Abstract
RTA (replication and transcription activator; also referred to as ORF50, Lyta, and ART), an immediate-early gene product of Kaposi's sarcoma-associated herpesvirus (KSHV)/human herpesvirus 8, disrupts latency and drives lytic replication. RTA activates the expression of polyadenylated nuclear (PAN) RNA (also known as T1.1 or nut-1) of KSHV. This novel noncoding PAN RNA is the most abundant lytic transcript of KSHV; therefore, studying PAN RNA expression serves as a model system for understanding how RTA transactivates target genes during lytic replication. The RTA-responsive element of the PAN promoter (pPAN RRE) was previously identified, and our data suggested direct binding of full-length RTA to the pPAN RRE. Here, we present a detailed analysis of specific interactions between RTA and the PAN promoter. We expressed and purified the DNA-binding domain of RTA (Rdbd) to near homogeneity and measured its affinity for the pPAN RRE. In electrophoretic mobility shift assays (EMSAs), the dissociation constant (Kd) of Rdbd on the pPAN RRE was determined to be approximately 8 × 10−9 M, suggesting a strong interaction between RTA and DNA. The specificity of RTA binding to the PAN promoter was confirmed with supershift assays. The Rdbd binding sequences on the PAN promoter were mapped within a 16-bp region of the pPAN RRE by methylation interference assays. However, the minimal DNA sequence for Rdbd binding requires an additional 7 bp on both sides of the area mapped by interference assays, suggesting that non-sequence-specific as well as sequence-specific interactions between RTA and DNA contribute to high-affinity binding. To better understand the molecular interactions between RTA and the PAN promoter, an extensive mutagenesis study on the pPAN RRE was carried out by using EMSAs and reporter assays. These analyses revealed base pairs critical for both Rdbd binding in vitro and RTA transactivation in vivo of the PAN promoter. The results from methylation interference, deletion analysis, and mutagenesis using EMSAs and reporter assays were closely correlated and support the hypothesis that RTA activates PAN RNA expression through direct binding to DNA.
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8, has been established as an etiological agent of all clinical forms of Kaposi's sarcoma (KS), including AIDS-associated KS, classic KS, endemic forms of KS, and renal transplant-related KS (1, 3, 20, 22, 31, 34, 38, 43, 47). KSHV is also associated with two AIDS-related lymphoproliferative diseases, primary effusion lymphoma (PEL) (6) and multicentric Castleman's disease (42). Sequence analysis of KSHV revealed that KSHV belongs to a gammaherpesvirus subfamily which also includes herpesvirus saimiri, murine gammaherpesvirus 68 (MHV-68), and Epstein-Barr virus (7, 35, 39).
Like other herpesviruses, KSHV exhibits two distinct phases of its life cycle: latency and lytic replication. Although latent infection with KSHV has been proposed to be critical for tumorigenesis, it is clear that lytic replication is also important in KSHV pathogenesis. Antibodies against lytic proteins are significantly elevated in the sera of patients developing KS (50). Furthermore, treatment with ganciclovir, a drug which inhibits lytic replication of herpesviruses, significantly reduced the incidence of KS development in AIDS patients at high risk for KS (30). Most tumor cells are latently infected with KSHV; however, the virus undergoes lytic replication in a small number of cells in the tumor lesion (15, 38, 43, 44, 47, 57). Upon reactivation of KSHV from latency to lytic replication, these cells express viral chemokines and proinflammatory cytokines, such as viral macrophage inflammatory proteins and viral interleukin-6 (vIL-6) (33, 43, 47). These lytic gene products may play a key role in disease progression in an autocrine or paracrine fashion (4, 16, 32). Therefore, it is important to study the molecular mechanisms of reactivation for a better understanding of KSHV pathogenesis.
KSHV RTA (replication and transcription activator; also referred to as ORF50, Lyta, and ART) is an immediate-early gene product encoded primarily by open reading frame 50 (ORF50), which is well conserved among all gammaherpesviruses (28, 48, 51, 53). RTA plays a central role in the switch of the viral life cycle from latency to lytic replication. Ectopic expression of RTA is sufficient to disrupt latency and activate lytic replication to completion in KSHV latently infected PEL cell lines (17, 27, 46). A C-terminal truncated form of RTA (amino acids 1 to 530) acted as a dominant-negative mutant and inhibited the ability of RTA to reactivate KSHV from latency (26). Thus, the expression of KSHV RTA is sufficient and necessary for viral reactivation. Similar results have also been obtained from studies on MHV-68 RTA (53). Furthermore, MHV-68 RTA was shown to play a critical role in de novo infection, as well as in reactivation (52). In Epstein-Barr virus (a gamma-1 herpesvirus), ZEBRA (also referred to as BZLF1, Zta, or Z) plays a key role in disruption of viral latency in synergy with RTA (9, 10, 24, 36, 54).
The RTA homologues from gammaherpesviruses function as transcriptional activators. RTA autostimulates its own expression (12, 17, 37, 53). The DNA-binding domain (DBD) of RTA is highly conserved, suggesting important interactions between RTA and target gene promoters. KSHV RTA transactivates the expression of several downstream genes, including polyadenylated nuclear (PAN) RNA, kaposin (K12), ORF57, K-bZIP (K8, the ZEBRA homologue of KSHV), thymidine kinase, K5, ORF6 (single-stranded DNA binding protein), ORF59 (DNA polymerase-associated processivity factor), K14 (vOX-2), viral G protein-coupled receptor, and vIL-6 (8, 14, 19, 21, 25, 26, 41, 55; H. Deng, M. J. Song, J. Chu, and R. Sun, submitted for publication). KSHV RTA also interacts with known cellular factors such as the cyclic AMP-responsive element binding protein-binding protein, histone deacetylase-1, and Oct-1 and with a novel cellular protein, MGC2663 (named KSHV RTA binding protein) (18, 40, 49). However, the detailed molecular mechanism by which RTA activates target genes during lytic replication remains unclear. To date, both direct and indirect mechanisms have been suggested for RTA transactivation (11, 25, 36, 40, 41).
PAN RNA (also known as T1.1 or nut-1) is an early lytic gene transcript of KSHV with no apparent capacity to encode a protein (45, 57). PAN RNA is an RNA polymerase II transcript that is polyadenylated and forms a ribonucleoprotein complex located in the nuclear speckles (45, 56). PAN RNA is the most abundant lytic transcript of KSHV. Its copy number was as high as 2.5 × 105 to 5.0 × 105 per cell in PEL cells after induction of lytic replication. The abundance is apparent, as it can be visualized in an ethidium bromide-stained agarose gel (41, 45). The function of PAN RNA has yet to be elucidated. However, activation of PAN RNA expression by RTA offers an excellent model system to understand how this viral transactivator regulates high levels of lytic gene expression upon reactivation.
A previous study revealed a very strong RTA-responsive element (RRE) located in the PAN promoter and suggested direct binding of RTA to this element (41). Here, we have undertaken a detailed analysis of interactions between RTA and the PAN promoter RRE (pPAN RRE). We have mapped the RTA-binding site and identified base pairs of the pPAN RRE that are essential for RTA binding in vitro as well as transactivation in vivo.
MATERIALS AND METHODS
Cell culture.
All cells were cultured at 37°C in the presence of 5% CO2. 293T (a human embryonic kidney cell line transformed with the E1 region of adenovirus and the simian virus 40 T antigen) cells were cultured in Dulbecco's modified Eagle's medium (Cellgro) containing 10% fetal bovine serum and antibiotics (50 U of penicillin and 50 μg of streptomycin per ml).
Plasmids.
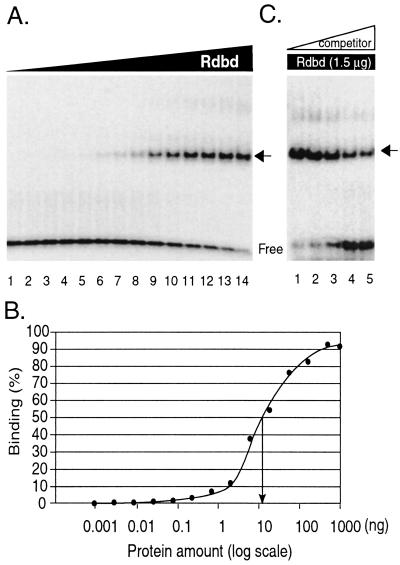
The luciferase reporter construct pLUC(−122) contains the PAN promoter region spanning nucleotides (nt) −122 to +14 amplified using a pair of primers, pan8b/KpnI and pan4/NheI, and cloned into a pGL3-basic plasmid (Promega, Madison, Wis.) as previously described (41). pLUC(−122) served as the wild-type template to generate a series of mutant constructs with two base-pair substitutions (see Fig. 6A and B), using PCR-directed site mutagenesis. For example, to construct the MJ75 mutant, which has the base pairs mutated at positions nt −75 and −74 in the PAN promoter (CC → AT), the distal end of the promoter region was amplified with the primers pan8b/KpnI and MJ75R (5′-CCACCCATTTTTATAAGCCACGCCCC-3′) and Pfuturbo DNA polymerase (Stratagene, La Jolla, Calif.). The proximal end of the promoter region was amplified with the primers MJ75F (5′-GGGGCGTGGCTTATAAAAATGGGTGG-3′) and pan4/NheI. In all sequences, uppercase letters represent viral sequences and mutated sequences are in boldface. These PCR products were subjected to a second round of PCR with pan8b/KpnI and pan4/NheI. The final PCR product was cloned into the pGL3-basic plasmid with KpnI and NheI cloning sites. A series of mutant constructs, MJ73 to MJ39 (see Fig. 6), were similarly prepared. The two base pair substitutions for each mutant construct were confirmed by sequencing. A mutant construct containing five base pair substitutions, MJmulti, was constructed as described above. The primer sequences used to generate the mutant constructs are listed in Table 1.
FIG. 6.
