substitution was scored as either evolutionarily conserved (EC) or evolutionarily unconserved (EU). We observed that 12 of the 35 (34%) EC variants resulted in decreased function compared with only four of the 45 (9%) EU variants (χ2 = 7.93, P = 0.047). Twelve of the 16 variants that resulted in loss of function were EC, giving a sensitivity of 75%. However, 23 of 64 variants that retained function also occurred at EC residues, giving a specificity of only 64%. In the second method, we used the prediction algorithm SIFT, which scores each variant as either “tolerant” or “intolerant” to the indicated substitution. Of the 80 single amino acid substitution variants, SIFT correctly predicted as “intolerant” 75% of the variants that decreased function and predicted as “tolerant” 75% of the variants that retained function. Overall, SIFT performed better, mispredicting only 20 variants (25%) compared with a misprediction of 27 variants (34%) by our EC/EU method.
Selection acts on variants with decreased function in cellular assays, and on variants that retain function in cellular assays but alter evolutionarily conserved residues
We plotted the fraction of variants with decreased function vs. allele frequency and compared that distribution with the distribution of the variants that retained function (Fig. 3A). Our results demonstrated that there was a significantly lower allele frequency distribution of variants with decreased function compared with those that retained function in cellular assays (Log-Rank test, P = 9.3 × 10-3). These data are consistent with decreased function variants being selected against. It is notable that all four ethnic populations had variants with decreased function. In fact, 17 of the 22 variants with decreased function (77%) were present in only a single ethnic or racial population sample. Fourteen of the 17 were singletons (only found on a single chromosome in our sample) and by definition, present in only one population. However, three were nonsingletons: OCT1-G465R (MAF = 0.04 in European Americans), OCTN2-F17L (MAF = 0.02 in Asian Americans), and CNT1-V385del (MAF = 0.03 in African Americans). The existence of functional variants common in some ethnicities and absent in others highlights the importance of considering race and ethnicity in human genetics, as different populations may carry a different set of deleterious polymorphisms, especially when the causative mutations are of low frequency (<10%) (Risch et al. 2002).
Figure 3.
Retention or loss of function in cellular assays predicts function in vivo. Variants were classified as having reduced function if they exhibited uptake values <60% of control. (A) Allele frequency distributions between variants that retained function vs. those that exhibited loss of function in cellular assays. The resulting curves were significantly different (Log-Rank test, P = 9.3 × 10-3), with a skew toward lower population allele frequencies for reduced-function variants. (B) Allele frequency distributions between variants at evolutionarily conserved (EC) positions that retained function vs. variants at evolutionarily unconserved (EU) positions that retained function. EC variants showed a significant shift toward lower allele frequencies (Log-Rank test, P = 0.02), even for variants that retained function in cellular assays.
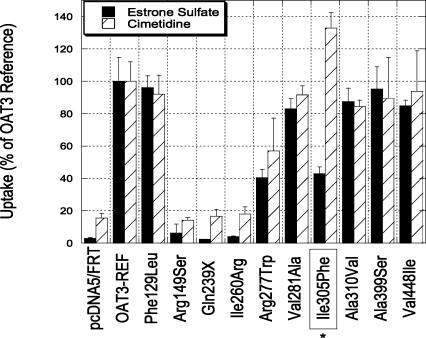
We then plotted the allele frequency distributions of the EC variants that retained function and the EU variants that retained function (Fig. 3B). Interestingly, the EC variants that retained function had an allele frequency distribution that was skewed toward lower frequencies and was significantly different from that of the EU variants that retained function (Log-Rank, P = 0.02). The data suggest that variants that appear to retain function in biochemical assays, but alter evolutionarily conserved residues, may affect some function important in organism fitness that is not measured in these assays. For example, this function may be an entirely different (i.e., nontransport) function mediated by the same gene, or may simply be the transport activity with respect to substrates that were not studied. Figure 4 shows one variant of OAT3, OAT3-I305F, which retained activity toward one substrate, the peptic ulcer drug, cimetidine, but had reduced activity toward the model substrate estrone sulfate, an endogenous steroid hormone.
Figure 4.
