Abstract
KLF4 is induced upon growth-arrest in vitro and during epithelial maturation in vivo, and is essential for proper cell fate specification of post-mitotic cells. In spite of a normal role in post-mitotic cells, expression is upregulated and constitutive in certain tumor types. KLF4 functions as an oncogene in vitro, and enforced expression in basal cells of mouse skin rapidly induces lesions similar to hyperplasia, dysplasia and squamous cell carcinoma (SCC). Here we used conditional expression to characterize early steps in KLF4-mediated tumor initiation. In contrast to SCC-like lesions that result when using a conditional, keratin 14 promoter-dependent strategy, lower conditional expression achieved using a MMTV promoter induced only epidermal cycling within morphologically normal skin, a process we termed occult cell turnover. Surprisingly, KLF4-induced hyperplastic lesions showed increased transgene-derived mRNA and protein in maturing, PCNA-negative cells, a property of endogenous KLF4. In contrast, hyperplastic lesions induced by GLI1, a control, showed uniform transgene expression. In KLF4-induced dysplasia and SCC the complementarity of KLF4 and PCNA was replaced by concordance of the two proteins. These studies show that KLF4 transcripts are normally suppressed in cycling cells in a promoter-independent fashion, consistent with a post-transcriptional control, and reveal loss of this control in the transition from hyperplasia to dysplasia. Like the mouse tumors, human cutaneous SCCs and adjacent dysplasias frequently showed maturation-independence of KLF4, with co-expression of KLF4 and PCNA. A smaller subset of human SCCs showed complementarity of KLF4 and PCNA, similar to hyperplastic mouse skin. The results identify parallels between a mouse model and human primary tumors, and show that successive increases of KLF4 in the nuclei of basal keratinocytes leads to occult cell turnover followed by hyperplasia, dysplasia, and invasive SCC.
Keywords: initiation, progression, squamous cell carcinoma, skin, animal model, KLF4, mRNA stability
Abbreviations: KLF4 kruppel-like factor 4, K14 keratin 14, MD maturation dependent, MI maturation independent, MIN maturation independent with nuclear localization, SCC squamous cell carcinoma, TRE tetracycline response element, DEJ dermo-epidermal junction
Introduction
Deregulation of tissue specific ‘gatekeeping’ pathways through genetic or epigenetic means is implicated in the early steps in human tumor progression.1–4 Typically, such pathways influence cell fate by modulating the activity of a transcription factor oncoprotein. Based upon its activity as an oncogene in vitro and its upregulation in certain tumor types, the zinc finger protein Kruppel-like factor 4 (KLF4 or GKLF) represents a candidate effector of the malignant phenotype in tumors such as squamous cell carcinoma (SCC) of the skin or oral cavity.5–9
Klf4 was identified by reduced-stringency hybridization to zinc finger consensus probes.10,11 Both mRNA in situ hybridization and immunostaining showed that KLF4 is most highly expressed in vivo in the post-mitotic, differentiating epithelial cells of the skin and gut, and similarly expressed in a maturation dependent (MD) fashion in other epithelial tissues.5,10–14 KLF4 transcripts are low or negative in the proliferation-competent basal keratinocytes of normal skin or the intestinal crypts, an unusual feature for an oncogene. The lower expression of Klf4 transcripts in proliferating cells is consistent with either transcriptional control or regulated mRNA turnover.11,15–17 However, the mechanism by which MD expression is established remains poorly understood.
Interestingly, the induction of KLF4 upon growth-arrest can be recapitulated in vitro. For example, Klf4 transcripts are induced in growth-arrested NIH3T3 fibroblasts following serum starvation.11 Likewise, other methods of cell cycle delay induce KLF4, such as DNA damage-induced activation of p53 and p21/Waf1.15 While several observations suggest a role for KLF4 as a growth suppressor in the gut,18,19 our expression cloning studies identified KLF4 as one of the most potent transforming activities in human oropharyngeal SCC cell lines.5 Using an E1a-immortalized epithelial reporter line termed RK3E, screening of cDNA expression libraries identified only two transforming activities, KLF4 and c-MYC, and these were repeatedly isolated. Consistent with transformation of these E1a-immortalized rat kidney cells, Klf4 was recently identified as an oncogene that requires deregulation of specific G1 cell cycle regulators such as Cyclin D1 or p21/Waf1 to induce transformation.9
Induction of KLF4 in the basal layer of mouse skin induced hyperplasia, dysplasia and lesions similar to cutaneous SCC within 2–28 days.7 In the current study, we characterized four steps in KLF4-mediated skin tumor initiation, and compared these mouse tumors with their human counterpart. The KLF4 transgene recapitulated the MD expression previously observed for endogenous KLF4. Thus, MD expression is a promoter-independent property of the KLF4 protein-coding region and flanking untranslated regions (UTRs), and may be mediated by a post-transcriptional mechanism. This mechanism appears critical to tumor progression, as KLF4 became maturation independent in both mouse and human skin tumors, with increased expression in the nuclei of dysplastic skin and SCC.
Materials and Methods
Plasmid constructs, cell culture, and immunoblot analysis
To construct pGFP-KLF4, an EcoRI-Asp718I fragment encoding human KLF4 was inserted in to pEGFP-C1 (Clontech).5 HEK293 cells were transfected using transIT-LT1 (Mirus), lysed in nonionic detergent, and extracts were quantitated using the Bradford assay (BioRad). Following SDS-PAGE and transfer to nitrocellulose, anti-KLF4 was applied at 1.3 μg/ml and detected using ECL (Amersham).7 MCF10A cells were obtained from the ATCC (Manassas, VA) and cultured as described.20
Generation and analysis of transgenic mice
For conditional expression of KLF4 or GLI1 in K14-positive keratinocytes, tetracycline response element (TRE)-KLF432831 or TRE-GLI1 transgenic males on an FVB/NJ background were crossed to X chromosome-linked, homozygous K14-reverse tetracycline-responsive transcriptional activator (K14-rtTA) females as described.7,21 To place KLF4 under control of the mouse mammary tumor virus (MMTV) promoter, TRE-KLF432831 animals were crossed to FVB/NJ animals transgenic for MMTV-rtTA (strain MTB).22
To induce KLF4, dox (2.0 mg/ml, Sigma) was administered in 5% sucrose water in amber bottles at 3 weeks of age and was changed three times per week. Where indicated, BrdU (Zymed #40286619, 1.0 ml/100 gr body mass) was injected into the peritoneum 2 hrs before sacrificing. Following fixation and embedding in paraffin, analysis of hematoxylin- and eosin-stained slides was performed by a pathologist (A.R.F.). The described microscopic results were observed in multiple sections taken from each of the 2–4 animals examined, suggesting that penetrance is either very high or complete.
mRNA in situ hybridization
mRNA in situ hybridization analysis of mouse skin cryosections was performed using digoxigenin-labeled transcripts as described.23 Templates for in vitro transcription were isolated from total mRNA of mouse NIH3T3 cells by RT-PCR. PCR products incorporated a T7 RNA polymerase binding site, and forward and reverse primers were:
KLF4-AS, 5’ GAATTCAGTATTTTTTACTTTTCA 3’, and
5’ GGATCCTAATACGACTCACTATAGGGAGACTGGTCTTCCCTCCCCC 3’;
KLF4-S, 5’ GGATCCTAATACGACTCACTATAGGGAGAGAATTCAGTATTTTTTACTTTTCA 3’, and
5’ CTGGTCTTCCCTCCCCC 3’;
TUBB1-AS, 5’ TTGTGAGAAAGGAAGCTGAG 3’, and
5’ GGATCCTAATACGACTCACTATAGGGAGATAGGTCACCGTAAGTGGGT 3’;
ITGA3-AS, 5’ CCCTCACGGCCCACAAG 3’, and
5’ GGATCCTAATACGACTCACTATAGGGAGAGCCCAGCTGGCACATGC 3’.