Binding of Rdbd to mutant oligonucleotides of the pPAN RRE in vitro. (A) A series of 2-bp mutations were introduced into the pPAN RRE from nt −75 to −38. The wild-type sequence is shown at the top. The same mutant sequences were incorporated into probes for EMSA as well as into reporter constructs. Probes used for EMSA contained the flanking sequences at both ends (Fig. 5A). (B) Single- and multiple-base-pair mutations introduced into the pPAN RRE. (C) EMSA of mutant probes of the pPAN RRE. Each mutant or wild-type (wt) probeCROSS-REF>was end labeled and incubated with 20 ng of Rdbd for EMSA. (D) Quantitative analysis of RTA binding in vitro to pPAN RRE mutant probes. Binding efficiency of each mutant probe was calculated as the ratio of bound to total, and the relative binding efficiencies are shown. Data are averages of the results from at least three different experiments with standard errors. (E) Critical base pairs of the pPAN RRE required for RTA binding in vitro. Closed ovals indicate base pairs with severe mutation effects (<30% of wild-type binding), and open ovals mark those with moderate effects (30 to 60% of wild-type binding).
TABLE 1.
Sequences of oligonucleotides used to construct mutants
| Name | Sequence (5′→3′)a |
|---|---|
| MJ75F | GGGGCGTGGCTTATAAAAATGGGTGG |
| MJ73F | GCGTGGCTTCCGCAAATGGGTGGC |
| MJ71F | GTGGCTTCCAACGATGGGTGGCATAC |
| MJ69F | GGCTTCCAAAACGGGGTGGCTAACC |
| MJ67F | GCTTCCAAAAATTTGTGGCTAACCTG |
| MJ65F | CTTCCAAAAATGGTGGGCTAACCTGTC |
| MJ63F | CCAAAAATGGGTTTCTAACCTGTCC |
| MJ61F | CAAAAATGGGTGGAGAACCTGTCCAAAATATG |
| MJ59F | ATGGGTGGCTCCCCTGTCCAAAATATG |
| MJ57F | GGGTGGCTAAAATGTCCAAAATATG |
| MJ55F | GGGTGGCTAACCGTTCCAAAATATG |
| MJ53F | GGTGGCTAACCTGGACAAAATATGG |
| MJ51F | GGCTAACCTGTCACAAATATGGGAAC |
| MJ49F | AACCTGTCCACCATATGGGAAC |
| MJ47F | CTAACCTGTCCAAACGATGGGAACACTGG |
| MJ45F | CCTGTCCAAAATCGGGGAACACTGGAG |
| MJ43F | CCTGTCCAAAATATTTGAACACTGGAG |
| MJ41F | CCAAAATATGGTCACACTGGAGATAAAAGG |
| MJ39F | CCAAAATATGGGTCAACTGAGATAAAAGG |
| MJmultiF | CGGGATCCGCTTCCAAAAATGGGTGTCTACCCGTGCCAAAATATGGGAACAGATCTTC |
| MJ75R | CCACCCATTTTTATAAGCCACGCCCC |
| MJ73R | GCCACCCATTTGCGGAAGCCACGC |
| MJ71R | GTTAGCCACCCATCGTTGGAAGCCAC |
| MJ69R | GGTTAGCCACCCCGTTTTGGAAGCC |
| MJ67R | CAGGTTAGCCACAAATTTTTGGAAGC |
| MJ65R | GACAGGTTAGCCCACCATTTTTGGAAG |
| MJ63R | GGACAGGTTAGAAACCCATTTTTGG |
| MJ61R | CATATTTTGGACAGGTTCTCCACCCATTTTTG |
| MJ59R | TTTTGGACAGGGGAGCCACCCATTTTTG |
| MJ57R | ATATTTTGGACATTTTAGCCACCC |
| MJ55R | CCCATATTTTGGAACGGTTAGCCAC |
| MJ53R | GTTCCCATATTTTGTCCAGGTTAGCC |
| MJ51R | GTGTTCCCATATTTGTGACAGGTTAG |
| MJ49R | GTTCCCATATGGTGGACAGGTTAG |
| MJ47R | CCAGTGTTCCCATCGTTTGGACAGGTTAG |
| MJ45R | CTCCAGTGTTCCCCGATTTTGGACAGG |
| MJ43R | CTCCAGTGTTCAAATATTGGACAGG |
| MJ41R | CCTTTTATCTCCAGTGTGACCATATTTTGG |
| MJ39R | CCTTTTATCTCAGTTGTCCCATATTTTG |
| MJmultiR | CCCATATTTTGGCACGGGTAGACACCCATTTTTG |
Mutations are in bold.
An RTA-expressing plasmid with a FLAG peptide sequence at the N terminus (pFLAG/RTA) contains the cDNA sequence of RTA ORF following the cytomegalovirus enhancer-promoter sequence as described previously (H. J. Brown, M. J. Song, H. Deng, T.-T. Wu, G. Cheng, and R. Sun, submitted for publication). For overexpression in bacteria, RTA (amino acids 1 to 691) and the Rdbd (amino acids 1 to 320) were PCR amplified from the cDNA clone of RTA tagged with FLAG, using the primers HB-FLAG (5′-agtctagagcctgcagccatggactacaaagac-3′) and TLS2 (5′-gaagatctctcgaggcacagacgcccttgcgaac-3′) for RTA and the primers Rdbd-1 (5′-gaagatctcccgggatggactacaaggacgacgatgacaag-3′) and Rdbd-2 (5′-gaagatctcgagcgggtctgtacccgatgggttaaag-3′) for Rdbd. The underlined nucleotides represent restriction enzyme sites. The PCR product for full-length RTA was digested with PstI and XhoI and blunt-ended with a fill-in reaction. This insert was cloned into a vector plasmid, pET11d (Novagen, Madison, Wis.), that was also blunt-ended after NcoI and BamHI digestion. The PCR product for Rdbd was digested with EcoRV and XhoI and cloned into pET22b(+), after the vector was digested with NdeI and XhoI. Rdbd expressed from pET22b was tagged with six histidine residues at the C terminus (Novagen).
Transfections.
For the luciferase reporter assays, 2.5 × 105 293T cells were transfected with 40 ng of pcDNA3 or pcDNA3/RTA and 200 ng of a PAN promoter reporter plasmid in 12-well plates by a calcium phosphate transfection method. Each transfection for reporter assays was performed in duplicate and included 10 ng of pRLCMV (Promega) as a control for transfection efficiencies, as well as 350 ng of a carrier DNA (plasmid DNA lacking any mammalian promoter-enhancer sequences). At 24 h posttransfection, cells were washed with 1× phosphate-buffered saline (PBS) and subjected to reporter assays.
To prepare nuclear extracts, 10 μg of either vector alone or pFLAG/RTA was transfected into 3 × 106 293T cells in a 100-mm plate by a calcium phosphate transfection method. Cells were harvested at 36 h posttransfection and washed with 1× PBS. Nuclear extracts were prepared according to the method of Dignam et al. (13).
Dual luciferase assay.
The dual luciferase reporter assay system (Promega) was used to test promoter activity. Transfected 293T cells in a 12-well plate were washed with 1× PBS and incubated with 250 μl of 1× passive lysis buffer provided by the manufacturer. Lysates were frozen, thawed once, and centrifuged at top speed in a microcentrifuge for 5 min. Supernatants of 293T cells were diluted 1/100 to obtain a reading in a linear range and assayed with an Optocomp I Luminometer (MGM Instruments, Hamden, Calif.). The reporter assays were carried out according to manufacturer's protocol for the dual luciferase reporter assay system (Promega).
Expression and purification of recombinant RTA and Rdbd proteins.
Full-length RTA tagged with a FLAG peptide at the N terminus was cloned into pET11d. Recombinant RTA protein was expressed in Escherichia coli and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Full-length RTA was purified from bacterial sonicates using a FLAG affinity column, as previously described (23).
Recombinant Rdbd protein tagged with a FLAG peptide at the N terminus and six histidine residues at the C terminus was expressed similarly to the full-length RTA protein. After 1 h of induction with 1 mM IPTG, cells were resuspended in a lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole [pH 8.0]). The Rdbd protein was purified using nickel-nitrilotriacetic acid (Ni-NTA) metal affinity chromatography (Qiagen, Valencia, Calif.). Briefly, the cell lysates were incubated with Ni-NTA agarose at 4°C for 4 h. The Ni-NTA agarose beads were vigorously washed with buffer (50 mM NaH2PO4, 300 mM NaCl, and 40 mM imidazole [pH 8.0]). Rdbd protein was then eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 100 mM imidazole [pH 8.0]). Protease inhibitor cocktail (Boehringer Mannheim, Mannheim, Germany) was used throughout the purification procedures.
Western analysis.
Nuclear extracts prepared from 293T cells transfected with either vector or pFLAG/RTA and purified RTA and Rdbd proteins were used for Western analysis. Protein samples (50 μg of nuclear extracts or 100 ng of purified recombinant proteins) were mixed with Laemmli buffer containing 0.25 M Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate (SDS), 5% β-mercaptoethanol (β-ME), 10% glycerol, and 0.002% bromophenol blue and separated by SDS-10% polyacrylamide gel electrophoresis (PAGE), along with a broad-range prestained protein molecular standard (Bio-Rad, Hercules, Calif.). Proteins were electrotransferred (Bio-Rad) onto nitrocellulose membranes (Amersham Pharmacia Biotech, Arlington Heights, Ill.). The membranes were blocked in 1× PBS containing 0.1% Tween 20 and 5% skim milk and incubated with either anti-FLAG monoclonal antibody (1:5,000) (Sigma, St. Louis, Mo.) or rabbit serum against RTA (1:2,000). Anti-RTA rabbit serum was generated by using purified Rdbd protein as an immunogen (Covance Research Products, Richmond, Calif.). The membranes were washed and incubated with secondary antibody, either anti-mouse immunoglobulin G conjugated with horseradish peroxidase (Amersham Pharmacia Biotech) or anti-rabbit immunoglobulin G conjugated with horseradish peroxidase (Amersham Pharmacia Biotech), respectively. The membranes were subjected to chemiluminescence detection (ECL+PLUS system; Amersham Pharmacia Biotech), and signals were detected with a STORM imaging system (Molecular Dynamics, Sunnyvale, Calif.).