Functional characterization of protein-altering variants of OAT3 (SLC22A8). Uptake of estrone sulfate and cimetidine in HEK-293 cells expressing reference OAT3 and OAT3 protein-altering variants. Uptake values are expressed as a percentage of reference OAT3. Each value represents mean ± SD from triplicate wells in a representative experiment. (*) The selectivity variant Ile305Phe.
Since the uptake of multiple substrates had been measured for variants of nine of the 11 transporters in our dataset, we calculated the fraction of variants that showed substrate-specific changes in uptake activity. Of the 58 variants for which multiple substrates had been assayed, eight (14%) showed substrate-specific differences (Table 3). The distribution of allele frequencies for those eight substrate-specificity variants was comparable to that of the entire dataset, and contained both rare (<1%) EC variants and common (>10%) EU variants. Notably, however, the allele frequency distribution of these specificity variants was significantly different from that of the reduced function variants, with the specificity variants having higher allele frequencies than variants that exhibited reduced activity toward the prototypical substrate (Log-Rank, P = 0.01).
Table 3.
Number of variants in nine SLC transporters that change substrate specificity
| Gene | Substrates tested (#) | Variants tested (#) | Number of variants with altered substrate specificity (%) |
|---|---|---|---|
| OCT2 | 6 | 4 | 3 |
| 7 | 5 | 0 | |
| OCTN1 | 2 | 6 | 1 |
| OCTN2 | 2 | 8 | 1 |
| OAT3 | 2 | 10 | 1 |
| CNT1 | 2 | 4 | 1 |
| CNT2 | 2 | 3 | 0 |
| 3 | 1 | 1 | |
| CNT3 | 2 | 7 | 0 |
| 5 | 3 | 0 | |
| ENT1 | 2 | 1 | 0 |
| 7 | 1 | 0 | |
| ENT2 | 4 | 5 | 0 |
| Total | 58 | 8 (14%) |
Discussion
Our study suggests that healthy human populations harbor a significant number of severely reduced function polymorphisms and rare variants. In a set of 88 protein-altering variants from 11 membrane transporter genes, we found that 14% of the polymorphic (allele frequency ≥1% in at least one ethnic population) variants had decreased transport function (see Fig. 2). We then examined the variants to identify any characteristics that could be used to predict a reduction in function. First, we found that mutations that alter more than a single amino acid (e.g., frameshift and nonsense mutations) all showed virtually complete loss of function. For the amino acid substitutions, we examined the magnitude of the chemical change, and found that there was a trend toward larger chemical changes in variants with decreased function compared with those that retained function. These data are consistent with Miller and Kumar who demonstrated that amino acid substitutions associated with disease had higher Grantham values (larger chemical changes) than amino acid substitutions across species (Miller and Kumar 2001).
We then assessed the ability of evolutionary conservation to predict effects on protein function. Previous studies have demonstrated that EC residues are under stronger purifying selection than EU residues, suggesting that variation at EC residues is more likely to affect protein function than variation at EU residues. For example, Miller and Kumar demonstrated that nonsynonymous variants associated with disease occur at EC sites more frequently than expected by chance (Miller and Kumar 2001). We found that a simple measure of evolutionary conservation using a small number of known orthologs does reasonably well at predicting decreased function variants, but less well at predicting variants that retain function. The prediction algorithm SIFT, which uses a much larger set of homologous sequences, provides a similar sensitivity for prediction of functional significance, but had higher specificity compared with the EC/EU method. These results suggest that evolutionary conservation across larger distances may more accurately predict effects on protein function, or, in other words, that knowledge of the degree of conservation allows for more specific prediction of functionally significant amino acid substitutions (Botstein and Risch 2003). New efforts at sequencing the genomes of additional species should therefore facilitate the improvement of predictive algorithms (Cooper et al. 2003).
We found that our measure of protein function in cellular assays correlated significantly with allele frequency, a measure of effect on human fitness. That is, variants with grossly impaired function in biochemical assays were present at lower allele frequencies than variants that retained function, consistent with the idea that biochemical assays should be performed as a confirmatory measure for variants found to be associated with a disease phenotype. However, we found that alleles that altered evolutionarily conserved amino acid residues, but retained apparently normal function in biochemical assays, were also under negative or purifying selection. This finding suggests that even direct biochemical assay of variant protein function is not perfectly predictive of function in vivo, and that evolutionary conservation contains residual information independent of loss/retention of function in biochemical assays. An implication of this is that a negative finding in a biochemical assay of a disease-associated polymorphism is not necessarily evidence against a role of that variant in the phenotype of interest. This may be particularly important to the genetics of complex disease, in which the contribution of any individual risk-conferring polymorphism is expected to be very small, and thus may not be detectable in cellular assays.