Hybridized transcripts were detected using enzyme-antibody conjugates and the alkaline phosphatase substrate, Fast Red.
Immunostaining and imaging
Indirect immunofluorescence of cultured cells and immunostaining of paraffin sections utilized anti-KLF47 at 3.6 μg/ml.8 For paraffin section, bound antibody was applied twice for 40 minutes each, and then detected using Envision Plus reagents and the chromogenic substrate diaminobenzidine (Dako). Normal rabbit immunoglobulin (Dako #X0903) was used as a negative control for KLF4. Staining for PCNA was previously described.7 BrdU was detected by immunostaining of paraffin sections as directed by Zymed (#93-3943). Frozen sections of mouse skin were fixed in neutral buffered formalin and stained with rabbit anti-Gli1 antibody or normal rabbit IgG at 0.5 μg/ml.21 Bound antibody was detected as described.8 Staining patterns were assessed by J.M.R. and A.R.F. Images were acquired on an Axioskop 2 plus microscope equipped with an AxioCam HRc digital camera and AxioVision Software (Zeiss, version 4.4).
Study population
Patients underwent Mohs micrographic surgery for skin lesions, including SCC or basal cell carcinoma (BCC), in the Department of Dermatology at the University of Alabama at Birmingham between September and November 2004. Normal skin was obtained at the time of wound repair after excision of SCC or BCC, and consisted of the excision of redundant skin, or was obtained separately from reduction mammoplasty specimens. Nondiagnostic tissue containing SCC, BCC, or normal tissue were placed immediately in neutral buffered zinc formalin (10% v/v) and then transferred to the Tissue Procurement Core Facility for processing. Histopathologic diagnoses were made by a board-certified dermatopathologist (James E. Elder, MD, Skin Pathology Associates Inc, Birmingham, AL). Statistical analyses utilized Fisher’s exact test, two-tailed. All procedures were approved by the Institutional Review Board at this institution.
Results
Characterization of KLF4 antibody
Anti-KLF4 is an immuno-affinity purified rabbit antibody to a nonconserved human peptide, and specifically stains mouse skin expressing a human KLF4 transgene.7 To further characterize the specificity of anti-KLF4, we performed immunoblot analysis of HEK293 cells following transient transfection of a green fluorescent protein (GFP)-KLF4 fusion vector (Fig. 1A). Anti-KLF4 recognized a single species with a molecular weight similar to the predicted size of the fusion protein. Compared with other cell lines tested, human mammary epithelial MCF10A cells exhibit more prominent KLF4 by Northern blot, and stained strongly with anti-KLF4 (Fig. 1B). No staining was observed using normal rabbit immunoglobulin at the same concentration (Fig. 1B, lower panels). Mixed staining of the cytoplasm and the nucleus is consistent with previous studies.8
Fig. 1.
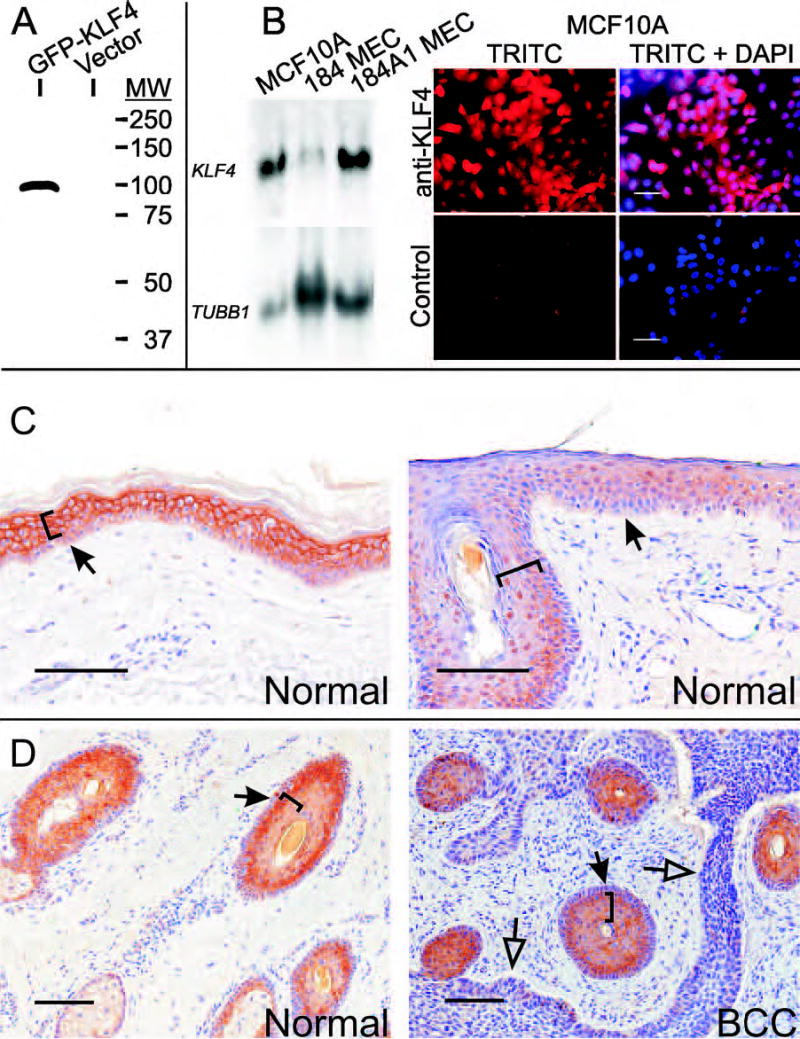
Characterization of anti-KLF4. A, HEK293 cells were transfected with the indicated plasmid and cell extracts were analyzed by immunoblot. Ponceau S stain of the filter demonstrated equal loading (not shown). B, Total RNA was isolated from the indicated cell lines and analyzed by Northern blot (left panel). A β-tubulin probe (TUBB1) served as a control for loading. To detect the endogenous protein, anti-KLF4 (upper panels) or normal rabbit immunoglobulin (lower panels) was applied to MCF10A cells. Bound antibody was detected using a red-fluorescent secondary antibody. Signal corresponding to KLF4 is shown either alone (TRITC) or else superimposed on the cell nuclei (TRITC+DAPI). Scale bars, 50μ. C–D, Tissue sections were stained with anti-KLF4. Bound antibody is indicated as a brown precipitate, and sections were counterstained blue using hematoxylin. C, Normal skin was obtained by reduction mammoplasty (left panel) or by excision of uninvolved skin adjacent to a SCC (right panel). D, Uninvolved adnexal epithelium in a patient with SCC (left panel) or stratified squamous epithelium and tumor cells in a patient with BCC (right panel). Brackets indicate cell layers with more prominent staining, solid arrows denote the dermo-epidermal junction (DEJ), and open arrows indicate the tumor-stromal border. Scale bars, 100μ.