EMSAs.
All double-stranded oligonucleotides used for electrophoretic mobility shift assays (EMSAs) were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.) and filled in with Klenow DNA polymerase (New England Biolabs) to obtain similar labeling efficiencies for each probe. Labeled probes were incubated with various amounts of either purified RTA or Rdbd protein for 30 min in a binding buffer [10 mM Tris-HCl (pH 7.5), 150 mM KCl, 5.7 mM MgCl2, 1 mM EDTA, 0.1 μg of poly(dI · dC), 5 μg of bovine serum albumin, 0.38 mM dithiothreitol, 0.38 mM phenylmethylsulfonyl fluoride, 50 mM β-ME, 5% glycerol]. The binding mixtures were loaded onto a 4.5% polyacrylamide gel in 1× TGE buffer (5 mM Tris, 190 mM glycine, and 1 mM EDTA [pH 8.3]) in the presence of 55 mM β-ME. The gel was run at 130 V at 4°C, dried, and exposed to a phosphorimager screen. Quantitative analysis of signals was performed with a STORM imaging system (Molecular Dynamics).
The labeled oligonucleotides pan1* (nt −78 to −37) and pan2* (nt −69 to −42) contain the PAN promoter sequence as well as a BamHI or BglII flanking sequence at the 5′ and 3′ ends, respectively (41). For supershift assays, the full-length RTA protein (60 ng) was incubated with increasing amounts (0, 1, and 3 μl) of anti-RTA rabbit serum for 20 min prior to the addition of pan2*. The purified Rdbd protein (20 ng) was incubated with pan1* for 30 min in the absence or presence of anti-FLAG antibody (2.2 μg; Sigma) or anti-RTA rabbit serum (1 μl).
To determine the apparent Kd, increasing amounts of purified Rdbd were incubated with pan1* (10−11 mol). Threefold serial dilutions from 500 ng of Rdbd were made, and final protein amounts per reaction were 0.0009, 0.0028, 0.0085, 0.025, 0.076, 0.229, 0.686, 2.06, 6.17, 18.5, 55.7, 167, and 500 ng. In addition, 1 μg of purified Rdbd was incubated with pan1*. To ensure that DNA was indeed limiting, EMSAs with various amounts of Rdbd were repeated with a fivefold-increased amount of the labeled probe (pan1*). This resulted in 50% binding of Rdbd at similar protein amounts, confirming that the DNA amount used in the initial titration was below the apparent Kd. The concentration of protein required to generate 50% binding is defined as the apparent Kd. The concentration of active protein needs to be determined to measure the true Kd (5). Once the apparent Kd was obtained, competition assays were performed to determine the true Kd. Purified Rdbd (1.5 μg) was incubated with 5, 10, 20, 40, or 60 pmol of the unlabeled oligonucleotide pan1.
For the mutagenesis study on Rdbd binding, a series of double-stranded oligonucleotides that contained two-base-pair, single-base-pair, or multiple-base-pair substitutions at various positions were generated (see Fig. 6A and B). The sequences of the oligonucleotides used in EMSAs are listed in Table 2. An oligonucleotide containing mutated sequences was annealed with either GSRRE rev or GSRRE rev2 to make double-stranded oligonucleotides. To construct deletion mutants, two oligonucleotides corresponding to each position were annealed. For example, 77/32 was generated from GSPAN(−77)F and GSPAN(−32)R.
TABLE 2.
Sequences of oligonucleotides used for EMSAs
| Name | Sequence (5′→3′)a |
|---|---|
| GSMJ75 | cgggatccGCTTATAAAAATGGGTGGCTAACCTGTCCAAAATATGGGAACAgatcttcg |
| GSMJ73 | cgggatccGCTTCCGCAAATGGGTGGCTAACCTGTCCAAAATATGGGAACAgatcttcg |
| GSMJ71 | cgggatccGCTTCCAACGATGGGTGGCTAACCTGTCCAAAATATGGGAACAgatcttcg |
| GSMJ69 | cgggatccGCTTCCAAAACGGGGTGGCTAACCTGTCCAAAATATGGGAACAgatcttcg |
| GSMJ67 | cgggatccGCTTCCAAAAATTTGTGGCTAACCTGTCCAAAATATGGGAACAgatcttcg |
| GSMJ65 | cgggatccGCTTCCAAAAATGGTGGGCTAACCTGTCCAAAATATGGGAACAgatcttcg |
| GSMJ63 | cgggatccGCTTCCAAAAATGGGTTTCTAACCTGTCCAAAATATGGGAACAgatcttcg |
| GSMJ61 | cgggatccGCTTCCAAAAATGGGTGGAGAACCTGTCCAAAATATGGGAACAgatcttcg |
| GSMJ59 | cgggatccGCTTCCAAAAATGGGTGGCTCCCCTGTCCAAAATATGGGAACAgatcttcg |
| GSMJ57 | cgggatccGCTTCCAAAAATGGGTGGCTAAAATGTCCAAAATATGGGAACAgatcttcg |
| GSMJ55 | cgggatccGCTTCCAAAAATGGGTGGCTAACCGTTCCAAAATATGGGAACAgatcttcg |
| GSMJ53 | cgggatccGCTTCCAAAAATGGGTGGCTAACCTGGACAAAATATGGGAACAgatcttcg |
| GSMJ51 | cgggatccGCTTCCAAAAATGGGTGGCTAACCTGTCACAAATATGGGAACAgatcttcg |
| GSMJ49 | cgggatccGCTTCCAAAAATGGGTGGCTAACCTGTCCACCATATGGGAACAgatcttcg |
| GSMJ47 | cgggatccGCTTCCAAAAATGGGTGGCTAACCTGTCCAAACGATGGGAACAgatcttcg |
| GSMJ45 | cgggatccGCTTCCAAAAATGGGTGGCTAACCTGTCCAAAATCGGGGAACAgatcttcg |
| GSMJ43 | cgggatccGCTTCCAAAAATGGGTGGCTAACCTGTCCAAAATATTTGAACAgatcttcg |
| GSMJ41 | cgggatccGCTTCCAAAAATGGGTGGCTAACCTGTCCAAAATATGGTCACAgatcttcg |
| GSMJ39 | cgggatccGCTTCCAAAAATGGGTGGCTAACCTGTCCAAAATATGGGACAAgatcttcg |
| GSMJ63S | cgggatccGCTTCCAAAAATTGGTTGCTAACCTGTCCAAAATATGGGAACAgatcttcg |
| GSMJ62S | cgggatccGCTTCCAAAAATGGGTGTCTAACCTGTCCAAAATATGGGAACAgatcttcg |
| GSMJ59S | cgggatccGCTTCCAAAAATGGGTGGCTCACCTGTCCAAAATATGGGAACAgatcttcg |
| GSMJ58S | cgggatccGCTTCCAAAAATGGGTGGCTACCCTGTCCAAAATATGGGAACAgatcttcg |
| GSMJ55S | cgggatccGCTTCCAAAAATGGGTGGCTAACCGGTCCAAAATATGGGAACAgatcttcg |
| GSMJ54S | cgggatccGCTTCCAAAAATGGGTGGCTAACCTTTCCAAAATATGGGAACAgatcttcg |
| GSMJ53S | cgggatccGCTTCCAAAAATGGGTGGCTAACCTGGCCAAAATATGGGAACAgatcttcg |
| GSMJ52S | cgggatccGCTTCCAAAAATGGGTGGCTAACCTGTACAAAATATGGGAACAgatcttcg |
| GSMJmulti | cgggatccGCTTCCAAAAATGGGTGTCTACCCGTGCCAAAATATGGGAACAgatcttcg |
| GSRRE rev | gctagatctGTT |
| GSRRE rev2 | cgaagatctGTT |
| GSPAN(−77)F | TCCAAAAATGGGTGGCTAACCTGT |
| GSPAN(−74)F | AAAAATGGGTGGCTAACCTGT |
| GSPAN(−71)F | AATGGGTGGCTAACCTGT |
| GSPAN(−32)R | TCCAGTGTTCCCATATTTTGGACAGGTTAGCCAC |
| GSPAN(−35)R | AGTGTTCCCATATTTTGGACAGGTTAGCCAC |
| GSPAN(−38)R | GTTCCCATATTTTGGACAGGTTAGCCAC |
| GSPAN(−41)R | CCCATATTTTGGACAGGTTAGCCAC |
| GSPAN(−44)R | ATATTTTGGACAGGTTAGCCAC |
| GSPAN(−47)R | TTTTGGACAGGTTAGCCAC |
| GSPAN(−50)R | TGGACAGGTTAGCCAC |
| GSPAN(−73)R | AAAATGGGTGGCTAACCTGT |
| GSPAN(−72)R | AAATGGGTGGCTAACCTGT |
| GSPAN(−45)R | TATTTTGGACAGGTTAGCCAC |
| GSPAN(−46)R | ATTTTGGACAGGTTAGCCAC |
Viral sequences are in capital letters; mutations are in bold.
Methylation interference assays.
pan1* was digested with either BglII or BamHI to generate a single-end-labeled probe for the forward or reverse strand. pan1* was incubated with 0.5% dimethyl sulfate (DMS) for 5 min in 200 μl of a reaction buffer (50 mM sodium cacodylate and 1 mM EDTA [pH 8]) at room temperature. The reaction was stopped with 40 μl of a stop buffer (1.5 M sodium acetate [pH 7] and 1.0 M β-ME). The concentration of DMS and duration of incubation were chosen after optimization to result in an average of one methylation per probe molecule. After purification, the modified probe was incubated with 125 ng of Rdbd protein for EMSA. Unbound and bound DNA fractions were purified from the gel with a QIAEX II DNA gel extraction system (Qiagen) and cleaved with 1 M piperidine at 90°C for 30 min. Cleaved DNA fragments were separated, along with a labeled 10-bp ladder (Life Technologies, Gaithersburg, Md.), on a 12% sequencing gel. The gel was dried and exposed to a phosphorimager screen. Quantitative analysis of signals was performed with a STORM imaging system (Molecular Dynamics).