Negative selection may act on variants that appear to “retain” function in cellular assays when those variants specifically alter occult functions of the protein that aren't measured in the assay or when small changes in protein function have large physiological consequences. For the membrane transport proteins in our study, possible occult functions include the transport of physiologically relevant substrates other than the model substrate. We examined this possibility by calculating the fraction of variants for which the transport of more than one substrate had been measured that showed substrate-specific changes in transport. Although relatively few variants (14%) showed substrate-specific changes in function, we likely underestimated the true fraction of these variants, since not all of the physiologically relevant substrates are known for each transporter and not all known substrates were tested. Variants with substrate-specific effects on function are probably not unique to membrane transporters, but common to all proteins that have multiple catalytic activities, multiple substrates, or multiple binding partners. This has important implications for pharmacogenetic association studies, since some of the protein variants that associate with variation in response to one drug may not associate with variation in the other drugs that interact with the same protein. Future biochemical assays of variant protein function should be interpreted with respect to how well the pertinent functions of the studied protein are known and how many of those functions are measured by the assay. Likewise, our best measure of evolutionary conservation (SIFT) failed to predict 25% of the reduced-function variants. Since measures of evolutionary conservation ignore species-specific physiology and are extremely sensitive to the availability of homologous sequence, they cannot substitute for direct measurement of protein function to predict and understand phenotypic diversity.
Early successes in pharmacogenetics (for example, the identification of the genetic determinants of polymorphism in debrisoquine metabolism [Gonzalez et al. 1988] or succinylcholine sensitivity [Lockridge 1990]), gave hope that the genetic determinants of drug-response phenotypes would be relatively easy to detect and verify. The underlying assumption of this optimism was that the genes involved in such nonessential functions as the elimination of xenobiotics would be under relatively little selective pressure, such that loss-of-function or severely reduced-function alleles would be expected to be found at high frequencies in healthy individuals. This assumption, that variation in “drug-response genes” is selectively neutral, may be incorrect. Experience has shown us that many genes that were discovered as drug-metabolizing enzymes or (as in this case) drug transport proteins are under significant negative selection. Whether this relates to possible homeostatic/physiological roles of these genes, or to an unrecognized importance of their protective effects in evolutionary fitness, is unknown. It is apparent, however, that high-frequency null alleles in drug-response genes have not often been observed. However, our identification of protein variants with substrate-specific changes in function suggests that there may exist variants in drug-response genes that may function normally with respect to endogenous or common environmental molecules, but show reduced function with respect to recently developed, clinically relevant drugs. These variants would be free of negative selection and could therefore reach high frequencies in healthy individuals. Indeed, of the small set of substrate selectivity variants identified in the current study, nearly half occurred at allele frequencies >10%. Thus, in addition to the reduced function variants identified here, these selectivity variants represent plausible candidates for association with drug-response phenotypes.
As we and others have seen, the relative frequency of deleterious mutations (the allelic spectrum) varies from gene to gene, and it may be that for many genes, association with a particular phenotype cannot be explained by one or a small number of high-frequency variants, even when the effect of that gene is significant and the phenotype is relatively common. A well-studied example is the association between the MC4R gene and obesity, in which no single variant occurs at a sufficiently high frequency to establish a significant association, yet the sum of deleterious variants of the MC4R gene has been shown consistently to be higher in obese individuals than in non-obese controls (Vaisse et al. 1998; Yeo et al. 1998 Hirschhorn and Altshuler 2002). A more recent example is the association of rare amino acid substitutions in several candidate genes (ABCA1, APOA1, and LCAT) with variation in plasma HDL cholesterol levels, in which various deleterious amino acid substitutions in these genes were found to be more common in individuals with low levels of HDL than in those with high HDL (Cohen et al. 2004). For pharmacogenetics, in cases similar to this, it may be possible to study the role of a candidate gene prospectively, by taking a “genotype-to-phenotype” approach. That is, identification of (rare) variants of severely reduced function, followed by phenotyping the (rare) individuals carrying these variants by drug administration. The knowledge that a particular variant alters function in cellular assays will greatly strengthen our confidence in positive associations found using such a strategy.