Regulation of subcellular localization in normal skin
Previously, Klf4 mRNA and protein were found to be expressed predominantly in the nucleus of suprabasal, postmitotic cells of mouse skin.10,12,14 To determine the pattern of KLF4 in human skin, we examined 6 samples of normal skin, including interfollicular skin (Fig. 1C) and follicle epithelium (Fig. 1D, left panel). In skin obtained from a mammoplasty specimen, staining was predominantly cytoplasmic in parabasal and more superficial epithelial cells, and was also present in the basal layer (Fig. 1C, left panel). All normal skin samples examined showed positive basal and suprabasal staining (e.g., Fig. 1, C–D, and see below). Although staining was often higher in suprabasal cells, multiple areas showed similar staining of basal and parabasal cells (e.g., Fig. 1C, right panel, at right of arrow).
Unlike Klf4 in mouse skin,12,14 the subcellular localization of human KLF4 varied during keratinocyte development. In morphologically normal skin several mm distant from SCC (Fig. 1C, right panel), staining became progressively nuclear as the cells matured, and then became negative as cells approached the corneal layer. Similar results were obtained in follicles and sebaceous glands (Fig. 1D, left panel). KLF4 was low or negative in the two BCCs examined in the current study, but was prominent in squamous epithelium interspersed among the BCC cells (Fig. 1D, right panel). These studies establish the KLF4 expression pattern in human skin, indicating that KLF4 is normally expressed in the basal cell layer, and that expression and nuclear localization are increased in maturing cells.
Conditional KLF4 expression leads to increased skin cell cycling without cell accumulation
Previously, we showed that dox induction of KLF4 in basal keratinocytes of K14-rtTA;TRE-KLF4 mice blocked the proliferation-differentiation switch. Strong TUNEL staining, observed at 48 hours, abated as the skin transitioned through phases of hyperplasia, dysplasia, and invasive SCC-like lesions over a period of 2-28 days.7 In contrast to the rapid onset phenotype using this conditional strategy, constitutive expression achieved using an MMTV-KLF4 transgene lead to slow onset of skin dysplasia and SCC-like lesions between 6 months and 2 years of age, depending upon the p53 status.7 Acceleration of tumorigenesis by p53 hemizygosity suggested that cell death may generally limit the outgrowth of lesions. However, skin dysplasia and SCC was observed with a penetrance close to 100% in K14-rtTA;TRE-KLF4 animals administered a wide range of dox, and no setting in which cell turnover was sufficient to limit lesion outgrowth was identified.
To further examine early steps in KLF4-induced skin neoplasia, and to better understand the long latency in MMTV-KLF4 mice,7 TRE-KLF4 mice were crossed to a well-characterized, MMTV-rtTA strain previously used to conditionally induce TRE-linked transgenes in breast epithelium.22 Administration of dox to these animals for 3 weeks yielded no discernable alteration of the skin or breast, as determined by gross examination and by hematoxylin-eosin (H&E) stain of paraffin sections (not shown). To detect more subtle alterations in cell growth control, we stained the skin of uninduced, bitransgenic animals (Control) and day 14 animals (+ Dox) with anti-KLF4 and anti-BrdU (Fig. 2, upper rows). In the absence of dox KLF4 staining was low or negative, representing either nonspecific staining or else low-level, dox-independent transgene expression. As expected, BrdU incorporation was prominent in the anagen-phase follicles of these control animals, but rare in interfollicular keratinocytes (Fig. 2). Following 14 days of dox treatment, bitransgenic animals had increased expression of KLF4 in basal keratinocytes of morphologically normal follicular and interfollicular skin. Incorporation of BrdU was striking in follicle cells and interfollicular keratinoctyes (Fig. 2). As a further control, K14-rtTA;TRE-KLF4 mice were analyzed in parallel, and showed a prominent dysplastic response, with high-levels of KLF4 in nuclei and increased expression of PCNA as previously reported (Fig. 2).7 Because the skin phenotype in these control animals was grossly apparent, BrdU was not utilized. Thus, low-level KLF4 transgene expression, achieved using a conditional, MMTV-based strategy, identifies an occult step in tumor progression in which the skin undergoes increased cell cycling without overt hyperplasia or dysplasia, indicating an increased rate of cell turnover.
Fig. 2.
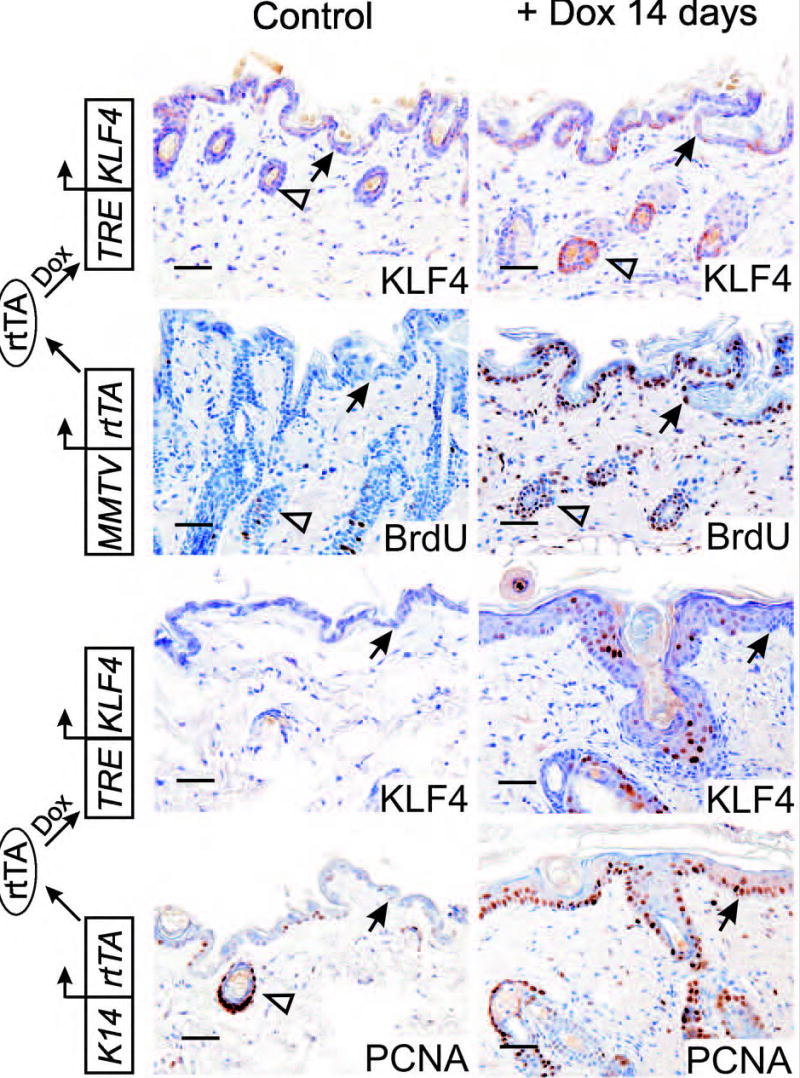
Low-level expression of KLF4 induces epithelial turnover without overt hyperplasia. Transgenic animals carrying the reverse tetracycline-responsive transcriptional activator (rtTA) under control of the MMTV promoter (i.e., MMTV-rtTA) were crossed with tetracycline response element (TRE)-KLF4 transgenic mice, and bitransgenic offspring were induced with dox (upper panels). Paraffin sections of skin were analyzed by immunostaining. Control animals were administered sucrose-water without dox. As an additional control, animals expressing KLF4 under control of a K14-rtTA transgene were induced and analyzed in parallel (lower panels). Closed arrows indicate the DEJ, and open arrows indicate follicle epithelium. Scale bars, 50μ.