RESULTS
Expression and purification of RTA and Rdbd.
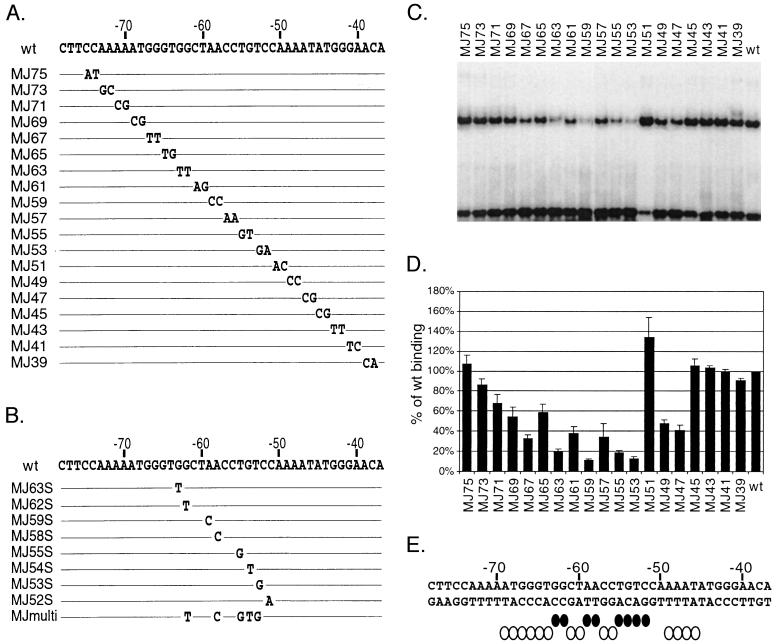
RTA has been shown to act as a strong transcription activator for several early and late transcripts of KSHV, including PAN RNA, ORF57, K-bZIP, kaposin, vIL-6, K14, viral G protein-coupled receptor, and thymidine kinase (14, 25, 41, 55; Deng et al., submitted). We previously undertook a deletion analysis of the PAN promoter and identified the RRE in a region between nt −69 and −38 of the PAN promoter. In addition, our data indicated RTA direct binding to this RRE, using bacterially expressed full-length RTA protein (amino acids 1 to 691) tagged with a FLAG peptide at the N terminus (Fig. 1A) (41). However, due to the limited expression level of full-length RTA protein in bacteria and difficulties in the purification process (i.e., degradation from the C terminus), we continued our studies with the N-terminal portion of RTA (amino acids 1 to 320). This region of RTA includes a DBD and a leucine zipper domain based on homology with other gammaherpesvirus RTA sequences. The Rdbd was tagged with a FLAG peptide at the N terminus and six histidine residues at the C terminus (Fig. 1A). Rdbd was purified to near homogeneity (Fig. 1B). The recombinant Rdbd migrated in SDS-PAGE at approximately 37 kDa, similar to its predicted molecular mass, 36.7 kDa. In addition, purified Rdbd was used to generate anti-RTA rabbit serum, which was utilized for Western analysis and EMSAs.
FIG. 1.
Expression of RTA and Rdbd. (A) Schematic diagram of recombinant RTA proteins. Full-length RTA protein (amino acids 1 to 691) was tagged with FLAG at the N terminus. Rdbd (amino acids 1 to 320) includes a putative DBD as well as a leucine zipper domain (LZ) tagged with FLAG at the N terminus and six histidine residues at the C terminus. AD, activation domain. (B) Representative Coomassie blue-stained gel of purified Rdbd protein. Rdbd protein was expressed in bacteria and purified with a Ni-NTA column. Eluates (E1 to E3) were analyzed by SDS-PAGE and stained with Coomassie blue. The arrow indicates purified Rdbd protein. M, molecular mass markers. (C) RTA and Rdbd expression in 293T cells and E. coli. Nuclear extracts were prepared from 293T cells transfected with either vector alone (lanes 1 and 5) or a FLAG-tagged RTA-expressing plasmid (pFLAG/RTA; lanes 2 and 6). Bacterially expressed recombinant full-length RTA (lanes 3 and 7) and Rdbd (lanes 4 and 8) were purified and separated on 10% gels with nuclear extracts from transfected 293T cells. Monoclonal anti-FLAG antibody (left) and polyclonal anti-RTA rabbit serum (right) were used for Western analysis.
It has been shown that RTA protein undergoes posttranslational modifications (i.e., phosphorylation) depending on the time after transfection (26; T. Symensma, unpublished data). To compare bacterially expressed proteins with the proteins expressed in eukaryotic cells, we transfected either a vector alone or a FLAG-tagged RTA-expressing plasmid (pFLAG/RTA) into 293T cells and prepared nuclear extracts at 36 h posttransfection. Two distinct bands were detected by Western analysis of nuclear extracts, using monoclonal anti-FLAG (Fig. 1C, lane 2), as well as polyclonal anti-RTA antiserum (Fig. 1C, lane 6). The apparent molecular masses of these bands are approximately 90 and 110 kDa. The 110-kDa species was proposed to be due mainly to a phosphorylated form of RTA (26). However, the role of phosphorylation in RTA function is not known. Western analysis of purified full-length RTA detected one major band at the same molecular mass as the unmodified form of RTA expressed in 293T cells (Fig. 1C, lanes 3 and 7), suggesting that bacterially expressed RTA, which lacks posttranslational modifications, is similar to the unmodified protein expressed in mammalian cells. Purified Rdbd was detected with anti-FLAG antibody and anti-RTA antibody at a molecular mass consistent with the predicted size (Fig. 1C, lanes 4 and 8).
Supershift of the protein-DNA complex.
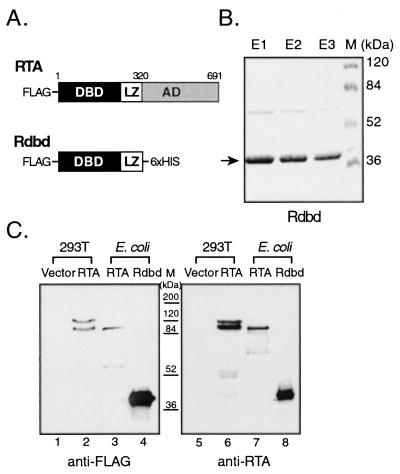
We previously showed that full-length purified RTA binds to a region of the PAN promoter in a dose-dependent and sequence-specific manner (41). Supershift of full-length RTA using anti-FLAG antibody was not possible in these experiments because the full-length RTA was purified from an affinity column by elution with an excess of FLAG peptides. However, availability of the antibody against the Rdbd made it possible to test the specificity of RTA binding to DNA.
Increasing amounts of anti-RTA rabbit serum (Fig. 2A) were incubated with full-length purified RTA prior to addition of pan2* (nt −69 to −42). Incubation with the anti-RTA antibody resulted in inhibition of protein-DNA complex formation, indicating the specificity of the RTA-DNA complex. Similar results were observed when purified Rdbd (20 ng) was incubated with pan1* (nt −78 to −37) in the presence of anti-RTA rabbit serum (Fig. 2B, lane 4). Incubation of Rdbd with anti-FLAG antibody yielded the supershift of the Rdbd-DNA complex (Fig. 2B, lane 3). We observed two different effects of the antibodies on the protein-DNA complex. When an antibody such as anti-RTA recognizes the DBD, the antibody may prevent DNA binding, leading to a reduction in the amount of protein-DNA complex. The tag peptide at the N terminus of the protein is unlikely to interfere with the binding of RTA, so that the anti-FLAG antibody may render the complex supershifted. These two results reinforce the evidence for RTA-specific binding to the pPAN RRE. Furthermore, incubation with irrelevant antibodies, such as anti-HA and antitrimethylguanosine and a polyclonal rabbit serum against an irrelevant protein, neither inhibited nor supershifted the Rdbd-DNA complex (data not shown). In addition, purified Rdbd was tested for its ability to bind various deletion probes of the pPAN RRE. EMSA results for the Rdbd protein with labeled double-stranded oligonucleotides, pan2* to pan6* (41), compared well with those obtained with full-length RTA (data not shown). Our results indicate that the purified recombinant Rdbd protein expressed in bacteria is functional in terms of DNA binding.
FIG. 2.
Specificity of the RTA-DNA and Rdbd-DNA complexes. (A) Supershift of the RTA-DNA complex. Full-length RTA protein (60 ng) was incubated with increasing amounts of anti-RTA rabbit serum (0, 1, and 3 μl in lanes 1 to 3, respectively), prior to the addition of an end-labeled probe (pan2*; nt −69 to −42). The arrow indicates the specific RTA-DNA complex. (B) Supershift of the Rdbd-DNA complex. An end-labeled probe (pan1*; nt −78 to −37) and 20 ng of purified Rdbd protein were incubated for EMSA in the absence (lane 2) or presence of an antibody against FLAG (lane 3) or RTA (lane 4). Closed and open arrowheads indicate Rdbd-DNA (lane 2) and shifted (lane 3) complexes.
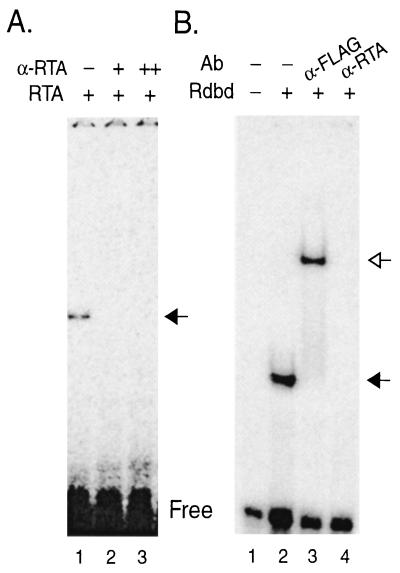
Determination of the Kd of Rdbd binding to the pPAN RRE.