Methods
Variant identification
The coding regions (all exons and 50-100 bp of flanking intronic region per exon) of 11 membrane transporter genes [SLC22A1 (OCT1), U77086; SLC22A2 (OCT2), X98333; SLC22A4 (OCTN1), NM_003059; SLC22A5 (OCTN2), NM_003060; SLC22A6 (OAT1), AF097490; SLC22A8 (OAT3), NM_004254; SLC28A1 (CNT1), U62968; SLC28A2 (CNT2), U84392; SLC28A3 (CNT3), AF305210; SLC29A1 (ENT1), U81375; SLC29A2 (ENT2), AF029358] were screened for polymorphism by denaturing HPLC or by direct sequencing of a large number of DNA samples collected from ethnically diverse populations. Set I genes (SLC22A1, SLC22A2, SLC28A1, SLC28A2, SLC29A1, and SLC29A2) were screened using ethnically identified DNA samples (100 African-Americans and 100 European-Americans) from the Coriell Institute; Set II genes (SLC22A4, SLC22A5, SLC22A6, SLC22A8, and SLC28A3) were screened using a cohort of individuals (80 African-Americans, 80 European-Americans, 60 Asian-Americans, 50 Mexican-Americans, and six Pacific Islanders) from the San Francisco Bay Area enrolled in the SOPHIE project (Studies of Pharmacogenetics in Ethnically Diverse Populations). Nucleotide diversity (π), which is the average proportion of nucleotide differences between all possible pairs of sequences in the sample, was used to estimate nucleotide diversity at synonymous sites (πS) and amino acid-altering or nonsynonymous sites (πNS) (Tajima 1989; Hartl and Clark 1997). All variants identified and their ethnic-specific allele frequencies have been deposited in the public databases PharmGKB (http://www.pharmgkb.org) and dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/).
Functional characterization
Uptake studies for OCT1, OCT2, OAT1, CNT1, CNT2, CNT3, and ENT2 were performed using Xenopus laevis oocytes as described previously (Leabman et al. 2002; Shu et al. 2003; Gray et al. 2004; Badagnani et al. 2005; Fujita et al. 2005; Owen et al. 2005). Studies of ENT1 were performed in Saccharomyces cerevisiae using cytotoxicity and cell growth assays as described previously (Osato et al. 2003). Studies of OAT3, OCTN1, and OCTN2 were performed by transient transfection of HEK-293 cells using the Lipofectamine 2000 reagent as per the manufacturer's protocol (Invitrogen). Reference and variant cDNA clones in the expression vector pcDNA5/FRT (OAT3 and OCTN1) or pcDNA3 (OCTN2) were used to transfect HEK-293 cells at 90% confluence in 24-well poly-D-lysine coated plates (BD Discovery Labware) using 1 μg of DNA and 3 μg of Lipofectamine 2000 per well. Cells were assayed for activity at 48 h post-transfection by measurement of initial rate uptake of radiolabeled probe substrates: 0.1 μM 3H-estrone-3-sulfate (OAT3), 10 μM 14C-tetraethylammonium (OCTN1), or 1 μM 3H-L-carnitine (OCTN2). For each transporter, variant cDNAs were constructed by site-directed mutagenesis of the reference sequence clone, defined as the most common amino acid sequence in our sample. Initial rate of uptake of radiolabeled probe compound (Fig. 1) was measured for each variant, and the results expressed as a percent of the uptake of the reference sequence clone after subtracting background uptake. Experiments were performed several times and the average values from multiple experiments were used in the analysis.
Data Analysis
Prediction of function in cellular assays
The rate of uptake of the radiolabeled probe compound for each variant as a percentage of the reference sequence clone was used as the measure of biochemical function of the variant. The distribution of this variable was multimodal, with some variants having biochemical function <55% of the control and other variants having function of >60% of the control in cellular assays. We used the value of 60% of the control uptake value as a cut-off point to demarcate reduced or loss-of-function variants from “normal” function variants.