Suppression of KLF4 transgene-derived protein levels in PCNA-positive, hyperplastic keratinocytes
KLF4 transcripts and protein were previously shown to be selectively expressed in post-mitotic cells in vivo, and induced upon growth-arrest of cultured cells in vitro.10–12,14,15 As a part of the study described above (Fig. 2), we isolated samples from K14-rtTA;TRE-KLF4 mice representing morphologically normal skin (Fig. 3A), hyperplasia (Fig. 3B), dysplasia (Fig. 3C), or SCC-like lesions (Fig. 3D). Using serial paraffin sections, we analyzed expression of PCNA and transgene-derived KLF4. To identify cells in which K14-rtTA may be expressed, we determined the expression pattern of endogenous K14, which is normally restricted to basal keratinocytes.
Fig. 3.
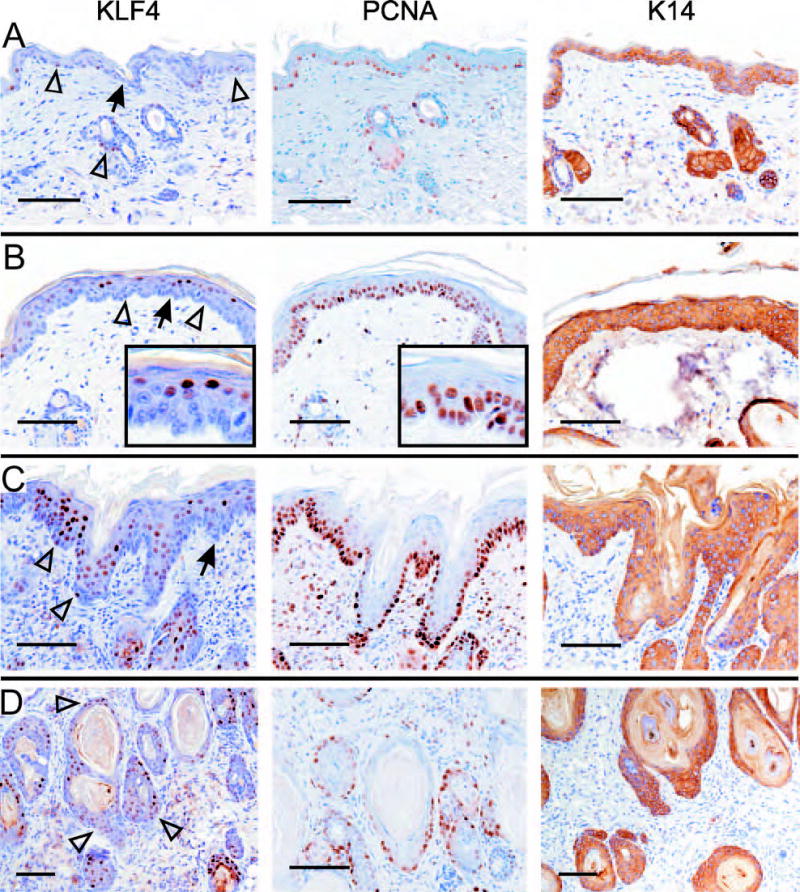
KLF4 and PCNA identify stages of tumor initiation. K14-rtTA;TRE-KLF4 bitransgenic mice were induced with dox for 14 days. The skin was sampled at multiple sites, fixed in paraffin, and tissue samples showing representative histologies were analyzed by immunostaining. After staining of serial sections with one of three different antibodies, images were recorded at a similar position on each sample. A, Lesion-adjacent skin exhibiting little or no morphologic abnormality. On normal skin analyzed in parallel, K14 staining was limited to basal keratinocytes, as expected (not shown). B, Hyperplasia. C, Dysplasia. D, SCC-like lesion. Panels A and C show adjacent areas of one tissue section, while B and D were sections taken from another mouse. Insets show the cell-type specific staining at increased magnification. Arrows indicate the DEJ, and open arrowheads indicate examples of KLF4-positive, basal keratinocytes.
Lesion-adjacent skin showed nuclear KLF4 in some basal and parabasal cells, but KLF4 was low or negative in most keratinocytes (Fig. 3A). In these samples, PCNA was increased compared to normal (see Fig. 2 above), particularly in the interfollicular skin. Whereas samples of normal skin showed the expected pattern of basal-specific K14 (not shown), this cytokeratin was uniformly expressed in lesion-adjacent skin, indicative of a block to differentiation.7 Thus, lesion-adjacent skin is abnormal at the molecular level, and perhaps similar to the MMTV-rtTA;TRE-KLF4 skin described above (Fig. 2).
Strikingly, hyperplastic skin showed prominent nuclear KLF4 in maturing, suprabasal cells, with only sporadic, low-level expression in basal and parabasal keratinocytes (Fig. 3B, left panel). Compared with lesion-adjacent skin, PCNA was increased in hyperplasia, and was complementary to KLF4 in a serial section (Fig. 3B, middle panel; and see insets). The preferential expression of KLF4 in PCNA-negative epithelial layers was in contrast to K14 staining, which was uniform throughout (Fig. 3B, right panel). This uniformity of K14 suggested that differential expression of TRE-KLF4 is not due to differential expression of K14-rtTA.
The complementarity of KLF4 and PCNA, observed consistently in hyperplastic lesions from multiple animals, was less evident in dysplastic lesions, where KLF4 was detected both in PCNA-positive regions of the basal layer and in more superficial cells (Fig. 3C). Unlike the uniform expression of K14, these lesions typically exhibited a gradient of KLF4, with increasing KLF4 and decreasing PCNA as cells matured.
In lesions more similar to SCC, the KLF4 gradient observed in dysplasia (Fig. 3C) was abolished, and KLF4 and PCNA were coexpressed, particularly in basal-like cells adjacent to the stroma (Fig. 3D). The marked variation of KLF4 expression during tumor initiation (Fig. 3A–D, left panels) was in contrast to the more consistent staining of PCNA (Fig. 3, middle panels) and K14 (Fig. 3, right panels) at each stage.
Differential expression of KLF4 transcripts in hyperplastic skin
In the studies describe above (Fig. 3B), we observed MD expression of KLF4 protein derived from a human cDNA transgene. This transgene, which lacks KLF4 promoter sequences, the introns, and part of the 3’ UTR, was sufficient to impose cell cycle-regulated expression of the human KLF4 protein. To determine if regulation occurred at the level of mRNA or protein, we used in situ hybridization to detect transcripts derived from the transgene and two endogenous genes, the basal cell marker integrin α3 (Itga3) and the housekeeping gene β-tubulin (Tubb1).
An anti-sense probe derived from the human KLF4 3’ UTR specifically labeled the upper cell layers that are Itga3-negative, as shown by analysis of serial sections (Fig. 4A, upper panels). In contrast, the Tubb1 probe labeled a nonadjacent section uniformly (Fig. 4A, lower right), and no signal was obtained using a KLF4 sense control probe (not shown). Independent induction/hybridization experiments using sections from at least 4 different animals yielded consistent results. As shown above for the protein (Fig. 3D), we previously showed that established SCC-like lesions exhibit uniform expression of KLF4 transgene-derived transcripts.7
Fig. 4.