We set out to determine the binding affinity of the Rdbd for the pPAN RRE. Specific binding of the Rdbd to DNA increased in a dose-dependent manner with increasing amounts of purified Rdbd protein, ranging from 0.9 pg to 1 μg in the presence of a limited amount (10−11 mol) of 32P-labeled pan1* (Fig. 3A). To determine the affinity of Rdbd for the pPAN RRE, we plotted the percentage binding as a function of protein amount expressed on a log scale (Fig. 3B). The amount of protein required for 50% binding was approximately 11 to 12 ng per 13-μl reaction mixture (Kd = 11.5 to 12.6 nM). Since RTA homologues of gammaherpesviruses are known to form a homodimer complex through the leucine zipper domain (29), we calculated Rdbd concentration as a dimer. This represents the apparent Kd, as it reflects total (i.e., active and inactive) protein concentration.
FIG. 3.
Determination of the Kd of the Rdbd binding to the pPAN RRE. (A) EMSAs of the pPAN RRE with serial dilutions of purified Rdbd protein (see Materials and Methods). A double-stranded oligonucleotide (pan1*; nt −78 to −37) containing the pPAN RRE was incubated with increasing amounts of Rdbd. The amount of Rdbd protein increased from ca. 1 pg (lane 1) to 1 μg (lane 14). The arrow indicates specific binding of Rdbd to DNA. (B) Efficiencies of binding of Rdbd to the pPAN RRE. The binding efficiency was calculated based on the ratio of the bound probe to the total and plotted as a function of protein amounts (in nanograms on the log scale). The Kd of RTA on the PAN promoter was estimated from the amount of protein resulting in 50% binding. The arrow indicates the amount of Rdbd for the apparent Kd. (C) EMSAs for competition assays. Purified Rdbd protein (1.5 μg) was incubated with pan1* in the presence of 5, 10, 20, 40, or 60 pmol of unlabeled competitor, pan1, per reaction (lanes 1 to 5). The competitor amount at 50% inhibition of Rdbd binding was determined and used to calculate the active protein amount. The arrow indicates the specific binding of Rdbd to DNA.
In order to determine the true Kd (i.e., the concentration of active protein required for 50% binding), competition assays were performed in the presence of an oversaturating amount of Rdbd (1.5 μg per reaction). When the protein concentration is well above the apparent Kd, this would lead to almost 100% binding of the probe. Increasing amounts of unlabeled cold competitor (pan1) were then added to the binding reaction mixture. When the competitor began to exceed the amount of active protein in the reaction, the labeled unbound probe was observed (Fig. 3C). The signal was quantitated and the percent unbound probe was plotted as a function of the amount of the cold competitor added to each mixture. The amount of competitor resulting in 50% competition is equivalent to twice the amount of active homodimeric protein. The amount of active protein present in 1.5 μg of purified Rdbd was estimated to be 14 pmol, whereas the amount of total protein was calculated to be 20 pmol. Hence, given the apparent Kd of 11.5 to 12.6 nM, the true Kd of Rdbd binding for the pPAN RRE is approximately 7.9 to 8.6 nM.
Mapping the RTA binding site in the pPAN RRE.
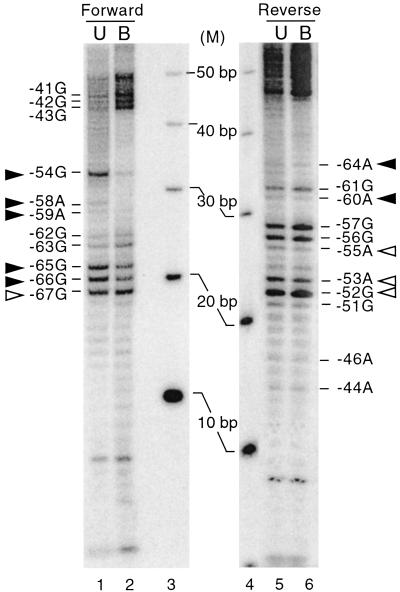
Direct binding of RTA to DNA has been suggested to be an important mechanism by which RTA activates expression of downstream genes, including PAN RNA, kaposin, ORF57, K-bZIP, and vIL-6 (14, 25, 41, 55; Deng et al., submitted). The sequence of the ORF57, the K-bZIP, and the vIL-6 promoters that confers RTA binding does not share any obvious homology to that of the PAN promoter, while the RRE of the PAN promoter shares significant homology to that of the kaposin promoter (25, 41). However, the RTA binding sequence within the promoter has not been determined in detail for these genes. To precisely map the RTA binding site on the PAN promoter, methylation interference assays were used for the following reason. The RTA binding site was narrowed down to a 28-bp DNA fragment, the region between nt −69 to −42 of the PAN promoter; therefore, it was necessary to use a high-resolution method to pinpoint the binding sequence. DNase I footprinting assays do not meet this criterion, because DNase I is a large protein that renders a protected region approximately 8 to 10 bp larger than the actual binding site (5). In contrast, the methylation interference assay is a high-resolution method for measuring the bases involved in sequence-specific recognition by proteins. This method uses a small molecule, DMS, to freely methylate nucleotides prior to incubation with a protein, in order to determine whether such modifications interfere with protein-DNA binding (2, 5).
pan1* was used to generate either a reverse (3′)- or forward (5′)-strand-labeled probe. DMS strongly modifies nucleotides with guanine in the major groove and to a lesser extent adenine in the minor groove. Both forward and reverse strands were analyzed to obtain comprehensive mapping of the pPAN RRE. These single-end-labeled probes were incubated with 0.5% DMS and subsequently with 125 ng of purified Rdbd. The unbound and bound probes were purified, digested with piperidine to cleave the modified bases, and resolved on a sequencing gel with an end-labeled 10-bp ladder (Fig. 4). The bands in the unbound fraction represent an enriched pool of probes with modified bases at the positions that interfere with protein binding, while those in the bound fraction contain modified bases that permit protein binding. The signal for each band was normalized with the total signal in each lane. The ratios of bound to unbound DNA were calculated. The result identified critical nucleotides involved in Rdbd binding: nt −67 to −64, nt −60 to −58, and nt −55 to −52. Therefore, it is likely that RTA interacts with the PAN promoter sequence from nt −67 to −52 and recognizes a subset of base pairs within this region.
FIG. 4.
Mapping the RTA binding sequence in pPAN RRE. Methylation interference assays of Rdbd binding to pPAN RRE were performed. 5′ (forward; lanes 1 and 2)- and 3′ (reverse; lanes 5 and 6)-end-labeled oligonucleotides were generated by cutting either end of pan1* with BamHI or BglII. These probes were incubated with DMS and used for EMSA with 125 ng of purified Rdbd. Purified unbound (U) and bound (B) DNA fragments were digested with piperidine, and digested DNA fragments were resolved in a sequencing gel with an end-labeled 10-bp ladder (M; lanes 3 and 4). The signal in each band was normalized with the total signal in each lane by phosphorimaging. The ratios of the bound to the unbound DNA were calculated. Closed triangles, base pairs which, when methylated, severely reduced Rdbd binding (<50%); open triangles, those with mild effects on Rdbd binding (50 to 70%).
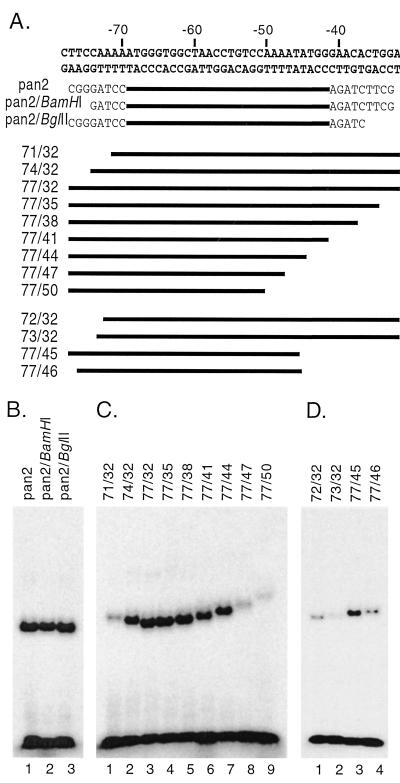
Minimal DNA length of the PAN promoter required for RTA binding.
Methylation interference assays demonstrated that base pairs in the PAN promoter from nt −67 to −52 interact with Rdbd. However, RTA may also require flanking regions for its optimal binding. To test this hypothesis and to determine the minimal DNA length required for RTA binding, a series of deletion probes was generated (Fig. 5A). In a previous deletion study, pan2* (nt −69 to −42) was the smallest fragment shown to be sufficient for RTA binding (41). This double-stranded oligonucleotide was end labeled and cut at the BamHI (5′) or BglII (3′) restriction site, resulting in a 3- or 4-bp deletion. In addition, a set of synthesized oligonucleotides were made with 3-bp serial deletions at the 5′ or 3′ end of the PAN promoter (nt −77 to −32). End-labeled deletion probes were incubated with purified Rdbd (20 ng) and separated on a native gel (Fig. 5B and C). The labeled probes, pan2/BamHI* and/BglII*, displayed Rdbd binding similar to that of pan2* (Fig. 5B). The majority of the serial deletion probes also maintained Rdbd binding similar to that of pan2*. However, the loss of three base pairs from nt −74 to −71 or from nt −44 to −47 of the promoter significantly reduced Rdbd binding (Fig. 5C, lanes 1 and 8).
FIG. 5.
Determination of minimal DNA length for RTA binding. (A) Sequences of deletion probes. pan2* was cut with either BamHI or BglII. Three-base-pair serial deletions in the region from nt −77 to −32 of the PAN promoter were made from either the 5′ or 3′ end. Based on the results from these probes, the second set of 1-bp-deletion probes was designed. The bold lines indicate a region corresponding to the PAN promoter sequences, and the flanking sequences including enzyme sites are indicated. (B) EMSA of pan2*, pan2/BamHI*, and pan2/BglII*. pan2* was digested with BamHI or BglII to generate 3-or 4-bp deletions at the 5′ or 3′ end, respectively. The probes were incubated with 20 ng of purified Rdbd and run on a native polyacrylamide gel. (C) EMSA of deletion probes. Each probe was end labeled and incubated with 20 ng of purified Rdbd. (D) EMSAs with the second set of deletion probes. Probes with 1-bp deletions in a region from nt −71 to −74 and from nt −44 to −47 were used for EMSAs.