Each amino acid substitution was then evaluated for characteristics that might be expected to aid in prediction of functional activity, i.e., evolutionary conservation, degree of chemical change, and location in the protein (transmembrane domain vs. intracellular or extracellular loop regions). The degree of chemical change for each amino acid substitution was scored using the substitution matrix of Grantham (1974). For evolutionary conservation, two methods were used to score the variants. In the first method, the human amino acid sequence of each of our 11 transporter genes was aligned with three to seven known vertebrate orthologs (chimp, dog, mouse, rat, pig, cow, chicken, and/or frog). Residues that were identical across 80% or more of the reference sequences of all comparator species were classified as evolutionarily conserved (EC); all other residues were classified as evolutionarily unconserved (EU). The second method utilized the prediction algorithm SIFT (for Sort Intolerant From Tolerant amino acid substitutions) (Ng and Henikoff 2001). In contrast to our alignments with only a few orthologs, the SIFT algorithm generates alignments with a large number of homologous sequences and assigns scores to each residue, ranging from zero to one. Scores close to zero indicate evolutionary conservation and intolerance to substitution, while scores close to one indicate tolerance to substitution. SIFT scores <0.05 are predicted by the algorithm to be intolerant or deleterious amino acid substitutions, whereas scores >0.05 are considered tolerant. SIFT analysis was performed by allowing the algorithm to search for homologous sequences (i.e., without inputting known homologs) and using the default settings (SWISS-PROT 45 and TrEMBL 28 databases, median conservation score 3.00, remove sequences >90% identical to query sequence). To determine the structural location of each variant, the secondary structure of the reference sequence of each protein was estimated by hydropathy analysis (Pepplot, GCG sequence analysis suite). Each variant was scored as occurring in either the transmembrane domain (TMD) or loop region of the protein. The variables scored were tested as predictors of functional activity in biochemical assays using χ2 test for association.
Prediction of in vivo function
Alleles that are deleterious in nature suffer strong selection pressure and are therefore more likely to be found at low frequency. The allele frequency of each variant was used as an estimator for the effect on human fitness of that allele. The probability that an SNP had a minor allele frequency greater than some frequency, x, was modeled. Plots of these probability curves were generated, stratifying over dichotomous variables (function/no function in cellular assays, evolutionary conservation, SIFT) to determine whether these variables were predictive of allele frequency distribution, and thus might be correlated with gene function in vivo. The curves generated were analogous to Kaplan-Meier survival curves with allele frequency replacing time. These curves were compared using the Log-Rank Test.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) GM61390 and GM36780.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4356206.
References
- Badagnani, I., Chan, W., Castro, R.A., Brett, C.M., Huang, C.C., Stryke, D., Kawamoto, M., Johns, S.J., Ferrin, T.E., Carlson, E.J., et al. 2005. Functional analysis of genetic variants in the human concentrative nucleoside transporter 3 (CNT3; SLC28A3). Pharmacogenomics J. 5 157-165. [DOI] [PubMed] [Google Scholar]
- Botstein, D. and Risch, N. 2003. Discovering genotypes underlying human phenotypes: Past successes for mendelian disease, future approaches for complex disease. Nat. Genet. 33 228-237. [DOI] [PubMed] [Google Scholar]
- Cargill, M., Altshuler, D., Ireland, J., Sklar, P., Ardlie, K., Patil, N., Lane, C.R., Lim, E.P., Kalayanaraman, N., Nemesh, J., et al. 1999. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22 231-238. [DOI] [PubMed] [Google Scholar]