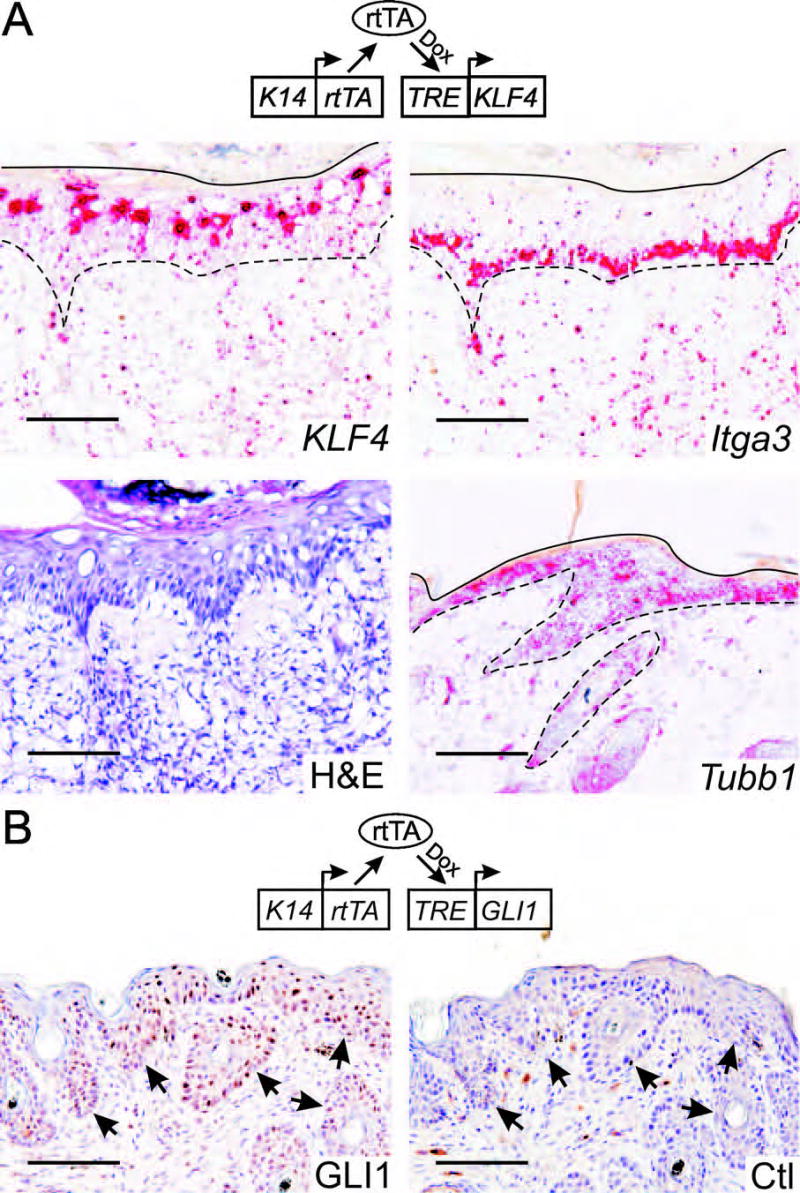
TRE-KLF4 and TRE-GLI1 transgene expression in hyperplastic skin. K14-rtTA;TRE-KLF4 (A) or K14-rtTA;TRE-GLI1 (B) bitransgenic mice were induced with dox, and cryosections of hyperplastic skin were analyzed. This 21 day induction was performed independently of the experiment shown in Fig. 3. A, mRNA in situ hybridization analysis using the indicated anti-sense probe. Positive staining is indicated as a red precipitate. Orientation lines correspond to the skin surface (solid lines) or to the DEJ (dashed lines). An H&E-stained cryosection is shown at bottom left. B, Immunostain of GLI1-induced hyperplastic lesions with GLI1 antibody. Normal rabbit immunoglobulin served as a control (Ctl). Staining by anti-GLI1 was low or negative on normal, interfollicular mouse skin (not shown). Arrows indicate the DEJ. Scale bars, 100μ.
MD expression of KLF4 transcripts could result from differential activity of the K14-rtTA transgene in dividing vs. maturing cells. To address this possibility, we utilized a different transgene, termed TRE-GLI1, in combination with the same K14-rtTA. When induced with dox, K14-rtTA;TRE-GLI1 mice develop well-characterized, hyperplastic lesions composed of BrdU-positive basal/parabasal cells that undergo a delayed proliferation-differentiation switch and upregulate K1 in BrdU-negative, superficial cells.21 To stain GLI1 in sections, we tested a new antibody that exhibits specificity by immunoblot.21
GLI1 staining of nuclei was uniform throughout the hyperplastic skin of K14-rtTA;TRE-GLI1 animals, without evidence of preferential staining of superficial cells (Fig. 4B). Normal rabbit antibody served as a control and did not stain. Analysis of normal mouse skin revealed little or no staining with this antibody in the interfollicular epithelium (not shown). These studies demonstrate that the MD expression of KLF4 protein in initiating lesions (Fig. 3B–C) is due to suppression of KLF4 transgene-derived transcripts in PCNA-positive cells, that suppression is specific to KLF4 transcripts, and that suppression is lost in KLF4-induced, SCC-like lesions.
Subtypes of SCC based upon KLF4 staining
We previously showed that anti-KLF4 stained two of five cases of human cutaneous SCC.7 To better determine the frequency and timing of KLF4 upregulation in SCC, we prospectively collected tissue corresponding to 48 new cases. 16 samples were dysplastic lesions termed actinic keratosis (AK), without adjacent SCC (Table 1, cases 1–16), and 32 were cases of SCC with or without adjacent AK (Table 1, cases 17–48).
Table 1.
KLF4 staining patterns in cutaneous SCC progression.
| Case# | Diagnosis | AK* | SCC* |
|---|---|---|---|
| 1 | AK | MIN | - |
| 2 | AK | Low/Neg. | - |
| 3 | AK | Low/Neg. | - |
| 4 | AK | MD | - |
| 5 | AK | MD | - |
| 6 | AK | MIM | - |
| 7 | AK | MIM | - |
| 8 | AK | MIC | - |
| 9 | AK | MIC | - |
| 10 | AK | Low/Neg. | - |
| 11 | AK | MIC | - |
| 12 | AK | MIM | - |
| 13 | AK | MIC | - |
| 14 | AK | MIC | - |
| 15 | AK | MIC | - |
| 16 | AK or SCCis | MIN | - |
| 17 | SCC | Low/Neg. | MD |
| 18 | SCCis | MD | Low/Neg. |
| 19 | SCC | MD | Low/Neg. |
| 20 | SCC | MD | Low/Neg. |
| 21 | SCC | MD | Low/Neg. |
| 22 | SCCis | MD | Low/Neg. |
| 23 | SCC | MD | MD |
| 24 | SCC | MD | MD |
| 25 | SCC | MD | MD |
| 26 | SCC | MD | MD |
| 27 | SCC | MIC | MIM |
| 28 | SCCis | MIC | MIM |
| 29 | SCCis | MIC | MIN |
| 30 | SCC | MIM | MIM |
| 31 | SCC | MIM | MIN |
| 32 | SCC | MIM | MIN |
| 33 | SCC | MIM | MIN |
| 34 | SCC | MIN | MIN |
| 35 | SCC | MIN | MIN |
| 36 | SCC | MIN | MIN |
| 37 | SCC | MIN | MIN |
| 38 | SCC | MIN | MIN |
| 39 | SCC | MIN | MIN |
| 40 | SCCis | MIN | MIN |
| 41 | SCC | - | Low/Neg. |
| 42 | SCC | - | Low/Neg. |
| 43 | SCC | - | MIM |
| 44 | SCC | - | MD |
| 45 | SCC | - | MIM |
| 46 | SCC | - | MIN |
| 47 | SCC | - | MIM |
| 48 | SCC | - | MIN |
A dash (-) indicates that no tissue with this histology was present. KLF4 expression patterns: MD – maturation dependent; MIC – maturation independent, cytoplasmic; MIM – maturation independent, mixed; MIN – maturation independent, nuclear; Low/Neg. – Low or negative expression.