A second set of probes with 1-bp deletions were also generated and tested to further define the minimal length of DNA required for RTA binding (Fig. 5A). Deletion from nt −74 to −73 at the 5′end reduced the Rdbd binding (Fig. 5D, lane 2). When one base pair was deleted (nt −45 to −46) at the 3′ end, Rdbd did not bind to the probe (Fig. 5D, lane 4). Therefore, the minimal DNA length for Rdbd binding is 30 bp, spanning the region from nt −74 to −45 of the promoter. Rdbd bound to probes 74/32 and pan2/BamHI* to a similar extent, suggesting that the base pairs between nt −74 and −70 at the 5′ end of the minimal DNA fragment (−74 to −45) are not sequence specific for RTA binding. Based on the results from methylation interference assays and deletion analysis, the 7-bp AT-rich sequences outside the mapped Rdbd binding sequences at both ends may play a role in stabilizing the protein-DNA complex rather than being involved in direct base pair recognition from the protein. The Rdbd binding sequence mapped by interference assays is consistent with this concept because base pairs at both the 5′and 3′ ends of the minimal DNA fragment are less important in sequence-specific recognition (Fig. 4) but are still essential for high-affinity binding (Fig. 5).
Mutagenesis study of the pPAN RRE. (i) Rdbd binding in vitro.
To gain more comprehensive information on the interactions between RTA and the PAN promoter and to examine the relationship between in vitro binding and in vivo transactivation by RTA, we carried out an extensive mutagenesis study of the PAN promoter. Two-base-pair substitutions were made sequentially across the PAN promoter from nt −75 to −38 (Fig. 6A). Since transversion mutations are known to introduce the most radical structural change possible at a particular position, a C/G base pair was replaced with A/T, G/C was replaced with T/A, A/T was replaced with C/G, and T/A was replaced with G/C. The mutated sequences were analyzed to ensure that no other known transcription factor binding sites would be introduced. The same mutant sequences were incorporated into probes for EMSA and into reporter constructs for transfection analysis.
We first examined the effect of pPAN RRE mutations on Rdbd DNA binding in vitro. The pan1* probe (nt −78 to −36) served as the wild type, and mutant probes shared the pPAN RRE and flanking sequences with the wild type except for the two mutated base pairs (Fig. 6A). Double-stranded wild-type and mutant probes were end labeled and incubated with 20 ng of purified Rdbd. A representative result is shown in Fig. 6C. Relative binding efficiencies of mutant probes were used for quantitative analysis of mutation effects (Fig. 6D). Mutations in the area from nt −69 to −46 significantly reduced Rdbd binding in vitro. Among these, mutations of base pairs at −63, −62, −59 to −54, −53, and −52 demonstrated less than 30% of wild-type binding activity and mutation of base pairs at −46 to −49, −55, −56, −61, −62, and −64 to −69 demonstrated 30 to 60% of wild-type binding activity (Fig. 6E). We noticed that mutations at −51 and −50 resulted in 34% higher binding activity than wild type. The difference in binding of this mutant was less than 40%, which we used as a cutoff for mutation effects. Nevertheless, this may suggest high flexibility of RTA in sequence recognition (see Discussion). However, this result clearly demonstrated that specific base pairs (−63G, −62G, −59A, −58A, −55C, −54T, −53G, and −52T) in the PAN promoter are critical for Rdbd binding in vitro. These results are consistent with the results of methylation interference assays and deletion analysis. In addition, the mutagenesis study implicated the importance of extra nucleotides at the 3′ end of the previously identified Rdbd binding site (nt −49 to −45 of the PAN promoter), suggesting that the 3′ AT-rich sequences may also play a role in sequence-specific recognition of RTA.
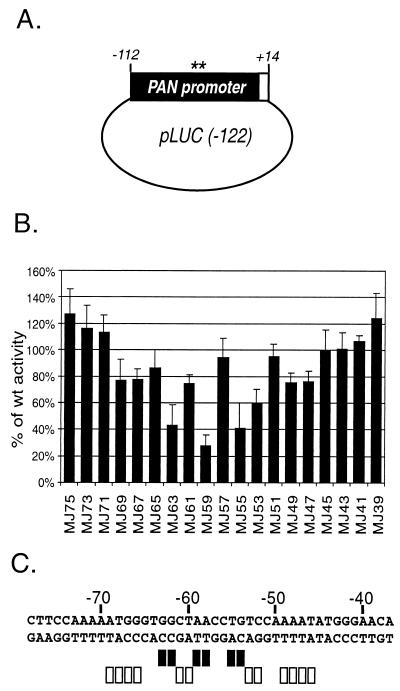
(ii) RTA transactivation in vivo.
We next sought to characterize the base pairs of the PAN promoter important in DNA binding, which affect RTA transactivation in vivo by introducing the same mutations into a reporter construct. pLUC(−122) includes 122 bp of the PAN promoter sequence with 14 bp of the 5′ transcribed region of the PAN RNA sequence cloned into a vector driving firefly luciferase. The same 2-bp mutations used for the Rdbd binding mutagenesis study were introduced into pLUC(−122) by PCR-directed site mutagenesis (Fig. 7A).
FIG. 7.
Transactivation of pPAN RRE mutant reporters by RTA in vivo. (A) Schematic diagram of the reporter construct used in the mutagenesis study. The wild-type construct, pLUC(−122), includes 122 bp of the PAN promoter sequence as well as 14 bp of the 5′ transcribed sequence of PAN RNA that was cloned into a pGL3-basic vector driving the expression of firefly luciferase. Asterisks indicate two base pairs replaced with transversion mutations in a region from nt −75 to −38 of the PAN promoter. (B) Quantitative analysis of RTA transactivation in vivo of the pPAN RRE mutant reporters. Mutant constructs (200 ng) were cotransfected into 293T cells with 40 ng of pcDNA3 or pcDNA3/RTA in the presence of a control vector (10 ng), pRLCMV, which constitutively expresses Renilla luciferase. At 24 h posttransfection, cells were harvested, and dual luciferase assays were performed. Firefly luciferase activity from reporter constructs was normalized to the corresponding Renilla luciferase activity. The normalized luciferase activity of pcDNA3-transfected cells was subtracted from that of pcDNA3/RTA-transfected cells to calculate promoter activity. The average of the results for each mutant reporter is shown relative to the wild-type promoter activity from at least three different duplicate transfections. (C) Base pairs critical for transactivation of the pPAN RRE by RTA in vivo. Closed rectangles represent base pairs with severe mutation effects (<50% of wild-type promoter activity), and open rectangles indicate moderate effects (50 to 80% of wild-type promoter activity).
Reporter constructs (200 ng) were transfected into 293T cells with 40 ng of either vector (pcDNA3) alone or an RTA expression plasmid (pcDNA3/RTA). Plasmid pRLCMV (10 ng) containing the coding sequence for Renilla luciferase under the control of a constitutively active human cytomegalovirus immediate-early enhancer-promoter was included in each transfection and served as an internal control for transfection efficiency. At 24 h posttransfection, cell lysates were assayed for both firefly and Renilla luciferase activities. Firefly luciferase activity was normalized with the corresponding Renilla luciferase activities in each transfection. The normalized luciferase activity of pcDNA3-transfected cells was subtracted from that of pcDNA3/RTA-transfected cells and used as the promoter activity (Fig. 7B). Compared with the promoter activity of the wild type, that of each mutant showed varied promoter activities. We considered mutants with less than 50% of wild-type promoter activity as having severe mutation effects and those with 50 to 80% of wild-type activity as having mild effects (Fig. 7C). This analysis demonstrated that nt −69 to −46 of the PAN promoter except nt −65, −64, −51, and −50 are important for promoter activation by RTA. The base pairs at −63G, −62G, −59A, −58A, −55T, and −54G were consistently found to be critical in mutant studies for in vivo reporter analysis as well as in vitro binding assays. A parallel comparison of reporter analysis with Rdbd binding analysis revealed a good correlation between the base pairs required for Rdbd binding in vitro and those required for transactivation by RTA in vivo.
Detailed analysis of the pPAN RRE in RTA binding in vitro and transactivation in vivo.
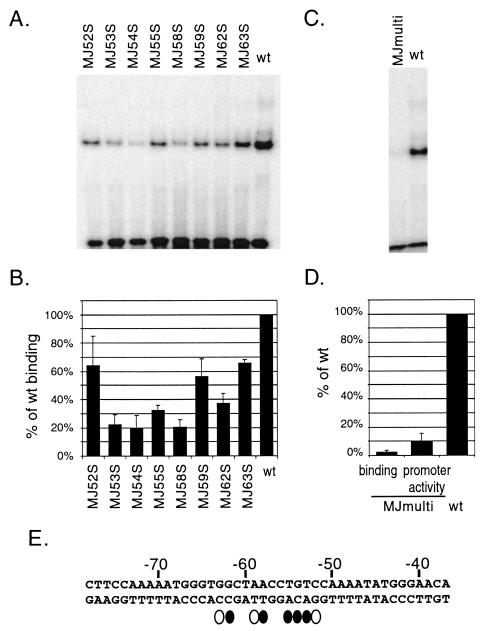
In order to determined which of the two base pairs mutated in each mutant probe or reporter construct contributed most to the reduced Rdbd binding or transactivation, we generated eight mutant probes with single base pair substitutions at nt −63G, −62G, −59A, −58A, −55C, −54T, −53G, and −52T (Fig. 6B). Single-base-pair mutant probes were tested by EMSAs performed in parallel with the wild type (Fig. 8A). Quantitative analysis of the mutants was performed as described earlier (Fig. 8B). Five of eight base pairs mutated at nt −62, −58, −55, −54, and −53 resulted in severely reduced binding of Rdbd (Fig. 8E). These single-base-pair mutations rendered the mutant probes fivefold less efficient than the wild-type probe, suggesting that these individual base pairs play a key role in interactions between RTA and the promoter.
FIG. 8.