- Chasman, D. and Adams, R.M. 2001. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: Structure-based assessment of amino acid variation. J. Mol. Biol. 307 683-706. [DOI] [PubMed] [Google Scholar]
- Cohen, J.C., Kiss, R.S., Pertsemlidis, A., Marcel, Y.L., McPherson, R., and Hobbs, H.H. 2004. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science 305 869-872. [DOI] [PubMed] [Google Scholar]
- Cooper, G.M., Brudno, M., Green, E.D., Batzoglou, S., and Sidow, A. 2003. Quantitative estimates of sequence divergence for comparative analyses of mammalian genomes. Genome Res. 13 813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, J.C., Wyckoff, G.J., and Wu, C.I. 2001. Positive and negative selection on the human genome. Genetics 158 1227-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, T., Brown, C., Carlson, E.J., Taylor, T., de la Cruz, M., Johns, S.J., Stryke, D., Kawamoto, M., Fujita, K., Castro, R., et al. 2005. Functional analysis of polymorphisms in the organic anion transporter, SLC22A6 (OAT1). Pharmacogenet. Genomics 15 201-209. [DOI] [PubMed] [Google Scholar]
- Gonzalez, F.J., Skoda, R.C., Kimura, S., Umeno, M., Zanger, U.M., Nebert, D.W., Gelboin, H.V., Hardwick, J.P., and Meyer, U.A. 1988. Characterization of the common genetic defect in humans deficient in debrisoquine metabolism. Nature 331 442-446. [DOI] [PubMed] [Google Scholar]
- Grantham, R. 1974. Amino acid difference formula to help explain protein evolution. Science 185 862-864. [DOI] [PubMed] [Google Scholar]
- Gray, J.H., Mangravite, L.M., Owen, R.P., Urban, T.J., Chan, W., Carlson, E.J., Huang, C.C., Kawamoto, M., Johns, S.J., Stryke, D., et al. 2004. Functional and genetic diversity in the concentrative nucleoside transporter, CNT1, in human populations. Mol. Pharmacol. 65 512-519. [DOI] [PubMed] [Google Scholar]
- Halushka, M.K., Fan, J.B., Bentley, K., Hsie, L., Shen, N., Weder, A., Cooper, R., Lipshutz, R., and Chakravarti, A. 1999. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat. Genet. 22 239-247. [DOI] [PubMed] [Google Scholar]
- Hartl, D.L. and Clark, A.G. 1997. Principles of population genetics. Sinauer Associates, Sunderland, MA.
- Hirschhorn, J.N. and Altshuler, D. 2002. Once and again-issues surrounding replication in genetic association studies. J. Clin. Endocrinol. Metab. 87 4438-4441. [DOI] [PubMed] [Google Scholar]
- Hirschhorn, J.N., Lohmueller, K., Byrne, E., and Hirschhorn, K. 2002. A comprehensive review of genetic association studies. Genet. Med. 4 45-61. [DOI] [PubMed] [Google Scholar]
- Howard, H.C., Mount, D.B., Rochefort, D., Byun, N., Dupre, N., Lu, J., Fan, X., Song, L., Riviere, J.B., Prevost, C., et al. 2002. The K-Cl cotransporter KCC3 is mutant in a severe peripheral neuropathy associated with agenesis of the corpus callosum. Nat. Genet. 32 384-392. [DOI] [PubMed] [Google Scholar]
- Leabman, M.K., Huang, C.C., Kawamoto, M., Johns, S.J., Stryke, D., Ferrin, T.E., DeYoung, J., Taylor, T., Clark, A.G., Herskowitz, I., et al. 2002. Polymorphisms in a human kidney xenobiotic transporter, OCT2, exhibit altered function. Pharmacogenetics 12 395-405. [DOI] [PubMed] [Google Scholar]
- Leabman, M.K., Huang, C.C., DeYoung, J., Carlson, E.J., Taylor, T.R., de la Cruz, M., Johns, S.J., Stryke, D., Kawamoto, M., Urban, T.J., et al. 2003. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc. Natl. Acad. Sci. 100 5896-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockridge, O. 1990. Genetic variants of human serum cholinesterase influence metabolism of the muscle relaxant succinylcholine. Pharmacol. Ther. 47 35-60. [DOI] [PubMed] [Google Scholar]
- Miller, M.P. and Kumar, S. 2001. Understanding human disease mutations through the use of interspecific genetic variation. Hum. Mol. Genet. 10 2319-2328. [DOI] [PubMed] [Google Scholar]