Unlike normal epithelium, in which expression and subcellular localization varied as cells matured (see above), most SCCs exhibited loss of this regulation. Maturation independent (MI) staining was usually associated with constitutive nuclear localization (MIN). Other staining patterns observed included maturation dependent (MD), and low or negative staining (Low/Neg.) (Fig. 5A). In total, MI staining was observed in 19 of 32 cases (59%) (Table 1), and 13 of these 19 cases showed MIN staining. In other cases both the cytoplasm and nucleus were stained (MI, mixed or MIM) or else cytoplasmic staining predominated (MI, cytoplasmic or MIC).
Fig. 5.
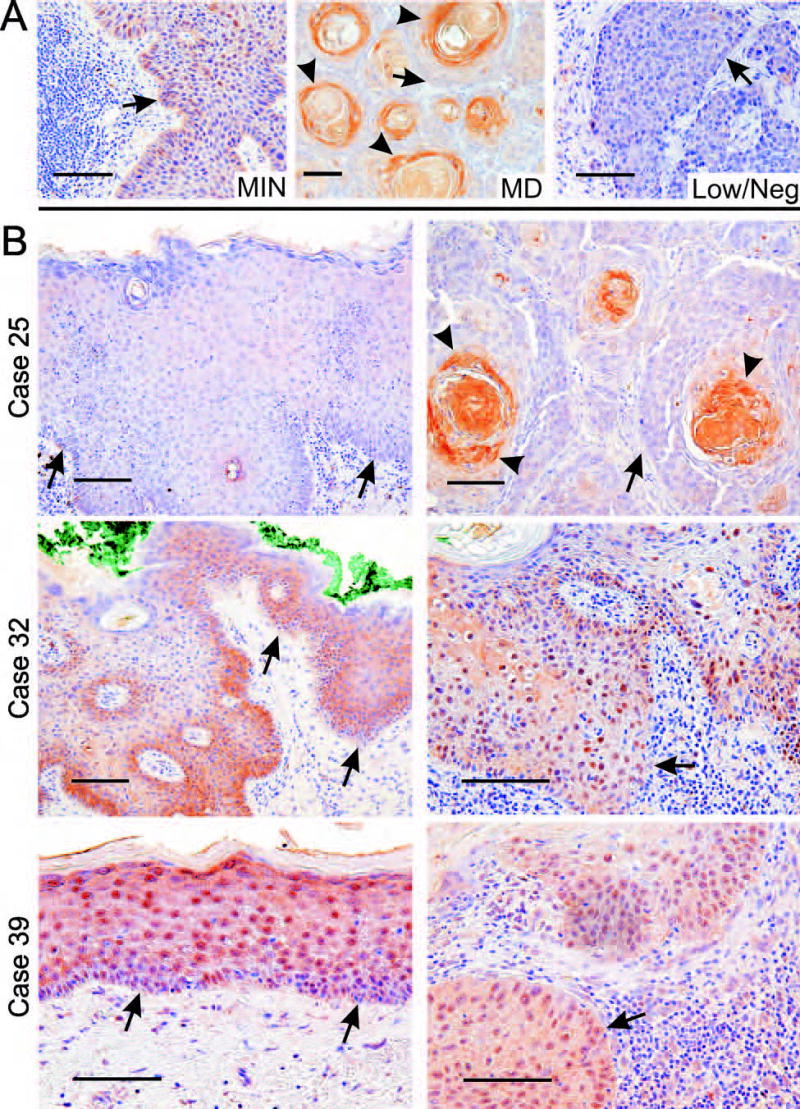
KLF4 expression in human cutaneous SCC. A, Tumors showing different KLF4 staining patterns (see Table 1). B, Comparison of staining in tumors with that of adjacent skin. Arrows indicate the DEJ in sections showing epidermis (left), or else the tumor-stromal interface for sections showing tumor cells (right). Arrowheads indicate maturing cells with expression of KLF4. Scale bars, 100μ.
Six cases (19%) were MD, showing prominent staining of KLF4 in maturing squamous cells within tumor islands (Fig. 5A, case 25, and Table 1). This pattern exaggerated that of normal skin, with low or even negative expression in basal-like tumor cells contacting the stroma. Finally, 7 cases of SCC (22%) exhibited Low/Neg. expression of KLF4.
MI expression and nuclear localization are sequential features of SCC progression
To better understand the temporal nature of KLF4 deregulation, we examined SCC and adjacent AK (Table 1, cases 17–40, Fig. 5B). Of 10 SCCs typed as Low/Neg. or MD, one of these patterns was likewise observed in the associated AK (Table 1, case 25, Fig. 5B). In contrast, 14 SCCs typed as MI showed similar staining in the adjacent AK (P<0.0001). The strong correlation of these adjacent lesions, and the frequent MI staining in AK lesions not associated with SCC (Table 1, cases 1–16) suggests that KLF4 expression patterns are established early in tumor progression.
As observed above in the mouse model, nuclear localization was often greater in human SCC than in the adjacent skin. Six of the 14 cases with MI staining in the AK lesion showed transition to a more nuclear pattern in the SCC on the same slide (Table 1, cases 27–30, 31–33; Fig. 5B, case 32). For another 7 cases, MIN staining was observed in both lesions (Table 1, cases 34–40; Fig. 5B, case 39), suggesting that constitutive nuclear localization can occur early in tumor progression.
Further support of a role for nuclear localization in progression was suggested by comparison of AK adjacent to SCC with cases containing AK in isolation (Table 1). For the 24 cases in which AK was found in association with SCC, MIN expression within the AK lesion was observed in 7 (29%). In contrast, of 15 cases of AK not associated with SCC, only 1 case had predominantly nuclear staining (6.7%; P=0.104). Interestingly, one additional case of AK that could not be definitively distinguished from SCC in situ (SCCis) also had a MIN expression pattern (Table 1, case 16).
To compare KLF4 and PCNA in human lesions, we stained serial sections (Fig. 6). PCNA staining was rare in the KLF4-positive interfollicular cells of normal skin (Fig. 6, Normal), but was positive in most cases of SCC, where it was usually co-expressed with KLF4 (Fig. 6, SCC-MIN). PCNA was generally higher in tumors compared with the adjacent skin or AK, but in several cases KLF4 and PCNA were co-expressed in the dysplastic skin adjacent to SCC (not shown). Such co-expression was likewise observed in one of two AK lesions that were not associated with SCC (Fig. 6, AK).
Fig. 6.