Detailed analysis of the pPAN RRE in RTA binding in vitro and transactivation in vivo. (A and C) EMSA of single (A)- and multiple (C)-base-pair mutants of the pPAN RRE. An equal molar amount of each mutant probe shown in Fig. 6B was assayed as described for Fig. 6C. (B) Quantitative analysis of single-base-pair mutations. The relative binding efficiencies of the mutant probes were calculated as described for Fig. 6D. (D) Effects of multiple-base-pair mutations on RTA binding in vitro and transactivation in vivo of the pPAN RRE. A mutant probe with multiple mutations (MJmulti) was used for EMSA, as described for Fig. 6C. A reporter construct containing the same set of multiple mutations was transfected into 293T cells, as described for Fig. 7B. Relative binding efficiency and promoter activity of MJmulti are presented in comparison with those of the wild type. (E) Base pairs critical for in vitro binding and in vivo transactivation of the pPAN RRE by RTA. Closed ovals indicate base pairs with severe mutation effects (<40% of wild-type activity), and open ovals mark those with moderate effects (40 to 70% of wild-type activity).
A mutant probe containing multiple base pair mutations (nt −62, −58, −55, −54, and −53) was tested to determine whether the mutations have an additive effect (Fig. 6B). Simultaneous mutation of the five base pairs completely abolished the in vitro binding of Rdbd (Fig. 8C). A reporter construct with the same mutation was generated and tested in 293T cells. The promoter activity of this mutant construct was decreased dramatically to 10% of wild-type promoter activity (Fig. 8D). Therefore, direct interactions of RTA with DNA at these critical positions are essential for its ability to activate the PAN promoter. These results support the hypothesis that direct DNA binding of RTA is a key mechanism by which the PAN promoter is activated.
DISCUSSION
It has been shown that the PAN promoter is strongly activated by RTA (41). Here, we defined the interactions between RTA and the PAN promoter. To better understand the interactions between RTA and DNA, we expressed and purified the Rdbd (1 to 320 amino acids). Purified full-length RTA and the truncated Rdbd proteins displayed specific DNA binding to the PAN promoter. The specificity of the interaction between RTA and DNA was shown by EMSAs using an excess of sequence-specific competitor as well as a nonspecific competitor. In addition, the RTA forms a stable complex with the pPAN RRE, even under stringent high-salt conditions (i.e., 300 mM KCl) (41). We have tested several RREs from other viral promoters, including kaposin, ORF57, K-bZIP, and vIL-6, all of which showed Rdbd- and RTA-specific binding in EMSAs (unpublished results). We confirmed the specificity of the interactions by using the anti-RTA antibody to inhibit complex formation and the anti-FLAG antibody to supershift the complex. In addition, sequence-specific binding of the Rdbd to DNA was tested by using an end-labeled oligonucleotide with nonrelated sequence (41), which resulted in no Rdbd-specific binding within the range of protein amounts used (data not shown). Therefore, purified RTA and Rdbd proteins expressed in bacteria bound to the target gene promoters in a sequence-specific manner, recapitulating how RTA may function as a transcriptional activator during viral lytic replication.
Among the RREs, the pPAN RRE and the kaposin RRE share a significant homology of 25 bp, including the same consecutive 16-bp sequence at the 5′-end side (5′-AAATGGGTGGCTAACCCCTACATAA-3′; the shared sequence is underlined), which corresponds to nt −71 to −47 of the PAN promoter. Previously, it was shown that the RRE of the ORF57 promoter (5′-AGTGTAACAATAATGTTCCCACGGC-3′) shares partial homology with that of the K-bZIP promoter (14, 25), which is capable of binding to RTA. However, the similarity of these two groups of the RRE sequences is not apparent, and a recently identified RRE from the vIL-6 promoter does not reveal any recognizable homology to these RREs (Deng et al., submitted), suggesting that RTA has high flexibility in terms of sequence recognition. It is of interest to understand how RTA binds to a variety of sequences with different affinities and controls the expression levels of various viral genes during lytic replication.
We have measured the Kd of Rdbd binding to the pPAN RRE. RTA is known to form a homodimer complex via a leucine zipper domain (29). The amount of active protein was calculated from the known concentration of the cold competitor. Given the oligomeric status of RTA, active Rdbd protein comprises 70% of the purified protein sample. The determined true Kd of RTA to the PAN promoter is in the nanomolar range, indicating a high-affinity interaction. In our study, more full-length RTA protein was required to show RTA binding than Rdbd. It is most likely because there is less active full-length protein present in the preparation than in the Rdbd preparation. We have noticed that the full-length RTA is much less stable than the DBD alone. It is also consistent with a general notion that the activation domain is less structured on its own unless bound by a coactivator. However, it is possible that the actual affinity of RTA for the PAN promoter in KSHV-infected cells may be even higher, due to cooperative binding between RTA and other viral or cellular factors. The abundance of PAN RNA expression may be attributed primarily to high-affinity binding of RTA to the PAN promoter during viral lytic replication.
We used three independent methods to characterize the interactions between RTA and the pPAN RRE in vitro and addressed its functional relevance in vivo. A summary of these results is presented in Fig. 9. The most comprehensive data on specific interactions between RTA and its responsive promoter were gained from the mutagenesis study on Rdbd binding in vitro. A group of base pairs located between nt −69 and −46 of the promoter (except nt −51 and −50) were shown to be critical for Rdbd binding. These base pairs were also found to be essential for promoter transactivation in reporter assays. Furthermore, the relative extent of mutation effects correlated between results from EMSAs and reporter assays. Methylation interference assays pinpointed fewer base pairs than the RTA recognition site, but these base pairs were also determined to be important by the mutagenesis study. For example, the promoter regions from nt −67 to −66, nt −60 to −58, and nt −55 to −52 were consistently shown to be critical for in vitro binding in all the assays performed. However, nt −51 and −50 were not shown to be sequence specific by any methods used. The minimal promoter region required for RTA binding in vitro includes all the base pairs found to be critical in other assays.
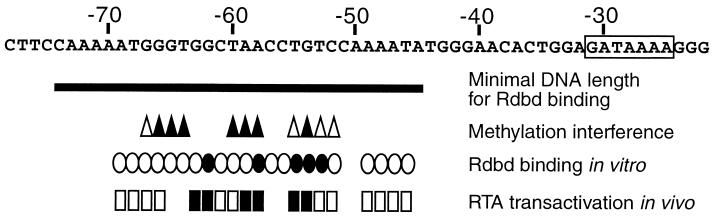
FIG. 9.
Summary of the interactions between RTA and the PAN promoter. The PAN promoter sequence from nt −78 to −22 is shown. A TATA box is outlined. Each symbol indicates a base pair identified as being important in each analysis. Triangles are used for methylation interference assays (Fig. 4), ovals are for Rdbd binding in vitro assayed by EMSA (Fig. 6 and 8), and rectangles are for RTA transactivation in vivo (Fig. 7). Closed symbols indicate base pairs of great importance, and open symbols mark those with mild effects. The minimal length of DNA required for RTA binding is indicated under the mapped region (Fig. 5).
Despite the strong correlation between results from the different assays, there were some discrepancies. Interference assays permit detection of nucleotides in close proximity to the protein, although they do not measure direct protein contacts with nucleotides. DMS methylates guanine residues at the N-7 position, which protrudes into the major groove, and also methylates adenines at the N-3 position, which protrudes into the minor groove. Although adenines can be detected in these assays, adenine modifications are much weaker than the guanidine modifications. Thus, the lower sensitivity for detection of adenines may account for different results for adenines of nt −49 to −46. These adenines were found to be important in the mutation studies on RTA binding; however, they were not identified as critical nucleotides by methylation interference assays. Deletion analysis to determine the minimal region required for RTA binding in vitro allows us to examine the overall boundary of interactions between RTA and DNA. In addition to sequence-specific interactions between a protein and bases in the major groove and/or the minor groove, nonspecific contacts of the protein with the phosphate or sugar backbone of DNA are known to contribute to the affinity of the protein for DNA. The presence of both specific and nonspecific interactions between RTA and DNA is consistent with our results showing that the minimal region for RTA binding spans a larger region than the critical base pairs identified by methylation interference assays or mutagenesis (Fig. 9). In our previous study, inverted and direct repeats in the pPAN RRE were proposed, and the RTA binding site was speculated to be within these repeats (41). However, our detailed analysis indicated that only a part of these repeats is important, and therefore, the significance of the palindromic sequences has yet to be defined. It is possible that these repeats might be involved in binding by cellular or viral factors other than RTA. Critical base pairs identified from mutagenesis studies on Rdbd binding in vitro were well matched with those important for RTA transactivation in vivo. However, RTA binding defects in EMSAs were more severe than transactivation defects in reporter assays. With a purified protein, EMSAs are designed to test the binding of only one factor to DNA, while multiple factors may contribute to promoter activity in reporter assays. In fact, the wild-type reporter construct pLUC(−122) contains a longer promoter than the minimal RTA-responsive reporter pLUC(−69). This might have increased the contribution from cellular factors, because deletion of 53 bp from nt −122 to −69 of the PAN promoter resulted in threefold drop in transactivation (41). Additional posttranslational modifications of RTA expressed in mammalian cells might also contribute to this difference. However, results of methylation interference assays, deletion analysis, and mutagenesis for Rdbd binding in vitro and RTA transactivation in vivo are consistent, strongly supporting our hypothesis that direct DNA binding of RTA is the key mechanism for activation of the PAN promoter.
In EMSAs, we observed different mobilities of the protein-DNA complexes when probes of different lengths were used (Fig. 5B and D). While the gel showed little differentiation in mobility of the free probes, the protein-DNA complexes migrated more slowly when the shorter probes were tested. This was not simply due to “smiling” of the gel, since it was observed repeatedly and since other lanes with different samples in the same gel did not exhibit extended tailing (data not shown). The contribution from molecular masses of the probes with different lengths cannot account for the slow migration of the complex with a short probe. Upon binding to DNA, many Dna-binding proteins undergo conformational changes. Thus, it is reasonable to conceive that longer probes may generate more stable and compacted protein conformation, leading to faster migration. A high-resolution X-ray crystal structure of RTA cocrystallized with or without DNA may allow us to visualize conformational changes of RTA and envision how RTA interacts with its target gene promoter during viral lytic replication. The present study, using chemical probing and mutagenesis, delineated the molecular interactions between RTA and the PAN promoter, which are also important for RTA transactivation in transient-transfection reporter assays. These data will also serve as a framework for interpreting future crystal structure data.