- Muller, T., Rahmann, S., and Rehmsmeier, M. 2001. Non-symmetric score matrices and the detection of homologous transmembrane proteins. Bioinformatics 17 S182-S189. [DOI] [PubMed] [Google Scholar]
- Ng, P.C. and Henikoff, S. 2001. Predicting deleterious amino acid substitutions. Genome Res. 11 863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 12 436-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2003. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31 3812-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, P.C., Henikoff, J.G., and Henikoff, S. 2000. PHAT: A transmembrane-specific substitution matrix. Predicted hydrophobic and transmembrane. Bioinformatics 16 760-766. [DOI] [PubMed] [Google Scholar]
- Osato, D.H., Huang, C.C., Kawamoto, M., Johns, S.J., Stryke, D., Wang, J., Ferrin, T.E., Herskowitz, I., and Giacomini, K.M. 2003. Functional characterization in yeast of genetic variants in the human equilibrative nucleoside transporter, ENT1. Pharmacogenetics 13 297-301. [DOI] [PubMed] [Google Scholar]
- Owen, R.P., Gray, J.H., Taylor, T.R., Carlson, E.J., Huang, C.C., Kawamoto, M., Johns, S.J., Stryke, D., Ferrin, T.E., and Giacomini, K.M. 2005. Genetic analysis and functional characterization of polymorphisms in the human concentrative nucleoside transporter, CNT2. Pharmacogenet. Genomics 15 83-90. [DOI] [PubMed] [Google Scholar]
- Pascual, J.M., Wang, D., Lecumberri, B., Yang, H., Mao, X., Yang, R., and De Vivo, D.C. 2004. GLUT1 deficiency and other glucose transporter diseases. Eur. J. Endocrinol. 150 627-633. [DOI] [PubMed] [Google Scholar]
- Patil, N., Berno, A.J., Hinds, D.A., Barrett, W.A., Doshi, J.M., Hacker, C.R., Kautzer, C.R., Lee, D.H., Marjoribanks, C., McDonough, D.P., et al. 2001. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294 1719-1723. [DOI] [PubMed] [Google Scholar]
- Ramensky, V., Bork, P., and Sunyaev, S. 2002. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 30 3894-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch, N.J. 2000. Searching for genetic determinants in the new millennium. Nature 405 847-856. [DOI] [PubMed] [Google Scholar]
- Risch, N., Burchard, E., Ziv, E., and Tang, H. 2002. Categorization of humans in biomedical research: Genes, race and disease. Genome Biol. 3 comment2007. [DOI] [PMC free article] [PubMed]
- Shu, Y., Leabman, M.K., Feng, B., Mangravite, L.M., Huang, C.C., Stryke, D., Kawamoto, M., Johns, S.J., DeYoung, J., Carlson, E., et al. 2003. Evolutionary conservation predicts function of variants in the human organic cation transporter, OCT1. Proc. Natl. Acad. Sci. 100 5902-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, J.C., Schneider, J.A., Tanguay, D.A., Choi, J., Acharya, T., Stanley, S.E., Jiang, R., Messer, C.J., Chew, A., Han, J.H., et al. 2001. Haplotype variation and linkage disequilibrium in 313 human genes. Science 293 489-493. [DOI] [PubMed] [Google Scholar]
- Sunyaev, S., Ramensky, V., and Bork, P. 2000. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 16 198-200. [DOI] [PubMed] [Google Scholar]
- Sunyaev, S., Ramensky, V., Koch, I., Lathe III, W., Kondrashov, A.S., and Bork, P. 2001. Prediction of deleterious human alleles. Hum. Mol. Genet. 10 591-597. [DOI] [PubMed] [Google Scholar]
- Sunyaev, S., Kondrashov, F.A., Bork, P., and Ramensky, V. 2003. Impact of selection, mutation rate and genetic drift on human genetic variation. Hum. Mol. Genet. 12 3325-3330. [DOI] [PubMed] [Google Scholar]
- Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisse, C., Clement, K., Guy-Grand, B., and Froguel, P. 1998. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat. Genet. 20 113-114. [DOI] [PubMed] [Google Scholar]
- Wang, Z. and Moult, J. 2001. SNPs, protein structure, and disease. Hum. Mutat. 17 263-270. [DOI] [PubMed] [Google Scholar]
- Yeo, G.S., Farooqi, I.S., Aminian, S., Halsall, D.J., Stanhope, R.G., and O'Rahilly, S. 1998. A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat. Genet. 20 111-112. [DOI] [PubMed] [Google Scholar]