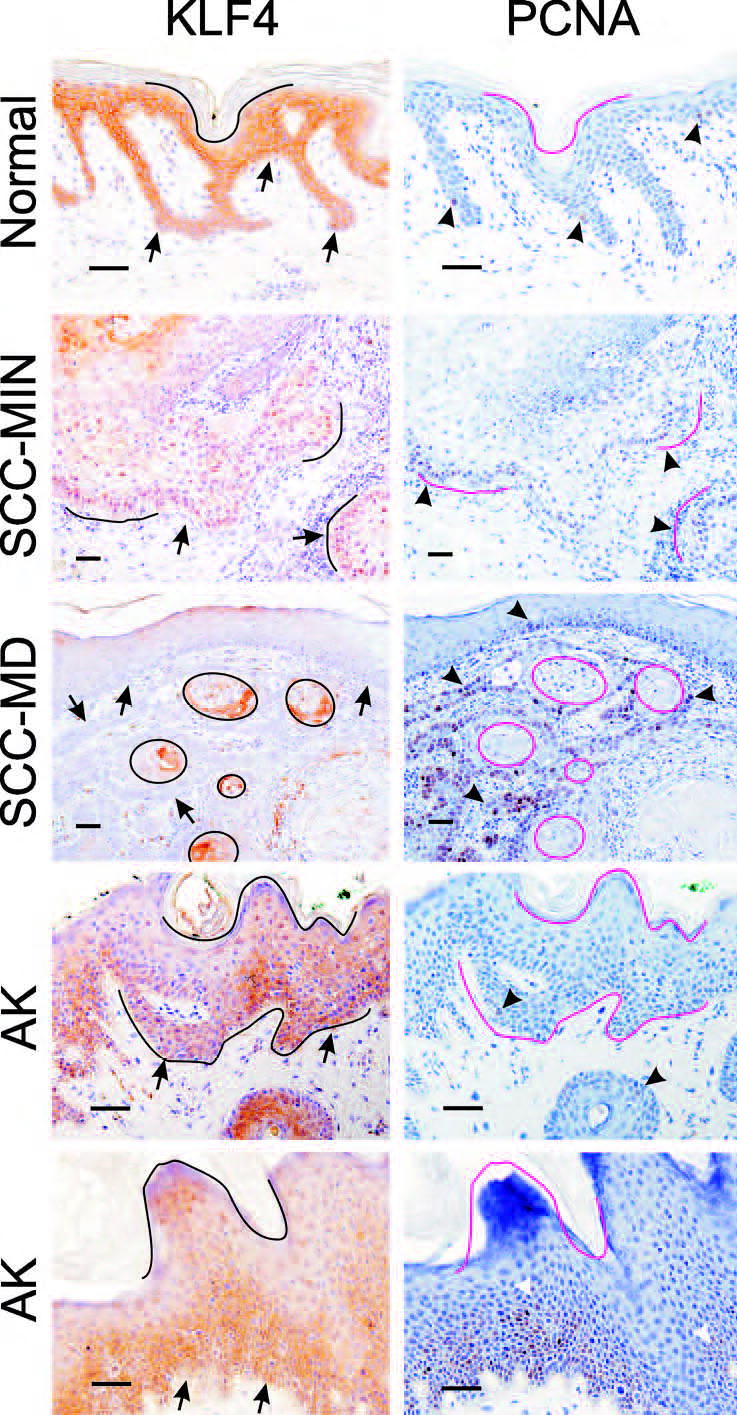
Co-localization of KLF4 and PCNA in human tissues. Serial sections of paraffin-embedded samples were analyzed by immunostaining. Identical orientation lines were placed on the matched images. Arrows indicate the DEJ or tumor-stromal borders, and arrowheads indicate examples of PCNA-positive cells. Scale bars, 50μ.
In contrast to the co-expression of KLF4 and PCNA in MI tumors, each of the two MD tumors that were analyzed exhibited strong expression of both markers, but in a complementary pattern (Fig. 6, SCC-MD). With respect to KLF4 regulation, these malignant tumors appear similar to hyperplastic mouse skin (Fig. 3B), indicating that the machinery that normally suppresses KLF4 in dividing cells may be intact in this subset of human SCCs.
Discussion
In the current study we used two conditional KLF4 expression strategies to identify stages in tumor initiation, and characterized lesions with distinct morphologies according to expression of KLF4 and PCNA. The phenotype resulting from deregulation of a TRE-KLF4 transgene was dependent upon the promoter used to drive expression of rtTA. Whereas K14-rtTA mice developed dysplasias and SCC-like lesions with prominent KLF4 expression, MMTV-rtTA mice exhibited modest expression of KLF4 in morphologically normal skin that was strongly positive for BrdU, indicating an increased rate of cell turnover. Although KLF4 induction in the skin results in prominent TUNEL staining,7 for unclear reasons TUNEL is prominent in the skin when proliferation and differentiation are both increased, as in psoriasis, even though apoptotic bodies are not evident by microscopy.24,25 As apoptotic bodies are rare in KLF4-induced lesions, increased TUNEL staining is consistent with cell turnover mediated by accelerated cell differentiation rather than apoptosis. Increased turnover within normal-appearing epithelium, or occult cell turnover, is a potential mechanism of tumor promoters such as phorbol ester, and may represent a common, cell death limited step in other settings of tumorigenesis. For example, expression of activated Smo in breast epithelium (MMTV-SmoM2) leads to prominent BrdU and Ki67 staining in the absence of overt hyperplasia or dysplasia, and with little evidence of increased apoptosis (personal communication, Michael T. Lewis, Baylor College of Medicine).26 Similarly, occult cell turnover might manifest in at-risk patients as prominent Ki67 staining within morphologically normal epithelium.
A surprising outcome of these studies was the MD expression of KLF4 in hyperplastic lesions, where KLF4 and PCNA were expressed in a complementary fashion. Such a pattern is distinct from the uniform expression of endogenous K14 or exogenous TRE-GLI1 that we observed within hyperplastic lesions. Furthermore, in developing the K14-rtTA strain that we used, Xie and colleagues conditionally expressed a TRE-erbB2 allele to induce hyperplastic lesions.27 By mRNA in situ hybridization, rtTA transcript levels were uniform within these lesions, and TRE-erbB2 was prominently expressed in all cell layers, with no evidence of preferential expression in post-mitotic cells. Therefore, a preponderance of evidence indicates that K14-rtTA expression and activity is cell cycle independent, and that the MD expression pattern of the KLF4 cDNA transgene is due to sequence elements within the protein coding region and/or the flanking UTRs.
Analysis of human skin provided further insight. We consistently observed prominent expression of KLF4 in all cell layers of normal human skin. Although certain samples showed more prominent suprabasal expression, we found no normal skin samples where expression was restricted to these cells. Taken together, our data suggest that KLF4 is transcribed in diverse cell types of the skin, perhaps even in a constitutive fashion, and that the transcripts are unstable in actively dividing cells. Support for this instability in human tissues was the complementary pattern of KLF4 and PCNA in MD SCCs.