In our effort to address the question of how KSHV achieves such a high level of PAN RNA expression during lytic replication, we have characterized the interactions between RTA and the PAN promoter and demonstrated functional correlation of these interactions. Since PAN RNA is the most abundant lytic transcript of KSHV, it may help us understand an effective mechanism by which RTA activates downstream viral genes. Therefore, a study of RTA activation of the PAN promoter serves as a good model system for investigating KSHV gene regulation by RTA.
Acknowledgments
We thank T. Symensma and M. Carey for critical reading and J. Chu and W. Aft for editing the manuscript. We thank George Miller for communication of unpublished results.
This work was supported by NIH grants CA83525, CA91791, and DE14153, the Stop Cancer Foundation (R.S.), and the Universitywide AIDS Research Program (H.J.B.). M.J.S. was supported by a UCLA Chancellor's Dissertation Year Fellowship.
REFERENCES
- 1.Alkan, S., D. S. Karcher, A. Ortiz, S. Khalil, M. Akhtar, and M. A. Ali. 1997. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus in organ transplant patients with immunosuppression. Br. J. Haematol. 96:412-414. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M. (ed.). 1998. Current protocols in molecular biology, vol. 2. John Wiley and Sons, Inc., New York, N.Y.
- 3.Boshoff, C., T. F. Schulz, M. M. Kennedy, A. K. Graham, C. Fisher, A. Thomas, J. O. McGee, R. A. Weiss, and J. J. O'Leary. 1995. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274-1278. [DOI] [PubMed] [Google Scholar]
- 4.Cannon, J. S., J. Nicholas, J. M. Orenstein, R. B. Mann, P. G. Murray, P. J. Browning, J. A. DiGiuseppe, E. Cesarman, G. S. Hayward, and R. F. Ambinder. 1999. Heterogeneity of viral IL-6 expression in HHV-8-associated diseases. J. Infect. Dis. 180:824-828. [DOI] [PubMed] [Google Scholar]
- 5.Carey, M., and S. T. Smale. 2000. Transcriptional regulation in eukaryotes: concepts, strategies, and techniques. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 6.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 7.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, and K. Yamanishi. 2000. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 74:8623-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, M. A., J. Leahy, and J. M. Hardwick. 1990. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivators. J. Virol. 64:313-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darr, C. D., A. Mauser, and S. Kenney. 2001. Epstein-Barr virus immediate-early protein BRLF1 induces the lytic form of viral replication through a mechanism involving phosphatidylinositol-3 kinase activation. J. Virol. 75:6135-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, H., A. Young, and R. Sun. 2000. Auto-activation of the rta gene of human herpesvirus-8./Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 81:3043-3048. [DOI] [PubMed] [Google Scholar]
- 13.Dignam, J. D., P. L. Martin, B. S. Shastry, and R. G. Roeder. 1983. Eukaryotic gene transcription with purified components. Methods Enzymol. 101:582-598. [DOI] [PubMed] [Google Scholar]
- 14.Duan, W., S. Wang, S. Liu, and C. Wood. 2001. Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch. Virol. 146:403-413. [DOI] [PubMed] [Google Scholar]
- 15.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ensoli, B., G. Barillari, and R. C. Gallo. 1992. Cytokines and growth factors in the pathogenesis of AIDS-associated Kaposi's sarcoma. Immunol. Rev. 127:147-155. [DOI] [PubMed] [Google Scholar]
- 17.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gwack, Y., H. Byun, S. Hwang, C. Lim, and J. Choe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haque, M., J. Chen, K. Ueda, Y. Mori, K. Nakano, Y. Hirata, S. Kanamori, Y. Uchiyama, R. Inagi, T. Okuno, and K. Yamanishi. 2000. Identification and analysis of the K5 gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 74:2867-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, Y. Q., J. J. Li, M. H. Kaplan, B. Poiesz, E. Katabira, W. C. Zhang, D. Feiner, and A. E. Friedman-Kien. 1995. Human herpesvirus-like nucleic acid in various forms of Kaposi's sarcoma. Lancet 345:759-761. [DOI] [PubMed] [Google Scholar]
- 21.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kedda, M. A., L. Margolius, M. C. Kew, C. Swanepoel, and D. Pearson. 1996. Kaposi's sarcoma-associated herpesvirus in Kaposi's sarcoma occurring in immunosuppressed renal transplant recipients. Clin. Transplant. 10:429-431. [PubMed] [Google Scholar]
- 23.Koh, S. S., C. S. Hengartner, and R. A. Young. 1997. Baculoviral transfer vectors for expression of FLAG fusion proteins in insect cells. BioTechniques 23:627-630. [DOI] [PubMed] [Google Scholar]
- 24.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and S. D. Hayward. 1990. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 28.Manet, E., H. Gruffat, B. M. C. Trescol, N. Moreno, P. Chambard, J. F. Giot, and A. Sergeant. 1989. Epstein-Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional trans-activators. EMBO J. 8:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manet, E., A. Rigolet, H. Gruffat, J. F. Giot, and A. Sergeant. 1991. Domains of the Epstein-Barr virus (EBV) transcription factor R required for dimerization, DNA binding and activation. Nucleic Acids Res. 19:2661-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, D. F., J. P. Dunn, J. L. Davis, J. S. Duker, R. E. Engstrom, Jr., D. N. Friedberg, G. J. Jaffe, B. D. Kuppermann, M. A. Polis, R. J. Whitley, R. A. Wolitz, and C. A. Benson. 1999. Use of the ganciclovir implant for the treatment of cytomegalovirus retinitis in the era of potent antiretroviral therapy: recommendations of the International AIDS Society-USA panel. Am. J. Ophthalmol. 127:329-339. [DOI] [PubMed] [Google Scholar]
- 31.Memar, O. M., P. L. Rady, and S. K. Tyring. 1995. Human herpesvirus-8: detection of novel herpesvirus-like DNA sequences in Kaposi's sarcoma and other lesions. J. Mol. Med. 73:603-609. [DOI] [PubMed] [Google Scholar]
- 32.Miles, S. A., O. Martinez-Maza, A. Rezai, L. Magpantay, T. Kishimoto, S. Nakamura, S. F. Radka, and P. S. Linsley. 1992. Oncostatin M as a potent mitogen for AIDS-Kaposi's sarcoma-derived cells. Science 255:1432-1434. [DOI] [PubMed] [Google Scholar]
- 33.Moore, P. S., C. Boshoff, R. A. Weiss, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 34.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 332:1181-1185. [DOI] [PubMed] [Google Scholar]
- 35.Moore, P. S., L. A. Kingsley, S. D. Holmberg, T. Spira, P. Gupta, D. R. Hoover, J. P. Parry, L. J. Conley, H. W. Jaffe, and Y. Chang. 1996. Kaposi's sarcoma-associated herpesvirus infection prior to onset of Kaposi's sarcoma. AIDS 10:175-180. [DOI] [PubMed] [Google Scholar]
- 36.Quinlivan, E. B., E. A. Holley-Guthrie, M. Norris, D. Gutsch, S. L. Bachenheimer, and S. C. Kenney. 1993. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 21:1999-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ragoczy, T., and G. Miller. 2001. Autostimulation of the Epstein-Barr virus BRLF1 promoter is mediated through consensus Sp1 and Sp3 binding sites. J. Virol. 75:5240-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed, J. A., R. G. Nador, D. Spaulding, Y. Tani, E. Cesarman, and D. M. Knowles. 1998. Demonstration of Kaposi's sarcoma-associated herpes virus cyclin D homolog in cutaneous Kaposi's sarcoma by colorimetric in situ hybridization using a catalyzed signal amplification system. Blood 91:3825-3832. [PubMed] [Google Scholar]
- 39.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song, M. J., H. J. Brown, T. T. Wu, and R. Sun. 2001. Transcription activation of polyadenylated nuclear RNA by RTA in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:3129-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 43.Staskus, K. A., R. Sun, G. Miller, P. Racz, A. Jaslowski, C. Metroka, H. Brett-Smith, and A. T. Haase. 1999. Cellular tropism and viral interleukin-6 expression distinguish human herpesvirus 8 involvement in Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. J. Virol. 73:4181-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun, R., S. F. Lin, L. Gradoville, and G. Miller. 1996. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 93:11883-11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Santen, V. L. 1993. Characterization of a bovine herpesvirus 4 immediate-early RNA encoding a homolog of the Epstein-Barr virus R transactivator. J. Virol. 67:773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitby, D., M. R. Howard, M. Tenant-Flowers, N. S. Brink, A. Copas, C. Boshoff, T. Hatzioannou, F. E. Suggett, D. M. Aldam, A. S. Denton, et al. 1995. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet 346:799-802. [DOI] [PubMed] [Google Scholar]
- 51.Whitehouse, A., I. M. Carr, J. C. Griffiths, and D. M. Meredith. 1997. The herpesvirus saimiri ORF50 gene, encoding a transcriptional activator homologous to the Epstein-Barr virus R protein, is transcribed from two distinct promoters of different temporal phases. J. Virol. 71:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, T. T., L. Tong, T. Rickabaugh, S. Speck, and R. Sun. 2001. Function of Rta is essential for lytic replication of murine gammaherpesvirus 68. J. Virol. 75:9262-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, T. T., E. J. Usherwood, J. P. Stewart, A. A. Nash, and R. Sun. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74:3659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, L., J. Chiu, and J. C. Lin. 1998. Activation of human herpesvirus 8 (HHV-8) thymidine kinase (TK) TATAA-less promoter by HHV-8 ORF50 gene product is SP1 dependent. DNA Cell Biol. 17:735-742. [DOI] [PubMed] [Google Scholar]
- 56.Zhong, W., and D. Ganem. 1997. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8). J. Virol. 71:1207-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 93:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]