This complementary pattern may result from post-transcriptional control of KLF4. The TRE-KLF4 transgene used in the current study7 retains all of the 5’ UTR previously isolated5 and over 500 bases of the 3’ UTR. Cell cycle-dependent, post-transcriptional regulation of mRNA stability is well-established for the histone mRNAs and for polyadenylated mRNAs of genes important in a wide variety of processes, including the mitogenic response, cell differentiation, and cell cycle regulation.28–30 Recently, micro RNAs (miRs) have been implicated in control of transcript stability, and computational studies feature KLF4 as a prominent candidate target of this regulation.31,32 For example, miR7 has two binding sites in KLF4, both of which are present in our transgene. Like KLF4, several miRs are prominently expressed in proliferating cells but downregulated during differentiation.33 We are currently using RK3E cells, in which endogenous KLF4 is induced upon growth arrest (not shown), to map RNA destabilizing elements. This regulation appears relevant to pathogenesis, as complementarity with PCNA was lost as lesions progressed to dysplasia and SCC. Although we suggest that a post-transcriptional mechanism suppresses KLF4, pre-mature transcriptional termination would also be consistent with our in vivo observations.
We observed clear parallels between KLF4-initiated lesions in mice and a majority of human cutaneous SCCs, identifying KLF4 as a candidate effector of SCC tumor progression in human skin. A better understanding of how KLF4 induces transformation in vitro and tumor initiation in vivo may provide new opportunities for clinical intervention in this common disease.
Acknowledgments
MMTV-rtTA transgenic mice were a kind gift of Lewis A. Chodosh. We thank James E. Elder for performing histopathologic diagnoses, and Eben L. Rosenthal for critical reading of the manuscript. Supported by American Cancer Society RSG-05-207-01-TBE (A.R.F., J.M.R.), NCI grants RO1-CA65686 (A.R.F., J.M.R.), RO1-CA094030 (C.C.H., J.M.R. and S.M. L.-R.), and P50-CA89019 (J.M.R. and S.M.L.-R.). The UAB Tissue Procurement Core Facility is supported by NCI Grant 5P50 CA13148.
References
- 1.Taipale J, Beachy PA. The Hedgehog and Wnt signaling pathways in cancer. Nature. 2001;411:349–54. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 3.Nusse R. Wnts and Hedgehogs: lipid-modified proteins and similarities in signaling mechanisms at the cell surface. Development. 2003;130:5297–305. doi: 10.1242/dev.00821. [DOI] [PubMed] [Google Scholar]
- 4.Pires-daSilva A, Sommer RJ. The evolution of signalling pathways in animal development. Nat Rev Genet. 2003;4:39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- 5.Foster KW, Ren S, Louro ID, Lobo-Ruppert SM, McKie-Bell P, Grizzle W, Hayes MR, Broker TR, Chow LT, Ruppert JM. Oncogene expression cloning by retroviral transduction of adenovirus E1a-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 1999;10:423–34. [PubMed] [Google Scholar]
- 6.Foster KW, Frost AR, McKie-Bell P, Lin CY, Engler JA, Grizzle WE, Ruppert JM. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–95. [PubMed] [Google Scholar]
- 7.Foster KW, Liu Z, Nail CD, Li X, Fitzgerald TJ, Bailey SK, Frost AR, Louro ID, Townes TM, Paterson AJ, Kudlow JE, Lobo-Ruppert SM, Ruppert JM. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene. 2005;24:1491–500. doi: 10.1038/sj.onc.1208307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandya AY, Talley LI, Frost AR, Fitzgerald TJ, Trivedi V, Chakravarthy M, Chhieng DC, Grizzle WE, Engler JA, Krontiras H, Bland KI, Lobuglio AF, Lobo-Ruppert SM, Ruppert JM. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin Cancer Res. 2004;10:2709–19. doi: 10.1158/1078-0432.ccr-03-0484. [DOI] [PubMed] [Google Scholar]
- 9.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol 2005; .. [DOI] [PubMed]
- 10.Garrett-Sinha LA, Eberspaecher H, Seldin MF, de Crombrugghe B. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem. 1996;271:31384–90. doi: 10.1074/jbc.271.49.31384. [DOI] [PubMed] [Google Scholar]
- 11.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–17. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–60. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 13.Panigada M, Porcellini S, Sutti F, Doneda L, Pozzoli O, Consalez GG, Guttinger M, Grassi F. GKLF in thymus epithelium as a developmentally regulated element of thymocyte-stroma cross-talk. Mechanisms of Development. 1999;81:103–13. doi: 10.1016/s0925-4773(98)00237-8. [DOI] [PubMed] [Google Scholar]
- 14.Jaubert J, Cheng J, Segre JA. Ectopic expression of kruppel like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development. 2003;130:2767–77. doi: 10.1242/dev.00477. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, Kaestner KH, Biggs JR, Kraft AS, Yang VW. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J Biol Chem. 2000;275:18391–8. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–50. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 17.Sancho E, Batlle E, Clevers H. Live and let die in the intestinal epithelium. Curr Opin Cell Biol. 2003;15:763–70. doi: 10.1016/j.ceb.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23:395–402. doi: 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katz JP, Perreault N, Goldstein BG, Actman L, McNally SR, Silberg DG, Furth EE, Kaestner KH. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128:935–45. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 20.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Deng W, Nail CD, Bailey SK, Kraus MH, Ruppert JM, Lobo-Ruppert SM. Snail induction is an early response to Gli1 that determines the efficiency of epithelial transformation. Oncogene 2005; .:in press [DOI] [PMC free article] [PubMed]
- 22.Gunther EJ, Belka GK, Wertheim GB, Wang J, Hartman JL, Boxer RB, Chodosh LA. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB J. 2002;16:283–92. doi: 10.1096/fj.01-0551com. [DOI] [PubMed] [Google Scholar]
- 23.Louro ID, Bailey EC, Li X, South LS, McKie-Bell PR, Yoder BK, Huang CC, Johnson MR, Hill AE, Johnson RL, Ruppert JM. Comparative gene expression profile analysis of GLI and c-MYC in an epithelial model of malignant transformation. Cancer Res. 2002;62:5867–73. [PubMed] [Google Scholar]
- 24.Gandarillas A. Epidermal differentiation, apoptosis, and senescence: common pathways? Exp Gerontol. 2000;35:53–62. doi: 10.1016/s0531-5565(99)00088-1. [DOI] [PubMed] [Google Scholar]
- 25.Gandarillas A, Goldsmith LA, Gschmeissner S, Leigh IM, Watt FM. Evidence that apoptosis and terminal differentiation of epidermal keratinocytes are distinct processes. Exp Dermatol. 1999;8:71–9. doi: 10.1111/j.1600-0625.1999.tb00350.x. [DOI] [PubMed] [Google Scholar]
- 26.Lewis MT, Veltmaat JM. Next stop, the twilight zone: hedgehog network regulation of mammary gland development. J Mammary Gland Biol Neoplasia. 2004;9:165–81. doi: 10.1023/B:JOMG.0000037160.24731.35. [DOI] [PubMed] [Google Scholar]
- 27.Xie W, Chow LT, Paterson AJ, Chin E, Kudlow JE. Conditional expression of the ErbB2 oncogene elicits reversible hyperplasia in stratified epithelia and up-regulation of TGFα expression in transgenic mice. Oncogene. 1999;18:3593–607. doi: 10.1038/sj.onc.1202673. [DOI] [PubMed] [Google Scholar]
- 28.Marzluff WF. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol. 2005;17:274–80. doi: 10.1016/j.ceb.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–50. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bevilacqua A, Ceriani MC, Capaccioli S, Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J Cell Physiol. 2003;195:356–72. doi: 10.1002/jcp.10272. [DOI] [PubMed] [Google Scholar]
- 31.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 32.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–78. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]


